Abstract
TCP proteins are plant-specific transcription factors involved in many different processes. Because of their involvement in a large number of developmental pathways, their roles have been investigated in various plant species. However, there are almost no studies of this transcription factor family in orchids. Based on the available transcriptome of the inflorescence of the orchid Orchis italica, in the present study we identified 12 transcripts encoding TCP proteins. The phylogenetic analysis showed that they belong to different TCP classes (I and II) and groups (PCF, CIN and CYC/TB1), and that they display a number of conserved motifs when compared with the TCPs of Arabidopsis and Oryza. The presence of a specific cleavage site for the microRNA miRNA319, an important post-transcriptional regulator of several TCP genes in other species, was demonstrated for one transcript of O. italica, and the analysis of the expression pattern of the TCP transcripts in different inflorescence organs and in leaf tissue suggests that some TCP transcripts of O. italica exert their role only in specific tissues, while others may play multiple roles in different tissues. In addition, the evolutionary analysis showed a general purifying selection acting on the coding region of these transcripts.
The TCP genes encode plant-specific transcription factors that regulate plant growth and development by controlling cell proliferation. Initially identified in angiosperms, members of the TCP family have also been found in green algae, mosses, ferns, lycophytes and gymnosperms, showing that their origin predates the emergence of land plants1. The members of this large family of proteins share the TCP domain, a non-canonical ∼60 amino acid basic helix-loop-helix (bHLH) motif involved in DNA binding, nuclear localization and protein-protein interactions2,3. The acronym TCP derives from the name of the proteins where this domain was initially described: TEOSINTE BRANCHED1 (TB1) from Zea mays4, CYCLOIDEA (CYC) from Antirrhinum majus5 and PROLIFERATING CELL FACTORS 1 and 2 (PCF1 and PCF2) from Oryza sativa6.
Based on the sequence differences in the TCP domain, the members of the TCP family can be classified into two subfamilies, class I (or PCF or TCP-P) and class II (or TCP-C)1,3,7. The main difference between these two groups is the presence of an additional short amino acid stretch (four residues) in the basic domain of the class II proteins. In addition, the consensus binding site on their DNA targets is different, even if overlapping: GGNCCCAC for class I and G(T/C)GGNCCC for class II proteins7,8. Outside the TCP domain, some members of the class II subfamily present an arginine-rich motif of 18–20 amino acids called the R domain, whose function is unknown2. The members of the class II subfamily are quite heterogeneous and can be further classified into two sub-clades: the CYC/TB1-like group, also known as the ECE group based on the presence of a glutamic acid-cysteine-glutamic acid stretch9, and the CIN-like group3. Both class I and class II members are present in some green algae, making it difficult to establish which TCP subfamily is more ancient. In contrast, the apparent absence of CYC/TB1-like genes (class II) in mosses and lycophytes supports the hypothesis that the CIN-like genes are more ancestral10.
The role of class I TCP genes during plant development is mainly to promote cell division, growth and differentiation, and most of them act in a redundant manner6,11. In contrast, class II genes are generally involved in the inhibition of cell growth and proliferation3, and their expression is strictly controlled at various transcriptional and post-transcriptional levels. For example, some angiosperm members of the CIN-like clade involved in leaf and flower morphogenesis are targeted by the microRNA miR31912,13,14,15,16. Recently, the role of TCP proteins has been demonstrated in plant immunity responses, through the interaction with pathogenic effectors17, and in mitochondrial metabolism, through the binding to the cis-acting regulatory element TGGGGCY18. Another relevant role of the TCP genes, in particular the members of the CYC/TB1-like clade, is the establishment of the floral monosymmetry through their interactions with MYB transcription factors and cell cycle proteins19,20,21. Zygomorphic flowers have evolved independently multiple times during the angiosperm evolution. The involvement of CYC/TB1-like genes in the evolution and maintenance of bilateral symmetry in dicots has been extensively analyzed5,22,23,24,25,26,27,28, whereas only a few studies are available for monocots29,30,31,32. Other members of the CYC/TB1-like genes, e.g. TB1 of Zea mays and its orthologs in dicots BRC1, are involved in the control of lateral branches33,34. Among monocots, the family Orchidaceae is characterized by highly specialized zygomorphic flowers, and to date, CYC/TB1-like genes have not been identified in orchids35.
The application of the next-generation sequencing techniques is producing an increasing amount of data that opens new opportunities to study the TCP genes at the genomic and transcriptomic level36,37,38,39 in non-model species, including orchids40,41,42. The family Orchidaceae is one of the most species-rich plant families including species with highly specialized morphological and physiological characteristics. Orchis italica (sub-family Orchidoideae, tribe Orchidinae) is an Eurasian orchid species. Its inflorescence is oval and dense, with numerous pink flowers that exhibit the typical organization of the orchid floral organs43. In brief, there are three sepals (outer tepals) and three petals distinguished in two inner lateral tepals and one inner median tepal (lip or labellum). The column is a fusion of male and female reproductive tissues, at the top of which are located the pollen grains. The maturation of the ovary, placed at the base of the column, is triggered by pollination44,45.
In the present paper, we report the transcriptome-wide identification of the TCP genes expressed in the inflorescence of Orchis italica based on the inflorescence transcriptome46 and miRNAome47 available for this wild Mediterranean species. We examined the expression patterns of the identified TCP transcripts and of the microRNA miR319 in different tissues and performed a comparative analysis to identify conserved motifs among the TCP proteins of O. italica, other orchid species and the model species, Arabidopsis thaliana and Oryza sativa.
Results and Discussion
Transcripts encoding TCP proteins in the inflorescence of O. italica
To identify transcripts encoding TCP proteins, the inflorescence transcriptome of O. italica46 was screened through a standalone TBLASTN search using as query the sequences of the TCP DNA-binding domain of A. thaliana and O. sativa. Eleven not-redundant transcripts (ranging from 850 to 2788 bp) present in the inflorescence transcriptome of O. italica contain a region encoding the TCP domain (Supplementary Data S1 and Figure S1) flanked downstream by at least 500 bp of additional sequence. This indicates that at least eleven distinct TCP genes are expressed in the inflorescence of O. italica, increasing the small number of known TCP genes of orchids (five in Phalaenopsis hybrid cultivar and one in Dendrobium hybrid cultivar)35. Surprisingly, a first phylogenetic analysis of these virtually translated sequences of O. italica and the TCP proteins of Arabidopsis thaliana and Oryza sativa revealed that none of the TCP transcripts present in the inflorescence transcriptome of O. italica belong to the class II CYC/TB1-like group, whose members are involved in the establishment of bilateral symmetry in numerous plant species26,28,48,49,50,51. The recent release of the assembled genome of the orchid species Phalaenopsis equestris42 provided for the first time the opportunity to check for the presence of TCP genes belonging to the class II CYC/TB1-like group within the genome of an orchid species. Using the TCP transcripts of O. italica as queries, standalone BLASTN and BLASTX searches of the assembled genome of P. equestris were conducted. In addition, the annotated sequences of the genome of P. equestris were checked to verify the presence of additional TCP genes. Excluding three sequences with an incomplete TCP domain, in the genome of P. equestris there are 17 TCP-encoding genes and among them three belong to the CYC/TB1-like group (Supplementary Table S1). To check for the presence of CYC/TB1-like sequences in O. italica, degenerate primers based on the CYC/TB1-like sequences of P. equestris were used to amplify the genomic DNA of O. italica. The single fragment of 378 bp obtained from this PCR amplification (named OitaTB1 and deposited in GenBank with the accession number KR858306) encodes the region from the TCP domain to the R domain. The online TBLAST search confirmed its homology with CYC/TB1-like genes. As expected, the attempts to obtain the full-length cDNA of this sequence starting from the RNA extracted from an inflorescence of O. italica failed. In fact, the absence of an assembled transcript in the inflorescence transcriptome of O. italica corresponding to OitaTB1 suggests that this gene is not expressed or it is expressed at very low levels and/or only in specific tissues of the inflorescence of O. italica at the stage of development examined. Further attempts to amplify the full-length cDNA of OitaTB1 starting from RNA extracted from various tissues of the inflorescence or from leaf tissue did not give positive results. However, the OitaTB1 sequence was used for all the subsequent analyses.
Phylogeny, conserved motifs and evolutionary analysis
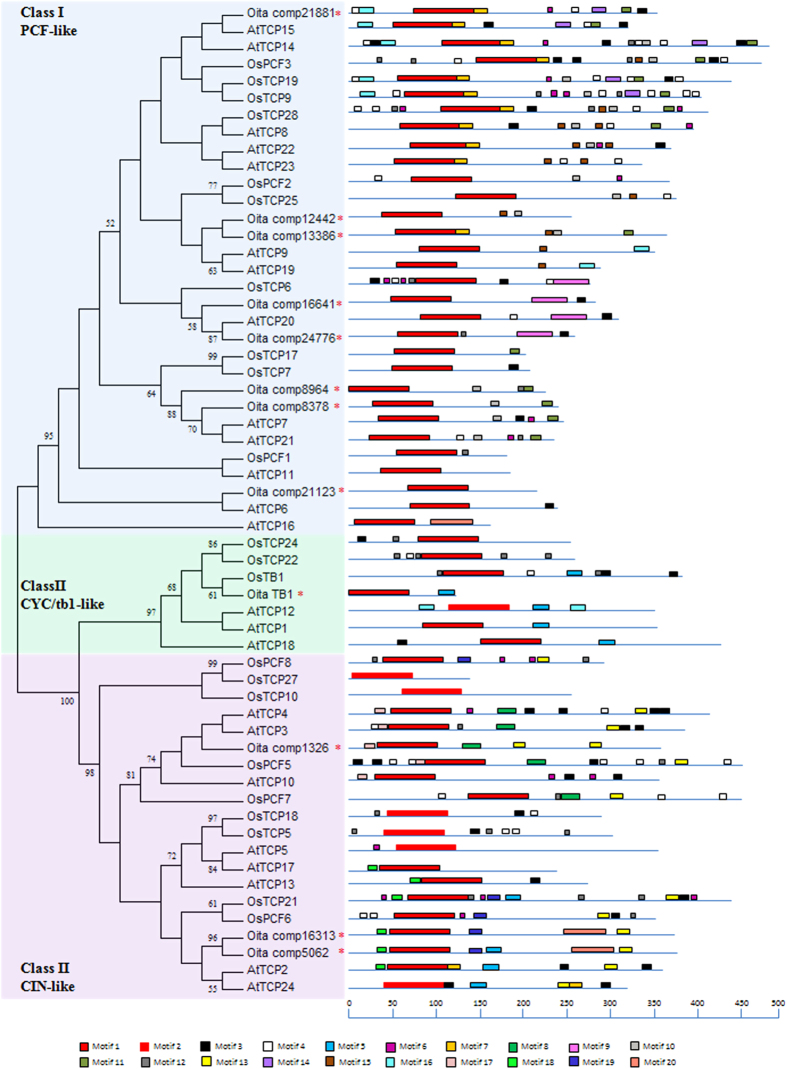
The Neighbor-Joining (NJ) tree obtained from the multiple amino acid alignment of the TCP domain encoded by the selected transcripts of O. italica (12 sequences), A. thaliana (24 sequences), and O. sativa (22 sequences) is shown in Fig. 1. A high bootstrap value (95%) supports the class I PCF-like group that includes eight TCP transcripts of O. italica. Similarly, the class II TCP group is statistically well supported (bootstrap value 100%). It is divided into the CIN-like group (bootstrap value 98%) that includes three TCP transcripts of O. italica and the CYC/TB1-like group (bootstrap value 97%) that includes OitaTB1.
Figure 1. The NJ tree and graphical view of the conserved domains of the TCP proteins examined.
. The NJ tree was obtained from the amino acid alignment of the TCP domain of Orchis italica, Arabidopsis thaliana and Oryza sativa (Supplementary Table S1). The bootstrap percentages >50% are shown next to the branches. The red asterisks indicate the sequences of O. italica. The graph of the conserved domains and the corresponding legend were obtained from the MEME search using the full length of the TCP proteins. The scale below the conserved domains indicates the amino acid position.
The generally dispersed positions of the different TCPs of O. italica among the branches of the tree confirm the hypothesis of the expansion of the TCP family before the divergence of the lineages examined, as observed in some dicot species37,38,39. The robustness of the NJ tree was confirmed by the topology of the Minimum Evolution and the Maximum Likelihood trees (Supplementary Figure S2 and S3, respectively), both showing branch patterns in agreement with the one obtained from the NJ analysis. In addition, the NJ tree inferred from the amino acid alignment of the TCP domains of Orchidaceae is in agreement with the classification of the TCP proteins (Supplementary Table S1 and Supplementary Figure S4). Similarly, the topology of the NJ tree obtained from the amino acid alignment of the TCP domains of O. italica, P. equestris, P. hybrid cultivar, D. hybrid cultivar, A. thaliana and O. sativa (Supplementary Figure S5) consistently distinguishes the different groups of the TCP proteins.
The amino acid sequences of the TCP proteins of O. italica, A. thaliana, and O. sativa were scanned to search for conserved shared domains using the motif-based sequence analysis tool MEME, and the results are shown in Fig. 1 and Supplementary Figure S6. The pattern distribution of the shared motifs is in general agreement with the phylogeny inferred from the alignment of the TCP domain. All the sequences share the TCP domain, included in Motif 1 and 2. The R domain (Motif 5) is shared by 9 sequences belonging to the class II TCP family, both CYC/TB1-like and CIN-like. Among them, two are sequences of O. italica, one CIN-like (comp5062) and one CYC/TB1-like (OitaTB1). Another interesting motif shared by 12 sequences of the CIN-like group (Motif 13) corresponds to the amino acid stretch encoded by the target site of the microRNA miR31912,13,14,15. This motif is present in three sequences of O. italica (comp5062, comp1326 and comp16313). Many other motifs are restricted to specific sub-groups of the tree, suggesting that they might contribute to the specific function of the different TCP proteins. For example, Motif 9 is present in OsTCP6, AtTCP20 and in two sequences of O. italica, comp16641 and comp24776; Motif 8 is shared by AtTCP3, AtTCP4, OsPCF5, OsPCF7 and comp1326. Scanning the identified consensus motifs against the database of protein domains PROSITE52 resulted in positive hits with the TCP domain or with other regions of the TCP proteins with unknown functions, or in no positive matches. In conclusion, excluding the TCP domain, the functions of the other conserved motifs identified are still unknown.
The estimates of the non-synonymous (Ka) and synonymous (Ks) substitution rates was performed on the putative orthologous TCP nucleotide sequences of O. italica and P. equestris to evaluate the evolutionary constraints acting on these sequences. The Ka/Ks ratio equal to 1 indicates neutral selection, while values lower or higher than 1 reveal purifying or positive selection, respectively53. The search for the best reciprocal BLASTP hits between the TCP sequences of O. italica and P. equestris gave significant scores for 10 pairs of sequences. The results of the evolutionary analysis are summarized in Table 1. Among the sequences examined, the mean Ka/Ks value is 0.222, and the pair-wise comparisons reveal that a strong purifying selection is acting on the nucleotide sequences encoding the TCP domain in O. italica and P. equestris. However, the comparison between the sequence comp16641 of O. italica and PEQU28429 of P. equestris resulted in a Ka/Ks value much higher than all the other comparisons and higher than 1, even though the p value was not significant. As shown in Fig. 1 and reported in Supplementary Table S1, the transcripts comp16641 of O. italica and PEQU28429 of P. equestris are related to the TCP factor AtTCP20 of A. thaliana. AtTCP20 belongs to the class I PCF-like group. It is expressed in different organs and developmental stages and is involved in cell division and elongation, which must be coordinated for normal plant development and differentiation54. The Ka/Ks value detected between the transcripts comp16641 of O. italica and PEQU28429 of P. equestris suggests a possible relaxation of the selective constraints acting on these sequences in orchids, which might drive the evolution of morphological innovations.
Table 1. The evolutionary analysis of ten putative TCP orthologs between O. italica and P. equestris.
| Sequence comparison | Ka | Ks | Ka/Ks | P-Value (Fisher) |
|---|---|---|---|---|
| comp24776_PEQU28429 | 0.03255 | 1.06064 | 0.030689 | 9.87E-18 |
| comp21123_PEQU21260 | 0.149384 | 0.922071 | 0.16201 | 2.50E-05 |
| comp8964_PEQU03822 | 0.07316 | 99 | 0.000739 | 0 |
| comp12442_PEQU09751 | 0.011686 | 1.45184 | 0.008049 | 1.18E-22 |
| comp13386_PEQU40981 | 0.022818 | 2.92432 | 0.007803 | 9.93E-30 |
| comp1326_PEQU06996 | 0.114233 | 1.6597 | 0.068828 | 9.26E-28 |
| comp5062_PEQU08153 | 0.073761 | 1.22877 | 0.060028 | 3.83E-19 |
| comp16313_PEQU19547 | 0.111968 | 3.73888 | 0.029947 | 3.01E-17 |
| comp16641_PEQU28429 | 4.05786 | 2.28581 | 1.77524 | 0.488247 |
| OitaTB1_PEQU11715 | 0.249986 | 3.26082 | 0.076664 | 1.61E-26 |
The non-synonymous (Ka) and synonymous (Ks) substitution rates are calculated with the Yang and Nielsen (YN) method.
MicroRNA target sites and expression analysis
To check for the presence of target sites of specific microRNAs (miRNAs) expressed in the inflorescence, the selected transcripts of O. italica encoding TCP-like proteins were scanned with the psRNATarget online tool55 using the inflorescence miRNAs of O. italica47 as queries. The results of this in silico analysis predicted for three different transcripts of O. italica belonging to the class II CIN-like group (comp5062, comp1326 and comp16313) the presence of a putative cleavage site for miR319, in agreement with the results obtained from the analysis of the shared conserved motifs. Phylogenetic analysis shows that comp5062 and comp16313 of O. italica are grouped with AtTCP2 and AtTCP24 of A. thaliana, comp1326 with AtTCP3 and AtTCP4 of A. thaliana and OsPCF5 of Oryza sativa (Fig. 1). All of these transcripts have been described as targets of miR319 and are involved in different developmental processes, from petal and leaf formation to senescence and response to different stresses12,13,14,15,16.
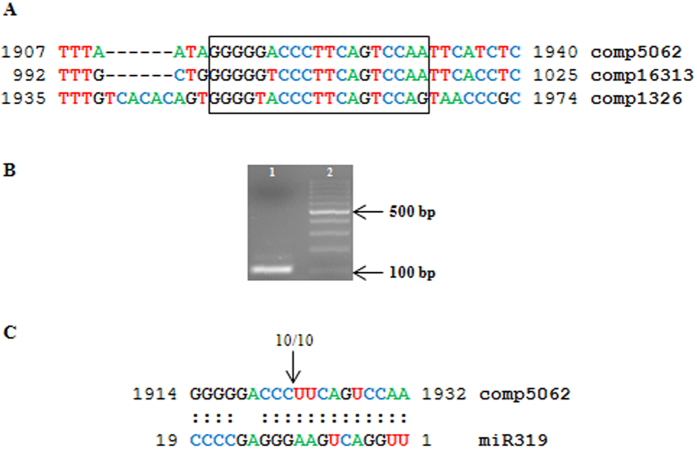
The validation of the miRNA cleavage was performed by conducting a modified 5′-RACE experiment. Cleavage was confirmed for the transcript comp5062 (Fig. 2B,C), whereas for the transcripts comp1326 and comp16316, no cleavage fragment of the expected size was detected. The apparent absence of the cleaving activity of miR319 on comp1326 and 16313 might be due to the sequence divergence in the upstream and downstream regions surrounding the predicted cleavage site.
Figure 2.
The cleavage site of miR319 on the TCP transcript of O. italica comp5062 . (A) The nucleotide alignment of the in silico predicted target site (highlighted in the box) of miR319 on three TCP transcripts of O. italica. Numbers refer to the nucleotide positions on the transcript. (B) Agarose gel electrophoresis of the modified 5′RACE experiment on the TCP transcript comp5062 (lane 1) and the 100 bp Ladder (Fermentas). (C) Alignment of the target site in the transcript comp5062 and the miR319. The arrow and the numbers above the sequences indicate the cleavage site and the number of sequenced clones that revealed the cleavage in that position, respectively.
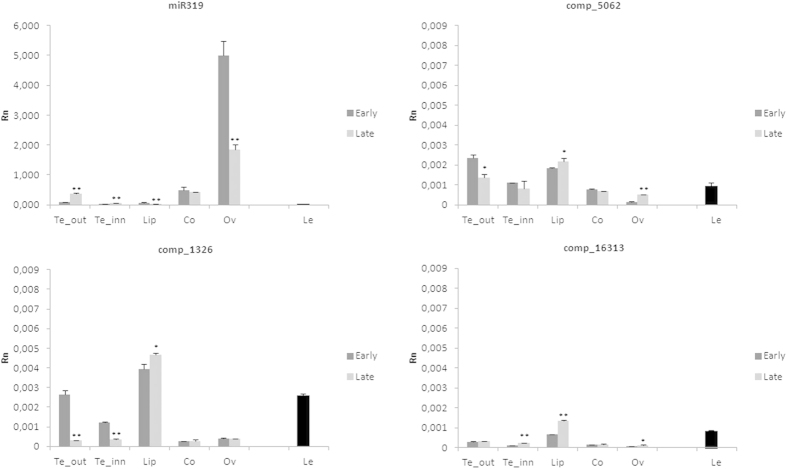
The analysis of the expression patterns is useful to hypothesize possible gene functions and overlapping expression profiles indicate probable functional redundancy. The expression analysis of miR319 and of its putative target transcripts in different tissues of the inflorescence of O. italica at two developmental stages (Fig. 3C) was evaluated by real time PCR. The early stage corresponds to floral buds with a diameter of approximately 9 mm (Fig. 3A) and the late stage to completely opened flowers after anthesis (Fig. 3B). In both stages, cell division is completed, and the flower organs are completely formed; however, in the early stage, cells are still elongating. Figure 4 shows the relative expression of the transcripts comp5062, comp1326, comp16316 and miR319 in the outer and inner tepal, lip, column, unpollinated ovary and in leaf tissue. The expression patterns of these three transcripts in the tissues of O. italica at the two different stages examined are similar but with different expression levels. In the inflorescence, they are mainly expressed in the organs of the perianth (tepals and lip) with some difference between the early and late stage. In the column and ovary, the expression of the three transcripts is generally lower than in the other tissues, including leaves. In contrast, the expression pattern of miR319 shows the opposite trend, with the highest expression levels observed in the column and ovary. The observed values of the relative expression of miR319 are negatively correlated with those of its three putative targets; however, the only statistically significant Pearson correlation coefficient (r = –0.61, p < 0.05) is relative to the transcript comp5062. This result, together with the failure to detect specific fragments cleaved by miR319 for the transcripts comp1326 and comp16316, suggests the existence of different regulators of the expression of these two TCP transcripts in the tissues of O. italica at the stages examined. Our results demonstrate for the first time the activity of miR319 on a TCP target in the floral tissues of a monocot species. In the model dicot species Arabidopsis, miR319 has been demonstrated to have a critical role in the development of petal and stamens, through the cleavage of a CIN-like TCP target15. The differences of the expression pattern of the transcript comp5062 in the inflorescence tissues of O. italica and its regulation by miR319 make this TCP transcript a good candidate gene involved in the development of the floral phenotype of O. italica and suggest a possible conserved function of miR319 in flower development of dicots and monocots.
Figure 3. The Mediterranean orchid Orchis italica.
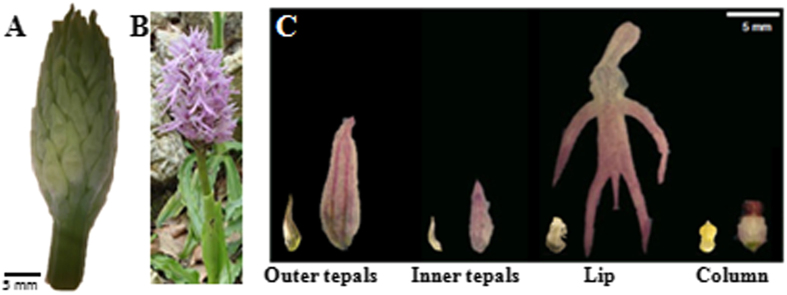
. (A) An inflorescence at the early stage; (B) An inflorescence at the late stage, after anthesis. (C) Tepals (outer and inner), lip and column collected from a single floret of the early (left) and late (right) inflorescence of O. italica.
Figure 4.
The relative expression of TCP transcripts comp5062, comp1326 and comp16313 and of miR319 in different floral tissues and in leaves of O. italica. The bars indicate standard deviation. The asterisks indicate statistically significant differences between the relative expression of the early and the late stages (*p < 0.05, **p < 0.01). Te_out, outer tepals; Te_inn, inner tepals; Lip, labellum; Co, column; Ov, ovary; Le, leaf. Rn, relative normalized expression.
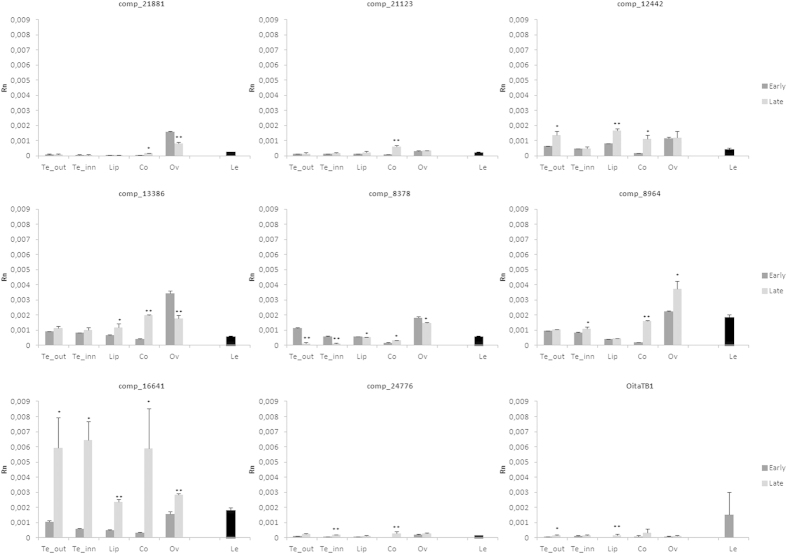
The different tissues of O. italica were examined to evaluate the expression pattern of the other identified transcripts that encode TCP proteins. The results reported in Fig. 5 show distinct expression profiles for some transcripts, suggesting their different roles in the various tissues and stages examined, while other transcripts share similar patterns. In particular, the transcript comp21881 is expressed almost exclusively in the ovary tissue and the transcript comp21123 in the column at the late stage, suggesting that they might have a specific function in these tissues. The transcript comp21881 is related to the gene AtTCP15 of Arabidopsis (Fig. 1), involved in many developmental pathways including gynoecium development56,57. The expression profile observed in O. italica suggests its functional conservation, at least for its possible role in the development of the female reproductive structures. The transcripts comp12442 and 13386, belonging to the same branch of the NJ tree, display generally overlapping profiles, being expressed in all the tissues examined, including leaves, with some differences in the ovary tissue. This suggests pleiotropic and redundant functions of these two transcripts in the tissues examined of O. italica, in agreement with the diffuse functional redundancy of the class I TCPs. The transcripts comp8378 and 8964 belong to the same branch of the NJ tree, and they display similar patterns at the early stage. In the late stage, differences are detectable in tepals and ovary, where the levels of comp8378 significantly decrease, whereas the levels of comp8964 increase markedly in column and ovary. These two transcripts are related to the genes AtTCP21 and AtTCP7 (Fig. 1). In Arabidopsis these two genes show not fully overlapping expression profiles and functions. AtTCP7 is mainly involved in leaf development and AtTCP21 is a component of the circadian clock58,59. The expression pattern of comp8378 and 8964 indicates their possible functional diversification also in O. italica. Despite the fact that the transcripts comp16641 and 24776 are included in the same branch of the NJ tree, their expression profiles are very different. The first is highly expressed in all the floral tissues at the late stage and in leaves; the latter is very weakly expressed in all the tissues examined, suggesting that its functions are performed in different organs and/or developmental stages. The transcript comp16641 shows the strongest variation of the expression profile between the two stages examined and, as previously discussed, it shows relaxation of the selective constraints acting on its coding region. These findings let hypothesize a possible sub- or neo-functionalization of comp16641 and suggest to investigate in more detail the possible role of this gene in the development and maintenance of floral structures in orchids. Finally, OitaTB1 is expressed at a very low level in the inflorescence tissues and at a slightly higher level in leaves. OitaTB1 seems to be phylogenetically close to OsTB1 of Oryza sativa (Fig. 1, Supplementary Figure S2 and S5), a negative regulator of lateral branching60. It would be of great interest to ascertain whether OitaTB1 is also involved in this developmental process in O. italica and in orchids in general.
Figure 5.
The relative expression of nine TCP transcripts in different floral tissues and in leaves of O. italica. The bars indicate standard deviation. The asterisks indicate statistically significant differences between the relative expression of the early and the late stages (*p < 0.05, **p < 0.01). Te_out, outer tepals; Te_inn, inner tepals; Lip, labellum; Co, column; Ov, ovary; Le, leaf. Rn, relative normalized expression.
Conclusions
This study has led to the identification of 12 transcripts encoding TCP proteins in the Mediterranean orchid O. italica. This number is lower than the number of the TCP genes of the model species A. thaliana (24) or O. sativa (22) because the approach used in the present study was focused on the TCPs expressed in the inflorescence tissues. Other TCP transcripts in O. italica are possibly expressed in tissues and/or developmental stages different from those examined. However, the number of transcripts identified in O. italica is not much different from the number annotated in the genome of the orchid P. equestris (17), suggesting that in orchids there could be fewer TCP genes than in Arabidopsis and Oryza. The analysis of the expression profiles of the TCP transcripts of O. italica shows that some of them are expressed in all the tissues and developmental stages examined, suggesting their potential involvement in multiple pathways, whereas others are restricted to specific tissues, indicating a possible more specialized role. In addition, the presence of a cleavage site for miR319 on one TCP transcript of O. italica supports the existence of an evolutionary conserved role of this miRNA in regulating the activity of specific TCP factors.
Methods
Isolation of the TCP genes expressed in the inflorescence of O. italica
The sequences of the TCP DNA-binding domain of Arabidopsis and Oryza (Pfam PF03634) were used as query to perform a standalone TBLASTN (e-value 1e-003) against the non-redundant sequences of the inflorescence transcriptome of O. italica46. In addition, the transcripts of the inflorescence transcriptome of O. italica annotated as TCP genes were selected. The transcripts of O. italica with significant hits from the TBLASTN search and those previously annotated as TCP genes were checked for the presence of an ORF that exceeded the TCP domain, excluding those shorter than 500 bp from further analyses.
The coding sequences (CDSs) and predicted proteins of the genome of the orchid Phalaenopsis equestris42 were downloaded from the FTP site ftp://ftp.genomics.org.cn/from_BGISZ/20130120/. The coding sequences annotated as TCP genes were selected (Supplementary Table S1) and used for the subsequent analyses.
Isolation of the class II CYC/TB1-like sequences of O. italica
To identify the nucleotide sequences coding for CYC/TB1-like proteins, the genomic DNA of O. italica was extracted from leaf tissue61. Using the three nucleotide sequences of P. equestris annotated as CYC/TB1-like proteins, a degenerate primer pair was designed to amplify the region from the TCP domain to the R domain (Supplementary Table S2). PCR amplification was conducted on 100 ng of genomic DNA of O. italica in a final volume of 50 μl. The reaction mixture contained 200 μM dNTPs, 0.4 μM of each primer, buffer 1X and 2.5 U HotMaster Taq polymerase (5Prime). The thermal cycle was the following: 94 °C for 3 min, 35 cycles of 94 °C for 30 sec, 54 °C for 20 sec, 65 °C for 2 min, followed by a final extension of 10 min at 65 °C.
The amplification product was cloned into the pGEM-T Easy vector (Promega), and the positive clones were sequenced using the plasmid primers T7 and SP6 and analyzed on an ABI 310 Automated Sequencer (Applied Biosystems).
Phylogenetic analysis and identification of conserved motifs
The amino acid sequences of the TCP proteins of Arabidopsis thaliana (24 accessions) and Oryza sativa (22 accessions) were downloaded from the Plant TFDB (Supplementary Table S1). Multiple sequence alignment of the TCP conserved domains of the downloaded sequences and the corresponding regions of the TCP proteins of O. italica obtained from the virtual translation of the selected transcripts was performed with MUSCLE62 and then manually adjusted. The Neighbor-Joining (NJ), Minimum Evolution (ME) and Maximum Likelihood (ML) trees were constructed using MEGA 6.0663 with 1000 bootstrap replicates. The amino acid sequences of the TCP proteins of the orchids Phalaenopsis hybrid cultivar (5 accessions) and Dendrobium hybrid cultivar (1 accession) were downloaded from GenBank (Supplementary Table S1). The TCP domains of the orchids O. italica, P. equestris, P. hybrid cultivar and D. hybrid cultivar were aligned, and the NJ, ME and ML trees were constructed as described above.
The whole amino acid sequences of the TCP proteins of O. italica, A. thaliana and O. sativa were scanned for the presence of shared conserved motifs using the online tool MEME64, setting the parameters to any number of repetitions, the optimum width from 4 to 70 and the maximum number of motifs to 20.
Evolutionary analysis
To identify the best reciprocal hits, a custom-made perl script was used to perform BLASTP search on the TCP sequences of O. italica and P. equestris. Based on the results obtained, pair-wise nucleotide alignment between the resulting orthologous nucleotide sequence encoding the TCP domain of O. italica and P. equestris was constructed and used to estimate the synonymous (Ks) and non-synonymous sites (Ka) substitution rates using the pair-wise approximate YN method65 implemented in the KaKs Calculator66.
Search for miRNA targets and cleavage validation
The psRNATarget online tool55 was used to search for the presence of microRNA targets within the TCP transcripts of O. italica, using as the query the microRNAs expressed in the inflorescence of O. italica47 and setting stringent search parameters (maximum expectation 0.0).
Based on the predicted position of the putative miRNA cleavage sites on the TCP transcripts of O.italica, specific reverse primers were designed, all of them annealing downstream from the predicted putative miRNA target, and used to verify the presence of the cleavage product by performing a modified 5′-RACE experiment67. In brief, total RNA was extracted from the inflorescence of O. italica before anthesis (~9 mm diameter) using the Trizol Reagent (Ambion). After DNase treatment, RNA was quantified using the spectrophotometer Nanodrop 2000 (Thermo Scientific). The 5′ adaptor of the RLM-RACE GeneRace kit (Invitrogen) was ligated to the 5′-terminus of the extracted RNA (500 ng) without any enzymatic treatment to remove the 5′ cap. The RNA was then reverse transcribed, and the cDNA was amplified using the GeneRace 5′ Primer and the TCP-specific reverse primers (Supplementary Table S2). A second PCR amplification was performed on 1 μl of the first reaction using the nested adaptor forward primer and the nested TCP-specific reverse primers (Supplementary Table S2). The amplification products were cloned into the pGEM-T Easy vector (Promega), and 10 clones were sequenced as described above.
The specimens of O. italica used in the present work are present in the orchids collection of the Botanical Garden of Naples (Italy).
Expression analysis
Total RNA was extracted from different tissues of O. italica following the protocol described above. In particular, outer and inner tepal, lip, column and unpollinated ovary were dissected from the inflorescence of O. italica before anthesis (defined as the early stage, ~9 mm diameter) and after anthesis (late stage). Although in the early stage, cell division has been completed and flower organs formed, cell elongation is still occurring. In addition, total RNA was extracted from leaf tissue. RNA (350 ng) was reverse transcribed using the Advantage RT-PCR kit (Clontech) and an oligo dT primer.
Using the 5.8 S RNA as the endogenous control gene, real time PCR experiments were conducted on 30 ng of the first strand cDNA from each tissue using the primer pairs specific for the TCP transcripts of O. italica (Supplementary Table S2). The reactions were performed using the SYBR Green PCR Master Mix (Life Technologies) in technical triplicates and biological duplicates, and the thermal cycle was followed by a melting curve step68.
Stem-loop real time PCR experiments were conducted to evaluate the expression pattern of the microRNA miR319 of O. italica in all the tissues examined69. In brief, total RNA (150 ng) from each tissue was separately reverse transcribed using the miR319 and the 5.8 S stem-loop primers (Supplementary Table S2). The first strand cDNA (5 ng) was then amplified in technical triplicates and biological duplicates using the miR319 and 5.8 S specific primers and the stem-loop universal primer (Supplementary Table S2).
The PCR efficiency (E) and the optimal threshold cycle (CT) for each well were calculated using the Real Time PCR Miner online tool70. The mean relative expression ratio (Rn) and standard deviation of the TCP transcripts and of the miR319 in the different tissues was calculated using the 5.8 S RNA as the endogenous control gene by applying the formula Rn = (1 + Etarget gene)−CT target gene/(1 + Econtrol gene)−CT control gene. Differences in relative expression levels of the TCP transcripts and of miR319 between and/or among the different tissues were assessed by the two-tailed t test and ANOVA followed by the Tukey HSD post hoc test. To exclude the presence of PCR artifacts, the real time PCR products of several samples were cloned and sequenced as described above.
Additional Information
How to cite this article: Paolo, S. D. et al. Analysis of the TCP genes expressed in the inflorescence of the orchid Orchis italica. Sci. Rep. 5, 16265; doi: 10.1038/srep16265 (2015).
Supplementary Material
Acknowledgments
The authors are grateful to Prof. S. Cozzolino for plant material and to Mr Enzo Iacueo for the technical support. This work was supported by the 2007 Regione Campania Grant L.R. N5/2002.
Footnotes
Author Contributions S.D.P. performed the experiments and participated to the data analysis. L.G. contributed to the interpretation of the data and critically read the manuscript. S.A. designed the experiments, analyzed the data and wrote the manuscript.
References
- Navaud O., Dabos P., Carnus E., Tremousaygue D. & Herve C. TCP transcription factors predate the emergence of land plants. J Mol Evol 65, 23–33, doi: 10.1007/s00239-006-0174-z (2007). [DOI] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J. & Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18, 215–222 (1999). [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M. & Cubas P. TCP genes: a family snapshot ten years later. Trends Plant Sci 15, 31–39, doi: 10.1016/j.tplants.2009.11.003 (2010). [DOI] [PubMed] [Google Scholar]
- Doebley J., Stec A. & Hubbard L. The evolution of apical dominance in maize. Nature 386, 485–488, doi: 10.1038/386485a0 (1997). [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L. & Coen E. Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799, doi: 10.1038/383794a0 (1996). [DOI] [PubMed] [Google Scholar]
- Kosugi S. & Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619, doi: 10.1105/tpc.9.9.1607 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S. & Ohashi Y. DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30, 337–348, doi: 1294 (2002). [DOI] [PubMed] [Google Scholar]
- Viola I. L., Uberti Manassero N. G., Ripoll R. & Gonzalez D. H. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem J 435, 143–155, doi: 10.1042/BJ20101019 (2011). [DOI] [PubMed] [Google Scholar]
- Howarth D. G. & Donoghue M. J. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc Natl Acad Sci USA 103, 9101–9106, doi: 10.1073/pnas.0602827103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer R. & Theissen G. MADS and more: transcription factors that shape the plant. Methods Mol Biol 754, 3–18, doi: 10.1007/978-1-61779-154-3_1 (2011). [DOI] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colon-Carmona A., Gutierrez R. A. & Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102, 12978–12983, doi: 10.1073/pnas.0504039102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N. et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39, 787–791 (2007). [DOI] [PubMed] [Google Scholar]
- Palatnik J. F. et al. Sequence and expression differences underlie functional specialization of arabidopsis microRNAs miR159 and miR319. Dev Cell 13, 115–125 (2007). [DOI] [PubMed] [Google Scholar]
- Schommer C. et al. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6, e230 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., King S. & Jack T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106, 22534–22539, doi: 10.1073/pnas.0908718106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M. & Luo H. Role of microRNA319 in creeping bentgrass salinity and drought stress response. Plant Signal Behav 9, e28700, doi: 28700 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J. A., Sun Y., Blair P. B. & Mukhtar M. S. TCP three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci 20, 238–245, doi: 10.1016/j.tplants.2015.01.005 (2015). [DOI] [PubMed] [Google Scholar]
- Giraud E. et al. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22, 3921–3934, doi: 10.1105/tpc.110.074518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J. C. & Hileman L. C. Developmental genetics of floral symmetry evolution. Trends Plant Sci 14, 147–154, doi: 10.1016/j.tplants.2008.12.005 (2009). [DOI] [PubMed] [Google Scholar]
- Hileman L. C. & Cubas P. An expanded evolutionary role for flower symmetry genes. J Biol 8, 90, doi: 10.1186/jbiol193 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman L. C. Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos Trans R Soc Lond B Biol Sci 369, doi: 10.1098/rstb.2013.0348 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C., Le Guilloux M., Jager M. & Charon C. Diversity and evolution of CYCLOIDEA-like TCP genes in relation to flower development in Papaveraceae. Plant Physiol 143, 759–772, doi: 10.1104/pp.106.090324 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsch A. & Gleissberg S. Diversification of CYCLOIDEA-like TCP genes in the basal eudicot families Fumariaceae and Papaveraceae s.str. Plant Biol (Stuttg) 8, 680–687, doi: 10.1055/s-2006-924286 (2006). [DOI] [PubMed] [Google Scholar]
- Preston J. C., Kost M. A. & Hileman L. C. Conservation and diversification of the symmetry developmental program among close relatives of snapdragon with divergent floral morphologies. New Phytol 182, 751–762, doi: 10.1111/j.1469-8137.2009.02794.x (2009). [DOI] [PubMed] [Google Scholar]
- Gao Q., Tao J. H., Yan D., Wang Y. Z. & Li Z. Y. Expression differentiation of CYC-like floral symmetry genes correlated with their protein sequence divergence in Chirita heterotricha (Gesneriaceae). Dev Genes Evol 218, 341–351, doi: 10.1007/s00427-008-0227-y (2008). [DOI] [PubMed] [Google Scholar]
- Chapman M. A. et al. Genetic analysis of floral symmetry in Van Gogh’s sunflowers reveals independent recruitment of CYCLOIDEA genes in the Asteraceae. PLoS Genet 8, e1002628, doi: 10.1371/journal.pgen.1002628 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang Y. & Luo D. LjCYC genes constitute floral dorsoventral asymmetry in Lotus japonicus. J Integr Plant Biol 52, 959–970, doi: 10.1111/j.1744-7909.2010.00926.x (2010). [DOI] [PubMed] [Google Scholar]
- Wang Z. et al. Genetic control of floral zygomorphy in pea (Pisum sativum L.). Proc Natl Acad Sci USA 105, 10414–10419, doi: 10.1073/pnas.0803291105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z. et al. RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol 149, 235–244, doi: 10.1104/pp.108.128231 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett M. E. & Specht C. D. Changes in expression pattern of the teosinte branched1-like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. Am J Bot 98, 227–243, doi: 10.3732/ajb.1000246 (2011). [DOI] [PubMed] [Google Scholar]
- Preston J. C. & Hileman L. C. Parallel evolution of TCP and B-class genes in commelinaceae flower bilateral symmetry. Evodevo 3, 6, doi: 10.1186/2041-9139-3-6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y. et al. Characterization of CYCLOIDEA-like genes in controlling floral zygomorphy in the monocotyledon Alstroemeria. Sci Hortic-Amsterdam 169, 6–13, doi: 10.1016/j.scienta.2014.01.046 (2014). [DOI] [Google Scholar]
- Doebley J., Stec A. & Gustus C. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141, 333–346 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poza-Carrion C., Aguilar-Martinez J. A. & Cubas P. Role of TCP Gene BRANCHED1 in the Control of Shoot Branching in Arabidopsis. Plant Signal Behav 2, 551–552 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon-Palomino M. & Trontin C. High time for a roll call: gene duplication and phylogenetic relationships of TCP-like genes in monocots. Ann Bot 107, 1533–1544, doi: 10.1093/aob/mcr059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S. et al. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J Exp Bot 64, 5673–5685, doi: 10.1093/jxb/ert337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R. et al. Genomewide analysis of TCP transcription factor gene family in Malus domestica. J Genet 93, 733–746 (2014). [DOI] [PubMed] [Google Scholar]
- Ma J. et al. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci Rep 4, 6645, doi: 10.1038/srep06645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parapunova V. et al. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol 14, 157, doi: 10.1186/1471-2229-14-157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedeek K. E. et al. Transcriptome and proteome data reveal candidate genes for pollinator attraction in sexually deceptive orchids. PLoS One 8, e64621, doi: 10.1371/journal.pone.0064621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W. C. et al. OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol 54, e7, doi: 10.1093/pcp/pcs187 (2013). [DOI] [PubMed] [Google Scholar]
- Cai J. et al. The genome sequence of the orchid Phalaenopsis equestris. Nat Genet 47, 65–72, doi: 10.1038/ng.3149 (2015). [DOI] [PubMed] [Google Scholar]
- Montieri S., Gaudio L. & Aceto S. Isolation of the LFY/FLO homologue in Orchis italica and evolutionary analysis in some European orchids. Gene 333, 101–109, doi: 10.1016/j.gene.2004.02.015 (2004). [DOI] [PubMed] [Google Scholar]
- Rudall P. J. & Bateman R. M. Roles of synorganisation, zygomorphy and heterotopy in floral evolution: the gynostemium and labellum of orchids and other lilioid monocots. Biol Rev Camb Philos Soc 77, 403–441 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang X. S. & O’Neill S. D. Ovary and gametophyte development are coordinately regulated by auxin and ethylene following pollination. Plant Cell 5, 403–418, doi: 10.1105/tpc.5.4.403 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paolo S., Salvemini M., Gaudio L. & Aceto S. De novo transcriptome assembly from inflorescence of Orchis italica: analysis of coding and non-coding transcripts. PLoS One 9, e102155, doi: 10.1371/journal.pone.0102155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aceto S. et al. The analysis of the Inflorescence miRNome of the Orchid Orchis italica Reveals a DEF-Like MADS-Box Gene as a New miRNA Target. PLoS One 9, e97839, doi: 10.1371/journal.pone.0097839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J. & Galego L. Flower symmetry and shape in Antirrhinum. Int J Dev Biol 49, 527–537, doi: 10.1387/ijdb.041967ja (2005). [DOI] [PubMed] [Google Scholar]
- Feng X. et al. Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA 103, 4970–4975, doi: 10.1073/pnas.0600681103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth D. G., Martins T., Chimney E. & Donoghue M. J. Diversification of CYCLOIDEA expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Ann Bot 107, 1521–1532, doi: 10.1093/aob/mcr049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour F. et al. Specific duplication and dorsoventrally asymmetric expression patterns of Cycloidea-like genes in zygomorphic species of Ranunculaceae. PLoS One 9, e95727, doi: 10.1371/journal.pone.0095727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. J. et al. New and continuing developments at PROSITE. Nucleic Acids Res 41, D344–347, doi: 10.1093/nar/gks1067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. & Bielawski J. P. Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15, 496–503, doi: 10.1016/S0169-5347(00)01994-7 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herve C. et al. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol 149, 1462–1477, doi: 10.1104/pp.108.126136 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X. & Zhao P. X. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39, W155–159, doi: 10.1093/nar/gkr319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberti-Manassero N. G., Lucero L. E., Viola I. L., Vegetti A. C. & Gonzalez D. H. The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J Exp Bot 63, 809–823, doi: 10.1093/jxb/err305 (2012). [DOI] [PubMed] [Google Scholar]
- Lucero L. E. et al. TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J, doi: 10.1111/tpj.12992 (2015). [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J. L., Breton G., Para A. & Kay S. A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485, doi: 10.1126/science.1167206 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martinez J. A. & Sinha N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front Plant Sci 4, 406, doi: 10.3389/fpls.2013.00406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T. et al. The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33, 513–520, doi: 10.1046/j.1365-313X.2003.01648.x (2003). [DOI] [PubMed] [Google Scholar]
- Doyle J. J. & Doyle J. L. A rapid DNA isolation procedure for small amounts of leaf tissue. Phytochem Bull 19, 11–15 (1987). [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797, doi: 10.1093/nar/gkh340 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30, 2725–2729, doi: 10.1093/molbev/mst197 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–208, doi: 10.1093/nar/gkp335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. & Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17, 32–43 (2000). [DOI] [PubMed] [Google Scholar]
- Zhang Z. et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4, 259–263, doi: 10.1016/S1672-0229(07)60007-2 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K. D. & Carrington J. C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056 (2002). [DOI] [PubMed] [Google Scholar]
- Salemme M., Sica M., Gaudio L. & Aceto S. Expression pattern of two paralogs of the PI/GLO-like locus during Orchis italica (Orchidaceae, Orchidinae) flower development. Dev Genes Evol 221, 241–246, doi: 10.1007/s00427-011-0372-6 (2011). [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E., Wu R., Wood M., Walton E. F. & Hellens R. P. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. & Fernald R. D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol 12, 1047–1064, doi: 10.1089/cmb.2005.12.1047 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.