Abstract
Some studies have demonstrated that familial non-medullary thyroid cancer (FNMTC) has a more aggressive clinical behavior compared to sporadic NMTC (SNMTC). However, FNMTC is difficult to differentiate from SNMTC by the morphology and immunohistochemistry. Although genes responsible for FNMTC were unclear, screening for rare germline mutations on known important tumor suppressor genes might offer more insights on predicting susceptibility to FNMTC. Here, a customized panel was designed to capture all exons of 31 cancer susceptive genes possibly related to FNMTC. Using next-generation sequencing we performed deep sequencing to achieve 500× coverage of the targeted regions. At the end 45 variants were identified in 29 of 47 familial patients and 6 of 16 sporadic patients. Notably, several germline mutations were found matching between paired FNMTC patients from the same family, including APC L292F and A2778S, BRAF D22N, MSH6 G355S and A36V, MSH2 L719F, MEN1 G508D, BRCA1 SS955S, BRCA2 G2508S, and a GNAS inframe insertion. We demonstrated a novel approach to help diagnose and elucidate the genetic cause of the FNMTC patients, and assess whether their family members are exposed to a higher genetic risk. The findings would also provide insights on monitoring the potential second cancers for thyroid cancer patients.
Thyroid cancer is the most common malignancy of endocrine organs, and its prevalence is increasing rapidly. Non-medullary thyroid cancer (NMTC) arising from the follicular thyroid epithelium cells accounts for more than 95% of all thyroid cancers1. About 5%–10% of NMTC patients have reported familial history, therefore named familial NMTC (FNMTC)2,3,4.
FNMTC is defined by the diagnosis of two or more first degree relatives affected by differentiated thyroid cancer of follicular cell origin. FNMTC can be divided into two groups based on clinical characteristics. The first includes familial tumor syndromes characterized by a preponderance of non-thyroidal tumors, for example, familial adenomatous polyposis (FAP), Gardner’s syndrome, Cowden’s disease and so on. The second group includes familial syndromes characterized by a preponderance of NMTC. Histologically, FNMTC includes papillary thyroid carcinoma (PTC), follicular thyroid carcinoma (FTC), and anaplastic thyroid carcinoma. However, FNMTC is difficult to differentiate from sporadic non-medullary thyroid cancer (SNMTC) because FNMTC is usually indistinguishable from sporadic follicular cell cancers either by morphological examination or immunohistochemical assay, which means that FNMTC cannot be diagnosed until at least one of the patient’s first degree relatives is affected by NMTC.
Some studies have demonstrated that compared to SNMTCs, FNMTCs have a more aggressive clinical behavior, as well as a worse prognosis5,6,7,8,9. Patients with FNMTC have an increased risk of multifocal disease, local invasion, increased local or regional recurrence, lymph node metastases, and intraglandular dissemination. Comparing the first and the second generation of FNMTC patients, the off-springs usually show an earlier age at disease onset and have more aggressive disease when compared with their parents10. Although the prognosis of FNMTC is poor compared to SNMTC, standardized treatment performed at early stage of disease is considerably effective to reduce the relapse and metastasis after surgery. However, since there has not been a sensitive and specific experimental approach available in clinic to screen for high genetic risks related to FNMTC, it’s difficult to identify susceptible FNMTC patients at an early stage. This could lead to insufficient treatment in clinic and higher risk of post-operation relapse for those FNMTC patients.
Reviews of various genetic studies suggest that inheritance of FNMTC is autosomal dominant and that the penetrance is incomplete9,11,12,13,14,15,16. In 1997, Bignell16 evaluated the contribution of MNG1 to FNMTC, studied 37 NMTC families and found that a small group of patients could be attributed to MNG1 (14q32). Canzian15 investigated a French pedigree with multiple cases of multinodular goiter and NMTC and reported the mapping of the responsible gene, named “TCO”. TCO was mapped to chromosome 19p13.2 by linkage analysis with a whole genome panel of microsatellite markers. McKay’s study11 found the existence of a susceptibility locus for FNMTC on chromosome 2q21 (NMTC1). Malchoff13 demonstrated that the incidence of an America family (five PTC and two papillary renal neoplasia, PRN) was linked to 1q21 (FPTC/PRN). In one study, the authors evaluated the roles of MNG1, TCO1, FPTC/PTEN, TSHR, and TRKA in FNMTC, and carried out a comprehensive mutation and linkage analysis of these genes in 22 families12. One family was linked to chromosome 19q13.2, confirming that TCO1 underlies a subset of familial non-medullary thyroid cancer. However, none of the LOD scores in this study actually achieved statistical significance, including for TCO1. Lack of successful identification of significant loci from these studies indicates that the genetic causality model for FNMTC might be complex. On the other hand, there are tumor suppressor genes whose germline mutations are known to correlate with some NMTC-related familial tumor syndromes, e.g. PTEN with Cowden Syndrome and WRN with Werner Syndrome17,18. Therefore, instead of the classical linkage analysis approach, screening for rare germline mutations on known important tumor suppressor genes might offer more insights on predicting susceptibility to FNMTC.
The key challenge on tumor suppressor genes mutation profiling is that the whole exons have to be sequenced, since most loss-of-function mutations do not demonstrate any “hot spot” patterns. Next-generation sequencing (NGS) offers simultaneous sequencing of thousands to millions of short nucleic acid sequences and may offer a cost-effective approach for parallel detection of multiple genetic alterations. It provides a clear advantage over the conventional sequencing technique, such as Sanger sequencing, by allowing simultaneous analysis of large regions of the genome and offering high sensitivity of mutation detection and quantitative assessment of mutant alleles. In this study, we evaluated capture-based targeted DNA sequencing as a new approach for testing a broad spectrum of point mutations (SNVs) and short insertion-deletions (Indels) possibly related to FNMTC. Targeted sequencing enables deep coverage of the interested genomic regions only with affordable cost per patient. Here, we assess the feasibility of such an approach in a NMTC-patient cohort and found a few mutational loci in FNMTC patients that haven’t been reported before.
Results
Quality assessment of the targeted sequencing data
We performed deep sequencing with the FNMTC susceptibility panel on two batches of samples to achieve average 500× coverage of the targeted regions. Among the 38 samples in the first batch we profiled, we obtained an average of 2.46 million reads for each sample (Fig. 1a). On average 98.9% of all reads can be mapped back to the genome and 54.3% of all reads are mapped to our designed target regions (Fig. 1b). This indicates a high capture efficiency of the probes. We then assessed the distribution of the coverage within the target regions. Figure 1c plots the histogram of the average coverage depth among the 38 samples across all target regions. The average coverage depth appears to be normally distributed, with a minimum of 157× and a maximum of 1505×. Figure 1d shows the coverage depth in each sample, where all samples have greater than 96% of all regions with coverage depth greater than 200×.
Figure 1. Quality assessment of the targeted sequencing data among 38 samples in first batch.
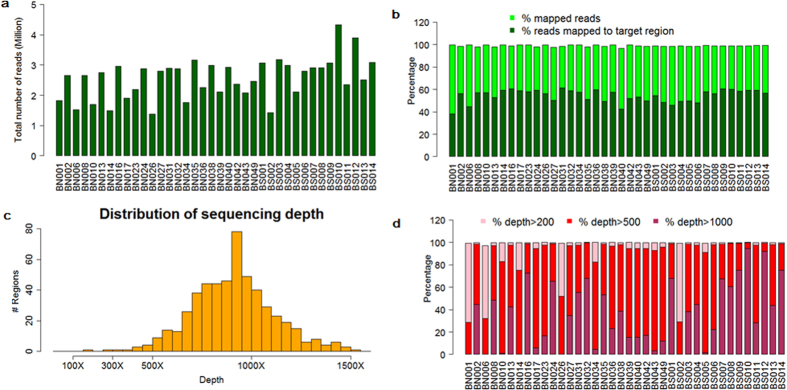
(a) Total number of reads in every sample (with an average of 2.46 million reads for each sample). (b) Percentage of all mapped reads and percentage of reads mapped to target regions for every sample. (c) The average distribution of sequencing depth over regions across all samples (the average coverage depth was well-distributed among regions, with a minimum of 157× and a maximum of 1505×). (d) The coverage depth in each sample (all samples have greater than 96% of all regions with coverage depth greater than 200×).
We then assessed the repeatability of probe capturing efficiency by measuring the correlation of coverage depth in each target region between different samples. Figure 2a shows a heatmap of such correlation between each pair of samples. Most pairs have a correlation coefficient greater than 0.8, indicating excellent repeatability of capturing efficiency of probes across different regions. The scatterplots of coverage depth across all regions between the most and least correlated pairs of samples are also shown (Fig. 2b,c). Sequencing data quality and region coverage reproducibility is similar among the second batch of samples (see Supplementary Fig. S1–2 online).
Figure 2. The correlation of coverage depth in each target region between 38 samples in first batch.
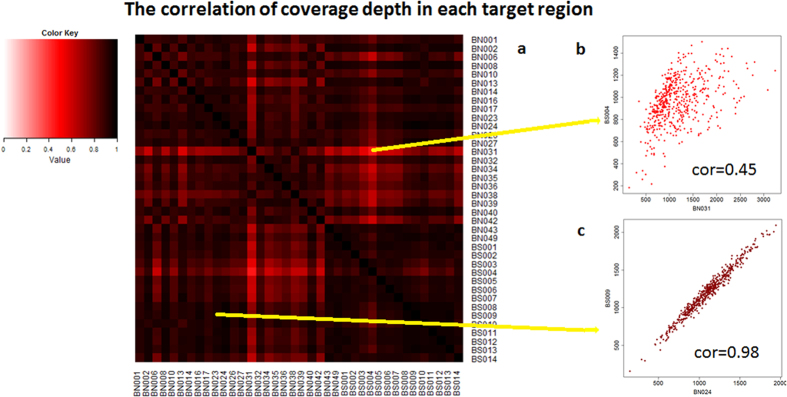
(a) A heatmap of such correlation between each pair of samples. (b) The scatterplot of coverage depth across all regions in the least correlated pair of samples. (c) The scatterplot of coverage depth across all regions in the most correlated pair of samples.
Germline mutation identification
We applied GATK 3.2 for genotyping the two batches of samples and identified 8462 single-nucleotide variants (SNVs) and 1699 insertion/deletion (INDELs) overall. The base substitution pattern is highly consistent across all samples (Fig. 3a), with an average transition-transversion (ti/tv) ratio of 2.55, similar to previously reported ti/tv ratio among exome regions. We then plotted the distribution of the minor allele frequencies (MAF) across all identified variants (Fig. 3b). The MAF demonstrates a clear bi-modal distribution, peaking at 0.5 and 1, a distribution expected for germline variants.
Figure 3. Base change distribution of all SNVs and distribution of MAF.
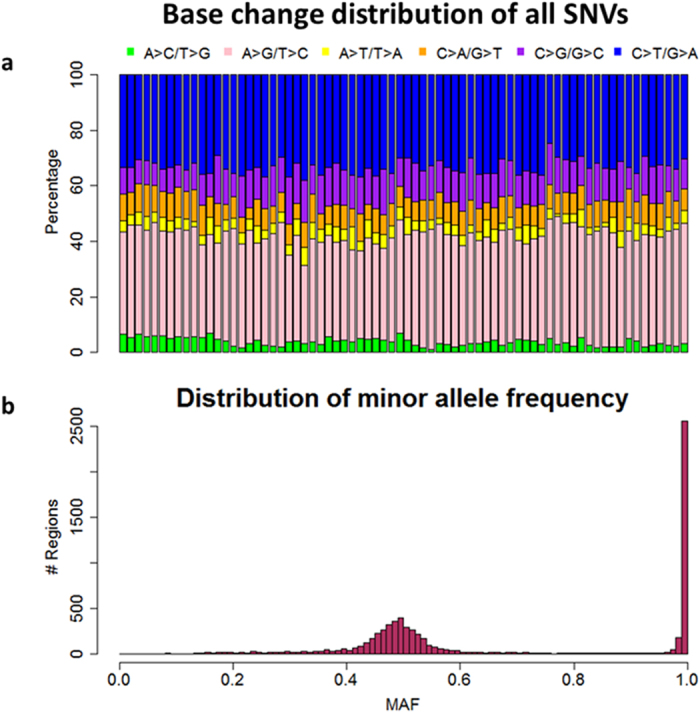
(a) Base substitution distribution of all single-nucleotide variants (SNVs) across all samples. (b) The distribution of the minor allele frequencies (MAF) across all identified variants.
In order to identify clinically meaningful germline variants, we applied filters to pick out non-synonymous high-confidence variants that were not reported previously in dbSNP or 1000 Genome Project as a common SNP (Fig. 4). Through the filtering steps, 45 variants were identified in the end and listed in Table 1, including 38 SNVs, one short 4-bp frame-shift deletion, one splice donor variant and five in-frame insertions or deletions. The MAF of these SNVs ranges from 43.4% to 60.0%, indicating that all of them are heterozygous germline mutations.
Figure 4. Process of mutation filtering.
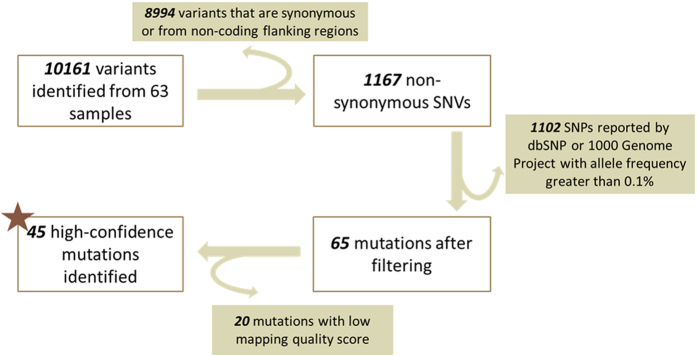
Among 10161 variants identified from 63 samples, there were 8994 variants that were synonymous or from non-coding flanking regions, 1102 SNPs reported by dbSNP or 1000 Genome Project with allele frequency greater than 0.1%, 20 mutations with low mapping quality score. After filtering out above variants 45 variants were identified in the end and listed in Table 1, including 38 SNVs and 7 indels. The MAF of these SNVs ranges from 43.4% to 60.0%, indicating that all of them are heterozygous germline mutations.
Table 1. List of the 45 high-confidence non-synonymous mutations identified among 63 NMTC patients.
| Family | Sample | Gene | Type | AA change | AF | GVGD | Clin-Var | ExAC ALL | ExAC EAS | SIFT_ score | Polyphen2 _HDIV _score | LRT_ score | Mutation Taster_ score | Mutation Assessor_ score | FATHMM_ score | Radial SVM_ score | LR_ score | Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BN01 | BN001 | APC | SNV | L292F | 45.3% | Class C0 | NA | . | . | 0.18 | 0.468 | 0.04 | 0.715 | 0.69 | −2.53 | 0.136 | 0.62 | 3 |
| BN01 | BN001 | EPCAM | SNV | T164I | 48.3% | Class C0 | US | 0 | 0 | 0.09 | 0.578 | 0.006 | 0.999 | 1.15 | −0.66 | −0.891 | 0.143 | 0 |
| BN01 | BN002 | APC | SNV | L292F | 48.5% | Class C0 | NA | . | . | 0.18 | 0.468 | 0.04 | 0.715 | 0.69 | −2.53 | 0.136 | 0.62 | 3 |
| BN01 | BN002 | MSH6 | SNV | L1081F | 49.4% | Class C0 | NA | . | . | 0.72 | 0.045 | 0.007 | 1 | 0.255 | −2.17 | −0.918 | 0.291 | 2 |
| BN03 | BN010 | BRAF | SNV | D22N | 48.9% | Class C0 | US | 0.0001 | 0 | 0.2 | 0.805 | 0.304 | 0.997 | 0 | −1.04 | −0.703 | 0.315 | 1 |
| BN03 | BN049 | BRAF | SNV | D22N | 49.8% | Class C0 | US | 0.0001 | 0 | 0.2 | 0.805 | 0.304 | 0.997 | 0 | −1.04 | −0.703 | 0.315 | 1 |
| BN04 | BN013 | ATM | SNV | C693Y | 45.9% | Class C0 | NA | . | . | 1 | 0 | 0.541 | 1 | −0.755 | −0.59 | −0.941 | 0.082 | 0 |
| BN05 | BN017 | APC | SNV | V1352A | 43.4% | Class C0 | US | 0 | 0.0005 | . | 0.595 | 0.002 | 1 | 0.975 | −2.5 | −0.25 | 0.483 | 2 |
| BN06 | BN023 | MSH6 | INDEL | L1358fs | 50.6% | NA | LB | 0.0023 | 0.032 | . | . | . | . | . | . | . | . | 0 |
| BN06 | BN024 | ATM | SNV | R2691C | 47.8% | Class C45 | US | 0.0001 | 0.0007 | 0.02 | 0.999 | 0.003 | 1 | 1.975 | −1.21 | 0.121 | 0.552 | 6 |
| BN09 | BN034 | MEN1 | SNV | A216V | 46.5% | Class C0 | NA | . | . | 0.23 | 0.053 | 0.3 | 1 | 0.805 | −5.8 | 0.824 | 0.897 | 3 |
| BN09 | BN035 | BRCA2 | SNV | V2503I | 47.7% | Class C0 | US | 0 | 0 | 1 | 0.001 | 0.002 | 1 | −1.845 | −1.84 | −0.728 | 0.12 | 1 |
| BN10 | BN036 | GNAS | SNV | P220A | 45.2% | Class C0 | NA | . | . | 0.15 | 0.005 | 0.002 | 1 | 0.695 | −3.65 | −0.328 | 0.677 | 3 |
| BN10 | BN036 | MSH6 | SNV | G355S | 48.7% | Class C0 | US | 0.0001 | 0.0012 | 0.69 | 0.015 | 0.042 | 1 | 0 | −1.81 | −0.815 | 0.309 | 2 |
| BN10 | BN038 | MSH6 | SNV | G355S | 51.1% | Class C0 | US | 0.0001 | 0.0012 | 0.69 | 0.015 | 0.042 | 1 | 0 | −1.81 | −0.815 | 0.309 | 2 |
| BN11 | BN039 | MSH2 | SNV | L719F | 47.8% | Class C0 | NA | . | . | 0 | 0.974 | 0.001 | 1 | 1.775 | −2.01 | 0.118 | 0.608 | 7 |
| BN11 | BN040 | MSH2 | SNV | L719F | 49.5% | Class C0 | NA | . | . | 0 | 0.974 | 0.001 | 1 | 1.775 | −2.01 | 0.118 | 0.608 | 7 |
| BN12 | BN043 | BRCA1 | SNV | R762S | 49.8% | Class C0 | US | 0.0001 | 0.0015 | 0 | 0.993 | 0.003 | 1 | 2.94 | −1.09 | −0.225 | 0.61 | 4 |
| BN13 | BN069 | EPCAM | SNV | R338G | 44.6% | Class C0 | NA | . | . | 0.01 | 0.999 | 0 | 0.905 | 2.015 | −0.83 | 0.007 | 0.504 | 7 |
| BN13 | BN070 | MSH6 | SNV | K854M | 46.4% | Class C65 | US | 0.0004 | 0.0015 | 0 | 1 | 0 | 1 | 2.71 | −2.83 | 0.822 | 0.845 | 7 |
| BN15 | BN053 | GNAS | SNV | R16C | 51.0% | Class C0 | NA | 0.0001 | 0.0022 | 0.01 | 0.916 | . | 1 | 1.39 | −2.42 | 0.042 | 0.482 | 4 |
| BN15 | BN053 | MEN1 | SNV | G508D | 56.0% | Class C0 | US | 0.0005 | 0.0073 | 0.55 | 0.059 | 0.527 | 1 | 0 | −5.73 | 0.53 | 0.844 | 3 |
| BN15 | BN054 | MEN1 | SNV | G508D | 55.9% | Class C1 | US | 0.0005 | 0.0073 | 0.55 | 0.059 | 0.527 | 1 | 0 | −5.73 | 0.53 | 0.844 | 3 |
| BN16 | BN052 | MSH6 | INDEL | NA | 54.0% | NA | US | 0.0013 | 0.0042 | . | . | . | . | . | . | . | . | 0 |
| BN17 | BN081 | BRCA1 | INDEL | SS955S | 49.6% | NA | NA | . | . | . | . | . | . | . | . | . | . | 0 |
| BN17 | BN081 | WRN | SNV | V755A | 52.3% | Class C0 | NA | . | . | 0.33 | 0.212 | 0.035 | 0.991 | 1.15 | 3.61 | −0.956 | 0.012 | 0 |
| BN17 | BN082 | BRCA1 | INDEL | SS955S | 50.0% | NA | NA | . | . | . | . | . | . | . | . | . | . | 0 |
| BN18 | BN084 | BRCA2 | SNV | F3328C | 48.7% | Class C15 | NA | 0 | 0 | 0.09 | 0.89 | 0.016 | 1 | 1.79 | 5.85 | −0.876 | 0.004 | 0 |
| BN19 | BN085 | BRCA2 | SNV | G2508S | 48.2% | Class C55 | US | 0.0001 | 0.0015 | 0.05 | 1 | 0 | 1 | 2.71 | −2.19 | 0.748 | 0.819 | 8 |
| BN19 | BN086 | APC | SNV | A2778S | 44.9% | Class C0 | US | 0.0002 | 0.0023 | . | 1 | 0 | 1 | 0.975 | −1.53 | −0.021 | 0.573 | 5 |
| BN19 | BN086 | BRCA2 | SNV | G2508S | 44.2% | Class C55 | US | 0.0001 | 0.0015 | 0.05 | 1 | 0 | 1 | 2.71 | −2.19 | 0.748 | 0.819 | 8 |
| BN19 | BN087 | APC | SNV | A2778S | 49.8% | Class C1 | US | 0.0002 | 0.0023 | . | 1 | 0 | 1 | 0.975 | −1.53 | −0.021 | 0.573 | 5 |
| BN21 | BN094 | TIMM44 | SNV | R277Q | 54.3% | Class C0 | NA | 0 | 0 | 0.01 | 0.997 | 0 | 1 | 3.155 | −1.66 | 0.572 | 0.679 | 8 |
| BN22 | BN095 | GNAS | INDEL | P459PDAPADPDSGAAR | 57.9% | NA | NA | . | . | . | . | . | . | . | . | . | . | 0 |
| BN22 | BN095 | MSH6 | SNV | A36V | 55.4% | Class C0 | US | 0.0002 | 0.0008 | 0.2 | 0.386 | 0.001 | 0.999 | 0.55 | −2.02 | −0.514 | 0.411 | 1 |
| BN22 | BN096 | GNAS | INDEL | P459PDAPADPDSGAAR | 60.0% | NA | NA | . | . | . | . | . | . | . | . | . | . | 0 |
| BN22 | BN096 | MSH6 | SNV | A36V | 51.1% | Class C0 | US | 0.0002 | 0.0008 | 0.2 | 0.386 | 0.001 | 0.999 | 0.55 | −2.02 | −0.514 | 0.411 | 1 |
| BS01 | BS001 | MSH2 | SNV | M261V | 43.5% | Class C0 | US | . | . | 1 | 0.097 | 0 | 1 | 0.805 | −2.32 | −0.748 | 0.334 | 3 |
| BS02 | BS002 | MSH2 | SNV | V78I | 50.3% | Class C25 | NA | 0 | 0.0002 | 0.03 | 0.998 | 0 | 1 | 2.38 | −2.56 | 0.783 | 0.82 | 8 |
| BS07 | BS007 | APC | SNV | L662I | 53.0% | Class C0 | NA | 0.0001 | 0.0008 | 0 | 0.995 | 0 | 1 | 2.045 | −0.31 | −0.372 | 0.384 | 5 |
| BS07 | BS007 | WT1 | SNV | S322N | 46.6% | Class C0 | NA | . | . | 0.38 | 0.001 | 0.054 | 1 | −0.69 | 3.41 | −1.054 | 0.039 | 1 |
| BS12 | BS012 | MSH6 | SNV | A53D | 44.0% | Class C0 | NA | . | . | 0.36 | 0.739 | 0.967 | 1 | 0.55 | −1.89 | −0.493 | 0.425 | 2 |
| BS12 | BS012 | MSH6 | INDEL | G56GPR | 44.7% | Class C0 | US | . | . | . | . | . | . | . | . | . | . | 0 |
| BS14 | BS014 | BRCA1 | SNV | A1368V | 45.4% | Class C0 | NA | . | . | 0.62 | 0.985 | 0.663 | 0.995 | −0.145 | −1.71 | −1.016 | 0.042 | 3 |
| BS17 | BS017 | ATM | SNV | I346N | 49.2% | Class C45 | NA | . | . | 0 | 0.938 | 0 | 0.999 | 1.845 | −0.35 | −0.373 | 0.311 | 3 |
AA: Amino Acid; NA: Not Available; US: Uncertain Significance; LB: Likely Benign. AF: Allele Frequency; Score: Number of algorithms predicting the variant to be “damaging”; BN: FNMTC samples; BS: SNMTC samples; 14 variants with score ≥ 4 were predicted to be damaging or deleterious by at least half of the predicting algorithms; More details are available in Supplementary Table S1 online.
We annotated all 45 variants with both the ExAC and ClinVar databases. Twenty-three out of the 37 variants (62%) from the familial group and 2 out of the 8 variants (25%) from the sporadic group are archived in the ExAC database. We examined the prevalence of all variants among both the whole ExAC population and the East Asian population only. Most archived variants have lower than 0.0001 minor allele frequency in both populations. Interestingly, 19 out of all 25 (78%) ExAC-archived variants show a greater prevalence among East Asian population than among the whole ExAC population. Twenty out of the 37 variants (54%) and 2 out of the 8 variants (25%) have at least one previous submission in the ClinVar databases. However, most of them have unclear clinical interpretation offered by ClinVar. None of the variants are archived as clearly pathogenic. One variant, MSH6 L1358fs, even though appearing to be a loss-of-function mutation, is archived as “likely benign”, probably because it is located at the very end of the MSH6 protein.
Since we could only find limited information about the detected variants from ExAC and ClinVar, in order to elucidate their possible impact on cancer susceptibility, we further annotated all variants with eight different tools, including SIFT, Polyphen2, LRT, MutationTaster, MutationAssessor, FATHMM, MetaSVM, and MetaLR, to predict the impact of the variants on protein function (Table 1; More details are available in Supplementary Table S1 online). Out of all 45 variants detected, 14 of them were predicted to be damaging or deleterious by at least half of the predicting algorithms. Most importantly, three matching variants within FNMTC families (BRCA2 G2508S, MSH2 L719F, and APC A2778S) were predicted to be damaging. The BRCA2 variant was also predicted to be highly likely to be damaging by GVGD (Class C55), further indicating possible contribution of these variants to the patients’ susceptibility (Table 1; More details are available in Supplementary Table S1 online).
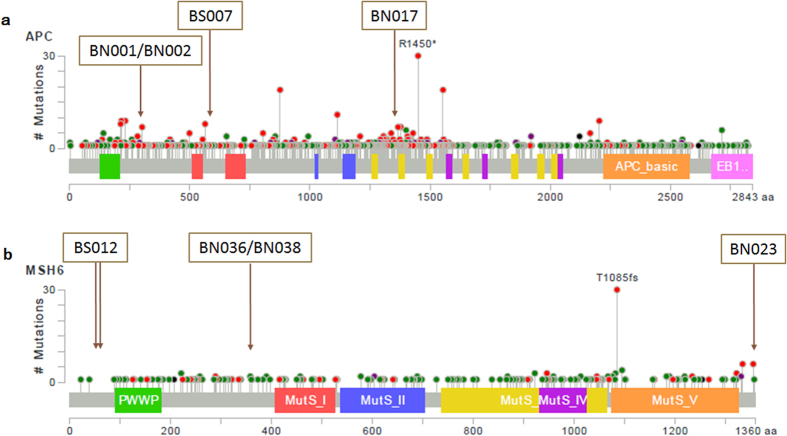
Mutations were detected among 13 out of the 31 genes profiled, including APC, MSH2, MSH6, ATM, BRAF, BRCA1, BRCA2, EPCAM, GNAS, MEN1, TIMM44, WRN and WT1. The distribution of mutational genes between the two groups was shown in Fig. 5. Twelve mutational genes were detected in FNMTC patients and six in SNMTC cases. The mutational spectrum of FNMTC was wider than that of SNMTC. The six most common genes detected in the two groups of patients were APC, BRCA1, BRCA2, GNAS, MSH2 and MSH6. Mutations detected in APC and MSH6 are overlaid on mutation distribution diagrams from TCGA (Fig. 6). Ten patients (8 FNMTC and 2 SNMTC) carry two germline mutations among all genes profiled, while all other patients carry one or none. We then compared the percentage of any germline mutation carriers between the two groups. Twenty-nine out of 47 (61.7%) among the familial group and 6 out of 16 (37.5%) among the sporadic group display a marginal but not statistically significant difference (OR = 2.64; p-value = 0.14), largely due to the limited sample size (Fig. 7). Notably, ten germline mutations from eight genes were found matching between paired FNMTC patients from the same family, including APC L292F and A2778S, BRAF D22N, MSH6 G355S and A36V, MSH2 L719F, MEN1 G508D, BRCA1 SS955S, BRCA2 G2508S, and a GNAS inframe insertion. The mutation percentage of these eight genes in FNMTC group was 53.2%, while 31.3% in SNMTC group (OR = 2.46; P = 0.16). It’s plausible that some of these variants might have contributed to these patients’ susceptibility to thyroid cancer.
Figure 5. The distribution of mutational genes between the two groups.
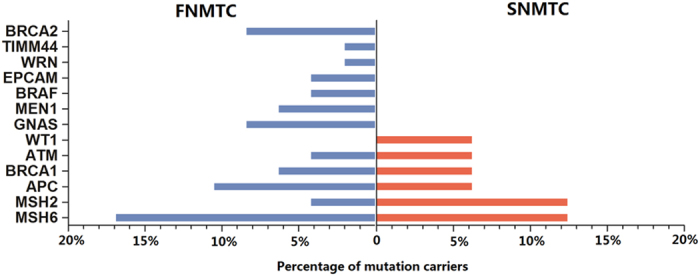
(12 mutational genes were detected in FNMTC patients and 6 in SNMTC cases; APC, BRCA1, BRCA2, GNAS, MSH2 and MSH6 were common genes detected in the two groups of patients).
Figure 6. Mutations detected in APC and MSH6 are overlaid on mutation distribution diagrams from TCGA.
(a) APC. (b) MSH6.
Figure 7. The rate of germline mutation carriers.
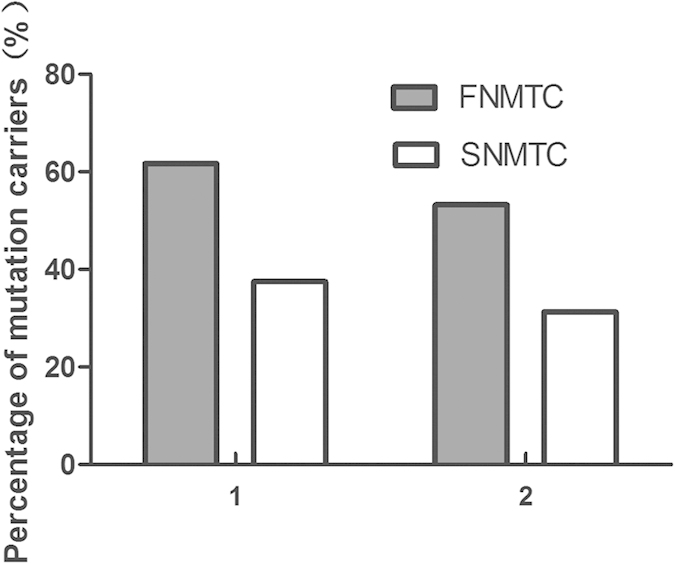
(“1” represents the rate of any germline mutation carriers in FNMTC group and in SNMTC group (61.7% vs. 37.5%); “2” represents the rate of mutations of eight genes with recurrent germline mutations detected (53.2% in FNMTC vs. 31.3% in SNMTC)).
Association of germline mutations and clinical-pathological characteristics
The clinical-pathological information of 61 patients in this research was collected and based on sequencing results, patients were divided into two groups: mutation positive and mutation negative. We compared the clinical features of mutation positive patients with that of negative patients (Table 2). There was a trend for a younger age in the positive group (mean 43.26 ± 9.66 vs. 47.19 ± 13.02, P = 0.18). No statistically significant differences were identified between the two groups on other factors, including female/male ratio (28: 7 vs. 20: 6, p = 0.77), tumor size (1.05 ± 0.50 vs. 1.01 ± 0.47, p = 0.73), the presence of multifocal disease (45.7% vs. 42.3%, p = 0.79) and the presence of bilateral disease (34.3% vs. 34.6%, p = 0.98). Interestingly, we found that central lymph nodes involvement occurred more frequently in the mutation-positive group than in the negative group (68.6% vs. 30.8%, p = 0.003), even though the treatment approach, such as extension of surgery, displayed no statistically significant difference between the two groups of patients.
Table 2. Association of germline mutations and clinical-pathological characteristics.
| Mutation + | Mutation − | p-value | |
|---|---|---|---|
| N | 35 | 26 | |
| Female: Male | 28:7 | 20:6 | 0.77 |
| Age | 43.26 ± 9.66 | 47.19 ± 13.02 | 0.18 |
| Tumor size | 1.05 ± 0.50 | 1.01 ± 0.47 | 0.73 |
| Multifocality | 16(45.7%) | 10(42.3%) | 0.79 |
| Bilaterality | 12(34.3%) | 8(34.6%) | 0.98 |
| Total thyroidectomy | 15(42.9%) | 12(46.2%) | 0.80 |
| Central compartment neck dessection | 34(97.1%) | 25(96.2%) | 0.83 |
| Extrathyroidal extension | 16(45.7%) | 14(53.9%) | 0.53 |
| Central lymph node involvement | 24(68.6%) | 8(30.8%) | 0.003 |
| Lateral lymph node involvement | 7(20.0%) | 6(23.1%) | 0. 77 |
Proportions were compared using Fisher’s exact test or Chi-square test, and continuous variables were compared using Student’s t-test or Mann Whitney U-test, as appropriate. A p-value < 0.05 was considered statistically significant.
Discussion
In this study, we developed a new approach for identifying potential genetic risk factors for FNMTC using next-generation sequencing. It uses a small amount of DNA and tests for a broad range of point mutations and indels simultaneously with high accuracy and sensitivity. The panel includes whole exon regions from 31 genes and one intronic region from RET for potential RET fusion profiling. Sequencing data showed comprehensive and evenly distributed coverage among different regions. With an average of ~500× coverage depth, more than 99% of all regions were covered by more than 200 reads. Furthermore, the high coverage distribution correlation between different batches of samples indicates a great reproducibility. Given such distribution, since germline mutations mostly possess an allele frequency around 50% or 100%, future application of this NGS panel for germline mutation detection would only need an average coverage depth of 50–100×, equalizing 17 MB sequencing data per sample, which would be very affordable for clinical application.
Our study has identified 37 variants among 29 (61.7%) NMTC patients with strong familial history and 8 variants among 6 (37.5%) NMTC patients without apparent familial history. Although statistical significance was not reached (p = 0.14), a notable OR of 2.64 was observed. It would have reached statistical significance if the sample size increased by 50% and prevalence in each group remained the same. It should also be noted that the co-occurrence pattern in many families with multiple NMTC cases might be attributed to environmental factors but not genetic risks.
Interestingly, when ignoring the familial pattern and simply comparing the clinical features between mutation positive patients with mutation negative patients, we found that the former group showed a trend for younger age and more frequent central lymph nodes involvement though they had no different in tumor size. This is consistent with previous studies, which suggested patients with FNMTC have an increased risk of lymph node metastases5,19,20. However, lymph node metastasis in central compartment could not be identified by preoperative examination easily. Therefore, central compartment neck dissection is needed for patients with such germline mutation or family history of thyroid cancer, at least central lymph nodes ipsilateral to the lesion should be removed.
The six commonly mutated genes detected in this study were APC, BRCA1, BRCA2, GNAS, MSH2 and MSH6. There are five patients in FNMTC group with APC mutations, four patients of whom belong to two families, indicating that the shared variants of APC may be associated with the incidence of NMTC. Defects in this gene cause familial adenomatous polyposis (FAP), an autosomal dominant pre-malignant disease that usually progresses to malignancy. An association between follicular adenoma and FAP has been described21,22,23. It had been reported that the cribriform-morular variant of papillary thyroid carcinoma was frequently seen in patients with FAP24 and thirty percent of thyroid carcinomas are diagnosed 4–12 years before the development of polyposis coli21. Although the siblings carrying APC mutation are follicular variant of papillary thyroid carcinoma in this study, we could not confirm whether they have adenomatous polyposis. Therefore regular colonic examination is recommended for these patients. In another family, the siblings have the same variant of BRAF D22N, different from the well-known BRAF V600E variant that is highly prevalent in papillary thyroid carcinoma. The highly mutated genes (KRAS, HRAS, NRAS, etc.) in somatic cells of thyroid cancer were rarely observed to mutate in patients’ blood in our study.
Matching mutations within family in MSH6 or MSH2 were detected in three other families. MSH6 and MSH2, together with MLH1 and PMS2, belong to DNA mismatch repair (MMR) genes, preventing mutational events through correction of mismatched bases during DNA replication. Germline mutations in MMR genes can give rise to the autosomal dominant condition, Lynch Syndrome (LS), previously referred to as hereditary nonpolyposis colon cancer (HNPCC). LS was characterized by increased lifetime risks for colorectal (40–80%), endometrial (25–60%), ovarian (4–24%), and gastric (1–13%) cancers25,26. During our follow-up with the tested patients, patient BN040, who carries the same MSH2 L719F mutation as her sibling, reported suffering from frequent diarrhea (3–4 times per day), and was recommended for further examination. We also learned that within family BN10, from which two family members were detected with MSH6 G355S in our study, four out of all five sisters suffer from thyroid cancer. Rein27 reported a 44-year-old woman from a Lynch syndrome, Amsterdam positive family who was referred for DNA testing. She had a recent history of a colorectal adenoma and an undifferentiated carcinoma of her thyroid and was shown to carry the truncating MSH2 mutation that was known to segregate in her family. Traditionally, thyroid cancer is not considered to be part of the Lynch syndrome tumour spectrum. However, Rein’s study demonstrated that the patient’s undifferentiated thyroid carcinoma showed complete loss of immunohistochemical expression of the MSH2 and MSH6 protein. In addition, Pande’s findings28 confirmed that a significant number of Lynch syndrome patients can present with thyroid carcinoma or other types of tumors as primary tumors that are not part of the Lynch syndrome spectrum. Our sequencing results showed germline mutations of some MMR genes detected in 37.1% (13/35) of all NMTC patients with a mutation. Therefore, the causality of MMR gene mutations on thyroid carcinoma calls for further investigation. Screening of colonic polyposis by total colonoscopy should be recommended for thyroid carcinoma patients carrying MMR gene mutation. Studies had demonstrated that annual surveillance of colon for MMR mutations patients would reduce the incidence of colon cancer and mortality29,30,31. Women carrying these germline mutations have dramatically elevated rates of gynecological cancer compared to women in the general population. They face a 40–60% lifetime risk of endometrial cancer and a 10–12% lifetime risk of ovarian cancer, compared to the general population risks of 3% and 1.4%, respectively32,33,34. Some authors recommend annual surveillance in this high-risk group of mutation carriers, including transvaginal ultrasonography, tumor marker CA125 blood tests and/or endometrial biopsy35,36. Schmeler’s findings37 suggest that prophylactic hysterectomy with bilateral salpingo-oophorectomy is an effective strategy for preventing endometrial and ovarian cancer in women with germline mutations related to the Lynch syndrome.
Previous studies38,39,40,41,42 have indicated that patients with thyroid cancer have an increased risk of developing a second cancer in all sites examined including salivary gland, kidney, prostate, skin, breast, brain, myeloma, leukemia, and non-Hodgkin lymphoma, compared to the general population. Fallah43 studied lifetime cumulative risk of thyroid cancer (CRTC) in 63495 first degree relatives of 11206 NMTC patients, Family history of PTC increased risk of non-thyroid endocrine cancers (such as parathyroid cancer) and non-melanoma skin cancer up to about two-fold in both sexes and kidney (1.4-fold) and prostate (1.2-fold) cancer in men. An additional advantage of this panel is that it might help to forecast the risk and the type of second tumor after diagnosis of primary thyroid cancer from the germline genetic variant that patient carries. For example, mutations in ATM, BRCA1 and BRCA2 increase the risk of breast cancer and ovarian cancer, while WT1 is commonly expressed in ovarian serous carcinomas and endometrial serous carcinoma and considered a diagnostic marker of these tumors40. MEN1 and RET are associated with multiple endocrine neoplasia type I and multiple endocrine neoplasia type II, respectively. During our follow-up with mutation-positive patients from this study, one SNMTC patient BS012 has most recently developed a 3-cm lump in her breast. She is undergoing further examination for diagnosis.
In summary, we demonstrated the feasibility and practicability of the application of next generation sequencing in screening for germline mutations on cancer-related genes among FNMTC patients. This method provides a novel approach for us to help diagnose and elucidate the genetic cause of the FNMTC patients, and assess whether their family members are exposed to a higher genetic risk. The findings would also provide insights on monitoring the potential second cancers for patients affected with thyroid cancer.
Methods and Materials
Ethics statement
Peripheral blood of all patients was obtained from Tumor Tissue Banking Facility of Tianjin Medical University Cancer Institute and Hospital and all participants provided written informed consent. This study was performed in accordance with the approved guidelines of Tianjin Medical University Cancer Institute and Hospital’s Ethics Committee.
Patients
FNMTC is defined by the diagnosis of two or more first-degree relatives affected by differentiated thyroid cancer of follicular cell origin. Therefore, in the present work, the criteria for eligibility of the FNMTC families were that two (or more) first-degree family members had to be affected with NMTC. In this study, total of 63 NMTC patients were recruited, including 47 cases with an apparent family history of thyroid cancer and 16 cases with no family history. The task of sequencing and analysis was completed in two times. In the first batch of samples, 24 FNMTC patients from 12 unrelated families and 14 patients with SNMTC were included. The ratio of female and male was 3:1 and the average age was 46 years. There were 23 FNMTC patients from 10 families and 2 SNMTC patients in the second batch which was the complement and verification for the first batch of samples. The ratio of female and male was 5.3:1 and the average age was 43 years. Histological types of all patients were papillary thyroid carcinoma. In addition, the clinic-pathological characteristics, including tumor size, extension of surgery, lymph node metastasis and extra-thyroidal extension, were compared between patients with or without actionable mutations. But the operations of two cases were not performed in our hospital, so the clinical-pathological information of them was absent.
Design of Gene Chip
One study suggested that premenopausal breast carcinoma may occur with a greater frequency than expected in NMTC patients44. It has been hypothesized that breast carcinoma is a side effect of 131I therapy for PTC45. Alternatively, breast and thyroid carcinoma, which both arise from an epithelial cell type, may share a common susceptibility factor, gene or otherwise. Consistent with this hypothesis, studies of malignancies in first-degree relatives of NMTC patients showed a greater frequency of breast cancer than expected among first-degree relatives of FNMTC patients46,47. Other malignancies potentially associated with PTC include kidney, ovarian, and right-sided colon cancer46. In order to expand the screening of susceptibility genes, the panel we designed includes candidate susceptibility genes and loci for hereditary thyroid cancer: TIMM44, DNMT1, SMARCA4, RET, MEN1 and possible susceptibility genes for breast cancer and ovarian cancer, as well as other previously known familial cancer or cancer-related syndromes: BRCA1, BRCA2, TP53, PTEN, STK11, APC, CDH1, MITF, VHL, WT1, SMAD4, MUTYH, MLH1, MSH2, MSH6, PMS2, EPCAM, ATM, GNAS1, PRKAR1A, WRN, which may also relate to the incidence of thyroid cancer.
In addition, recent studies of rare hereditary lung cancer showed that carrying known disease-relevant somatic mutations (e.g. EGFR T790M) as germline might be another important mechanism of hereditary cancer syndromes48,49. Therefore, our panel also included important genes with somatic mutations found previously in thyroid cancer, multiple endocrine adenoma, parathyroid carcinoma and pheochromocytoma, for example, RET fusion, BRAF, KRAS, HRAS, NRAS, PTEN, and PIK3CA. Overall, the panel included whole exon regions from 31 genes and one intronic region from RET for potential RET fusion profiling.
Targeted DNA sequencing
To design the capture probe baits and prepare the SureSelect reagents, 170 kb human genomic sequence from 526 target regions were submitted to the Agilent eArray platform and manufactured by Agilent.
Peripheral blood was collected from 47 FNMTCs patients and 16 SNMTCs respectively. DNA was extracted from peripheral blood leukocytes using standard protocols. DNA of the 63 samples were extracted (QIAamp DNA blood mini kit), and the concentration of the DNA samples were measured by Qubit dsDNA assay. The gDNA quality was then assessed to make sure A260/A280 is within the range of 1.8 to 2.0. Shearing fragmentation by sonication (covaris M220) was then conducted, followed by end repair, phosphorylation and adaptor ligation. Fragments of size 200–400 bp were selected by bead (Agencourt AMPure XP Kit), followed by hybridization with the capture probes baits, hybrid selection with magnetic beads, and PCR amplification. A bioanalyzer high sensitivity DNA assay was then used to assess the quality and size range. Indexed samples were pooled to be loaded onto the flow cells for sequencing on a Miseq (Illumina, Inc., USA) with 150-cycle pair-end reads.
Sequence data analysis
Sequence data were mapped to the human genome (hg19) using BWA aligner 0.7.10. PCR duplicate reads were removed before base substitution detection. Local alignment optimization and variant calling and annotation were performed using GATK 3.2. DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3.
Statistical analysis
Analysis was performed for patients’ characteristics between the mutation positive group and negative group and the percentage of mutation carriers in FNMTC group was compared with that of SNMTC control. Proportions were compared using Fisher’s exact test or Chi-square test, and continuous variables were compared using Student’s t-test or Mann Whitney U-test, as appropriate. A p-value < 0.05 was considered statistically significant.
Additional Information
How to cite this article: Yu, Y. et al. Targeted DNA Sequencing Detects Mutations Related to Susceptibility among Familial Non-medullary Thyroid Cancer. Sci. Rep. 5, 16129; doi: 10.1038/srep16129 (2015).
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81472580, No. 81272282), Key Projects in the National Science & Technology Pillar Program (No. 2013ZX09303001, No. 2015BAI12B15), Natural Science Foundation of Tianjin (No. 13JCQNJC10200), Tianjin Municipal Commission of Science & Technology Key Research Program (No. 13ZCZCSY20300).
Footnotes
Author Contributions J.Y. and M.G. conceived the research and supervised the research activities. Y.Y., L.D., D.L., S.C., Z.W., X.Z., Y.C. and L.H. performed research, analyzed and interpreted the data. Y.Y., L.D. and S.C. wrote the paper. All authors discussed the results and contributed to the refinement of the paper.
References
- Schlumberger M. J. Papillary and Follicular Thyroid Carcinoma. N Engl J Med. 338, 297–306 (1998). [DOI] [PubMed] [Google Scholar]
- Goldgar D. E., Easton D. F., Cannon-Albright L. A. & Skolnick M. H. Systematic Population-Based Assessment of Cancer Risk in First-Degree Relatives of Cancer Probands. J Natl Cancer Inst. 86, 1600–1608 (1994). [DOI] [PubMed] [Google Scholar]
- Ron E. et al. A Population-Based Case-Control Study of Thyroid Cancer. J Natl Cancer Inst. 79, 1–12 (1987). [PubMed] [Google Scholar]
- Stoffer S. S., Van Dyke D. L., Bach J. V., Szpunar W. & Weiss L. Familial Papillary Carcinoma of the Thyroid. Am J Med Genet. 25, 775–782 (1986). [DOI] [PubMed] [Google Scholar]
- Mazeh H. et al. In Patients with Thyroid Cancer of Follicular Cell Origin, a Family History of Nonmedullary Thyroid Cancer in One First-Degree Relative is Associated with More Aggressive Disease. Thyroid. 22, 3–8 (2012). [DOI] [PubMed] [Google Scholar]
- Triponez F. et al. Does Familial Non-Medullary Thyroid Cancer Adversely Affect Survival? World J Surg. 30, 787–793 (2006). [DOI] [PubMed] [Google Scholar]
- Uchino S. et al. Familial Nonmedullary Thyroid Carcinoma Characterized by Multifocality and a High Recurrence Rate in a Large Study Population. World J Surg. 26, 897–902 (2002). [DOI] [PubMed] [Google Scholar]
- Alsanea O. et al. Is Familial Non-Medullary Thyroid Carcinoma More Aggressive than Sporadic Thyroid Cancer? A Multicenter Series. Surgery. 128 1043–1050, 1050–1051 (2000). [DOI] [PubMed] [Google Scholar]
- Loh K. C. Familial Nonmedullary Thyroid Carcinoma: A Meta-Review of Case Series. Thyroid. 7, 107–113 (1997). [DOI] [PubMed] [Google Scholar]
- Capezzone M. et al. Familial Non-Medullary Thyroid Carcinoma Displays the Features of Clinical Anticipation Suggestive of a Distinct Biological Entity. Endocr Relat Cancer. 15, 1075–1081 (2008). [DOI] [PubMed] [Google Scholar]
- McKay J. D. et al. Localization of a Susceptibility Gene for Familial Nonmedullary Thyroid Carcinoma to Chromosome 2Q21. Am J Hum Genet. 69, 440–446 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan S. et al. A Comprehensive Analysis of MNG1, TCO1, fPTC, PTEN, TSHR, and TRKA in Familial Nonmedullary Thyroid Cancer: Confirmation of Linkage to TCO1. J Clin Endocrinol Metab. 86, 3701–3704 (2001). [DOI] [PubMed] [Google Scholar]
- Malchoff C. D. et al. Papillary Thyroid Carcinoma Associated with Papillary Renal Neoplasia: Genetic Linkage Analysis of a Distinct Heritable Tumor Syndrome. J Clin Endocrinol Metab. 85, 1758–1764 (2000). [DOI] [PubMed] [Google Scholar]
- Malchoff C. D. et al. Familial Papillary Thyroid Carcinoma is Genetically Distinct From Familial Adenomatous Polyposis Coli. Thyroid. 9, 247–252 (1999). [DOI] [PubMed] [Google Scholar]
- Canzian F. et al. A Gene Predisposing to Familial Thyroid Tumors with Cell Oxyphilia Maps to Chromosome 19P13.2. Am J Hum Genet. 63, 1743–1748 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell G. R. et al. Familial Nontoxic Multinodular Thyroid Goiter Locus Maps to Chromosome 14Q but Does Not Account for Familial Nonmedullary Thyroid Cancer. Am J Hum Genet. 61, 1123–1130 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw D. et al. Germline Mutations of the PTEN Gene in Cowden Disease, an Inherited Breast and Thyroid Cancer Syndrome. Nat Genet. 16, 64–67 (1997). [DOI] [PubMed] [Google Scholar]
- Huang S. et al. The Spectrum of WRN Mutations in Werner Syndrome Patients. Hum Mutat. 27, 558–567 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenbrand A. et al. Familial Nonmedullary Thyroid Carcinoma-Clinical Relevance and Prognosis. A European Multicenter Study. ESES Vienna Presentation. Langenbecks Arch Surg. 395, 851–858 (2010). [DOI] [PubMed] [Google Scholar]
- Sippel R. S., Caron N. R. & Clark O. H. An Evidence-Based Approach to Familial Nonmedullary Thyroid Cancer: Screening, Clinical Management, and Follow-Up. World J Surg. 31, 924–933 (2007). [DOI] [PubMed] [Google Scholar]
- Plail R. O., Bussey H. J., Glazer G. & Thomson J. P. Adenomatous Polyposis: An Association with Carcinoma of the Thyroid. Br J Surg. 74, 377–380 (1987). [DOI] [PubMed] [Google Scholar]
- Harach H. R., Williams G. T. & Williams E. D. Familial Adenomatous Polyposis Associated Thyroid Carcinoma: A Distinct Type of Follicular Cell Neoplasm. Histopathology. 25, 549–561 (1994). [DOI] [PubMed] [Google Scholar]
- Soravia C. et al. Familial Adenomatous Polyposis-Associated Thyroid Cancer: A Clinical, Pathological, and Molecular Genetics Study. Am J Pathol. 154, 127–135 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetta F. et al. Germline Mutations of the APC Gene in Patients with Familial Adenomatous Polyposis-Associated Thyroid Carcinoma: Results From a European Cooperative Study. J Clin Endocrinol Metab. 85, 286–292 (2000). [DOI] [PubMed] [Google Scholar]
- Aarnio M. et al. Cancer Risk in Mutation Carriers of DNA-mismatch-repair Genes. Int J Cancer. 81, 214–218 (1999). [DOI] [PubMed] [Google Scholar]
- Bonadona V. et al. Cancer Risks Associated with Germline Mutations in MLH1, MSH2, and MSH6 Genes in Lynch Syndrome. JAMA. 305, 2304–2310 (2011). [DOI] [PubMed] [Google Scholar]
- Stulp R. P. et al. Thyroid Cancer in a Patient with a Germline MSH2 Mutation. Case Report and Review of the Lynch Syndrome Expanding Tumour Spectrum. Hered Cancer Clin Pract. 6, 15–21 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande M. et al. Cancer Spectrum in DNA Mismatch Repair Gene Mutation Carriers: Results From a Hospital Based Lynch Syndrome Registry. Fam Cancer. 11, 441–447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvinen H. J. et al. Controlled 15-Year Trial On Screening for Colorectal Cancer in Families with Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 118, 829–834 (2000). [DOI] [PubMed] [Google Scholar]
- de Vos T. N. C. W. et al. Surveillance for Hereditary Nonpolyposis Colorectal Cancer: A Long-Term Study On 114 Families. Dis Colon Rectum. 45, 1588–1594 (2002). [DOI] [PubMed] [Google Scholar]
- Dove-Edwin I., Sasieni P., Adams J. & Thomas H. J. Prevention of Colorectal Cancer by Colonoscopic Surveillance in Individuals with a Family History of Colorectal Cancer: 16 Year, Prospective, Follow-Up Study. BMJ. 331, 1047 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch H. T. & de la Chapelle A. Hereditary Colorectal Cancer. N Engl J Med. 348, 919–932 (2003). [DOI] [PubMed] [Google Scholar]
- Weissman S. M. et al. Genetic Counseling Considerations in the Evaluation of Families for Lynch Syndrome–A Review. J Genet Couns. 20, 5–19 (2011). [DOI] [PubMed] [Google Scholar]
- Watson P. et al. The Risk of Extra-Colonic, Extra-Endometrial Cancer in the Lynch Syndrome. Int J Cancer. 123, 444–449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindor N. M. et al. Recommendations for the Care of Individuals with an Inherited Predisposition to Lynch Syndrome: A Systematic Review. JAMA. 296, 1507–1517 (2006). [DOI] [PubMed] [Google Scholar]
- McCann G. A. & Eisenhauer E. L. Hereditary Cancer Syndromes with High Risk of Endometrial and Ovarian Cancer: Surgical Options for Personalized Care. J Surg Oncol. 111, 118–124 (2015). [DOI] [PubMed] [Google Scholar]
- Schmeler K. M. et al. Prophylactic Surgery to Reduce the Risk of Gynecologic Cancers in the Lynch Syndrome. N Engl J Med. 354, 261–269 (2006). [DOI] [PubMed] [Google Scholar]
- Kim C. et al. The Risk of Second Cancers After Diagnosis of Primary Thyroid Cancer is Elevated in Thyroid Microcarcinomas. Thyroid. 23, 575–582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. H. et al. Second Primary Malignancies Following Thyroid Cancer: A Population-Based Study in Taiwan. Eur J Endocrinol. 169, 577–585 (2013). [DOI] [PubMed] [Google Scholar]
- Oakley G. M. et al. Increased Melanoma Risk in Individuals with Papillary Thyroid Carcinoma. JAMA Otolaryngol Head Neck Surg. 140, 423–427 (2014). [DOI] [PubMed] [Google Scholar]
- Hsu C. H. et al. Co-Occurrence of Second Primary Malignancy in Patients with Thyroid Cancer. QJM. 107, 643–648 (2014). [DOI] [PubMed] [Google Scholar]
- Van Fossen V. L., Wilhelm S. M., Eaton J. L. & McHenry C. R. Association of Thyroid, Breast and Renal Cell Cancer: A Population-Based Study of the Prevalence of Second Malignancies. Ann Surg Oncol. 20, 1341–1347 (2013). [DOI] [PubMed] [Google Scholar]
- Fallah M. et al. Risk of Thyroid Cancer in First-Degree Relatives of Patients with Non-Medullary Thyroid Cancer by Histology Type and Age at Diagnosis: A Joint Study From Five Nordic Countries. J Med Genet. 50, 373–382 (2013). [DOI] [PubMed] [Google Scholar]
- Vassilopoulou-Sellin R., Palmer L., Taylor S. & Cooksley C. S. Incidence of Breast Carcinoma in Women with Thyroid Carcinoma. Cancer-Am Cancer Soc. 85, 696–705 (1999). [DOI] [PubMed] [Google Scholar]
- Chen A. Y. et al. The Development of Breast Carcinoma in Women with Thyroid Carcinoma. Cancer-Am Cancer Soc. 92, 225–231 (2001). [DOI] [PubMed] [Google Scholar]
- Hemminki K., Eng C. & Chen B. Familial Risks for Nonmedullary Thyroid Cancer. J Clin Endocrinol Metab. 90, 5747–5753 (2005). [DOI] [PubMed] [Google Scholar]
- Pal T. et al. Increased Risk for Nonmedullary Thyroid Cancer in the First Degree Relatives of Prevalent Cases of Nonmedullary Thyroid Cancer: A Hospital-Based Study. J Clin Endocrinol Metab. 86, 5307–5312 (2001). [DOI] [PubMed] [Google Scholar]
- Gazdar A. et al. Hereditary Lung Cancer Syndrome Targets Never Smokers with Germline EGFR Gene T790M Mutations. J Thorac Oncol. 9, 456–463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H. et al. Novel Germline Mutation in the Transmembrane Domain of HER2 in Familial Lung Adenocarcinomas. J Natl Cancer Inst. 106, t338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.