Abstract
Many nonmodel species exemplify important biological questions but lack the sequence resources required to study the genes and genomic regions underlying traits of interest. Reef-building corals are famously sensitive to rising seawater temperatures, motivating ongoing research into their stress responses and long-term prospects in a changing climate. A comprehensive understanding of these processes will require extending beyond the sequenced coral genome (Acropora digitifera) to encompass diverse coral species and related anthozoans. Toward that end, we have assembled and annotated reference transcriptomes to develop catalogs of gene sequences for three scleractinian corals (Fungia scutaria, Montastraea cavernosa, Seriatopora hystrix) and a temperate anemone (Anthopleura elegantissima). High-throughput sequencing of cDNA libraries produced ∼20–30 million reads per sample, and de novo assembly of these reads produced ∼75,000–110,000 transcripts from each sample with size distributions (mean ∼1.4 kb, N50 ∼2 kb), comparable to the distribution of gene models from the coral genome (mean ∼1.7 kb, N50 ∼2.2 kb). Each assembly includes matches for more than half the gene models from A. digitifera (54–67%) and many reasonably complete transcripts (∼5300–6700) spanning nearly the entire gene (ortholog hit ratios ≥0.75). The catalogs of gene sequences developed in this study made it possible to identify hundreds to thousands of orthologs across diverse scleractinian species and related taxa. We used these sequences for phylogenetic inference, recovering known relationships and demonstrating superior performance over phylogenetic trees constructed using single mitochondrial loci. The resources developed in this study provide gene sequences and genetic markers for several anthozoan species. To enhance the utility of these resources for the research community, we developed searchable databases enabling researchers to rapidly recover sequences for genes of interest. Our analysis of de novo assembly quality highlights metrics that we expect will be useful for evaluating the relative quality of other de novo transcriptome assemblies. The identification of orthologous sequences and phylogenetic reconstruction demonstrates the feasibility of these methods for clarifying the substantial uncertainties in the existing scleractinian phylogeny.
Keywords: coral, phylogenomics, nonmodel system, database
Transcriptome sequencing provides a rapid and cost-effective approach for gene discovery in nonmodel organisms. Analysis of transcriptomes from a diverse range of invertebrates such as sponges (Riesgo et al. 2014; Conaco et al. 2012), ctenophores (Ryan et al. 2013), annelids (Riesgo et al. 2012), and mollusks (Riesgo et al. 2012; Kocot et al. 2011) has enhanced comparative and evolutionary studies of metazoans. Quantitative analysis of these sequences (RNA-Seq) has become the method of choice to profile genome-wide transcription levels. This technique provides an unbiased approach to discovering functional processes through identification and quantification of differentially expressed genes between phenotypic states including experimental treatments (Meyer et al. 2011), tissue types (Siebert et al. 2011), and developmental stages (Graveley et al. 2011).
Genomic and transcriptomic resources have been developed for a variety of species within the phylum Cnidaria (Moya et al. 2012; Barshis et al. 2013; Fuchs et al. 2014; Helm et al. 2013; Lehnert et al. 2012; Meyer et al. 2009, 2011; Polato et al. 2011; Shinzato et al. 2014; Soza‐Ried et al. 2010; Traylor-Knowles et al. 2011; Wenger and Galliot 2013; Sun et al. 2013; Meyer and Weis 2012; Lehnert et al. 2014), a diverse group of evolutionarily and ecologically significant species that range from hydroids (Class Hydrozoa) and jellyfish (Class Medusozoa) to sea anemones and corals (Class Anthozoa). Cnidarians are among early-diverging or basal metazoans and occupy a key position as a sister taxon to the bilaterians (Dunn et al. 2008). Many cnidarians play an important role in marine trophic cascades due to their mutualistic relationship with dinoflagellate species of the genus Symbiodinium that reside inside of cnidarian host cells. This relationship is based on nutritional exchange in which Symbiodinium spp. provide the cnidarian host with products from photosynthesis in return for inorganic nutrients and a stable, high-light environment (Davy et al. 2012). The paramount examples of this partnership are the reef-building corals, which form the trophic and structural foundation of productive and biodiverse coral reef ecosystems. Anthropogenic stressors, especially those associated with global climate change, are gravely threatening these reef ecosystems, including the corals themselves (Douglas 2003; Weis and Allemand 2009). Insight into the molecular mechanisms that underlie coral-dinoflagellate symbioses and their stress response to environmental perturbation is critical for future management and conservation of coral reef ecosystems.
To date, there are three publically available sequenced genomes from the Anthozoa: the symbiotic coral Acropora digitifera (Shinzato et al. 2011), symbiotic sea anemone Aiptasia sp. (Baumgarten et al. 2015) and the nonsymbiotic sea anemone, Nematostella vectensis (Putnam et al. 2007). These genomes have provided insight into the genomic complexity of cnidarians, furthering studies of gene evolution and function across basal metazoans (Poole and Weis 2014; Putnam et al. 2007; Shinzato et al. 2011; Marlow et al. 2009; Ryan et al. 2006; Hamada et al. 2013; Shinzato et al. 2012a, b; Wood-Charlson and Weis 2009; Dunn et al. 2008). Comparison of these genomes has revealed putative symbiosis-associated genes that may function in the onset and maintenance of cnidarian-dinoflagellate symbiosis (Meyer and Weis 2012). Annotated de novo transcriptomes, generated using next generation sequencing (NGS) [expressed sequence tags (ESTs), 454 pyrosequencing and Illumina HiSeq technologies], have been published for eight genera of anthozoans (Polato et al. 2011; Kenkel et al. 2013; Meyer et al. 2009; Traylor-Knowles et al. 2011; Lehnert et al. 2012; Pratlong et al. 2015; Shinzato et al. 2014; Vidal-Dupiol et al. 2013). These resources have been used in a variety of contexts, including the study of gene family evolution (Poole and Weis 2014), symbiosis-enhanced gene expression (Lehnert et al. 2014), and responses to environmental stressors such as elevated seawater temperature (Meyer et al. 2011; Kenkel et al. 2013), bacterial infection (Closek et al. 2014), and CO2-driven changes in seawater pH (Vidal-Dupiol et al. 2013). These studies are adding to earlier generation omics studies [EST studies, subtractive hybridization and cDNA microarrays (Meyer and Weis 2012)] and are providing information on the mechanisms of cnidarian-dinoflagellate symbiosis and coral bleaching, a stress response that results from the breakdown of the partnership (Weis 2008; Davy et al. 2012). Expression studies are therefore contributing not only to our basic understanding of cellular processes in cnidarians but also to our ability to link molecular responses with phenotypic change due to environmental perturbation.
The available anthozoan resources are limited in taxonomic diversity and dominated by a few genera from a narrow geographic range (Meyer and Weis 2012). In addition, many resources are from aposymbiotic (lacking dinoflagellate symbionts) samples or nonsymbiotic species, which limits the study of interplay between the two partners. One goal of this work is to increase the number and diversity of anthozoan resources for comparative, phylogenetic, and functional analyses.
In this study, we present transcriptomes from four anthozoans: the sea anemone Anthopleura elegantissima (Brandt 1835) and the corals Fungia scutaria (Lamarck 1801), Montastraea cavernosa (Linnaeus 1767), and Seriatopora hystrix (Dana 1846) in varying symbiotic states, life history stages, and geographic locations (Table 1). These species are of particular interest to investigations into the molecular mechanisms associated with the onset, maintenance, and breakdown of cnidarian-dinoflagellate symbioses. We highlight how these transcriptomes can be used in applications ranging from targeted gene searches to orthologous group predictions and phylogenomic analysis. In addition, we outline a method to screen for cross-contamination between sequencing libraries that can be broadly applied to other transcriptome studies.
Table 1. Collection sites, life history stages, and symbiotic states of the four anthozoans used for transcriptome assembly.
| Organism | Collection Site | Developmental Stage | Symbiotic State |
|---|---|---|---|
| Anthopleura elegantissima | Seal Rock, Oregon | Adult | Aposymbiotic |
| Fungia scutaria | Coconut Island, Hawaii | Larval | Aposymbiotic |
| Montastraea cavernosa | Florida Keys, Florida | Adult | Symbiotic |
| Seriatopora hystrix | Nanwan Bay, Taiwan | Adult | Symbiotic |
Materials and Methods
Sample collection and RNA extraction
All four anthozoan species examined in this study engage in symbiosis with Symbiodinium spp., and therefore RNA extractions typically include contributions from the dinoflagellate symbionts at some level. Here, two samples (M. cavernosa and S. hystrix) were collected from symbiotic specimens and two samples (F. scutaria and A. elegantissima) were collected from nominally aposymbiotic stages or specimens (Table 1). Larvae of F. scutaria were reared in filtered seawater at the Hawaii Institute of Marine Biology following fertilization and development and remained symbiont-free during development (Schnitzler and Weis 2010). The aposymbiotic specimen of A. elegantissima was collected in that condition in the field.
Total RNA was extracted from S. hystrix, F. scutaria, and A. elegantissma using the following methods. S. hystrix tissue was stored in RNAlater Stabilization Solution (Qiagen, CA) and RNA was extracted using the RNeasy Mini Kit (Qiagen, CA) according to the manufacturer’s protocol. Whole animal specimens of A. elegantissima (aposymbiotic) and F. scutaria (larvae) were collected, frozen in liquid nitrogen, and stored at −80°. RNA was extracted using a combination of the TRIzol RNA isolation protocol (Life Technologies, CA) and RNeasy Mini Kit (Qiagen, CA). The TRIzol protocol was used for initial steps up to and including the chloroform extraction. Following tissue homogenization, an additional centrifugation step was performed at 12,000 × g for 10 min to remove tissue debris. After the chloroform extraction, the aqueous layer was combined with an equal volume of 100% EtOH and the RNeasy Mini Kit was used to perform washes following the manufacturer’s protocol.
A core sample of M. cavernosa was collected, frozen in liquid nitrogen, and stored at −80°. Total RNA from M. cavernosa was extracted following a modified TRIzol protocol with a 12-M LiCl precipitation (Mazel et al. 2003). Briefly, the coral fragment was vortexed in TRIzol reagent for 15 min and then processed according to the manufacturer’s instructions through phase separation. To precipitate RNA, 0.25 ml of isopropanol and 0.25 ml of a high-salt solution (0.8 M sodium citrate and 1.2 M NaCl) per 1 ml of TRIzol used was added to the aqueous supernatant. The addition of the high-salt solution removes proteoglycan and polysaccharide contaminants. The solution was incubated at room temperature for 10 min and then centrifuged at 12,000 × g for 10 min at 4°. After centrifugation, the standard TRIzol protocol was followed through the ethanol wash. To remove PCR inhibitors of an unknown nature that are frequently encountered in coral samples, RNA was precipitated by adding an equal volume of 12 M LiCl and then was incubated for 30 min at −20°. The sample was centrifuged at 12,000 × g for 15 min at room temperature and washed with 75% EtOH (1 ml per 1 ml of TRIzol), followed by centrifugation at 7500 × g for 5 min at room temperature. The supernatant was removed and the RNA pellet was air-dried.
The extracted total RNA from each sample was DNase-treated using a TURBO DNA-Free Kit (Ambion, CA) to remove genomic DNA contamination. RNA quantity and quality were assessed using the NanoDrop ND-1000 UV-Vis Spectrophotometer (Thermo Scientific, MA) and gel electrophoresis.
Preparation of sequencing libraries
Polyadenylated RNA was purified from 10 µg of total RNA using the Magnetic mRNA Isolation Kit (New England Biolabs, MA). First strand cDNA was synthesized using ProtoScript M-MuLV FS-cDNA Synthesis Kit (New England Biolabs, MA) according to the manufacturer’s protocol and modified oligonucleotides in Supporting Information, Table S1. Second strand synthesis was performed by incubating first-strand cDNA with 1× NEBNext Second Strand Synthesis Buffer (New England Biolabs, MA), 0.2 mM dNTPs, 15 units of Escherichia coli DNA ligase (New England Biolabs, MA), 75 units of E. coli DNA polymerase I (New England Biolabs, MA), and 3 units of RNase H (New England Biolabs, MA) for 2 hr at 16°. cDNA was purified using the GeneJet PCR Purification Kit (Fermentas, MA) and then fragmented using NEBNext dsDNA Fragmentase (New England Biolabs, MA) according to the manufacturer’s protocol, with the addition of 5 mM MgCl2 and 1 mg ml−1 BSA (New England Biolabs, MA). Fragmented cDNA was purified and the ends were repaired using NEB Quick Blunting Kit (New England Biolabs, MA) according to manufacturer’s protocol. The product was purified and A-tailed in a reaction with nuclease-free water, 1× NEB Standard Taq buffer (New England Biolabs, MA), 1 mM dATP, and 2 units of NEB Standard Taq (New England Biolabs, MA) at 68° for 2 hr. Tailed templates were ligated to double stranded adaptors prepared with oligonucleotides from the Illumina Customer Sequence Letter (version August 12, 2014, Illumina 2014; Table S1). Purified, tailed cDNA was combined with T4 DNA ligase buffer (New England Biolabs, MA), T4 DNA ligase (New England Biolabs, MA), and the double stranded adaptors, and the solution was incubated at 12° for at least 6 hr. Ligation products were purified and then amplified using custom sample-specific barcodes (“indices”) designed with a 3-bp minimum Hamming distance based on Illumina barcodes (Illumina 2014) (Table S1). PCR included template cDNA, Phusion Taq polymerase buffer (Thermo Scientific, MA), dNTPs, 5′ Illumina “i5” barcoding oligo, 3′ Illumina “i7” multiplex oligonucleotide, and Phusion High Fidelity Taq polymerase (Thermo Scientific, MA). Reactions were incubated at 98° for 30 sec, followed by 17–21 cycles of the following: 98° for 10 sec, 63° for 30 sec, and 72° for 1.5 min. Reactions were amplified for the minimum cycle number required to produce a visible product on a 1% agarose gel. PCR products were size-selected by excising the 350–550 bp fraction from a 2% agarose gel. Finally, size-selected sequencing libraries were extracted using the E.Z.N.A. Gel Extraction Kit (Omega Bio-Tek, GA).
Sequencing, processing, and assembly
cDNA libraries were sequenced on Illumina HiSeq 2000 at University of Oregon’s Genomics Core Facility (Eugene, OR). All cDNA libraries were pooled on a single lane to produce 100-bp paired-end reads. Raw sequences were filtered using custom Perl scripts to remove uninformative (matching adaptors in Table S1, or poly-A tail) and low-quality reads (>20 positions with quality scores <20) (Meyer et al. 2011). All custom scripts used in this study are available online at GitHub (https://github.com/Eli-Meyer). The high-quality filtered reads were then assembled using default settings in Trinity v2.0.2, a de Bruijn graph-based assembler that uses paired-end data to reconstruct transcripts and group these into components intended to represent the collection of transcripts originating from a single gene (Grabherr et al. 2011).
Functional annotation
To develop these assemblies as resources for functional studies, we assigned putative gene names and functional categories [Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)] to assembled transcripts based on sequence comparisons with online databases. All sequence comparisons were conducted using BLAST+ from National Center for Biotechnology Information (NCBI) (Package version 2.2.29) (Altschul et al. 1990). Gene names were assigned by comparing transcript sequences against UniProt protein sequence databases (SwissProt and TREMBL) using BLASTx with an expect value (E value) cutoff of 10−4. Each transcript was assigned a gene name based on its best match, excluding matches with uninformative names (e.g., uncharacterized, unknown, or hypothetical). GO terms describing biological processes, molecular functions, and cellular components were assigned to each transcript based on GO-UniProt associations of its best match downloaded from the Gene Ontology website (Ashburner et al. 2000). KEGG orthology terms were assigned from single-directional best hit BLAST searches of each transcriptome on the KEGG Automatic Annotation Server (Moriya et al. 2007).
Reference transcriptome databases
To enhance the utility of these resources for the coral research community, we have also developed searchable databases and made these publicly available on the author’s laboratory website hosted at Oregon State University (http://people.oregonstate.edu/∼meyere/index.html). Databases were produced using the open-source SQLite software library and can be queried directly using a publicly accessible web form. To demonstrate the utility of our searchable databases for rapidly identifying genes of interest, we searched each database for a few genes previously studied in cnidarians, including a cell adhesion molecule (sym32) (Reynolds et al. 2000), a cysteine biosynthesis enzyme [cystathionine β-synthase, (Cbs)] (Shinzato et al. 2011), and a fluorescent protein (GFP) (Mazel et al. 2003; Shinzato et al. 2012b; Smith-Keune and Dove 2008). For comparison with these simple text searches, we also conducted a more comprehensive search for each gene based on reciprocal BLAST. Representative sequences for each gene were obtained from the UniProt database (version 2014_09, downloaded October 20, 2014), and searched against each assembly using tBLASTn (bit-score ≥45). The matching transcripts were then reciprocally compared against UniProt using BLASTx. Reciprocal matches were evaluated at the level of gene names; transcripts identified by searching with a target gene (e.g., B5T1L4, GFP from Acropora millepora) were accepted if they reciprocally matched a different gene with corresponding annotation (e.g., Q9U6Y6, a GFP gene from Anemonia manjano).
Evaluating gene content and completeness of assembly
An ideal reference transcriptome would include all genes present in the genome of an organism, but low or tissue-specific expression can lead to incomplete sampling of genes during cDNA library preparation. To evaluate the gene representation of our assemblies, we searched each assembly for sequence similarity with a core set of conserved eukaryotic genes (CEGMA) (Parra et al. 2007) and with gene models from sequenced anthozoan genomes: the coral A. digitifera (OIST: adi_v1.0.1) (Shinzato et al. 2011) and the anemone N. vectensis (assembly version: Nemve1) (Putnam et al. 2007). Sequence comparisons were conducted using NCBI’s BLASTx (Altschul et al. 1990) and bit-scores ≥50 were considered significant.
An ideal transcriptome assembly would also include complete transcripts as contiguous sequences or contigs, but variation in coverage and sequence characteristics lead to fragmented assemblies consisting of partial transcripts. To evaluate the effectiveness of our assemblies in reconstructing complete transcripts, we calculated the Ortholog Hit Ratio (OHR), a metric ranging from 0 to 1 that indicates the proportion of each gene included in the assembled transcript (O’Neil et al. 2010). Each assembly was compared to gene models from the N. vectensis genome using BLASTx to identify orthologs. We calculated OHR first with a relatively stringent approach (OHRHITS) as the proportion of each N. vectensis gene included within local alignments with assembled transcripts (high-scoring segment pairs in BLASTx output). Because this approach excludes divergent regions, we calculated OHR with an alternative and more inclusive approach (OHRORF) as the ratio of the transcript’s longest ORF (in the BLASTx-defined reading frame) relative to the length of its corresponding N. vectensis protein. When multiple transcripts matched a single gene, we considered only the longest OHR. Distributions of maximum OHR scores and summary statistics were examined to evaluate the completeness of each assembly.
Screening for biological contamination
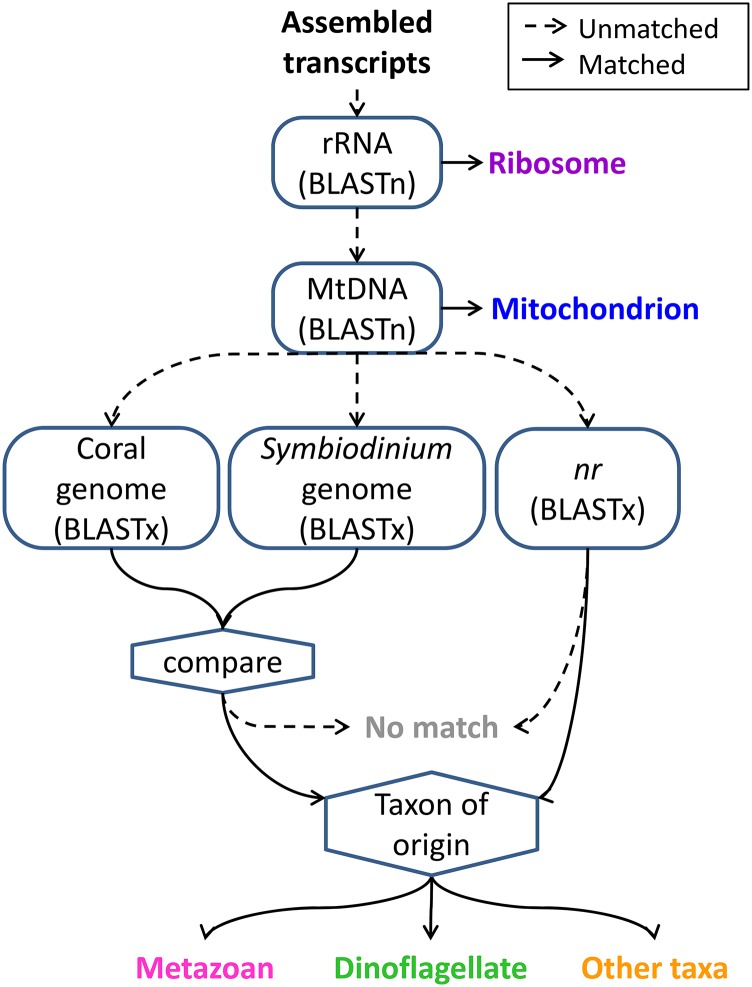
All species used in this study engage in symbiotic associations with intracellular dinoflagellate symbionts; therefore, RNA extracted from these specimens is expected to include contributions from both animal hosts and dinoflagellate symbionts. To evaluate these contributions we conducted a series of sequence comparisons aiming to identify the taxonomic origin of each transcript (Figure 1). Transcripts were compared with a series of sequence databases using BLAST v.2.2.29 with a bit-score threshold of 45. To identify transcripts derived from rRNA, each assembly was compared with cnidarian rRNA sequences using BLASTn. N. vectensis sequences were chosen for this purpose because they represent the most complete cnidarian sequences in the SILVA rRNA database (SILVA: ABAV01023297, ABAV01023333) (Quast et al. 2013). Transcripts were compared with a cnidarian mitochondrial genome using BLASTn; for this analysis, we chose the complete mitochondrial genome from Acropora tenuis (NCBI: NC_003522.1) (van Oppen et al. 2002). To identify the taxonomic origin of each transcript, sequences were compared with the NCBI nonredundant (nr) protein database (downloaded March 12, 2014) using BLASTx (E value ≤10−5) (Altschul et al. 1990). To avoid errors that might arise from the scarcity of cnidarian and dinoflagellate sequences in these databases, transcripts were compared with gene models from Symbiodinium minutum (clade B) (OIST: symbB.v1.2.augustus.prot) and A. digitifera (OIST: adi_v1.0.1_prot) using BLASTx. The taxonomic origin of each sequence was categorized as follows. First, transcripts matching rRNA or mitochondrial sequences were assigned to those categories. Transcripts matching Symbiodinium genes more closely than coral genes that did not return a metazoan hit as their best match in nr were assigned to the dinoflagellate category. Transcripts matching coral genes more closely than Symbiodinium genes that also matched metazoan records or lacked matches in nr were categorized as metazoan. Transcripts that showed conflicting results (metazoan in one database but nonmetazoan in the other) were categorized as “unknown.” Transcripts lacking any match to either coral or Symbiodinium genes were assigned based on taxonomic annotation of the best match in nr, if available. This series of decisions made it possible to classify each transcript based on origin (ribosomal, mitochondrial, other metazoan, dinoflagellate, or other taxa, which includes prokaryotes, uncertain, or no match).
Figure 1.
Annotation pipeline used to classify origins of each assembled transcript. A series of sequence comparisons was performed comparing each transcript against N. vectensis rRNA (SILVA: ABAV01023297, ABAV01023333), A. tenuis mitochondrial DNA (NCBI: NC_003522.1), A. digitifera and S. minutum gene models, and the NCBI nonredundant protein database (bit-score threshold of 45 for small databases; E value threshold of 10−5 for large databases). Transcripts were assigned to categories by evaluating their similarity to each database in the order shown (see Materials and Methods for details).
Screening for cross-contamination
During preliminary analysis of the transcriptome assemblies, we observed a few orthologs with unexpectedly high sequence similarity (>99%) among species.
Because cross-contamination could realistically occur at several different stages during multiplex library preparation and sequencing, we tested for evidence of cross-contamination in our transcriptome assemblies and developed a pipeline to eliminate contaminating sequences. To evaluate the extent of cross-contamination in our libraries, we mapped the cleaned reads used to produce each assembly against that assembly using the Trinity utility align_and_estimate_abundance.pl (Haas et al. 2013). We then compared all transcriptome libraries sequenced and prepared together using BLASTn to identify nearly identical sequences present in multiple assemblies (bit-score ≥100). This analysis identified many sequences occurring in multiple assemblies, which were highly abundant in one sample (consistent with this being their true origin) but were very low in abundance (<10-fold lower) in other assemblies (consistent with cross-contamination). To evaluate the level of sequence similarity expected among anthozoan transcriptomes, for comparison with the similarity observed among our assemblies, we compared publicly available transcript assemblies produced independently in different labs [Pocillopora damicornis, (Traylor-Knowles et al. 2011); A. digitifera, (Shinzato et al. 2011); and A. millepora, (Meyer et al. 2009)]. To eliminate putative cross-contaminants identified in our assemblies, we first compared assemblies using BLASTn to identify highly similar sequences (bit-score ≥100). We then estimated the abundance of each transcript in each assembly by mapping and counting reads from each library against the assembly produced from those reads using the Trinity utility align_and_estimate_abundance.pl. To identify and remove sequences that might result from cross-contamination, we categorized each transcript based on sequence similarity and relative expression in all other assemblies. Any transcripts with nearly identical matches in more than one assembly were assigned to the assembly in which each was most abundant if the sequence was at least 10-fold more abundant in that library than any others. Alternatively, transcripts found at comparable levels (<10-fold difference) in multiple assemblies were flagged as “unknown origin” and excluded from further analysis.
Development of SSR markers
Simple sequence repeats (SSRs), also known as microsatellites, are sequences with repetitive 2–5 base pairs of DNA. These molecular markers have been widely used for studies of genome mapping, genetic linkage, and population structure. Although SSRs have largely been replaced with sequencing-based approaches for single nucleotide polymorphism (SNP) genotyping, in some situations they may still be the most practical option. To demonstrate the utility of transcriptome assemblies for SSR marker development and to identify SSR markers for the four species described here, we used a pipeline we have previously described for identifying SSRs in coral sequence data (Davies et al. 2013). In brief, sequences containing repetitive regions (≥30 bp, ≤15% deviation from perfect repeat structure, ≥30 bp flanking regions) were identified using RepeatMasker v3.2.9 (Smit et al. 1996) and then assembled using CAP3 to eliminate redundancy (Huang and Madan 1999). Target sequences were further screened for redundancy using BLASTn (Altschul et al. 1990) to identify unique targets within each repeat type (e.g., AT, CCG, etc.). Finally, primer sequences flanking these SSRs were developed using Primer3 (Rozen and Skaletsky 1999), targeting regions 150–500 bp with 45–65% GC content.
Identification of orthologous groups
To facilitate comparative studies of cnidarian gene sequences, and to demonstrate the utility of our transcriptome assemblies for phylogenetic analysis, we identified orthologous groups among the four transcriptomes generated in this study. We also compared these with sequence resources from other cnidarians and basal metazoans, including a marine sponge Amphimedon queenslandica (Srivastava et al. 2010), the hydrozoan Hydra magnipapillata (Chapman et al. 2010), the schyphozoan Aurelia aurita (Fuchs et al. 2014), and a variety of other anthozoans including Aiptasia pallida (Lehnert et al. 2012), N. vectensis (Putnam et al. 2007), A. digitifera (Shinzato et al. 2011), Porites astreoides (Kenkel et al. 2013), P. damicornis (Vidal-Dupiol et al. 2013), Stylophora pistillata (Karako-Lampert et al. 2014), Orbicella faveolata (formerly belonging to the genus Montastraea) (Budd and Stolarski 2011; DeSalvo et al. 2008), and Pseudodiploria strigosa (Table S2). These resources varied in the types of sequencing technologies used to create them, and this resulted in differing degrees of assembly completeness, ranging from whole genomes to EST libraries (Table S2). All resources were converted into candidate protein coding sequences using the package TransDecoder (transdecoder.sourceforge.net) that identifies open reading frames. Protein sequences were then processed with FastOrtho (enews.patricbrc.org/fastortho), an OrthoMCL-based program (Li et al. 2003) that performs an all-by-all BLAST of the input sequences (E value cutoff ≤10−5) and clusters orthologous groups with the Markov Cluster algorithm (Van Dongen 2000).
Phylogenetic analysis
The four transcriptomes from this study and other sequence resources were used to infer phylogenetic relationships from commonly used markers and newly identified orthologs. The mitochondrial gene cytochrome c oxidase 1 (COI) has been used to reconstruct the most comprehensive phylogeny of corals (Anthozoa, Scleractinia) (Kitahara et al. 2010), and mitochondrial sequences are commonly used to infer evolutionary relationships of the Cnidaria (Kitahara et al. 2010; Bridge et al. 1992; Kayal et al. 2013). Recent findings suggest, however, that a concatenated set of NADH dehydrogenase genes (ND 2, 4, and 5), called ND supergene, outperforms COI in metazoan datasets including in anthozoans (Havird and Santos 2014).
To investigate the effect of increased gene sampling on phylogenetic inferences, we compared phylogenetic trees constructed based on the widely used marker COI, the ND supergene, and the set of orthologs identified from a comparison of our transcriptomes with other cnidarian sequence resources. All taxa used in searches for orthologous groups were included and A. queenslandica served as the outgroup. The Transdecoder catalog of proteins for each organism was made into a local BLAST database. Then, the mitochondrial protein sequences of COI, ND2, ND4, and ND5 were found from BLASTx searches against our local databases, UniProt database, or NCBI database (Table S3 and Table S4). In some cases, mitochondrial genes were not recovered from the local protein databases, but they were found by tBLASTx to the original resources. These transcripts were instead translated using Expasy Translate Tool (http://web.expasy.org/translate/) under the “invertebrate mitochondrial” genetic code. Proteins sequences for COI, ND2, ND4, and ND5 were aligned using MAFFT v6.864b (Katoh et al. 2002). In some cases, the mitochondrial sequences were fragmented within a single database or recovered from two separate databases (Table S3 and Table S4). These fragments were aligned and manually combined to increase total alignment positions. Individual MAFFT alignments of ND2, ND4, and ND5 were concatenated into a single matrix in Mesquite (v. 3.02) (Maddison and Maddison 2015). Protein alignments of COI and the ND genes were run through ProtTest server (http://darwin.uvigo.es/software/prottest_server.html) (Abascal et al. 2005) to select the appropriate substitution rate model based on AIC and BIC criterion. Phylogenetic trees were constructed using maximum likelihood (ML) in RAxML v8.0.26 (Stamatakis 2014) under the MTZOA+G+F model (Rota-Stabelli et al. 2009). Optimal topology was selected based on ML scores from 500 replicate trees. Nodal support was assessed from 500 bootstrap replicates.
For phylogenomic reconstruction, the computational pipeline PhyloTreePruner (Kocot et al. 2013) was applied to orthologous groups with a minimum amino acid length of 100 from the 15 taxa identified in Table S2. PhyloTreePruner is a phylogenetic approach used to refine orthologous groups identified in programs like OrthoMCL by removing predicted paralogs resulting from gene duplication or splice variants through single gene-tree evaluation (Kocot et al. 2013). First, each group of orthologs was aligned using MAFFT v6.864b with 1000 iterations. Ambiguous or uninformative positions were removed from the alignment using Gblocks v0.91b (Castresana 2000). Then, single-gene ML trees for each group inferred with FastTree2 (Price et al. 2010) were screened for paralogy with PhyloTreePruner and the longest sequence for each taxon was retained. The pruned orthologous groups were then merged into a single matrix using FASconCAT v1.0 (Kück and Meusemann 2010). To examine the impact of missing data on tree topology, two trees were constructed. In the conservative tree, 14–15 taxa were sampled per ortholog for a total of 397 groups (73,833 unique alignment positions). The relaxed tree allowed more missing data, requiring only at least 10 taxa sampled per ortholog for a total of 2896 groups (535,413 unique alignment positions). For each dataset, ML trees were inferred with RAxML v8.0.26 using the WAG+GAMMA+F substitution model (Whelan and Goldman 2001). Topology for each tree was selected from 100 replicate trees, and nodal support values are based on 100 and 500 bootstrap replicates in the conservative and relaxed trees, respectively.
Data availability
The data sets supporting the results of this article are available from the Sequence Read Archive at NCBI (Accession number: SRP063463), the Dryad Digital Repository (doi: 10.5061/dryad.3f08f), and the author’s website (http://people.oregonstate.edu/∼meyere/index.html).
Results and Discussion
Sequencing and de novo assembly
The four libraries described here were sequenced on Illumina HiSeq 2000 (each occupying 1/6th of a lane), yielding on average 26.3 million paired reads per library (range, 21.2–30.3; Table S5). A fraction of these (22% on average; range 14–28%) were removed during quality and adaptor filtering prior to assembly. Assembly of the remaining high-quality reads produced on average ∼170,000 transcripts. This is substantially higher than the number of genes in sequenced cnidarian genomes (23,677 in A. digitifera, 27,273 in N. vectensis), which likely results from redundancy, fragmentation in the assemblies, and biological contamination. Assemblies included many small contigs (on average, 47% were <400 bp) that were unlikely to provide significant matches, so for analyses based on sequence homology we considered only contigs ≥400 bp (average n = 91,792). For these core transcriptome datasets used for downstream analyses, the average length ranged from 1.1 to 1.7 kb and N50 ranged from 1.4 to 2.7 kb. These are slightly shorter than the expected size distribution for a complete cnidarian transcriptome (e.g., average ∼1700 and N50 ∼2200 bp transcripts in the A. digitifera genome), suggesting incomplete assemblies. Assembly statistics of the four transcriptome references developed in this study are broadly comparable to previously published anthozoan transcriptomes (Moya et al. 2012; Shinzato et al. 2011, 2014; Abascal et al. 2005; Traylor-Knowles et al. 2011; Lehnert et al. 2012).
Completeness of transcriptomes
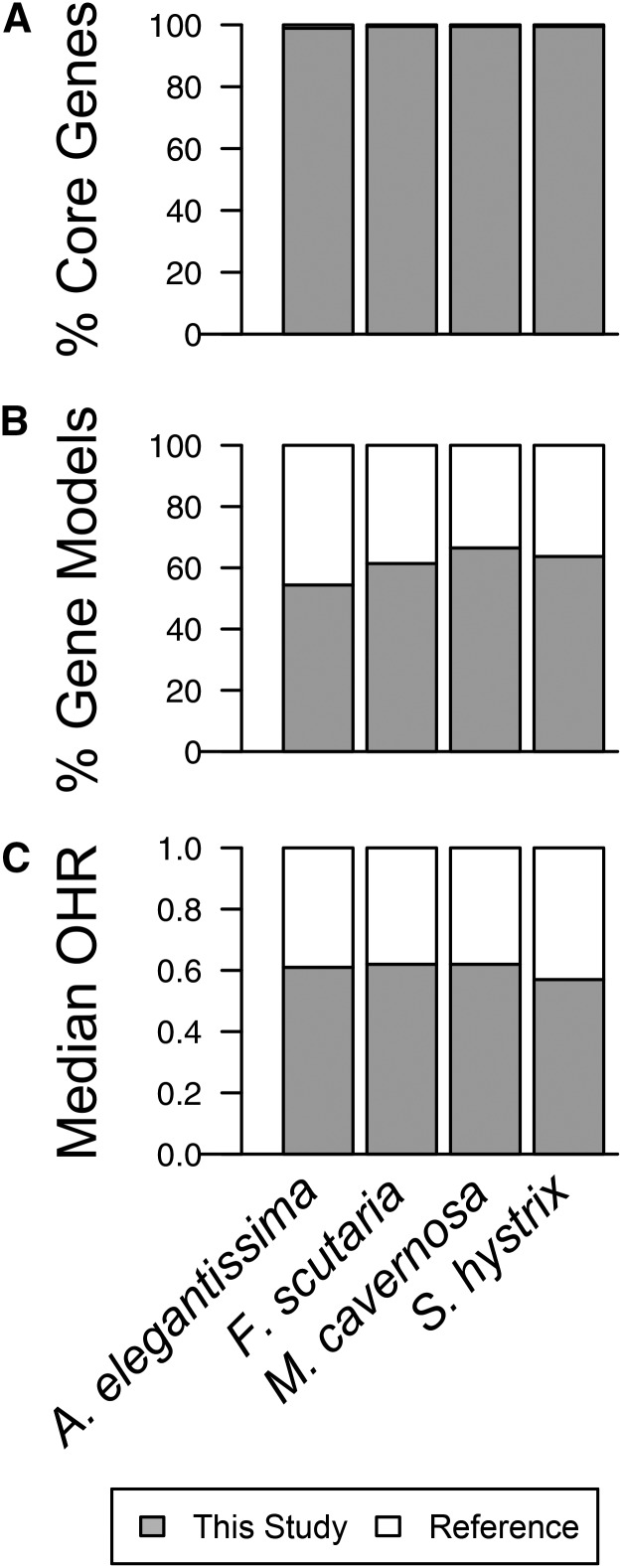
To evaluate the completeness of the transcriptome assemblies from the perspective of gene content, we conducted sequence comparisons with conserved eukaryotic genes and gene models from sequenced relatives. The core eukaryotic genes (CEGMA) (Parra et al. 2007) are expected to be expressed in most eukaryotes (Nakasugi et al. 2013; Sanders et al. 2014) and are widely used to estimate transcriptome completeness. Sequence comparisons revealed matches for 453 (98.9%) of these conserved genes in A. elegantissma and 456 (99.5%) in F. scutaria, M. cavernosa, and S. hystrix (Figure 2A). For a more comprehensive view of gene representation, the transcriptomes were compared with gene models from sequenced relatives (the coral A. digitifera and the anemone N. vectensis). This analysis identified matches for more than 14,000 gene models in each genome (BLASTx, bit-score ≥50): 54–67% of gene models in A. digitifera (Figure 2B) and 48–49% in N. vectensis. This is comparable to the level of sequence similarity observed among anthozoans with completed genomes. BLASTp comparisons of predicted proteins from the A. digitifera and N. vectensis genomes using the same thresholds recover 35% and 42% of genes in the other genome. This is substantially lower than the optimistic estimates of representation based on CEGMA, perhaps reflecting essential functions and constitutive expression of these highly conserved genes. Comparisons with gene models of closely related taxa appear to provide a more conservative estimate of gene representation in transcriptome assemblies.
Figure 2.
Three metrics used to evaluate gene representation and assembly of complete transcripts in de novo transcriptome assemblies. (A) Percent of core eukaryotic genes (CEGMA) identified in each assembly. (B) Percent of A. digitifera gene models with significant matches in each assembly. (C) Median proportion of each N. vectensis protein aligned with transcripts in each assembly (OHRHITS). Gray = our transcriptome assembly compared to the respective reference for each analysis.
To evaluate the effectiveness of our assemblies in reconstructing complete transcripts, we calculated ortholog hit ratios (OHR) for each final assembly. This method estimates the amount of a de novo transcript contained in the best ortholog from a reference genome (O’Neil et al. 2010), ranging from 1 (for complete transcripts) to 0 (for transcript fragments). We calculated OHR based on sequence comparisons with N. vectensis gene models using two approaches. First, a relatively stringent analysis based on the proportion of each N. vectensis gene included in regions of local similarity (OHRHITS) produced median OHR of 63.8, 64.7, 65.7, and 58.0% for A. elegantissma, F. scutaria, M. cavernosa, and S. hystrix, respectively (Figure 2C). A more inclusive analysis based on the longest ORF (in BLAST defined frame) produced similar estimates (median OHRORF: 67.4, 75.8, 77.2, and 60.3%, respectively). Each assembly included more than 5000 reasonably complete transcripts spanning at least 75% of the corresponding N. vectensis gene (range, 5262–6725). Overall, these comparisons with existing cnidarian sequence resources quantify the representation and completeness of our assemblies and provide a framework for comparison with other de novo assemblies. These estimates compare favorably with previous transcriptome completeness estimates for cnidarians (Sanders et al. 2014) and several invertebrates (O’Neil and Emrich 2013; Riesgo et al. 2012) using similar methods.
Annotation of transcriptomes
Transcripts were annotated using BLAST homology searches against the UniProt databases. Approximately one-third of all transcripts matched records in UniProt (range, 30–40%) (Table S5). The relatively low fraction of sequences annotated is attributable in part to sequence lengths: on average, 21% of transcripts <400 bp in length were annotated as compared with 42% of transcripts 400–1000 bp in length and 78% of transcripts >1000 bp. Even among the longest transcripts (>1 kb), a substantial number of sequences lacked annotated matches in UniProt (range, 6647–12,090 sequences per assembly). This highlights the well-known bias in taxonomic composition of existing databases and the value of ongoing gene sequencing in underrepresented metazoan taxa for public sequence databases.
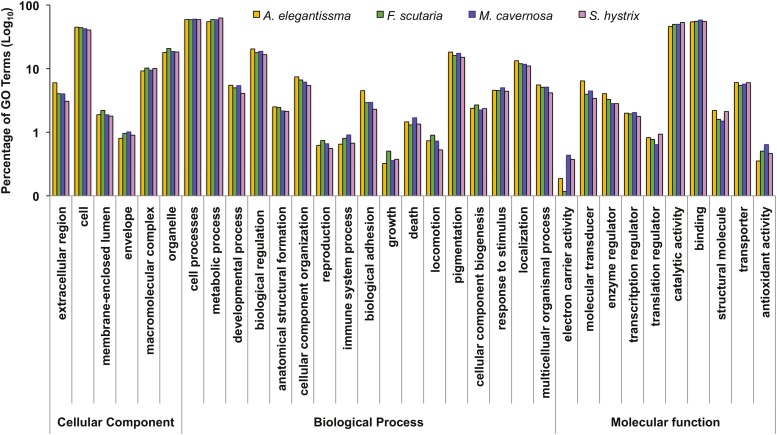
To categorize the biological functions inferred from sequence similarity, Gene Ontology (GO) terms were assigned to transcripts matching GO-annotated records in the UniProt database. This process identified functional annotation for 77% of transcripts with BLAST matches, providing tentative gene identities for a large number of sequences in each assembly (range, 32,299–47,547 transcripts; Table S5). Figure 3 shows the distribution of functional categories across the four transcriptomes, visualized using the Web Gene Ontology Annotation Plotting (WEGO) application. The GO terms were broadly distributed across the three domains and the percentages of sequences mapped to a given sub-ontology were highly similar for all species and comparable to other invertebrate transcriptomes (Riesgo et al. 2012; O’Neil et al. 2010; Lehnert et al. 2012; Moya et al. 2012; Polato et al. 2011; Shinzato et al. 2014; Stefanik et al. 2014; Traylor-Knowles et al. 2011). The similarities in functional distributions of assemblies prepared from diverse species, developmental stages, and symbiotic states highlight the constitutive expression of a broad set of genes in cnidarian transcriptomes. These core genes should facilitate comparative transcriptome studies by increasing the overlap among incomplete libraries.
Figure 3.
Distribution of functional categories (GO terms) in each transcriptome assembly. The percentage of transcripts with GO annotation for each category under the three main ontology domains was calculated for each assembly.
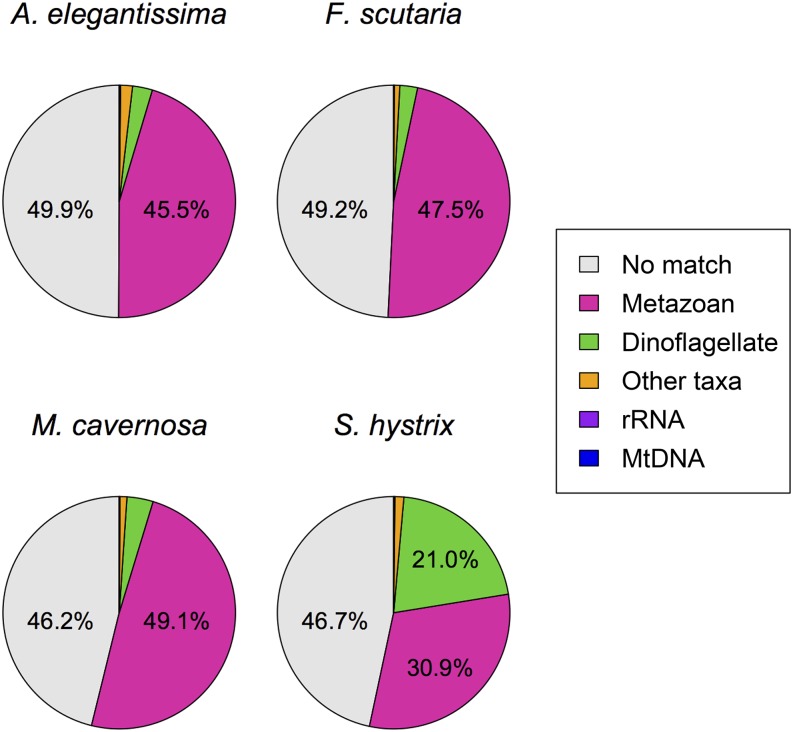
To determine taxonomic origin for each transcript, we conducted a series of BLAST searches and filtering steps outlined in Figure 1. Because our assemblies were produced from symbiotic and aposymbiotic specimens, the transcriptomes contain genes not only from anthozoans but also from their associated microbial community. To investigate the relative contributions of these sources we classified each transcript based on sequence similarity (Figure 1). These analyses confirmed that metazoan sequences comprised the majority of each library as expected. Fortunately, only a small fraction of transcripts were derived from organelles (mitochondria and ribosomes): on average, 212 transcripts (range, 127–284) in each assembly matched rRNA (N. vectensis) and 30 transcripts (range, 16–54) matched the mitochondrial genome (A. tenuis). Notably, almost half of the transcripts in each assembly (range, 46.2–49.9%) lacked matches to coral or Symbiodinium spp. genes, or NCBI’s nr database (Figure 4), a range that is consistent with results from other anthozoan transcriptomes (Sun et al. 2013; Karako-Lampert et al. 2014; Polato et al. 2011; Traylor-Knowles et al. 2011). These "unknown" transcripts may represent lineage-specific genes ("taxonomically restricted genes") that require further characterization. Comparison with NCBI’s nr database revealed that the majority of sequences with matches in one or more databases (59–95%) matched a metazoan sequence better than any other taxon, suggesting they originated from the animal host rather than from dinoflagellate or prokaryotic symbionts. A negligible fraction of transcripts in each assembly (0.8–1.7%) was assigned to the “Other taxa” category, most of which matched either coral or Symbiodinium genes but were classified as “unknown” because of conflicting results in the nr search (e.g., transcripts that matched Symbiodinium more closely than coral but whose best matches in nr were from metazoans).
Figure 4.
Predicted taxonomic origin of transcriptomes based on homology searches with BLAST. The percent of transcripts that were assigned to rRNA (purple), mtDNA (blue), dinoflagellate (green), metazoan (pink), other taxa (orange), and no match (gray) are shown.
The contribution of algal symbionts varied widely across samples. In nominally aposymbiotic samples of F. scutaria and A. elegantissma, 2.6% of transcripts on average were classified as dinoflagellate in origin (Figure 4), which may have resulted either from unexpected presence of symbionts at low abundance in these samples or from genes lacking orthologs in the A. digitifera reference. The symbiotic samples from S. hystrix, in contrast, showed a comparable abundance of transcripts classified as metazoan (61,369) and dinoflagellate in origin (41,724). Surprisingly, the M. cavernosa library that was similarly prepared from a symbiotic sample showed only 7278 transcripts from dinoflagellates (Figure 4). This striking contrast in Symbiodinium contributions from symbiotic specimens may have arisen from differing methods of RNA extraction. For S. hystrix, tissue was airbrushed off the coral skeleton directly into RNAlater Stabilization Solution (Qiagen, CA), followed by complete tissue homogenization. In contrast, the M. cavernosa fragment was simply vortexed to disrupt tissue, without physical homogenization. Our findings suggest that omitting physical homogenization during lysis can minimize symbiont contamination for studies aiming to focus on the cnidarian host, whereas studies investigating both components may benefit from thorough homogenization during extraction. The gene names, functional categories, and putative origin of each transcript are annotated in Table S6, Table S7, Table S8, and Table S9.
Gene searches of the database
The resulting annotations and sequences are available in a set of searchable databases hosted by Oregon State University (http://people.oregonstate.edu/∼meyere/index.html). To illustrate the utility of databases for cnidarian researchers targeting specific genes, we compared the effectiveness of simple text searches of the databases with reciprocal BLAST (RB) analysis, a more comprehensive approach that requires additional work by the end-user. Text searches targeting a few selected genes [cell adhesion molecule sym32, green fluorescent protein (GFP), and cystathionine β-synthase (Cbs)] produced comparable results as RB searches (Table S10). Text searches are obviously sensitive to query phrasing; the query “fluorescent” retrieves 51 putative GFP homologs, and functionally related synonyms (“GFP”, “chromoprotein”) retrieved an additional 10. Interestingly, the Cbs homologs identified in nominally aposymbiotic samples (A. elegantissima and F. scutaria) showed greater sequence similarity with Symbiodinium gene models than coral (A. digitifera) and were classified as dinoflagellate in our assignment procedure (Figure 1), whereas Cbs homologs in symbiotic samples (M. cavernosa and S. hystrix) included both metazoan and dinoflagellate transcripts. This unexpected observation of apparently dinoflagellate homologs of Cbs in nominally aposymbiotic samples is noteworthy because of their variable distribution among corals and possible roles in coral nutritional dependency on symbiosis (Shinzato et al. 2011). While this finding could be explained by undetected Symbiodinium harbored in these putatively aposymbiotic samples, the uncertainty introduced by these observations suggests that studies investigating the diversity of Cbs homologs across corals may require additional data (e.g., in situ hybridization) to confirm transcript origins. Overall, the close agreement between rigorous computational searches and simple text searches in these examples illustrates the utility of our searchable online databases for rapidly identifying genes of interest in reference transcriptome assemblies.
Novel SSR markers
Simple sequence repeats (SSRs or microsatellites) have been widely used to study genetic diversity, hybridization events, population structure, and connectivity in anthozoans (Concepcion et al. 2010; Fernandez-Silva et al. 2013; Selkoe and Toonen 2006; Ruiz-Ramos and Baums 2014), and can directly influence phenotypic traits by altering DNA replication, translation, and gene expression (Ruiz-Ramos and Baums 2014). SSR markers can be readily identified from de novo assemblies of NGS data and emerge as a side benefit in transcriptome assembly projects conducted for other purposes. We identified and developed primers for 52, 49, 73, and 75 candidate SSR markers in A. elegantissma, F. scutaria, M. cavernosa, and S. hystrix, respectively. Primer pairs for each species are listed in Table S11. For three of the species studied here, varying numbers of SSR markers are already available. Previous studies of S. hystrix have developed 10 SSR markers (Maier et al. 2001; Underwood et al. 2006) to study habitat partitioning within a single reef (Bongaerts et al. 2010), dispersal and recruitment patterns across multiple reefs (van Oppen et al. 2008; Kininmonth et al. 2010), and population changes associated with bleaching events (Underwood et al. 2007). Candidate SSR markers have been identified in F. scutaria (n = 118) from the coral host and dinoflagellate symbionts (Concepcion et al. 2010). SSR markers previously developed in M. cavernosa (Shearer et al. 2005; Serrano et al. 2014) have been used to investigate the population connectivity across depth and geographic distance (Serrano et al. 2014). The candidate SSR markers identified in this study provide additional markers for future studies along similar lines. To our knowledge, SSR markers have not been previously developed in A. elegantissima. Although the population structure of the host has not been described, analysis of their dinoflagellate symbionts revealed highly structured populations across their geographic range (Sanders and Palumbi 2011). The markers developed in this study for A. elegantissima provide tools to investigate population structure of the host across a similar range.
Orthologous groups and phylogenomic reconstructions
With the increasing availability of transcriptomes and genomes, these datasets can now be mined to discover novel phylogenetic markers within Anthozoa and across the Cnidaria to resolve taxonomic uncertainties. Phylogenetic reconstruction of anthozoans has presented challenges because analyses based on morphology, life history, and molecular sequences have failed to adequately delineate taxonomic boundaries or evolutionary relationships (Daly et al. 2003). To date, molecular phylogenies for anthozoans have been based on one or a small number of markers including nuclear ribosomal 28S and 18S genes (Daly et al. 2003; Berntson et al. 1999), β-tubulin (Fukami et al. 2008), mitochondrial 16S (Daly et al. 2003), cytochrome b (Fukami et al. 2008), and COI (Kitahara et al. 2010; Fukami et al. 2008). Interestingly, mitochondrial sequences in anthozoans have extremely low mutation rates compared to the bilaterians and are therefore highly conserved, allowing for robust comparisons across distantly related taxa (van Oppen et al. 2002; Galtier et al. 2009). Therefore, the mitochondrial gene COI has been used recently to define evolutionary relationships among scleractinian corals (Kitahara et al. 2010; Fukami et al. 2008; Budd and Stolarski 2011) and to support the distinction of robust corals from the complex corals (Romano and Palumbi 1996).
One disadvantage to single gene phylogenetic inferences is that they suffer from weak phylogenetic signals, sensitivity to hidden paralogy, and spurious tree artifacts (Philippe et al. 2004). Despite these potential limitations, single gene trees have advanced the field of cnidarian systematics. However, polyphyly remains a problem among several anthozoan families when using both maximum likelihood and Bayesian analyses (Fukami et al. 2008; Budd and Stolarski 2011), which has led to recent shifts in taxonomic classification (Budd and Stolarski 2011). To expand beyond previous single-gene approaches, we performed phylogenomic analyses incorporating the four new transcriptomes and other available "omic" resources. By simultaneously increasing taxon and gene sampling, phylogenetic inference is expected to improve (Philippe et al. 2004) and may help resolve some of the challenges in reconstructing the evolutionary relationships of the Anthozoa and, more broadly, phylum Cnidaria.
For phylogenomic analysis, transcripts larger than 400 bp were converted to protein with TransDecoder and clustered into orthologous groups using FastOrtho. The number of assigned orthologous groups ranged from 14,144 to 21,147 for the four transcriptomes (Figure S1). Comparison of all four resulted in 6560 shared orthologs (Figure S1). The three coral species shared 2045 orthologs not found in anemones and the two most closely related corals (M. cavernosa and F. scutaria) shared 1682 orthologs absent from the other assemblies. By incorporating 11 additional taxa for phylogenomic analysis (Table S2), 443 orthologs were identified between all taxa. After setting a minimum protein length (100 amino acids), these orthologs were refined using the PhyloTreePruner analysis pipeline (Kocot et al. 2013). Filtering resulted in the identification of 397 orthologs for ≥14 taxa. These were used to construct a phylogenetic tree we termed “conservative” because loci with any missing data were excluded (Table S12).
Missing data are a commonly encountered problem in phylogenomic analyses, from either reduced transcript length or gene absence from a transcriptome (Philippe et al. 2004; Kocot et al. 2013; Roure et al. 2013). However, the sensitivity of phylogenetic inference to incomplete datasets is still under investigation, with mixed results from phylogenomic analyses on large but patchy supermatrices (Roure et al. 2013; Philippe et al. 2004). Because the resources in this study used for ortholog identification differed in completeness, ranging from EST libraries to complete genomes (Table S2), we tested the influence of missing data on our phylogenetic reconstruction. To investigate this, we lowered the required number of taxa per orthologous group to ≥10, which identified 2897 orthologs (Table S12). This second set was used to create the “relaxed” phylogeny, so called because loci with some missing data were included.
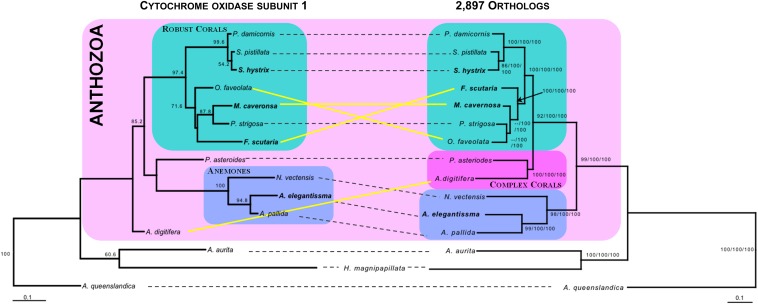
Both maximum likelihood phylogenomic analyses reconstructed identical and strongly supported topologies (bootstrap = 100; Figure S2), demonstrating that our phylogenetic inference was insensitive to missing data (Figure 5). However, the relationship of the corals in the family Faviidae, containing M. cavernosa, P. strigosa, and O. faveolata, varied among the COI, ND supergene, and phylogenomic analyses. The mitochondrial ND supergene identified by Havird and Santos (2014) produced a phylogenetic tree nearly synonymous with the accepted cnidarian taxonomic relationships and phylogenomic analyses from this study (Kitahara et al. 2010), except for the placement of the M. cavernosa as sister taxon to O. faveolata and P. strigosa. The analysis of single gene COI resulted in a discordant phylogenetic topology (Figure 5), failing to reconstruct the complex coral clade (P. astreoides and A. digitifera), which was recovered by ND supergene, relaxed and conserved trees (Figure 5, Figure S2). In the COI tree, the placement of the F. scutaria, from the family Fungiidae, as sister taxon to P. strigosa and M. cavernosa from the family Faviidae, instead of O. faveolata is incongruent with current taxonomic placement (Figure 5) (Kitahara et al. 2010; Budd and Stolarski 2011). Furthermore, while the phylogenomic analyses placed O. faveolata as sister to P. strigosa with strong support (bootstrap = 100), this relationship was not recovered in either mitochondrial phylogeny (Figure S2). Overall, the tree topology from the phylogenomic analyses is consistent with accepted evolutionary relationships within Anthozoa (Budd and Stolarski 2011; Fukami et al. 2008; Kitahara et al. 2010).
Figure 5.
Discordance in maximum likelihood phylogenetic reconstruction of COI compared to a combined phylogeny of concatenated ND (2, 4, and 5) genes and two phylogenomic trees. The COI phylogeny is presented on the left and the combined phylogeny is presented on the right. Topology for the ND mitochondrial set, relaxed and conservative phylogenomic trees were nearly identical. Therefore, nodal support is summarized on the relaxed tree (right). Bootstrap support at the nodes from left to right represents ND gene set/relaxed/conservative. If topologies differed in the summary tree, then the nodal support is presented - - as next to the node. Yellow solid lines connect taxon with different positions and/or relationships between the two trees, whereas black dashed lines connect those with the same position and/or relationship. Reconstructions of groups in the class Anthozoa based on Kitahara et al. (2010) are highlighted in boxes: teal= robust corals; dark pink = complex corals; and light blue = anemones. The names of species used in this study are emphasized by bold font. Scale bars indicate the amino acid replacements per site.
Conclusion
The annotated transcriptome assemblies developed in this study provide useful resources for genomic research in anthozoan species for which sequences resources were previously lacking. The searchable databases developed from these assemblies make it possible to rapidly identify genes of interest from each species. Our ortholog analysis demonstrates the feasibility of phylogenetic inference in corals using transcriptome assemblies from diverse stages and symbiotic states, highlighting a promising path toward resolving major uncertainties in the existing phylogeny of scleractinians. Future studies will benefit from the growing body of anthozoan sequence resources, including the four assemblies contributed in this study.
Supplementary Material
Acknowledgments
We acknowledge Dr. Christine Schnitzler and the labs of Dr. Tung-Yung Fan and Dr. Andrew Baker for assistance with sample collection. In addition, we thank Sarah Guermond and Emily Weiss for assistance in sample preparation and analysis. Research funding was provided by Oregon State University, Department of Integrative Biology. Publication of this article in an open access journal was funded by the Oregon State University Libraries & Press Open Access Fund. E.M., S.K., and A.P. conceived the investigation. S.K., C.C., and A.P. performed library preparation and sequencing. C.C., A.P., and E.M. assembled and annotated the transcriptomes. S.K. performed computational analyses related to transcriptome completeness, GO annotation, and phylogenetics. C.C. complied transcriptome statistics. C.C. and S.K. performed targeted gene searches. E.M. performed cross-contamination screens, identified SSR markers, and provided bioinformatic expertise. S.K., C.C., and E.M. made significant contributions to the preparation of the manuscript. All authors revised and approved the final manuscript. The authors declare no competing interests.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.020164/-/DC1
Accession Numbers: Raw data: NCBI’s SRA, accession SRP063463. Assemblies: DRYAD digital repository, doi: 10.5061/dryad.3f08f
Communicating editor: A. Gasch
Literature Cited
- Abascal F., Zardoya R., Posada D., 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene ontology: tool for the unification of biology. Nature Genetics 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Barshis D. J., Ladner J. T., Oliver T. A., Seneca F. O., Traylor-Knowles N., et al. , 2013. Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. USA 110: 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S., Simakov O., Esherick L. Y., Liew Y. J., Lehnert E. M., et al. , 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. USA 112: 11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berntson E. A., France S. C., Mullineaux L. S., 1999. Phylogenetic relationships within the class Anthozoa (phylum Cnidaria) based on nuclear 18S rDNA sequences. Mol. Phylogenet. Evol. 13: 417–433. [DOI] [PubMed] [Google Scholar]
- Bongaerts P., Riginos C., Ridgway T., Sampayo E. M., van Oppen M. J. H., et al. , 2010. Genetic divergence across habitats in the widespread coral Seriatopora hystrix and its associated Symbiodinium. PLoS One 5: e10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge D., Cunningham C. W., Schierwater B., DeSalle R., Buss L. W., 1992. Class-level relationships in the phylum Cnidaria: evidence from mitochondrial genome structure. Proc. Natl. Acad. Sci. USA 89: 8750–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd A. F., Stolarski J., 2011. Corallite wall and septal microstructure in scleractinian reef corals: comparison of molecular clades within the family Faviidae. J. Morphol. 272: 66–88. [DOI] [PubMed] [Google Scholar]
- Castresana J., 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chapman J. A., Kirkness E. F., Simakov O., Hampson S. E., Mitros T., et al. , 2010. The dynamic genome of Hydra. Nature 464: 592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Closek C. J., Sunagawa S., DeSalvo M. K., Piceno Y. M., DeSantis T. Z., et al. , 2014. Coral transcriptome and bacterial community profiles reveal distinct Yellow Band Disease states in Orbicella faveolata. ISME J. 8: 2411–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C., Neveu P., Zhou H., Arcila M. L., Degnan S. M., et al. , 2012. Transcriptome profiling of the demosponge Amphimedon queenslandica reveals genome-wide events that accompany major life cycle transitions. BMC Genomics 13: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion G., Polato N., Baums I., Toonen R., 2010. Development of microsatellite markers from four Hawaiian corals: Acropora cytherea, Fungia scutaria, Montipora capitata and Porites lobata. Conserv. Genet. Resour. 2: 11–15. [Google Scholar]
- Daly M., Fautin D. G., Cappola V. A., 2003. Systematics of the Hexacorallia (Cnidaria: Anthozoa). Zool. J. Linn. Soc. 139: 419–437. [Google Scholar]
- Davies S. W., Rahman M., Meyer E., Green E. A., Buschiazzo E., et al. , 2013. Novel polymorphic microsatellite markers for population genetics of the endangered Caribbean star coral, Montastraea faveolata. Mar. Biodivers. 43: 167–172. [Google Scholar]
- Davy S. K., Allemand D., Weis V. M., 2012. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76: 229–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSalvo M., Voolstra C., Sunagawa S., Schwarz J., Stillman J., et al. , 2008. Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Mol. Ecol. 17: 3952–3971. [DOI] [PubMed] [Google Scholar]
- Douglas A. E., 2003. Coral bleaching—how and why? Mar. Pollut. Bull. 46: 385–392. [DOI] [PubMed] [Google Scholar]
- Dunn C. W., Hejnol A., Matus D. Q., Pang K., Browne W. E., et al. , 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: 745–749. [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva I., Whitney J., Wainwright B., Andrews K. R., Ylitalo-Ward H., et al. , 2013. Microsatellites for next-generation ecologists: a post-sequencing bioinformatics pipeline. PLoS One 8: e55990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs B., Wang W., Graspeuntner S., Li Y., Insua S., et al. , 2014. Regulation of polyp-to-jellyfish transition in Aurelia aurita. Curr. Biol. 24: 263–273. [DOI] [PubMed] [Google Scholar]
- Fukami H., Chen C. A., Budd A. F., Collins A., Wallace C., et al. , 2008. Mitochondrial and nuclear genes suggest that stony corals are monophyletic but most families of stony corals are not (Order Scleractinia, Class Anthozoa, Phylum Cnidaria). PLoS One 3: e3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., Jobson R. W., Nabholz B., Glémin S., Blier P. U., 2009. Mitochondrial whims: metabolic rate, longevity and the rate of molecular evolution. Biol. Lett. 5: 413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., et al. , 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., et al. , 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M., Shoguchi E., Shinzato C., Kawashima T., Miller D. J., et al. , 2013. The complex NOD-like receptor repertoire of the coral Acropora digitifera includes novel domain combinations. Mol. Biol. Evol. 30: 167–176. [DOI] [PubMed] [Google Scholar]
- Havird J. C., Santos S. R., 2014. Performance of single and concatenated sets of mitochondrial genes at inferring metazoan relationships relative to full mitogenome data. PLoS One 9: e84080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm R. R., Siebert S., Tulin S., Smith J., Dunn C. W., 2013. Characterization of differential transcript abundance through time during Nematostella vectensis development. BMC Genomics 14: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Madan A., 1999. CAP3: a DNA sequence assembly program. Genome Res. 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina, 2014 Illumina customer sequence letter. Illumina, Inc., San Diego. [Google Scholar]
- Karako-Lampert S., Zoccola D., Salmon-Divon M., Katzenellenbogen M., Tambutté S., et al. , 2014. Transcriptome analysis of the scleractinian coral Stylophora pistillata. PLoS One 9: e88615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K. I., Miyata T., 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E., Roure B., Philippe H., Collins A. G., Lavrov D. V., 2013. Cnidarian phylogenetic relationships as revealed by mitogenomics. BMC Evol. Biol. 13: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenkel C., Meyer E., Matz M., 2013. Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Mol. Ecol. 22: 4322–4334. [DOI] [PubMed] [Google Scholar]
- Kininmonth S., van Oppen M. J. H., Possingham H. P., 2010. Determining the community structure of the coral Seriatopora hystrix from hydrodynamic and genetic networks. Ecol. Modell. 221: 2870–2880. [Google Scholar]
- Kitahara M. V., Cairns S. D., Stolarski J., Blair D., Miller D. J., 2010. A comprehensive phylogenetic analysis of the Scleractinia (Cnidaria, Anthozoa) based on mitochondrial CO1 sequence data. PLoS One 5: e11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocot K. M., Cannon J. T., Todt C., Citarella M. R., Kohn A. B., et al. , 2011. Phylogenomics reveals deep molluscan relationships. Nature 477: 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocot K. M., Citarella M. R., Moroz L. L., Halanych K. M., 2013. PhyloTreePruner: a phylogenetic tree-based approach for selection of orthologous sequences for phylogenomics. Evol. Bioinform. Online 9: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kück P., Meusemann K., 2010. FASconCAT: Convenient handling of data matrices. Mol. Phylogenet. Evol. 56: 1115–1118. [DOI] [PubMed] [Google Scholar]
- Lehnert E. M., Burriesci M. S., Pringle J. R., 2012. Developing the anemone Aiptasia as a tractable model for cnidarian-dinoflagellate symbiosis: the transcriptome of aposymbiotic A. pallida. BMC Genomics 13: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert, E. M., M. E. Mouchka, M. S. Burriesci, N. D. Gallo, J. A. Schwarz et al., 2014 Extensive differences in gene expression between symbiotic and aposymbiotic Cnidarians. G3 (Bethesda) 4: 277–295. [DOI] [PMC free article] [PubMed]
- Li L., Stoeckert C. J., Roos D. S., 2003. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W. P., Maddison D. R., 2015. Mesquite: a modular system for evolutionary analysis. v3.02. http://mesquiteproject.org [Google Scholar]
- Maier E., Tollrian R., Nürnberger B., 2001. Development of species-specific markers in an organism with endosymbionts: microsatellites in the scleractinian coral Seriatopora hystrix. Mol. Ecol. Notes 1: 157–159. [Google Scholar]
- Marlow H. Q., Srivastava M., Matus D. Q., Rokhsar D., Martindale M. Q., 2009. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev. Neurobiol. 69: 235–254. [DOI] [PubMed] [Google Scholar]
- Mazel C. H., Lesser M. P., Gorbunov M. Y., Barry T. M., Farrell J. H., et al. , 2003. Green-fluorescent proteins in Caribbean corals. Limnol. Oceanogr. 48: 402–411. [Google Scholar]
- Meyer E., Weis V. M., 2012. Study of cnidarian-algal symbiosis in the “omics” age. Biol. Bull. 223: 44–65. [DOI] [PubMed] [Google Scholar]
- Meyer E., Aglyamova G. V., Wang S., Buchanan-Carter J., Abrego D., et al. , 2009. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics 10: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E., Aglyamova G. V., Matz M. V., 2011. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol. Ecol. 20: 3599–3616. [DOI] [PubMed] [Google Scholar]
- Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M., 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35(Suppl 2): W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A., Huisman L., Ball E., Hayward D., Grasso L., et al. , 2012. Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2‐driven acidification during the initiation of calcification. Mol. Ecol. 21: 2440–2454. [DOI] [PubMed] [Google Scholar]
- Nakasugi K., Crowhurst R. N., Bally J., Wood C. C., Hellens R. P., et al. , 2013. De Novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One 8: e59534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil S., Emrich S., 2013. Assessing De Novo transcriptome assembly metrics for consistency and utility. BMC Genomics 14: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil S., Dzurisin J., Carmichael R., Lobo N., Emrich S., et al. , 2010. Population-level transcriptome sequencing of nonmodel organisms Erynnis propertius and Papilio zelicaon. BMC Genomics 11: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Korf I., 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Philippe H., Snell E. A., Bapteste E., Lopez P., Holland P. W., et al. , 2004. Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol. Biol. Evol. 21: 1740–1752. [DOI] [PubMed] [Google Scholar]
- Polato N. R., Vera J. C., Baums I. B., 2011. Gene discovery in the threatened elkhorn coral: 454 sequencing of the Acropora palmata transcriptome. PLoS One 6: e28634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole A. Z., Weis V. M., 2014. TIR-domain-containing protein repertoire of nine anthozoan species reveals coral–specific expansions and uncharacterized proteins. Dev. Comp. Immunol. 46: 480–488. [DOI] [PubMed] [Google Scholar]
- Pratlong M., Haguenauer A., Chabrol O., Klopp C., Pontarotti P., et al. , 2015. The red coral (Corallium rubrum) transcriptome: a new resource for population genetics and local adaptation studies. Mol. Ecol. Resour. 15: 1205–1215. [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P., 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N. H., Srivastava M., Hellsten U., Dirks B., Chapman J., et al. , 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317: 86–94. [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., et al. , 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds W. S., Schwarz J. A., Weis V. M., 2000. Symbiosis-enhanced gene expression in cnidarian-algal associations: cloning and characterization of a cDNA, sym32, encoding a possible cell adhesion protein. Comp. Biochem. 126: 33–44. [DOI] [PubMed] [Google Scholar]
- Riesgo A., Andrade S. C., Sharma P., Novo M., Perez-Porro A., et al. , 2012. Comparative description of ten transcriptomes of newly sequenced invertebrates and efficiency estimation of genomic sampling in non-model taxa. Front. Zool. 9: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesgo A., Farrar N., Windsor P. J., Giribet G., Leys S. P., 2014. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol. Biol. Evol. 31: 1102–1120. [DOI] [PubMed] [Google Scholar]
- Romano S. L., Palumbi S. R., 1996. Evolution of scleractinian corals inferred from molecular systematics. Science 271: 640–642. [Google Scholar]
- Rota-Stabelli O., Yang Z., Telford M. J., 2009. MtZoa: A general mitochondrial amino acid substitutions model for animal evolutionary studies. Mol. Phylogenet. Evol. 52: 268–272. [DOI] [PubMed] [Google Scholar]
- Roure B., Baurain D., Philippe H., 2013. Impact of missing data on phylogenies inferred from empirical phylogenomic data sets. Mol. Biol. Evol. 30: 197–214. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 1999 Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ramos D., Baums I., 2014. Microsatellite abundance across the Anthozoa and Hydrozoa in the phylum Cnidaria. BMC Genomics 15: 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J. F., Burton P. M., Mazza M. E., Kwong G. K., Mullikin J. C., et al. , 2006. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 7: R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J. F., Pang K., Schnitzler C. E., Nguyen A.-D., Moreland R. T., et al. , 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342: 1232592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J. G., Palumbi S. R., 2011. Populations of Symbiodinium muscatinei show strong biogeographic structuring in the intertidal anemone Anthopleura elegantissima. Biol. Bull. 220: 199–208. [DOI] [PubMed] [Google Scholar]
- Sanders S., Shcheglovitova M., Cartwright P., 2014. Differential gene expression between functionally specialized polyps of the colonial hydrozoan Hydractinia symbiolongicarpus (Phylum Cnidaria). BMC Genomics 15: 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzler C. E., Weis V. M., 2010. Coral larvae exhibit few measurable transcriptional changes during the onset of coral-dinoflagellate endosymbiosis. Mar. Genomics 3: 107–116. [DOI] [PubMed] [Google Scholar]
- Selkoe K. A., Toonen R. J., 2006. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol. Lett. 9: 615–629. [DOI] [PubMed] [Google Scholar]
- Serrano X., Baums I. B., O’Reilly K., Smith T. B., Jones R. J., et al. , 2014. Geographic differences in vertical connectivity in the Caribbean coral Montastraea cavernosa despite high levels of horizontal connectivity at shallow depths. Mol. Ecol. 23: 4226–4240. [DOI] [PubMed] [Google Scholar]
- Shearer T. L., Gutiérrez-Rodríguez C., Coffroth M. A., 2005. Generating molecular markers from zooxanthellate cnidarians. Coral Reefs 24: 57–66. [Google Scholar]
- Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., et al. , 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476: 320–323. [DOI] [PubMed] [Google Scholar]
- Shinzato C., Hamada M., Shoguchi E., Kawashima T., Satoh N., 2012a The repertoire of chemical defense genes in the coral Acropora digitifera genome. Zoolog. Sci. 29: 510–517. [DOI] [PubMed] [Google Scholar]
- Shinzato C., Shoguchi E., Tanaka M., Satoh N., 2012b Fluorescent protein candidate genes in the coral Acropora digitifera genome. Zoolog. Sci. 29: 260–264. [DOI] [PubMed] [Google Scholar]
- Shinzato C., Inoue M., Kusakabe M., 2014. A snapshot of a coral “holobiont”: a transcriptome assembly of the scleractinian coral, Porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS One 9: e85182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert S., Robinson M. D., Tintori S. C., Goetz F., Helm R. R., et al. , 2011. Differential gene expression in the siphonophore Nanomia bijuga (Cnidaria) assessed with multiple next-generation sequencing workflows. PLoS One 6: e22953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A., R. Hubley, and P. Green, RepeatMasker Open-3.0. http://www.repeatmasker.org
- Smith-Keune C., Dove S., 2008. Gene expression of a green fluorescent protein homolog as a host-specific biomarker of heat stress within a reef-building coral. Mar. Biotechnol. (NY) 10: 166–180. [DOI] [PubMed] [Google Scholar]
- Soza‐Ried J., Hotz‐Wagenblatt A., Glatting K. H., del Val C., Fellenberg K., et al. , 2010. The transcriptome of the colonial marine hydroid Hydractinia echinata. FEBS J. 277: 197–209. [DOI] [PubMed] [Google Scholar]
- Srivastava M., Simakov O., Chapman J., Fahey B., Gauthier M. E. A., et al. , 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466: 720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanik D. J., Lubinski T. J., Granger B. R., Byrd A. L., Reitzel A. M., et al. , 2014. Production of a reference transcriptome and transcriptomic database (EdwardsiellaBase) for the lined sea anemone, Edwardsiella lineata, a parasitic cnidarian. BMC Genomics 15: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Chen Q., Lun J. C., Xu J., Qiu J.-W., 2013. PcarnBase: Development of a transcriptomic database for the brain coral Platygyra carnosus. Mar. Biotechnol. (NY) 15: 244–251. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein et al, 2000. Gene Ontology: tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traylor-Knowles N., Granger B. R., Lubinski T. J., Parikh J. R., Garamszegi S., et al. , 2011. Production of a reference transcriptome and transcriptomic database (PocilloporaBase) for the cauliflower coral, Pocillopora damicornis. BMC Genomics 12: 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood J. N., Souter P. B., Ballment E. R., Lutz A. H., van Oppen M. J. H., 2006. Development of 10 polymorphic microsatellite markers from herbicide-bleached tissues of the brooding pocilloporid coral Seriatopora hystrix. Mol. Ecol. Notes 6: 176–178. [Google Scholar]
- Underwood J. N., Smith L. D., van Oppen M. J. H., Gilmour J. P., 2007. Multiple scales of genetic connectivity in a brooding coral on isolated reefs following catastrophic bleaching. Mol. Ecol. 16: 771–784. [DOI] [PubMed] [Google Scholar]
- Van Dongen S., 2000. Graph clustering by flow simulation, University of Utrecht, The Netherlands. [Google Scholar]
- van Oppen M. J. H., Catmull J., McDonald B. J., Hislop N. R., Hagerman P. J., et al. , 2002. The mitochondrial genome of Acropora tenuis (Cnidaria; Scleractinia) contains a large group I intron and a candidate control region. J. Mol. Evol. 55: 1–13. [DOI] [PubMed] [Google Scholar]
- van Oppen M. J. H., Lutz A., De’ath G., Peplow L., Kininmonth S., 2008. Genetic traces of recent long-distance dispersal in a predominantly self-recruiting coral. PLoS One 3: e3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Dupiol J., Zoccola D., Tambutté E., Grunau C., Cosseau C., et al. , 2013. Genes related to ion-transport and energy production are upregulated in response to CO2-driven pH decrease in corals: New insights from transcriptome analysis. PLoS One 8: e58652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis V. M., 2008. Cellular mechanisms of Cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 211: 3059–3066. [DOI] [PubMed] [Google Scholar]
- Weis V. M., Allemand D., 2009. What determines coral health? Science 324: 1153–1155. [DOI] [PubMed] [Google Scholar]
- Wenger Y., Galliot B., 2013. RNAseq vs. genome-predicted transcriptomes: a large population of novel transcripts identified in an Illumina-454 Hydra transcriptome. BMC Genomics 14: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S., Goldman N., 2001. A general empirical model of protein evolution derived from multiple protein families using a Maximum-Likelihood approach. Mol. Biol. Evol. 18: 691–699. [DOI] [PubMed] [Google Scholar]
- Wood-Charlson E. M., Weis V. M., 2009. The diversity of C-type lectins in the genome of a basal metazoan, Nematostella vectensis. Dev. Comp. Immunol. 33: 881–889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the results of this article are available from the Sequence Read Archive at NCBI (Accession number: SRP063463), the Dryad Digital Repository (doi: 10.5061/dryad.3f08f), and the author’s website (http://people.oregonstate.edu/∼meyere/index.html).