Abstract
The phytohormone abscisic acid (ABA) induces accumulation of reactive oxygen species (ROS), which can disrupt seed dormancy and plant development. Here, we report the isolation and characterization of an Arabidopsis thaliana mutant called ars1 (aba and ros sensitive 1) that showed hypersensitivity to ABA during seed germination and to methyl viologen (MV) at the seedling stage. ARS1 encodes a nuclear protein with one zinc finger domain, two nuclear localization signal (NLS) domains, and one nuclear export signal (NES). The ars1 mutants showed reduced expression of a gene for superoxide dismutase (CSD3) and enhanced accumulation of ROS after ABA treatment. Transient expression of ARS1 in Arabidopsis protoplasts strongly suppressed ABA-mediated ROS production. Interestingly, nuclear-localized ARS1 translocated to the cytoplasm in response to treatment with ABA, H2O2, or MV. Taken together, these results suggest that ARS1 modulates seed germination and ROS homeostasis in response to ABA and oxidative stress in plants.
Keywords: abiotic stress, abscisic acid, Arabidopsis, C2H2 zinc finger, reactive oxygen species, redox
Introduction
The phytohormone abscisic acid (ABA) regulates important physiological processes including embryogenesis, seed dormancy, vegetative growth, and abiotic stress responses (Cutler et al., 2010; Raghavendra et al., 2010). ABA signaling is associated with the accumulation of intracellular reactive oxygen species (ROS), which initiates diverse signal transduction processes such as gene expression, enzyme activation, and programmed cell death (Neill et al., 2002; Wasilewska et al., 2008). ROS such as hydrogen peroxide (H2O2), superoxide anion (O2-), singlet oxygen (1O2), and hydroxyl radical (OH-) form as toxic byproducts of metabolic processes, including photosynthesis, dark respiration, and photorespiration, as well as under abiotic stress conditions; ROS also act as important signaling molecules under optimal growth conditions (Mittler et al., 2004; Kim et al., 2008; Foyer and Shigeoka, 2011).
Reactive oxygen species, as key endogenous messengers, play a crucial role in the complex ABA signaling network (Wang and Song, 2008). This network involves diverse regulators, such as NADPH oxidases, SNF1-related protein kinases (SnRK), type-2C/A protein phosphatases (PP2C), calcineurin B-like (CBL) interacting protein kinases (CIPK), calcium-dependent protein kinases, and mitogen-activated protein kinases (MAPK). The ABA receptors RCAR/PYR1/PYL (Regulatory Components of ABA-receptor/Pyrabactin resistant Protein/PYR-like protein) perceive ABA, bind to ABA, and interact with a group of PP2Cs (Ma et al., 2009; Park et al., 2009). In the absence of ABA, PP2Cs interact with SnRK2/OST1 (OPEN STOMATA1) and dephosphorylate SnRK2 to inactive its kinase activity. When ABA binds to its receptors, SnRK2 is activated, via the lack of PP2C function (Umezawa et al., 2009). Activated SnRK2 phosphorylates and regulates various downstream target proteins, including guard cell ion channels, NADPH oxidase, and transcription factors (Kwak et al., 2003; Sato et al., 2009; Yoshida et al., 2010; Brandt et al., 2012). Among the SnRK2 targets, NADPH oxidases (respiratory burst oxidase homologues, RBOHs) localize in the plasma membrane and the phosphorylation of RBOHs by SnRK2/OST1 plays a major role in triggering ROS production in plants (Kwak et al., 2003). In addition, RBOHF is phosphorylated by the CBL/CBL9-CIPK26 complex and mediates ROS production (Drerup et al., 2013).
Regulation of ROS-generating and ROS-scavenging systems maintains the delicate balance of ROS (Foyer and Noctor, 2009). Non-enzymatic antioxidants detoxify singlet oxygen and hydroxyl radical; by contrast, antioxidant enzymes including superoxide dismutases (SOD), catalases (CAT), and ascorbate peroxidases (APX) detoxify H2O2 (Op den Camp et al., 2003; Gadjev et al., 2006; Gechev et al., 2006; Foyer and Noctor, 2009). SOD catalyzes the dismutation of O2- to H2O2; CAT and APX directly react with H2O2 to form water and oxygen (Mittler et al., 2004).
Recently, ROS have been implicated in mediating complex, systemic signaling in plant cells. These ROS signals may function alone or interact with other molecules, including plant hormones, Ca2+ signals, proteins, and RNA (Karpiński et al., 2013; Shah et al., 2014; Wang et al., 2014). In addition, ABA and ROS may act together in regulating systemic responses to abiotic stress (Suzuki et al., 2013). In this study, we isolated Arabidopsis ARS1 (ABA AND ROS SENSITIVE 1) using a genetic screening system and showed that ARS1 is essential for seed germination and maintenance of ROS homeostasis in plants challenged with ABA or oxidative stress. We also demonstrated that ARS1 translocates from the nucleus to the cytoplasm in response to ABA or oxidative stress. We report that ARS1 functions as a positive regulator countering ABA to break seed dormancy and maintaining ROS homeostasis in response to ABA and oxidative stress.
Materials and Methods
Plant Materials and Growth Conditions
The activation T-DNA vector pSKI015 was used to generate an insertion mutant population (T1) in the Arabidopsis thaliana C24 RD29A::LUC (WT) background, based on Basta herbicide selection. Plants were grouped into 10-line pools, and T2 progenies were screened for mutants that exhibited ABA hypersensitivity compared to WT. T-DNA insertion mutants of ARS1 (At3g02860), ars1-2 (SALK_009596), ars1-3 (SALK_030445), and ars1-4 (SALK_126300), were obtained from the Arabidopsis Biological Resource Center (ABRC). Seeds were surface-sterilized and sown onto MS medium [1/2 Murashige and Skoog (MS) salts, 1.5% sucrose and 0.6% agar, pH 5.7] with or without ABA (as indicated in figures). Plants were grown in a growth chamber with a cycle of 16 h light and 8 h dark at 22°C
Thermal Asymmetric Interlaced PCR Analysis
DNA flanking the left border of the inserted T-DNA in the ars1-1 mutant was isolated by thermal asymmetric interlaced PCR (TAIL-PCR). The entire isolated fragment was sequenced. The primers used in the TAIL-PCR were specific primers for the T-DNA left border (LB1, LB2, and LB3) and degenerate primers (DP1, DP2, and DP3; Supplementary Table S1). The nucleotide sequence of the PCR product was determined and subjected to BLASTn analysis.
Determination of Transcript Levels
Total RNA was isolated from 10-d-old seedlings using the RNeasy kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. Isolated total RNA was treated with DNase I (Qiagen, Valencia, CA, USA) to remove genomic DNA contamination. The first-strand cDNA was synthesized using 2 μg total RNA with a cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA) in a 20-μl reaction volume, and subjected to PCR for examination of gene expression. The specific primers were designed according to the sequence of ARS1. TUBULIN2 was used as a control in the experiment. The primers used for the RT-PCR analysis are listed in Supplementary Table S1.
For quantitative RT-PCR (qRT-PCR) analysis, the first-strand cDNA was synthesized using 2 μg total RNA with a cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA). The QuantiSpeed SYBR No-Rox Mix (PhileKorea, Seoul, Korea) was used for qRT-PCR as follows: 50°C for 10 min, 95°C for 2 min, and 40 cycles of 95°C for 5 s, and 60°C for 30 s. TUBULIN2 was used for RNA normalization. The relative expression levels of all samples were automatically calculated using CFX Manager program (Bio-Rad, Hercules, CA, USA) and carried out in three biological replicates. The primers used for the qRT-PCR analysis are listed in Supplementary Table S1.
Protoplast Transient Expression Analysis
The cDNA encoding ARS1 was isolated from a cDNA library by PCR. The PCR product was confirmed by nucleotide sequencing and was inserted into XbaI and BamHI sites of the sGFP vector (kindly provided by Inhwan Hwang, POSTEC, Korea) to create chimeric GFP-fusion constructs under the control of the 35S promoter (Supplementary Table S1). The sGFP plasmid vector is a pUC-based vector containing CaMV35S-sGFP-NOS3 for protoplast expression.
Protoplast isolation from Arabidopsis leaves and transformation into protoplasts was as described in Jin et al. (2001). Expression of the fusion constructs was monitored at various time points after transformation and images were captured with a Zeiss Axioplan fluorescence microscope (Carl Zeiss Co., Jena, Germany). The filter sets used were: XF116 (exciter, 474AF20; dichroic, 500DRLP; and emitter, 605DF50) and XF137 (exciter, 540AF30; dichroic, 500DRLP; and emitter, 585ALP; Omega, Inc., Brattleboro, VT) for GFP and RFP, respectively. Data were then processed using Adobe Photoshop software (Adobe System, Mountainview, CA, USA) and presented in pseudo-color format.
Detection of ROS in Protoplasts
To measure intracellular ROS levels, an aliquot of protoplast suspension (∼2 × 105⋅ml-1) was incubated with 5 μM 2,7-dichlorohydrofluoroscein diacetate (DCFH-DA, Molecular Probes, Eugene, OR, USA) for 5 min and 20 μM dihydrorhodamine123 (Rh123, Molecular Probes, Eugene, OR, USA) for 15 min and were observed under a Zeiss Axioplan fluorescence microscope using XF116 (DCFH-DA; exciter, 474AF20; dichroic, 500DRLP; and emitter, 605DF50) and XF33/E (Rh123; exciter, 535DF35; dichroic, 570DRLP; emitter, 605DF50; Omega, Inc., Brattleboro, VT, USA).
Histochemical Detection of O2-
NBT (nitro blue tetrazolium; Sigma–Aldrich, Saint Louis, MO, USA) staining was used to detect O2- accumulation in tissues. O2- was visualized as a dark blue formazan compound within tissues. Seven-day-old seedlings were immersed in 50 mM potassium phosphate buffer (pH 7.8) containing 0.1% NBT and 10 mM sodium azide and incubated for 2 h in the dark. Chlorophyll was removed from the seedlings prior to imaging by infiltrating them with lacto-glycerol-ethanol (1: 1: 4 volume) and boiling for 5 min (Bournonville and Díaz-Ricci, 2011).
Measurement of H2O2
H2O2 was measured in tissues using the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen/Molecular Probes, Eugene, OR, USA) following the manufacturer’s instructions. Fluorescence was determined by excitation at 530 nm and emission at 590 nm. H2O2 concentration was calculated based on a standard curve and expressed as H2O2 per fresh weight (Bournonville and Díaz-Ricci, 2011).
Results
Isolation and Identification of the ABA-Hypersensitive ars1 Mutant
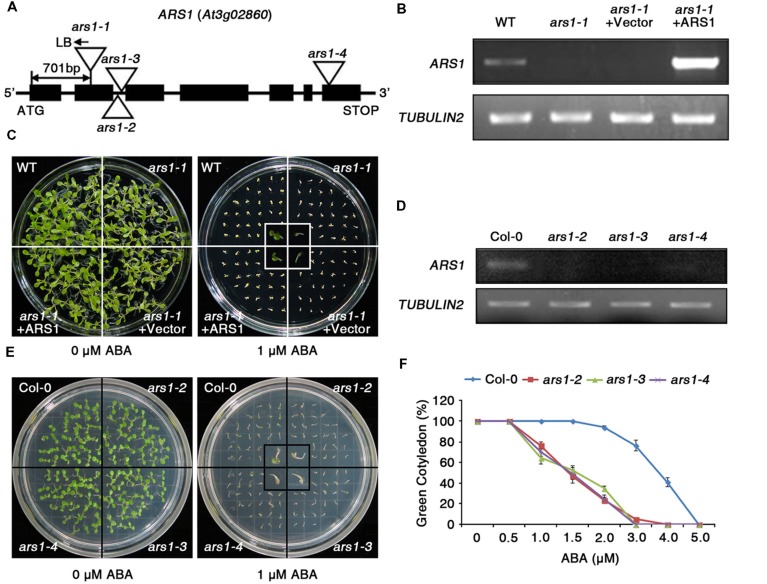
The RD29Apro::LUC transgene has been widely adapted to screen for ABA- or abiotic stress-responsive mutants from large populations of Arabidopsis C24 ecotype plants with T-DNA insertions (Ishitani et al., 1997; Zhu et al., 2008). Here, the T2 progeny were screened for enhanced sensitivity of seed germination to ABA compared to wild type (WT, C24 ecotype), and the phenotypes were further identified in T3 progeny. The ars1-1 mutant displayed hypersensitivity to ABA with reduced seed germination and retarded emergence of green cotyledons compared to WT (Supplementary Figure S1). Using TAIL-PCR, we found that the ars1-1 mutant had a T-DNA insertion in ARS1 (At3g02860), located in the second exon, 701 bp upstream of the ATG translation start site. Genotypic analysis showed the T-DNA insertion in ARS1 segregated with the ars1-1 mutant phenotype, as all tested F2 homozygotes exhibited identical phenotypes (n = 100, data not shown).
ARS1 encodes a zinc ion-binding protein (Figure 1A), but its biological functions have not been reported yet. To elucidate the functional roles of ARS1 in the ABA response, we generated a construct overexpressing ARS1 under the control of the cauliflower mosaic virus 35S promoter, and transformed this construct into the ars1-1 mutant (ars1-1+ARS1). RT-PCR analysis revealed that the ars1-1+ARS1 plants had high levels of ARS1 transcripts compared to WT, but ARS1 transcripts were absent in both ars1-1 and ars1-1 harboring the empty vector (ars1-1+Vector) (Figure 1B). In addition, ars1-1+ARS1 plants showed rescue of the ABA hypersensitivity of ars1-1 and displayed WT phenotypes in seed germination, while ars1-1+Vector showed germination defects similar to those of ars1-1 (Figure 1C).
FIGURE 1.
The absence of ARS1 leads to ABA-hypersensitive seed germination. (A) T-DNA insertions in the ars1 mutant alleles. (B) RT-PCR analysis of ARS1 expression in WT, ars1-1, ars1-1+Vector, and ars1-1+ARS1 plants. TUBULIN2 serves as a control for RNA integrity. (C) Comparison of seed germination between the WT, ars1-1, ars1-1+Vector, and ars1-1+ARS1 plants exposed to 0 or 1 μM ABA. The photograph shows Arabidopsis seedlings after 5 days of ABA treatment. (D) Expression of ARS1 in WT Col-0 and ars1 allelic mutants determined by RT-PCR. TUBULIN2 serves as a control for RNA integrity. (E) Comparison of seed germination between the WT Col-0 and ars1 mutants exposed to 0 or 1 μM ABA. The photograph shows Arabidopsis seedlings after 5 days of ABA treatment. (F) Quantification of green cotyledons in WT Col-0 and ars1 mutants grown on various concentrations of ABA for 5 days. The data represent the means ± SE of three independent experiments, with 50 seeds per experiment.
To identify the roles of ARS1 in response to ABA in multiple alleles, we obtained three additional, different T-DNA alleles, ars1-2, ars1-3, and ars1-4, in the Arabidopsis Col-0 background (Figure 1A). ARS1 transcripts were absent in ars1-2 and ars1-3 plants compared to Col-0 plants, but were barely detectable in ars1-4 (Figure 1D). All ars1 allelic mutants in the Col-0 background also showed an ABA-sensitive phenotype with retarded emergence of green cotyledons compared to Col-0 plants (Figures 1E,F). Root length also provided a measure of ABA sensitivity (Supplementary Figure S2). Four-day-old plants grown on MS medium were transferred to MS medium containing ABA, and root growth was monitored 11 days later. At 30 and 40 μM ABA, root growth in ars1-2 and ars1-3 plants lacking ARS1 transcripts was significantly lower compared to that in Col-0 plant; however ars1-4 mutants showed no significant differences from Col-0. Although the ABA-dependent root growth phenotypes of ars1-4 marginally differed from the germination phenotypes, the data are consistent with the differences in expression of ARS1 in those mutants, suggesting that ARS1 may also affect post-germination plant growth (Figure 1D, Supplementary Figure S2). These results indicate that ARS1 is essential for breaking seed dormancy in germination, which is inhibited mainly by ABA.
Characterization of ARS1
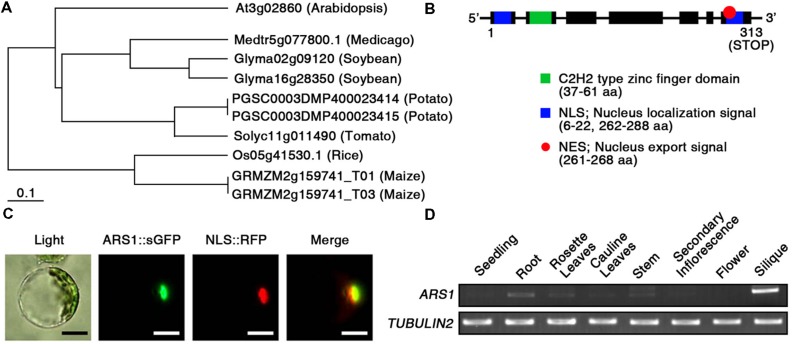
To predict the potential function of ARS1 in plants, we compared the protein sequences of Arabidopsis ARS1 and its homologs in other plants, retrieved by BLAST-P search. Phylogenetic analysis using the BAR Expressolog Tree program1 revealed that ARS1 had the highest sequence similarity (53.1%) to a zinc finger protein in Medicago truncatula, and also shared 32.4–49.9% similarity with its homologs in plant species such as Glycine max, Solanum tuberosum, Lycopersicon esculentum, Oryza sativa, and Zea mays (Figure 2A). The tree clearly divided ARS1 homologs from dicots or monocots into different branches. However, their biological functions remain elusive in plants. ARS1 encoded a 313-amino acid protein with predicted molecular mass of 35.2 kDa2 (Figure 2B). ARS1 protein possesses a nuclear localization signal (NLS; 6–22 a.a.), a C2H2-type zinc finger domain (37–61 a.a.) in the N-terminal region, a nuclear export signal (NES; 261–268 a.a.), and another NLS (262–288 a.a.) in the C-terminus. C2H2 zinc finger proteins are classified three sets (A, B, and C), and set C is sub-classified into three distinguishable subsets such as C1, C2, and C3 (Englbrecht et al., 2004). We found that ARS1 belongs to subfamily of C3 subset (Supplementary Figure S3). It indicates that ARS1 as a C2H2 zinc finger protein may be located either in the nucleus or cytoplasm and may translocate under certain conditions. To reveal the intracellular localization of ARS1, we used protoplast transient expression co-transforming an ARS1::sGFP construct and a chimeric construct containing the NLS domain fused to RFP (NLS::RFP) as a nuclear marker (Lee et al., 2001). As shown in Figure 2C, the intracellular distribution of green and red fluorescent signals overlapped, indicating that ARS1 localizes to the nucleus. We also examined the expression of ARS1 transcripts in different organs by RT-PCR analysis (Figure 2D). Interestingly, ARS1 transcripts accumulated to high levels in silique and root, which show strong effects of ABA. ARS1 expression was slightly higher in rosette leaves and stem than in highly proliferating organs such as seedlings, secondary inflorescences, and flowers.
FIGURE 2.
Characterization of ARS1. (A) Phylogenetic tree of ARS1 homologs, generated using the BAR Expressolog Tree program. (B) ARS1 encodes an unknown protein with two NLS domains, an NES domain, and a C2H2 type zinc-finger domain. (C) Intracellular localization of the ARS1 protein in the nucleus. Protoplasts prepared from Arabidopsis seedlings were co-transformed with ARS1::sGFP and NLS::RFP. The transformed protoplasts were examined by fluorescence microscopy 12 h after transformation. Green and red images are GFP and RFP signals, respectively. Bars indicate 20 μm. (D) RT-PCR analysis of ARS1 expression in Arabidopsis tissues. TUBULIN2 serves as a control for RNA integrity.
ARS1 Represses ABA-Induced ROS Production
Abscisic acid shows a strong relationship with abiotic stress tolerance, especially with drought tolerance via regulation of stomatal closure to reduce water loss (Lim et al., 2015). Thus, we first examined water loss, but found that water loss assays showed no significant differences between Col-0 and ars1-2 plants in the absence or presence of ABA (Supplementary Figure S4).
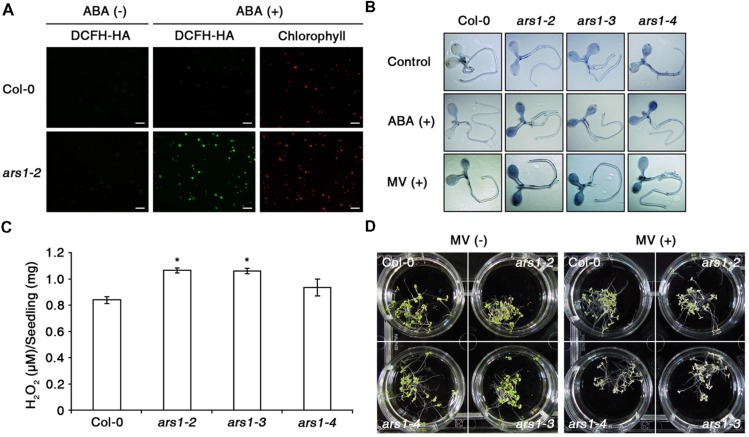
Next, we examined ABA-induced ROS production in Col-0 and ars1-2 protoplasts using an ROS-sensitive, cell-permeable fluorescent dye, 2′,7′-dichlorofluorescin diacetate (DCFH-DA) (Wang and Joseph, 1999; Pei et al., 2000). As shown in Figure 3A, the fluorescent signals increased slightly in response to ABA treatment in Col-0 plants, consistent with previous reports (Wang and Song, 2008). Interestingly, the signals increased dramatically in ars1-2 plants exposed to ABA. In addition, ABA-induced ROS production in ars1-3 and ars1-4 was similar to that in ars1-2 mutants (data not shown). We further examined ABA- or MV- induced ROS induction in planta by staining the superoxide anion with nitroblue tetrazolium (NBT) (Figure 3B). Both ABA and MV increased the dark purple staining, indicating increased superoxide levels in all ars1 mutant plants, while those in Col-0 plants remained consistent in the absence or presence of ABA or MV. Interestingly, ars1 mutants showed more staining than Col-0 plants, even in the absence of treatments, indicating that the absence of ARS1 may increase ROS levels in plants. To confirm the ROS accumulation in ars1 mutants, we compared hydrogen peroxide contents of Col-0 and ars1 mutants (Figure 3C). As shown in Figure 3B, hydrogen peroxide contents in all ars1 mutants were higher than those in Col-0 plants, even in the absence of treatments. The ars1-2 and ars1-3 mutants showed significantly higher ROS than ars1-4, which is also consistent with the different levels of ARS1 transcripts among ars1 mutants (Figure 1D). Furthermore, ars1 mutants displayed MV-sensitive phenotypes, showing bleaching of leaves compared to Col-0 (Figure 3D).
FIGURE 3.
ARS1 represses ABA-induced ROS production. (A) Representative images of ROS production indicated by the fluorescent dye DCFH-DA. Protoplasts from 3 week-old-seedlings of WT Col-0 and ars1-2 mutants were treated with ABA (100 μM) for 1 h. Bars indicate 100 μm. (B) WT Col-0 and ars1 mutants seedlings under ABA (100 μM) and MV (5 μM) treatment stained with NBT. The dark blue color indicates insoluble formazan deposits that are produced when NBT reacts with superoxide. (C) Internal H2O2 production assays in WT Col-0 and ars1 mutants. Experiments were performed twice, and approximate fluorescence was measured by excitation at 530 nm and emission at 590 nm. Asterisks represent significant differences from the WT (∗p-value ≤0.05, Student’s t-test). (D) Photographs of seedlings that were grown on MS medium for 4 days, and transferred to MS medium with or without MV (5 μM) for another 3 days.
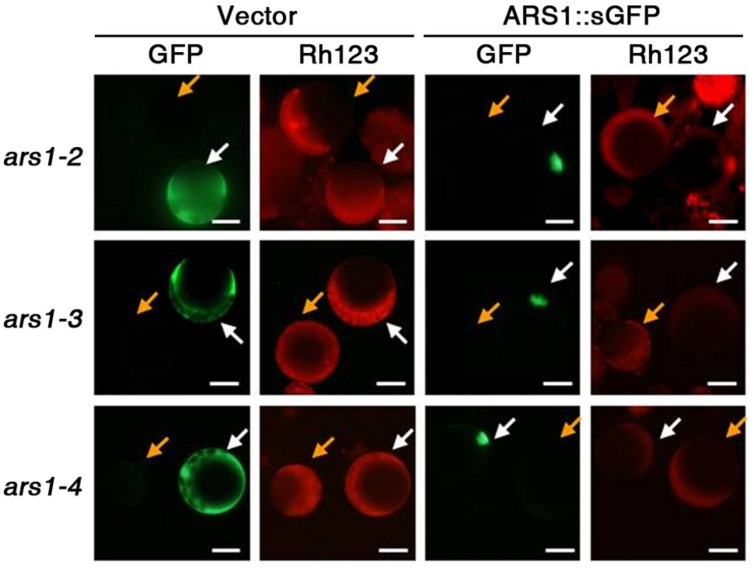
To determine whether ARS1 directly affects ABA-induced ROS production, we carried out complementation tests in protoplasts using the fluorescent dye dihydrorhodamine123 (Rh123) to monitor ROS production. Rh123 becomes the fluorescent chromophore Rh123 upon oxidation by ROS (Schulz et al., 1996). Empty GFP vector and ARS1::sGFP constructs were independently transformed into protoplasts isolated from the ars1 mutants, and approximately 40% of the protoplasts showed expression of both constructs (data not shown). After treatment with ABA for 1 h and then with Rh123 for 15 min, we found that ARS1::sGFP clearly targeted to the nucleus but the empty vector produced a GFP signal in the cytoplasm of protoplasts from all ars1 mutants (Figure 4). In addition, the fluorescent signals from Rh123 were greatly reduced in all protoplasts transformed with ARS1::sGFP compared to those transformed with empty vector (Figure 4). These results suggest that ARS1 acts positively in repressing ABA-induced ROS production.
FIGURE 4.
ARS1 inhibits ABA-induced ROS production. Protoplasts were isolated from ars1 mutants seedlings transformed with the empty sGFP vector (Vector) or ARS1::sGFP. Twelve hours after transformation, protoplasts were treated with ABA (100 μM) for 1 h and stained with dihydrorhodamine123 (Rh123) for 15 min. The images are green (GFP) and red (Rh123) fluorescence images of one aliquot of protoplasts. Bars indicate 20 μm. Yellow arrows point to the non-expressing protoplasts, white arrows point to the protoplasts expressing sGFP and ARS1::sGFP.
ARS1 Deficiency Reduces Expression of SOD
To identify how ARS1 regulates ABA responses, we first used qRT-PCR to analyze the transcript levels of ARS1 in response to ABA. ARS1 transcripts slightly increased (1.2-fold induction) in response to ABA treatments for 1 and 3 h (Supplementary Figure S5A). Englbrecht et al. (2004) found that C2H2 zinc finger proteins act as transcriptional regulators in conserved biological processes in response to stress. Accordingly, we examined the expression of RD29A genes in ars1 mutants to explore the possible role of ARS1 in the ABA signaling pathway. RD29A expression is highly induced by ABA as a stress-responsive marker gene but does not change in response to H2O2 treatment (Trouverie et al., 2008). However, RD29A transcript accumulation did not show any significant differences in the ars1 mutants compared to that in Col-0 plants either in the absence or presence of ABA (Supplementary Figure S5B). Thus, ARS1 activity may not be necessary for regulation at the transcriptional level in the ABA signaling pathway.
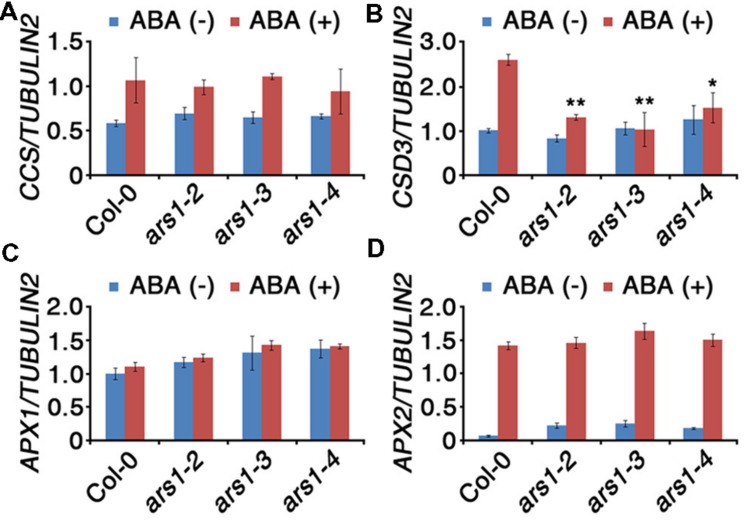
Based on the effect of ARS1 on ABA-induced ROS production, we investigated the transcripts encoding ROS-scavenging enzymes in ars1 mutants (Figure 5). SOD catalyze the dismutation of O2- to O2 and H2O2, which is subsequently reduced to water by CAT and APX (Mittler et al., 2004). We used qRT-PCR to measure the transcript levels of two SOD genes (CCS and CSD3) and two APX genes (APX1 and APX2) in the absence or presence of ABA. Transcripts of CCS (COPPER/ZINC SUPEROXIDE DISMUTASE), APX1, and APX2 did not show any significant differences between Col-0 and ars1 mutants and even in response to ABA treatment (Figures 5A,C,D). However, transcripts of CSD3, encoding a copper/zinc superoxide dismutase 3, significantly decreased in all ars1 mutants in response to ABA treatment (Figure 5B). These results indicate that ARS1 represses ABA-induced ROS accumulation via inhibiting SOD transcripts, especially CSD3.
FIGURE 5.
Expression of ROS-scavenging genes in response to ABA treatment. Total RNA was isolated from 10-d-old seedlings of WT and ars1 mutants with or without ABA (100 μM) treatment for 3 h. Relative transcript levels of CCS (A), CSD3 (B), APX1 (C), and APX2 (D) in Col-0 and ars1 mutants determined by qRT-PCR. Transcript levels were normalized to those of TUBULIN2. Bars represent mean ± SD of three biological replicates with three technical replicates each. Asterisks represent significant differences from the Col-0 (∗; 0.01 < p-value ≤ 0.05, ∗∗; p-value ≤ 0.01, Student’s t-test).
ARS1 Translocates from the Nucleus to the Cytoplasm in response to ABA and Oxidative Stress
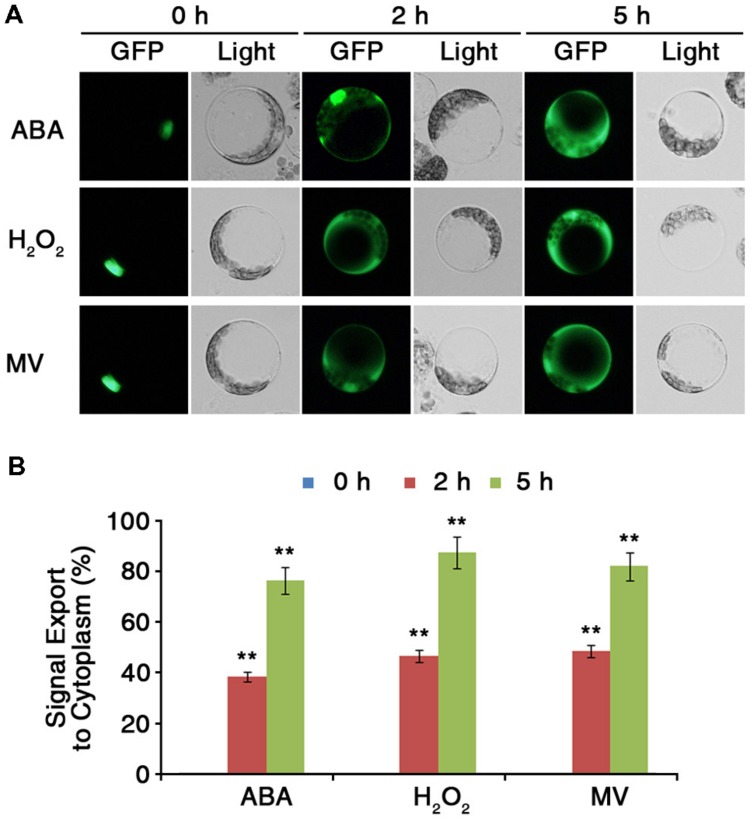
Protein sequence analysis revealed that ARS1 has a putative NES motif at its C terminus (Figure 2B). The presence of the putative NES signal suggested that ARS1 could be exported from the nucleus in response to certain stress conditions and led us to investigate the changes of subcellular localization of ARS1 in Arabidopsis cells. To analyze the subcellular localization of ARS1 under stress conditions, we first transiently expressed ARS1::sGFP in Arabidopsis (Col-0) protoplast cells. Twelve hours after transformation of ARS1::sGFP into protoplasts, we treated the protoplasts with ABA, H2O2, or methyl viologen (MV) as ROS triggers. In the absence of stressors, ARS1 clearly localized to the nucleus (Figures 2C and 6A, Supplementary Figure S5). Surprisingly, nuclear-localized ARS1 was abundantly translocated to the cytoplasm in response to ABA, H2O2, and MV. (Figure 6, Supplementary Figure S6). These translocation patterns of ARS1 in protoplasts markedly increased as the stress duration increased up to 5 h (Figure 6B). These results indicate that the putative NES motif is likely important for ARS1 function in stress responses. Together, this evidence suggests that ARS1 exists as an inactive form in the nucleus, but changes its localization to the cytoplasm as a result of ROS signals induced by ABA and other stresses to repress ABA/stress-induced ROS production in plant cells.
FIGURE 6.
ARS1 is translocated from the nucleus to the cytoplasm in response to ABA and oxidative stresses. (A) Protoplasts were isolated from leaves of 3-week-old WT Col-0 plants transformed with ARS1::sGFP. Twelve hours after transformation, protoplasts were treated with ABA (100 μM), H2O2 (1.5 mM) or MV (3 μM) for the indicated times. Bars indicate 20 μm. (B) Percentage of ARS1 signal exported to the cytoplasm in response to ABA, H2O2, and MV shown in (A). Bars represent mean ± SD of three biological replicates with three technical replicates each (n = 200). Asterisks represent significant differences from the 0 h(∗∗p-value ≤0.01, Student’s t-test).
Discussion
Seed germination involves extensive crosstalk between phytohormones and secondary messengers. ABA and gibberellin (GA) act antagonistically in germinating seeds, and secondary messengers such as ROS and Ca2+ might be also responding differently in germination. In dormant seeds, high levels of ABA in seed coats repress germination via expression of the DELLA gene RGL2 and the ABA biosynthesis gene ABA1, thus triggering induction of ABI5 (Lee et al., 2010). Seed dormancy can be broken by environmental cues such as exposure to cold, oxygen, and water (Finch-Savage and Leubner-Metzger, 2006). In germinating seeds, endogenous ROS and cytosolic Ca2+ concentrations increase to promote germination, counteracting the effects of ABA. Exogenous ROS and Ca2+ treatments also enhance seed germination (El-Maarouf-Bouteau and Bailly, 2008; Kong et al., 2015). However, either low or excess ROS delay or inhibit germination, indicating that ROS homeostasis, termed the “oxidative window of germination,” is essential for breaking seed dormancy (Bailly et al., 2008). In this study, we identified ARS1, encoding an uncharacterized zinc finger domain protein, as essential for seed germination to escape ABA-induced dormancy in Arabidopsis (Figures 1 and 2, Supplementary Figure S1). Interestingly, ars1 mutants displayed higher ROS accumulation either in the absence or presence of ABA, indicating that ARS1 may act to repress ROS production (Figures 3 and 4). Furthermore, this finding also suggests that ARS1 may regulate the “oxidative window of germination” to prevent excess ROS accumulation during the period of breaking seed dormancy. Using in silico analysis, we found 211 C2H2-type zinc finger proteins that constitute the most abundant family of putative transcriptional regulators in Arabidopsis3. Several of these proteins act in abiotic stress responses, in particular ABA or oxidative stress signaling. SAZ (SA- AND ABA-DOWNREGULATED ZINC FINGER GENE), ZFP3 (ZINC FINGER PROTEIN 3), AZF1 (ARABIDOPSIS C2H2 ZINC FINGER PROTEIN 1), and AZF2 act as negative regulators in ABA signaling during seed germination and early seedling development (Jiang et al., 2008; Kodaira et al., 2011; Joseph et al., 2014). SAP12 (STRESS-ASSOCIATED PROTEIN 12) contains two AN1 zinc fingers and conformational changes due to redox states of cysteine residues located between the zinc finger structures modulate its activity (Ströher et al., 2009). ZAT10 (ZINC-FINGER OF ARABIDOPSIS 10) is phosphorylated by MPK3 and MPK6 and is involved in ROS-dependent ABA signaling (Mittler et al., 2006). The high conservation of functional motifs among the C2H2 zinc finger proteins suggests that these proteins may have similar molecular functions with respect to transcriptional regulation in diverse biological processes.
Reactive oxygen species act as important signaling molecules and control various biological processes including germination, growth, development, and abiotic stress responses. Diverse abiotic stresses lead to production of toxic levels of ROS, which causes oxidative damage to organelles such as chloroplasts and mitochondria in plant cells (Bailey-Serres and Mittler, 2006; Foyer and Noctor, 2009). To balance the accumulation of toxic ROS, plants have efficient, well-conserved mechanisms for the removal of ROS from cells, including both enzymatic and non-enzymatic ROS scavenging antioxidant systems (Mittler et al., 2004; Bailey-Serres and Mittler, 2006; Foyer and Noctor, 2009). For instance, SOD, CAT, and APX serve as ROS-scavenging enzymes. Abiotic stresses as well as ABA induce production of ROS such as oxygen radicals and hydrogen peroxide via plasma membrane-localized NADPH oxidases (Kwak et al., 2003). ROS accumulation in the ars1 mutant plants may be caused by repressed expression of the SOD gene CDS3 (Figure 5). Our results indicate that the function of ARS1 in stress tolerance may be associated with the regulation of antioxidant ability. However, the direct mechanisms by which ARS1 regulates CDS3 expression are still elusive. To better understand the mechanisms, future research should examine the interaction between ARS1, ROS scavenging systems, and antioxidant enzyme activity.
Reactive oxygen species are generated in various subcellular organelles including chloroplasts, mitochondria, and peroxisomes, and they trigger changes in the nuclear transcriptome during stress (Apel and Hirt, 2004; Suzuki et al., 2012). ROS signaling occurs via interlinked exchanges between two distinct pathways: retrograde (organelle-to-nucleus) and anterograde (nucleus-to-organelle) signaling, which might be involved in acclimation, adaptation, or resistance against stresses (Suzuki et al., 2012). Disruption of ROS homeostasis can occur in chloroplasts or mitochondria, from which signals are transmitted to the nucleus via retrograde signaling cascades. We found that ARS1 in the nucleus was translocated to the cytoplasm upon exposure to ABA or oxidative stress, presumably in response to ROS signals (Figure 6). We cannot conclude that a protein is involved in retrograde regulation just because it exists in two different locations or relocalizes to another subcellular compartment depending on conditions. However, ARS1 translocates from the nucleus to cytoplasm in an ROS-dependent manner (Figure 6). This suggests that ARS1 may affect ROS-dependent anterograde signaling between the nucleus and cytoplasm under stress conditions or in response to ABA.
Conclusion
The phytohormone ABA regulates important physiological processes, and is closely related with the accumulation of intracellular ROS to transfer signals triggered in diverse physiological and environmental cues. In this study, we isolated and designated an ars1 mutant from large populations of Arabidopsis with T-DNA insertions as an ABA hypersensitive mutant. We identified that ARS1 encodes a C2H2 type zinc finger domain protein and may play as a positive regulator for seed germination and maintenance of ROS homeostasis in response to ABA and oxidative stress, which trigger the induction of toxic ROS in cells, via the regulation of a gene for superoxide dismutase (CSD3). Interestingly, we also demonstrated that nuclear-localized ARS1 is translocated to the cytoplasm in response to ABA or oxidative stress. Translocation of ARS1 induced by ROS signals may help clarify the role of ROS-dependent anterograde signaling pathways that underlie plant stress responses. Taken together, our results suggest that ARS1 is essential to modulate seed germination and ROS homeostasis in response to ABA and oxidative stress in Arabidopsis.
Author Contributions
DB, J-YC, and D-JY designed the experiments. DB performed most of the experiments, and J-YC and D-JY wrote the manuscript. DK, SL, and MK discussed and commented on the results and manuscripts. SK, BP, H-JL, HH, and HC performed some of the experiments. D-JY provided funding for research work as corresponding author.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Research Foundation funded by the Korean Government (MSIP; 2013R1A2A1A01005170) and the Next-Generation BioGreen21 Program (SSAC, grant#: PJ011051), Rural Development Administration Republic of Korea; as well as the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2013R1A1A2011511) to DB.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00963
References
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Mittler R. (2006). The roles of reactive oxygen species in plant cells. Plant Physiol. 141 311 10.1104/pp.104.900191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C., El-Maarouf-Bouteau H., Corbineau F. (2008). From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. C. R. Biol. 331 806–814. 10.1016/j.crvi.2008.07.022 [DOI] [PubMed] [Google Scholar]
- Bournonville C. F., Díaz-Ricci J. C. (2011). Quantitative determination of superoxide in plant leaves using a modified NBT staining method. Phytochem. Anal. 22 268–271. 10.1002/pca.1275 [DOI] [PubMed] [Google Scholar]
- Brandt B., Brodsky D. E., Xue S., Negi J., Iba K., Kangasjärvi J., et al. (2012). Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. U.S.A. 109 10593–10598. 10.1073/pnas.1116590109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61 651–679. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- Drerup M. M., Schlücking K., Hashimoto K., Manishankar P., Steinhorst L., Kuchitsu K., et al. (2013). The Calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant. 6 559–569. 10.1093/mp/sst009 [DOI] [PubMed] [Google Scholar]
- El-Maarouf-Bouteau H., Bailly C. (2008). Oxidative signaling in seed germination and dormancy. Plant Signal. Behav. 3 175–182. 10.4161/psb.3.3.5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englbrecht C. C., Schoof H., Böhm S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5:39 10.1186/1471-2164-5-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage W. E., Leubner-Metzger G. (2006). Seed dormancy and the control of germination. New Phytol. 171 501–523. 10.1111/j.1469-8137.2006.01787.x [DOI] [PubMed] [Google Scholar]
- Foyer C. H., Noctor G. (2009). Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11 861–905. 10.1089/ars.2008.2177 [DOI] [PubMed] [Google Scholar]
- Foyer C. H., Shigeoka S. (2011). Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 155 93–100. 10.1104/pp.110.166181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T. S., Laloi C., Minkov I. N., Shulaev V., et al. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141 436–445. 10.1104/pp.106.078717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev T. S., Van Breusegem F., Stone J. M., Denev I., Laloi C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28 1091–1101. 10.1002/bies.20493 [DOI] [PubMed] [Google Scholar]
- Ishitani M., Xiong L., Stevenson B., Zhu J. K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9 1935–1949. 10.1105/tpc.9.11.1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C. J., Aono M., Tamaoki M., Maeda S., Sugano S., Mori M., et al. (2008). SAZ, a new SUPERMAN-like protein, negatively regulates a subset of ABA-responsive genes in Arabidopsis. Mol. Genet. Genomics 279 183–192. 10.1007/s00438-007-0306-1 [DOI] [PubMed] [Google Scholar]
- Jin J. B., Kim Y. A., Kim S. J., Lee S. H., Kim D. H., Cheong G. W., et al. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph M. P., Papdi C., Kozma-Bognár L., Nagy I., López-Carbonell M., Rigó G., et al. (2014). The Arabidopsis ZINC FINGER PROTEIN3 interferes with abscisic acid and light signaling in seed germination and plant development. Plant Physiol. 165 1203–1220. 10.1104/pp.113.234294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpiński S., Szechyńska-Hebda M., Wituszyńska W., Burdiak P. (2013). Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 36 736–744. 10.1111/pce.12018 [DOI] [PubMed] [Google Scholar]
- Kim C., Meskauskiene R., Apel K., Laloi C. (2008). No single way to understand singlet oxygen signalling in plants. EMBO Rep. 9 435–439. 10.1038/embor.2008.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaira K. S., Qin F., Tran L. S., Maruyama K., Kidokoro S., Fujita Y., et al. (2011). Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiol. 157 742–756. 10.1104/pp.111.182683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D., Ju C., Parihar A., Kim S., Cho D., Kwak J. M. (2015). Arabidopsis glutamate receptor homolog3.5 modulates cytosolic Ca22+ level to conteract effect of abscisic acid in seed germination. Plant Physiol. 167 1630–1642. 10.1104/pp.114.251298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J. M., Mori I. C., Pei Z. M., Leonhardt N., Torres M. A., Dangl J. L., et al. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22 2623–2633. 10.1093/emboj/cdg277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. P., Piskurewicz U., Turečková V., Strnad M., Lopez-Molina L. (2010). A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. U.S.A. 107 19108–19113. 10.1073/pnas.1012896107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. J., Kim D. H., Kim Y. W., Hwang I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13 2175–2190. 10.2307/3871501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. W., Baek W., Jung J., Kim J. H., Lee S. C. (2015). Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 16 15251–15270. 10.3390/ijms160715251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324 1064–1068. 10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- Mittler R., Kim Y., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., et al. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580 6537–6542. 10.1016/j.febslet.2006.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 490–498. 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Neill S., Desikan R., Hancock J. (2002). Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 5 388–395. 10.1016/S1369-5266(02)00282-0 [DOI] [PubMed] [Google Scholar]
- Op den Camp R. G., Przybyla D., Ochsenbein C., Laloi C., Kim C., Danon A., et al. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15 2320–2332. 10.1105/tpc.014662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324 1068–1071. 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z. M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406 731–734. 10.1038/35021067 [DOI] [PubMed] [Google Scholar]
- Raghavendra A. S., Gonugunta V. K., Christmann A., Grill E. (2010). ABA perception and signalling. Trends Plant Sci. 15 395–401. 10.1016/j.tplants.2010.04.006 [DOI] [PubMed] [Google Scholar]
- Sato A., Sato Y., Fukao Y., Fujiwara M., Umezawa T., Shinozaki K., et al. (2009). Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 424 439–448. 10.1042/BJ20091221 [DOI] [PubMed] [Google Scholar]
- Schulz J. B., Weller M., Klockgether T. (1996). Potassium deprivation-induced apoptosis of cerebellar granule neurons: a sequential requirement for new mRNA and protein synthesis, ICE-like protease activity, and reactive oxygen species. J. Neurosci. 16 4696–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D., Mahajan N., Sah S., Nath S. K., Paudyal B. (2014). Oxidative stress and its biomarkers in systemic lupus erythematosus. J. Biomed. Sci. 21:23 10.1186/1423-0127-21-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ströher E., Wang X. J., Roloff N., Klein P., Husemann A., Dietz K. J. (2009). Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in Arabidopsis thaliana. Mol. Plant. 2 357–367. 10.1093/mp/ssn084 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Koussevitzky S., Mittler R., Miller G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35 259–270. 10.1111/j.1365-3040.2011.02336.x [DOI] [PubMed] [Google Scholar]
- Suzuki N., Miller G., Salazar C., Mondal H. A., Shulaev E., Cortes D. F., et al. (2013). Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell 25 3553–3569. 10.1105/tpc.113.114595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouverie J., Vidal G., Zhang Z., Sirichandra C., Madiona K., Amiar Z., et al. (2008). Anion channel activation and proton pumping inhibition involved in the plasma membrane depolarization induced by ABA in Arabidopsis thaliana suspension cells are both ROS dependent. Plant Cell Physiol. 49 1495–1507. 10.1093/pcp/pcn126 [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106 17588–17593. 10.1073/pnas.0907095106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., El-Shetehy M., Shine M. B., Yu K., Navarre D., Wendehenne D., et al. (2014). Free radicals mediate systemic acquired resistance. Cell Rep. 7 348–355. 10.1016/j.celrep.2014.03.032 [DOI] [PubMed] [Google Scholar]
- Wang H., Joseph J. A. (1999). Quantifying cellular oxidative stress by dichlorofluorescein assay using microplate reader. Free Radic. Biol. Med. 27 612–616. 10.1016/S0891-5849(99)00107-0 [DOI] [PubMed] [Google Scholar]
- Wang P., Song C. P. (2008). Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol. 178 703–718. 10.1111/j.1469-8137.2008.02431.x [DOI] [PubMed] [Google Scholar]
- Wasilewska A., Vlad F., Sirichandra C., Redko Y., Jammes F., Valon C., et al. (2008). An update on abscisic acid signaling in plants and more. Mol. Plant. 1 198–217. 10.1093/mp/ssm022 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61 672–685. 10.1111/j.1365-313X.2009.04092.x [DOI] [PubMed] [Google Scholar]
- Zhu J., Jeong J. C., Zhu Y., Sokolchik I., Miyazaki S., Zhu J. K., et al. (2008). Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. U.S.A. 105 4945–4950. 10.1073/pnas.0801029105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.