Abstract
A long awn is one of the distinct morphological features of wild rice species. This organ is thought to aid in seed dispersal and prevent predation by animals. Most cultivated varieties of Oryza sativa and Oryza glaberrima, however, have lost the ability to form long awns. The causal genetic factors responsible for the loss of awn in these two rice species remain largely unknown. Here, we evaluated three sets of chromosome segment substitution lines (CSSLs) in a common O. sativa genetic background (cv. Koshihikari) that harbor genomic fragments from Oryza nivara, Oryza rufipogon, and Oryza glaberrima donors. Phenotypic analyses of these libraries revealed the existence of three genes, Regulator of Awn Elongation 1 (RAE1), RAE2, and RAE3, involved in the loss of long awns in cultivated rice. Donor segments at two of these genes, RAE1 and RAE2, induced long awn formation in the CSSLs whereas an O. sativa segment at RAE3 induced long awn formation in O. glaberrima. These results suggest that the two cultivated rice species, O. sativa and O. glaberrima, have taken independent paths to become awnless.
Keywords: awn, domestication, Asian rice, African rice, CSSLs
The awn, a typical feature of Poaceae, is a needle-like organ extending from the tip of the lemma and is considered to be a modified leaf blade (Dahlgren et al. 1985). This bristly, barbed extension of the spikelet facilitates seed dispersal by attaching the seed to animal fur and deters seed predation by birds and mammals (Grundbacher 1963). In some cases, such as in wild tetraploid wheat, the movement of awns may even propel the seed into soil (Elbaum et al. 2007; Elbaum et al. 2008). Despite the important roles of grass awns under wild conditions, long and barbed awns hinder manual harvesting under agricultural conditions and have largely been avoided during artificial selection of rice by humans (Takahashi 1955). In contrast to barley awns, which are capable of photosynthesis during grain-filling, rice awns lack chlorenchyma and cannot contribute to photosynthesis (Grundbacher 1963; Kjack and Witters 1974; Takahashi et al. 1986; Tatsumi and Kawano 1972). Consistent with that observation, removal of awns has been shown to have a negligible effect on rice grain maturation (Tsudamori 1933) and, consequently, cultivated rice varieties may have become awnless to enhance ease of harvest without adverse effects on yield.
The genus Oryza has two independently domesticated species: cultivated Asian rice (Oryza sativa) and cultivated African rice (Oryza glaberrima) (Khush 1997). Domestication of O. sativa from its wild progenitor, Oryza rufipogon, is thought to have started ∼8000 years ago, and there is evidence of a polyphyletic origin of O. sativa (Yamanaka et al. 2002; Cheng et al. 2003; Fuller et al. 2010; Huang et al. 2012). In contrast to its Asian cultivated counterpart, O. glaberrima was domesticated from Oryza barthii in West Africa more recently, ∼3500 years ago (Linares 2002; Li et al. 2011). Despite the independent domestication histories of O. sativa and O. glaberrima, most varieties of both species are awnless, whereas their ancestral species, O. rufipogon and O. barthii, possess long awns (Chang et al. 1977). Past genetic studies have identified multiple awn-related quantitative trait loci (QTL) in rice (Sato et al. 1996; Xiong et al. 1999; Lorieux et al. 2000; Thomson et al. 2003; Yoshimura et al. 2010); however, only one gene has been cloned. This gene, named An-1, is a bHLH transcription factor located on chromosome 4, which was identified using an interspecific cross between O. rufipogon and O. sativa (Luo et al. 2013). The study showed that the O. sativa allele of An-1 has lost its prolonged expression at the distal end of the lemma compared to the O. rufipogon allele. This expression change confers the awnless phenotype in grains of O. sativa. While a recent mutant study revealed two additional developmental genes, DL and OsETT2, that also affect awn morphology (Toriba and Hirano 2014), the regulatory mechanism of long awn formation is still largely unknown and the question of how O. sativa and O. glaberrima lost their awns remains.
In this study, three sets of chromosome segment substitution lines (CSSLs) in a common O. sativa genetic background were used to identify three long awn-inducing loci. The three loci, Regulator of Awn Elongation 1 (RAE1), RAE2, and RAE3, may be involved in the loss of long awns during the domestication of O. sativa and O. glaberrima. Our data suggest that O. sativa lost the function of RAE1 and RAE2, whereas O. glaberrima achieved an awnless phenotype through mutation(s) in RAE3 while maintaining functional alleles at both RAE1 and RAE2. This report is the first account of two closely related crop species converging on a single morphological feature through mutations in different genes.
Materials and Methods
Plant materials
O. sativa ssp. japonica cv. Koshihikari (hereinafter referred to as Koshihikari), O. glaberrima Acc IRGC104038, and three sets of chromosome segment substitution lines (CSSLs) were used for phenotypic analyses to identify loci that control long awn formation. The three CSSL populations (WBSLs, RSLs, and GLSLs) share Koshihikari as the recurrent parent but differ in their donor parents: WBSLs contain Oryza nivara Acc W0054 donor introgressions; RSLs contain O. rufipogon Acc W0106 donor introgressions (Furuta et al. 2014); and GLSLs contain O. glaberrima Acc IRGC104038 donor introgressions (Shim et al. 2010). These plant materials were grown either in the greenhouses of the Laboratory of Plant Molecular Biosystems or under natural conditions in the research field of Nagoya University, Togo, Aichi, Japan. Seedlings were first grown in the greenhouse for 30 d and then transplanted in the field.
Measurement of yield related traits and awn frequency per panicle
Panicles of the CSSLs were harvested after seed maturation. We measured the following yield-related and awn traits: panicle length; number of primary branches; number of seeds per panicle; seed length; seed width; and awn frequency per panicle. Any seed with an awn longer than 3 mm (measured by a ruler) was considered awned. Awn frequency per panicle was calculated as the number of awned seeds per panicle divided by the total number of seeds per panicle. Seed length and width were measured using a scanned image analyzing software called SmartGrain (Tanabata et al. 2012).
Linkage analysis and fine mapping of RAE1 and RAE3
To map RAE1, ∼8000 F2 plants were produced by crossing GLSL-13 with Koshihikari. For RAE3, we used the progenies of 54 BC4F1 derived from a cross between O. glaberrima Acc IRGC104038 as the recurrent female parent and O. sativa cv. Taichung65 as the donor male parent. To develop the BC4F1 populations, F1 plants derived from a cross between O. glaberrima and Taichung65 were backcrossed successively to O. glaberrima four times. The 54 BC4F2 populations comprising approximately 100 plants each were subjected to the linkage analysis between awn formation and the genotypes of the mapping population. Each mapping population was genotyped using SSR and insertion/deletion (indel)-based markers developed to target the putative gene location (see Supporting Information, Table S1 for primer sequences). Genomic DNA from the mapping population was extracted using the TPS method (Hattori et al. 2007). SSRs and indel markers were amplified using standard PCR protocols and run on a 3% agarose gels containing ethidium bromide.
Sequence analysis of RAE1
A BAC clone library for O. glaberrima Acc IRGC104038 was provided by Honda Research Institute (HRI) in Kisarazu, Chiba, Japan. The BAC clone (HWC026-A20) carrying the RAE1 candidate locus was screened from the BAC clone library using the flanking markers previously used for fine mapping of RAE1. Sequencing of the BAC clone was performed by shotgun sequence method using an ABI3700 capillary DNA sequencer (Applied Biosystems). Base-calling and assembly were performed using Sequencher sequence analysis software (HITACHI SOFT). Comparative analysis between sequences within the candidate region for RAE1 derived from the BAC clone and the corresponding sequences in Nipponbare [Rice Annotation Project Database, (RAP-DB); IRGSP-1.0] was performed using ClustalW ver. 2.0 set at the default settings (Larkin et al. 2007).
Transformation constructs for complementation and overexpression
To develop a subclone for the complementation test of RAE1, the DNA of BAC clone HWC026-A20 was partially digested with HindIII to generate genomic fragments containing the RAE1 candidate locus. The binary vector, pYLTAC7 (Liu et al. 1999), was digested with HindIII and dephosphorylated by calf intestine alkaline phosphatase (CIAP) treatment. The BAC DNA fragments were ligated to the digested pYLTAC7 using Takara DNA Ligation Kit LONG (TAKARA), following the manufacturer’s protocol. The recombinant plasmids were transformed into Escherichia coli strain DH10B by electroporation and plated on LB medium with kanamycin. Three recombinant plasmids containing (1) the entire genomic sequence of Os04g0350700, (2) the entire sequence of Os04g0351333, and (3) the coding region of Os04g0351333 and its downstream noncoding region were obtained. To produce overexpression lines, the open reading frame (ORF) of Os04g0350700 was obtained from the full-length cDNA of Os04g0350700 using the SMARTer RACE cDNA Amplification kit (TAKARA) and cloned into pENTR/D-TOPO (Invitrogen). By LR recombination reaction (Invitrogen), the ORF was transferred into pGWB502 and fused with the CaMV35S promoter (Nakagawa et al. 2007). All constructs were introduced into Agrobacterium tumefaciens, strain EHA105, by electroporation (Hood and Helmer 1986) and used to transform the awnless cultivar cv. Nipponbare following the methods of Hiei et al. (1994).
RNA extraction and RAE1 expression analysis
Immature lemmas were collected from panicles during the booting stage, whereas mature lemmas were collected from panicles during the heading stage of Koshihikari and GLSL-13. Total RNA was extracted from each sample using the RNeasy Plant Mini Kit (QIAGEN) and treated with RNase-Free DNase set (QIAGEN) to remove genomic DNA contamination. Omniscript RT Kit (QIAGEN) was used for first strand cDNA synthesis from each of the extracted RNA samples.
StepOne Real-Time PCR System (Applied Biosystems) was used to analyze the relative expression levels of RAE1 in Koshihikari and GLSL-13. Reaction mixtures for real-time PCR were prepared following the manual of Power SYBR Green PCR Master Mix (Applied Biosystems). Relative expression levels were calculated by dividing RAE1 expression levels by UBQ expression levels. The following primers were used for PCR of RAE1 and UBQ: forward primer 5′-ATCCTCCTCTTCACGGCTTCTA-3′ and reverse primer 5′-CGTATGTACAGAAGGAGAGGTCG-3′ for RAE1 and forward primer 5′-ACACGGTTCAACAACATCCA-3′ and reverse primer 5′-GATCAAGAACTAGAGCGTCA-3′ for UBQ.
Data availability
The plant materials used in the present study are available upon request. Figure S1 shows the comparison of amino acid sequences of RAE/An-1 between O. glaberrima and O. sativa. Table S1 provides the sequences of primers used in this study.
Results
Two loci cause awnless phenotype in O. sativa
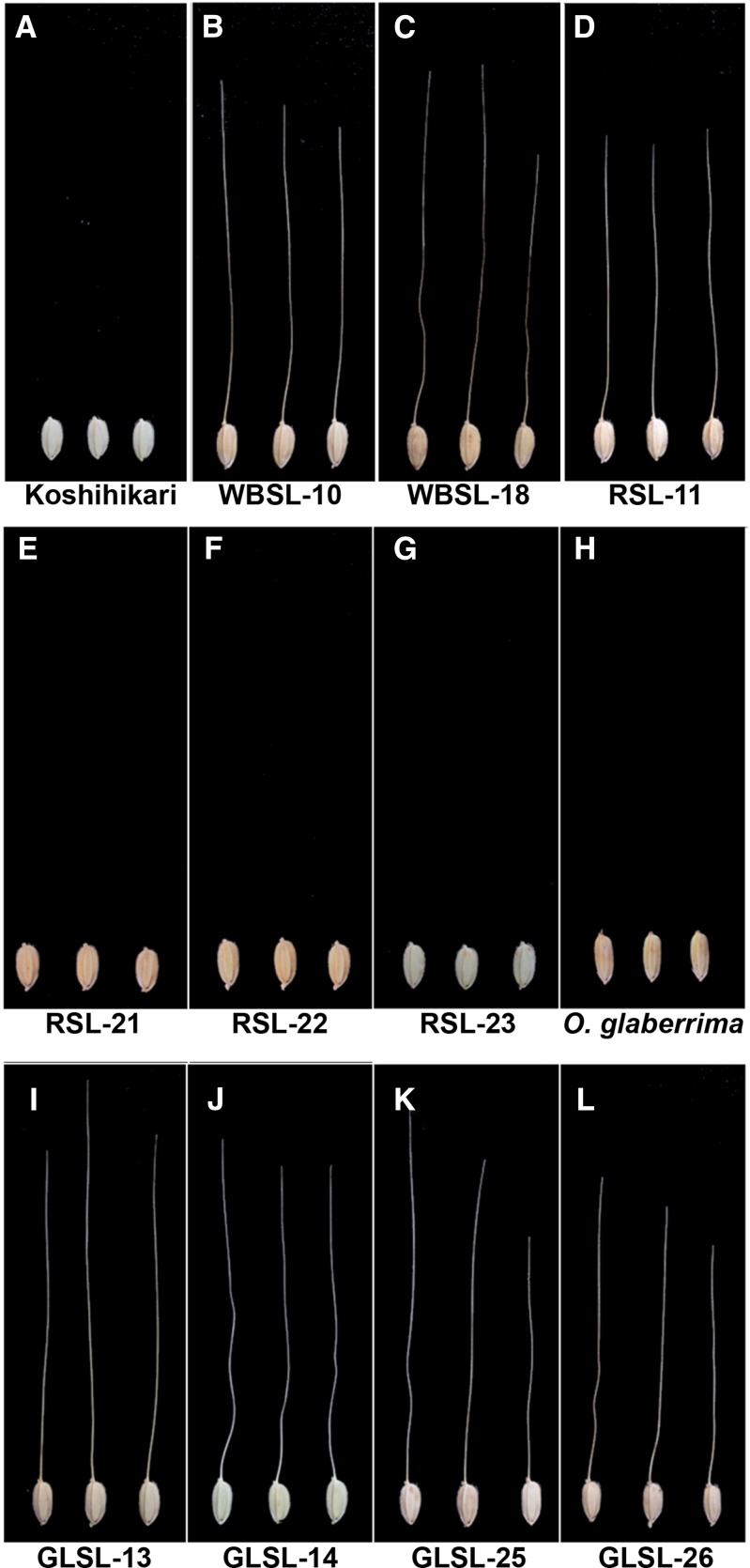
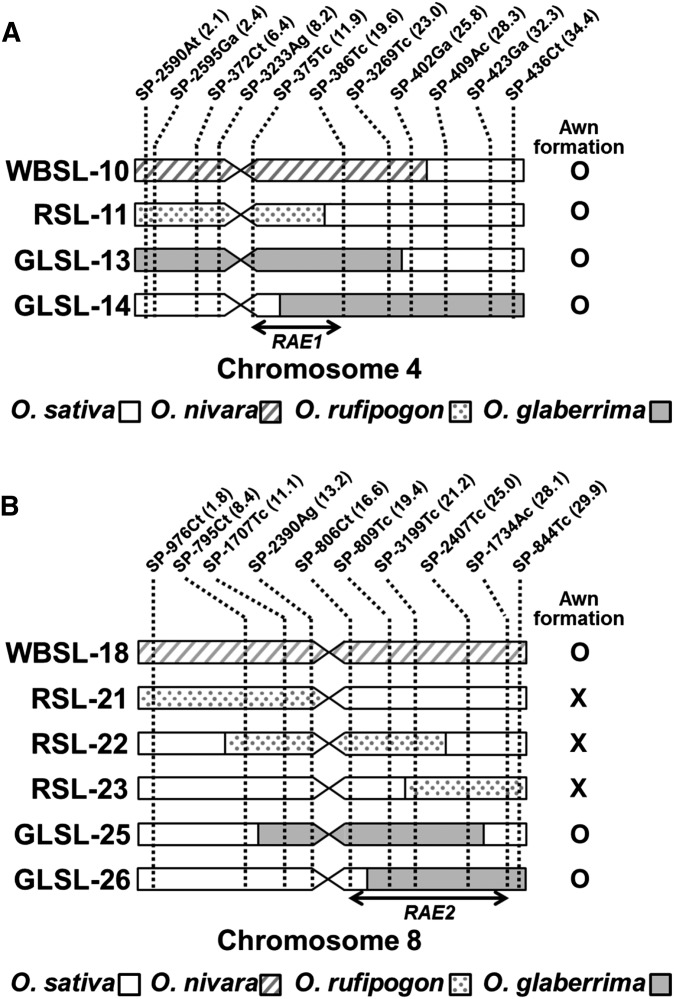
The CSSL population WBSLs were evaluated phenotypically for the presence of awns, and two awned lines were identified, WBSL-10 and WBSL-18 (Figure 1, A–C). Because the recurrent parent, Koshihikari, is an awnless variety, the formation of awns in WBSL-10 and WBSL-18 must be due to the substituted segments derived from O. nivara. WBSL-10 carries a ∼25-Mb donor segment on the end of the short arm of chromosome 4 (Figure 2A), whereas WBSL-18 has an entirely substituted chromosome 8 from O. nivara (Figure 2B). This result reveals that O. nivara contains two functional genes located on chromosomes 4 and 8, and either of which can independently induce awn formation in Koshihikari. Thus, we conclude that the functionality of at least two genes has been lost in O. sativa, resulting in an awnless phenotype. We named the causal genes on chromosomes 4 and 8, RAE1 and RAE2, respectively.
Figure 1.
Long awn formation in CSSLs. Seed’s morphology of Koshihikari (A), WBSL-10 (B), WBSL-18 (C), RSL-11 (D), RSL-21(E), RSL-22 (F), RSL-23 (G), O. glaberrima (H), GLSL-13 (I), GLSL-14 (J), GLSL-25 (K), and GLSL-26 (L). Koshihikari and O. glaberrima do not form long awns. However, the CSSLs except for RSL-21, RSL-22, and RSL-23 have long awns at the distal end of lemmas. WBSLs: CSSLs harboring O. nivara chromosome segments in the genetic background of O. sativa ssp. japonica cv. Koshihikari. RSLs: CSSLs harboring O. rufipogon chromosome segments in the genetic background of O. sativa ssp. japonica cv. Koshihikari. GLSLs: CSSLs harboring O. glaberrima chromosome segments in the genetic background of O. sativa ssp. japonica cv. Koshihikari.
Figure 2.
Graphical genotypes of the CSSLs forming long awns. Graphical genotypes of the CSSLs harboring substituted chromosome segments on chromosome 4 (A) and chromosome 8 (B). Chromosome segments of O. sativa cv. Koshihikari are represented by white boxes, whereas substituted segments from O. nivara, O. rufipogon, and O. glaberrima are indicated as shaded, dotted, and gray boxes, respectively. Presence and absence of long awn in each CSSL are indicated as O and X, respectively. Markers used for the development of the CSSLs are shown with their physical positions (Mb) (indicated in parenthesis) above the chromosome images. Candidate regions of RAE1 and RAE2 are indicated by double-headed arrows.
To confirm our findings, we evaluated the awn phenotype in a second set of interspecific CSSLs, the RSLs. The RSL population also had a Koshihikari background but harbored O. rufipogon donor segments (Furuta et al. 2014). As expected, we found a long awn forming line, RSL-11 (Figure 1D). RSL-11 has a chromosome segment derived from O. rufipogon that spans ∼19 Mb on the end of the short arm of chromosome 4 (Figure 2A). However, in contrast to the results from the WBSL evaluation, we did not observe the induction of awns due to introgressions from the donor parent on chromosome 8: lines RSL-21, RSL-22, and RSL-23 were awnless, although they collectively possess donor segments covering the entire chromosome 8 (Figure 1, E–G; Figure 2B). This suggested a dysfunctional RAE2 allele in the O. rufipogon donor. Evaluation of WBSLs and RSLs showed that RAE1 and RAE2 can independently induce the formation of long awns and that the awnless phenotype in O. sativa resulted from mutations in both RAE1 and RAE2. A previous study by Luo et al. (2013) identified An-1 in chromosome 4, a transcription factor that induces awn formation in rice. The candidate locus of RAE1 identified in this study using O. nivara and O. rufipogon CSSLs includes the An-1 locus, suggesting that RAE1 and An-1 might possibly be identical.
Genetic independence of awnless phenotype in O. sativa and O. glaberrima
Most O. sativa and O. glaberrima varieties share the awnless phenotype (Figure 1, A and H), despite their independent domestication histories in Asia and Africa, respectively. To verify the functionality of RAE1 and RAE2 in O. glaberrima, we evaluated a third set of introgression lines, 34 GLSLs, for the presence of awns. GLSLs carry O. glaberrima chromosomal segments in a Koshihikari background (Shim et al. 2010). We identified four GLSLs with awns (GLSL-13, GLSL-14, GLSL-25, and GLSL-26) despite being derived from two awnless parents (Figure 1, A and I–L).
To investigate whether RAE1 and RAE2 were functional in these awned GLSLs, we examined the four lines for overlap with donor introgressions in the awned RSLs and WBSLs. We found a common 7.7-Mb region (11.9 Mb–19.6 Mb) on the proximal region of the long arm of chromosome 4 in WBSL-10, RSL-11, GLSL-13, and GLSL-14 (Figure 2A). We also discovered a shared donor region spanning 11.5 Mb in the awned lines WBSL-18, GLSL-25, and GLSL-26 on chromosome 8 (Figure 2B). These results implied that although the O. glaberrima donor is phenotypically awnless (Figure 1H), its genome harbors functional copies of RAE1 and RAE2 and is able to induce long awns in Koshihikari. At the same time, it is also implied that awnlessness in O. glaberrima arose from mutation(s) in gene(s) other than RAE1 and RAE2. Our findings suggested that O. sativa and O. glaberrima have independently obtained the awnless phenotype through mutations in different genes.
Fine mapping and identification of the causal gene for RAE1
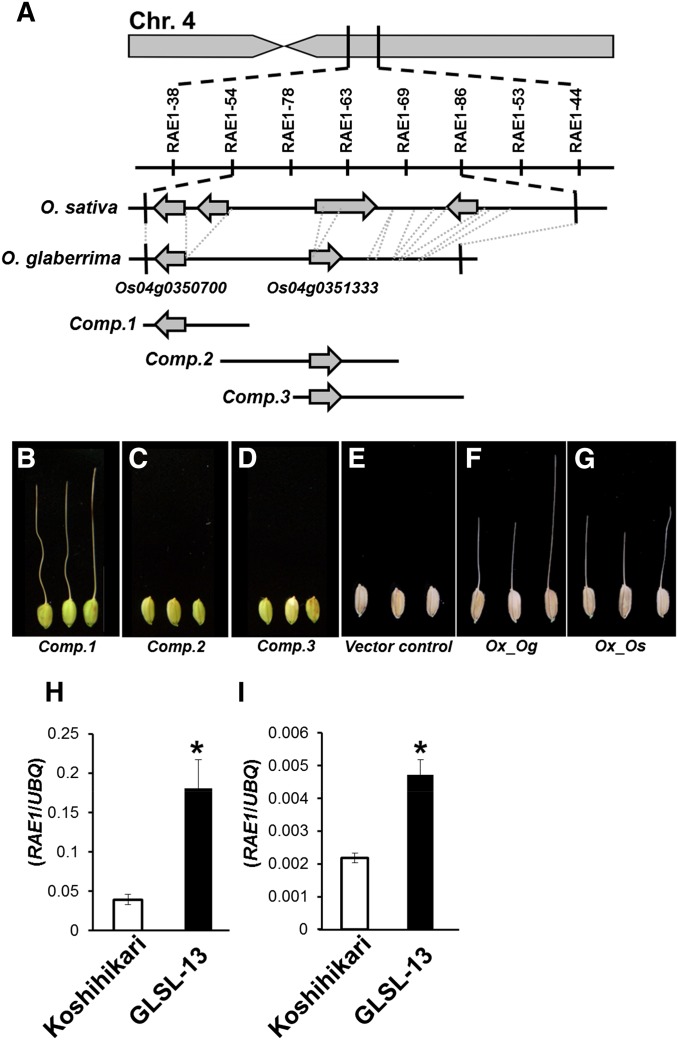
Our study revealed that O. glaberrima also has functional RAE1 on chromosome 4, even though this species do not form long awn. To clarify whether the causal gene for RAE1 in O. glaberrima is exactly identical to An-1 or not, we performed fine mapping for RAE1 in O. glaberrima. We used a population of ∼8000 F2 plants created from a cross between Koshihikari and GLSL-13, the introgression line containing a functional RAE1 from O. glaberrima. We successfully mapped the RAE1 location to a 60.4-kb region flanked by indel-based markers, RAE1-54 (16.73 Mb) and RAE1-86 (16.79 Mb), located on the long arm of chromosome 4 (Figure 3A). Sequence alignment revealed that the 60.4-kb candidate region for RAE1 in O. sativa corresponded to a 41.1-kb region in O. glaberrima, which contained six large insertion/deletion regions. Our data showed that the O. glaberrima genome has only two candidate genes in this region, Os04g0350700, which was reported as An-1, and Os04g0351333 (Figure 3A) (Luo et al. 2013).
Figure 3.
Positional cloning, complementation, and expression analysis of RAE1. (A) RAE1 was mapped at the proximal part of the long arm of chromosome 4. This region has four annotated genes in O. sativa, whereas O. glaberrima only has two (Os04g0350700 and Os04g0351333) of four genes in the corresponding region. (B–G) Complementation test and overexpression analysis were performed using awnless O. sativa cv. Nipponbare. Long awn formations were observed in the transgenic line of Comp.1 harboring the entire genomic fragment of Os04g0350700 derived from O. glaberrima (B), whereas Comp.2 and Comp.3 did not show awn formation (C and D). Overexpression of O. glaberrima allele (Ox_Og) (F) and Koshihikari allele (Ox_Os) (G) of Os04g0350700 induced long awn formations in Nipponbare, whereas vector control showed no awn (E). (H and I) Expression analysis of Os04g0350700 in immature lemma (H) and mature lemma (I) collected from GLSL-13 and Koshihikari. Mean values of three biological replicates are shown. An asterisk indicates the statistical significance at P < 0.01 in Student’s t-test.
To identify which of the two genes caused long awn formation, we performed a complementation test. The candidate region was subcloned from the BAC clone HWC026-A20 in three fragments, namely Comp.1, Comp.2, and Comp.3 (Figure 3A). Of the three fragments, only Comp.1 induced long awn formation in the transformants (Figure 3, B–D), implicating Os04g0350700 as the causal gene for RAE1. Thus, these results verified that RAE1 is identical to An-1 and that the O. glaberrima allele of RAE1/An-1 is functional for long awn formation.
RAE1/An-1 has a 792-bp ORF and encodes 263 amino acids of a bHLH-type transcription factor of group 1A1, classified based on conserved motifs capable of binding E-box and G-box domains (Li et al. 2006) (Figure S1). We identified six SNPs and two indels in the RAE1/An-1 coding region based on sequence comparison between O. glaberrima and O. sativa cv. Nipponbare (IRGSP-1.0). Although no polymorphism was located within the bHLH-conserved sequence, five of the eight mutations caused changes in the amino acid sequence (Figure S1). Additionally, the putative regulatory region upstream of the coding region in O. sativa contained a 4.4-kb transposable element (TE) insertion, along with numerous smaller indels, the same as with the case of O. rufipogon allele of RAE1/An-1 described in the previous study for An-1 (Luo et al. 2013).
Although sequence comparison between O. glaberrima and O. sativa RAE1/An-1 alleles did not directly reveal which mutation(s) was responsible for the awnless phenotype in O. sativa, we hypothesized that the TE insertion in O. sativa might disrupt normal gene expression and performed additional experiments to test this. We produced overexpression lines of RAE1/An-1 using alleles from both O. glaberrima and Koshihikari transformed into Nipponbare, a temperate japonica variety that harbors the same TE insertion found in Koshihikari. The overexpression lines of RAE1/An-1 using both the O. glaberrima and Koshihikari alleles induced long awns in the Nipponbare background (Figure 3, E–G), proving that the Koshihikari RAE1/An-1 indeed retains molecular function despite its awnless phenotype. Additionally, we compared RAE1/An-1 expression in GLSL-13 and Koshihikari and found significantly increased expression in the introgression line vs. the recurrent parent in the immature and mature spikelet, respectively (Figure 3, H and I). These data confirm that it is an alteration of RAE1/An-1 expression, rather than a structural change, that causes Koshihikari’s awnless phenotype. In the An-1 work, the TE insertion at the promoter region of An-1 has been suggested as the possible causal mutation for the loss of its expression (Luo et al. 2013). Therefore, the presence of the TE insertion in both Koshihikari and Nipponbare support that temperate japonica varieties of O. sativa might become awnless due to altered RAE1 cis regulation.
Novel awn-inducing locus causes loss of long awns in O. glaberrima
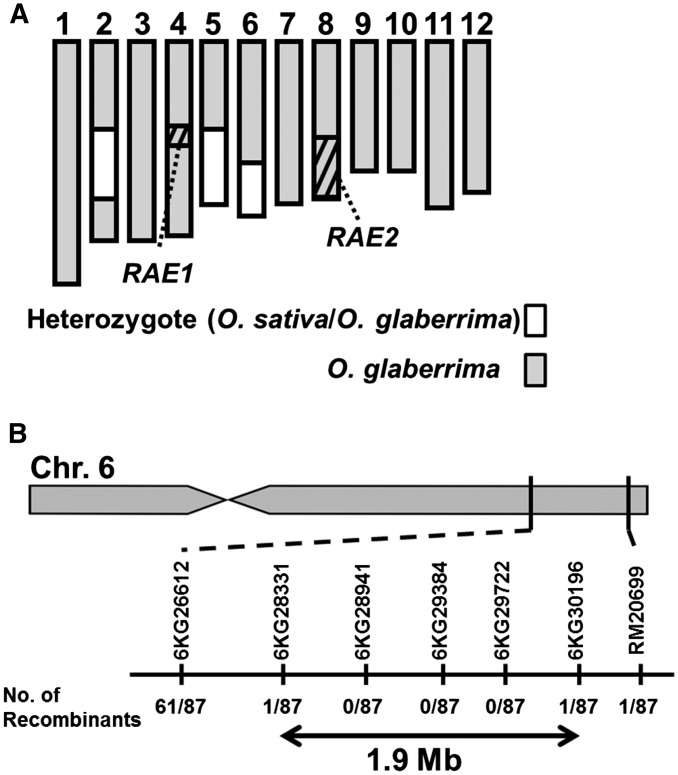
Phenotypic analysis of the GLSLs showed that O. glaberrima had no long awns, despite retaining functional copies of both RAE1 and RAE2. This finding suggested that another gene(s) was responsible for the loss of long awns in O. glaberrima. To identify this causal locus, we performed linkage analysis using progenies of the BC4F1 population derived from a cross between two awnless varieties: O. sativa cv. Taichung 65 × O. glaberrima (Acc IRGC104038), with O. glaberrima as the recurrent parent. In this BC4F1 population, we expect an average of 94.75% of the genome to be fixed for the O. glaberrima genotype and 6.25% to be heterozygous and able to segregate in the next generation. We evaluated progeny from 54 BC4F1 lines to identify families that were fixed for functional alleles at both RAE1/An-1 and RAE2 (O. glaberrima segments) but that still segregated phenotypically for long awns. We identified one such family of 87 BC4F2 plants that segregated phenotypically in a 3:1 ratio (61 long awned individuals vs. 26 awnless individuals). The parental BC4F1 plant of this population harbored heterozygous segments on chromosomes 2, 5, and 6 (Figure 4A).
Figure 4.
Location of causal locus for the loss of long awns in O. glaberrima. (A) The graphical genotype of the parental BC4F1 plant that showed segregation of long awn phenotype in its progenies. Gray boxes indicate homozygous regions derived from O. glaberrima, whereas heterozygous segments of O. sativa and O. glaberrima are indicated in white. Shaded boxes in chromosomes 4 and 8 represent the location of RAE1/An-1 and the candidate region of RAE2, respectively. RAE1 and RAE2 regions have been fixed as the O. glaberrima homozygote in the BC4F1. (B) RAE3 was roughly mapped at a 1.9-Mb region in chromosome 6 flanked by the genotype markers, 6KG28331 and 6KG30196. Numbers below the marker names indicate the number of recombinants.
We further analyzed the genotypes of these segments using the SSR markers RM341 (chromosome 2), RM6346 (chromosome 5), and RM20699 (chromosome 6). Among the 61 long awned individuals, RM341 and RM6346 allele frequencies were consistent with the theoretical segregation ratio for an F2 population; P values were 0.347 for RM341 and 0.975 for RM6346 in a chi-square test for 1:2:1 ratio (Table 1). In contrast, segregation of genotypes at RM20699 was highly distorted from the theoretical 1:2:1 ratio (P value = 2.94×10−5) (Table 1). Similar trends were observed in the 26 awnless plants (Table 1). From these results, we concluded that the genetic factor responsible for the awned phenotype in these BC4F2 plants was located at or near RM20699 on chromosome 6. The presence of a homozygous O. glaberrima segment around RM20699 eliminated awn formation, whereas heterozygous or homozygous O. sativa segments across that region induced long awn formation. Thus, a recessive allele found in O. glaberrima suppressed awn formation in this domesticated species, but the introduction of a functional O. sativa allele at this locus restored long awn formation. We named the causal gene on chromosome 6 regulating awn formation in O. glaberrima RAE3.
Table 1. Linkage between long awn phenotype and genotype.
| Segregation of Genotypes at the Three Markers in the Plants Forming Long Awn | ||||
|---|---|---|---|---|
| O. sativa | Hetero | O. glaberrima | Total no. of plant | |
| RM341 (Chr. 2) | 11 | 31 | 19 | 61 |
| RM6346 (Chr. 5) | 15 | 30 | 16 | 61 |
| RM20699 (Chr. 6) | 18 | 43 | 0 | 61 |
| Segregation of Genotypes at the Three Markers in the Plants Forming No Awn | ||||
|---|---|---|---|---|
| O. sativa | Hetero | O. glaberrima | Total no. of plant | |
| RM341 (Chr. 2) | 4 | 16 | 6 | 26 |
| RM6346 (Chr. 5) | 4 | 10 | 12 | 26 |
| RM20699 (Chr. 6) | 0 | 1 | 25 | 26 |
Each number indicates the number of plants that are homozygous for the O. sativa allele and heterozygous or homozygous for the O. glaberrima allele at specific markers.
Further detailed mapping was performed using genotypic markers located around RM20699 (Figure 4B), which narrowed down the candidate region to 1.9 Mb (marker 6KG28331 to marker 6KG30196) (Figure 4B). Although fine mapping is required to identify the causal gene(s) underlying RAE3, our results indicate that we have identified a novel locus controlling long awn formation that is responsible for the loss of long awns in O. glaberrima, and that this locus is independent of the RAE1/An-1 and RAE2 loci controlling awn formation in O. sativa.
Effects of awn formation on yield related traits
To investigate the effects of awn formation on yield related traits, we measured panicle length, number of primary branches, number of seeds, seed length, and seed width in Koshihikari and long-awned CSSLs (Table 2). We found that GLSL13, 14, and 26 showed a significant decrease in the number of seeds per panicle (Table 2). In GLSL-13, there was a significant increase of seed length, suggesting a trade-off between length and number of seeds. This result is consistent with the previous report for An-1 (Luo et al. 2013). However, from our work, we did not observe a trade-off in number of seeds vs. seed length in CSSLs in two of the lines (GLSL-14 and GLSL-25). It is possible that other genes derived from the donor parents affected the traits we examined and masked the effects of long awn formation on yield-related traits. Interestingly, awn frequency per panicle exhibited significant differences among the CSSLs (Table 2). One striking example can be seen by comparing GLSL-13 with GLSL-14, where significantly different awn frequencies were observed despite the fact that both carry functional alleles at RAE1/An-1. This observation suggests the possibility that awn frequency is regulated independently from awn formation.
Table 2. Yield related traits and awn frequency per panicle in CSSLs.
| Lines | Panicle length (mm) | No. of primary branch | No. of seeds | Seed length (mm) | Seed width (mm) | Awn frequency per panicle† |
|---|---|---|---|---|---|---|
| Koshihikari | 217.80±5.63 | 11.40±1.14 | 159.80±11.37 | 6.03±0.34 | 3.21±0.14 | 0.00±0.00a |
| RSL-11 | 200.20±13.54 | 10.80±0.84 | 147.40±12.05 | 6.05±0.36 | 3.19±0.16 | 64.43±15.40b |
| WBSL-10 | 200.00±12.81 | 9.80±0.45 | 146.60±12.42 | 6.04±0.32 | 3.11±0.14* | 91.10±5.89c |
| WBSL-18 | 184.00±16.06* | 13.60±1.82 | 167.80±15.74 | 6.22±0.28 | 3.04±0.12* | 91.30±6.09c |
| GLSL-13 | 208.60±17.74 | 9.60±0.55 | 121.20±19.38* | 6.59±0.32* | 3.23±0.13 | 56.29±9.41b |
| GLSL-14 | 169.60±8.91* | 10.60±2.07 | 110.80±18.57* | 5.78±0.31* | 3.10±0.13* | 32.94±7.89d |
| GLSL-25 | 155.25±19.70* | 9.00±0.82 | 68.50±22.49* | 6.43±0.30* | 3.27±0.11 | 92.34±2.67c |
| GLSL-26 | 193.80±6.53 | 12.60±1.14 | 141.20±7.09* | 6.02±0.31 | 3.09±0.17* | 83.62±6.17c |
P<0.05 in t-test with Holm’s adjustment comparing with Koshihikari.
Mean ± sd followed by different letters indicate significantly different pairs detected in pairwise t-test with Holm’s adjustment (P<0.05).
Discussion
The absence of long awns is one factor that helps distinguish the cultivated rice species, O. sativa and O. glaberrima, from their wild progenitors, O. rufipogon and O. barthii. While long, barbed awns serve to deter seed predation and enable seed dissemination in the wild, it is likely that they were actively selected against during domestication to facilitate harvesting and postharvest processing.
In this study, we demonstrate that an Asian rice cultivar, O. sativa cv. Koshihikari, is awnless due to two dysfunctional genes (RAE1 and RAE2), whereas an African rice cultivar, O. glaberrima Acc IRGC104038, is awnless due to a third gene (RAE3). This discovery suggests that different awn loci were the targets of selection during the domestication of O. sativa and O. glaberrima, despite the phenotypic similarity. Previous studies have investigated genes involved in the domestication of O. glaberrima (Gross et al. 2010; Sanyal et al. 2010; Wang et al. 2014), but they focused on domestication genes that were already well-characterized in O. sativa (e.g., Rc, qSh1, Sd1, Dep1, OsSh1, and Sh4) and concluded that convergent selection on common genes via independent mutations gave rise to similar phenotypes in Asian and African rice cultivars (Gross et al. 2010; Wang et al. 2014). In our study, we demonstrate that O. sativa and O. glaberrima have independently acquired the same phenotype, awnless seeds, via selection on different genes, namely RAE1 and RAE2 in O. sativa and RAE3 in O. glaberrima. This is the first report showing that selection on independent mutations in different genes conferred the same phenotype in two closely related cultivated crop species.
Using our introgression lines, we found that introduction of either a functional RAE1 or a functional RAE2 allele could induce awn formation in cv. Koshihikari, a temperate japonica cultivar of O. sativa. This leads us to ask what phenotype might have been under selection by humans during the domestication of O. sativa if selection for a dysfunctional allele at only one of the loci already produces awnless rice. One possible explanation is that RAE1 and RAE2 have pleiotropic effects on other biological processes that may have been the targets of selection during domestication. Luo et al. (2013) reported pleiotropic effects of the awn gene, An-1 (equivalent to RAE1 in this study), using NILs in an indica background. In that study, NIL-An-1, containing a functional An-1 allele derived from O. rufipogon, exhibited a reduction in seed number per panicle and increased seed length compared with the indica recurrent parent, Guangluai4. Although significant, the difference in seed number between NIL-An-1 and Guangluai4 was only an average of 10 seeds per panicle. In our study, we observed trade-offs between seed length and seed number in one CSSL, whereas others showed no correlation between seed length and seed number.
Another explanation is that both functional and dysfunctional RAE2 alleles already existed as standing variation in the wild, making it likely that RAE1 was the target of direct selection during domestication (in at least one subpopulation of O. sativa whose ancestor carried a functional allele of RAE2). Subsequent recombination would easily have brought RAE1 and RAE2 together. We see hints of this in the wild donors of the CSSLs used in this study; the O. nivara parent used in the WBSL evaluation carried a functional allele at RAE2, whereas the O. rufipogon parent that was used in the RSL evaluation did not. More detailed genome-wide analyses using diverse cultivars and wild relatives will be required to determine the frequencies of both RAE1 and RAE2 alleles in O. sativa and its wild relatives, and will allow us to explore hypotheses about which gene was selected on first and unveil the processes leading to the awnless phenotype during domestication of O. sativa.
Our finding that a mutation(s) in RAE3 causes the loss of long awns in O. glaberrima, which carries functional alleles of both RAE1 and RAE2, suggests that RAE3 functions as a common upstream or downstream factor of both RAE1 and RAE2, interacting epistatically with RAE1 and RAE2. Evaluation of GLSL-13 and GLSL-14 demonstrated that the introduction of functional alleles of RAE1 can induce long awns in a genotype carrying dysfunctional alleles at RAE2, and evaluation of GLSL-25 and GLSL-26 demonstrated that functional alleles at RAE2 in combination with dysfunctional alleles at RAE1 also induces long awn formation. In O. glaberrima, despite the presence of functional alleles at both RAE1 and RAE2, the long awned phenotype was eliminated by a mutation in RAE3. Isolation and molecular characterization of RAE3 will open the door to a better understanding of the regulatory mechanism(s) underlying long awn formation in rice.
Previous genetic studies identified five loci associated with awn formation. These included: (1) An8, located on the long arm of chromosome 4; (2) An6, located on the long arm of chromosome 8; (3) An7 located on the short arm of chromosome 5; (4) An9 located on the short arm of chromosome 1; and (5) An10 on the long arm of chromosome 1 (Yoshimura et al. 2010). All were identified using introgression lines harboring genomic segments derived from Oryza glumaepatula and Oryza meridionalis. An8 colocates on chromosome 4 with RAE1/An-1, and An6 colocates on chromosome 8 with RAE2, reported here. In the genetic material used in our study, we did not observe long awns in the CSSLs harboring substituted segments in the regions corresponding to An7, An9, and An10. It is possible that O. glumaepatula and O. meridionalis have functional alleles of An7, An9, and An10, whereas O. sativa, O. rufipogon, O. nivara, and O. glaberrima have lost them.
Our observations about differing frequencies of awns on panicles of the CSSLs support the hypothesis that multiple genes affect awn development in rice. Thus, we conclude that awn formation is regulated by a complex network of interacting genes, despite its simple morphology. As a phenotype, awn formation offers an interesting model for studying plant morphogenesis and development, as well as crop evolution and domestication. Further in-depth molecular analysis will be required to unravel the fine-tuned genetic control and pleiotropic impacts of awn development in rice.
Supplementary Material
Acknowledgments
This work was supported mainly by the Program for the Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry, and in part by Grants in Aid for Scientific Research 22119007 from the Ministry of Education, Culture, Sports, Science, by the Japan Science and Technology (JST) Agency-Japan International Cooperation Agency within the framework of the Science and Technology Research Partnership for Sustainable Development (SATREPS), by the Core Research for Evolutional Science and Technology, JST, and by the National Science Foundation (USA) through a graduate research fellowship (to D.W.) and a Plant Genome Research Program Grant (#1026555).
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.020834/-/DC1
Communicating editor: M. Warburton
Literature Cited
- Chang, T. T., A. P. Marciano, and G. C. Loresto, 1977 Morpho-agronomic variousness and economic potentials of Oryza glaberrima and wild species in the genus Oryza. In Meeting on African Rice Species, ORSTOM-IRAT, Paris, pp. 67–75. [Google Scholar]
- Cheng C., Motohashi R., Tsuchimoto S., Fukuta Y., Ohtsubo H., et al. , 2003. Polyphyletic origin of cultivated rice: based on the interspersion pattern of SINEs. Mol. Biol. Evol. 20: 67–75. [DOI] [PubMed] [Google Scholar]
- Dahlgren R., Clifford H. T., Yeo P. F., 1985. The families of the monocotyledons: Structure, evolution and taxonomy, Springer, New York. [Google Scholar]
- Elbaum R., Zaltzman L., Burgert I., Fratzl P., 2007. The role of wheat awns in the seed dispersal unit. Science 316: 884–886. [DOI] [PubMed] [Google Scholar]
- Elbaum R., Gorb S., Fratzl P., 2008. Structures in the cell wall that enable hygroscopic movement of wheat awns. J. Struct. Biol. 164: 101–107. [DOI] [PubMed] [Google Scholar]
- Fuller D., Sato Y., Castillo C., 2010. Consilience of genetics and archaeobotany in the entangled history of rice. Archaeol. Anthropol. Sci. 2: 115–131. [Google Scholar]
- Furuta T., Uehara K., Angeles-Shim R. B., Shim J., Ashikari M., et al. , 2014. Development and evaluation of chromosome segment substitution lines (CSSLs) carrying chromosome segments derived from Oryza rufipogon in the genetic background of Oryza sativa L. Breed. Sci. 63: 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B. L., Steffen F. T., Olsen K. M., 2010. The molecular basis of white pericarps in African domesticated rice: novel mutations at the Rc gene. J. Evol. Biol. 23: 2747–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundbacher F. J., 1963. The physiological function of the cereal awn. Bot. Rev. 29: 366–381. [Google Scholar]
- Hattori Y., Miura K., Asano K., Yamamoto E., Mori H., et al. , 2007. A Major QTL Confers Rapid Internode Elongation in Response to Water Rise in Deepwater Rice. Breed. Sci. 57: 305–314. [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T., 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Hood E., Helmer G., 1986. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J. Bacteriol. 168: 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Kurata N., Wei X., Wang Z. X., Wang A., et al. , 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.-G., Hirano Y., Fukaki H., Yanai Y., Tasaka M., et al. , 1999. Complementation of plant mutants with large genomic DNA fragments by a transformation-competent artificial chromosome. Proc. Natl. Acad. Sci. USA 96: 6535–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush G. S., 1997. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35: 25–34. [PubMed] [Google Scholar]
- Kjack J. L., Witters R. S., 1974. Physiological activity of awns in isolines of Atlas barley. Crop Sci. 14: 243–248. [Google Scholar]
- Larkin M., Blackshields G., Brown N., 2007. Clustal W and Clustal X version 2.0. Bioinfomatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li X., Duan X., Jiang H., Sun Y., Tang Y., 2006. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141: 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. M., Zheng X. M., Ge S., 2011. Genetic diversity and domestication history of African rice (Oryza glaberrima) as inferred from multiple gene sequences. Theor. Appl. Genet. 123: 21–31. [DOI] [PubMed] [Google Scholar]
- Linares O., 2002. African rice (Oryza glaberrima): history and future potential. Proc. Natl. Acad. Sci. USA 99: 16360–16365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux M., Ndjiondjop M., Ghesquière A., 2000. A first interspecific Oryza sativa× Oryza glaberrima microsatellite-based genetic linkage map. Theor. Appl. Genet. 100: 593–601. [Google Scholar]
- Luo J., Liu H., Zhou T., Gu B., 2013. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25: 3360–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Suzuki T., Murata S., Nakamura S., Hino T., et al. , 2007. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100. [DOI] [PubMed] [Google Scholar]
- Sanyal A., Ammiraju J.S.S., Lu F., Yu Y., Rambo T., Currie J., Kollura K., Kim H. , Chen J., Ma J., et al. , 2010. Orthologous comparisons of the Hd1 region across genera reveal Hd1 gene lability within diploid Oryza species and disruptions to microsynteny in Sorghum. Mol. Biol. Evol. 27: 2487–2506. [DOI] [PubMed] [Google Scholar]
- Sato S., Setsuji I., Masaki S., Choyu S., 1996. Genetic studies on an awnness gene An-4 on chr 8 in rice, Oryza sativa L. Breed. Sci. 46: 321–327. [Google Scholar]
- Shim R. A., Angeles E. R., Ashikari M., Takashi T., 2010. Development and evaluation of Oryza glaberrima Steud. chromosome segment substitution lines (CSSLs) in the background of O. sativa L. cv. Koshihikari. Breed. Sci. 60: 613–619. [Google Scholar]
- Takahashi R., 1955. The origin and evolution of cultivated barley. Adv. Genet. 7: 227–266. [Google Scholar]
- Takahashi N., Alwan H., Alterfa H., Sato T., 1986. Significant role of awn in rice plants (1): A survey of agricultural value of rice awn. Reports of the Institute for Agricultural Research Tohoku University 35: 21–31. [Google Scholar]
- Tanabata T., Shibaya T., Hori K., Ebana K., Yano M., 2012. SmartGrain: high-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 160: 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi J., Kawano K., 1972. Suitou no noge ni tsuite (about the awn in rice). Nippon sakumotsu-gakkai toukai-shibu kenkyu-happyou kougai (Research reports of Tokai branch of crop science society of Japan) 56: 11–15. [Google Scholar]
- Thomson M., Tai T., McClung A., 2003. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar. Theor. Appl. Genet. 107: 479–493. [DOI] [PubMed] [Google Scholar]
- Toriba T., Hirano H.-Y., 2014. The DROOPING LEAF and OsETTIN2 genes promote awn development in rice. Plant J. 77: 616–626. [DOI] [PubMed] [Google Scholar]
- Tsudamori M., 1933. Seishoku-kikan to shiteno ine noge no kachi (Values of the rice awn as a reproductive organ). Nissakuki 5: 380–390. [Google Scholar]
- Wang M., Yu Y., Haberer G., Marri P. R., Fan C., et al. , 2014. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46: 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Liu K., Dai X., Xu C., Zhang Q., 1999. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 98: 243–251. [Google Scholar]
- Yamanaka S., Nakamura I., Nakai H., Sato Y., 2002. Dual origin of the cultivated rice based on molecular markers of newly collected annual and perennial strains of wild rice species, Oryza nivara and O. rufipogon. Genet. Resour. Crop. Ev. 50: 529–538. [Google Scholar]
- Yoshimura A., Nagayama H., Kurakazu T., 2010. Introgression lines of rice (Oryza sativa L.) carrying a donor genome from the wild species, O. glumaepatula Steud. and O. meridionalis Ng. Breed. Sci. 60: 597–603. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The plant materials used in the present study are available upon request. Figure S1 shows the comparison of amino acid sequences of RAE/An-1 between O. glaberrima and O. sativa. Table S1 provides the sequences of primers used in this study.