Abstract
Liver cirrhosis (LC) is a chronic illness caused by inflammatory responses and progressive fibrosis. Globally, the most common causes of chronic liver disease include persistent alcohol abuse, followed by viral hepatitis infections and nonalcoholic fatty liver disease. However, regardless of the etiological factors, the susceptibility and degree of liver damage may be influenced by genetic polymorphisms that are associated with distinct ethnic and cultural backgrounds. Consequently, metabolic genes are influenced by variable environmental lifestyle factors, such as diet, physical inactivity, and emotional stress, which are associated with regional differences among populations. This Topic Highlight will focus on the genetic and environmental factors that may influence the metabolism of alcohol and nutrients in the setting of distinct etiologies of liver disease. The interaction between genes and environment in the current-day admixed population, Mestizo and Native Mexican, will be described. Additionally, genes involved in immune regulation, insulin sensitivity, oxidative stress and extracellular matrix deposition may modulate the degree of severity. In conclusion, LC is a complex disease. The onset, progression, and clinical outcome of LC among the Mexican population are influenced by specific genetic and environmental factors. Among these are an admixed genome with a heterogenic distribution of European, Amerindian and African ancestry; a high score of alcohol consumption; viral infections; a hepatopathogenic diet; and a high prevalence of obesity. The variance in risk factors among populations suggests that intervention strategies directed towards the prevention and management of LC should be tailored according to such population-based features.
Keywords: Genomic medicine, Polymorphisms, Viral hepatitis, Obesity, Nonalcoholic steatohepatitis, Risk factors
Core tip: Liver cirrhosis is a global health problem. The onset, progression, and clinical outcome of liver cirrhosis are influenced by several hereditary and lifestyle factors. Worldwide, interactions between genetic and environmental factors involved in liver cirrhosis may be associated with ethnic-based variations. Globally, in populations with admixed genomes, such as the Mexican population, individualized medicine represents a new challenge for hepatologists and gastroenterologists.
INTRODUCTION
Liver cirrhosis (LC) is a chronic illness caused by multiple, long-term injuries to the liver[1]. Globally, LC represents the 14th leading cause of mortality[2] but is the 12th leading cause of mortality in the United States[1] and the 4th leading cause in Central Europe[3] and Mexico[4]. In general, the main etiologies of LC are chronic alcohol abuse, followed by viral infections (hepatitis C and B viruses) and nonalcoholic fatty liver disease (NAFLD), including nonalcoholic steatohepatitis (NASH)[5]. Other causes that occur in a lesser extent are autoimmune hepatitis, obstructive cholestasis, hereditary hemochromatosis, alpha-1-antitrypsin deficiency, Wilson’s disease and drug toxicity[6].
Liver cirrhosis is characterized by the degeneration and necrosis of hepatocytes, the replacement of liver parenchyma with fibrotic tissues and regenerative nodules and loss of liver function[7]. The impaired liver architecture induces intrahepatic vascular distortion and portal hypertension, which manifests in major complications, such as ascites, upper gastrointestinal bleeding, jaundice and hepatic encephalopathy[8]. However, regardless of the etiological factors, the susceptibility and degree of liver damage may be influenced by genetic polymorphisms that are associated with particular ethnic backgrounds[9]. Consequently, genes involved in various metabolic pathways are influenced by distinct environmental lifestyle factors, such as diet, physical inactivity, and emotional stress[10].
Thus, this Topic Highlight will focus on the genetic and environmental factors that may influence the metabolism of alcohol and nutrients in the setting of distinct etiologies of liver disease. Additionally, genes involved in immune regulation, insulin sensitivity, oxidative stress and extracellular matrix deposition may modulate the degree of severity. The interaction between genes and environment in the current-day admixed population, Mestizo and Native Mexican, will be described. The inheritance of a heterogenic distribution of European, Amerindian and African ancestry in this population[11,12] may play a central role in the clinical outcome of liver disease in conjunction with specific environmental factors.
CELLULAR AND MOLECULAR MECHANISMS INVOLVED IN THE PATHOGENESIS OF LIVER CIRRHOSIS
Hepatic fibrogenesis is a key factor for the progression of liver damage as a result of an excessive healing response triggered by acute and chronic liver injury associated with the continuous deposition of extracellular matrix (ECM), mainly fibrillary collagen[13]. The accumulation of ECM is due to both increased synthesis and decreased degradation that contributes to the loss of hepatocyte microvilli and endothelial fenestrations, resulting in LC and liver failure[14,15]. Hepatic fibrosis is driven by a population of activated fibrogenic cells known as myofibroblasts (MFs)[16]. The hepatic stellate cells (HSCs) are the primary source of MFs, orchestrating the deposition of ECM in normal and fibrotic liver, and activation of the immune response[17]. However, recent research has revealed that several cell types contribute to MF formation, including portal fibroblasts and bone marrow-derived mesenchymal cells[18].
The clinical assessment of cirrhosis and the staging of fibrosis have given rise to a number of non-invasive techniques that have been validated as surrogate tests for liver biopsy, such as the most recently developed transient elastography (FibroScan; Echosens, Paris, France). By this method, shear wave velocity correlates with liver stiffness, which is staged from F0 to F4, regardless of the etiological agent of liver fibrosis (Figure 1)[1].
Figure 1.
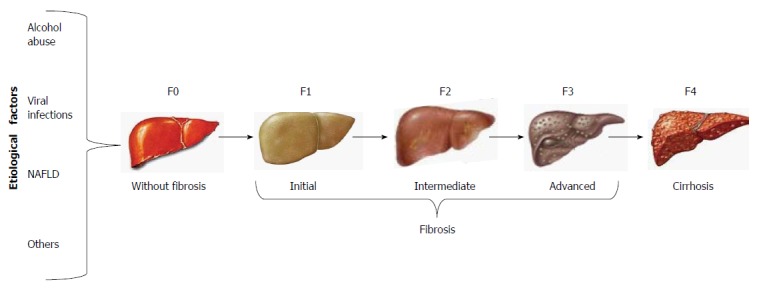
Stages of liver fibrosis. Liver fibrosis may be evaluated by liver biopsy and non-invasive methods. Regardless of etiological factors, liver fibrosis encompasses 3 stages until the development of cirrhosis. NAFLD: Nonalcoholic fatty liver disease.
Regarding the cellular and molecular mechanisms of LC, activation of HSCs involves two main stages (Figure 2). The first stage of initiation renders the cells responsive to diverse agents of liver injury, which results from the paracrine stimulation of neighboring cells, such as hepatocytes, circulating leukocytes, platelets and endothelial and Kupffer cells (KCs). Such stimuli include reactive oxygen species (ROS) and inflammatory cytokines. Others effectors are transcriptional factors, such as the three types of peroxisome proliferator-activated receptors (PPAR-α, -β and -γ), c-Myb, nuclear factor kappa B (NF-κB), Kruppel-like factor 6 (KLF6) and the CCAAT/enhancer binding protein-β (C/EBPβ). In addition, growth factors, mainly transforming growth factor beta 1 (TGF-β1), platelet-derived growth factor (PDGF), epidermal growth factor (EGF) and insulin-like growth factor 1 (IGF-1), play an important role in the activation of HSCs[19,20].
Figure 2.
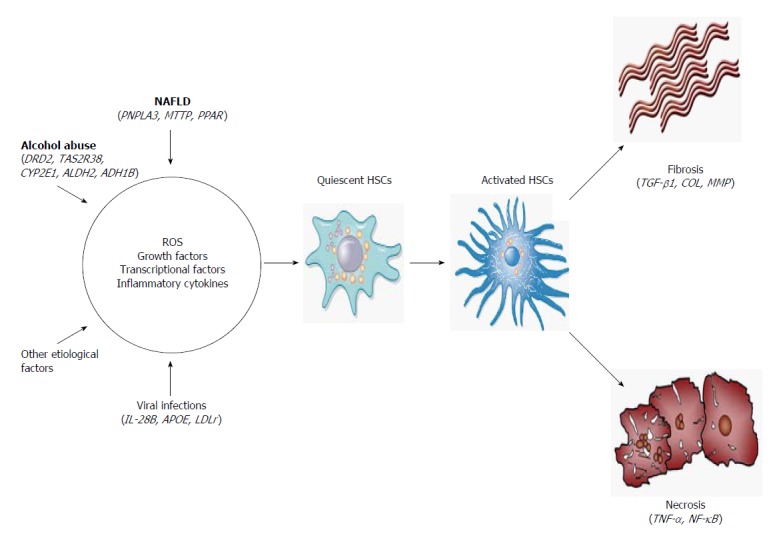
Hepatic fibrogenesis. Different etiological factors induce production of several stimuli to HSCs activation. Activated HSCs promote fibrosis and necrosis of hepatocytes. These pathogenic processes can be modulated by genetic polymorphisms involved in each stage of the pathophysiological process. HSC: Hepatic stellate cells; DRD2: Dopamine receptor D2; TAS2R38: Bitter taste receptor; CYP2E1: Cytochrome P450, family 2, subfamily E, polypeptide 1; ALDH2: Aldehyde dehydrogenase 2 family; ADH1B: Alcohol dehydrogenase class I, beta polypeptide; PNPLA3: Patatin-like phospholipase 3; MTTP: Microsome triglyceride transfer protein; PPAR-γ2: Peroxisome proliferator-activated receptors; IL-28B: Interleukin-28B; APOE: Apolipoprotein E; LDLr: Low-density lipoprotein receptor; TGF-β1: transforming growth factor beta 1; COL: Collagenases; MMP: Matrix metalloproteinase; TNF-α: Tumor necrosis factor alpha; NF-κB: Nuclear factor kappa B; ROS: Reactive oxygen species.
Once the HSCs are activated, the perpetuation stage encompasses phenotypic changes in cell biology, such as proliferation, contractility, fibrogenesis, matrix degradation, chemotaxis, retinoid loss, cytokine release and white blood cell chemoattraction. These events are regulated in an autocrine and paracrine manner by proinflammatory, pro-fibrogenic and pro-mitogenic signals to exacerbate the accumulation of ECM[20]. Finally, if the causal agent is eliminated, liver injury may be resolved. In this case, activated HSCs limit the fibrogenic response through the up-regulation of genes involved in apoptosis, enhanced immune surveillance and the increased secretion of ECM-degrading enzymes[21].
GENETIC, METABOLIC AND ENVIRONMENTAL FACTORS INVOLVED IN LIVER DISEASE
Alcoholic liver disease
Overconsumption of alcohol is directly associated with liver disease and affects millions of individuals worldwide[22]. It is estimated that a consumption of > 40 g of alcohol/d in men and > 20 g of alcohol/d in women over a period of 10 years significantly increases the risk of LC[23]. Alcohol-induced liver disease pathogenesis involves alcohol-metabolizing enzymes that alter the levels of acetaldehyde and ROS; these enzymes are known to have variable allelic distribution worldwide (Table 1)[24]. High levels of ROS diminish antioxidant activity, leading to oxidative damage of proteins, lipids, and DNA, which serve as antigens for eliciting the host immune response[25]. In addition, oxidative stress can modify intracellular signaling cascades related to an inflammatory state[26]. The resulting oxidative damage and subsequent mitochondrial and autophagy dysfunction lead to energy depletion, the accumulation of cytotoxic mediators and cell death[27]. Furthermore, acetaldehyde promotes bacterial lipopolysaccharide (LPS) translocation from the gut to the liver, which in turn stimulates Kupffer cells to release proinflammatory cytokines[28]. Additionally, acetaldehyde alters lipid homeostasis by disrupting lipoprotein transportation from the liver, decreasing β-oxidation of fatty acids and increasing its biosynthesis; resulting in hepatic steatosis[29]. Moreover, acetaldehyde is involved in fibrogenesis because this metabolite stimulates the synthesis of collagen and ECM components through the activation of the TGF-β1/SMAD3 signaling pathway[30]. Finally, acetaldehyde and ROS can form DNA adducts that are prone to mutagenesis and carcinogenesis[31].
Table 1.
Genetic polymorphisms associated with alcohol dependence, alcohol abuse and liver disease
| Gene | Risk allele | Association | Population | Ref. |
| Alcohol-metabolizing enzymes | ||||
| CYP2E1 | CYP2E1*c2 | Higher susceptibility to LC; decompensated liver function | Mexican (Mestizo), West Mexico | [110] |
| ADH1B | ADH1B*2 | Higher risk to LC | Japanese | [111] |
| ADH1B | ADH1B*1 | Alcohol dependence | European, Asian | [112-114] |
| ALDH2 | ALDH2*1 | Higher susceptibility to LC | Japanese | [111] |
| Alcohol dependence genes | ||||
| DRD2 | Taq I A1 | Alcohol dependence | European, East Asian | [115] |
| TAS2R38 | AVV haplotype | Higher alcohol intake | Mexican, (Mestizo), West Mexico | [39] |
| Lipid metabolism | ||||
| APOE | APOE*2 | Hypertriglyceridemia and increased development of early LC | Mexican (Mestizo), West Mexico | [36] |
| FABP2 | Ala54 | Earlier onset of LC | Mexican (Mestizo), West Mexico | [116] |
| PNPLA3 | M148 | Alcoholic liver disease and clinically evident LC | Mixed European and Native American, Mexico City | [117,118] |
| PPAR-γ2 | Ala12 | Increased risk to develop severe steatohepatitis and fibrosis | German | [119] |
| Immune response | ||||
| TNF-α | -238 A | Higher prevalence of LC | Spanish | [120] |
| NF-ΚB | ATTG deletion | Higher prevalence of LC | Spanish | [121] |
| CXCL1 | rs4074 A | Higher prevalence of LC | German | [122] |
| CD14 | -159 T | Advanced liver disease, hepatitis and especially with LC | Finnish | [123,124] |
LC: Liver cirrhosis.
Furthermore, in alcoholic liver disease (ALD), additional genetic factors involved in alcohol dependence and alcohol abuse have been intensively studied. Among these factors are the dopamine receptor D2 (DRD2); the bitter taste receptor (TAS2R38); and the liver enzymes alcohol dehydrogenase class I, beta polypeptide (ADH1B), cytochrome P450, family 2, subfamily E, polypeptide 1 (CYP2E1) and aldehyde dehydrogenase 2 family (ALDH2). Moreover, polymorphisms of lipid metabolizing genes, such as apolipoprotein E (APOE), fatty acid-binding protein 2 (FABP2), patatin-like phospholipase domain- containing 3 (PNPLA3) and peroxisome proliferator-activated receptor γ2 (PPAR-γ2) have been associated with the severity of ALD. Others genes related to the process are inflammatory tumor necrosis factor alpha (TNF-α), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), CXC chemokine ligand 1 (CXCL1) and CD14 endotoxin receptor (CD14) (Table 1).
Additionally, environmental cultural factors involved in ALD may vary among populations. In Spain, drinking moderate amounts of red wine is part of the traditional Mediterranean diet and has been associated with reduced all-cause mortality[32]. Mexico has a strong Spanish cultural influence; however, the Mexican population did not inherit this Mediterranean alcohol drinking pattern. Instead, the Mexican population has a pattern of alcohol consumption that differs from what has been reported in other regions worldwide[24]. Specifically, in West Mexico, young people consume 80 to 360 g of alcohol (mainly beer) over the weekends in a period of 10 years. In the next decade, alcohol consumption eventually increases from 360 to 640 g of alcohol per day, including beer and a combination of beer and distilled beverages, such as tequila. Finally, 3-5 years later, the amount of alcohol increases up to 720 g daily, mainly tequila[24,33], until LC clinically manifests[34,35]. This pattern of alcohol consumption is consistent with the age-related onset of alcohol-induced LC previously reported[36]. The average peak age for LC onset has been reported to be 45 years; however, one group reported an average age of 30 years, which may be the earliest onset ever reported most likely due to genetic susceptibility[36]. Genetic polymorphisms in the CYP2E1, APOE, and FABP2 genes have been associated with the early onset of LC in the population of West Mexico (Table 1). However, the distribution of these genetic polymorphisms has demonstrated ethnic differences. Interestingly, the APOE*2 risk allele is highly frequent in the Caucasian population[12], whereas the CYP2E1*c2 risk allele prevails among Amerindians[37]. Other factors may affect the development of alcoholic liver damage, including dose, duration and type of alcohol consumption; drinking pattern; gender; obesity; iron overload; and diet[24]. Consistently, patients with chronic alcohol abuse had a higher intake of cholesterol, sodium and simple carbohydrates than other groups with liver disease, which may accelerate hepatocellular injury[38].
Furthermore, certain polymorphisms that are associated with Amerindian ancestry may have placed the current-day Mestizo population at a higher risk of genetic susceptibility to alcoholism. The novel AVV haplotype of TAS2R38, which has not been identified in other populations, was associated with alcohol abuse in the population of West Mexico[39]. Globally, Mexico has the highest frequency of the allele A1 in DRD2, which has been correlated with alcohol addiction[24]. The identification of these factors has contributed to an integrative understanding of ALD in Mexico. However, further large-scale studies that involve distinct ethnic groups are needed to determine how genetics may influence the onset and progression of LC.
VIRAL HEPATITIS
Hepatitis C virus
The hepatitis C virus (HCV) is a hepatotropic single-stranded RNA virus of the Flaviviridae family[40]. HCV infection is one of the most common causes of chronic liver disease worldwide, with an estimated prevalence of 3% or 170 million infected people[41]. Latin America has one of the lowest prevalences (1.23%); however, this prevalence varies between different regions[42]. In Mexico, 400000 to 1400000 individuals are infected with HCV. Of these individuals, 200000 to 700000 present with active viremia[43]. HCV has been classified into six main genotypes with a heterogeneous global distribution. HCV genotypes 4, 5 and 6 are common in Africa and South East Asia, whereas genotypes 2 and 3 are more frequent in European countries[44]. However, the most predominant HCV genotypes in Mexico are 1a and 1b[45], which have been associated with increased resistance to treatment with pegylated interferon and ribavirin. These HCV endemic differences are explained in part by the prevalent routes of transmission in each region. In West Mexico, the predominant risk factors for HCV transmission are blood transfusions from infectious donors and surgeries with contaminated surgical instruments[46]. In contrast, in North America, injection drug use is the major route of HCV infection[47].
Approximately 18%-34% of patients with acute HCV infection undergo spontaneous clearance through a sustained, vigorous and virus-specific CD4+ T-cell response in peripheral blood[48]. However, in cases of persistent HCV infection, the virus evades the host innate immune response[49]. This condition may potentially increase the risk of progression to hepatic fibrosis, cirrhosis and hepatocellular carcinoma (HCC) over 20-30 years of HCV infection[50]. HCV is a non-cytopathic virus; therefore, the related liver damage is mainly caused by the host immune response. The potential mechanisms of LC include portal lymphoid infiltration, focal and bridging necrosis, and degenerative lobular lesions, which may be amplified by the accumulation of specific T cells that are recruited by adhesion molecules and chemokine expression[51].
Studies in West Mexico have found that cytokines influence the extent of HCV development in patients infected with genotype 1a. Our data reveal that chronic infection results in an increased secretion of interleukin 8 (IL-8) and the chemokine (C-C motif) ligand 5 (CCL5), whereas patients who spontaneously cleared HCV exhibited augmented levels of interleukin 1 alpha (IL-1α), interleukin 13 (IL-13), interleukin 15 (IL-15), TNF-α, TGF-β1 and the chemokine (C-C motif) ligand 8 (CCL8)[52]. This finding correlates with studies conducted in regions where the virus is endemic and suggests that variations in the profile of cytokines involved in the immune response may contribute to either the ability to clear HCV or to the long-term persistence of HCV and thus the potential development of LC[53].
Chronic HCV infection causes numerous pathogenic changes in the liver, including iron overload, steatosis, insulin resistance, the induction of endoplasmic reticulum stress, the unfolded protein response, oxidative stress, mitochondrial dysfunction and altered growth control[54-56]. Other factors are significantly associated with the progression of fibrosis in chronic HCV carriers. These factors include the duration of infection, advanced age, male sex, heavy alcohol use (> 50 g/d), HIV coinfection, diabetes and a low CD4 count[57-59]. Moreover, physical inactivity, obesity, and related lipid alterations may play a critical role in the progression of fibrosis[42]. Recently, we found that approximately 65% of HCV-infected patients in West Mexico have a sedentary lifestyle, and approximately 70% of these patients are overweight or obese[38]. Because overnutrition may influence the course of infection, the analysis of distinct cohorts, including obese and non-obese individuals, should be conducted to characterize the influence of this factor on LC development.
In addition to environmental and viral features, host genetic factors are important contributors to the modulation of HCV outcomes[42]. Diverse polymorphisms in genes involved in lipid metabolism have been described, such as apolipoprotein B (APOB), APOE, LDL receptor (LDLr), microsomal triglyceride transfer protein (MTTP) and PNPLA3. Polymorphisms in several immune regulatory genes have been demonstrated to influence HCV outcomes, including CXCL1, interleukin 28B (IL-28B), TGF-β1 and TNF-α; and matrix metalloproteinase 1 (MMP-1), 3 (MMP-3) and 9 (MMP-9). Additionally, genetic polymorphisms that have been implicated in the metabolism of homocysteine (methylenetetrahydrofolate reductase, MTHFR), iron (hemochromatosis gene, HFE), and vitamin D (vitamin D receptor, VDR) have been reported (Table 2). Finally, a meta-analysis demonstrated that two polymorphisms in IL-28B (rs12979860 CC and rs8099917 TT) were strong predictors of sustained virological response in patients on pegylated interferon and ribavirin treatment[60].
Table 2.
Genetic polymorphisms associated with the outcomes of hepatitis C virus infection
| Gene | Risk allele | Association | Population | Ref. |
| Lipid metabolism | ||||
| APOB | -516 C | Increased susceptibility of HCV infection | Chinese | [125] |
| APOE | APOE*3 | Viral persistence | Northern European | [126] |
| LDLr | rs2738459 C, rs2569540 G, rs1433099 A, rs11672123 A | Higher viral load in genotypes 1 and 4 | Spanish | [127] |
| MTTP | -493 T | Higher degree of steatosis, HCV RNA serum levels and hepatic fibrosis | Native Italian | [128,129] |
| PNPLA3 | M148 | Higher risk for steatosis and fibrosis progression | European: Belgian, German and French | [130] |
| Inmune response mediators | ||||
| CXCL1 | rs4074 A | Higher risk for LC | German | [131] |
| IL-28B | rs12979860 T | Higher risk for LC and HCC | Native Italian and Chinese | [132,133] |
| TGF-β1 | -509 T | Higher risk for LC and HCC | Egyptian | [134] |
| TNF-α | -308 A | Higher risk for LC and HCC | Egyptian | [134] |
| Fibrogenesis | ||||
| MMP-1 | -1607 2G | Higher prevalence of LC | Japanese | [135] |
| MMP-3 | -1171 5A | Lower age at LC diagnosis and a higher Child-Pugh score | Japanese | [135] |
| MMP-9 | -1562 C | Higher prevalence of LC | Japanese | [135] |
| Nutrient metabolism | ||||
| MTHFR | 677 T | Hyperhomocysteinemia and higher degree of steatosis and fibrosis | Italian | [136] |
| HFE | 63 D | Higher likelihood of LC | Taiwanese | [137] |
| VDR | CAA haplotype (rs1544410 C, rs7975232 A, rs731236 A) | Higher fibrosis progression and LC | Swiss | [138] |
LC: Liver cirrhosis; HCV: Hepatitis C virus; HCC: Hepatocellular carcinoma.
Hepatitis B virus
The hepatitis B virus (HBV) is a noncytopathic, hepatotropic virus of the Hepadnaviridae family[61]. HBV infection is a public health problem, in which nearly 400 million people suffer chronic HBV infection[62]. In Mexico, epidemiological studies have indicated that at least 15 million adults may have been exposed to HBV during their lifetime[63], and up to 300000 individuals may be active carriers[64]. Globally, geographical areas with a high endemicity of HBV infection include Asian countries, regions in South America with indigenous peoples and Alaska[65,66]. In contrast, Mexico is considered a region of low endemicity[43], which is associated with predominant HBV genotypes H and G[67], low hepatitis B surface antigen (HBsAg) seroprevalence[64], and low viral loads[63] but a high prevalence of occult B infection (OBI) among native Mexican groups (Nahuas and Huichol)[68]. OBI is diagnosed based on a negative serum hepatitis B surface antigen (HBsAg) test, the presence of HBV DNA in the liver and very low HBV DNA levels in serum[69]. Therefore, given these features, the detection of HBV infection in Mexico may be underestimated in the setting of HBV-induced chronic liver disease.
The outcome of HBV infection is the result of complex interactions between HBV and the host immune system[53,70]. A study of cytokine sera profiles in native Mexican groups revealed that OBI patients displayed increased interleukin 2 (IL-2) secretion and a characteristic inflammatory profile (reduction in IL-8 and TNF-α, and increased levels of TGF-β1)[71]. This finding correlates with the accepted role of TGF-β1 in the progression of liver disease[72], whereas the high secretion of IL-2 suggests that OBI may be modulated by IL-2. Experimental data indicate that cytotoxic T cells are the main cell type responsible for the inhibition of viral replication and for hepatocyte lysis[73].
Moreover, HBV genotypes influence the natural course of infection and related liver damage. Regarding the distribution of HBV genotypes, HBV genotype H in native and Mestizo Mexicans and genotype F in native Latin Americans are linked to a less severe natural course of disease and a faster resolution of infection[74]. This finding may be due to a high degree of immune adaptation to the virus in these populations. Nonetheless, in Mestizo Americans, genotype F is commonly associated with acute and chronic liver disease with a tendency toward HCC[74].
Additionally, HBV is associated with metabolic alterations in host cells, which lead to hepatic steatosis, oxidative stress, inflammation, and carcinogenesis. The main mechanisms include the transcriptional up-regulation of genes involved in the biosynthesis of lipids, phosphatidylcholine, and hexosamine; as well as modifications in cell proliferation and differentiation. Together, such changes promote HBV replication and liver damage[75]. Other factors, such as alcohol abuse (> 60 g/d) for at least 10 years, central obesity, insulin resistance, diabetes and environmental hepatotoxins, including tobacco smoke and aflatoxins, may increase the progression rate of liver injury in chronic HBV-infected patients[76]. In addition, genetic polymorphisms modulate the outcome of HBV infection. These polymorphisms include interleukin 10 (IL-10), IL-28B, TGF-β1 and the collagenases, type I, alpha 1 (COL1A1) and type III, alpha 1 (COL3A1) (Table 3).
Table 3.
Genetic polymorphisms associated with the outcomes of hepatitis B virus infection
| Gene | Risk allele | Association | Population | Ref. |
| Immune response mediators | ||||
| IL-10 | -592 C | Significant increased risk of LC | Asian | [139] |
| IL-28B | rs12979860 C | Increased risk for developing LC | Asian | [140] |
| TGF-β1 | 10 T | Higher prevalence of LC | Chinese | [141] |
| TGF-β1 | -509 C | Higher susceptibility to LC | Chinese | [142] |
| Fibrogenesis | ||||
| COL1A1 | TC haplotype (-1997 T, -1363 C) | Higher prevalence of LC | Chinese | [143] |
| COL3A1 | rs3106796 A | Higher prevalence of chronic hepatitis, LC and HCC | Koreans | [144] |
LC: Liver cirrhosis; HCC: Hepatocellular carcinoma.
Other viral hepatitis
Recently, we described a high seroprevalence of the hepatitis E virus (HEV) in serum samples from cirrhotic patients in whom no etiological agent was found[43]. Consequently, this finding suggests a potential role of HEV in LC development and is consistent with the description of HEV-related cirrhosis in immunocompromised patients[77]. Further epidemiological studies are warranted among the Mexican population to elucidate the plausible influence of HEV in liver disease.
NAFLD
Obesity is considered the main factor associated with multiple metabolic disorders, such as NAFLD, which comprises simple steatosis, NASH, cirrhosis and HCC[78,79]. Hepatic steatosis is defined by the presence of cytoplasmic triglyceride (TG) droplets in more than 5% of hepatocytes as a result of an imbalance between lipid input and output[80,81]. The abnormal accumulation of TGs in the liver induces lipotoxicity, endoplasmic reticulum (ER) stress, mitochondrial dysfunction, and inflammation, which all result in fibrosis (Figure 3)[82,83]. Conventionally, international guidelines and intervention strategies have been proposed for the treatment of NAFLD/NASH. However, because disparities in genetic and environmental factors among populations were not considered, the application of these strategies may not be feasible for all populations worldwide. Therefore, intervention strategies for the prevention and management of such obesity-related diseases should be based on a regionalized genome-based diet by focusing on the specific ancestry of each population and the convenience of consuming traditional ethnic food[11].
Figure 3.
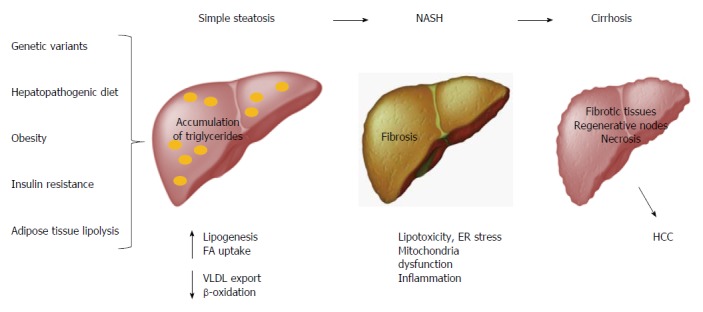
Nonalcoholic fatty liver disease pathogenesis. The spectrum of NAFLD includes simple steatosis, NASH, LC and even HCC. Risk factors such as obesity, hepatopathogenic diet, insulin resistance and adipose tissue lipolysis lead to accumulation of triglycerides. This abnormality can stimulate lipotoxicity, ER stress, mitochondria dysfunction and inflammation, promoting fibrosis. Chronic fibrogenesis causes histological changes in the liver that lead to LC, which in turn may evolve to HCC. NAFLD: Nonalcoholic fatty liver disease; NASH: Nonalcoholic steatohepatitis; LC: Liver cirrhosis; HCC: Hepatocellular carcinoma; FA: Fatty acids; VLDL: Very low-density lipoprotein; ER: Endoplasmic reticulum.
Currently, Mexico is one of the countries with the highest number of obese individuals, which is closely related to dietary factors and physical inactivity[84]. The diet in West Mexico is hepatopathogenic because of its high content of fat derived mainly from the habitual consumption of meat, fried foods and sausages[11,38]. Interestingly, we have observed that a genetic variant in the fat taste CD36 receptor may play an important role in this condition[85]. High-fat diets are associated with an increase in body weight, the upregulation of blood glucose levels, the progression of steatosis, and marked inflammation of the liver[86]. Moreover, more than 70% of the Mexican population frequently consumes soda, which increases fructose input[38]. Fructose promotes steatosis, insulin resistance, necroinflammation, and fibrosis[87-89]. In contrast, our diet is deficient in n-3 polyunsaturated fatty acids (PUFA) and antioxidants, which favors oxidative stress and lipid alterations[90]. Finally, inadequate dietary behaviors, such as missing breakfast and a lack of feeding schedules, are common among the Mexican population[38] and have been linked to obesity[91].
Other interactions between genes and environmental lifestyle factors that favor changes in the body mass index have been studied in West Mexico. Dietary modifications consisting of a lower intake of saturated fat (< 7%) significantly decreased body mass index, waist circumference, waist-to-hip ratio, and reactive C protein in FABP2 Thr54 allele carriers compared to Ala54 allele carriers[92]. Additionally, recent studies in an obese cohort demonstrated that the expression of adiponectin levels may be modulated by changes in lifestyle regardless of the presence of an adiponectin (ADIPOQ) 11391G/A polymorphism[93].
However, despite the strong correlation between obesity and NAFLD, approximately 5% of morbidly obese patients do not develop NAFLD[94]. This finding supports the role of genetic factors in the development of NAFLD and NASH (Table 4). These factors may include polymorphisms in genes that affect lipid metabolism, such as apolipoprotein C3 (APOC3), PNPLA3, MTTP, and phosphatidylethanolamine N-methyltransferase (PEMT). Polymorphisms in genes that affect insulin sensitivity, such as ADIPOQ, adiponectin receptors 1 and 2 (ADIPOR 1 and ADIPOR 2), PPAR-γ, PPAR-γ coactivator 1α gene (PPARGC1A) and PPAR-α, may play a role in the development of NAFLD and NASH. Additionally, regulatory genes of oxidative stress, including glutamate cysteine ligase (GCLC), inducible nitric oxide synthase (NOS2), and manganese superoxide dismutase (SOD2), have been implicated. Genes related to immune regulation, including signal transducer and activator of transcription 3 (STAT3), TNF-α, IL-8 and interleukin-6 (IL-6); as well as MTHFR and HFE have been described. Finally, other less common genes that have been associated with the development of NAFLD and NASH include ATP-binding cassette subfamily C member 2 (ABCC2, also known as multidrug resistance protein 2, MRP2) and angiotensin II type I receptor (AGTR1)[95].
Table 4.
Genetic polymorphisms associated with the development of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis
| Gene | Risk allele | Association | Population | Ref. |
| Lipid metabolism | ||||
| APOC3 | 482 T, 455 C | Higher fasting plasma triglyceride concentration and higher prevalence of NAFLD | Asian Indian | [145] |
| PNPLA3 | M148 | Increased hepatic fat levels, hepatic inflammation and fibrosis in NAFLD and NASH patients | Hispanic, African American, European American, Finnish, Argentinean, Italian | [146-150] |
| MTTP | -493 G | Higher intrahepatic triglycerides content. Higher incidence and progression of NASH | French, Japanese | [151,152] |
| PEMT | M175 | Higher prevalence of NAFLD and NASH | Hispanic, African American, European American, Asian | [153,154] |
| Insulin resistance/sensitivity | ||||
| ADIPOQ | 45 T, 276 T | Higher prevalence of NAFLD. Lower postprandial adiponectin and higher postprandial triglyceride, VLDL, and FFA in NASH patients | Italian | [155] |
| 45 G, 276 G | Higher prevalence of NAFLD, severe fibrosis and insulin resistance in females | Japanese | [156] | |
| ADIPOR1 | -8503 A, -1927 C | Lower insulin sensitivity and higher liver fat | German | [157] |
| ADIPOR2 | rs767870 T | Increased hepatic fat and biochemical surrogates of NAFLD | Finnish | [158] |
| PPAR-γ | 161 T | Higher susceptibility of NAFLD | Chinese | [159] |
| PPARGC1A | rs2290602 T | Higher occurrence of NAFLD | Japanese | [160] |
| PPAR-α | Val227 | Higher prevalence of NAFLD and anthropometrical indicators of obesity | Chinese | [161] |
| Oxidative stress | ||||
| GCLC | -129 T | Higher prevalence of NASH | Brazilian | [162] |
| NOS2 | rs1060822 T | Higher fibrosis index in NAFLD patients | Japanese | [163] |
| SOD2 | 1183 T | Higher prevalence of NASH | Japanese | [152] |
| Immune response mediators | ||||
| STAT3 | rs6503695 T, rs9891119 A | Higher prevalence of NAFLD | Argentinean | [164] |
| TNF-α | -238 A | Higher prevalence of NAFLD and NASH | Italian, Chinese | [165,166] |
| IL-8 | -251 A | Disease progression in NASH | Turkish | [167] |
| IL-6 | -174 C | Higher risk for NAFLD and NASH | Italian | [168] |
| MTHFR | 1298 C, 677 C | Higher prevalence of NASH | Turkish | [169] |
| HFE | 282 Y | More hepatic fibrosis in NASH patients. Higher prevalence of NAFLD | Australian | [170-172] |
| ABCC2/MRP2 | rs17222723 T, rs8187710 A | NAFLD disease severity | Argentinean | [173] |
| AGTR1 | rs3772633 G, rs3772627 C, rs3772622 A | Higher prevalence of NASH | Japanese | [174] |
NAFLD: Nonalcoholic fatty liver disease; NASH: Nonalcoholic steatohepatitis.
HCC
Globally, HCC is the most common form of liver cancer, the sixth most prevalent cancer and the third most frequent cause of cancer-related death[96]. Nearly 75% of HCC cases are attributed to chronic HBV and HCV infections in high endemic regions[97]. Therefore, the particular characteristics of these viruses and the genetic structure and environmental factors that prevail in each population may explain the epidemiological differences in HCC worldwide. Regions with higher incidence rates of this malignant disease include sub-Saharan Africa, Eastern Asia, followed by Southern European countries[32]. However, HCC is rare in other geographical areas, such as North and South America, Northern Europe, Oceania, and Mexico[98]. The low incidence of HBV-related HCC among the Mexican population is associated with a low steady HBsAg seroprevalence[64]. Furthermore, because Mexican patients with alcoholic cirrhosis are young, death occurs earlier due to long-term complications. This fact hinders the study of the association between alcohol consumption and HCC in Mexico because the average age of death from HCC is at least one or two decades after the clinical diagnosis of LC[98]. Similarly, epidemiological studies have indicated a positive correlation between liver cancer and the consumption of several foodstuffs contaminated with aflatoxins and fumonisins, including maize, cereals, ground nuts and tree nuts[99]. These mycotoxins are potent liver carcinogens through the formation of pro-mutagenic DNA adducts. Nonetheless, the process of “nixtamalization” in the elaboration of Mexican tortillas since pre-Hispanic times[100] inactivates up to 95% of aflatoxins in corn (maize), which may exert a protective effect against the development of HCC[101].
EPIGENETIC FACTORS
Epigenetic changes affect gene expression without altering the underlying DNA sequence[102]. These changes include DNA methylation, histone modifications, chromatin remodeling, and microRNAs (miRs), which are essential for the proper maintenance of cellular homeostasis[103]. However, such processes are regulated by environmental factors, including diet, alcohol, drugs, exercise and stress[104]. Growing evidence suggests that epigenetic alterations are correlated with a wide range of chronic disorders, including liver diseases. Diverse miRs have been implicated in the development and progression of NAFLD by disrupting lipid and glucose metabolism, insulin resistance, the unfolded protein response, mitochondrial damage, ER stress, oxidative stress, cellular differentiation, inflammation and apoptosis[105,106]. Moreover, the reduction of histone expression promotes the differentiation of HSCs to myofibroblasts, thereby enhancing the fibrogenesis process[107]. Interestingly, alcohol directly stimulates changes in chromatin structure, which induces the trans-differentiation of HSCs and the increased expression of ECM proteins[108]. Global DNA hypomethylation and the dysregulated expression of non-coding RNAs and epigenetic regulatory genes have been found to trigger carcinogenesis of hepatocytes[109]. Taken together, these findings support the need to evaluate epigenetic factors in distinct populations and their relationship to the severity of liver damage.
CONCLUSION
LC is a complex disease. The onset, progression, and clinical outcome of LC are influenced by genetic polymorphisms that are associated with particular ethnic backgrounds and their interaction with environmental factors, such as lifestyle. Currently, chronic alcohol abuse is the main etiological factor of LC in Mexico, which is associated mainly with cultural and genetic aspects. To date, HCV genotypes 1a and 1b favor chronicity, liver damage and therapeutic failure among Mexicans. In contrast, the more favorable natural course of HBV infection in the native Mexican population may be linked to an evolutionary immune adaptation to HBV genotype H. Finally, the hepatopathogenic diet and the high prevalence of obesity in our population contribute to the onset and progression of NAFLD. These findings highlight the influence of genetic and environmental factors in the development of LC, which may be different than those reported in other regions of the world. Therefore, these factors represent a challenge for hepatologists and gastroenterologists in Mexico and worldwide because they require the incorporation of personalized genomic medicine into current medical practices. However, the development of intervention strategies according to individual or population-based features will have a substantial impact on the prevention, management and treatment of LC.
Footnotes
Supported by Promep-University of Guadalajara (UDG-CA-478, to Panduro A).
Conflict-of-interest statement: The authors declare no conflicts of interest.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: May 6, 2015
First decision: July 13, 2015
Article in press: September 14, 2015
P- Reviewer: Niu zs, Ruiz-Margain A S- Editor: Ma YJ L- Editor: A E- Editor: Zhang DN
References
- 1.Sharma S, Khalili K, Nguyen GC. Non-invasive diagnosis of advanced fibrosis and cirrhosis. World J Gastroenterol. 2014;20:16820–16830. doi: 10.3748/wjg.v20.i45.16820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsochatzis EA, Bosch J, Burroughs AK. Liver cirrhosis. Lancet. 2014;383:1749–1761. doi: 10.1016/S0140-6736(14)60121-5. [DOI] [PubMed] [Google Scholar]
- 3.Blachier M, Leleu H, Peck-Radosavljevic M, Valla DC, Roudot-Thoraval F. The burden of liver disease in Europe: a review of available epidemiological data. J Hepatol. 2013;58:593–608. doi: 10.1016/j.jhep.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Mokdad AA, Lopez AD, Shahraz S, Lozano R, Mokdad AH, Stanaway J, Murray CJ, Naghavi M. Liver cirrhosis mortality in 187 countries between 1980 and 2010: a systematic analysis. BMC Med. 2014;12:145. doi: 10.1186/s12916-014-0145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838–851. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarrilli F, Elce A, Scorza M, Giordano S, Amato F, Castaldo G. An update on laboratory diagnosis of liver inherited diseases. Biomed Res Int. 2013;2013:697940. doi: 10.1155/2013/697940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou WC, Zhang QB, Qiao L. Pathogenesis of liver cirrhosis. World J Gastroenterol. 2014;20:7312–7324. doi: 10.3748/wjg.v20.i23.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefton HB, Rosa A, Cohen M. Diagnosis and epidemiology of cirrhosis. Med Clin North Am. 2009;93:787–99, vii. doi: 10.1016/j.mcna.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Dongiovanni P, Anstee QM, Valenti L. Genetic predisposition in NAFLD and NASH: impact on severity of liver disease and response to treatment. Curr Pharm Des. 2013;19:5219–5238. doi: 10.2174/13816128113199990381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roman S, Panduro A. Genomic medicine in gastroenterology: A new approach or a new specialty? World J Gastroenterol. 2015;21:8227–8237. doi: 10.3748/wjg.v21.i27.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roman S, Ojeda-Granados C, Ramos-Lopez O, Panduro A. Genome-based nutrition: an intervention strategy for the prevention and treatment of obesity and nonalcoholic steatohepatitis. World J Gastroenterol. 2015;21:3449–3461. doi: 10.3748/wjg.v21.i12.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aceves D, Ruiz B, Nuño P, Roman S, Zepeda E, Panduro A. Heterogeneity of apolipoprotein E polymorphism in different Mexican populations. Hum Biol. 2006;78:65–75. doi: 10.1353/hub.2006.0021. [DOI] [PubMed] [Google Scholar]
- 13.Iwaisako K, Brenner DA, Kisseleva T. What’s new in liver fibrosis? The origin of myofibroblasts in liver fibrosis. J Gastroenterol Hepatol. 2012;27 Suppl 2:65–68. doi: 10.1111/j.1440-1746.2011.07002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novo E, Cannito S, Paternostro C, Bocca C, Miglietta A, Parola M. Cellular and molecular mechanisms in liver fibrogenesis. Arch Biochem Biophys. 2014;548:20–37. doi: 10.1016/j.abb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000;275:2247–2250. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- 16.Mallat A, Lotersztajn S. Cellular mechanisms of tissue fibrosis. 5. Novel insights into liver fibrosis. Am J Physiol Cell Physiol. 2013;305:C789–C799. doi: 10.1152/ajpcell.00230.2013. [DOI] [PubMed] [Google Scholar]
- 17.Lee UE, Friedman SL. Mechanisms of hepatic fibrogenesis. Best Pract Res Clin Gastroenterol. 2011;25:195–206. doi: 10.1016/j.bpg.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elpek GÖ. Cellular and molecular mechanisms in the pathogenesis of liver fibrosis: An update. World J Gastroenterol. 2014;20:7260–7276. doi: 10.3748/wjg.v20.i23.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hui AY, Friedman SL. Molecular basis of hepatic fibrosis. Expert Rev Mol Med. 2003;5:1–23. doi: 10.1017/S1462399403005684. [DOI] [PubMed] [Google Scholar]
- 20.Friedman SL. Mechanisms of disease: Mechanisms of hepatic fibrosis and therapeutic implications. Nat Clin Pract Gastroenterol Hepatol. 2004;1:98–105. doi: 10.1038/ncpgasthep0055. [DOI] [PubMed] [Google Scholar]
- 21.Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruha R, Dvorak K, Petrtyl J. Alcoholic liver disease. World J Hepatol. 2012;4:81–90. doi: 10.4254/wjh.v4.i3.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Shea RS, Dasarathy S, McCullough AJ. Alcoholic liver disease. Hepatology. 2010;51:307–328. doi: 10.1002/hep.23258. [DOI] [PubMed] [Google Scholar]
- 24.Roman S, Zepeda-Carrillo EA, Moreno-Luna LE, Panduro A. Alcoholism and liver disease in Mexico: genetic and environmental factors. World J Gastroenterol. 2013;19:7972–7982. doi: 10.3748/wjg.v19.i44.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cederbaum AI, Lu Y, Wu D. Role of oxidative stress in alcohol-induced liver injury. Arch Toxicol. 2009;83:519–548. doi: 10.1007/s00204-009-0432-0. [DOI] [PubMed] [Google Scholar]
- 26.Medina J, Moreno-Otero R. Pathophysiological basis for antioxidant therapy in chronic liver disease. Drugs. 2005;65:2445–2461. doi: 10.2165/00003495-200565170-00003. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gustot T, Lemmers A, Moreno C, Nagy N, Quertinmont E, Nicaise C, Franchimont D, Louis H, Devière J, Le Moine O. Differential liver sensitization to toll-like receptor pathways in mice with alcoholic fatty liver. Hepatology. 2006;43:989–1000. doi: 10.1002/hep.21138. [DOI] [PubMed] [Google Scholar]
- 29.Liu J. Ethanol and liver: recent insights into the mechanisms of ethanol-induced fatty liver. World J Gastroenterol. 2014;20:14672–14685. doi: 10.3748/wjg.v20.i40.14672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ceni E, Mello T, Galli A. Pathogenesis of alcoholic liver disease: role of oxidative metabolism. World J Gastroenterol. 2014;20:17756–17772. doi: 10.3748/wjg.v20.i47.17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 32.Gea A, Bes-Rastrollo M, Toledo E, Garcia-Lopez M, Beunza JJ, Estruch R, Martinez-Gonzalez MA. Mediterranean alcohol-drinking pattern and mortality in the SUN (Seguimiento Universidad de Navarra) Project: a prospective cohort study. Br J Nutr. 2014;111:1871–1880. doi: 10.1017/S0007114513004376. [DOI] [PubMed] [Google Scholar]
- 33.Campollo O, Martínez MD, Valencia JJ, Segura-Ortega J. Drinking patterns and beverage preferences of liver cirrhosis patients in Mexico. Subst Use Misuse. 2001;36:387–398. doi: 10.1081/ja-100102632. [DOI] [PubMed] [Google Scholar]
- 34.Bastidas-Ramirez BE, Nuño-Gonzalez P, Vivas-Arceo C, Sánchez-Orozco LV, Panduro A. Albumin mRNA in peripheral white blood cells of cirrhotic patients with a superimposed alcoholic hepatitis is associated to fatal outcome. Hepatol Res. 2002;24:265. doi: 10.1016/s1386-6346(02)00109-2. [DOI] [PubMed] [Google Scholar]
- 35.Nuño-González P, Ruíz-Madrigal B, Bastidas-Ramírez BE, Martínez-López E, Segura JE, Panduro A. Expression of apolipoprotein AI mRNA in peripheral white blood cells of patients with alcoholic liver disease. Biochim Biophys Acta. 2005;1740:350–356. doi: 10.1016/j.bbadis.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Hernández-Nazará ZH, Ruiz-Madrigal B, Martínez-López E, Roman S, Panduro A. Association of the epsilon 2 allele of APOE gene to hypertriglyceridemia and to early-onset alcoholic cirrhosis. Alcohol Clin Exp Res. 2008;32:559–566. doi: 10.1111/j.1530-0277.2007.00607.x. [DOI] [PubMed] [Google Scholar]
- 37.Gordillo-Bastidas E, Panduro A, Gordillo-Bastidas D, Zepeda-Carrillo EA, García-Bañuelos JJ, Muñoz-Valle JF, Bastidas-Ramírez BE. Polymorphisms of alcohol metabolizing enzymes in indigenous Mexican population: unusual high frequency of CYP2E1*c2 allele. Alcohol Clin Exp Res. 2010;34:142–149. doi: 10.1111/j.1530-0277.2009.01075.x. [DOI] [PubMed] [Google Scholar]
- 38.Ramos-Lopez O, Roman S, Ojeda-Granados C, Sepúlveda-Villegas M, Martínez-López E, Torres-Valadez R, Trujillo-Trujillo E, Panduro A. Patrón de ingesta alimentaria y actividad física en pacientes hepatópatas en el Occidente de México. Rev Endocrinol Nutr. 2013;21:7–15. [Google Scholar]
- 39.Ramos-Lopez O, Roman S, Martinez-Lopez E, Gonzalez-Aldaco K, Ojeda-Granados C, Sepulveda-Villegas M, Panduro A. Association of a novel TAS2R38 haplotype with alcohol intake among Mexican-Mestizo population. Ann Hepatol. 2015;14:729–734. [PubMed] [Google Scholar]
- 40.Zaltron S, Spinetti A, Biasi L, Baiguera C, Castelli F. Chronic HCV infection: epidemiological and clinical relevance. BMC Infect Dis. 2012;12 Suppl 2:S2. doi: 10.1186/1471-2334-12-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 42.Fierro NA, Gonzalez-Aldaco K, Torres-Valadez R, Martinez-Lopez E, Roman S, Panduro A. Immunologic, metabolic and genetic factors in hepatitis C virus infection. World J Gastroenterol. 2014;20:3443–3456. doi: 10.3748/wjg.v20.i13.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panduro A, Escobedo Meléndez G, Fierro NA, Ruiz Madrigal B, Zepeda-Carrillo EA, Román S. [Epidemiology of viral hepatitis in Mexico] Salud Publica Mex. 2011;53 Suppl 1:S37–S45. [PubMed] [Google Scholar]
- 44.Irshad M, Ansari MA, Singh A, Nag P, Raghvendra L, Singh S, Badhal SS. HCV-genotypes: a review on their origin, global status, assay system, pathogenecity and response to treatment. Hepatogastroenterology. 2010;57:1529–1538. [PubMed] [Google Scholar]
- 45.Panduro A, Roman S, Khan A, Tanaka Y, Kurbanov F, Martinez-Lopez E, Campollo O, Hernandez-Nazara Z, Mizokami M. Molecular epidemiology of hepatitis C virus genotypes in west Mexico. Virus Res. 2010;151:19–25. doi: 10.1016/j.virusres.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Muñoz-Espinosa LE, Trujillo-Trujillo ME, Martínez-Macías RF, Panduro A, Rivas-Estilla AM, Fierro NA, Silvera-Linares AL, Torres-Valadez R, Cordero-Pérez P, González-Aldaco K, et al. Increase of drug use and genotype 3 in HCV-infected patients from Central West and Northeast Mexico. Ann Hepatol. 2015;14:642–651. [PubMed] [Google Scholar]
- 47.Alter MJ. HCV routes of transmission: what goes around comes around. Semin Liver Dis. 2011;31:340–346. doi: 10.1055/s-0031-1297923. [DOI] [PubMed] [Google Scholar]
- 48.Westbrook RH, Dusheiko G. Natural history of hepatitis C. J Hepatol. 2014;61:S58–S68. doi: 10.1016/j.jhep.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 49.Oshiumi H, Matsumoto M, Seya T. [Chronic hepatitis C virus infection attenuates host antiviral innate immune response] Nihon Rinsho. 2015;73:234–238. [PubMed] [Google Scholar]
- 50.Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol. 2013;10:553–562. doi: 10.1038/nrgastro.2013.107. [DOI] [PubMed] [Google Scholar]
- 51.Pawlotsky JM. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 2004;12:96–102. doi: 10.1016/j.tim.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 52.Fierro NA, González-Aldaco K, Torres-Valadez R, Trujillo-Trujillo ME, Roman S, Trujillo-Ochoa JL, Panduro A. Spontaneous hepatitis C viral clearance and hepatitis C chronic infection are associated with distinct cytokine profiles in Mexican patients. Mem Inst Oswaldo Cruz. 2015;110:267–271. doi: 10.1590/0074-02760140377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fierro NA, Castro-Garcia FP, Panduro A. Rethinking cytokine function during hepatitis A and hepatitis C infections. Adv Biosci Biotechnol. 2013;4:13–18. [Google Scholar]
- 54.Isom HC, McDevitt EI, Moon MS. Elevated hepatic iron: a confounding factor in chronic hepatitis C. Biochim Biophys Acta. 2009;1790:650–662. doi: 10.1016/j.bbagen.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Adinolfi LE, Restivo L, Zampino R, Lonardo A, Loria P. Metabolic alterations and chronic hepatitis C: treatment strategies. Expert Opin Pharmacother. 2011;12:2215–2234. doi: 10.1517/14656566.2011.597742. [DOI] [PubMed] [Google Scholar]
- 56.Ivanov AV, Bartosch B, Smirnova OA, Isaguliants MG, Kochetkov SN. HCV and oxidative stress in the liver. Viruses. 2013;5:439–469. doi: 10.3390/v5020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Torres M, Poynard T. Risk factors for liver fibrosis progression in patients with chronic hepatitis C. Ann Hepatol. 2003;2:5–11. [PubMed] [Google Scholar]
- 58.Monto A, Patel K, Bostrom A, Pianko S, Pockros P, McHutchison JG, Wright TL. Risks of a range of alcohol intake on hepatitis C-related fibrosis. Hepatology. 2004;39:826–834. doi: 10.1002/hep.20127. [DOI] [PubMed] [Google Scholar]
- 59.Chiquete E, Ochoa-Guzmán A, García-Lamas L, Anaya-Gómez F, Gutiérrez-Manjarrez JI, Sánchez-Orozco LV, Godínez-Gutiérrez SA, Maldonado M, Román S, Panduro A. Hepatitis C virus infection and type 2 diabetes mellitus in Mexican patients. Rev Med Inst Mex Seguro Soc. 2012;50:481–486. [PubMed] [Google Scholar]
- 60.Chen Y, Xu HX, Wang LJ, Liu XX, Mahato RI, Zhao YR. Meta-analysis: IL28B polymorphisms predict sustained viral response in HCV patients treated with pegylated interferon-α and ribavirin. Aliment Pharmacol Ther. 2012;36:91–103. doi: 10.1111/j.1365-2036.2012.05131.x. [DOI] [PubMed] [Google Scholar]
- 61.Glebe D, Bremer CM. The molecular virology of hepatitis B virus. Semin Liver Dis. 2013;33:103–112. doi: 10.1055/s-0033-1345717. [DOI] [PubMed] [Google Scholar]
- 62.Ioannou GN. Chronic hepatitis B infection: a global disease requiring global strategies. Hepatology. 2013;58:839–843. doi: 10.1002/hep.26516. [DOI] [PubMed] [Google Scholar]
- 63.Panduro A, Maldonado-Gonzalez M, Fierro NA, Roman S. Distribution of HBV genotypes F and H in Mexico and Central America. Antivir Ther. 2013;18:475–484. doi: 10.3851/IMP2605. [DOI] [PubMed] [Google Scholar]
- 64.Roman S, Panduro A, Aguilar-Gutierrez Y, Maldonado M, Vazquez-Vandyck M, Martinez-Lopez E, Ruiz-Madrigal B, Hernandez-Nazara Z. A low steady HBsAg seroprevalence is associated with a low incidence of HBV-related liver cirrhosis and hepatocellular carcinoma in Mexico: a systematic review. Hepatol Int. 2009;3:343–355. doi: 10.1007/s12072-008-9115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Datta S. An overview of molecular epidemiology of hepatitis B virus (HBV) in India. Virol J. 2008;5:156. doi: 10.1186/1743-422X-5-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liaw YF, Chu CM. Hepatitis B virus infection. Lancet. 2009;373:582–592. doi: 10.1016/S0140-6736(09)60207-5. [DOI] [PubMed] [Google Scholar]
- 67.Roman S, Panduro A. HBV endemicity in Mexico is associated with HBV genotypes H and G. World J Gastroenterol. 2013;19:5446–5453. doi: 10.3748/wjg.v19.i33.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roman S, Tanaka Y, Khan A, Kurbanov F, Kato H, Mizokami M, Panduro A. Occult hepatitis B in the genotype H-infected Nahuas and Huichol native Mexican population. J Med Virol. 2010;82:1527–1536. doi: 10.1002/jmv.21846. [DOI] [PubMed] [Google Scholar]
- 69.Pollicino T, Raimondo G. Occult hepatitis B infection. J Hepatol. 2014;61:688–689. doi: 10.1016/j.jhep.2014.04.036. [DOI] [PubMed] [Google Scholar]
- 70.Chen J, Yuan Z. Interplay between hepatitis B virus and the innate immune responses: implications for new therapeutic strategies. Virol Sin. 2014;29:17–24. doi: 10.1007/s12250-014-3412-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fierro NA, Roman S, Realpe M, Hernandez-Nazara Z, Zepeda-Carrillo EA, Panduro A. Multiple cytokine expression profiles reveal immune-based differences in occult hepatitis B genotype H-infected Mexican Nahua patients. Mem Inst Oswaldo Cruz. 2011;106:1007–1013. doi: 10.1590/s0074-02762011000800018. [DOI] [PubMed] [Google Scholar]
- 72.Dooley S, ten Dijke P. TGF-β in progression of liver disease. Cell Tissue Res. 2012;347:245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Busca A, Kumar A. Innate immune responses in hepatitis B virus (HBV) infection. Virol J. 2014;11:22. doi: 10.1186/1743-422X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roman S, Jose-Abrego A, Fierro NA, Escobedo-Melendez G, Ojeda-Granados C, Martinez-Lopez E, Panduro A. Hepatitis B virus infection in Latin America: a genomic medicine approach. World J Gastroenterol. 2014;20:7181–7196. doi: 10.3748/wjg.v20.i23.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H, Zhu W, Zhang L, Lei H, Wu X, Guo L, Chen X, Wang Y, Tang H. The metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatment. Sci Rep. 2015;5:8421. doi: 10.1038/srep08421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fattovich G, Bortolotti F, Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol. 2008;48:335–352. doi: 10.1016/j.jhep.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 77.Kamar N, Dalton HR, Abravanel F, Izopet J. Hepatitis E virus infection. Clin Microbiol Rev. 2014;27:116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raszeja-Wyszomirska J, Lawniczak M, Marlicz W, Miezyńska-Kurtycz J, Milkiewicz P. [Non-alcoholic fatty liver disease--new view] Pol Merkur Lekarski. 2008;24:568–571. [PubMed] [Google Scholar]
- 79.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koo SH. Nonalcoholic fatty liver disease: molecular mechanisms for the hepatic steatosis. Clin Mol Hepatol. 2013;19:210–215. doi: 10.3350/cmh.2013.19.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berlanga A, Guiu-Jurado E, Porras JA, Auguet T. Molecular pathways in non-alcoholic fatty liver disease. Clin Exp Gastroenterol. 2014;7:221–239. doi: 10.2147/CEG.S62831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Neuschwander-Tetri BA. Hepatic lipotoxicity and the pathogenesis of nonalcoholic steatohepatitis: the central role of nontriglyceride fatty acid metabolites. Hepatology. 2010;52:774–788. doi: 10.1002/hep.23719. [DOI] [PubMed] [Google Scholar]
- 83.Ramadori P, Kroy D, Streetz KL. Immunoregulation by lipids during the development of non-alcoholic steatohepatitis. Hepatobiliary Surg Nutr. 2015;4:11–23. doi: 10.3978/j.issn.2304-3881.2015.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rtveladze K, Marsh T, Barquera S, Sanchez Romero LM, Levy D, Melendez G, Webber L, Kilpi F, McPherson K, Brown M. Obesity prevalence in Mexico: impact on health and economic burden. Public Health Nutr. 2014;17:233–239. doi: 10.1017/S1368980013000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramos-Lopez O, Panduro A, Martinez-Lopez E, Fierro NA, Ojeda-Granados C, Sepulveda-Villegas M, Roman S. Genetic variant in the CD36 Gene (rs1761667) is associated with higher fat intake and high serum cholesterol among the population of West Mexico. J Nutr Food Sci. 2015;5:353. [Google Scholar]
- 86.Yasutake K, Kohjima M, Nakashima M, Kotoh K, Nakamuta M, Enjoji M. Nutrition therapy for liver diseases based on the status of nutritional intake. Gastroenterol Res Pract. 2012;2012:859697. doi: 10.1155/2012/859697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nseir W, Nassar F, Assy N. Soft drinks consumption and nonalcoholic fatty liver disease. World J Gastroenterol. 2010;16:2579–2588. doi: 10.3748/wjg.v16.i21.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Basaranoglu M, Basaranoglu G, Sabuncu T, Sentürk H. Fructose as a key player in the development of fatty liver disease. World J Gastroenterol. 2013;19:1166–1172. doi: 10.3748/wjg.v19.i8.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moore JB, Gunn PJ, Fielding BA. The role of dietary sugars and de novo lipogenesis in non-alcoholic fatty liver disease. Nutrients. 2014;6:5679–5703. doi: 10.3390/nu6125679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Conlon BA, Beasley JM, Aebersold K, Jhangiani SS, Wylie-Rosett J. Nutritional management of insulin resistance in nonalcoholic fatty liver disease (NAFLD) Nutrients. 2013;5:4093–4114. doi: 10.3390/nu5104093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yasutake K, Kohjima M, Kotoh K, Nakashima M, Nakamuta M, Enjoji M. Dietary habits and behaviors associated with nonalcoholic fatty liver disease. World J Gastroenterol. 2014;20:1756–1767. doi: 10.3748/wjg.v20.i7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martinez-Lopez E, Garcia-Garcia MR, Gonzalez-Avalos JM, Maldonado-Gonzalez M, Ruiz-Madrigal B, Vizmanos B, Hernandez-Nazara Z, Roman S, Panduro A. Effect of Ala54Thr polymorphism of FABP2 on anthropometric and biochemical variables in response to a moderate-fat diet. Nutrition. 2013;29:46–51. doi: 10.1016/j.nut.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 93.Garcia-Garcia MR, Morales-Lanuza MA, Campos-Perez WY, Ruiz-Madrigal B, Maldonado-Gonzalez M, Vizmanos B, Hernandez-Cañaveral I, Yañez-Sanchez I, Roman S, Panduro A, et al. Effect of the ADIPOQ Gene -11391G/A Polymorphism Is Modulated by Lifestyle Factors in Mexican Subjects. J Nutrigenet Nutrigenomics. 2014;7:212–224. doi: 10.1159/000371801. [DOI] [PubMed] [Google Scholar]
- 94.Ong JP, Elariny H, Collantes R, Younoszai A, Chandhoke V, Reines HD, Goodman Z, Younossi ZM. Predictors of nonalcoholic steatohepatitis and advanced fibrosis in morbidly obese patients. Obes Surg. 2005;15:310–315. doi: 10.1381/0960892053576820. [DOI] [PubMed] [Google Scholar]
- 95.Hooper AJ, Adams LA, Burnett JR. Genetic determinants of hepatic steatosis in man. J Lipid Res. 2011;52:593–617. doi: 10.1194/jlr.R008896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 97.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pujol FH, Roman S, Panduro A, Navas MC, Lampe E. Hepatocellular Carcinoma in Latin America. In: Chemin S, editor. Hepatocellular Carcinoma: A Global Challenge. Nova Biomedical; 2012. pp. 56–68. [Google Scholar]
- 99.Ok HE, Kim HJ, Shim WB, Lee H, Bae DH, Chung DH, Chun HS. Natural occurrence of aflatoxin B1 in marketed foods and risk estimates of dietary exposure in Koreans. J Food Prot. 2007;70:2824–2828. doi: 10.4315/0362-028x-70.12.2824. [DOI] [PubMed] [Google Scholar]
- 100.Roman S, Fierro NA, Moreno-Luna LE, Panduro A. Hepatitis B virus genotype H and environmental factors associated to the low prevalence of hepatocellular carcinoma in Mexico. J Cancer Ther. 2013;4:2A. [Google Scholar]
- 101.Anguiano-Ruvalcaba GL, Verver y Vargas-Cortina A, Guzmán-de Peña D. [Inactivation of aflatoxin B1 and aflatoxicol through traditional “nixtamalización” of corn and their regeneration by acidification of corn dough] Salud Publica Mex. 2005;47:369–375. doi: 10.1590/s0036-36342005000500007. [DOI] [PubMed] [Google Scholar]
- 102.Choi SW, Friso S. Epigenetics: A New Bridge between Nutrition and Health. Adv Nutr. 2010;1:8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee JH, Friso S, Choi SW. Epigenetic mechanisms underlying the link between non-alcoholic fatty liver diseases and nutrition. Nutrients. 2014;6:3303–3325. doi: 10.3390/nu6083303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lo CL, Zhou FC. Environmental alterations of epigenetics prior to the birth. Int Rev Neurobiol. 2014;115:1–49. doi: 10.1016/B978-0-12-801311-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li YY. Genetic and epigenetic variants influencing the development of nonalcoholic fatty liver disease. World J Gastroenterol. 2012;18:6546–6551. doi: 10.3748/wjg.v18.i45.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun C, Fan JG, Qiao L. Potential epigenetic mechanism in non-alcoholic Fatty liver disease. Int J Mol Sci. 2015;16:5161–5179. doi: 10.3390/ijms16035161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao Q, Qin CY, Zhao ZH, Fan YC, Wang K. Epigenetic modifications in hepatic stellate cells contribute to liver fibrosis. Tohoku J Exp Med. 2013;229:35–43. doi: 10.1620/tjem.229.35. [DOI] [PubMed] [Google Scholar]
- 108.Page A, Paoli PP, Hill SJ, Howarth R, Wu R, Kweon SM, French J, White S, Tsukamoto H, Mann DA, et al. Alcohol directly stimulates epigenetic modifications in hepatic stellate cells. J Hepatol. 2015;62:388–397. doi: 10.1016/j.jhep.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ozen C, Yildiz G, Dagcan AT, Cevik D, Ors A, Keles U, Topel H, Ozturk M. Genetics and epigenetics of liver cancer. N Biotechnol. 2013;30:381–384. doi: 10.1016/j.nbt.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 110.García-Bañuelos J, Panduro A, Gordillo-Bastidas D, Gordillo-Bastidas E, Muñoz-Valle JF, Gurrola-Díaz CM, Sánchez-Enríquez S, Ruiz-Madrigal B, Bastidas-Ramírez BE. Genetic polymorphisms of genes coding to alcohol-metabolizing enzymes in western Mexicans: association of CYP2E1*c2/CYP2E1*5B allele with cirrhosis and liver function. Alcohol Clin Exp Res. 2012;36:425–431. doi: 10.1111/j.1530-0277.2011.01617.x. [DOI] [PubMed] [Google Scholar]
- 111.Yokoyama A, Mizukami T, Matsui T, Yokoyama T, Kimura M, Matsushita S, Higuchi S, Maruyama K. Genetic polymorphisms of alcohol dehydrogenase-1B and aldehyde dehydrogenase-2 and liver cirrhosis, chronic calcific pancreatitis, diabetes mellitus, and hypertension among Japanese alcoholic men. Alcohol Clin Exp Res. 2013;37:1391–1401. doi: 10.1111/acer.12108. [DOI] [PubMed] [Google Scholar]
- 112.Whitfield JB. Alcohol dehydrogenase and alcohol dependence: variation in genotype-associated risk between populations. Am J Hum Genet. 2002;71:1247–150; author reply 1247-150;. doi: 10.1086/344287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. 2009;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van Beek JH, Willemsen G, de Moor MH, Hottenga JJ, Boomsma DI. Associations between ADH gene variants and alcohol phenotypes in Dutch adults. Twin Res Hum Genet. 2010;13:30–42. doi: 10.1375/twin.13.1.30. [DOI] [PubMed] [Google Scholar]
- 115.Munafò MR, Matheson IJ, Flint J. Association of the DRD2 gene Taq1A polymorphism and alcoholism: a meta-analysis of case-control studies and evidence of publication bias. Mol Psychiatry. 2007;12:454–461. doi: 10.1038/sj.mp.4001938. [DOI] [PubMed] [Google Scholar]
- 116.Salguero ML, Leon RE, Santos A, Roman S, Segura-Ortega JE, Panduro A. The role of FABP2 gene polymorphism in alcoholic cirrhosis. Hepatol Res. 2005;33:306–312. doi: 10.1016/j.hepres.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 117.Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet. 2010;42:21–23. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 118.Falleti E, Fabris C, Cmet S, Cussigh A, Bitetto D, Fontanini E, Fornasiere E, Bignulin S, Fumolo E, Bignulin E, et al. PNPLA3 rs738409C/G polymorphism in cirrhosis: relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int. 2011;31:1137–1143. doi: 10.1111/j.1478-3231.2011.02534.x. [DOI] [PubMed] [Google Scholar]
- 119.Rey JW, Noetel A, Hardt A, Canbay A, Alakus H, Zur Hausen A, Dienes HP, Drebber U, Odenthal M. Pro12Ala polymorphism of the peroxisome proliferator-activated receptor γ2 in patients with fatty liver diseases. World J Gastroenterol. 2010;16:5830–5837. doi: 10.3748/wjg.v16.i46.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pastor IJ, Laso FJ, Romero A, González-Sarmiento R. -238 G& gt; A polymorphism of tumor necrosis factor alpha gene (TNFA) is associated with alcoholic liver cirrhosis in alcoholic Spanish men. Alcohol Clin Exp Res. 2005;29:1928–1931. doi: 10.1097/01.alc.0000187595.19324.ca. [DOI] [PubMed] [Google Scholar]
- 121.Marcos M, Pastor I, González-Sarmiento R, Laso FJ. A functional polymorphism of the NFKB1 gene increases the risk for alcoholic liver cirrhosis in patients with alcohol dependence. Alcohol Clin Exp Res. 2009;33:1857–1862. doi: 10.1111/j.1530-0277.2009.01023.x. [DOI] [PubMed] [Google Scholar]
- 122.Nischalke HD, Berger C, Lutz P, Langhans B, Wolter F, Eisenhardt M, Krämer B, Kokordelis P, Glässner A, Müller T, et al. Influence of the CXCL1 rs4074 A allele on alcohol induced cirrhosis and HCC in patients of European descent. PLoS One. 2013;8:e80848. doi: 10.1371/journal.pone.0080848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Järveläinen HA, Orpana A, Perola M, Savolainen VT, Karhunen PJ, Lindros KO. Promoter polymorphism of the CD14 endotoxin receptor gene as a risk factor for alcoholic liver disease. Hepatology. 2001;33:1148–1153. doi: 10.1053/jhep.2001.24236. [DOI] [PubMed] [Google Scholar]
- 124.Meiler C, Muhlbauer M, Johann M, Hartmann A, Schnabl B, Wodarz N, Schmitz G, Scholmerich J, Hellerbrand C. Different effects of a CD14 gene polymorphism on disease outcome in patients with alcoholic liver disease and chronic hepatitis C infection. World J Gastroenterol. 2005;11:6031–6037. doi: 10.3748/wjg.v11.i38.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu C, Zhang R, Liu D, Mukhtar MM, Liu W, Peng G, Wang K, Hao Q, Xu Y, Liu F, et al. Association of functional polymorphism of ApoB promoter with hepatitis C virus infection. Clin Chim Acta. 2009;401:124–127. doi: 10.1016/j.cca.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 126.Price DA, Bassendine MF, Norris SM, Golding C, Toms GL, Schmid ML, Morris CM, Burt AD, Donaldson PT. Apolipoprotein epsilon3 allele is associated with persistent hepatitis C virus infection. Gut. 2006;55:715–718. doi: 10.1136/gut.2005.079905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Caruz A, Neukam K, Rivero-Juárez A, Herrero R, Real LM, Camacho A, Barreiro P, Labarga P, Rivero A, Pineda JA. Association of low-density lipoprotein receptor genotypes with hepatitis C viral load. Genes Immun. 2014;15:16–24. doi: 10.1038/gene.2013.56. [DOI] [PubMed] [Google Scholar]
- 128.Zampino R, Ingrosso D, Durante-Mangoni E, Capasso R, Tripodi MF, Restivo L, Zappia V, Ruggiero G, Adinolfi LE. Microsomal triglyceride transfer protein (MTP) -493G/T gene polymorphism contributes to fat liver accumulation in HCV genotype 3 infected patients. J Viral Hepat. 2008;15:740–746. doi: 10.1111/j.1365-2893.2008.00994.x. [DOI] [PubMed] [Google Scholar]
- 129.Mirandola S, Bowman D, Hussain MM, Alberti A. Hepatic steatosis in hepatitis C is a storage disease due to HCV interaction with microsomal triglyceride transfer protein (MTP) Nutr Metab (Lond) 2010;7:13. doi: 10.1186/1743-7075-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Trépo E, Pradat P, Potthoff A, Momozawa Y, Quertinmont E, Gustot T, Lemmers A, Berthillon P, Amininejad L, Chevallier M, et al. Impact of patatin-like phospholipase-3 (rs738409 C& gt; G) polymorphism on fibrosis progression and steatosis in chronic hepatitis C. Hepatology. 2011;54:60–69. doi: 10.1002/hep.24350. [DOI] [PubMed] [Google Scholar]
- 131.Nischalke HD, Berger C, Luda C, Müller T, Berg T, Coenen M, Krämer B, Körner C, Trebicka J, Grünhage F, et al. The CXCL1 rs4074 A allele is associated with enhanced CXCL1 responses to TLR2 ligands and predisposes to cirrhosis in HCV genotype 1-infected Caucasian patients. J Hepatol. 2012;56:758–764. doi: 10.1016/j.jhep.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 132.Fabris C, Falleti E, Cussigh A, Bitetto D, Fontanini E, Bignulin S, Cmet S, Fornasiere E, Fumolo E, Fangazio S, et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. J Hepatol. 2011;54:716–722. doi: 10.1016/j.jhep.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 133.Suo GJ, Zhao ZX. Association of the interleukin-28B gene polymorphism with development of hepatitis virus-related hepatocellular carcinoma and liver cirrhosis: a meta-analysis. Genet Mol Res. 2013;12:3708–3717. doi: 10.4238/2013.September.19.1. [DOI] [PubMed] [Google Scholar]
- 134.Radwan MI, Pasha HF, Mohamed RH, Hussien HI, El-Khshab MN. Influence of transforming growth factor-β1 and tumor necrosis factor-α genes polymorphisms on the development of cirrhosis and hepatocellular carcinoma in chronic hepatitis C patients. Cytokine. 2012;60:271–276. doi: 10.1016/j.cyto.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 135.Okamoto K, Mimura K, Murawaki Y, Yuasa I. Association of functional gene polymorphisms of matrix metalloproteinase (MMP)-1, MMP-3 and MMP-9 with the progression of chronic liver disease. J Gastroenterol Hepatol. 2005;20:1102–1108. doi: 10.1111/j.1440-1746.2005.03860.x. [DOI] [PubMed] [Google Scholar]
- 136.Adinolfi LE, Ingrosso D, Cesaro G, Cimmino A, D’Antò M, Capasso R, Zappia V, Ruggiero G. Hyperhomocysteinemia and the MTHFR C677T polymorphism promote steatosis and fibrosis in chronic hepatitis C patients. Hepatology. 2005;41:995–1003. doi: 10.1002/hep.20664. [DOI] [PubMed] [Google Scholar]
- 137.Mah YH, Kao JH, Liu CJ, Chen CL, Chen PJ, Lai MY, Chen DS. Prevalence and clinical implications of HFE gene mutations (C282Y and H63D) in patients with chronic hepatitis B and C in Taiwan. Liver Int. 2005;25:214–219. doi: 10.1111/j.1478-3231.2005.01055.x. [DOI] [PubMed] [Google Scholar]
- 138.Baur K, Mertens JC, Schmitt J, Iwata R, Stieger B, Eloranta JJ, Frei P, Stickel F, Dill MT, Seifert B, et al. Combined effect of 25-OH vitamin D plasma levels and genetic vitamin D receptor (NR 1I1) variants on fibrosis progression rate in HCV patients. Liver Int. 2012;32:635–643. doi: 10.1111/j.1478-3231.2011.02674.x. [DOI] [PubMed] [Google Scholar]
- 139.Jin XY, Wang YQ, Yan T, Wang J, Qing S, Ding N, Tian H, Zhao P. Interleukin-10 gene promoter polymorphism and susceptibility to liver cirrhosis. Hepatogastroenterology. 2014;61:442–446. [PubMed] [Google Scholar]
- 140.Xia P, Zhou M, Dong DS, Xing YN, Bai Y. Association of polymorphisms in interleukin-18 and interleukin-28B genes with outcomes of hepatitis B virus infections: a meta-analysis. Tumour Biol. 2014;35:1129–1137. doi: 10.1007/s13277-013-1151-y. [DOI] [PubMed] [Google Scholar]
- 141.Yang ZX, Wang H, Gao CF, Xu LL, Zhao WJ. [Effect of polymorphism of transforming growth factor beta1 gene on HBV-induced liver cirrhosis] Zhonghua Yi Xue Zazhi. 2005;85:1021–1026. [PubMed] [Google Scholar]
- 142.Li H, Wu HL, Lv H, Wei HS, Wang HB, Wang PL. [The association of TGF beta1 and AT1R gene polymorphisms with hereditary susceptibility and clinical phenotype of HBV-induced liver cirrhosis] Zhonghua Yi Xue Yi Chuan Xue Zazhi. 2007;24:298–301. [PubMed] [Google Scholar]
- 143.Zhao YP, Wang H, Fang M, Ji Q, Yang ZX, Gao CF. Study of the association between polymorphisms of the COL1A1 gene and HBV-related liver cirrhosis in Chinese patients. Dig Dis Sci. 2009;54:369–376. doi: 10.1007/s10620-008-0340-9. [DOI] [PubMed] [Google Scholar]
- 144.Lee SK, Yi CH, Kim MH, Cheong JY, Cho SW, Yang SJ, Kwack K. Genetic association between functional haplotype of collagen type III alpha 1 and chronic hepatitis B and cirrhosis in Koreans. Tissue Antigens. 2008;72:539–548. doi: 10.1111/j.1399-0039.2008.01144.x. [DOI] [PubMed] [Google Scholar]
- 145.Petersen KF, Dufour S, Hariri A, Nelson-Williams C, Foo JN, Zhang XM, Dziura J, Lifton RP, Shulman GI. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med. 2010;362:1082–1089. doi: 10.1056/NEJMoa0907295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kotronen A, Johansson LE, Johansson LM, Roos C, Westerbacka J, Hamsten A, Bergholm R, Arkkila P, Arola J, Kiviluoto T, et al. A common variant in PNPLA3, which encodes adiponutrin, is associated with liver fat content in humans. Diabetologia. 2009;52:1056–1060. doi: 10.1007/s00125-009-1285-z. [DOI] [PubMed] [Google Scholar]
- 148.Sookoian S, Castaño GO, Burgueño AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894–903. doi: 10.1002/hep.23759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Valenti L, Al-Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, Nobili V, Mozzi E, Roviaro G, Vanni E, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 151.Bernard S, Touzet S, Personne I, Lapras V, Bondon PJ, Berthezène F, Moulin P. Association between microsomal triglyceride transfer protein gene polymorphism and the biological features of liver steatosis in patients with type II diabetes. Diabetologia. 2000;43:995–999. doi: 10.1007/s001250051481. [DOI] [PubMed] [Google Scholar]
- 152.Namikawa C, Shu-Ping Z, Vyselaar JR, Nozaki Y, Nemoto Y, Ono M, Akisawa N, Saibara T, Hiroi M, Enzan H, et al. Polymorphisms of microsomal triglyceride transfer protein gene and manganese superoxide dismutase gene in non-alcoholic steatohepatitis. J Hepatol. 2004;40:781–786. doi: 10.1016/j.jhep.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 153.Song J, da Costa KA, Fischer LM, Kohlmeier M, Kwock L, Wang S, Zeisel SH. Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD) FASEB J. 2005;19:1266–1271. doi: 10.1096/fj.04-3580com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dong H, Wang J, Li C, Hirose A, Nozaki Y, Takahashi M, Ono M, Akisawa N, Iwasaki S, Saibara T, et al. The phosphatidylethanolamine N-methyltransferase gene V175M single nucleotide polymorphism confers the susceptibility to NASH in Japanese population. J Hepatol. 2007;46:915–920. doi: 10.1016/j.jhep.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 155.Musso G, Gambino R, De Michieli F, Durazzo M, Pagano G, Cassader M. Adiponectin gene polymorphisms modulate acute adiponectin response to dietary fat: Possible pathogenetic role in NASH. Hepatology. 2008;47:1167–1177. doi: 10.1002/hep.22142. [DOI] [PubMed] [Google Scholar]
- 156.Tokushige K, Hashimoto E, Noto H, Yatsuji S, Taniai M, Torii N, Shiratori K. Influence of adiponectin gene polymorphisms in Japanese patients with non-alcoholic fatty liver disease. J Gastroenterol. 2009;44:976–982. doi: 10.1007/s00535-009-0085-z. [DOI] [PubMed] [Google Scholar]
- 157.Stefan N, Machicao F, Staiger H, Machann J, Schick F, Tschritter O, Spieth C, Weigert C, Fritsche A, Stumvoll M, et al. Polymorphisms in the gene encoding adiponectin receptor 1 are associated with insulin resistance and high liver fat. Diabetologia. 2005;48:2282–2291. doi: 10.1007/s00125-005-1948-3. [DOI] [PubMed] [Google Scholar]
- 158.Kotronen A, Yki-Järvinen H, Aminoff A, Bergholm R, Pietiläinen KH, Westerbacka J, Talmud PJ, Humphries SE, Hamsten A, Isomaa B, et al. Genetic variation in the ADIPOR2 gene is associated with liver fat content and its surrogate markers in three independent cohorts. Eur J Endocrinol. 2009;160:593–602. doi: 10.1530/EJE-08-0900. [DOI] [PubMed] [Google Scholar]
- 159.Hui Y, Yu-Yuan L, Yu-Qiang N, Wei-Hong S, Yan-Lei D, Xiao-Bo L, Yong-Jian Z. Effect of peroxisome proliferator-activated receptors-gamma and co-activator-1alpha genetic polymorphisms on plasma adiponectin levels and susceptibility of non-alcoholic fatty liver disease in Chinese people. Liver Int. 2008;28:385–392. doi: 10.1111/j.1478-3231.2007.01623.x. [DOI] [PubMed] [Google Scholar]
- 160.Yoneda M, Hotta K, Nozaki Y, Endo H, Uchiyama T, Mawatari H, Iida H, Kato S, Hosono K, Fujita K, et al. Association between PPARGC1A polymorphisms and the occurrence of nonalcoholic fatty liver disease (NAFLD) BMC Gastroenterol. 2008;8:27. doi: 10.1186/1471-230X-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen S, Li Y, Li S, Yu C. A Val227Ala substitution in the peroxisome proliferator activated receptor alpha (PPAR alpha) gene associated with non-alcoholic fatty liver disease and decreased waist circumference and waist-to-hip ratio. J Gastroenterol Hepatol. 2008;23:1415–1418. doi: 10.1111/j.1440-1746.2008.05523.x. [DOI] [PubMed] [Google Scholar]
- 162.Oliveira CP, Stefano JT, Cavaleiro AM, Zanella Fortes MA, Vieira SM, Rodrigues Lima VM, Santos TE, Santos VN, de Azevedo Salgado AL, Parise ER, et al. Association of polymorphisms of glutamate-cystein ligase and microsomal triglyceride transfer protein genes in non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2010;25:357–361. doi: 10.1111/j.1440-1746.2009.06001.x. [DOI] [PubMed] [Google Scholar]
- 163.Yoneda M, Hotta K, Nozaki Y, Endo H, Tomeno W, Watanabe S, Hosono K, Mawatari H, Iida H, Fujita K, et al. Influence of inducible nitric oxide synthase polymorphisms in Japanese patients with non-alcoholic fatty liver disease. Hepatol Res. 2009;39:963–971. doi: 10.1111/j.1872-034X.2009.00539.x. [DOI] [PubMed] [Google Scholar]
- 164.Sookoian S, Castaño G, Gianotti TF, Gemma C, Rosselli MS, Pirola CJ. Genetic variants in STAT3 are associated with nonalcoholic fatty liver disease. Cytokine. 2008;44:201–206. doi: 10.1016/j.cyto.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 165.Valenti L, Fracanzani AL, Dongiovanni P, Santorelli G, Branchi A, Taioli E, Fiorelli G, Fargion S. Tumor necrosis factor alpha promoter polymorphisms and insulin resistance in nonalcoholic fatty liver disease. Gastroenterology. 2002;122:274–280. doi: 10.1053/gast.2002.31065. [DOI] [PubMed] [Google Scholar]
- 166.Hu ZW, Luo HB, Xu YM, Guo JW, Deng XL, Tong YW, Tang X. Tumor necrosis factor--alpha gene promoter polymorphisms in Chinese patients with nonalcoholic fatty liver diseases. Acta Gastroenterol Belg. 2009;72:215–221. [PubMed] [Google Scholar]
- 167.Cengiz M, Yasar DG, Ergun MA, Akyol G, Ozenirler S. The role of interleukin-6 and interleukin-8 gene polymorphisms in non-alcoholic steatohepatitis. Hepat Mon. 2014;14:e24635. doi: 10.5812/hepatmon.24635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Carulli L, Canedi I, Rondinella S, Lombardini S, Ganazzi D, Fargion S, De Palma M, Lonardo A, Ricchi M, Bertolotti M, et al. Genetic polymorphisms in non-alcoholic fatty liver disease: interleukin-6-174G/C polymorphism is associated with non-alcoholic steatohepatitis. Dig Liver Dis. 2009;41:823–828. doi: 10.1016/j.dld.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 169.Sazci A, Ergul E, Aygun C, Akpinar G, Senturk O, Hulagu S. Methylenetetrahydrofolate reductase gene polymorphisms in patients with nonalcoholic steatohepatitis (NASH) Cell Biochem Funct. 2008;26:291–296. doi: 10.1002/cbf.1424. [DOI] [PubMed] [Google Scholar]
- 170.George DK, Goldwurm S, MacDonald GA, Cowley LL, Walker NI, Ward PJ, Jazwinska EC, Powell LW. Increased hepatic iron concentration in nonalcoholic steatohepatitis is associated with increased fibrosis. Gastroenterology. 1998;114:311–318. doi: 10.1016/s0016-5085(98)70482-2. [DOI] [PubMed] [Google Scholar]
- 171.Bonkovsky HL, Jawaid Q, Tortorelli K, LeClair P, Cobb J, Lambrecht RW, Banner BF. Non-alcoholic steatohepatitis and iron: increased prevalence of mutations of the HFE gene in non-alcoholic steatohepatitis. J Hepatol. 1999;31:421–429. doi: 10.1016/s0168-8278(99)80032-4. [DOI] [PubMed] [Google Scholar]
- 172.Valenti L, Dongiovanni P, Fracanzani AL, Santorelli G, Fatta E, Bertelli C, Taioli E, Fiorelli G, Fargion S. Increased susceptibility to nonalcoholic fatty liver disease in heterozygotes for the mutation responsible for hereditary hemochromatosis. Dig Liver Dis. 2003;35:172–178. doi: 10.1016/s1590-8658(03)00025-2. [DOI] [PubMed] [Google Scholar]
- 173.Sookoian S, Castaño G, Gianotti TF, Gemma C, Pirola CJ. Polymorphisms of MRP2 (ABCC2) are associated with susceptibility to nonalcoholic fatty liver disease. J Nutr Biochem. 2009;20:765–770. doi: 10.1016/j.jnutbio.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 174.Yoneda M, Hotta K, Nozaki Y, Endo H, Uchiyama T, Mawatari H, Iida H, Kato S, Fujita K, Takahashi H, et al. Association between angiotensin II type 1 receptor polymorphisms and the occurrence of nonalcoholic fatty liver disease. Liver Int. 2009;29:1078–1085. doi: 10.1111/j.1478-3231.2009.01988.x. [DOI] [PubMed] [Google Scholar]


