Abstract
Since its discovery in 1989, efforts to grow clinical isolates of the hepatitis C virus (HCV) in cell culture have met with limited success. Only the JFH-1 isolate has the capacity to replicate efficiently in cultured hepatoma cells without cell culture-adaptive mutations1-3. We hypothesized that cultured cells lack one or more factors required for the replication of clinical isolates. To identify the missing factors, we transduced Huh-7.5 human hepatoma cells with a pooled lentivirus-based human cDNA library, transfected with HCV subgenomic replicons lacking adaptive mutations, and selected for stable replicon colonies. This led to the identification of a single cDNA, SEC14L2, whose expression allowed RNA replication of all HCV genotypes in several hepatoma cell lines. This effect was dose-dependent, and required the continuous presence of SEC14L2. Full-length HCV genomes also replicated and produced low levels of infectious virus. Remarkably, SEC14L2-expressing Huh-7.5 cells also supported HCV replication following inoculation with patient sera. Mechanistic studies suggest that SEC14L2 promotes HCV infection by enhancing vitamin E-mediated protection against lipid peroxidation. This sets the stage for development of in vitro replication systems for all HCV isolates, and provides an attractive platform to dissect the mechanisms by which cell culture-adaptive mutations act.
Hepatitis C virus (HCV) is a leading cause of liver disease worldwide, with an estimated global burden of 185 million chronic infections4. Studies of this important human pathogen have historically been hampered by the lack of a cell culture system that supports replication of clinical isolates. Inoculation of primary human hepatocytes or hepatoma cell lines with serum from HCV infected patients leads to extremely little, if any, replication. In addition, molecularly cloned HCV genomes that are infectious in experimentally infected chimpanzees fail to establish infection in Huh-7 derived human hepatoma cells5-8. Drug-selectable subgenomic replicons derived from these genomes must acquire mutations to replicate in cell culture1,2. This replication block could be due to the presence of inhibitory factor(s) that limit replication and/or the lack of essential host factor(s).
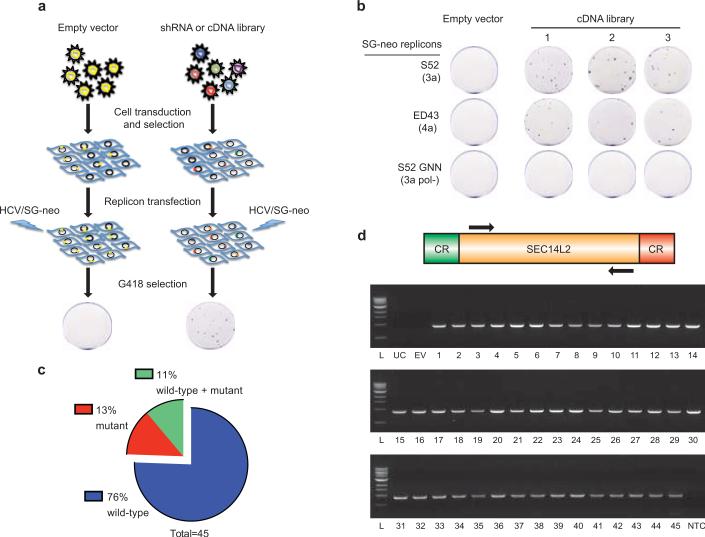
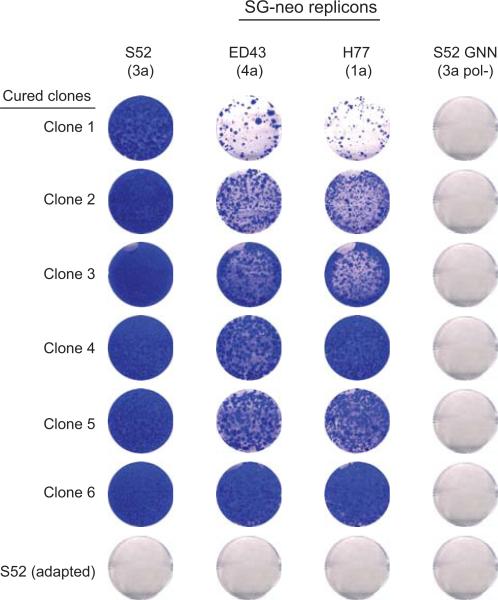
To overcome the block, we transduced Huh-7.5 cells with lentivirus libraries expressing either shRNAs or cDNAs. The transduced cell populations were then electroporated with in vitro transcripts of wild-type G418-selectable genotype 3a and 4a HCV subgenomic replicons, which in unaltered Huh-7.5 cells require at least two adaptive mutations for replication (Fig. 1a). While the shRNA library did not yield any hits, the cDNA library produced numerous colonies (Fig. 1b). The vast majority of these colonies (34 of 45) harbored replicons with the parental sequence (Fig. 1c); mutations were present in the remaining colonies but none corresponded to known adaptive changes. To confirm their ability to support non-adapted replicons, cell colonies were treated with anti-HCV compounds to clear replicating viral genomes and then re-transfected with a second set of wild-type replicons. Selection with G418 resulted in the production of numerous G418-resistant colonies. In contrast, no colonies were produced from control cells cured of cell culture-adapted replicons (Extended Data Fig. 1).
Figure 1. cDNA screening of Huh-7.5 cells identifies SEC14L2 as a critical host factor for HCV RNA replication.
(a) Illustration of the shRNA or cDNA screen. (b) Three Huh-7.5 cell populations independently transduced with a pooled lentiviral cDNA library and a population of empty vector-expressing control cells was electroporated with the indicated wild-type HCV replicons or a replication-defective S52 GNN replicon. After 3 weeks of G418 selection, the resulting cell colonies were stained with crystal violet. (c) HCV NS3-NS5B region from 45 cell colonies was amplified by RT-PCR and subjected to direct sequencing. Shown are the % colonies harboring wild-type, mutant, or a mixture of wild-type and mutant sequences. (d) The SEC14L2 cDNA was amplified from 45 colonies (see methods). CR, constant region; L, ladder; UC, untransduced cells; EV, empty vector-transduced cells; NTC, no template control.
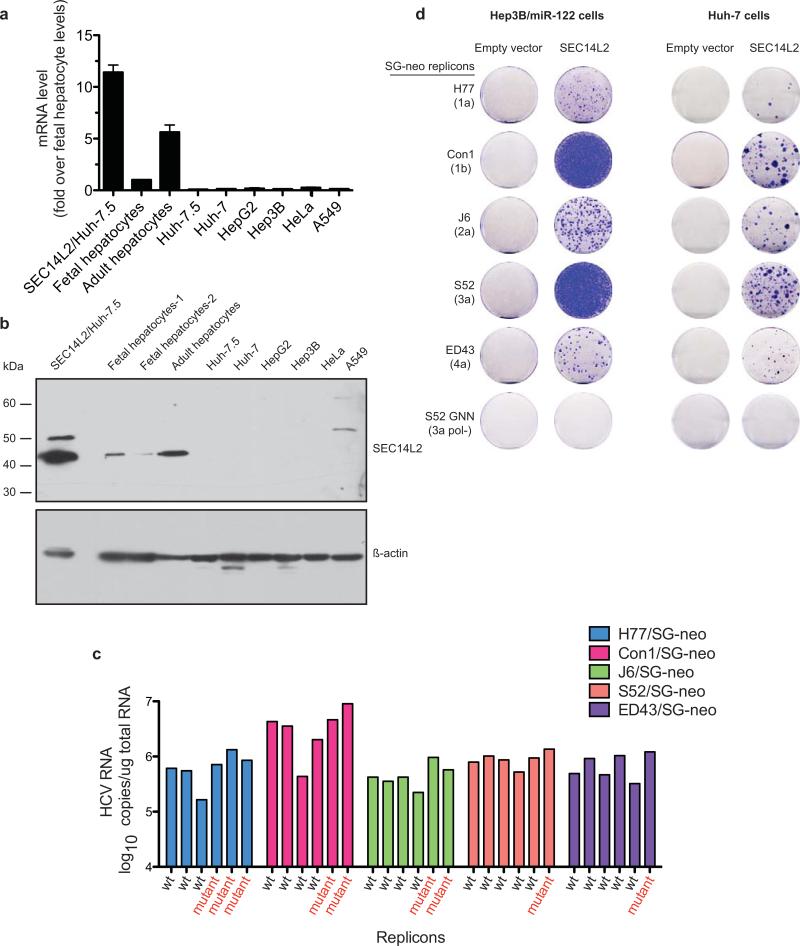
PCR amplification and sequence analysis of integrated cDNAs from 45 colonies revealed the same gene product, SEC14L2 (Fig. 1d). SEC14L2, also known as c22orf6, supernatant protein factor 1 (SPF1), or tocopherol-associated protein 1 (TAP1), is a cytosolic lipid-binding protein family member9,10, and is ubiquitously expressed in human tissues11. SEC14L2 RNA and protein could not be detected in human hepatoma and non-hepatoma cell lines. However, primary human hepatocytes, both from fetal and adult sources, expressed readily detectable levels (Extended Data Fig. 2a, b).
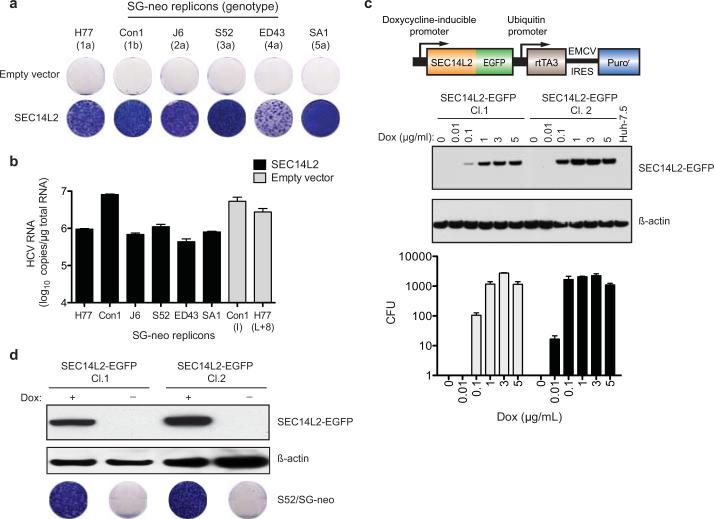
To confirm that SEC14L2 is necessary and sufficient for HCV RNA replication, we generated Huh-7.5 cells stably expressing SEC14L2 (SEC14L2/Huh-7.5) and transfected them with a panel of wild-type replicons from HCV genotypes 1a, 1b, 2a, 3a, 4a, and 5a. Selection with G418 yielded large numbers of colonies (Fig. 2a), with the majority harboring replicons with the parental sequences (Extended Data Fig. 2c). HCV RNA levels in these colonies were comparable to cell culture-adapted replicons, suggesting high levels of replication (Fig. 2b and Extended Data Fig. 2c). The effect was not cell line specific, since SEC14L2 expression in Huh-7 and Hep3B/miR122 cells also rendered them permissive for HCV replication, albeit to lower levels (Extended Data Fig. 2d).
Figure 2. SEC14L2 allows replication of wild-type HCV subgenomic replicons.
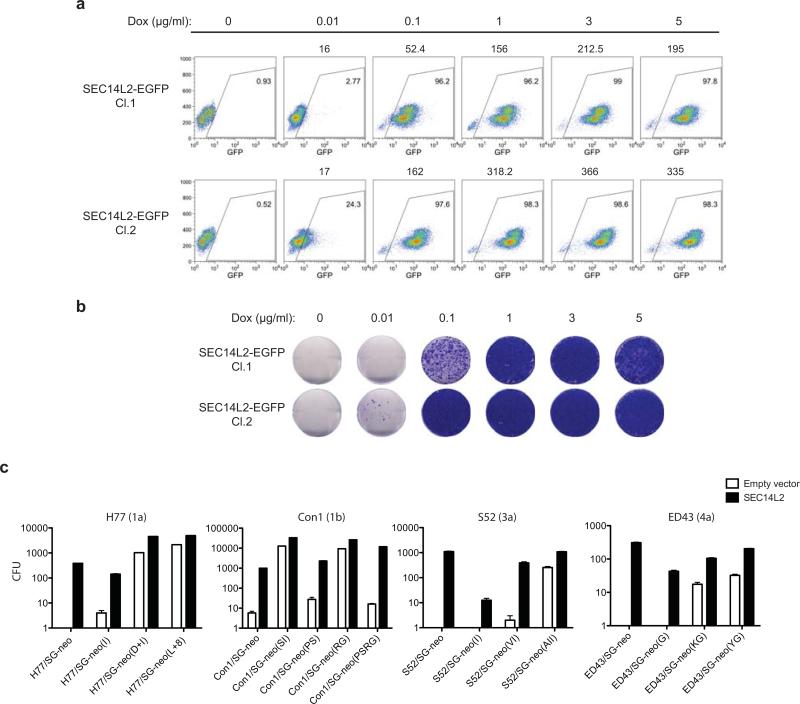
(a) Cell colonies obtained for the indicated replicons in Huh-7.5 cells transduced with empty vector or SEC14L2. (b) HCV RNA levels for wild-type replicons in SEC14L2-expressing cells (black bars) and two cell culture-adapted replicons in control cells (grey bars) are shown. (c) Schematic representation of the doxycycline-inducible SEC14L2-EGFP expression plasmid (top panel). Dose-dependent effect of doxycycline on SEC14L2-EGFP expression (middle panel) and S52/SG-neo replication (bottom panel) is shown in two single cell clones. CFU, colony forming units/100,000 transfected cells. (d) The colonies obtained for 1 μg/ml doxycycline in c were pooled and passaged in G418-free medium in the presence or absence of doxycycline. After 2 weeks, SEC14L2 protein was undetectable in cells passaged without doxycycline (top panel). These cells died when subjected to G418 selection (bottom panel). Bar graphs show means±SD from at least duplicate experiments.
To examine the temporal and dose-dependent relationship between SEC14L2 expression and HCV permissiveness in Huh-7.5 cells, we established a doxycycline-inducible expression system. Two cell clones with varying levels of SEC14L2 expression were tested for replicon colony formation. The number of G418-resistant colonies correlated with the level of SEC14L2, indicating a dose-dependent effect (Fig. 2c and Extended Data Fig. 3a, b). Withdrawal of doxycycline led to the loss of colonies, suggesting that continuous expression of SEC14L2 is needed to maintain HCV replication (Fig. 2d).
In contrast to wild-type HCV replicons, cell culture-adapted replicons showed a mild to moderate response to SEC14L2 expression (Extended Data Fig. 3c), indicating that cell culture adaptive mutations partially circumvent the need for SEC14L2. This notion is further supported by the fact that wild-type replicons in the presence of SEC14L2 yielded almost as many colonies as were obtained for cell culture-adapted replicons in the absence of SEC14L2. Similarly, replication of the JFH-1 subgenomic replicon, which does not require adaptive mutations, was not enhanced by SEC14L2 (data not shown).
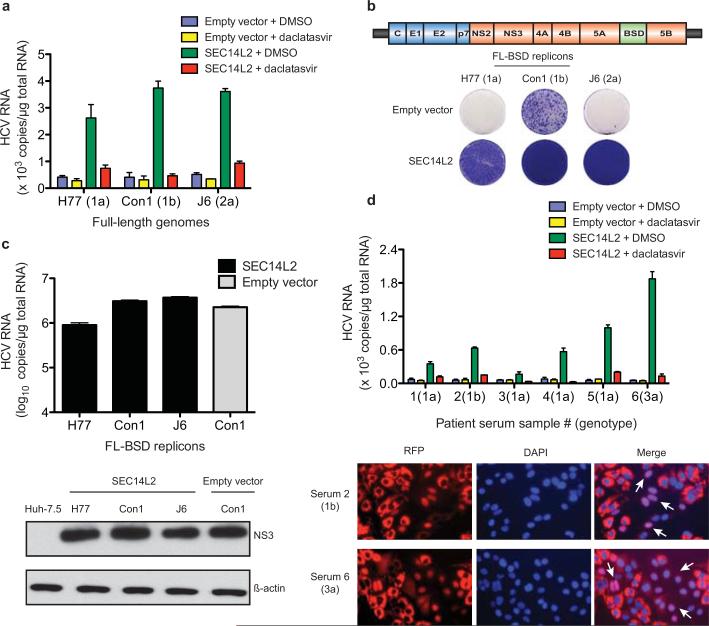
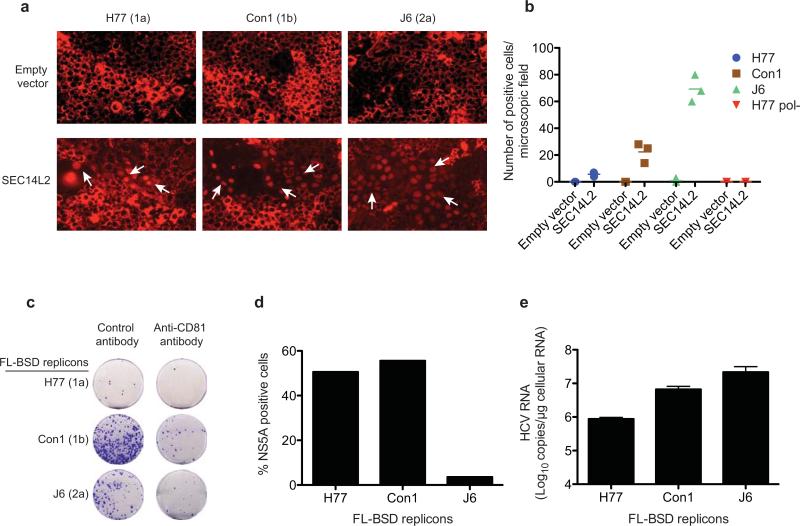
To examine replication of full-length HCV genomes, we took two approaches. First, we performed transient replication assays in Huh-7.5 cells stably expressing an HCV infection-dependent fluorescence relocalization (HDFR) reporter12, with or without SEC14L2. HCV RNA replication in HDFR cells results in nuclear localization of otherwise mitochondrially anchored RFP. When in vitro transcribed full-length HCV RNA genomes were electroporated into SEC14L2-expressing HDFR cells (SEC14L2/HDFR), varying numbers of positive cells were visible after 6 days (Extended Data Fig. 4a, b). These cells had readily detectable levels of HCV RNA that could be suppressed by treatment with daclatasvir, a specific antiviral targeting NS5A (Fig. 3a), confirming authentic HCV replication. As expected, no evidence of viral replication was seen in control HDFR cells.
Figure 3. SEC14L2 allows replication of full-length HCV genomes.
(a) HCV RNA levels 6 days after electroporation of the indicated full-length HCV genomes in HDFR reporter cells transduced with empty vector or SEC14L2. DMSO or daclatasvir (10 nM) was added 3 days prior to cell harvest. (b) Cell colonies obtained for the indicated blasticidin-selectable, full-length (FL-BSD) HCV genomes in Huh-7.5 cells transduced or not to express SEC14L2. (c) The cell colonies obtained in b were pooled and HCV RNA and protein abundance was determined by qRT-PCR (top panel) and immunoblotting with anti-NS3 antibody (bottom panel). (d) Direct infection of SEC14L2-expressing HDFR reporter cells with 6 serum samples from HCV patients. The top panel shows HCV RNA levels. The bottom panel shows the nuclear localization of RFP in individual cells infected with serum 2 (genotype 1b) and serum 6 (genotype 3a). White arrows indicate cells with nuclear RFP. Bar graphs show means±SD from at least duplicate experiments.
For stable replication assays, we inserted a blasticidin S deaminase (BSD) cassette into full-length HCV genomes and introduced them into SEC14L2/Huh-7.5 cells by electroporation. Selection with blasticidin resulted in large numbers of colonies that were positive for viral RNA and protein (Fig. 3b, c). Inoculation of naïve SEC14L2/Huh-7.5 cells with culture medium from these colonies and subsequent selection with blasticidin yielded varying numbers of colonies that were positive for viral RNA and protein, indicating production of infectious virus. Incubation of cells with anti-CD81 antibody prior to inoculation reduced colony formation (Extended Data Fig. 4c-e). Thus, SEC14L2 supported transient and stable replication of HCV genomes lacking adaptive mutations.
Past attempts to infect cultured cells with clinical HCV isolates have largely remained unsuccessful. Patient sera are infectious when inoculated into chimpanzees or human liver-chimeric mice13,14, but do not result in productive infection in hepatoma cells. To test if SEC14L2 facilitates infection with HCV from infected sera, we inoculated SEC14L2/HDFR cells with six high-titer HCV sera of genotypes 1a, 1b and 3a. Large numbers of cells with nuclear RFP were observed for genotype 3a, followed by genotype 1b (Fig. 3d, bottom panel). All serum samples yielded detectable levels of HCV RNA that were sensitive to treatment with daclatasvir (Fig. 3d, top panel). In contrast, control cells did not support HCV RNA replication.
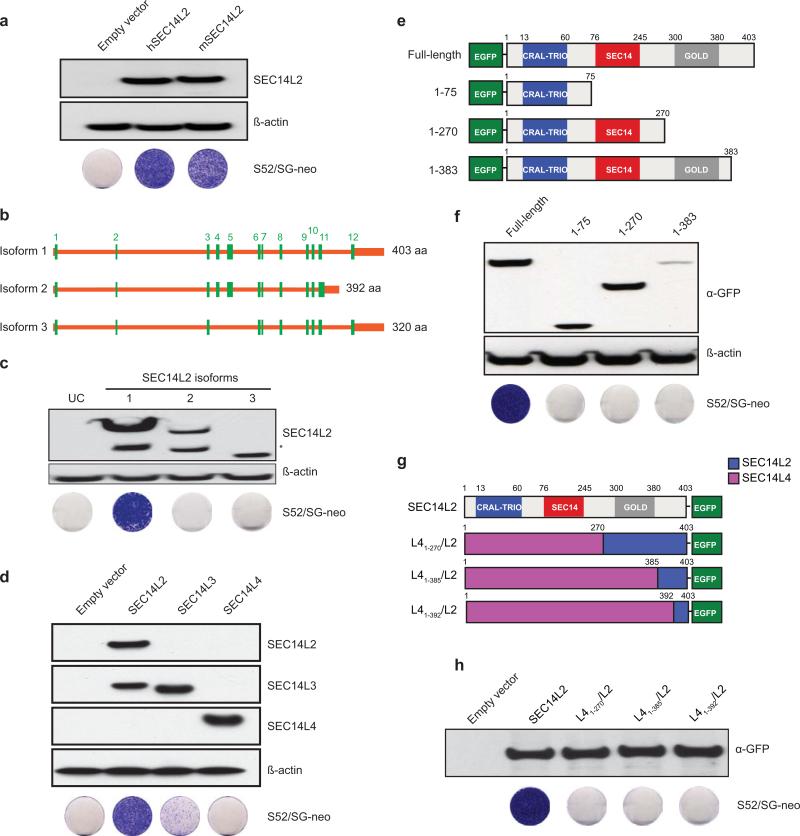
A number of experiments were undertaken to examine SEC14L2-specific determinants important for its HCV replication enhancing effects. Cells expressing murine SEC14L2 were highly permissive to wild-type HCV (Extended Data Fig. 5a). Naturally occurring splice isoforms of SEC14L2 encoding shorter than the full-length proteins, did not facilitate HCV replication (Extended Data Fig. 5b, c). Two other SEC14L2-related proteins have been described in humans: SEC14L3 and SEC14L411, none of which could be detected in Huh-7.5 cells (Extended Data Fig. 5d). While SEC14L3 allowed low level HCV replication, SEC14L4 had no effect (Extended Data Fig. 5d).
SEC14L2 has three domains: an N-terminal CRAL-TRIO domain (aa 12-60) that binds small lipophilic molecules15, a SEC14-like domain (aa 76-245) that contains a hydrophobic ligand-binding pocket, and a C-terminal GOLD domain (aa 300-380) that is postulated to facilitate protein-protein interactions. To test the importance of these domains in facilitating HCV replication, we constructed various deletion mutants (Extended Data Fig. 5e) and SEC14L2/L4 chimeras (Extended Data Fig. 5g). Neither the deletion mutants nor any of the chimeric constructs conferred HCV permissiveness to Huh-7.5 cells (Extended Data Fig. 5f, h), suggesting that multiple SEC14L2-specific determinants are required for HCV replication.
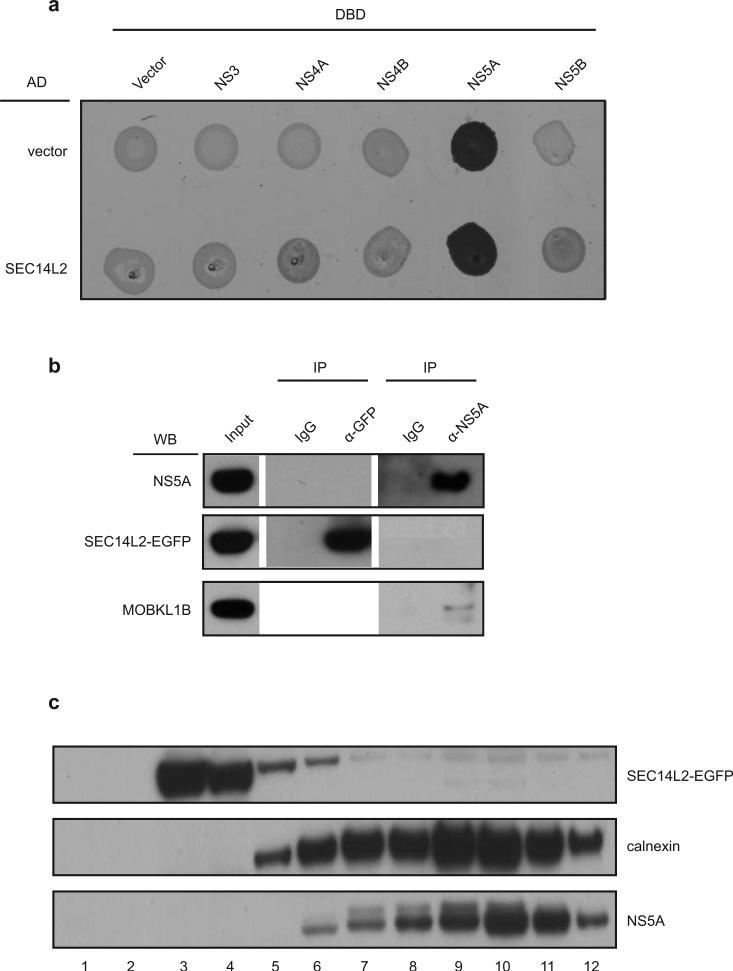
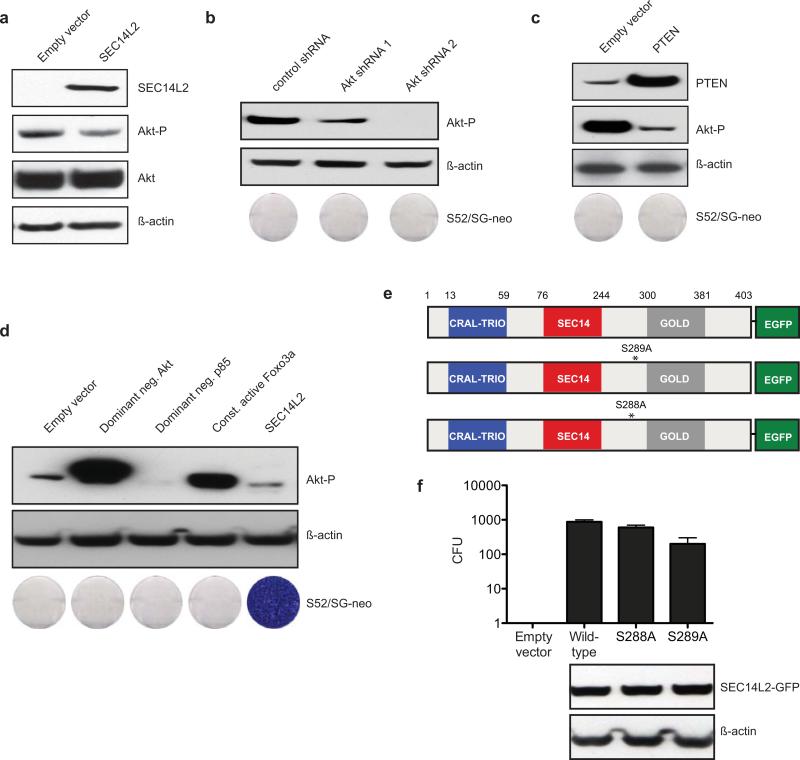
In terms of mechanism, yeast two-hybrid, co-immunoprecipitation and subcellular fractionation did not reveal a physical interaction between SEC14L2 and HCV replicase components (Extended Data Fig. 6a-c). Since SEC14L2 is implicated in the regulation of the PI3K/Akt signaling pathway11,16, cholesterol synthesis17,18, and vitamin E metabolism16,19, we hypothesized that modulation of these pathways might be involved in facilitating HCV RNA replication. As previously reported for prostate cancer cells16, SEC14L2 expression mildly reduced Akt phosphorylation in Huh-7.5 cells (Extended Data Fig. 7a); yet suppression of PI3K/Akt pathway by SEC14L2-independent methods failed to support HCV replication (Extended Data Fig. 7b-d).
SEC14L2 expression transiently stimulates cholesterol synthesis in rat hepatoma cells18. However, we did not see increased cholesterol levels in Huh-7.5 cells stably expressing SEC14L2 (data not shown). Furthermore, a SEC14L2 mutant lacking cholesterolgenic activity18 efficiently supported HCV replication (Extended Data Fig. 7e, f). In contrast, a SEC14L2 deletion mutant capable of stimulating cholesterolgenic enzymes in rat hepatoma cells failed to confer HCV permissiveness to Huh-7.5 cells (1-383 mutant18 in Extended Data Fig. 5e, f). Taken together, SEC14L2 phenotype cannot be attributed to stimulation of the cholesterol synthesis pathway.
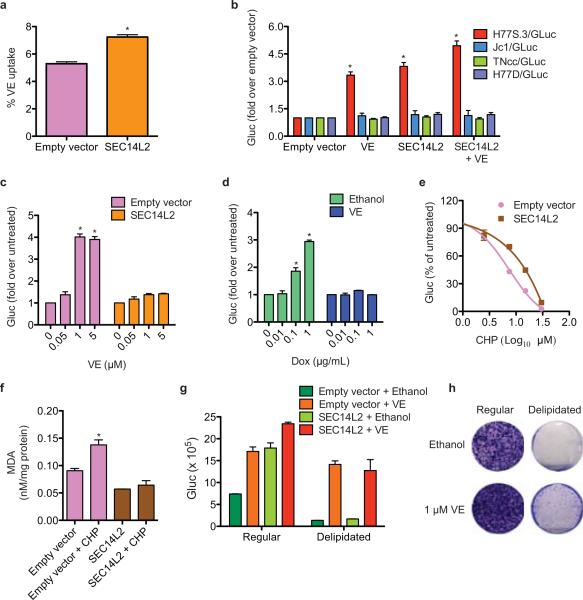
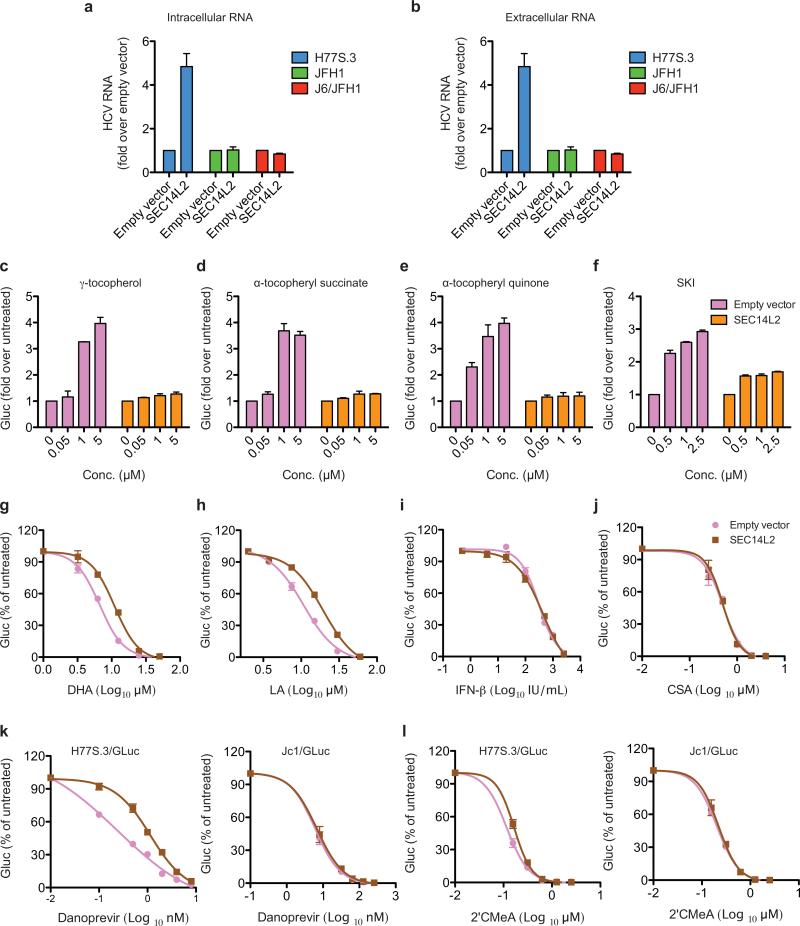
We next examined the role of SEC14L2 as a vitamin E (VE; tocopherol) binding protein16,19. Lipid-soluble antioxidants such as VE inhibit lipid peroxidation and enhance HCV replication20. As described for prostate cancer cells16, SEC14L2 enhanced the accumulation of VE in Huh-7.5 cells (Fig. 4a). Interestingly, HCV isolates resistant to lipid peroxidation (JFH-1, Jc1, H77D, TNcc) and not enhanced by VE20 were insensitive to SEC14L2 expression. In contrast, replication of a lipid peroxidation-sensitive clone, H77S.320, was enhanced to similar levels by VE and SEC14L2 (Fig. 4b and Extended Data Fig. 8a, b). SEC14L2 and VE did not show an additive effect on H77S.3 replication (Fig. 4c,d), suggesting an overlapping mechanism. The replication enhancing effect of other lipophilic anti-oxidants was also markedly suppressed in the presence of SEC14L2 (Extended Data Fig. 8c-f).
Figure 4. SEC14L2 promotes HCV replication by enhancing the antioxidant effect of VE.
(a) Effect of SEC14L2 expression on VE accumulation in Huh-7.5 cells. Results represent the mean±SD from 8 adjacent wells treated with radioactively labeled VE for 4 h. (b) Effect of VE (1 μM) and SEC14L2 on replication of the indicated HCV genomes. (c) Dose-response effects of VE on replication of H77S.3/GLuc in empty vector- and SEC14L2-expressing Huh-7.5 cells. (d) Dose-response effects of SEC14L2 expression on H77S.3/GLuc replication in SEC14L2-EGFP Cl. 1 cells (described in Fig. 2c) in presence or absence of VE. (e) Dose-response effect of cumene hydroperoxide (CHP), a lipophilic oxidant, on replication of H77S.3/GLuc in empty vector- and SEC14L2-expressing Huh-7.5 cells. (f) Intracellular abundance of malondialdehyde (MDA), an end product of lipid peroxidation, in empty vector and SEC14L2-expressing Huh-7.5 cells incubated for 16 h with 15 μM CHP. (g) H77S.3/GLuc replication and (h) S52/SG-neo colony formation in empty vector and SEC14L2-expressing Huh-7.5 cells grown in regular or delipidated medium. Since delipidated serum is suboptimal for hepatoma cell proliferation, the colonies obtained were small and slow growing. Results represent means±SD from 2 (d,f) or 3 (b,c,e,g) biological replicates. * P < 0.01 by one-tailed, Mann-Whitney test (a,b,c,d) or one-tailed, paired t-test (f).
SEC14L2 also reduced the inhibitory effect of lipophilic oxidants, but not that of interferon-alpha or cyclosporine A, on HCV replication (Fig. 4e and Extended Data Fig. 8g-j), indicating selective protection against lipid peroxidation. Consistent with this hypothesis, SEC14L2 expression lowered the intracellular abundance of malondialdehyde, a secondary product of lipid peroxidation (Fig. 4f). Furthermore, SEC14L2 phenocopied the reported effects of other lipid peroxidation inhibitors such as VE and sphingosine kinase inhibitor (SKI)20 by enhancing the 50% effective concentration (EC50) of NS3 protease (Danoprevir) and NS5B polymerase (2’CMeA) inhibitors against H77S.3, but not against lipid peroxidation-resistant Jc1 (Extended Data Fig. 8k-l).
SEC14L2 no longer supported HCV replication when cells were cultured in medium containing FBS that had been depleted of lipid-soluble materials including VE (delipidated FBS) (Fig. 4g, h), an effect that was reversed by VE supplementation (Fig. 4h), suggesting that antioxidative effect of SEC14L2 is mediated, at least in part, through VE. Since delipidated FBS lacks many other lipids, we cannot entirely rule out the involvement of other potential SEC14L2 ligands in HCV replication. Recently, sphingosine kinase-2 (SPHK2) was also shown to promote lipid peroxidation and inhibit replication of non-JFH1 isolates20. However, neither mRNA nor protein levels of SPHK2 were diminished by SEC14L2 expression. In addition, inhibition of SPHK2 with sphingosine kinase inhibitor (SKI) or by stable expression of dominant-negative SPHK2G212D21 did not allow HCV RNA replication in Huh-7.5 cells (data not shown), suggesting that the SEC14L2 effect is not mediated by inhibiting SPHK2.
Collectively, these data imply that SEC14L2 promotes HCV replication by enhancing VE-mediated inhibition of lipid peroxidation. As expected, RNA viruses insensitive to lipid peroxidation20 did not respond to SEC14L2 expression (Extended Data Fig. 9). Interestingly, although VE supplementation significantly enhanced SEC14L2-mediated replicon colony formation (Extended Data Fig. 10), it failed to support colony formation on its own (data not shown), suggesting that SEC14L2 might have additional functions. SEC14L2-mediated enrichment of VE at specific sites may be required to support stable HCV replication, but further studies will be required to elucidate the precise mechanism by which SEC14L2 facilitates persistent replication of wild-type HCV.
This study represents an important step forward in developing a culture system where HCV can be propagated without the need for adaptive mutations. All human hepatoma cell lines that we tested became permissive to wild-type HCV genomes when engineered to express SEC14L2. Most remarkably, these cells also supported HCV replication directly from patient sera. The low levels of transient replication observed in these cells may reflect inherently low replication potential of the wild-type HCV isolates or lack of additional host factors in Huh-7.5 cells. Moreover, although shRNA screening did not yield hits in our hands, the expression of restriction factors cannot be entirely ruled out. SEC14L2-expressing Huh-7.5 cells provide an excellent platform to test these possibilities. We anticipate that the ability of SEC14L2 to enable replication of non-adapted HCV in cell culture will open up new avenues for studying HCV biology, including drug resistant clinical variants that are emerging even in the new era of highly effective antiviral therapies22.
Methods
HCV constructs
H77/SG-neo (genotype 1a), Con1/SG-neo (genotype 1b), S52/SG-neo (genotype 3a), ED43/SG-neo (genotype 4a), and SA1/SG-neo (genotype 5a) have been described1,2,23,24. To generate J6/SG-neo, the region of J6 full-length genome encoding the structural proteins was replaced with a cassette containing the NPTII gene and the EMCV IRES. Con1/SG-Feo, J6/SG-Feo, S52/SG-Feo, ED43/SG-Feo, and SA1/SG-Feo were generated by replacing the NPTII gene of neo replicons with a chimeric gene encoding a fusion protein of firefly luciferase and NPTII, as described elsewhere2. A high copy number derivative of full-length H77 (pUC/H77FLX) and its replication-defective mutant (pUC/H77FL-X pol-, referred to here as H77/AAG) have been described25. Molecular clones of Con1 and J6 have been previously described and shown to be infectious in chimpanzee8,26. We generated a high copy number derivative of Con1 consensus cDNA in pUC19 vector backbone, pUC/Con1FL-X, that has a 5’ T7 promoter and a unique XbaI run off site for production of full-length RNA transcripts. To generate blasticidin-selectable full-length replicons from these clones (H77/FLBSD, Con1/FL-BSD, and J6/FL-BSD), we cloned a blasticidin s deaminase (bsd) gene flanked by the NS3/4A protease cleavage site between NS5A and NS5B. The cell culture-adapted subgenomic replicons from genotype 1a ((H/SG-neo(I), H/SG-neo(D+I), H/SG-neo(L+8)), 1b ((Con1/SG-neo(SI), Con1/SG-neo(PS), Con1/SG-neo(RG), Con1/SG-neo(PSRG)), 3a ((S52/SG-neo(I), S52/SG-neo(VI), S52/SG-neo(AII)), and 4a ((ED43/SG-neo(G), ED43/SG-neo(KG), ED43/SG-neo(YG)) have been previously described2,27,28. H77S.3, H77S.3/GLuc2A (referred to here as H77S.3/GLuc), H77D/GLuc, and TNcc/GLuc were a kind gift from Dr. Stanley M. Lemon20,29. JFH-1, J6/JFH1, and Jc1FLAG2(p7-nsGLuc2A) (referred to here as Jc1/GLuc) are previously described30-32.
Lentiviral pseudoparticles
To generate pseudoparticles for delivery of cDNA library or individual ORFs, 4 × 106 Lenti-X 293T cells (Clontec) in 10-cm dishes were co-transfected with plasmids encoding (1) pLOC proviruses (2) HIV-1 gag-pol/delta neo and (3) VSV-G in a ratio of 1/0.8/0.2, respectively. To generate pseudoparticles for delivery of shRNA library or individual shRNAs, 4 × 106 Lenti-X 293T cells were co-transfected with plasmids encoding (1) pGIPZ proviruses (2) pMD.G and (3) pCMVR8.91 in a ratio of 1/0.5/0.5. A total of 12 μg RNA was transfected using 24 μl jetPEI DNA transfection reagent (Polyplus Transfection, France). Transfections were carried out for 6 h, followed by a medium change to DMEM containing 10% FBS. Culture medium was collected at 48 h and 72 h, pooled, filtered through 0.45 μm filter, and stored at −80°C. Target cells seeded into 6-well plates at a density of 4.5 × 105 cells/well were transduced with pseudoparticles by spinoculation at 1000 x g for 45 min at 37°C in a medium containing 10%FBS, 20 mM HEPES, and 4 μg/ml polybrene.
shRNA/cDNA screening
For shRNA screening, Huh-7.5 cells were transduced with the pooled pGIPZ lentiviral shRNA library targeting 18,000 genes with ~ 60,000 shRNA. Briefly, Huh-7.5 cells (1.8 × 107) were infected to achieve an average representation of 100 cells per shRNA. Transductions were carried out at a low transduction efficiency of 0.3 to reduce the probability of multiple integrants per cell. After 48 h, shRNA-containing cells were selected with puromycin (5 μg/ml) for 7 days. We prepared three independent cell populations transduced with the shRNA library and one cell population transduced with the control shRNA. For cDNA screening, Huh-7.5 cells were transduced with a pool of pLOC lentiviruses containing ~ 7000 sequence-verified human cDNAs. Briefly, Huh-7.5 cells (2.1 × 106) were transduced to achieve a low transduction efficiency of 0.3 and an average representation of 100 cells per cDNA. After 48 h, cDNA-expressing cells were selected with blasticidin (5 μg/ml) for 10 days. We generated three independent cell populations transduced with the cDNA-containing lentiviruses and one control cell population transduced with the RFP-containing lentiviruses. The puromycin- and blasticidin-resistant cell populations were electroporated with the wild-type subgenomic replicons derived from genotype 3a (S52/SG-neo), genotype 4a (ED43/SG-neo), or as a negative control with a replication defective RNA (S52/SG-neo GNN), and selected with G418 (500 μg/ml) for 3 weeks.
Identification of integrated cDNAs
Genomic DNA was extracted from 45 replicon colonies using DNeasy Blood and Tissue Kit (Qiagen), following the manufacturer's instructions. Twenty ng DNA was subjected to PCR amplification using Expand High Fidelity PCR System (Roche) and a pair of primers complementary to the universal sequences flanking cDNAs (Forward primer 5’-CTCCATAGAAGACACCGAC-3’, reverse primer 5’- GGTTCATTAGCTAGCAACCACT-3’). Touchdown PCR conditions were used to avoid non-specific bands: Initial denaturation at 94°C for 2 min, 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at temperature starting from 62°C and decreasing by 0.5°C every cycle until it reached 56°C, and extension for 4 min at 72°C. The resulting PCR products were resolved on 1% agarose gel. Surprisingly, despite earlier transduction at low multiplicity, multiple bands were observed for most colonies. However, 3 colonies yielded a single PCR product encoding the same gene: SEC14L2. We then designed SEC14L2-specific primers spanning exon-exon junctions to examine the integration of this cDNA in the remaining 42 cell colonies.
Cells and reagents
Huh-7, Huh-7.5, Hep3B/miR12233, 293T, and BHK cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 0.1 mM nonessential amino acids (NEAA), at 37°C in a humidified atmosphere containing 5% CO2. All cells used in this study were tested for and found free of mycoplasma contamination. Medium containing 500 μg/ml G418 (Sigma Aldrich, MO) was used to select and maintain cells harboring selectable HCV subgenomic replicons. Arrayed precision lentiORFs were purchased from GE Healthcare (http://dharmacon.gelifesciences.com/mammalian-orf/precision-lentiorfs/) and pooled as previously described34. The GIPZ Human Whole Genome shRNA library (# RHS6037) was also from GE Healthcare. The following primary antibodies were used in this study: SEC14L2 mouse monoclonal antibody clone 4H2 (Origene, MD), SEC14L2 rabbit polyclonal antibody (Acris Antibodies, CA), SEC14L3 rabbit polyclonal antibody (Abcam, MA), SEC14L4 rabbit polyclonal antibody (Creative BioMart, NY), HCV NS5A mouse monoclonal antibody clone 9E1031, HCV NS3 mouse monoclonal antibody clone 2E3 (a gift from BioFront Technologies, FL), GFP rabbit polyclonal antiserum35, pan-Akt rabbit monoclonal antibody clone C67E7 (Cell Signaling Technology), Phospho-Akt (Ser473) rabbit monoclonal antibody clone D9E (Cell Signaling Technology), PTEN mouse monoclonal antibody clone 6H2.1 (Millipore, MA), CD81 human antibody clone JS-81 (BD Biosciences, CA), calnexin rabbit polyclonal antibody (Abcam, MA), and ß-actin HRP-conjugated mouse monoclonal antibody clone AC-40 (Sigma). α-tocopherol, γ-tocopherol, tocopherol succinate, docohexaenoic acid, and 5’ azacytidine were purchased from Sigma-Aldrich. D-alpha tocopheryl quinone, and cumene hydroperoxide were from Santa Cruz Biotechnology. Linoleic acid was from Cayman Chemical. 2’CMeA and daclatasvir were from Merck Research Laboratories (PA, USA) and Selleck Chemicals (TX, USA), respectively. [14C]RRR-α-tocopherol was a generous gift from Danny Manor (Case Western Reserve University, OH).
Plasmids
Human SEC14L2 or RFP was cloned in pLOC plasmid (http://lentivirus.imb.uq.edu.au/downloads/pdfs/LOC_Technical_Manual.pdf) to obtain pSEC14L2/BlastR or pRFP/BlastR, respectively. BlastR gene from pSEC14L2/BlastR and pRFP/BlastR was replaced with PuroR to obtain the puromycin-selectable plasmids, pSEC14L2/PuroR and pRFP/PuroR, respectively. pSEC14L2-EGFP/BlastR was generated by removing the TurboGFP tag from pSEC14L2/BlastR and fusing the C-terminal end of SEC14L2 in-frame with EGFP. Plasmids containing dominant negative Akt1 (pLNCX myr HA Akt1; Addgene plasmid 9005), dominant negative p85 subunit of PI3K (pWZL-neo delta-p85; Addgene plasmid 10888), SPHK2 (pDONR223-SPHK2; Addgene plasmid 23714), and constitutively active FOXO3a (HAFOXO3a TM; Addgene plasmid 1788) were purchased from Addgene. Dominant-negative SPHK2G212D was generated by PCR-based mutagenesis to substitute aspartic acid for glycine at amino acid position 212. Expression plasmids for SEC14L4 and mouse SEC14L2 were from Origene. The SEC14L3 expression plasmid was obtained from CCSB-Broad lentiviral expression library (Thermo Scientific). The open reading frames (ORFs) from these plasmids were transferred to pLOC lentiviral vector (Thermo Scientific) for stable expression in Huh-7.5 cells. Akt shRNA plasmids (clones NM_005163.1628s1c1 and NM_005163.x-642s1c1) were purchased from Sigma Aldrich. The lentivirus packaging HIV-1 gag-pol plasmid was sequenced by the primer-walking method and modified by removing the NPTII gene to obtain a neomycin-sensitive derivative, HIV-1 gag-pol/delta neo.
Doxycycline-inducible SEC14L2 expression system
Tet-On pTRIPZ vector (http://dharmacon.gelifesciences.com/uploadedfiles/resources/ptripz-induciblelentiviral-manual.pdf) was engineered to express the EGFP or SEC14L2-EGFP fusion protein under doxycycline-inducible promoter, resulting in the generation of pEGFPind and pSEC14L2-EGFPind, respectively. Lentivirus pseudoparticles were generated as described above and used to infect Huh-7.5 cells at a low multiplicity of 0.3 to reduce the probability of getting multiple integrants per cell. The untransduced cell population was eliminated by selection with puromycin (5 μg/ml) for 7 days. To obtain single cell clones, GFP-negative cells were sorted by flow cytometry at one cell per well in 96-well plates and grown to confluence. After testing multiple cell clones for absence of background GFP expression and responsiveness to doxycycline, 2 cell clones, SEC14L2-EGFP Cl. 1 and SEC14L2-EGFP Cl. 2 were selected for subsequent experiments.
Viruses and plaque formation assays
The following reporter viruses were used for FACS analysis: YFV-Venus12 (derived from YF17D-5’C25Venus2AUbi), DENV-GFP36 (generated from IC30P-A, a full-length infectious clone of strain 16681), SINV-GFP37 (derived from pS300/pS300-GFP), and RRV-GFP38 (generated from the T48 strain of RRV). Plaque formation assays were performed by using the following non-reporter viruses: YFV (17D)39 and DENV (16681)40. Infectious titers of YFV were determined by plaque assay on 80-90% confluent monolayers of Huh-7.5 cells in 6-well plates. Briefly, cells were incubated with 10-fold serial dilutions (prepared in DMEM containing 10% FBS) for 1 h at 37°C, and overlaid with 1.2% Avicel (FMC BioPolymer, PA) in DMEM containing 10% FBS. After 96h, the monolayers were fixed with 7% formaldehyde and plaques visualized by crystal violet staining. DENV titers were determined by plaque assay on 80-90% confluent monolayers of BHK cells in 12-well plates. Briefly, cells were incubated with 10-fold serial dilutions (prepared in FBS-free MEM) for 2 h at 32°C, and overlaid with 2.25% Avicel in MEM containing 5% FBS. Cells were further incubated for 5 days at 37°C, followed by fixation with 7% formaldehyde and staining with crystal violet.
HCV RNA synthesis and transfection of cultured cells
Plasmids carrying full-length or subgenomic HCV genomes were linearized and subjected to in vitro transcription using the T7 RiboMAX Express Large Scale RNA Production System (Promega, WI). Complete removal of template DNA was ensured by an additional on-column treatment with RNase-free DNase and the quality of in vitro transcribed RNA was examined by agarose gel electrophoresis. RNA transcripts were introduced into cells either by electroporation using a BTX ElectroSquarePorator as described previously2, or by transfection using TransIT-mRNA transfection kit (Mirus). For transfection, cells plated in 12-well plates were grown overnight to approximately 80% confluency. The culture medium was replaced with a fresh complete growth medium 30 min before transfection. The transfection complexes, prepared by adding 1 μg of HCV RNA, 2 μl of mRNA boost reagent and 2 μl of transfection reagent to 100 μl Opti-MEM in sterile tubes, were incubated for 3 min at room temperature and added drop wise to each well.
Colony formation assay
Colony formation efficiency was measured as described previously2. To test replicon colony formation in cells containing doxycycline-inducible SEC14L2, cells were treated with various concentrations of doxycycline for 24 h before transfection with S52/SG-neo. Forty-eight hours later, cells were subjected to G418 selection and fed with fresh G418 and doxycycline every 2 days. The resulting cell colonies were stained with crystal violet.
Luciferase assays
Cells transfected with HCV RNA encoding Gaussia luciferase (GLuc) were exposed to compounds 6 h after transfection. The culture medium harvested between 48-72 h after adding compounds was prepared by using 1X Renilla luciferase assay lysis buffer (Promega) and amounts of secreted GLuc were measured with the Renilla luciferase assay system (Promega) and Synergy NEO HTS Multi-Mode Microplate Reader (BioTek). To test the dose-response effects of SEC14L2 expression on H77S.3/GLuc replication in presence and absence of VE, SEC14L2-EGFP Cl. 1 cells were treated for 24 h with ethanol (vehicle) or 1 μM VE in presence of the indicated concentrations of doxycycline. Cells were then transfected with H77S.3/GLuc RNA for 6 h and incubated with VE and doxycycline for another 72 h before measuring the amounts of secreted GLuc.
HCV replication in delipidated medium
Huh-7.5 cells transduced with empty vector or SEC14L2 were passaged 3 times in the culture medium supplemented with 10% of regular or delipidated FBS. These cells were then transfected with H77S.3/GLuc or S52/SG-neo for 6 h, followed by medium change to ethanol- or VE-supplemented medium (1 μM). H77S.3/GLuc replication was examined by measuring the secreted GLuc activity at 72 h. For S52/SG-neo colony formation, the transfected cells were subjected to G418 selection at 48 h after transfection and fed with fresh G418 every 2 days. After 3 weeks of selection, the resulting cell colonies were stained with crystal violet.
Inoculation of cells with HCV infected sera
Six high-titer serum samples (5.98 × 106- 3.2 × 107 IU/ml), four of genotype 1a and one of each of genotypes 1b and 3a were obtained from HCV patients at the Mount Sinai Hospital, as approved by the Institutional Review Board of the Mount Sinai Hospital. Informed consent was obtained from all patients. For infection, 25 μl of patient serum diluted in 500 μl of DMEM containing 10% FBS, 1X NEAA, and 50 ng/ml EGF was added to cells grown overnight in 24-well plates. After overnight incubation with serum, cells were rinsed twice with 1X PBS and fed with 500 μl fresh medium containing 10% FBS, 1X NEAA, and 50 ng/ml EGF. Four days later, the cells were exposed to DMSO or daclatasvir (10 nM) for 48 h, after which they were harvested for qPCR or fixed with 4% PFA/PBS to test the nuclear localization of RFP.
Construction of SEC14L2 isoforms 2 and 3
SEC14L2 has three splice isoforms. Isoform 1, identified in the screen, is 403 aa long, comprised of 12 exons. Isoform 2 is 392 aa that shares the N-terminal 360 amino acids with isoform 1 but alternative splicing at exon 10 generates a unique C-terminus. Isoform 3 is generated by skipping of exons 4 and 5, resulting in a 320 aa protein. We used isoform 1 as a PCR template to construct the other two isoforms. Isoform 2 was generated by multiple rounds of PCR with the following primers: 1st round (Forward primer 5’-GCTCGGATCCACTAGTCCAGTG-3’, reverse primer 5’-AGGCAGAGATACTTACAGATGCCAGGATCAC-3’), 2nd round (Forward primer 5’-GCTCGGATCCACTAGTCCAGTG-3’, reverse primer 5’-TTCAAGGCATTGCCAAGGCAGAGATACTTAC-3’), 3rd round ((Forward primer 5’-GCTCGGATCCACTAGTCCAGTG-3’, reverse primer 5’-GAAAGCTGGACATGGGGCTTCAAGGCATTGC-3’ ), 4th round (Forward primer 5’-GCTCGGATCCACTAGTCCAGTG-3’, reverse primer 5’-GAGGAACCTCACAGGCAGAAAGCTGGACATG-3’), 5th round (Forward primer 5’-GCTCGGATCCACTAGTCCAGTG-3’, reverse primer 5’-AAAAATCCATGGAGGAAGAGGAACCTCACAG-3’ ), and 6th round (Forward primer 5’-GCTCGGATCCACTAGTCCAGTG-3’, reverse primer 5’-TAGCTAGCTCATTAACACTCAGAGCCAAAAATCCATGGAG-3’). The resulting PCR product was digested with BamHI and NheI and replaced with the corresponding fragment of pLOC-SEC14L2 isoform 1. To generate Isoform 3, the coding region for amino acids 55-140 of isoform 1 was deleted by a two-step PCR. In the first step, PCR was carried out with primers 5’-GCTCGGATCCACTAGTCCAGTG-3’/5’-CTTCCTCCCCAACTTCCGGAGCATGGCCT-3’ and 5’-AGGCCATGCTCCGGAAGTTGGGGAGGAAG-3’/5’-TAGCTAGCTCATTATTTCGGGGTGCCTGC-3’. Since the 5’ end of the internal reverse primer was complementary to the 5’ end of the internal forward primer, the two PCR products obtained in the first step were pieced together by a second round of PCR with primers 5’-GCTCGGATCCACTAGTCCAGTG-3’ and 5’- TAGCTAGCTCATTATTTCGGGGTGCCTGC-3’. The resulting PCR product was digested with BamHI and NheI and used to replace the corresponding fragment of pLOC-SEC14L2 isoform 1.
Yeast two-hybrid assays
Yeast two-hybrid binding assays were performed as described previously41, using the Matchmaker GAL4 Yeast Two Hybrid 3 system (Clontech, Mountain View, CA). SEC14L2 was cloned into pGADT7 (activation domain) vector (Clontech) using 5’ EcoRI and 3’ BamHI cloning sites. HCV nonstructural protein encoding genes were cloned into pGBKT7 (DNA binding domain) vector (Clontech) as described previously41. Saccharomyces cerevisiae AH-109 was co-transformed with SEC14L2 and individual viral gene containing plasmids and selected by culturing on Sabourand dextrose broth plates in the absence of Leu and Trp for 3 days at 30°C. For testing protein-protein interactions, ~100 colonies were pooled, resuspended and selected on Sabourand dextrose broth plates (minus Leu, minus Trp, minus Ade, minus His) for 3 days at 30°C.
Co-immunoprecipitation assay
SEC14L2-EGFP/Huh-7.5 cells (1 × 107) harboring wild-type Con1/SG-neo were harvested, washed 3 times with 1 ml of ice-cold phosphate-buffered saline (PBS), and suspended in 0.5 ml of lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM MgCl2 and 1% Triton X-100) supplemented with EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland), DNase I (50 U/ml) and RNase A (10 μg/ml). Lysates were clarified by centrifugation (16,000x g, 10 min, 4°C) and incubated with the mouse monoclonal anti-NS5A antibody (9E10)31 or the rabbit polyclonal anti-GFP antibody42 captured on Dynabeads® Protein A (Invitrogen, Carlsbad, CA) for 2 h at 4°C. The beads were then washed 3 times in lysis buffer and the bound proteins were released by boiling in SDS-PAGE loading buffer and analyzed by immunoblotting.
Subcellular fractionation
SEC14L2/Huh-7.5 cells (2 × 107) harboring Con1/SG-neo were washed twice with PBS, scraped into PBS, and pelleted by centrifugation at 1000x g for 5 min at 4°C. Cells were resuspended in 150 mM sucrose containing 10 mM HEPES and protease inhibitor cocktail without EDTA (Roche Diagnostic, Manheim, Germany) and lysed by cell disruption with a Parr Bomb at 900 psi. Nuclei and unbroken cells were removed by centrifugation at 1000x g for 10 min at 4°C. The supernatant was carefully removed and overlaid onto a discontinuous sucrose gradients (460 μl of 56% sucrose, 920 μl of 50% sucrose, 1.38 ml of 45% sucrose, 2.3 ml of 40% sucrose, 2.3 ml of 35% sucrose, 1.38 ml of 30% sucrose, 460 μl of 20% sucrose, and 1ml of 8% sucrose containing 10 mM HEPES, protease inhibitor and trypsin inhibitor) and centrifuged at 200,000x g for 18 h at 4°C in a SW41Ti rotor. 12 fractions were collected from the top and SEC14L2, calnexin, and HCV NS5A were detected by immunoblotting.
Vitamin E uptake assay
Stable Huh-7.5 cell populations expressing SEC14L2 or RFP were seeded into 24-well plates at a density of 5 × 104 cells/well and grown overnight. [14C]RRR-α-tocopherol pre-mixed with FBS was then added to the culture medium. After 4 h of incubation at 37°C, the media was removed, the cells washed with DMEM and then lysed in 20 mM HEPES (pH 7.4), 1 mM EDTA, 150 mM NaCl, 1% NP40, 20 mM sodium fluoride, 20 mM β-glycerophosphate, 1 mM sodium vanadate, and 200 μM PMSF. The radioactivity in cell lysate and culture medium was measured in a scintillation counter, and vitamin E (VE) accumulation was calculated as follows:
The values obtained were then normalized to the total protein and presented as % VE uptake.
Measurement of lipid peroxidation
1.5 × 106 cells seeded into 100 mm dishes were treated overnight with cumene hydroperoxide (15 μM) and malondialdehyde (MDA) levels were measured by a previously described classical method43. Briefly, cell monolayers were washed with 1X PBS, scrapped into 1X PBS and pelleted by centrifugation at 1000x g for 5 min at 4°C. The cell pellet was resuspended in 110 μl PBS and incubated on ice for 30 min. 10 μl of cell suspension was taken for protein measurement and the rest was used for MDA quantification. Briefly, 100 μl of cell suspension was mixed with 100 μl of 10% SDS, followed by addition of freshly made 100 μl of 0.75% Thiobarbituric acid, 30% trichloroacetic acid, and 0.5N HCl and incubated at 95°C for 15 min. The reaction was stopped by incubating the mixture on ice for 15 min, followed by centrifugation for 5 min at 13,500x g. 100 μl of each sample was transferred to a clear bottom black 96-well microplate. The fluorescence at 550 excitation/530nm was measured by reference to a standard curve prepared with 0 to 10 μM MDA.
Statistical analysis
No statistical methods were used to predetermine sample size. Statistical significance was tested using non-parametric, one-tailed Mann-Whitney test (for two-way comparisons) and one-tailed paired t-test (for multiple comparisons) in Prism v5.0 (GraphPad). A value of p < 0.05 was considered significant.
Extended Data
Extended Data Figure 1. Cured replicon cell clones supported replication of wild-type HCV.
Six of the 45 cell colonies obtained from cDNA screen and one control cell clone, S52 (adapted) that carried a cell culture-adapted HCV subgenomic replicon S52/SG-neo(AII), were treated for 15 days with a combination of an HCV NS5A inhibitor (daclatasvir, 1 nM) and an NS5B polymerase inhibitor (2’CMeA, 1 μM) to eliminate the replicating viral genomes. These cells were then transfected with the indicated wild-type HCV replicons and selected with G418 for 3 weeks. The resulting cell colonies were stained with crystal violet. Replication defective replicon, S52 GNN, with a catalytically inactive mutation in the NS5B polymerase served as a negative control.
Extended Data Figure 2. The ectopic expression of SEC14L2 confers HCV permissiveness to human hepatoma cells.
(a,b) SEC14L2 is not expressed in most cell lines. (a) SEC14L2 mRNA levels were measured in the indicated cells by qPCR, and the values were normalized to those of the housekeeping gene, RPS11. Shown is the fold difference from fetal hepatocytes. The results are plotted as mean±SEM of 2 different cell stocks. (b) SEC14L2 protein expression in the indicated cells was determined by immunoblotting with SEC14L2 rabbit polyclonal antibody. ß-actin is included as a loading control. (c) SEC14L2-expressing Huh-7.5 cells were transfected with the indicated wild-type subgenomic replicons. After 4 weeks of selection, HCV RNA was sequenced in 6 individual colonies from each of the replicons. HCV RNA levels in each of these colonies were determined by qRT-PCR. (d) Hep3B/miR-122 and Huh-7 cells transduced with SEC14L2 or empty vector were transfected with in vitro transcribed RNA from the indicated wild-type replicons and selected for 3 weeks with G418 (500 μg/ml). The resulting cell colonies were stained with crystal violet. S52 GNN with a catalytically inactive mutation in NS5B polymerase was included as a negative control.
Extended Data Figure 3. Effect of SEC14L2 expression on the replication of wild-type and cell culture-adapted HCV subgenomic replicons.
(a,b) SEC14L2 expression enhances replication of wild-type HCV RNA in a dose-dependent manner. (a) Huh-7.5 cells were transduced with lentiviruses encoding SEC14L2-EGFP fusion protein under a doxycycline-inducible promoter and flow cytometry was performed to obtain single cell clones. Two cell clones selected for downstream analysis were treated with the indicated concentrations of doxycycline for 24 h, followed by flow cytometry to determine the number of EGFP positive cells. Mean fluorescence intensities (MFI) of EGFP are shown at the top of each box. (b) S52/SG-neo colony formation in cells described in a. The results were confirmed by 2 independent transfections. (c) SEC14L2 expression enhances replication of cell culture-adapted HCV replicons to varying extents. Colony formation efficiency of the indicated subgenomic replicons in empty vector- and SEC14L2-expressing Huh-7.5 cells is plotted as CFU/100,000 transfected cells. Results represent mean±SEM from 2 independent transfections.
Extended Data Figure 4. SEC14L2 expression enables replication of wild-type full-length HCV genomes and production of low levels of infectious virus particles.
(a) HDFR reporter cells transduced with SEC14L2 or empty vector were transfected with in vitro transcribed RNA from H77, Con1, and J6 full-length genomes. Live cell images were captured 6 days after transfection. White arrows indicate the cells with nuclear RFP. (b) The numbers of cells exhibiting nuclear RFP were counted in 3 random microscopic fields at day 6 post-transfection. H77 pol-, with a catalytically inactive mutation in NS5B polymerase, was used as a negative control. (c,d,e) Infectious virus particles are produced from SEC14L2/Huh-7.5 cells harboring selectable full-length HCV genomes. (c) Huh-7.5 cells stably expressing SEC14L2 were incubated at 37°C for 30 min with anti-CD81 antibody or control IgG, and inoculated with the culture medium from SEC14L2/Huh-7.5 cells harboring blasticidin-selectable, full-length (FL-BSD) H77, Con1, and J6 genomes, described in Fig. 3b. After 72 h, selection with blasticidin (2.5 μg/ml) was imposed and the colonies obtained after 3 weeks were stained with crystal violet. (d) The cell colonies obtained in c were pooled and stained with anti-NS5A antibody (9E10 clone). The percentages of positive cells from 2 independent experiments are plotted. As previously described44, 9E10 antibody did not detect NS5A from the J6 isolate. (e) The cell colonies obtained in c were pooled and HCV RNA levels were measured by qPCR. The values are plotted as mean±SEM of 2 independent experiments.
Extended Data Figure 5. Only full-length SEC14L2 supports replication of wild-type HCV.
(a) Huh-7.5 cells stably expressing human or murine SEC14L2 (93% identity) were lysed and protein expression was confirmed by immunoblotting. These cells were then transfected with S52/SG-neo and selected with G418. The resulting cell colonies were stained with crystal violet. (b,c) Only isoform 1 of SEC14L2 supports HCV RNA replication. The SEC14L2 gene is comprised of 12 exons and results in 3 alternatively spliced transcript variants encoding 3 different protein isoforms. Isoform 1 was identified in cDNA screening. (b) Schematic representation of SEC14L2 isoforms. Coding exons are shown as green blocks and the amino acid length of each protein is shown on right. (c) Cell lysates from Huh-7.5 cells stably expressing 3 SEC14L2 isoforms were analyzed by 4-12% SDS-PAGE and immunoblotting was performed with SEC14L2 mouse monoclonal antibody. The bands highlighted with asterisks might be a cleavage product of SEC14L2. UC, untransduced cells. These cells were transfected with S52/SG-neo and selected with G418 for 3 weeks. The resulting cell colonies were stained with crystal violet. (d) SEC14L3 and SEC14L4 (86% and 80% amino acid similarity to SEC14L2, respectively)11 are not expressed in Huh-7.5 cells. Cell lysates from Huh-7.5 cells stably expressing SEC14L2, SEC14L3, or SEC14L4 were analyzed by 4-12% SDS-PAGE followed by immunoblotting with the indicated antibodies (The signal generated by SEC14L3 antibody in SEC14L2-expressing cells most likely reflects the cross-reactivity of SEC14L3 antibody). These cells were then tested for their ability to support replication of S52/SG-neo. (e,f) Deletion mutants of SEC14L2 do not support HCV RNA replication. (e) Schematic representation of N-terminal EGFP-tagged SEC14L2 deletion mutants. (f) Cell lysates from Huh-7.5 cells stably expressing the full-length SEC14L2 and the deletion mutants were analyzed by 4-12% SDS-PAGE followed by immunoblotting with the indicated antibodies. These cells were then transfected with S52/SG-neo and selected with G418 for 3 weeks. The resulting cell colonies were stained with crystal violet. (g,h) Since N-terminal deletion mutants of SEC14L2 were unstable in Huh-7.5 cells (they formed protein aggregates), we generated chimeric constructs by fusing C-terminal ends of SEC14L2 with the corresponding N-terminal sequences from SEC14L4 and tested their ability to support HCV replication. (g) Schematic representation of C-terminal EGFP-tagged chimeric constructs. (h) Huh-7.5 cells stably expressing the indicated chimeric constructs were lysed and protein expression was confirmed by immunoblotting with anti-GFP antibody. The cells were then transfected with S52/SG-neo and selected with G418 for 3 weeks.
Extended Data Figure 6. SEC14L2 does not interact with HCV non-structural proteins under the tested conditions.
(a) Yeast two-hybrid assay was performed to examine direct interaction between SEC14L2 and HCV non-structural proteins. SEC14L2-AD (GAL4 activation domain) fusion or control AD vector was co-expressed with DBD (DNA binding domain) fusion of the individual HCV non-structural proteins or control DBD vector and tested for positive yeast two-hybrid interactions under selective nutritional conditions (lacking leucine, tryptophan, histidine, and adenosine). The strong signal obtained for NS5A most likely reflects the intrinsic trans-activating activity of NS5A45. (b) Co-immunoprecipitation did not reveal binding of SEC14L2 with HCV NS5A protein. Since two-hybrid assay (panel a) did not yield unambiguous results on interaction between SEC14L2 and NS5A, we employed co-immunoprecipitation assay to probe binding between these proteins. Huh-7.5 cells stably expressing SEC14L2-EGFP and harboring wild-type Con1/SG-neo replicon were lysed and subjected to immunoprecipitation with anti-NS5A antibody, anti-EGFP antibody or control IgGs. The bound proteins were analyzed by immunoblotting. Endogenous MOBKL1B, a previously described binding partner of the NS5A protein41, served as a positive control. (c) Subcellular fractionation of SEC14L2-EGFP/Huh-7.5 cells harboring wild-type Con1/SG-neo replicon did not show significant co-fractionation between SEC14L2 and HCV NS5A protein.
Extended Data Figure 7. SEC14L2 does not facilitate HCV RNA replication by modulating PI3K/Akt or cholesterol synthesis pathways.
(a,b,c,d) Down-regulation of PI3K/Akt pathway does not support HCV replication. (a) Akt phosphorylation in control and SEC14L2-expressing Huh-7.5 cells was analyzed by immunoblotting; ß-actin is included as a loading control. (b) Stable Knockdown of Akt in Huh-7.5 cells with 2 different shRNAs did not facilitate G418 resistant colony formation by S52/SG-neo. (c) Huh-7.5 cells were transduced to stably express PTEN, a negative regulator of PI3K pathway. Despite decreased Akt phosphorylation, these cells did not support colony formation by S52/SG-neo. (d) Suppression of PI3K pathway in Huh-7.5 cells by stable expression of a dominant negative Akt, a dominant negative p85 subunit of PI3K, or a constitutively active FOXO3a did not render them permissive to HCV replication. Dominant negative Akt gets phosphorylated, but since it is kinase-dead, it cannot initiate the downstream signaling. Interestingly, increased Akt phosphorylation was seen in Huh-7.5 cells expressing constitutively active FOXO3a, suggesting a potential feed back mechanism. (e,f) A SEC14L2 mutant (S289A) lacking the cholesterolgenic activity supported HCV replication. (e) Schematic representation of carboxy-terminal EGFP-tagged SEC14L2 point mutants. S288A was generated as a negative control. (f) The colony formation efficiency of S52/SG-neo (plotted as CFU/100,000 transfected cells) and the expression levels of SEC14L2 mutants in Huh-7.5 are shown.
Extended Data Figure 8. SEC14L2 expression masks the effects of lipophilic oxidants and anti-oxidants on H77S.3/GLuc replication.
(a,b) SEC14L2 enhances transient replication of H77S.3, but not that of JFH-1 or J6/JFH1. Empty vector- and SEC14L2-expressing Huh-7.5 cells were electroporated with the indicated viral RNAs lacking GLuc insertions. (a) Intracellular and (b) extracellular RNA levels were measured 6 days after electroporation. Results are plotted as fold change from empty vector control. (c,d,e,f) The pro-viral effects of various lipophilic antioxidants on H77S.3/GLuc replication were suppressed in the presence of SEC14L2. (c) γ-tocopherol, (d) α-tocopheryl succinate, (e) α-tocopheryl quinone, and (f) sphingosine kinase inhibitor (SKI) were added to Huh-7.5 cells 20 h before transfection with H77S.3/GLuc. Transfections were carried out in the fresh medium lacking these compounds. 6 h after transfection, cells were again fed with each compound and the GLuc expression was measured at 72 h post-transfection. The results are presented as fold change from untreated cells. (g,h,i,j,k,l) SEC14L2 expression suppressed the inhibitory effect of lipophilic oxidants and direct-acting antivirals, but not that of the HCV host factor inhibitors, on H77S.3/GLuc replication. (g) Docosahexaenoic acid (oxidant), (h) Linoleic acid (oxidant), (i) IFN-ß, (j) CSA (cyclosporine A), (k) Danoprevir (NS3 protease inhibitor), and (l) 2’CMeA (NS5B polymerase inhibitor) were added to Huh-7.5 cells 6 h after transfection with H77S.3/GLuc (and Jc1/GLuc in case of Danoprevir and 2’CMeA) and the secreted GLuc activity at 72 h post-transfection was measured. The results are plotted as % inhibition relative to the untreated cells. All results in this figure represent mean±SD of 2 replicate experiments.
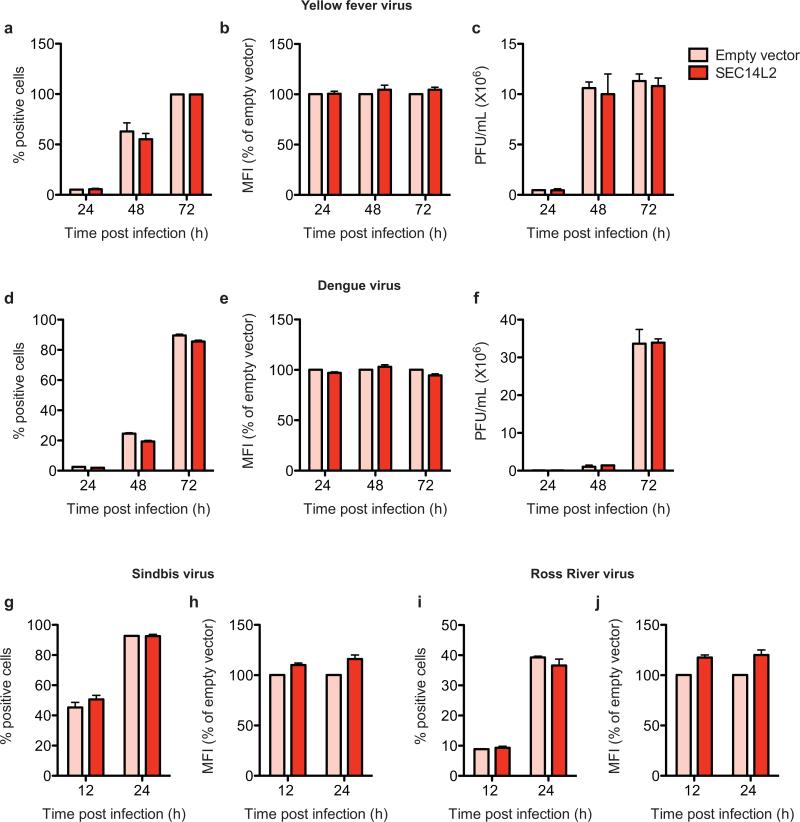
Extended Data Figure 9. SEC14L2 expression does not enhance replication of lipid peroxidation-resistant RNA viruses.
(a-f) Flaviviruses, such as Yellow fever virus (YFV) and dengue virus (DENV) do not respond to SEC14L2 expression. (a,b,d,e) Empty vector- and SEC14L2-expressing Huh-7.5 cells were infected with (a,b) YFV-venus or (d,e) DENV-GFP at a multiplicity of infection (MOI) of 0.01 and 0.1, respectively. The cells were harvested at the indicated times post infection followed by FACS analysis. (a,d) The number of yellow (YFV-venus) and green (DENV-GFP) cells are plotted as % positive cells and (b,e) the mean fluorescence intensities are presented as % relative to empty vector. (c,f) Empty vector- and SEC14L2-expressing Huh-7.5 cells were inoculated with (c) YFV (17D) or (f) DENV (serotype 2 strain 16681) at an MOI of 0.5 and 0.1, respectively. The culture medium was collected at the indicated times post infection and the infectious virus titers were determined by a plaque formation assay on Huh-7.5 cells (YFV-venus) or BHK cells (DENV). The results are presented as plaque-forming units (PFU)/ml. (g-j) Alphaviruses, such as Sindbis virus (SINV) and Ross River virus (RRV) are insensitive to SEC14L2 expression. Empty vector- and SEC14L2-expressing Huh-7.5 cells were infected with (g,h) SINV-GFP or (I,j) RRV-GFP (T48 strain) at an MOI of 0.05. The cells were harvested at the indicated times post infection and FACS analysis was carried out to determine the number of green cells and the mean fluorescence intensities. All results in this figure represent mean±SEM of 2 replicate experiments.
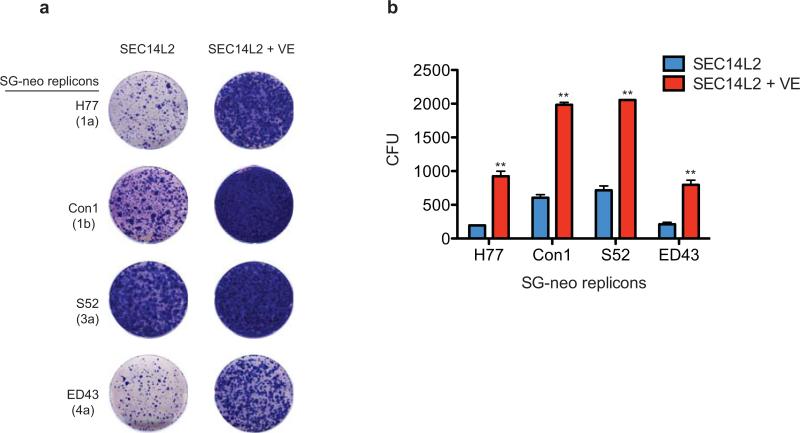
Extended Data Figure 10. VE increased SEC14L2-mediated replicon colony formation.
(a) VE (α-tocopherol: 1 μM) was added to Huh-7.5 cells 20 h before transfection with the indicated wild-type HCV subgenomic replicons. Transfections were carried out for 6 h in the fresh medium lacking VE, followed by medium change to VE-containing medium. After 48 h, cells were subjected to G418 selection and fed with fresh G418 and VE every 2 days. The resulting cell colonies were stained with crystal violet. Shown are the results of one of the 3 independent experiments. (b) Colony formation efficiency of the indicated subgenomic replicons was measured in SEC14L2- expressing cells in the absence or presence of 1 μM VE. The results are plotted as mean±SEM of CFU/100,000 transfected cells from 3 independent transfections. ** P < 0.005 by two-tailed, paired t-test.
Acknowledgments
We thank Y. Matsuura (Osaka University, Japan) for HEP3B/miR-122 cells, J. Bukh (University of Copenhagen, Denmark) for pJ6CF and pTNcc/GLuc, S. M. Lemon and D. Yamane (The University of North Carolina at Chapel Hill, NC) for pH77S.3/GLuc, pH77S.3, and pH77D/GLuc, and Danny Manor (Case Western Reserve University, OH) for [14C]RRR-α-tocopherol. We also thank E. Castillo and A. Webson for laboratory assistance, H. H. Hoffman and L. Andrus for technical input, and W. M. Schneider, M.M. Li, and M. R. MacDonald for critical reading of the manuscript. This work was supported in part by the National Institutes of Health, NCI grant R01CA057973, NIAID grants R01AI072613 and R01AI099284 (to CMR), NIDDK grant R01DK090317 and NIDA grant DA031095 (to ADB), and by a Helmsley Postdoctoral Fellowship for Basic and Translational Research on Disorders of the Digestive System at The Rockefeller University (to MS). The Greenberg Medical Research Institute, the Starr Foundation, the Ronald A. Shellow, M.D. Memorial Fund and anonymous donors provided the additional funding (to CMR).
Footnotes
Contributions
M.S. and C.M.R. designed the project, analyzed results, and wrote the manuscript. M.S., U.A., H.Y.C. and C.E. performed the experimental work. A.D.B. and J.M.S. contributed reagents and advice.
Competing financial interests
The authors declare the following conflicts of interest, which are managed under university policy: C.M.R. has equity in Apath, LLC, which holds commercial licenses for the Huh-7.5 cell line and the HCVcc cell culture system.
Contributor Information
Mohsan Saeed, Center for the Study of Hepatitis C, Laboratory of Virology and Infectious Disease, The Rockefeller University, New York, NY 10065, USA.
Ursula Andreo, Center for the Study of Hepatitis C, Laboratory of Virology and Infectious Disease, The Rockefeller University, New York, NY 10065, USA.
Hyo-Young Chung, Center for the Study of Hepatitis C, Laboratory of Virology and Infectious Disease, The Rockefeller University, New York, NY 10065, USA.
Christine Espiritu, Center for the Study of Hepatitis C, Laboratory of Virology and Infectious Disease, The Rockefeller University, New York, NY 10065, USA.
Andrea D. Branch, Division of Liver Diseases, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA
Jose M. Silva, Department of Pathology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA
Charles M. Rice, Center for the Study of Hepatitis C, Laboratory of Virology and Infectious Disease, The Rockefeller University, New York, NY 10065, USA
References
- 1.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 2.Saeed M, et al. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrobial agents and chemotherapy. 2012;56:5365–5373. doi: 10.1128/AAC.01256-12. doi:10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saeed M, et al. Replication of hepatitis C virus genotype 3a in cultured cells. Gastroenterology. 2013;144:56–58. e57. doi: 10.1053/j.gastro.2012.09.017. doi:10.1053/j.gastro.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi: 10.1002/hep.26141. doi:10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 5.Gottwein JM, et al. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. Journal of virology. 2010;84:5277–5293. doi: 10.1128/JVI.02667-09. doi:10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murayama A, et al. The NS3 helicase and NS5B-to-3'X regions are important for efficient hepatitis C virus strain JFH-1 replication in Huh7 cells. Journal of virology. 2007;81:8030–8040. doi: 10.1128/JVI.02088-06. doi:10.1128/JVI.02088-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murayama A, et al. RNA polymerase activity and specific RNA structure are required for efficient HCV replication in cultured cells. PLoS pathogens. 2010;6:e1000885. doi: 10.1371/journal.ppat.1000885. doi:10.1371/journal.ppat.1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukh J, et al. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14416–14421. doi: 10.1073/pnas.212532699. doi:10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen-Baume V, Segui B, Cockcroft S. Current thoughts on the phosphatidylinositol transfer protein family. FEBS letters. 2002;531:74–80. doi: 10.1016/s0014-5793(02)03412-9. [DOI] [PubMed] [Google Scholar]
- 10.Aravind L, Neuwald AF, Ponting CP. Sec14p-like domains in NF1 and Dbl-like proteins indicate lipid regulation of Ras and Rho signaling. Current biology : CB. 1999;9:R195–197. doi: 10.1016/s0960-9822(99)80127-4. [DOI] [PubMed] [Google Scholar]
- 11.Kempna P, et al. Cloning of novel human SEC14p-like proteins: ligand binding and functional properties. Free radical biology & medicine. 2003;34:1458–1472. doi: 10.1016/s0891-5849(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 12.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nature biotechnology. 2010;28:167–171. doi: 10.1038/nbt.1604. doi:10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farci P, et al. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Washburn ML, et al. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology. 2011;140:1334–1344. doi: 10.1053/j.gastro.2011.01.001. doi:10.1053/j.gastro.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panagabko C, et al. Ligand specificity in the CRAL-TRIO protein family. Biochemistry. 2003;42:6467–6474. doi: 10.1021/bi034086v. doi:10.1021/bi034086v. [DOI] [PubMed] [Google Scholar]
- 16.Ni J, et al. Tocopherol-associated protein suppresses prostate cancer cell growth by inhibition of the phosphoinositide 3-kinase pathway. Cancer research. 2005;65:9807–9816. doi: 10.1158/0008-5472.CAN-05-1334. doi:10.1158/0008-5472.CAN-05-1334. [DOI] [PubMed] [Google Scholar]
- 17.Mokashi V, Singh DK, Porter TD. Supernatant protein factor stimulates HMG-CoA reductase in cell culture and in vitro. Archives of biochemistry and biophysics. 2005;433:474–480. doi: 10.1016/j.abb.2004.10.002. doi:10.1016/j.abb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Mokashi V, Porter TD. Supernatant protein factor requires phosphorylation and interaction with Golgi to stimulate cholesterol synthesis in hepatoma cells. Archives of biochemistry and biophysics. 2005;435:175–181. doi: 10.1016/j.abb.2004.11.030. doi:10.1016/j.abb.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 19.Neuzil J, Dong LF, Wang XF, Zingg JM. Tocopherol-associated protein-1 accelerates apoptosis induced by alpha-tocopheryl succinate in mesothelioma cells. Biochemical and biophysical research communications. 2006;343:1113–1117. doi: 10.1016/j.bbrc.2006.03.052. doi:10.1016/j.bbrc.2006.03.052. [DOI] [PubMed] [Google Scholar]
- 20.Yamane D, et al. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med. 2014;20:927–935. doi: 10.1038/nm.3610. doi:10.1038/nm.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshimoto T, et al. Positive modulation of IL-12 signaling by sphingosine kinase 2 associating with the IL-12 receptor beta 1 cytoplasmic region. Journal of immunology. 2003;171:1352–1359. doi: 10.4049/jimmunol.171.3.1352. [DOI] [PubMed] [Google Scholar]
- 22.Poveda E, et al. Update on hepatitis C virus resistance to direct-acting antiviral agents. Antiviral research. 2014;108:181–191. doi: 10.1016/j.antiviral.2014.05.015. doi:10.1016/j.antiviral.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 24.Wose Kinge CN, et al. Hepatitis C virus genotype 5a subgenomic replicons for evaluation of direct-acting antiviral agents. Antimicrobial agents and chemotherapy. 2014;58:5386–5394. doi: 10.1128/AAC.03534-14. doi:10.1128/AAC.03534-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liehl P, et al. Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat Med. 2014;20:47–53. doi: 10.1038/nm.3424. doi:10.1038/nm.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanagi M, Purcell RH, Emerson SU, Bukh J. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology. 1999;262:250–263. doi: 10.1006/viro.1999.9889. doi:10.1006/viro.1999.9889. [DOI] [PubMed] [Google Scholar]
- 27.Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. Journal of virology. 2001;75:1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. doi:10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. Journal of virology. 2003;77:3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimakami T, et al. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology. 2011;140:667–675. doi: 10.1053/j.gastro.2010.10.056. doi:10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. doi:10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. doi:10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 32.Marukian S, et al. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48:1843–1850. doi: 10.1002/hep.22550. doi:10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukuhara T, et al. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with hepatitis C virus. Journal of virology. 2012;86:7918–7933. doi: 10.1128/JVI.00567-12. doi:10.1128/JVI.00567-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Barrueco R, Marshall N, Silva JM. Pooled shRNA screenings: experimental approach. Methods in molecular biology. 2013;980:353–370. doi: 10.1007/978-1-62703-287-2_21. doi:10.1007/978-1-62703-287-2_21. [DOI] [PubMed] [Google Scholar]
- 35.Yi Z, et al. Identification and characterization of the host protein DNAJC14 as a broadly active flavivirus replication modulator. PLoS pathogens. 2011;7:e1001255. doi: 10.1371/journal.ppat.1001255. doi:10.1371/journal.ppat.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoggins JW, et al. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14610–14615. doi: 10.1073/pnas.1212379109. doi:10.1073/pnas.1212379109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suthar MS, Shabman R, Madric K, Lambeth C, Heise MT. Identification of adult mouse neurovirulence determinants of the Sindbis virus strain AR86. Journal of virology. 2005;79:4219–4228. doi: 10.1128/JVI.79.7.4219-4228.2005. doi:10.1128/JVI.79.7.4219-4228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shabman RS, et al. Differential induction of type I interferon responses in myeloid dendritic cells by mosquito and mammalian-cell-derived alphaviruses. Journal of virology. 2007;81:237–247. doi: 10.1128/JVI.01590-06. doi:10.1128/JVI.01590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bredenbeek PJ, et al. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. The Journal of general virology. 2003;84:1261–1268. doi: 10.1099/vir.0.18860-0. [DOI] [PubMed] [Google Scholar]
- 40.Kinney RM, et al. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53. Virology. 1997;230:300–308. doi: 10.1006/viro.1997.8500. doi:10.1006/viro.1997.8500. [DOI] [PubMed] [Google Scholar]
- 41.Chung HY, Gu M, Buehler E, MacDonald MR, Rice CM. Seed sequence-matched controls reveal limitations of small interfering RNA knockdown in functional and structural studies of hepatitis C virus NS5A-MOBKL1B interaction. Journal of virology. 2014;88:11022–11033. doi: 10.1128/JVI.01582-14. doi:10.1128/JVI.01582-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Molecular & cellular proteomics : MCP. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. doi:10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Pan M, et al. Lipid peroxidation and oxidant stress regulate hepatic apolipoprotein B degradation and VLDL production. The Journal of clinical investigation. 2004;113:1277–1287. doi: 10.1172/JCI19197. doi:10.1172/JCI19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheel TK, et al. Analysis of functional differences between hepatitis C virus NS5A of genotypes 1-7 in infectious cell culture systems. PLoS pathogens. 2012;8:e1002696. doi: 10.1371/journal.ppat.1002696. doi:10.1371/journal.ppat.1002696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maqbool MA, et al. Regulation of hepatitis C virus replication by nuclear translocation of nonstructural 5A protein and transcriptional activation of host genes. Journal of virology. 2013;87:5523–5539. doi: 10.1128/JVI.00585-12. doi:10.1128/JVI.00585-12. [DOI] [PMC free article] [PubMed] [Google Scholar]