Abstract
Chikungunya virus (CHIKV) is a re-emerging alphavirus which causes severe and prolonged arthralgic febrile illness. The recent global spread of the virus and lack of approved therapeutic options makes it imperative to gain greater insight into the molecular mechanisms underlying CHIKV pathogenesis, in particular host factors recruited by the virus. In the current study, we identify sphingosine kinase 2 (SK2) as a CHIKV host factor co-localized with the viral replication complex (VRC) during infection. SK2 was demonstrated to co-localize with viral RNA and nonstructural proteins. Targeted impairment of SK2 expression or function significantly inhibited CHIKV infection. Furthermore, affinity purification-mass spectrometry studies revealed that SK2 associates with a number of proteins involved in cellular gene expression specifically during viral infection, suggesting a role in replication. Collectively these results identify SK2 as a novel CHIKV host factor.
Keywords: chikungunya virus, nsP3, sphingosine kinase 2, viral replication complex
Introduction
Chikungunya virus (CHIKV) is a mosquito-borne pathogen of the genus alphavirus within the family Togaviridae. The virus is responsible for a disease characterized by high fever and joint pain, which can potentially be recurring for years.1,2,3,4 Recently, CHIKV has emerged as a global health concern having made its way to the western hemisphere.5 Despite all of this there remain no effective therapeutic options available. Greater understanding of the mechanisms underlying pathogenesis will be useful in the design and development of effective CHIKV therapeutics.
Recent studies have aided our understanding of the mechanisms underlying CHIKV pathogenesis and also host restriction factors to infection, including proteins such as bone marrow stromal antigen 2 (BST-2) and interferon-stimulated gene 15 (ISG15).6,7,8,9 Proteomic studies examining either vector or mammalian host proteins have provided new insights into the biological processes modulated during the course of virus infection.10,11,12,13 Similarly, studies identifying viral host interaction partners have also provided greater understanding.14,15,16,17,18 Still there remains much to be understood about viral pathogenesis, including the cellular factors that are recruited early during infection, as they may provide novel targets for therapeutic intervention.
The sphingosine kinases (SKs), SK1 and SK2, are lipid kinases that catalyze the production of sphingosine-1 phosphate (S1P) from sphingosine.19 The SK/S1P pathway has been demonstrated to play critical roles in a variety of cellular processes, including cell growth, differentiation, and host immunity.20,21 Not surprisingly the SK/S1P pathway has been shown to play a role during viral infections as well. Studies on respiratory syncytial virus (RSV) revealed a positive link between SK activity and S1P production during infection.22 SK1 has been reported to be a key modulator of viral infections.23 For example, recent reports have shown that increased expression of SK1 is associated with increased susceptibility to influenza A and measles virus (MV) infection.24,25,26 SK1 has also been shown to play a negative role during dengue virus (DENV) and bovine viral diarrhea virus (BVDV) infection.27,28 In sharp contrast, very little is known about SK2 function, in particular as it relates to viral infection. Two recent reports however indicate that SK2 similar to SK1 plays a role during viral infection. The first study revealed that SK2 is associated with maintenance of Kaposi's sarcoma-associated herpesvirus (KSHV) latency, while the second study demonstrated that lipid peroxidation regulated in part through SK2, can restrict hepatitis C virus (HCV) replication.29,30 Taken together these studies clearly demonstrate a role for SK2 during viral infection.
In the current study, we observe that knockdown of SK2 or small molecule inhibitor treatment significantly impaired CHIKV infection. Furthermore, we show that CHIKV infection induced co-localization of SK2 with viral RNA and viral proteins indicating an association with the viral replication complex (VRC). In over-expression studies, nsP3 alone was found to co-localize with SK2, providing mechanistic insight into SK2 association with the VRC. Finally, affinity purification-mass spectrometry (AP-MS) studies revealed that during infection SK2 is associated with a number of proteins that function in gene expression, suggesting a role during infection. Taken together, we identify SK2 as an essential host factor during CHIKV infection.
Materials and methods
Cell culture
HeLa, U2OS, HepG2, MCF10A, and BHK-21 cells were all purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies, Carlsbad, CA, USA) supplemented with 10% inactivated fetal bovine serum (FBS) (GE Healthcare, Piscataway, NJ, USA). Human skeletal muscle cells (SkMCs) (Lonza, Walkersville, MD, USA) were cultured in skeletal muscle basal medium (SkBM) (Lonza, Walkersville, MD, USA) supplemented with SingleQuots Kit (Lonza, Walkersville, MD, USA) which consisted of 0.5 mL human epidermal growth factor (hEGF), 0.5 mL fetuin, 5.0 mL bovine serum albumin (BSA), 0.5 mL dexamethasone, 5.0 mL insulin, and 0.5 mL gentamicin/amphotericin-B.
Plasmids, compounds, and viruses
CHIKV plasmids GFP-nsP1/2/3/4 and GFP-fused capsid were kindly provided by Dr Subhash G Vasudevan (Duke-NUS Graduate Medical School, Singapore). The pCMV6/Myc-DDK SK2 was purchased from OriGene (Rockville, MD, USA). SKI-II and S1P were purchased from Tocris and Cayman Chemical, respectively. ABC294640 was kindly provided by Dr Charles D Smith (MUSC, Charleston, SC, USA). CHIKV (clone 181/25) was propagated from a USAMRIID collection.
RNA interference
Small interfering RNAs (siRNAs) targeting SK1 (siSK1) (5′-AAC UAC UUC UGG AUG GUC ATT-3′) and SK2 (siSK2) (5′-CAC UGC CCU CAC CUG UCU GTT-3′) were purchased from Life Technologies (Carlsbad, CA, USA) and Qiagen (Germantown, MD, USA) respectively. Additionally, the scramble siRNA controls were purchased from Life Technologies (Carlsbad, CA, USA) and Qiagen (Germantown, MD, USA). Cells were transfected with 15 nM of siSK1 or 50 nM siSK2 using HiPerFect (Qiagen, Germantown, MD, USA) according to the manufacturer's instructions. Knockdown of SK1 and SK2 was confirmed by quantitative real-time polymerase chain reaction (qRT-PCR).
Quantitative real-time polymerase chain reaction
Cells were lysed in either RLT buffer (Qiagen, Germantown, MD, USA) or supernatants prepared in Trizol reagent (Life Technologies, Carlsbad, CA, USA). Cell lysates were prepared using the RNeasy Kit (Qiagen, Germantown, MD, USA). In these experiments, 50 ng of isolated RNA was prepared for analysis using the Taqman RNA-to-Ct 1-Step Kit (Life Technologies, Carlsbad, CA, USA) and analyzed on an ABI PRISM 7900HT Sequence Detection System. SK2 and SK1 transcript levels were measured using inventoried Taqman probes (Life Technologies, Carlsbad, CA, USA). Relative transcript levels were normalized using the peptidylprolyl isomerase B (PPIB) gene as a control. To determine viral RNA copy numbers in the supernatant a MagMax 96 RNA Extraction Kit (Life Technologies, Carlsbad, CA, USA) was used and qRT-PCR assays were performed using the RNA UltraSenseTM One-step Kit (Life Technologies, Carlsbad, CA, USA) with custom virus-specific TaqMan prime/probe sets (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Samples were analyzed as mentioned above on the ABI PRISM 7900HT. Serial 10-fold dilutions of the assayed (102–107 copies) virus was used as standards.
Immunofluorescence experiments
For high-content imaging the cells were fixed in formalin and subsequently blocked in PBS containing 3% bovine serum albumin (PBS-3% BSA) for 1 h then stained with a rabbit polyclonal antibody that detects the CHIKV E2 glycoprotein (IBT Bioservices, Gaithersburg, MD, USA). Cell nuclei and cytoplasm were labeled with Hoechst 33342 (Life Technologies, Carlsbad, CA, USA) and HCS CellMask Red (Life Technologies, Carlsbad, CA, USA), respectively. High-content quantitative imaging data were acquired and analyzed on an Opera Confocal Plate-reader Model 3842 (Quadruple Excitation High Sensitivity (QEHS), PerkinElmer, Boston, MA, USA). Image analysis was accomplished using standard Acapella scripts. Percentage inhibition was calculated using mean percentage infection from infected, untreated cells as a positive control. For all other imaging studies with the exception of the 5-ethyl uridine (EU) labeling study, cells were fixed in ice-cold methanol, blocked in PBS-3%BSA. SK2 was detected with a polyclonal SK2 antibody (ECM biosciences, Versailles, KY, USA). CHIKV was detected with a mouse monoclonal antibody 2D21-1. G3BP1 was detected with a mouse monoclonal antibody (BD Biosciences, San Jose, CA, USA) and viral double-stranded RNA (dsRNA) was detected using the mouse monoclonal antibody J2 (Scicons, Szirák, Hungary). CHIKV nsP2 was detected with a mouse monoclonal antibody kindly provided by Dr Marc Lecuit. Alexa 488-conjugated donkey anti-mouse secondary antibody, Alexa 568-conjugated donkey anti-rabbit antibody or Alexa 647-conjugated donkey anti-goat antibody (Life Technologies, Carlsbad, CA, USA) were used to visualize primary antibodies. Confocal images were collected with a Leica TCS-SP5 confocal/multiphoton microscope.
For EU labeling studies, HeLa cells were infected with CHIKV at a multiplicity of infection (MOI) of 5. At 4-h postinfection, the cells were treated with actinomycin D (ActD) (Sigma, St. Louis, MO, USA) then subsequently incubated with EU (ClikIT; Life Technologies, Carlsbad, CA, USA) and processed according to the manufacturer's instructions. Labeled RNA was detected by click chemistry using an Alexa Fluor 488-coupled azide.31 Confocal images were collected as mentioned above.
Western blot
HeLa cells were left mock infected or infected with CHIKV at a MOI of 5 for 24 h. Thirty minutes prior to infection, the cells were left untreated or treated with 50, 100, or 500 nM of S1P (Cayman Chemical, Ann Arbor, MI, USA). At 24-h postinfection, the cells were lysed in lysis buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.25% Na-deoxycholate, 1 mM ethylenediaminetetraacetic acid, protease inhibitor (Complete; Roche, Nutley, NJ, USA), and PhosSTOP phosphatase inhibitors (Roche, Nutley, NJ, USA) and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis followed by Western blot using the antibody against CHIKV E2 (IBT Bioservices, Gaithersburg, MD, USA) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Abcam, Cambridge, UK).
AP-MS experiments
293T cells were transfected with pCMV6/myc-DDK-SK2 or pCMV6 (empty vector) using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). At 24-h post-transfection the cells were either mock infected or infected with CHIKV at a MOI of 5 for 24 h. As a control pCMV6/myc-DDK-SK2 transfected cells were mock infected, and as an additional control, cells were transfected with an empty vector and also infected. After 24 h, the cell lysates were prepared in lysis buffer (same as above). Myc-DDK-SK2 complexes were precipitated using a myc monoclonal antibody cross-linked to agarose beads (Sigma, St. Louis, MO, USA). The precipitated material was analyzed by SDS-PAGE analysis followed by Coomassie staining (Thermo Fisher Scientific, Middletown, VA, USA) and prepared for LC-MS/MS analysis. Instrumentation consisted of an Ultimate 3000 Nano-LC connected to a Thermo Scientific LTQ Orbitrap Elite using a data-dependent method consisting of a 30 000 resolution MS1 scan followed by 15 ms/ms rapid scans of the highest intensity ions. Searches were performed with MASCOT 2.4 using the SwissProt_2014_01 database with human taxonomy specified. The false discovery rate was set at 0.5%. Mass tolerances were 10 ppm for the MS1 scan and 200 ppm for all ms/ms scans.
Analysis of protein complexes
For analysis of SK2 specific interaction partners during CHIKV infection a P-value of 0.01 was used to determine the probability of a SK2-associated protein being specific. The inter-connectivity and the immediate interacting partners of the 126 proteins identified were evaluated by applying publicly available binary human protein–protein interaction (PPI) data derived from Reactome (http://reactome.org), BIND (http://www.bind.ca), MINT (http://mint.bio. uniroma2.it/mint/Welcome.do), HPRD (http://www.hprd.org), CORUM (http://mips.helmholtz-muenchen.de/genre/proj/corum/index.html).32,33,34,35,36 For networks too complex for visual interpretation, Molecular Complex Detection (MCODE) analysis was applied to identify densely connected network components.32 All network visualization was based on Cytoscape (version 2.8.0).37 Our implementation of the MCODE analysis was based on the Cytoscape MCODE plug-in source code.
Results
Inhibition of SK2 impairs CHIKV infection
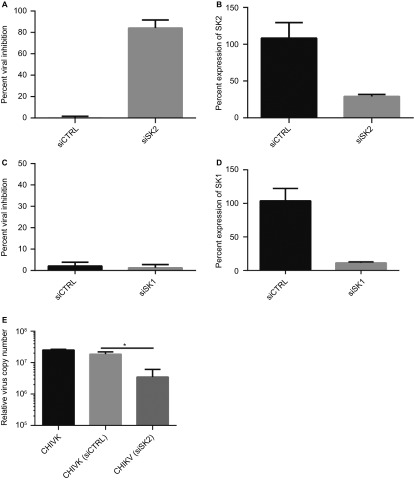
A primary siRNA screen using a human kinome library indicated that SK2 was required for CHIKV infection (data not shown). This was of interest in light of recent studies that have shown that SK1 exhibits pro-viral functions to enhance replication of influenza A and MV,25,26 and more recent studies showing that SK2 is involved in viral infections as well.29,30 To determine whether SKs indeed affect CHIKV infection we performed a series of knockdown experiments using siRNAs against SK1 or SK2 and utilized high-content imaging to detect viral infection. Knockdown of SK2 significantly inhibited CHIKV infection (Figure 1A); in contrast, knockdown of SK1 had no impact on virus infection (Figure 1C). In both experiments knockdown of SK gene expression was confirmed (Figures 1B and 1D). Next it was of interest to monitor virus production after SK2 knockdown. Knockdown of SK2 resulted in a significant reduction in CHIKV viral RNA in the supernatant of infected cells (Figure 1E). Taken together these experiments confirm that knockdown of SK2 expression significantly inhibited CHIKV infection.
Figure 1.
Inhibition of SK2 expression inhibits CHIKV infection. (A and C) U2OS cells were transfected with a siRNA control (siCTRL) (50 nM) (A), SK2 (50 nM) siRNA (A), siCTRL (15 nM) (C) or SK1 siRNA (15 nM) (C). At 48-h post-transfection, knockdown was confirmed for SK2 (B) and SK1 (D) by qRT-PCR. In parallel the cells were infected with CHIKV at a MOI of 5 and at 48-h postinfection the cells were fixed and analyzed by high-content confocal microscopy. (E) HeLa cells were untreated or transfected with siCTRL or siSK2 then infected with CHIKV, as previously mentioned. At 48-h postinfection, supernatant was collected and viral RNA copy number determined by qRT-PCR analysis. Values are means + SEM (n = 4). *P <0.0005.
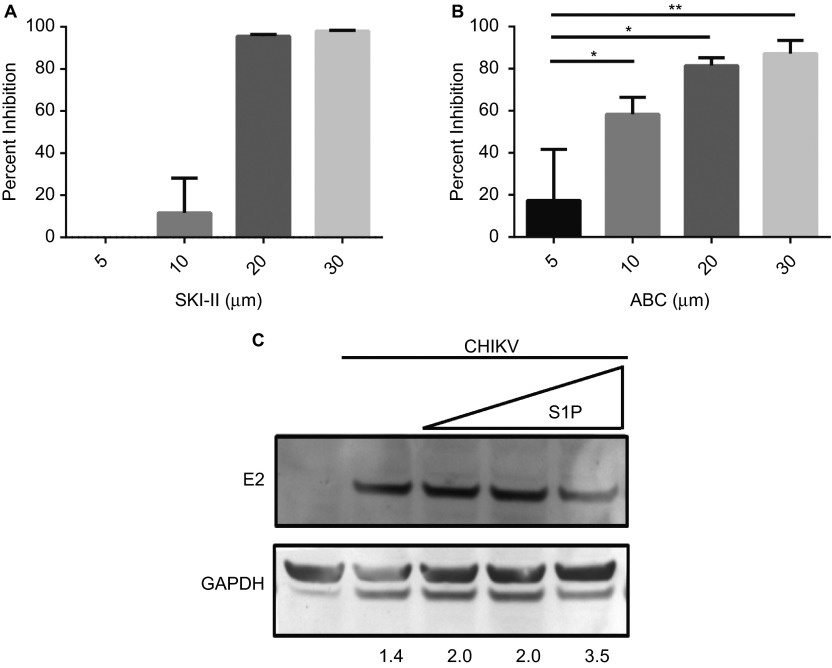
To chemically confirm the results of the previous experiments, we utilized the well-characterized SK-specific inhibitor, sphingosine kinase inhibitor II (SKI-II).38,39 HepG2 cells were pretreated with increasing concentrations of SKI-II (5, 10, 20, 30 µM), then subsequently infected. Treatment with SKI-II (between 20 µM and 30 µM) resulted in a significant reduction in CHIKV infection (Figure 2A). It is interesting to note, SKI-II was similarly shown to inhibit Influenza A infection at 30 µM.25 SKI-II is a dual SK inhibitor able to inhibit both SK1 and SK2.40 Because CHIKV infection was shown to be dependent on SK2 it was of interest to use a SK2-specific inhibitor. For this, we used the SK2-specific inhibitor ABC294640 (ABC).40,41 HepG2 cells were pretreated with increasing concentrations of ABC then infected with virus. ABC pretreatment potently inhibited CHIKV infection in a dose-dependent manner (Figure 2B). Comparatively, virus inhibition by ABC at 10 µM was greater than that observed for SKI-II at the same concentration. Since the enzymatic function of SK2 similar to SK1 is to catalyze the production of S1P from sphingosine, and our data thus far indicate SK2 is positively required for CHIKV infection, we wanted to determine if treatment with exogenous S1P would similarly impact virus infection. Pretreatment with increasing concentration of S1P had only a modest effect on viral protein expression (Figure 2C). These results demonstrate that targeting SK2 significantly inhibits CHIKV infection, while targeting the S1P system by the addition of exogenous S1P did not appear to affect viral protein production.
Figure 2.
SK2 inhibition impairs CHIKV infection. (A and B) HepG2 cells were pretreated for 2 h with 5, 10, 20, or 30 µM of SKI-II (A) or ABC (B) then infected with CHIKV at an MOI of 5. At 48-h postinfection, cells were fixed and analyzed for infection by high-content confocal microscopy. Values represent mean + SEM (n = 3). *P < 0.05, **P < 0.01 (C) HeLa cells were pretreated with either 50, 100, or 500 nM S1P for 15 min then infected with CHIKV at a MOI if 5 for 24 h. Cell lysates were harvested and subjected to Western blot analysis to detect the E2 viral glycoprotein and GAPDH, the relative intensity for each band is shown below.
SK2 is re-localized during virus infection
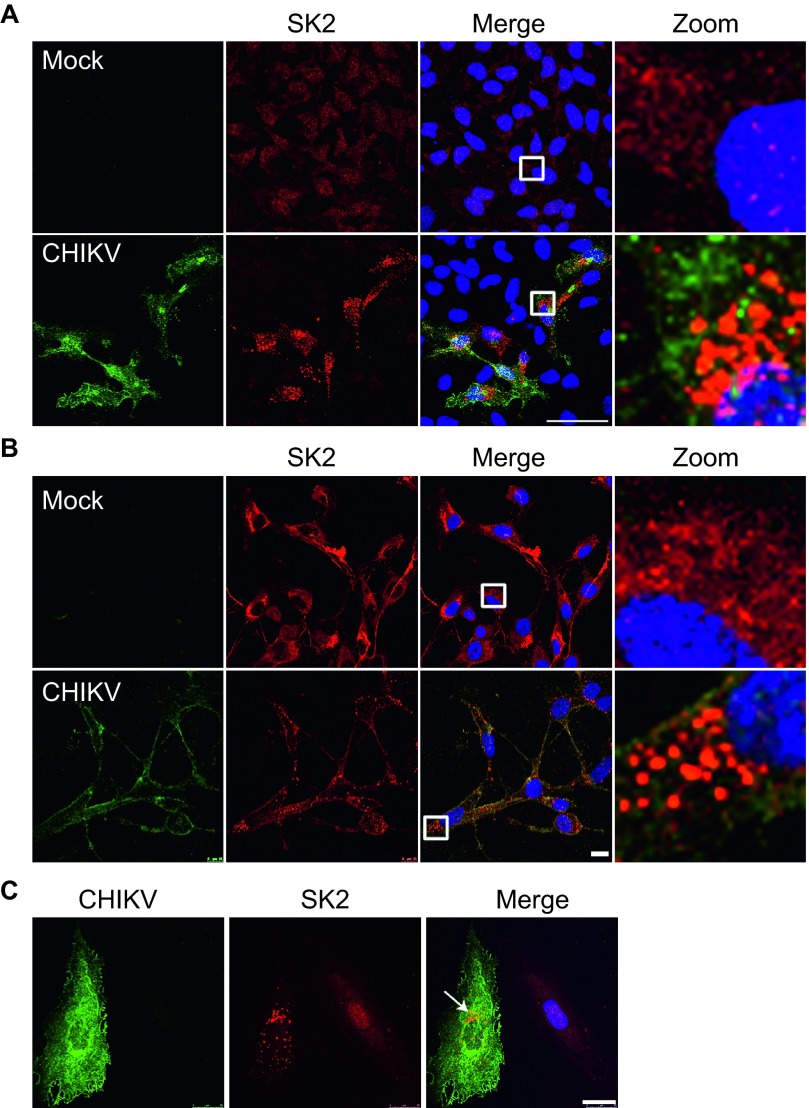
The requirement of SK2 during infection prompted us to examine the subcellular localization of SK2. Unlike SK1 which is localized in the cytoplasm, SK2 contains both a nuclear localization signal (NLS) and a nuclear export signal (NES),42,43 as a result its localization can vary in a cell-type dependent manner.44 In uninfected HeLa cells SK2 was observed to be localized in the nucleus with weak diffuse cytoplasmic staining. This observation is in line with previous reports.43 Strikingly, upon CHIKV infection SK2 was re-localized to distinct puncta in the cytoplasm (Figure 3A). We next wanted to confirm this observation in primary human skeletal muscle cells (hSkMCs) which are known to be targets of CHIKV infection.45,46 In contrast to HeLa cells, SK2 in uninfected hSkMCs was observed to be diffuse in the cytoplasm (Figure 3B, mock infected). Nevertheless, upon infection similar to HeLa cells SK2 was re-localized in a similar punctate staining pattern (Figure 3B). Additionally, this distinct re-localization of SK2 during CHIKV infection was confirmed in MCF10A cells (Figure 3C). Taken together these data show that SK2 is re-localized to distinct structures in the cytoplasm upon virus infection, further supporting a role for the kinase during infection.
Figure 3.
SK2 is re-localized during CHIKV infection. HeLa (A), hSKMC (B), or MCF10A (C) cells were uninfected or infected with CHIKV at a MOI of 5 for 24 h. The viral E2 protein (green), SK2 (red), and Hoechst nuclear stain (blue) were visualized by confocal microscopy. Scale bar = 50 µm (A), 10 µm (B), 25 µm (C).
SK2 is required early during CHIKV infection
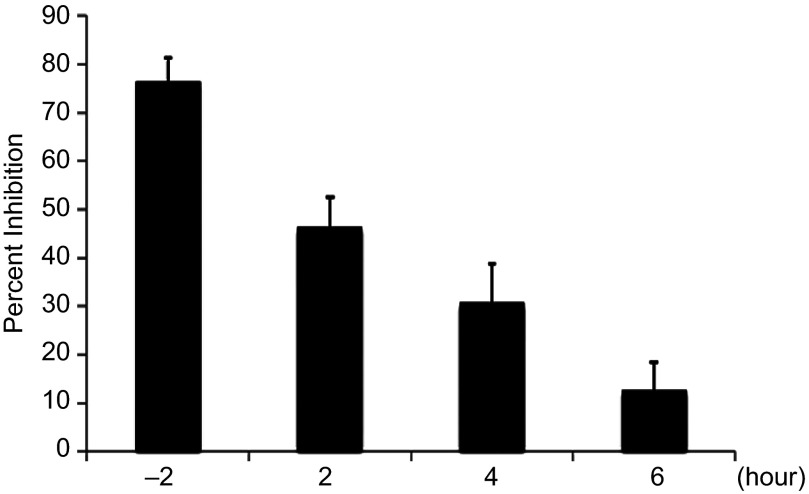
It was next of interest to determine the function of SK2 during viral infection. To perform in-depth analysis of what stage/s in the viral life cycle SK2 is playing a role we conducted a series of pseudo type studies. Pretreatment with ABC or siRNA-mediated knockdown of SK2 did not impair viral entry (data not shown), indicating that under these conditions SK2 is not essential for viral entry. Next, time of addition studies were conducted in which cells were either pretreated 2 h prior to infection or treated with ABC at several different times postinfection. As expected from our earlier studies, administration of ABC 2 h prior to infection resulted in strong inhibition of infection (∼80%), however when the compound was added postinfection it was unable to inhibit virus infection (Figure 4). The inability of ABC to inhibit CHIKV infection when added several hours postinfection along with the pseudo type studies suggests that SK2 is required early during infection post-entry.
Figure 4.
SK2 is required for an early step in virus infection. HeLa cells were treated with 20 µM of ABC at 2 h prior to infection or, 2-, 4-, or 6-h postinfection. The cells were then infected at a MOI of 5 for 48 h then infection analyzed by high-content confocal microscopy.
Identification of novel SK2 interaction partners during CHIKV infection
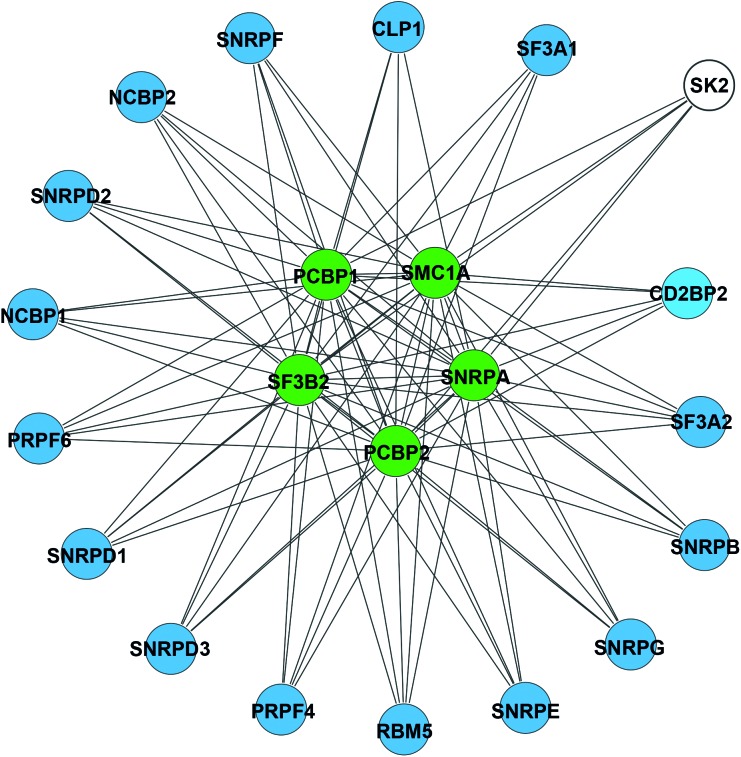
SK2 has previously been shown to interact with extracellular signal-regulated kinase (ERK) 1, eukaryotic elongation factor 1A and histone deacetylase (HDAC) 1 and 2.47,48,49 Association with HDAC1 and 2 revealed a novel role of SK2 in epigenetic regulation of gene expression.47 To better understand the role of SK2 during CHIKV infection AP-MS experiments were conducted to identify SK2 interaction partners specifically during viral infection. A total of 126 SK2-interacting proteins were present only in the presence of viral infection. Subsequently, a predictive PPI network was constructed to gain insights toward the interconnectivity and the immediate interacting partners. The PPI network revealed five enriched core factors that were previously characterized to be involved in mRNA processing and gene expression. These factors include splice factor 3b subunit 2 (SF3B2), structural maintenance of chromosome 1A (SMC1A), small nuclear ribonucleoprotein polypeptide A (SNRPA), poly(rC)-binding protein (PCBP) 1 and 2. Although not detected to directly interact with SK2, 16 additional factors (that are involved in mRNA processing and gene expression) were demonstrated to interact with the core factors (Figure 5). This suggests a potential role of SK2 in modulating gene expression during CHIKV infection.
Figure 5.
Identification of SK2 interaction partners during CHIKV infection. Network interaction map showing SK2 interactions gene expression network clusters. Network plots were generated using the Cytoscape 3 network analysis tool. Color codes represent, SK2 (white), proteins that directly interact with SK2 by AP-MS (green), secondary interactions of proteins associated with mRNA processing and gene expression (blue).
SK2 co-localizes with viral RNA
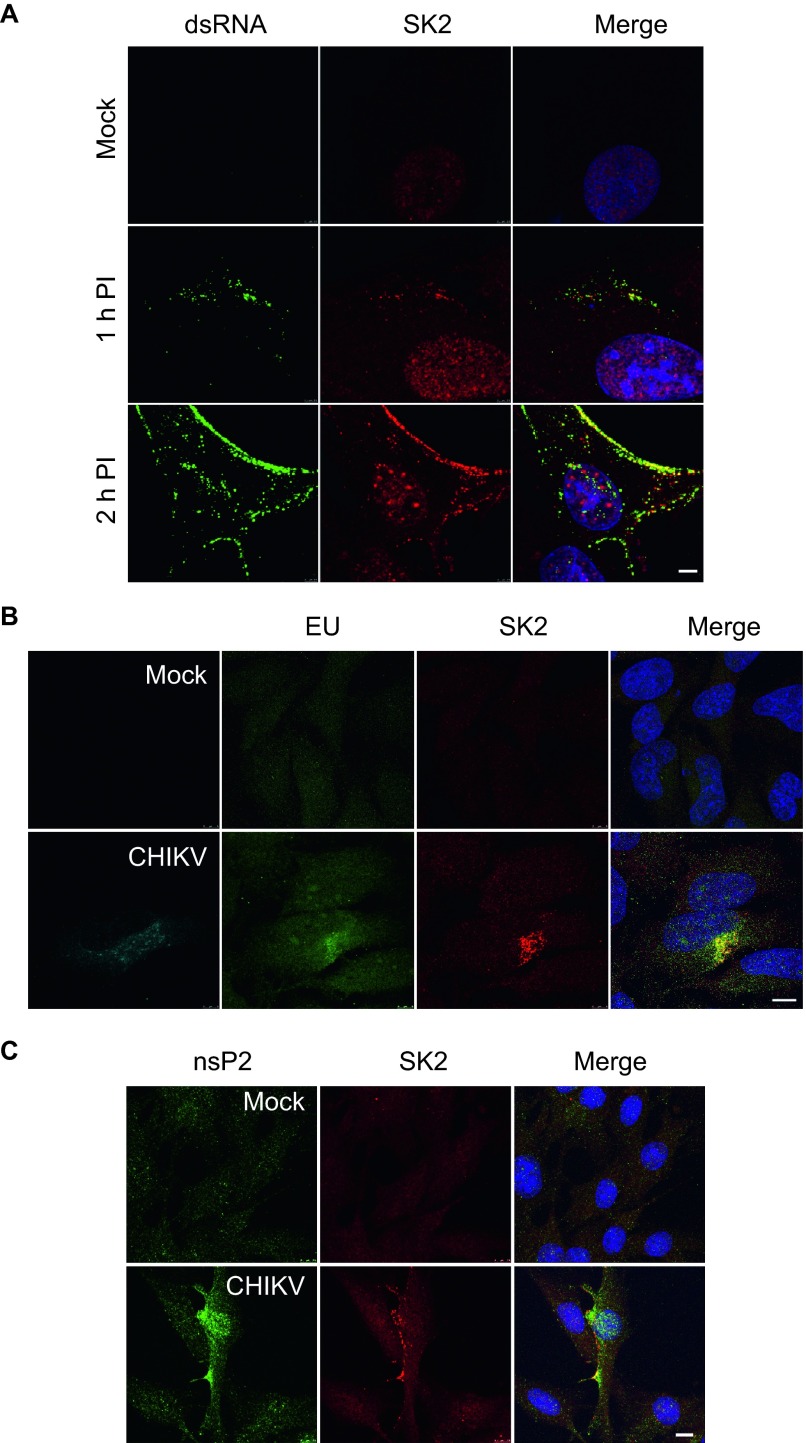
The result of the mass spec experiments indicated that SK2 may be involved in viral replication. To address a potential role in replication we examined the localization of SK2 during infection, in particular compared to the VRC. For these studies we used BHK-21 cells because this cell line has been used extensively for studying early events in alphavirus replication. BHK-21 cells were infected for 1 h or 2 h with CHIKV at a MOI of 5 or mock infected. At 1-h postinfection and more clearly at 2-h postinfection, staining for dsRNA demonstrated that virus-specific dsRNA was readily detected at the plasma membrane as well as in the cytoplasm. Localization of CHIKV VRCs at the plasma membrane is in line with prior studies demonstrating plasma membrane localization of alphavirus VRCs early during infection.50,51 In strong support of role in replication, SK2 was co-localized with the dsRNA-positive foci at 1 h, and more definitively at 2 h (Figure 6A). To further confirm that SK2 was co-localized with the VRC, HeLa cells were mock infected or infected with CHIKV for 4 h then treated with ActD to inhibit cellular transcription. Following 1-h incubation, RNA was labeled with EU. In ActD-treated, mock infected EU-labeled cells, only a diffuse background signal for EU could be detected (Figure 6B, upper panels). Strikingly, in infected cells as evidenced by viral E2 protein positive staining, a positive EU signal was detected and co-localized with SK2 (Figure 6B, lower panels). In the absence of ActD, EU-labeled cellular RNA was predominantly found localized in the nucleus and no RNA was detected in cells that did not receive EU (data not shown). Finally, to confirm co-localization of SK2 with a virally expressed protein, we examined localization of nsP2 2-h postinfection. At 2-h postinfection, nsP2 could be readily detected in both the cytoplasm and nucleus as previously reported.52 Similar to the dsRNA studies, nsP2 was found readily co-localized with SK2 during CHIKV infection (Figure 6C). These data demonstrate that SK2 is co-localized with VRC components during infection and suggest a role for the kinase in viral replication.
Figure 6.
SK2 associates with the CHIKV RNA and nsP2 during infection. (A) BHK-21 cells were infected with CHIKV at a MOI of 5 for 1 or 2 h or mock infected, then fixed and stained for dsRNA (green), SK2 (red), and Hoechst nuclear stain (blue) and visualized by confocal microscopy. Scale bars = 2.5 µm. (B) HeLa cells were mock or infected with CHIKV at a MOI of 5. At 4-h postinfection, the cells were treated with ActD then subsequently treated with 5-EU. Labeled RNA was subsequently detected with an Alexa Fluor 488-coupled azide. Labeled RNA (green), viral E2 protein (cyan), SK2 (red), Hoechst nuclear stain (blue) were visualized by confocal microscopy. Scale bars = 5 µm. (C) BHK-21 cells were either mock infected or infected with CHIKV at an MOI of 10 for 2 h. The cells were fixed and immune-stained for nsP2 (green), SK2 (red), and Hoechst nuclear stain (blue). Scale bars = 7.5 µm.
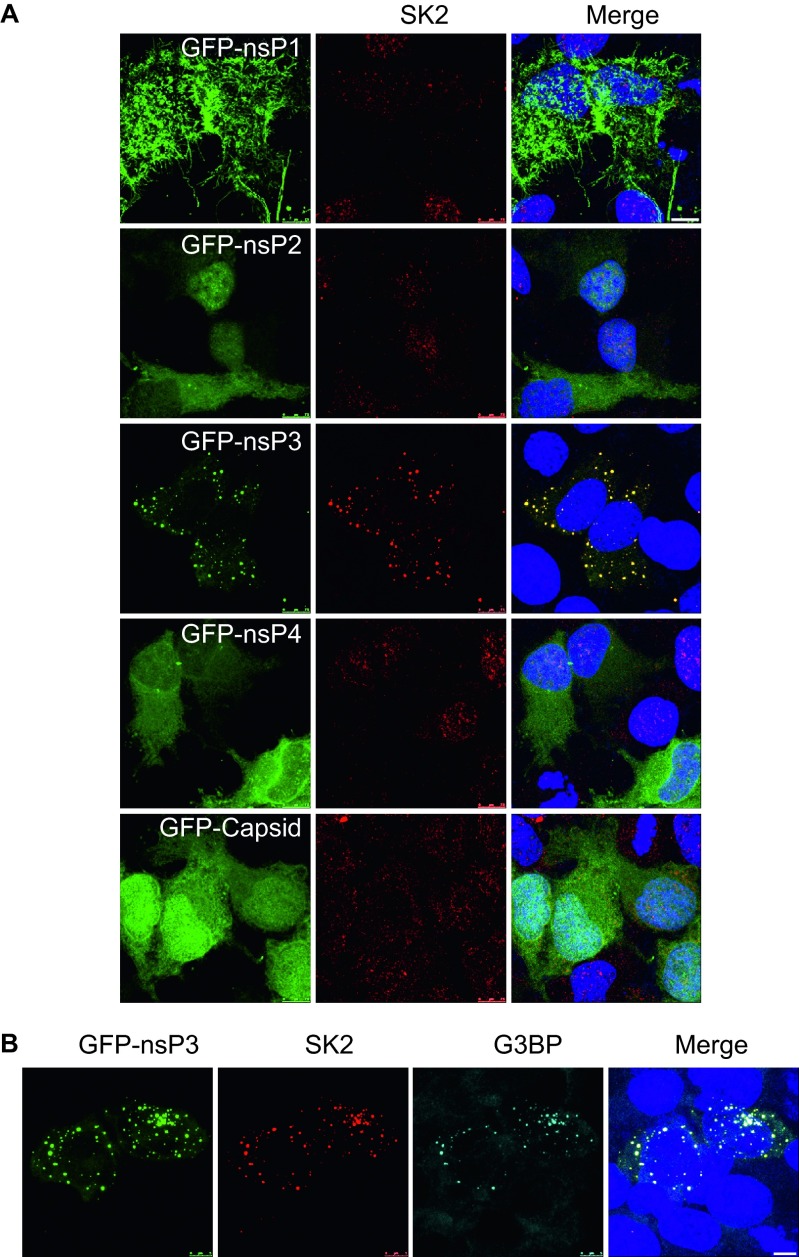
CHIKV nsP3 recruits SK2 to the viral replication complex
Host factor recruitment to sites of viral replication is often determined by specific associations with viral proteins. Therefore we wanted to determine if there is a CHIKV protein required for re-localization of SK2 to the VRC. To investigate this, HeLa cells were transfected with GFP-fused nonstructural proteins, nsP1-, nsP2-, nsP3-, nsP4-, or GFP-fused capsid. At 24-h post-transfection, the cells were analyzed for SK2 and GFP localization. Transfection of the GFP-nsP3 alone consistently induced re-localization of SK2 (Figure 7A). The observed localization of SK2 in the presence of GFP-nsP3 alone was similar to that seen during CHIKV infection, demonstrating that nsP3 of CHIKV is required for the recruitment of SK2 to the VRC. NsP3 mediated recruitment of SK2 identifies an additional function of the enigmatic nsP3 protein. Recent studies have shown that CHIKV nsP3 recruits G3BP1 during infection.16,17 We next wanted to determine if SK2 is co-localized with the nsP3-G3BP1 complex. Over-expression of GFP-nsP3 resulted in co-localization of both SK2 and G3BP1 (Figure 7B), suggesting that nsP3 recruits these two proteins to the same complex. As a result we have identified a novel association between CHIKV nsP3 and SK2 that is likely required for the recruitment of SK2 to the VRC. We also demonstrated that SK2 was recruited to nsP3-G3BP1 complexes.
Figure 7.
CHIKV nsP3 co-localizes with SK2. (A and B) HeLa cells were transfected with the indicated expression plasmids for GFP-fused CHIKV nsP1, nsP2, nsP3, nsP4, and capsid. At 24-h post-transfection, the cells were fixed and immunostained for GFP-fused viral proteins (green) SK2 (red) and Hoechst nuclear stain (blue). (B) GFP-nsP3 transfected cells were co-stained for SK2 (red), G3BP (cyan), and Hoechst nuclear stain (blue). Scale bars = 7.5 µm (A), 5 µm (B).
Discussion
In the current study, we identified SK2 as a novel CHIKV host factor. SK2 was found to co-localize with viral RNA and nonstructural proteins during infection and in transfection studies, respectively. These data suggest a role for the kinase in CHIKV replication and virus propagation. Interestingly, this is similar to the observed role of SK1 in the positive regulation of MV and influenza A virus replication.25,26 Very little is known about SK2 involvement in viral infection. A recent report indicates SK2 plays a role in KSHV latency an another reported showed that HCV replication can be restricted by lipid peroxidation regulated in part by SK2.29,30 This report combined with our current findings suggests the SK2 similar to SK1 likely plays a role in a number of viral infections.
SK1 and SK2 function as key regulators in sphingosine metabolism, phosphorylating sphingosine to S1P. As a result, these kinases are involved in regulation of a number of cellular and physiological processes, including viral infection. Viral entry into cells can be influenced by membrane sphingolipid compositions. As an example, HCV and HIV have been shown to be inhibited by ceramide.53,54 In contrast, alphavirus infection has long been positively associated with sphingolipids. Semliki Forest virus (SFV) fusion requires sphingolipids and Sindbis virus (SINV) entry has been associated with the release of the sphingolipid ceramide, a process leading to apoptosis.55,56 Because SK1 and SK2 can regulate the balance of sphingolipid levels these kinases likely play a key role in cell entry. However, in the current study SK2 did not appear to be required for entry.
SK1 has recently emerged as a host factor involved in the modulation of several viral infections, having been shown to play both a positive and negative regulatory role.23 The nonstructural protein 3 (NS3) from BVDV inhibits the catalytic activity of SK1, this inhibition had a beneficial impact on BVDV replication.28 Similarly, SK1 activity is reduced during DENV infection indicating SK1 likely plays a negative role during DENV infection as well.57 For some other viruses, including RSV, human cytomegalovirus (HCMV), MV, and influenza A virus, SK1 activity appears to positively impact virus infection.25,26,58,59 The differential role of SK1 during viral infection highlights the diverse strategies used by viruses to modulate the host cell machinery during infection. Our study presented several lines of evidence which strongly suggest that SK2 plays a positive role during CHIKV infection. Knockdown experiments demonstrated that SK2 expression is important for productive CHIKV infection (Figure 1A). Virus infection as detected by expression of the E2 viral glycoprotein was significantly reduced in SK2 knockdown-infected cells. Similarly, SK2 knockdown resulted in a significant reduction in CHIKV RNA in supernatant. In these experiments, SK1 knockdown did not impact CHIKV infection (Figure 1C). Importantly, complimentary studies with SK inhibitor SKI-II and the SK2-specific inhibitor ABC resulted in significant inhibition of virus infection, further confirming the siRNA experiments (Figures 2A and 2B). These results indicate that SK2 inhibitors should be explored as novel antivirals against CHIKV.
Of the two SKs, SK1 is by far the most studied; therefore, it is difficult to delineate precise functional differences between the two kinases. One clear difference with functional consequences is the subcellular localization of the two proteins. SK1 is localized in the cytoplasm and is recruited to the plasma membrane to mediate pro-survival and pro-proliferative signaling.60 By comparison SK2 contains both a NLS and a NES,42,43 indicating that the protein can readily shuttle between the nuclear and cytoplasmic compartments. In the current study, SK2 displayed predominantly nuclear localization in uninfected HeLa, MCF10A and BHK-21 cells, and cytoplasmic localization in hSkMCs. Upon infection in all cell types SK2 reproducibly re-localized to distinct cytoplasmic puncta (Figures 3A–3C). It is interesting to note that in these studies SK2 localization after CHIKV infection resembled that of studies showing co-localization between CHIKV nsP3 and G3BP1,16,17 suggesting a role for nsP3 in SK2 localization. Indeed, over-expression of nsp3 alone induced SK2 re-localization similar to that of viral infection. Moreover, SK2 was found to re-localize to nsP3-G3BP1-positive foci. SINV nsP3 has been demonstrated to form different complexes, including dsRNA-rich complexes that are indicative of a function in viral replication.61 The observed co-localization of SK2 with nsP3, along with experiments demonstrating that SK2 readily co-localizes with viral RNA early during infection, strongly suggests SK2 is associated with nsP3 in VRCs and moreover suggest a role for SK2 in viral replication. Further studies will be needed to confirm a specific role for SK2 during replication.
Our AP-MS studies revealed that SK2 associated with a number of proteins involved in mRNA processing and gene expression specifically during CHIKV infection. SK2 was found to associate with a core network consisting of SMC1A, SNRPA, PCBP1, PCBP2, and SF3B2. When this interaction network was expanded to include secondary interactions (i.e. proteins that interact with SK2 through interactions with the core network), we identified a further network of 16 total proteins involved in mRNA processing and gene expression that each individually interacted with all five proteins in our core network. These data along with a prior study identifying role for SK2 in gene expression suggested SK2 may play a similar role during viral infection.47 Further studies will be needed to fully determine a link between SK2 association with these proteins and viral replication. Nevertheless, association of SK2 with these cellular factors involved in mRNA processing and gene expression, specifically during viral infection, is strongly suggestive of a role in replication. Interestingly, although we were unable to identify individual viral proteins in the AP-MS studies, the viral polyprotein was reproducibly identified (data not shown), further supporting a role for SK2 in the VRC.
In our current study, we cannot eliminate the likelihood that SK2-mediated generation of S1P and in turn S1P function during CHIKV is critical for replication. Exogenously administered S1P did not significantly affect viral protein production, suggesting S1P signaling through its receptors may not affect CHIKV replication. A similar observation was reported for influenza A and MV where over-expression of SK1 enhanced viral infection; however, treatment with S1P did not significantly affect virus replication.24,26 This does not however eliminate the possibility that intracellular S1P can have an impact on virus replication. Further investigation to delineate a potential role for S1P in CHIKV infection will be needed. The kinase activity of SK2 was not addressed in this study and requires exploration. Specifically, is SK2 kinase activity required by CHIKV in the VRC? We observed that treatment with the SK2 inhibitor ABC severely reduced CHIKV infection. ABC targets SK2 kinase activity suggesting that this activity is likely required by CHIKV.38
In recent years there have been a number of proteomic studies aimed at elucidating the molecular mechanism underlying CHIKV replication and pathogenesis.9,10,11,12,13,14 While these studies have provided great insight much remains to be understood, in particular, essential host factors utilized by the virus during infection. In the current study, we identify a role for SK2 in viral replication. Further studies will provide greater insight into the precise function of SK2 during infection. The identification of this new cellular factor required by CHIKV provides the basis for the development of novel host-based therapeutic targets against the virus.
Acknowledgments
We thank the Dr Marc Lecuit (Institut Pasteur, Paris, France), Dr Subhash G Vasudevan (Duke-NUS Graduate Medical School, Singapore), Dr Charles D Smith (MUSC, Charleston, SC, USA), and Dr Michael Farzan (The Scripps Research Institute, Jupiter, FL, USA) for providing the nsP2 antibody, GFP-nsP/capsid plasmids, ABC294640 and MLV-GFP pseudotype plasmids respectively. We also thank Julie Tran and Dr Veronica Soloveva (USAMRIID, Frederick, MD, USA) for technical assistance and Dr Steven Bradfute (UNM, Albuquerque, NM, USA) for critical review of this manuscript. These studies were in part supported by Defense Threat Reduction Agency, Joint Science and Technology Office. The opinions, interpretations, conclusions, and recommendation in this report are not necessarily endorsed by the US Army.
References
- 1Brighton SW, Prozesky OW, de la Harpe AL. Chikungunya virus infection. A retrospective study of 107 cases. S Afr Med J 1983; 63: 313–315. [PubMed] [Google Scholar]
- 2Kennedy AC, Fleming J, Solomon L. Chikungunya viral arthropathy: a clinical description. J Rheumatol 1980; 7: 231–236. [PubMed] [Google Scholar]
- 3Mohan A, Kiran DH, Manohar IC et al. Epidemiology, clinical manifestations, and diagnosis of Chikungunya fever: lessons learned from the re-emerging epidemic. Indian J Dermatol 2010; 55: 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Schilte C, Staikowsky F, Couderc T et al. Chikungunya virus-associated long-term arthralgia: a 36-month prospective longitudinal study. PLoS Negl Trop Dis 2013; 7: e2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Weaver SC. Arrival of chikungunya virus in the new world: prospects for spread and impact on public health. PLoS Negl Trop Dis 2014; 8: e2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Jones PH, Maric M, Madison MN et al. BST-2/tetherin-mediated restriction of chikungunya (CHIKV) VLP budding is counteracted by CHIKV non-structural protein 1 (nsP1). Virology 2013; 438: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Schilte C, Couderc T, Chretien F et al. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J Exp Med 2010; 207: 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Werneke SW, Schilte C, Rohatgi A et al. ISG15 is critical in the control of Chikungunya virus infection independent of UbE1L mediated conjugation. PLoS Pathog 2011; 7: e1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Lum FM, Ng LF. Cellular and molecular mechanisms of chikungunya pathogenesis. Antiviral Res 2015; 120: 165–174. [DOI] [PubMed] [Google Scholar]
- 10Abere B, Wikan N, Ubol S et al. Proteomic analysis of chikungunya virus infected microglial cells. PLoS One 2012; 7: e34800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Dhanwani R, Khan M, Alam SI et al. Differential proteome analysis of Chikungunya virus-infected new-born mice tissues reveal implication of stress, inflammatory and apoptotic pathways in disease pathogenesis. Proteomics 2011; 11: 1936–1951. [DOI] [PubMed] [Google Scholar]
- 12Tchankouo-Nguetcheu S, Khun H, Pincet L et al. Differential protein modulation in midguts of Aedes aegypti infected with chikungunya and dengue 2 viruses. PLoS One 2010; 5: pii: e13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Thio CL, Yusof R, Abdul-Rahman PS et al. Differential proteome analysis of chikungunya virus infection on host cells. PLoS One 2013; 8: e61444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Bourai M, Lucas-Hourani M, Gad HH et al. Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component. J Virol 2012; 86: 3121–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Dudha N, Rana J, Rajasekharan S et al. Host-pathogen interactome analysis of Chikungunya virus envelope proteins E1 and E2. Virus Genes 2015; 50: 200–209. [DOI] [PubMed] [Google Scholar]
- 16Fros JJ, Domeradzka NE, Baggen J et al. Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci. J Virol 2012; 86: 10873–10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Panas MD, Ahola T, McInerney GM. The C-terminal repeat domains of nsP3 from the Old World alphaviruses bind directly to G3BP. J Virol 2014; 88: 5888–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Scholte FE, Tas A, Albulescu IC et al. Stress granule components G3BP1 and G3BP2 play a proviral role early in Chikungunya virus replication. J Virol 2015; 89: 4457–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol 2003; 4: 397–407. [DOI] [PubMed] [Google Scholar]
- 20Maceyka M, Harikumar KB, Milstien S et al. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell Biol 2012; 22: 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Spiegel S, Milstien S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat Rev Immunol 2011; 11: 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Monick MM, Cameron K, Powers LS et al. Sphingosine kinase mediates activation of extracellular signal-related kinase and Akt by respiratory syncytial virus. Am J Respir Cell Mol Biol 2004; 30: 844–852. [DOI] [PubMed] [Google Scholar]
- 23Carr JM, Mahalingam S, Bonder CS et al. Sphingosine kinase 1 in viral infections. Rev Med Virol 2013; 23: 73–84. [DOI] [PubMed] [Google Scholar]
- 24Seo YJ, Blake C, Alexander S et al. Sphingosine 1-phosphate-metabolizing enzymes control influenza virus propagation and viral cytopathogenicity. J Virol 2010; 84: 8124–8131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Seo YJ, Pritzl CJ, Vijayan M et al. Sphingosine kinase 1 serves as a pro-viral factor by regulating viral RNA synthesis and nuclear export of viral ribonucleoprotein complex upon influenza virus infection. PLoS One 2013; 8: e75005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Vijayan M, Seo YJ, Pritzl CJ et al. Sphingosine kinase 1 regulates measles virus replication. Virology 2014; 450–451: 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Carr JM, Kua T, Clarke JN et al. Reduced sphingosine kinase 1 activity in dengue virus type-2 infected cells can be mediated by the 3′ untranslated region of dengue virus type-2 RNA. J Gen Virol 2013; 94: 2437–2448. [DOI] [PubMed] [Google Scholar]
- 28Yamane D, Zahoor MA, Mohamed YM et al. Inhibition of sphingosine kinase by bovine viral diarrhea virus NS3 is crucial for efficient viral replication and cytopathogenesis. J Biol Chem 2009; 284: 13648–13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Yamane D, McGivern DR, Wauthier E et al. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med 2014; 20: 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Dai L, Plaisance-Bonstaff K, Voelkel-Johnson C et al. Sphingosine kinase-2 maintains viral latency and survival for KSHV-infected endothelial cells. PLoS One 2014; 9: e102314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci USA 2008; 105: 15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003; 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Ceol A, Chatr Aryamontri A, Licata L et al. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res 2010; 38: D532–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Croft D, O'Kelly G, Wu G et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res 2011; 39: D691–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Peri S, Navarro JD, Amanchy R et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res 2003; 13: 2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Ruepp A, Waegele B, Lechner M et al. CORUM: the comprehensive resource of mammalian protein complexes – 2009. Nucleic Acids Res 2010; 38: D497–D501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Shannon P, Markiel A, Ozier O et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38French KJ, Schrecengost RS, Lee BD et al. Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer Res 2003; 63: 5962–5969. [PubMed] [Google Scholar]
- 39Orr Gandy KA, Obeid LM. Targeting the sphingosine kinase/sphingosine 1-phosphate pathway in disease: review of sphingosine kinase inhibitors. Biochim Biophys Acta 2013; 1831: 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Gao P, Peterson YK, Smith RA et al. Characterization of isoenzyme-selective inhibitors of human sphingosine kinases. PLoS One 2012; 7: e44543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41French KJ, Zhuang Y, Maines LW et al. Pharmacology and antitumor activity of ABC294640, a selective inhibitor of sphingosine kinase-2. J Pharmacol Exp Ther 2010; 333: 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Ding G, Sonoda H, Yu H et al. Protein kinase D-mediated phosphorylation and nuclear export of sphingosine kinase 2. J Biol Chem 2007; 282: 27493–27502. [DOI] [PubMed] [Google Scholar]
- 43Igarashi N, Okada T, Hayashi S et al. Sphingosine kinase 2 is a nuclear protein and inhibits DNA synthesis. J Biol Chem 2003; 278: 46832–46839. [DOI] [PubMed] [Google Scholar]
- 44Neubauer HA, Pitson SM. Roles, regulation and inhibitors of sphingosine kinase 2. FEBS J 2013; 280: 5317–5336. [DOI] [PubMed] [Google Scholar]
- 45Couderc T, Chretien F, Schilte C et al. A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog 2008; 4: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Ozden S, Huerre M, Riviere JP et al. Human muscle satellite cells as targets of Chikungunya virus infection. PLoS One 2007; 2: e527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Hait NC, Allegood J, Maceyka M et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 2009; 325: 1254–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Hait NC, Bellamy A, Milstien S et al. Sphingosine kinase type 2 activation by ERK-mediated phosphorylation. J Biol Chem 2007; 282: 12058–12065. [DOI] [PubMed] [Google Scholar]
- 49Leclercq TM, Moretti PA, Vadas MA et al. Eukaryotic elongation factor 1A interacts with sphingosine kinase and directly enhances its catalytic activity. J Biol Chem 2008; 283: 9606–9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Frolova EI, Gorchakov R, Pereboeva L et al. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol 2010; 84: 11679–11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Kujala P, Ikaheimonen A, Ehsani N et al. Biogenesis of the Semliki Forest virus RNA replication complex. J Virol 2001; 75: 3873–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Rikkonen M, Peranen J, Kaariainen L. Nuclear and nucleolar targeting signals of Semliki Forest virus nonstructural protein nsP2. Virology 1992; 189: 462–473. [DOI] [PubMed] [Google Scholar]
- 53Finnegan CM, Rawat SS, Puri A et al. Ceramide, a target for antiretroviral therapy. Proc Natl Acad Sci USA 2004; 101: 15452–15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Voisset C, Lavie M, Helle F et al. Ceramide enrichment of the plasma membrane induces CD81 internalization and inhibits hepatitis C virus entry. Cell Microbiol 2008; 10: 606–617. [DOI] [PubMed] [Google Scholar]
- 55Jan JT, Chatterjee S, Griffin DE. Sindbis virus entry into cells triggers apoptosis by activating sphingomyelinase, leading to the release of ceramide. J Virol 2000; 74: 6425–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56Nieva JL, Bron R, Corver J et al. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J 1994; 13: 2797–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57Wati S, Rawlinson SM, Ivanov RA et al. Tumour necrosis factor alpha (TNF-alpha) stimulation of cells with established dengue virus type 2 infection induces cell death that is accompanied by a reduced ability of TNF-alpha to activate nuclear factor kappaB and reduced sphingosine kinase-1 activity. J Gen Virol 2011; 92: 807–818. [DOI] [PubMed] [Google Scholar]
- 58Machesky NJ, Zhang G, Raghavan B et al. Human cytomegalovirus regulates bioactive sphingolipids. J Biol Chem 2008; 283: 26148–26160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59Rana J, Sreejith R, Gulati S et al. Deciphering the host-pathogen protein interface in chikungunya virus-mediated sickness. Arch Virol 2013; 158: 1159–1172. [DOI] [PubMed] [Google Scholar]
- 60Pitson SM, Xia P, Leclercq TM et al. Phosphorylation-dependent translocation of sphingosine kinase to the plasma membrane drives its oncogenic signalling. J Exp Med 2005; 201: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61Gorchakov R, Garmashova N, Frolova E et al. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J Virol 2008; 82: 10088–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]