Abstract
The necrotrophic fungus Parastagonospora nodorum is an important pathogen of one of the world’s most economically important cereal crops, wheat (Triticum aestivum L.). P. nodorum produces necrotrophic protein effectors that mediate host cell death, providing nutrients for continuation of the infection process. The recent discovery of pathogen effectors has revolutionized disease resistance breeding for necrotrophic diseases in crop species, allowing often complex genetic resistance mechanisms to be broken down into constituent parts. To date, three effectors have been identified in P. nodorum. Here we use the effector, SnTox1, to screen 642 progeny from an eight-parent multiparent advanced generation inter-cross (i.e., MAGIC) population, genotyped with a 90,000-feature single-nucleotide polymorphism array. The MAGIC founders showed a range of sensitivity to SnTox1, with transgressive segregation evident in the progeny. SnTox1 sensitivity showed high heritability, with quantitative trait locus analyses fine-mapping the Snn1 locus to the short arm of chromosome 1B. In addition, a previously undescribed SnTox1 sensitivity locus was identified on the long arm of chromosome 5A, termed here QSnn.niab-5A.1. The peak single-nucleotide polymorphism for the Snn1 locus was converted to the KASP genotyping platform, providing breeders and researchers a simple and cheap diagnostic marker for allelic state at Snn1.
Keywords: fungal protein effectors, diagnostic genetic markers, plant disease resistance breeding, multiparent genetic mapping populations, high-density crop genotyping, MPP, Multiparent Advanced Generation Inter-Cross (MAGIC), multiparental populations
The necrotrophic wheat pathogen Parastagonospora (synonym: Septoria, Stagonospora, Phaeosphaeria) nodorum causes the disease Septoria nodorum blotch (SNB), responsible for significant wheat (Triticum aestivum L.) yield losses in Europe, Australasia, United States, North America, and North Africa (Friesen et al. 2005; Oliver et al. 2012). Necrotrophic pathogens derive their energy from dead host plant cells or tissues, in contrast to biotrophs, which obtain their energy from living cells. Recent work indicates that necrotrophic pathogens produce numerous proteinaceous and metabolite molecules, previously known as host-specific (or selective) toxins, now known as effectors, that facilitate pathogen entry by eliciting a necrosis response in the host (Oliver and Solomon 2010). Where the host plant is sensitive to the effector, the resulting cell death provides a rich nutrient source, which promotes necrotrophic infection (Vincent et al. 2012). This process is known as effector-triggered susceptibility. In P. nodorum, an inverse gene-for-gene interaction operates in which mutations in corresponding effector sensitively loci results in insensitivity and host resistance (Friesen et al. 2007). The identification and characterization of effectors in P. nodorum (Friesen et al. 2007; Oliver and Solomon 2010) and other pathogen species represents a paradigm shift within the field of phytopathology, providing tools with which to break down quantitative host sensitivity phenotypes into constitutive components.
The first necrotrophic protein effector to be isolated was PtrToxA from the wheat tan spot pathogen, Pyrenophora tritici-repentis (Ballance et al. 1989; Tómas et al. 1990), which causes necrosis in wheat lines carrying susceptible alleles at the Tsn1 locus (Thomas et al. 1990; Faris et al. 1996). Sequencing of the P. nodorum genome (Hane et al. 2007) identified a predicted protein almost identical in sequence to PtrToxA. This gene, termed SnToxA, encodes a 13-kDa protein containing two cysteine residues and an RGD-containing vitronectic-like motif (Ballance et al. 1996; Ciuffetti et al. 1997) and is thought to have been transferred from P. nodorum to P. tritici-repentis by lateral gene transfer (Friesen et al. 2006; Stukenbrock and McDonald 2007). ToxA from both species cause necrosis only in wheat lines carrying dominant sensitivity alleles at the Tsn1 locus (Friesen et al. 2006; Faris et al. 2010). Tsn1 has been cloned recently and encodes a predicted protein encoding domains commonly found in plant disease resistance genes: a serine/threonine protein kinase domain, a nucleotide-binding site (NBS), and leucine-rich repeat (LRR) domains (Faris et al. 2010). Although a small number of varieties have been identified with point mutations in Tsn1 that result in ToxA insensitivity (Faris et al. 2010), the vast majority of insensitive wheat varieties screened to date carry a deletion of Tsn1, allowing the development of a diagnostic molecular marker for allelic state at the locus.
After the identification of SnToxA, additional P. nodorum effectors have been isolated. The first of these was SnTox3, which encodes a 230 amino acid cysteine rich pro-protein, containing a predicted signal sequence and prosequence of 20 and ∼30 amino acids, respectively. SnTox3 causes necrosis in wheat lines carrying sensitive alleles at the Snn3 locus on chromosome 5BS (Liu et al. 2012). The SnTox3−Snn3 interaction has been shown to account for a significant proportion of disease phenotype in adult plants segregating for sensitivity alleles at Snn3, Tsn1, and Snn2, although only using P. nodorum isolates lacking SnToxA (Friesen et al. 2008), indicating the SnToxA-Tsn1 interaction is epistatic to the SnTox3−Snn3 interaction. SnTox1 is the most recent P. nodorum effector to be identified. It encodes a protein of 10.33 kDa and causes necrosis on wheat lines carrying dominant sensitive alleles at the Snn1 locus (Liu et al. 2012). SnTox1 was identified by screening a list of candidate genes identified in the P. nodorum genome has possessing characteristics common to small, secreted effector-like proteins (Liu et al. 2012). Candidates were expressed in a yeast expression system and used for infiltration into diagnostic wheat lines. SnTox1 was identified as the protein that specifically caused necrosis in wheat lines carrying Snn1, with its identity confirmed using transgenic approaches in virulent and avirulent P. nodorum strains (Liu et al. 2012). After cleavage of the 17 amino acid predicted signal peptide, the mature SnTox1 protein is 100 amino acids long and contains 16 cysteine residues, a common feature among avirulence effectors (Liu et al. 2012).
SnTox1 is widely prevalent in P. nodorum and found to be present in 85% of a global collection of 777 isolates, whereas it was absent in all isolates collected from wild grasses that are avirulent on wheat (Liu et al. 2012; McDonald et al. 2013). Although the molecular basis of necrosis-induced effector-triggered susceptibility is largely unknown, hallmarks of programmed cell death have been observed for SnTox1 in susceptible wheat, including oxidative burst, DNA laddering, and pathogenesis-related gene expression (Liu et al. 2012). SnTox2 is a small secreted peptide of ∼7 kDa that causes necrosis on wheat lines carrying the Snn2 gene on chromosome 2DS (Friesen et al. 2007). Compatible SnTox2-Snn2 and SnToxA-Tsn1 interactions are additive in their effects for susceptibility (Friesen et al. 2007), showing that in contrast to the classical gene-for-gene model, the presence of two effector-host sensitivity gene interactions results in more disease than one interaction alone (Friesen et al. 2008).
In view of the complex nature of host resistance and the significant genotype by environment (G × E) interaction (Friesen et al. 2008), screening segregating wheat populations with P. nodorum effectors has the potential to rapidly increase basal levels of host resistance to both pathogens by selection of effector insensitive lines for cultivar development and to discover (and remove) the corresponding host sensitivity loci. To date, only the Tsn1 host sensitivity locus has been cloned, with Snn1 (Liu et al. 2004; Reddy et al. 2008), Snn2 (Friesen et al. 2007), and Snn3-B1 (Zhang et al. 2011) genetically mapped to chromosomes 1BS, 2DS, and 5BS, respectively. In this study, we use SnTox1 to screen a recently completed eight-parent wheat multiparent advanced generation inter-cross (MAGIC) population (Mackay et al. 2014) genotyped with a 90,000 feature single-nucleotide polymorphism (SNP) array, fine-mapping the Snn1 locus to chromosome 1BS. The peak marker for Snn1 was converted to the KASP genotyping platform, providing a cheap and flexible genetic marker for breeders and researchers.
Materials and Methods
Wheat germplasm and high-density genotyping
The eight-parent MAGIC population described by Mackay et al. (2014) was used for all phenotypic screening. Briefly, the complete population consists of >1000 lines generated from three rounds of inter-crossing between eight elite United Kingdom wheat varieties (Alchemy, Brompton, Claire, Hereward, Rialto, Robigus, Soissons, Xi19) followed by five rounds of selfing to remove heterozygosity. Here, a subset of 642 lines (see Supporting Information, Table S1) were screened for SnTox1 sensitivity. Genotype data were generated using the wheat Illumina 90,000 SNP array (Wang et al. 2014), as described by Mackay et al. (2014). Genotyping was provided as a service by the Department of Primary Industries (Victorian AgriBiosciences Center, Bundoora, Australia). SNPs were called using GenomeStudio software (Illumina) using the clusterfiles developed by Wang et al. (2014). SNPs for downstream analyses were selected on the basis of genotyping success rate (>0.91) and minor allele frequency (>0.05). All genotype data are available from http://www.niab.com/magic/. In total, 15,514 polymorphic markers were used for quantitative trait locus (QTL) mapping (listed in Table S2). Genotype calling for selected SNPs was checked manually with GenomeStudio. Additionally, for SNPs not included in the initial analysis due to failure to automatically call genotypes, but predicted to map to the short arms of the group 1 chromosomes from BLASTn analysis to flow sorted wheat chromosomes (http://urgi.versailles.inra.fr/; The International Wheat Genome Sequencing Consortium 2014), genotypes were called manually with GenomeStudio. Updated allelic bins were defined with manually defined x and y coordinates, allowing for more accurate calling. A subset of 12 of these markers, selected by high linkage disequilibrium (D′ >0.94 and R2 >0.6) to the peak marker, were included in a second QTL scan.
SnTox1 protein production and MAGIC phenotyping
SnTox1 was heterologously expressed in Pichia pastoris, as previously described (Tan et al. 2014). The Pichia culture filtrate containing SnTox1 desalted in 10 mM sodium phosphate buffer (pH 7), filter-steralized, used undiluted. Empty vector culture filtrate was used as a control. Effectors were stored in a refrigerator at 4° on ice until used. MAGIC lines were grown in 96-well trays filled with medium grade compost and germinated in a heated and lit glasshouse held at 20° day, 17° night with a 16-hr photoperiod. Each line was represented by three replicates. All lines and replicates were allocated well positions with a randomized design. Outer cells of each tray were sown with discard to help reduce edge effects. Seedlings were grown for 2 wk, after which the third leaf from each plant was infiltrated with SnTox1 using a 1-mL syringe without needle, and the extent of infiltration along the leaf marked with a nontoxic pen. In addition, negative water infiltration controls were included for every MAGIC line. One week after infiltration, infiltrated leaves were scored visually for necrosis based on the protocol previously described by Waters et al. (2011), modified to a 0 to 4 scoring scale, where 0 = complete insensitivity and 4 = extensive necrosis (Figure 1).
Figure 1.
Score protocol for P. nodorum effector sensitivity. All scores were made on the infiltrated region delineated by indelible nontoxic marker pen as follows: 0: no visible effect, 1: very slight chlorosis, 2: fully chlorotic, 3: chlorosis and, or slight necrosis, 4: fully necrotic.
Statistical analyses and bioinformatics
SnTox1 phenotypic data were analyzed using REML as implemented in GenStat version 14 (VSN International, Hemel Hempstead, UK). Line means estimated as fixed effects were used in subsequent QTL analyses. Heritability among MAGIC line means was estimated as (variance among MAGIC lines estimated as random effects) / (variance among MAGIC lines + average variance of differences among MAGIC lines). QTL analyses were undertaken as described in Mackay et al. (2014) using the R package lme4 (Bates et al. 2013). Although the MAGIC population was designed to produce a population with uniform kinship relationships, some structure remains. As the full pedigree of the population is known, this was accounted for using a mixed model with variance components to account for each stratum (between funnels and between outcrossed plants within funnels). The full MAGIC population pedigree is available from http://www.niab.com/magic/. Marker-trait associations were considered significant above the Bonferroni corrected P = 0.05 threshold (-log10P = 5.49). SNPs were ordered in the resulting Manhattan plots according to the consensus genetic map generated with the wheat 90k SNP chip (Wang et al. 2014). For three significant SNPs, genetic position on the consensus map was amended by identification of their corresponding most highly correlated SNPs using Pearson correlation coefficient. Available genomic sequence flanking each SNP were used for BLASTn analysis against rice (Oryza sativa L. ssp. japonica cv. Nipponbare) coding regions (CDS) (MSU Osa1 assembly v6.1, http://rice.plantbiology.msu.edu/), brachypodium (Brachypodium distachyon accession BD21) gene models (assembly v1.0, using sequence data produced by the US Department of Energy Joint Genome Institute, http://modelcop.org/, MIPS/JGI v1.2 annotation), and hexaploid wheat cv. ‘Chinese Spring’ genome survey sequence (GSS) from flow-sorted chromosome arms (http://urgi.versailles.inra.fr/; The International Wheat Genome Sequencing Consortium 2014), as described by Wang et al. (2014). Gene predictions were undertaken on wheat genome sequence contigs using FGENESH (Solovyev et al. 2006). Interspecies orthology was determined by BLAST, with an e-value threshold of <1e-4.
Development of KASP markers
Selected SNPs were converted to the Kompetitive Allele-Specific PCR (KASP) genotyping platform (LGC Genomics), based on single-plex technology that employs a universal fluorescent reporting system in conjunction with allele-specific primers. SNP flanking sequences (Wang et al. 2014) were used for BLASTn searches of the genome survey sequence for wheat cultivar ‘Chinese Spring’ (The International Wheat Genome Sequencing Consortium 2014; available via http://wheat-urgi.versailles.inra.fr/). To identify putative homeologous copies on the A, B, and D wheat subgenomes, the highest matching sequence contigs were used as inputs for gene predictions using FGENESH (http://www.softberry.com/) using the ‘Triticum aestivum’ model. Predicted gene models for each homeologous set were aligned with ClustalW (Thompson et al. 1994) and manually edited with GENEDOC v2.6 (http://www.nrbsc.org/gfx/genedoc/). Homeologue-specific nucleotides flanking each targeted SNP were highlighted for incorporation into KASP primer design. Genomic DNA from the eight MAGIC parents and a subset of the progeny (see Table S1) was extracted using a modified Tanksley protocol (Fulton et al. 1995). DNA concentrations were determined using a Nanodrop 200 spectrophotometer (Thermo Scientific), and diluted to a final concentration of 10 ng/µL with sterile distilled water. KASP genotype data were returned from the service provider as .csv files, analyzed with SNP Viewer v.1.99 (http://lgcgenomics.com/), and compared with the corresponding SNP calls from the 90,000 SNP array.
Data availability
MAGIC germplasm is available from NIAB on request. ISelect 90k SNP genotype data is available from NIAB online at www.niab.com/magic/. SnTox1 sensitivity scores for the MAGIC lines screened are available in Table S1. The genetic markers used for QTL mapping, and their genetic map positions, are listed in Table S2.
Results
SnTox1 phenotyping
SnTox1 sensitivity was assessed on 642 MAGIC lines, with line means adjusted for block and position effects (see Table S1 and Figure 1). Parental lines exhibited varying SnTox1 sensitivity, with adjusted mean values ranging from 0.03 (Rialto) to 2.36 (Soissons). Analysis of progeny sensitivities identified transgressive segregation, with values ranging from -0.37 to 4.00, and a mean of 1.47. SnTox1 sensitivity of line means was found to be highly heritable, at 56.0%.
QTL mapping of SnTox1 sensitivity
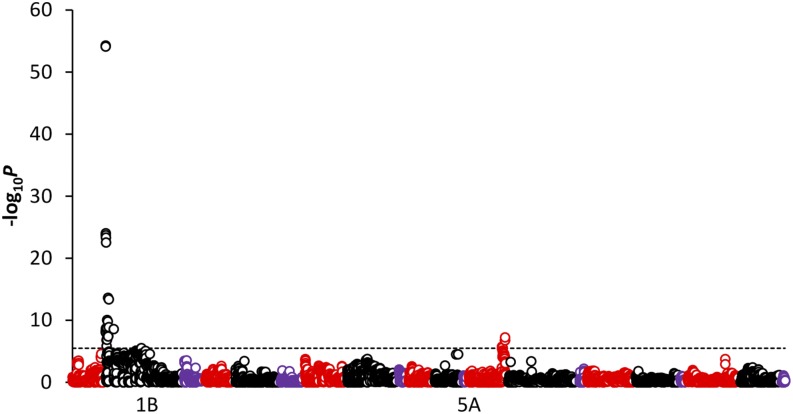
Using 15,514 SNPs across 642 MAGIC lines, QTL mapping of SnTox1 sensitivity in the MAGIC population identified 42 significant (-log10P > 5.49) SNPs (Table 1, Figure 2, and File S1). Of these, 29 genetically mapped to chromosome 1B, 11 to 5A, and 2 were unassigned. By far the strongest QTL was located on chromosome 1B, with the highest marker-trait associations identified for SNPs Excalibur_c21898_1423 (-log10P = 55.29) and BS00093078_51 (-log10P = 54.12), which cosegregated at 8.36 cM. The peak marker(s) explained 25.9% of the variation in line means for SnTox1 sensitivity, with box-plots of genotype at Excalibur_c21898_1423 vs. host sensitivity clearly indicating phenotypic partitioning according to genotype (Figure 3). The second genetic locus was identified by SNP BS00068108_51 (-log10P = 7.25) on the long arm of chromosome 5A, termed here QSnn.niab-5A.1 following Graingenes annotation (www.graingenes.org/), and explained 4.1% of the variation. Modeled together, the two peak SNPs at Snn1 and QSnn.niab-5A.1 explained 28.7% of the variation in line means. Of the six significant SNPs that lacked a genetic map position in the consensus map, all but BS00022296_51 could be tentatively assigned to chromosome arms on the basis of high linkage disequilibrium with the peak Snn1 marker (D′ > 0.94) or by BLASTn hits to wheat genome survey sequence generated from flow-sorted chromosome arms (Table 1). To enrich the 1BS QTL with additional markers, previously uncalled or unusable (due to quality control parameters) SNPs predicted to map to the short arm of the group 1 chromosomes by BLASTn analysis to the wheat reference genome sequence were recalled manually. Twelve markers in high linkage disequilibrium (D′ >0.94, R2 >0.6) with the peak markers (Excalibur_c21898_1423, BS00093078_51) were recovered: BobWhite_c4303_524, BS00011824_51, BS00015608_51, BS00022296_51, BS00026180_51a, BS00030768_51, BS00071333_51, BS00076192_51b, BS00111170_51b, Excalibur_c35316_388a, Jagger_c5878_119, and Kukri_c37738_417. Inclusion of these additional SNPs within subsequent QTL analyses (total number of markers = 15,526) found them to be highly significant (-log10P ≥ 42.26), although not more significant than the two best markers identified from the automated 90k SNP chip calls.
Table 1. Wheat genetic markers significantly associated (-log10P ≥ 5.49) with TOX1 sensitivity in the eight-parent MAGIC population (total number of significant SNPs = 54).
| Marker Name | Sig.-log10 P | Ta chr, cMa | Rice Homologb | % IDb | Brachy Homologc | % IDc | Ta GSS BLAST Hitd | % IDd | Gene Modelse |
|---|---|---|---|---|---|---|---|---|---|
| Excalibur_c21898_1423 | 55.29 | 1B, 8.361 | Os02g18940 (Os05g01040) | 89 | (2g40070) | . | 1BS_3482116 | 100 | 3 |
| BS00093078_51 | 54.12 | 1B, 8.361 | Os05g01090 (Os05g01110) | 73 | 2g40120 (2g40040) | 78 | 1BS_3479333 | 99 | 3 |
| BS00026180_51af | 51.61 | 1B, 8.361 | . | . | . | . | 1BS_3448531 | 98 | 1 |
| IAAV5782 | 38.60 | U | Os11g44580 | 68 | . | . | 1BS_3443196 | 100 | 1 |
| Kukri_c37738_417f | 38.44 | 1B, 9.679 | . | . | . | . | 1DS_108919 | 96 | |
| BobWhite_c4303_524f | 38.18 | 1B, 9.679g | Os05g01250 | 94 | 2g39980 | 97 | 1DS_1882757 | 99 | |
| BS00011824_51f | 37.87 | U | . | . | 2g40046 | 92 | 1DS_1907090 | 100 | |
| BS00030768_51f | 37.81 | 1B, 19.79 | . | . | . | . | 1BS_345816 | 99 | 3 |
| BS00022296_51f | 37.27 | U | . | . | 2g39247 | . | 1AS_1838762 | 100 | 1 |
| BS00076192_51bf | 35.50 | 1B, 60.62 | . | . | 4g09944 | . | 1BS_3456702 | 100 | 2 |
| BS00111170_51bf | 35.29 | 1B, 9.68 | . | . | . | . | 1BS_3454955 | 99 | 1 |
| Excalibur_c22958_433 | 24.95 | 1B, 8.361g | . | . | . | . | 2BL_8088363 | 99 | |
| Jagger_c5878_119f | 24.19 | 1B, 8.361 | Os05g01060 (Os05g10150) | 85 | 2g40100 (2g40090) | 85 | 1BS_3451992 | 100 | 3 |
| Kukri_c44369_131 | 24.04 | 1B, 8.361 | Os02g18940 (Os05g01040) | 93 | 2g40077 | 89 | 1BS_3482116 | 100 | 3 |
| BS00071333_51f | 24.02 | 1B, 8.361 | . | . | . | . | 1BS_2548463 | 99 | 1 |
| Excalibur_c35316_388af | 24.02 | U | . | . | 2g01490 | 85 | 1BS_3478312 | 99 | 2 |
| BS00015608_51f | 24.02 | U | . | . | 1g44420 | 86 | 1AS_3265927 | 100 | 1 |
| BS00022504_51 | 23.73 | 1B, 8.361 | (Os05g01230 Os05g01240) | . | (2g39980*) | . | 1BS_3475480 | 99 | 7 |
| RAC875_c24163_155 | 23.32 | 1B, 8.361 | Os04g30200 | 80 | . | . | 3DL_6837169 | 96 | |
| BS00082565_51 | 22.55 | 1B, 9.679 | (Os05g01280) | . | (2g03650) | . | 1BS_3483503 | 99 | 4 |
| TA003422-0757 | 14.03 | 1B, 41.057 | . | . | . | . | 1BS_3464287 | 100 | 3 |
| BS00011695_51 | 13.63 | 1B, 41.057 | (Os05g01780, Os05g01790) | . | 2g39347 (2g39340) | 70 | 1BS_3440902 | 99 | 3 |
| BobWhite_c14271_1379 | 13.39 | 1B, 43.858 | . | . | . | . | 5BS_1482084 | 99 | |
| Excalibur_c16851_835 | 10.05 | 1B, 31.04 | Os05g51630 | 90 | 2g39230 | 92 | 1BS_3480523 | 99 | 7 |
| IAAV4194 | 9.83 | 1B, 31.094 | Os05g02010 | 92 | 2g39150 (2g38750) | 91 | 1BS_3479684 | 100 | 2 |
| BS00071161_51 | 8.92 | 1B, 43.858 | (Os05g01020) | . | (2g40046) | . | 1BS_3450821 | 99 | 6 |
| BS00022429_51 | 8.77 | 1B, 30.338 | . | . | 4g14110 | 80 | 1BS_3484197 | 99 | 7 |
| Excalibur_c10657_796 | 8.70 | 1B, 8.361 | Os05g01060 | 89 | 2g40100 | 92 | 1DS_1904229 | 100 | |
| BS00022180_51 | 8.60 | 1B, 43.858g | (Os05g01090, Os05g01110) | . | 2g40120 (2g40040) | 74 | 1BS_3479333 | 99 | 3 |
| BobWhite_c5793_372h | 8.36 | 1B, 9.679 | Os10g02990 | . | . | . | 1BS_1682563 | 99 | 1 |
| BS00050522_51 | 8.11 | 1B, 5.275 | (Os05g01030) | . | . | . | 1BS_3482114 | 99 | 2 |
| BS00022482_51 | 8.04 | 1B, 9.679 | (Os05g01280) | . | . | . | 1BS_3483503 | 99 | 4 |
| Kukri_rep_c106834_139 | 8.04 | 1B, 9.679 | (Os05g01230 Os05g01240) | 84 | (2g39990, 2g40000) | . | 1BS_3475480 | 98 | 7 |
| BS00022505_51 | 7.90 | 1B, 9.679 | (Os05g01230 Os05g01240) | 86 | 2g39990 (2g40000) | 86 | 1BS_3475480 | 99 | 7 |
| BS00067961_51i | 7.79 | U | . | . | 4g05910 | 77 | 1BS_3432531 | 99 | 2 |
| Tdurum_contig47083_278 | 7.51 | 1B, 41.213 | (Os05g01550) | 97 | 2g39700 | 74 | 1BS_3435018 | 98 | 5 |
| BS00070706_51 | 7.39 | 1B, 41.213 | (Os05g01550) | . | (2g39700) | . | 1BS_3435018 | 99 | 5 |
| BS00068108_51 | 7.25 | 5A, 144.263 | . | . | . | . | 5AL_1712099 | 99 | |
| CAP7_c3299_342 | 6.88 | 1B, 18.741 | . | . | . | . | 1AS_1937927 | 97 | |
| BS00105846_51 | 6.87 | 1B, 28.761 | Os05g01710 | 91 | 5g01710 | 91 | 1BS_3398629 | 99 | 4 |
| Excalibur_c42255_425 | 6.79 | 5A, 144.137 | . | 85 | 3g09740 | 82 | 5AL_2807342 | 100 | |
| Excalibur_c77910_272 | 6.79 | 5A, 144.137 | . | . | . | . | 5AL_2685434 | 99 | |
| CAP7_c3299_316 | 6.78 | 1B, 18.741 | . | . | . | . | 1AS_1937927 | 97 | |
| Excalibur_c30569_384 | 5.86 | 1B, 21.045 | . | . | . | . | 1BS_3484442 | 99 | 7 |
| wsnp_Ku_rep_c71232_70948744 | 5.73 | 5A, 125.683 | Os03g02970 | 88 | 1g77087 | 95 | 5AL_2752914 | 99 | |
| Kukri_c75091_220 | 5.73 | 5A, 125.682 | . | . | . | . | 5AL_2752914 | 100 | |
| RAC875_c32639_395 | 5.73 | 5A, 125.682 | Os03g02980 | 87 | 1g77080 | 77 | 5AL_2670848 | 100 | |
| wsnp_Ex_c23795_33033150 | 5.73 | 5A, 125.682 | Os03g02970 | 86 | 1g77087 | 90 | 5AL_2752914 | 100 | |
| wsnp_Ex_c23795_33033959 | 5.73 | 5A, 125.682 | Os03g02970 | 86 | 1g77087 | 93 | 5AL_2752914 | 99 | |
| Kukri_c75091_154 | 5.73 | 5A, 125.682 | . | . | . | . | 5AL_2752914 | 99 | |
| Tdurum_contig78972_316 | 5.68 | U | (Os05g02020) | . | (2g38970) | . | 1BS_3460466 | 98 | 1 |
| BS00022299_51 | 5.63 | 5A, 124.588 | . | . | . | . | . | . | |
| wsnp_Ex_c23795_33033010 | 5.61 | 5A, 125.682 | Os03g02970 | 86 | 1g60100 | 82 | 5AL_2752914 | 100 | |
| RAC875_rep_c70803_1442 | 5.53 | 1B, 66.0715 | Os10g36060 | 89 | 3g30277 | 93 | 1BL_3907000 | 100 | 1 |
Both mapped and unmapped markers are included. MAGIC, multiparent advanced generation inter-cross; SNP, single-nucleotide polymorphism; Sig, significance; Ta, Triticum aestivum; Chr, chromosome; CDS, coding sequence; GSS, genome survey sequence; EST, expressed sequence tag, LD, linkage disequilibrium.
Wheat genetic map position according to Wang et al. (2014).
Best rice homolog, based on BLASTn searches of flanking SNP sequence vs. rice CDS. Rice genes agreeing with established colinearity with wheat chromosome 1B are indicated in bold. Genes in parentheses list colinear rice orthologs of predicted wheat gene models identified on wheat 1BS GSS contigs. Percentage identity (% ID) indicated.
Best brachypodium homolog, based on BLASTn searches of flanking SNP sequence vs. brachypodium CDS. Brachypodium genes agreeing with established colinearity with wheat chromosome 1B are indicated in bold. Genes in parentheses list colinear brachypodium orthologs of predicted wheat gene models identified on wheat 1BS GSS contigs. All brachypodium genes are prefixed by “Bradi.”
Best hit to wheat genome sequence, based on BLASTn searches of flanking SNP sequence vs. wheat GSS of flow-sorted chromosomes.
Number of FGENESH predicted genes on wheat GSS contig.
Re-called SNPs in high LD with Excalibur_c21898_1423, as measured by R2 (>0.6), D1 (>0.94).
Genetic map position amended (see File S1).
Marker from the same gene found by Reddy et al. (2008) to be proximal to Snn1 (based on EST BF293222). Wheat contigs predicted to possess NBS-LRR genes are highlighted in bold.
Classified as unmapped here, see Text S1 in File S1.
Figure 2.
Identification of SnTox1 sensitivity quantitative trait locus in the multiparent advanced generation inter-cross (i.e., MAGIC) population. Significant single-nucleotide polymorphism associations (-log10P ≥ 5.49, indicated by the dashed line) were identified on chromosomes 1B and 5A. Unmapped markers are not shown. Additional markers identified in the region by manual re-scoring (see Materials and Methods) and used in a second association scan are not shown here, but are listed in Table 1.
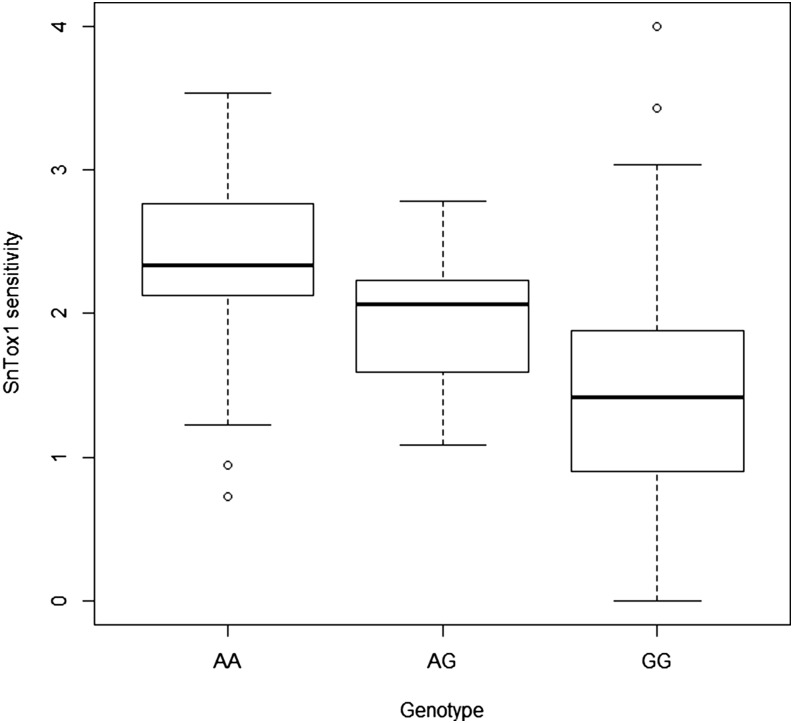
Figure 3.
Box plot for peak marker Excalibur_c21898_1423 on chromosome 1BS, contrasting allelic state (homozygous AA, heterozygous AG, and homozygous GG genotypes) with SnTox1 sensitivity (y-axis, adjusted means rescaled to a 0 to 4 scale).
To attempt to identify additional genetic loci controlling SnTox1 sensitivity, QTL analyses were repeated with the peak 1B marker (Excalibur_c21898_1423) as a covariate. These analyses failed to identify any additional QTL (Figure S1), indicating only two major genetic loci controlling SnTox1 sensitivity were segregating in the MAGIC population. It is of course possible that additional loci with smaller effects were segregating, but were not detected.
Comparative analysis of the Snn1 region
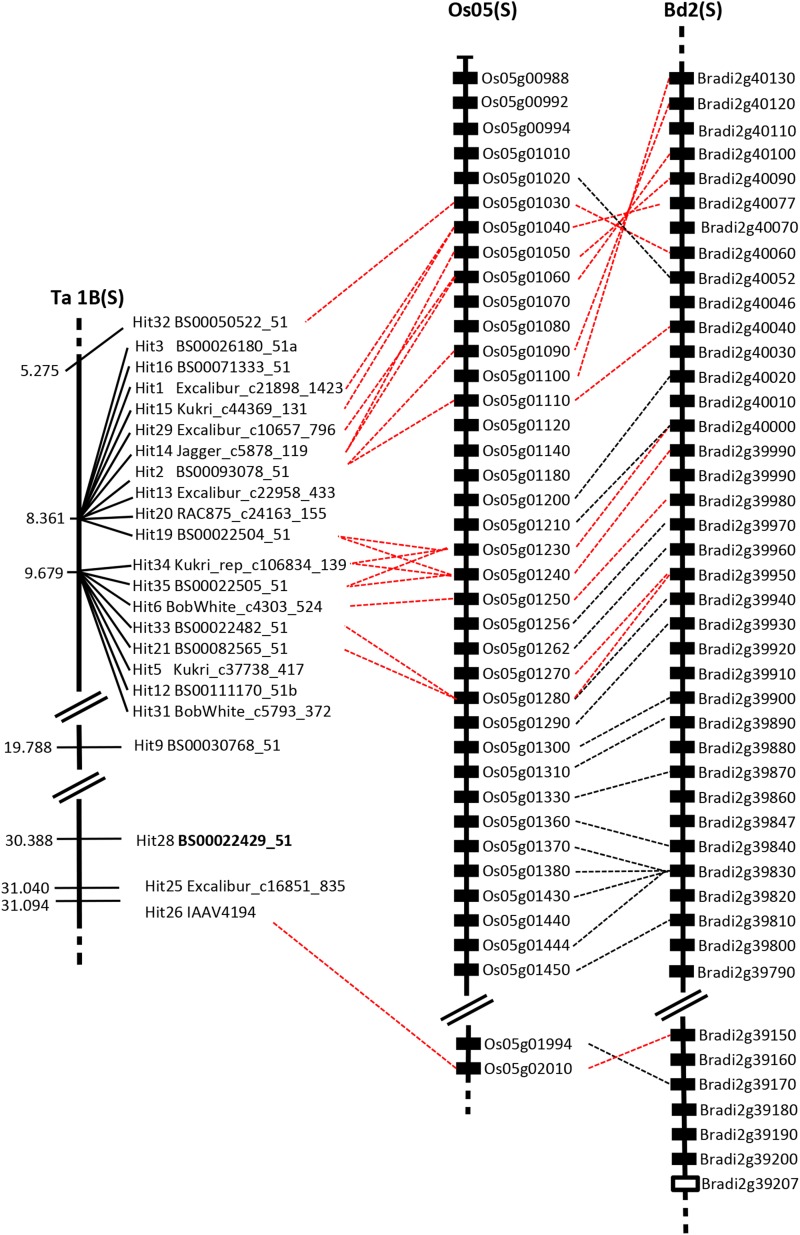
To investigate the possible gene content within the Snn1 region, markers mapped to the region were used to identify the region of the short arm of rice chromosome Os05, and long arm of brachypodium chromosome 4, known to display macro-colinearity with wheat 1BS (Peng et al. 2004; International Brachypodium Initiative 2010). Snn1 previously has been mapped to a 4.7-cM interval between restriction fragment length polymorphism marker ksuD14 and the 1BS telomere (Liu et al. 2004; 2012). KsuD14 lies within a sequenced wheat contig on the short arm of the 1D homeologous chromosome (GenBank accession AF532104S1), predicted to contain four genes, including the cloned resistance gene Lr21 (Huang et al. 2004). However, BLASTn analysis of the four predicted CDS to rice did not identify orthologous genes on the colinear region of rice chromosome Os05. Genomic sequence flanking the Snn1 region SNPs identified here were used as BLASTn queries against all rice and brachypodium gene models (Table 1). To further aid comparative analyses, predicted gene content of wheat GSS contigs identified as possessing Snn1 region SNPs were determined, and the resulting gene models used to identify rice and brachypodium orthologs (Table 1). In total, 19 wheat SNPs/contigs identified the following orthologs in the two sequenced grass species investigated: (1) Rice: 16 orthologous rice genes in the colinear region of Os05, spanning a region from LOC_Os05g02010 (based on orthology with wheat marker IAAV4194) to the short arm telomere. This delimited a physical region of 574 kb in rice, containing 84 predicted genes and 21 transposable elements (see Table S3). No NBS-LRR genes were present in this rice interval nor were any genes homologous to the cloned wheat SnToxA effector sensitivity locus, Tsn1. (2) Brachypodium: 16 orthologous brachypodium genes in the colinear region of Bradi2g, spanning a region from Bradi2g38970 to Bradi2g40120 (orthologous to wheat SNPs IAAV4194 and BS00093078_51, respectively). This delimited a physical interval of 884 kb in brachypodium, predicted to contain 118 genes. No genes homologous to Tsn1 were identified in this region. Although two NBS-LRR genes were found (Bradi2g38987, Bradi2g39207), the use of CDS from these gene as a queries for BLASTn searches of the wheat genome identified best hits on chromosomes 1D (IWGSC_GSS_1899471) and 7A (IWGSC_GSS_4256951), respectively. No NBS-LRR genes were found in brachypodium within the region colinear with the highest associated wheat SNPs (hits 1 to 21, see Figure 4). In wheat, predicted NBS-LRR genes were identified on wheat GSS contigs 1BS_3464287 (anchored by SNP TA003422-0757, 1BS at 41.057 cM), 1BS_3484197 (SNP BS00022429_51, 1BS at 30.338 cM), 1BS_3443196 (SNP IAAV5782, 1BS based on BLASTn of wheat GSS), 1AS_1838762 (SNP BS00022296_51, unmapped but allocated to group 1 chromosomes by BLAST), 1BS_3454955 (SNP BS00111170_51b, 1BS at 9.68 cM), 1BS_2548463 (SNP BS00071333_51, 1BS at 8.361 cM), and 1BS_3484442 (Excalibur_c30569_384, 1BS 1t 5.86 cM) (Table 1). Two additional NBS-LRR genes have been reported as being located in the Snn1 region (BE498831 and BF474204) (Reddy et al. 2008), which correspond to GSS contigs 1BS_3440935 and 1BS_ 3412727, respectively. However, none of the SNPs on the 90k array for which genotype data were returned originated from these contigs, indicating at least nine NBS-LRR genes are located within the region.
Figure 4.
Significant wheat genetic markers (between 0 and 9.7 cM on the short arm of chromosome 1B), and colinearity with the physical maps of rice and brachypodium. Red dashed lines link wheat genes with ortholgous in rice and brachypodium. Black dashed lines link rice and brachypodium orthologs. Brachypodium NBS-LRR genes are indicated in white. The wheat SNP located on a sequence contig predicted to contain an NBS-LRR gene is highlighted in bold. Chromosome arm is indicated: S (short), L (long).
Development of KASP markers for Snn1
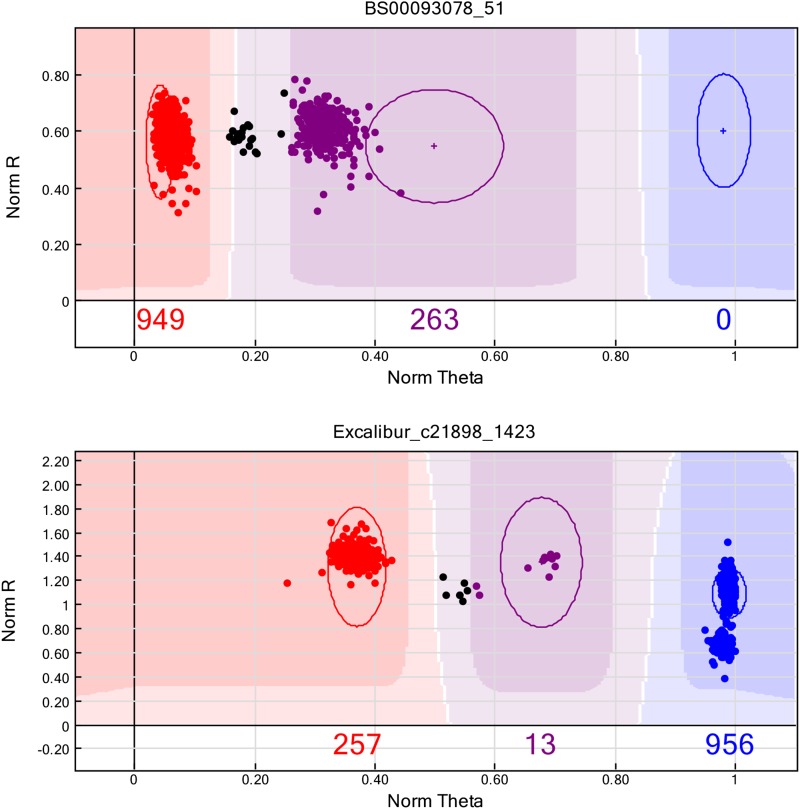
Markers BS00093078_51 (classified as dominant using the default SNP calling parameters) and Excalibur_c21898_1423 (codominant) were found to be highly significant (-log10P = 55.29 and 54.12, respectively). However, manual inspection of SNP calls showed that BS00093078_51 to possess a cluster of heterozygotes, expected in this MAGIC population, as some heterozygosity will remain, depending on how advanced recombinant inbred line development was at the time on genotyping (Figure 5). This suggests confounding of allele calling in the original panel of wheat accessions used to define the calling parameters for the SNP chip, possibly due to additional amplification from non-target homeologues. Thus, visual inspection indicates that BS00093078_51 is more accurately classified as a co-dominant marker. SNP Excalibur_c21898_1423 shows the clustering pattern expected from a co-dominant marker in the absence of amplification from non-target homeologues. Accordingly, both SNPs were selected for attempted conversion to KASP markers. BS00093078_51 was found to be transferable to the KASP system, and was validated on a panel of 95 wheat varieties (see Figure S2 and Table S4).
Figure 5.
Allele calling for the two peak Snn1 SNPs in the 90k SNP dataset, using GenomeStudio software. SNPs BS00093078_51 (top) and Excalibur_c21898_1423 (bottom) were classified as dominant and codominant, respectively, following the standard calling procedures. Visual inspection of BS00093078_51 shows clustering of AA homozygotes (red) and AB heterozygotes (purple), ‘no call’ genotypes (black) located between the AA and AB clusters, and an absence of BB genotypes. These ‘no call’ genotypes represent true heterozygotes, based on the observation that both the BB and AB clouds have two sub-clusters.
Discussion
The use of effectors vs. genetic markers for disease resistance breeding
The P. nodorum-wheat pathosystem was until recently thought to be due to the interaction of a suite of nonspecific toxins and cell wall degrading enzymes (Solomon et al. 2006; Liu et al. 2009). Wheat resistance to P. nodorum was quantitative, and controlled largely by numerous, weak and environment-specific QTL (Xu et al. 2004). The recent identification of necrotrophic effectors in P. nodorum provides tools with which to break down host resistance into its constituent parts, and so help study the disease and help develop resistant varieties. P. nodorum effectors have already been used by breeders to screen for insensitive germplasm within their breeding programs (Tan et al. 2014), resulting in an estimated saving of AUD $50m in Australia alone. However, the ability to develop genetic markers diagnostic for allelic state at the corresponding host sensitivity loci provides additional benefits. First, it allows workflows to be streamlined, as wheat breeders now routinely use genetic markers, allowing host sensitivity markers to be easily incorporated into current practices. Second, as chromosomal regions in close physical linkage with host sensitivity loci may harbor undesired allelic variants, the use of genetic markers allow precise manipulation of other loci on the same chromosome arm. Ultimately, the identification of the genes underlying host sensitivity loci will allow molecular and biochemical characterization of host-pathogen interactions. Such knowledge will help advance the pace of genomics-informed disease resistance breeding in wheat and other crops.
Genetic mapping of sensitivity to SnTox1 using MAGIC
The hexaploid wheat genome is large and complex (2n = 6X = 42, total size = 16 GB), complicating fine-scale genetic analyses. However, numerous genomic tools are now emerging in wheat, including genome survey sequence and dense SNP chips, as used in this study. In addition, recent developments in genetic mapping population design widen the opportunities for genetic analyses in crops. MAGIC is one of a number of multi-parent population designs now emerging in plant sciences (see the recent Multiparent Populations Collection in the journals Genetics and G3; http://www.genetics.org/site/misc/multiparental_populations.xhtml). The benefit of such populations is that they allow the use of linkage and association genetic methodologies for genetic analysis, while avoiding the difficulties imposed by highly structured association mapping populations (Mackay and Powell 2007). MAGIC populations are ideally suited to overcome many of the current barriers to genetic trait dissection in wheat, due to the high levels of genetic diversity captured (via multiple founders) and genetic recombination (via multiple rounds of inter-crossing) (Cavanagh et al. 2008). To date, only two wheat MAGIC populations are publicly available: the eight-parent winter United Kingdom wheat (Mackay et al. 2014) and the four-parent spring Australian wheat (Huang et al. 2012) populations. Here we use the United Kingdom MAGIC population genotyped with a 90,000 feature SNP array to fine-map the wheat Snn1 locus. Adjusted mean SnTox1 sensitivity in the parental lines ranged from 0.03 (Rialto), to 2.36 (Xi19). Genotypic scores at the three most significant SNPs discriminate between Xi19 and Soissions on the one hand, and the other six founders on the other (Table S5). We also identified SNPs uniquely tagging Soissions (Excalibur_c10657_796) and uniquely tagging Xi19 (Kukri_c37738_417), which map within a 1.3-cM interval. Both of these are highly significant (-log10P ≥ 8.11), with their joint effects accounting for a similar proportion of the total sum of squares (SS) compared with the peak SNP, Excalibur_c21898_1423 (29% compared to 26%). Therefore, SnTox1 sensitivity is introduced in the population via Xi19 and Soissons. The Xi19 QTL is the larger effect, accounting for 20% of the SS. Given the previous reports of a single major genetic locus, Snn1, controlling SnTox1 sensitivity at this location (Liu et al. 2012; Reddy et al. 2008), it is likely to represent a single genetic locus. However, in the absence of further information we cannot unambiguously state that these are independent alleles at the same QTL.
In addition to Snn1, we identified a second SnTox1 sensitivity locus on chromosome 5A. Previous analysis of SnTox1 and culture filtrates from SnTox1 knock out mutants has identified a sensitivity locus within a >10-cM interval on chromosome 4B in the ITMI bi-parental mapping population (Liu et al. 2012). However, no 4B QTL was identified in this study. Similarly, the 5A QTL QSnn.niab-5A.1 identified here has not been previously reported. Although the reasons for these differences could be numerous, it is most likely due to the different nature of the germplasm resources used: the ITMI population was constructed from a cross between a Mexican spring variety and a synthetic hexaploid wheat (Van Deynze et al. 1995), whereas elite winter-sown United Kingdom germplasm was used to construct the MAGIC population investigated here.
Comparative mapping of the Snn1 locus
Although genome survey sequence is available, wheat does not contain a completed physical map. This is largely due to the extremely large size of the wheat genome (16 Gb) in comparison to other sequenced cereal species (e.g., rice 430 Mb, brachypodium 255 Mb). However, conservation of gene content and order between members of the Poaceae has meant that colinearity is commonly used as a tool for inferring possible gene content in wheat and other large genome cereal species (e.g., Cockram et al. 2010a,b). Colinearity between wheat 1BS and rice (Os05g) and brachypodium (Bd2) was found to be variable across the Snn1 locus. Nevertheless, based either on homology of the gene in which the genotyped SNP originated, or on additional genes on the same genomic survey sequence contig as the SNP, 16 of the 32 most significant markers possessed orthologs on the colinear regions of the rice genome, with 18 possessing orthologous brachypodium genes. Colinearity between rice and brachypodium showed good overall levels of microcolinearity. In the regions colinear with the most significant wheat SNPs and associated contigs (between 5.275 and 8.361 cM), 62% of rice genes were conserved in brachypodium, whereas 68% of brachypodium genes were conserved in rice. Additionally, a small local inversion between these two species was observed, predicted to involve the wheat region carrying the peak SNP. We note that the rice region involved is within 60 kb of the chromosome Os05 short arm telomere, indicating this rearrangement could have occurred as a result of the macrochromosomal rearrangements that resulted in different chromosome numbers between rice and brachypodium. Thus, while colinearity with sequenced grass species will clearly be of use in map-based cloning of Snn1, differences in gene content mean that sequenced physical wheat maps of the region for cultivars showing SnTox1 sensitivity will be essential. This was found to be the case in the cloning of Tsn1, which is encoded by a gene absent from the colinear region of rice and other sequenced cereal species (Faris et al. 2010). We note that SNP marker IAAV5782 (-log10P = 38.60) originates from the same wheat contig as a marker (BE498831) previously shown to cosegregate with Snn1 (Reddy et al. 2008). As the peak SNP identified in this study was far more highly associated than IAAV5782 (-log10P = 55.29 vs. 38.60, respectively), this illustrates the additional power available in the MAGIC population, despite it not having been constructed specifically to investigate SnTox1 sensitivity.
Effector sensitivity vs. adult plant susceptibility
SNB is a complex disease. Although sensitivity to SnToxA has been shown to be a strong predictor of field SNB incidence, poor correlation between SnTox1 and SnTox3 sensitivity and disease susceptibility has previously been reported (Waters et al. 2011; Oliver et al. 2009). We have hypothesized that there are many other undiscovered effectors, and that interactions between these in establishing SNB have not been fully documented (Tan et al. 2014). Despite this, the role of Snn1 in conferring resistance SNB recently has been confirmed at the seedling and adult stages (Phan et al. 2015). Infection of a double haploid wheat mapping population segregating for Snn1 with P. nodorum SN15 identified Snn1 as a major SNB QTL at both growth stages, based on the severity of tissue necrosis. Furthermore, Snn1 was also confirmed as a major SnTox1 sensitivity QTL in the same population. These results demonstrate that the SnTox1-Snn1 interaction contributes to SNB disease incidence both at seedling and adult stages, and illustrate the utility of developing and using Snn1 linked markers for marker assisted selection.
Diagnostic markers for Snn1
Misclassification of SNP marker BS00093078_51 as dominant using the standard SNP calling protocol was likely due to a combination of the following reasons: (1) coamplification occurred from one or more of the nontarget homeologues in the MAGIC lines; (2) allele bin creation was constructed using a training set composed of a large and diverse set of germplasm, including varieties from around the world, as well as synthetic, tetraploid and diploid wheats. Thus, where nontarget homeologues were not amplified in the training set, a SNP allele calling bin would have been created (in this example allele call BB). In such instances, the heterozygote bin would include heterozygotes (AB), as well as homozygotes + nontarget homeologue (BBBB) and heterozygotes + nontarget homeologue (ABBB). Thus, allele calling would be confounded, as was observed here. This observation has important implications for all those using the array, as it will increase the number and accuracy of markers available for genetic and genomic studies. Indeed, marker enrichment around the Snn1 locus was found to be possible in this study following such an approach.
KASP has become the platform of choice for wheat breeding companies, and is in common use in the cereal research community (e.g., Cockram et al. 2012; Lister et al. 2013; Mackay et al. 2014). Only one (BS00093078_5) of the two Snn1 SNPs successfully converted to the KASP platform, due to a combination of the need to incorporate homeologue-specific nucleotides in the primers, and the constraints of effective primer design. The successfully converted KASP marker was shown to be co-dominant, allowing discrimination of heterozygotes from homozygotes, an important feature for effective use in wheat breeding and research. The development of co-dominant KASP markers closely linked to Snn1 provides a cheap, flexible and robust way for selecting for SnTox1 insensitivity in wheat germplasm.
Supplementary Material
Acknowledgments
The development of the MAGIC population was supported by the United Kingdom Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/E007260/1. The CCDM is supported by the Australian Grains Research and Development Corporation. A. S.’s placement in the United Kingdom was funded via an Erasmus studentship fund. Joint coordination and planning of project activities by J. C. and R. P. O. was aided by networking activities funded under the COST Action ‘SUSTAIN’.
J. C., K. G., G. R., R. P. O., and I. M. designed the research; A. S., J. C., R. K., G. R., and T. B. performed research; J. C., K. G., and I. M. analyzed the data; J. C., K.-C. T., E. F., H. P. P., R. P. O., N. G., and I. M. provided project resources; J. C. wrote the manuscript; and all authors edited and approved the manuscript.
Footnotes
Communicating editor: E. Huang
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.021584/-/DC1
Literature Cited
- Ballance G. M., Lamari L., Bernier C. C., 1989. Purification and characterization of a host-selective necrosis toxin from Pyrenophora tritici-repentis. Physiol. Mol. Plant Pathol. 35: 203–213. [Google Scholar]
- Ballance, G. M., L. Lamari, and C. C. Nermoer, 1996 Cloning expression and occurrence of the gene encoding the Ptr necrosis toxin from Pyrenophora tritici-repentis. Mol Plant Pathol. Available at: http://www.bspp.org.uk/mppol/1996/1209ballance/. Accessed October 1, 2015.
- Bates, D., M. Maechler, B. Bolker, and S. Walker, 2013 Lme4: Linear mixed-effects models using Eigen and S4. R package version 1.0–5. Available at: http://CRAN.R-project.org/package=lme4/. Accessed October 1, 2015.
- Cavanagh C., Morell M., Mackay I., Powell W., 2008. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11: 1–7. [DOI] [PubMed] [Google Scholar]
- Ciuffetti L. M., Tuori R. P., Gaventa J. M., 1997. A single gene encodes a selective toxin causal to the development of tan spot of wheat. Plant Cell 9: 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J., Howells R. M., O’Sullivan D. M., 2010a Segmental chromosomal duplications harbouring group IV CONSTANS-like genes in cereals. Genome 53: 231–240. [DOI] [PubMed] [Google Scholar]
- Cockram J., White J., Zuluaga D. L., Smith D., Comadran J., et al. , 2010b Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proc. Natl. Acad. Sci. USA 107: 21611–21616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J., Jones H., Norris C., O’Sullivan D. M., 2012. Evaluation of diagnostic molecular markers for DUS phenotypic assessment in the cereal crop, barley (Hordeum vulgare ssp. vulgare L.). Theor. Appl. Genet. 125: 1735–1749. [DOI] [PubMed] [Google Scholar]
- Faris J. D., Anderson J. A., Francl L. J., Jordahl J. G., 1996. Chromosomal location of a gene conditioning insensitivity in wheat to a necrosis-inducing culture filtrate from Pyrenophora tritici-repentis. Phytopathology 86: 459–463. [Google Scholar]
- Faris J. D., Zhang Z., Lu H., Lu S., Reddy L., et al. , 2010. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA 107: 13544–13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen T. L., Ali S., Klein K. K., Rasmussen J. B., 2005. Population genetic analysis of a global collection of Pyrenophora tritici-repentis, causal agent of tan spot of wheat. Phytopathology 95: 1144–1150. [DOI] [PubMed] [Google Scholar]
- Friesen T. L., Stukenbrock E. H., Liu Z., Meinhardt S., Ling H., et al. , 2006. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38: 953–956. [DOI] [PubMed] [Google Scholar]
- Friesen T. L., Meinhardt S. W., Faris J. D., 2007. The Stagonospora nodorum‐wheat pathosystem involves multiple proteinaceous host‐selective toxins and corresponding host sensitivity genes that interact in an inverse gene‐for‐gene manner. Plant J. 51: 681–692. [DOI] [PubMed] [Google Scholar]
- Friesen T. L., Zhang Z., Solomon P. S., Oliver R. P., Faris J. D., 2008. Characterization of the interaction of a novel Stagonospora nodorum host-selective toxin with a wheat susceptibility gene. Plant Physiol. 146: 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton T. M., Chunwongse J., Tanksley S. D., 1995. Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 13: 207–209. [Google Scholar]
- Hane J. K., Lowe R. G. T., Solomon P. S., Tan K.-C., Schoch C. L., et al. , 2007. Dothideomycete-plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell 19: 3347–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Brooks S. A., Li W., Fellers J. P., Trick H. N., et al. , 2004. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 164: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B. E., George A. W., Forrest F. L., Killian A., Hayden M. J., et al. , 2012. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol. J. 10: 826–839. [DOI] [PubMed] [Google Scholar]
- The International Brachypodium Initiative , 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- The International Wheat Genome Sequencing Consortium (IWGSC) , 2014. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 6194. [DOI] [PubMed] [Google Scholar]
- Lister D., Jones H., Jones M. K., O’Sullivan D. M., Cockram J., 2013. Analysis of DNA polymorphism in historic barley material: validation of the KASP SNP genotyping platform. Taxon 62: 779–789. [Google Scholar]
- Liu Z. H., Faris J. D., Meinhardt S. W., Ali S., Rasmussen J. B., et al. , 2004. Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host-selective toxin produced by Stagonospora nodorum. Phytopathology 94: 1056–1060. [DOI] [PubMed] [Google Scholar]
- Liu Z., Faris J. D., Oliver R. P., Tan K.-C., Solomon P. S., et al. , 2009. SnTox3 acts in effector triggered susceptibility to induce disease on wheat carrying the Snn3 gene. PLoS Pathog. 5: e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Zhang Z., Faris J. D., Oliver R. P., Syme R., et al. , 2012. The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harbouring Snn1. PLoS Pathog. 8: e1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I., Powell W., 2007. Methods for linkage disequilibrium mapping in plants. Trends Plant Sci. 12: 57–63. [DOI] [PubMed] [Google Scholar]
- Mackay I., Bansept-Basler P., Barber T., Bentley A. R., Cockram J., et al. , 2014. An eight-parent multiparent advanced generation intercross population for winter-sown wheat: creation, properties and validation. G3 (Bethesda) 4: 1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M. C., Oliver R. P., Friesen T. L., Brunner P. C., McDonald B. A., 2013. Global diversity and distribution of three necrotrophic effectors in Phaeosphaeria nodorum and related species. New Phytol. 199: 241–251. [DOI] [PubMed] [Google Scholar]
- Oliver R. P., Solomon P. S., 2010. New developments in pathogenicity and virulence of necrotrophs. Curr. Opin. Plant Biol. 13: 415–419. [PubMed] [Google Scholar]
- Oliver R. P., Rybak K., Solomon P. S., Ferguson-Hunt M., 2009. Prevalence of ToxA-sensitive alleles of the wheat gene Tsn1 in Australian and Chinese wheat cultivars. Crop Pasture Sci. 60: 348–352. [Google Scholar]
- Oliver R. P., Friesen T. L., Faris J. D., Solomon P. S., 2012. Stagonospora nodorum: from pathology to genomics and host resistance. Annu. Rev. Phytopathol. 50: 23–43. [DOI] [PubMed] [Google Scholar]
- Peng J. H., Zadeh H., Lazo G. R., Gustafson J. P., Chao S., et al. , 2004. Chromosome bin map of expressed sequence tags in homoeologous group 1 of hexaploid wheat and homoeology with rice and Arabidopsis. Genetics 168: 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, H. T. T., K. Rybak, E. Furuki, K. Chalmers, D. Mather et al., 2015 Modelling effector-receptor interactions and susceptibility to Septoria nodorum blotch in wheat. Presented at Plant and Animal Genome XXIII, January 10−14, 2015, San Diego, CA.
- Reddy L., Friesen T. L., Meinhardt S. W., Chao S., Faris J. D., 2008. Genomic analysis of the Snn1 locus on wheat chromosome arm 1BS and the identification of candidate genes. Plant Genome 1: 55–66. [Google Scholar]
- Solomon P. S., Wilson T. J., Rybak K., Parker K., Lowe R. G. T., et al. , 2006. Structural characterization of the interaction between Triticum aestivum and the dothideomycete pathogen Stagonospora nodorum. Eur. J. Plant Pathol. 114: 275–282. [Google Scholar]
- Solovyev V., Kosarev P., Seledsov I., Vorobyev D., 2006. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 7(Suppl. 1): S10.1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock E. H., McDonald B. A., 2007. Geographic variation and positive diversifying selection in the host-specific toxin SnToxA. Mol. Plant Pathol. 8: 321–332. [DOI] [PubMed] [Google Scholar]
- Tan K.-C., Walters O. D. C., Rybak K., Antoni E., Furuki E., et al. , 2014. Sensitivity to three Parastagonospora nodorum necrotrophic effectors in current Australian wheat cultivars and the presence of further fungal effectors. Crop Pasture Sci. 65: 150–158. [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J., 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tómas A., Geng G. H., Reeck G. R., Bockus W. W., Leach J. E., 1990. Purification of a cultivar-specific toxin from Pyrenophora tritici-repentis, causal agent of tan spot of wheat. Mol. Plant Microbe Interact. 3: 221–224. [Google Scholar]
- Van Deynze A. E., Dubcovsky J., Gill K. S., Nelson J. C., Sorrells M. E., et al. , 1995. Molecular-genetic maps for group 1 chromosomes of Triticeae species and their relation to chromosomes in rice and oat. Genome 38: 45–59. [DOI] [PubMed] [Google Scholar]
- Vincent D., Du Fall L. A., Livk A., Mathesius U., Lipscombe R. J., et al. , 2012. A functional genomics approach to dissect the mode of action of the Stagonospora nodorum effector protein SnTOXA in wheat. Mol. Plant Pathol. 13: 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wong D., Forrest K., Allen A., Chao S., et al. , 2014. Characterization of polyploid wheat genome diversity using a high-density 90000 single nucleotide polymorphism array. Plant Biotechnol. J. 12: 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters O. D. C., Lichtenzveig J., Rybak K., Friesen T. L., Oliver R. P., 2011. Prevalence and importance of sensitivity to the Stagonospora nodorum necrotrophic effector SnTox3 in current Western Australian wheat cultivars. Crop Pasture Sci. 62: 556–562. [Google Scholar]
- Xu S. S., Friesen T. L., Mujeeb-Kazi A., 2004. Seedling resistance to tan spot and Stagonospora nodorum blotch in synthetic hexaploid wheats. Crop Sci. 44: 2238–2245. [Google Scholar]
- Zhang Z., Friesen T. L., Xu S. S., Shi G., Liu Z., et al. , 2011. Two putatively homoeologous wheat genes mediate recognition of SnTox3 to confer effector-triggered susceptibility to Stagonospora nodorum. Plant J. 65: 27–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MAGIC germplasm is available from NIAB on request. ISelect 90k SNP genotype data is available from NIAB online at www.niab.com/magic/. SnTox1 sensitivity scores for the MAGIC lines screened are available in Table S1. The genetic markers used for QTL mapping, and their genetic map positions, are listed in Table S2.