SUMMARY
Expanded GGGGCC nucleotide repeats within the C9ORF72 gene are the most common genetic mutation associated with both amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Sense and antisense transcripts of these expansions are translated to form five dipeptide repeat proteins (DRPs). We employed primary cortical and motor neuron cultures, live-cell imaging, and transgenic fly models and found that the arginine-rich dipeptides, in particular Proline-Arginine (PR), are potently neurotoxic. Factors that anticipated their neurotoxicity included aggregation in nucleoli, decreased number of processing bodies, and stress granules formation, implying global translational dysregulation as path accountable for toxicity. Nuclear PR aggregates were also found in human-induced motor neurons and postmortem spinal cord tissues from C9ORF72 ALS and ALS/FTD patients. Intronic G4C2 transcripts, but not loss of C9ORF72 protein, are also toxic to motor and cortical neurons. Interestingly, G4C2 transcript-mediated neurotoxicity synergizes with that of PR aggregates, suggesting convergence of mechanisms.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS), a motor neuron degenerative disease (Kiernan et al., 2011), and frontotemporal dementia (FTD), a presenile onset dementia characterized by selective degeneration of frontal and temporal lobes (Warren et al., 2013), appear to be manifestations of the same clinico-pathological spectrum (Byrne et al., 2012; Elamin et al., 2013; Phukan et al., 2012; Phukan et al., 2007). The recent identification of aberrant GGGGCC (G4C2) intronic repeat expansions in the C9ORF72 gene (DeJesus-Hernandez et al., 2011; Renton et al., 2011) as the most common cause for both ALS and FTD have further emphasized this notion (Majounie et al., 2012). C9ORF72 patients carry from tens to hundreds of G4C2 repeats, while the majority of unaffected individuals have no more than 2 to 25 repeats (Rutherford et al., 2012). Correlation between these expanded repeats and severity of clinical manifestations has yet to be established.
Repeat expansions in genes cause many hereditary diseases in humans (La Spada & Taylor, 2010). The mechanisms by which G4C2 repeat expansions cause neurodegeneration are under intense investigations. Decreased C9ORF72 mRNA expression levels were detected in C9-ALS/FTD patient lymphoblasts (DeJesus-Hernandez et al., 2011). This led to the hypothesis that the intronic expansions could cause down-regulation of the C9ORF72 protein, which could then be responsible for neurodegeneration. Support for this loss-of-function hypothesis were first reported in zebrafish (Ciura et al., 2013), although confirmation is still lacking in mammalian models.
Lines of evidence supporting a gain-of-toxic-function hypothesis of pathogenesis have recently started to emerge. For instance, sense and antisense G4C2 repeat RNA transcripts accumulate in nuclear foci in neurons of different CNS areas of C9-ALS/FTD patients (Zu et al., 2013); antisense oligonucleotides against C9ORF72 transcripts reduced RNA-binding proteins sequestration and increased glutamate sensitivity of neurons derived from induced pluripotent stem cells (iPSCs) of C9-ALS patients (Donnelly et al., 2013); and G4C2 repeat expansions adopt stable G-quadruplex motifs (Reddy et al., 2013), which sequester ribonucleoproteins critical for cell survival (Haeusler et al., 2014). Furthermore, repeat-associated, non-ATG initiated (RAN) translation has been reported in several nucleotide repeat disorders (Zu et al., 2011). RAN-translated proteins from sense and anti-sense transcripts were reported accumulating in C9-ALS/FTD tissues (Ash et al., 2013; Mori et al., 2013), implying a pathogenic role for these proteins. Indeed, some of these RAN-translated proteins were recently described to cause toxicity (Kwon et al., 2014; Mizielinska et al., 2014; Zhang et al., 2014). All of the different and potentially toxic instances appear not to be mutually exclusive, as evidence for their co-existence was found in postmortem patient samples (Gendron et al., 2013a). However, it remains challenging to tease out their individual contributions and mechanisms of toxicity.
We were able to independently model these different C9ORF72 pathogenic instances. By transfecting cortical and motor neurons either with constructs engineered by a randomized codon strategy to express C9RAN proteins avoiding GC repeat sequences, or constructs encoding intronic expanded G4C2 sequences that do not initiate RAN translation, and constructs that efficiently knock down C9ORF72, we deciphered by means of time-lapse live-cell imaging their respective impact on neuronal viability. We also established transgenic drosophila models of C9RAN proteins. We found that one of the antisense C9RAN proteins, the Proline-Arginine dipeptide (PR), is potently neurotoxic when expressed in vivo and neurons with nuclear PR aggregates have a much higher risk to undergo degeneration. In addition, induced motor neuron (iMNs) derived from C9-ALS patients showed intranuclear PR+ aggregates, increased number of extracellular PR aggregates in the dish, and dramatic decrease in survival compared to controls. Nuclear PR aggregates were also found in human spinal cord tissues from C9ORF72 ALS/FTD patients. Interestingly, we found that poly-PR peptides aggregate in nucleoli, causing enlargement of the nucleolus, cell stress responses, and ultimately cell death.
In PR-expressing neurons, death is preceded by a decreased number of RNA-processing bodies (P-bodies) and by the appearance of cytosolic stress granules. These findings suggest that nuclear aggregation of PR peptides initiates a cascade of events leading to neurodegeneration via global perturbation in RNA processing and post-transcriptional regulation of gene expression.
RESULTS
RAN translated poly-PR proteins are neurotoxic
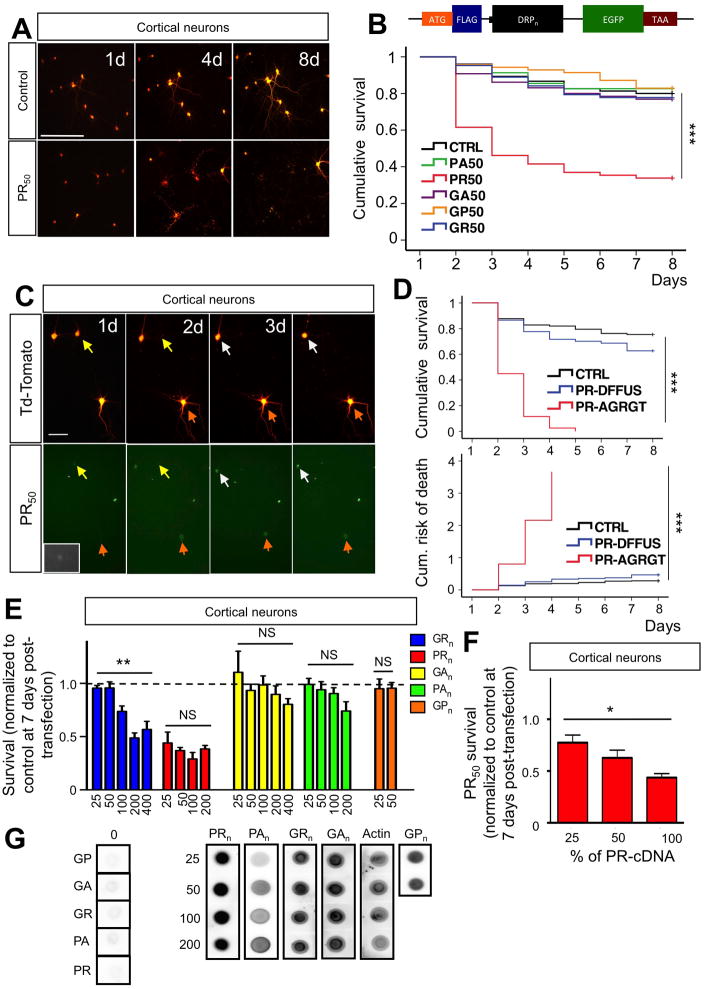
In vivo, pathogenic intronic G4C2 repeat expansions undergo RAN translation on both sense and antisense transcripts using all reading frames, giving rise to five dipeptide repeat proteins (DRPs) of presumably variable length (antisense: poly-PR and poly-PA; sense: poly-GA and poly-GR; both sense: poly-GP) (Ash et al., 2013; Gendron et al., 2013b; Mori et al., 2013; Zu et al., 2013). DRPs form aggregates in C9-patients in different neurons including those not affected by disease (Gendron et al., 2013a; Zu et al., 2013). It remains an open question whether and which DRPs initiate toxicity in neurons. To decipher the neurotoxic profiles of the various DRPs, we transfected mouse motor neuron like NSC34 cells, as well as primary cortical and motor neurons, with DRP-encoding constructs of different lengths (25–400 dipeptide-long sequences) tagged with GFP to aid identification of transfected cells. Constructs were designed to express DRPs in an ATG-dependent translation and with a randomized codon strategy to ensure expression of only the chosen DRP sequence at the desired length. This approach allowed us to engineer stable constructs containing moderate GC content encoding up to 400 dipeptide-long repeats for GR and GA, 200 for PR and PA, 50 for GP and to avoid potential toxicity induced by G4C2 repeats expansion at transcript level (Figure S1).
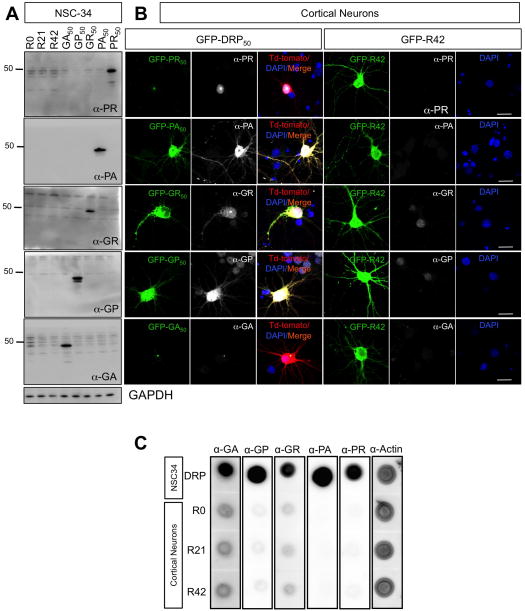
Proper expression of DRP constructs in NCS34 cells was assessed by western blot analysis using anti-DRPs (Ash et al., 2013) (Figure S2A) and GFP antibodies. Notably, the arginine-rich dipeptides, PR and GR, were consistently expressed at somewhat lower levels, whereas expression levels of the other DRPs were comparable (Figure S2B). Confocal microscopy analysis showed distinct PR and GR nuclear aggregates, whereas GA and PA expressing cells revealed predominant cytoplasmic aggregates. Conversely, GP displayed diffused localization (Figure S2C). The different DRPs had analogous localization patterns when transfected in cortical and motor neurons (Figure S2D, E).
We next sought to evaluate toxicity by longitudinal, live-cell imaging that gauged survival of DRP-expressing neurons. Mature cortical neurons (DIV10)(Lesuisse and Martin, 2002) were co-transfected with GFP tagged DRPs-constructs and Td-Tomato reporter driven by the synapsin promoter. This aided live visualization of transfected neurons (Figure S3A, B, C). GFP+ and Td-tomato+ cortical neurons were followed at 24-hour intervals, up to several days post-transfection.
Expression of PR50 dramatically decreased cortical neuron survival (Figure 1A), while the other four DRPs did not (Figure 1B). Most of PR50-mediated cell death occurred in the first 72 hours post-transfection. Interestingly, image analysis revealed that the majority of neurons harboring PR nuclear aggregates died within 24–48 hours after aggregates formed, while neurons featuring diffused pattern of PR expression survived (Figure 1C). This was confirmed by Kaplan-Meier survival and risk-of-death analysis performed on cohorts of neurons displaying either nuclear aggregates or diffused localization (Figure 1D). PR neurotoxicity did not significantly vary with increased length of repeats (Figure 1E), and the effect appeared to be concentration-dependent (Figure 1F). Clear length-dependent neurotoxicity was instead seen for poly-GR. Conversely, poly-PA, GA and GP were not significantly, or just marginally (poly-PA), toxic at the longest lengths tested. Expression levels were comparable among the DRPs of different lengths (Figure 1G). PR50 was also toxic to primary motor neurons. However, these neurons were equally susceptible to expression of GR50 (Figure S4A, B) which aggregated in the nucleus, similarly to PR50 although less aggressively (Figure S2, S4B). Notably, hippocampal neurons, which are marginally affected in C9-FTD (Warren et al., 2013), were also vulnerable to PR toxicity, but to a much lesser extent than cortical neurons and despite comparable expression levels, assessed as intensity of GFP fluorescence signal (Figure S4C, D, E). Localization pattern of PR and extent of its toxicity did not depend on the presence of the GFP tag in the dipeptide sequence as the expression of untagged PR50 displayed similar toxic and localization profile as the GFP-tagged dipeptide (Figure S4F, G, H). In contrast to the toxic effect of PR50 on both cortical and motor neurons, we found that SOD1-G93A displayed selective toxicity towards motor neurons (Figure S4I, J). This observation is consistent with the fact that the SOD1-G93A mutation causes pure ALS with no FTD symptoms.
Figure 1. Neurotoxicity of C9RAN poly-dipeptides.
(A) Representative live-cell images of cortical neurons co-transfected at DIV10 with control-GFP + Td-tomato plasmids (4:1) or PR50-GFP + Td-Tomato (4:1). The same neurons were imaged for 9 days (8 days post-transfection) at 24-hour intervals. Calibration bar is 100 μm. (B) Kaplan-Meier survival analysis of cortical neurons co-transfected with constructs encoding different DRPs + Td-Tomato. At least 80 neurons were followed/group; n=5 experiments; ***P<0.001. The insert shows the schematic of the construct. The sequence of the insert is the following: ATG-DYKDDK-KLGR-DPR50-GYRARIHYSSVVEFM-EGFP-TAA. The insert was subcloned into pcDNA3.1 vector and its expression driven by the CMV promoter. The DRP sequence has been constructed with a randomized codon strategy. (C) PR50 aggregates formation preceded neuronal death. Representative images show a group of cortical neurons expressing both PR50 (GFP tagged) and Td-tomato. Neurons in which PR50 formed aggregates underwent cell death within 24–36 hours (yellow arrows, day 1–2; white arrows, day 3–4). Conversely, neurons in which PR50 stayed in a diffused, not aggregated pattern (orange arrow and insets) outlived the neurons with PR aggregates. Calibration bar is 50 μm. (D) Cortical neurons transfected with PR50 that formed aggregates exhibited decreased survival and increased risk of death compared to those that didn’t form detectable PR50 aggregates (***p<0.001). PR50 transfected neurons with no detectable aggregates showed no difference in survival and risk of death compared to control transfected neurons (P = 0.08). (E) Length-dependent toxicity of DRPs in cortical neurons. Poly-GR displayed clear length-dependent toxicity (**P<0.01; one-way ANOVA), whereas the effect of poly-PR already peaked at 25 repeats, displaying no significant length-dependency (one-way ANOVA). (F) Cortical neuron toxicity of PR50 diminished as amount of transfected cDNA was reduced suggesting a dose-dependency in the effect. Maximum amount of cDNA-PR50 transfected was 0.8 μg/well (100%). Total amount of DNA transfected was kept constant across groups (1 μg/well). *P<0.05, **P<0.01, ***P<0.001. (G) NSC34 cells were transfected with indicated constrcuts. Expression levels of different lengths of DRP constructs were assessed using dot blots. Actin was used as a loading control. Comparable expression levels were observed for DRPs of different lengths.
Neuronal death by intronic G4C2-containing transcripts
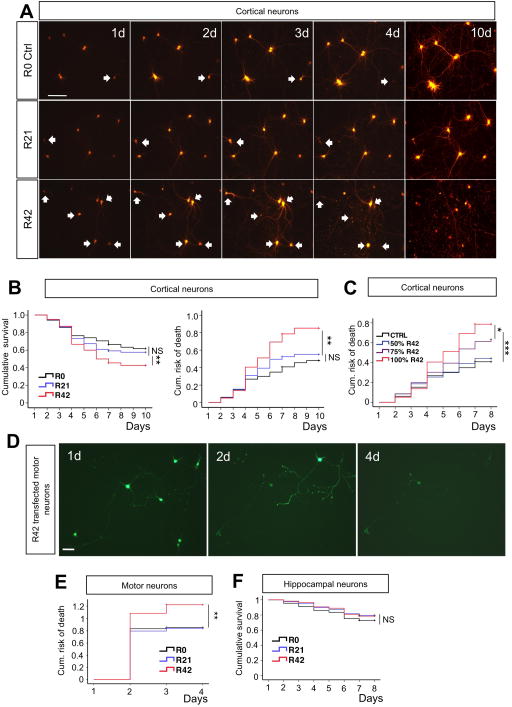
It was recently shown that expanded G4C2 repeats, cloned in a 5′ UTR sequence, are toxic to neuronal cell lines and zebrafish embryos (Lee et al., 2013). A still open question is whether G4C2 repeat expansions, harbored in an intronic sequence, cause neurodegeneration independently of the C9ORF72 gene sequence. We generated plasmids containing 0, 21 and 42 G4C2 repeats (R0, R21, R42) in an artificial intronic sequence inserted between two EGFP exons (Figure S5A, B) and transfected primary cortical neurons. These neurons could be readily identified for live-cell imaging by the concomitant Td-tomato and GFP fluorescence, as shown above (Figure S5C). GFP expression was confirmed by western blot analysis of lysates of R0–42 transfected NSC34 cells probed with an anti-GFP antibody (Figure S5D). Expression of the G4C2 transcripts was verified by FISH analysis (Figure S5E), which revealed prevalent nuclear localization of the G4C2 RNA transcript signal in cell transfected with R21 and R42 but not with R0.
Longitudinal live-cell imaging analysis showed that R42 transfected cortical neurons survived significantly less than R0 or R21-expressing neurons (Figure 2A). The risk of death increased ~2-fold (Figure 2B). R42 construct displayed dose-dependent toxicity with a threshold effect seen at 50% of the amount of transfected cDNA (Figure 2C). R42-expressing motor neurons had higher risk of death compared to R21 and controls (Figure 2D, E), whereas survival of hippocampal neurons was not affected by the R42 transcript (Figure 2F).
Figure 2. Intronic G4C2 repeats are directly toxic to primary neurons.
(A) Representative time-lapse images of cortical neurons co-transfected with Td-Tomato and R0, R21 or R42 constructs (1:4 ratio; 1 μg cDNA/well total). Neurons that went on to die are pointed by arrows. More death events were observed in neurons transfected with R42. Calibration bar is 100 μm. (B, C) Kaplan-Meier survival and Cox proportional hazards analysis of cortical neurons transfected with R0, R21 or R42 showed that expanded intronic G4C2 repeats caused cell death and significantly increased the risk of death. Neurotoxicity of G4C2 decreased with reduced amount of R42 cDNA transfected into neurons, suggesting dose-dependency. Note that 50% of R42 cDNA (or 0.4 μg) is not sufficient to cause toxicity. Total amount DNA used in transfection remained the same across groups. At least 80 neurons were followed/group; n=3 experiments. *P<0.05, **P<0.01, ***P<0.001. (D) Representative live-cell images based on GFP fluorescence at day 1, 2 and 4 post-transfection of R42 construct in motor neurons show clear motor neuron loss. Calibration bar is 50 μm. (E) Risk-of-death analysis of motor neurons transfected at DIV5 with R0–42 constructs. At least 40 motor neurons are imaged/group; n = 3 experiments. **P<0.01. (F) Kaplan-Meier survival analysis showed that hippocampal neurons were not sensitive to expanded intronic G4C2 repeats. At least 80 hippocampal neurons were imaged/group; n = 3 experiments. P = 0.337
G4C2 repeats neurotoxicity is independent on RAN translation
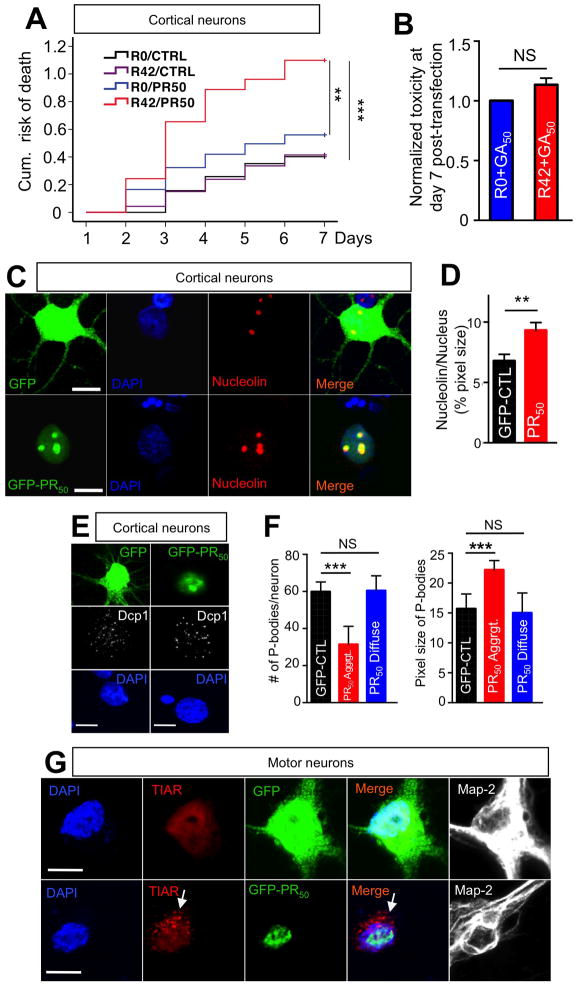
RNA sequence made of 38 G4C2 repeats can initiate RAN translation when expressed in HEK cells in a repeat length-dependent manner (Mori et al., 2013). To determine whether R42 neurotoxicity in our culture systems was mediated by RAN translation, we performed western blot analysis on lysates of R0–42 transfected motor neuron-like NSC34 cells using anti-DRPs antibodies. We found no evidence for formation of any of the five possible DRPs (Figure 3A). This was further confirmed in cortical neurons transfected with R42 by immunocytochemistry and dot-blot analysis (Figure 3B, C). While intronic R42 repeats do not produce detectable DRPs, we found that the concomitant expression of a sub-toxic concentration of R42 repeat transcripts (50% of the dose normally used) greatly enhanced the damaging effect of a marginally toxic concentration (50% of the dose) of PR50 on cortical neurons (Figure 4A). On the other hand, the prevalently cytosolic expression of GA aggregates in combination with R42 did not produce the same synergistic effect (Figure 4B).
Figure 3. DRPs were not detected in cells transfected with R42 repeat transcript.
(A) NSC34 cells were transfected with the indicated constructs. Efficiency of transfection is ~ 90% for all groups. Homogenates were probed with anti-DRPs antibodies. GAPDH reactivity indicated equal total protein loading across the lanes. Immunoblots showed no detectable DRPs in NSC34 cells transfected with R0, R21 and R42 constructs. (B) Cortical neurons were transfected with the indicated constructs. Immunofluorescence analysis suggested that DRPs did not form in neurons transfected with R42 constructs. Nuclei were stained with DAPI (blue), green represents GFP-DRPs, red represents Td-Tomato, which was cotransfected with GFP-DRP constructs, and white represents the respective DRPs. Calibration bar is 20 μm. (C) Primary cortical neurons were transfected with R0, R21or R42 constructs and collected 72h post-transfection. NSC34 cells transfected with DRP constructs were used as positive controls. Equal amounts of protein were loaded and actin was used as a loading control. Dot blot analysis suggests that no DRPs were generated from cortical neurons expressing R0, R21 and R42.
Figure 4. PR50 nuclear aggregates co-localize with nucleolin and mediate cellular stress responses.
(A) Cortical neurons transfected with a combination of R42 and PR50 constructs (0.4 μg R42 + 0.4 μg PR50 + 0.2 μg Td-tomato; total 1 μg/well cDNA) displayed significant increase in cumulative risk of death compared to the other three possible combinations, suggesting R42 and PR50 increased neural toxicity synergistically. The cDNA concentration for each toxic species is 50% of the concentration used when R42 or PR50 were transfected alone in other experiments. At least 80 neurons were imaged/group; n=3 experiments. **P<0.01; ***P<0.001. (B) The combination of GA50 and R42 did not have synergistic effects. P=0.08. (C) Immunofluorescence analysis of cortical neurons transfected with PR50 revealed that PR50 nuclear aggregates co-localized with nucleolin in the nucleolus (bottom panels), and PR50 aggregates induced (D) dispersal of nucleolin with increase in nucleolus size. DAPI: Blue; GFP control or PR50: green; Nucleolin: red. Calibration bar is 10 μm. (E) Neurons transfected with PR50 showed decreased number and increased size of P-bodies. Representative images of GFP control or PR50 transfected cortical neurons. DAPI: blue; Dcp1: white; GFP control or PR50: green. DCP1 is the mRNA-decapping enzyme 1A, constituent of P-bodies. Calibration bar is 10 μm. (F) Quantification of number and size of P-bodies in control and PR50 transfected cortical neurons displaying diffused and aggregated PR50. Diffused PR-staining was previously associated to neuronal survival. At least 20 neurons/group were counted per experiment; n=3. (***P<0.001; two-tailed t-test). Calibration bar is 10 μm. (G) Stress granules formed only in motor neurons in which PR50 aggregated (arrow). DAPI: blue; TIAR: red; GFP control or PR50: green; MAP-2: white. TIAR = TIA1 (cytotoxic granule-associated RNA binding protein-like 1). Calibration bar is 10 μm.
Recently, nucleolar stress was proposed as a potentially toxic mechanism of the G-quadruplexes formed by G4C2 transcripts (Haeusler et al., 2014). Interestingly, PR aggregates co-localized with nucleolin, a constituent protein of nucleolus. PR-containing nucleoli appeared larger in size with dispersal of nucleolin staining, suggesting PR-mediated dynamic nucleolar stress response (Figure 4C, 4D, S6A, S6B). PR aggregates co-localized with fibrillarin, another protein constituent of nucleolus (Ochs et al., 1985), confirming their nucleolar partition (Figure S6C, D). Similar aggregation in nucleoli was seen for GR50, even though a good portion of them were still retained in the cytosol (Figure S6E). We observed that neurons with nuclear PR aggregates had a consistent reduction in the number of cytosolic processing bodies (P-bodies) (Figure 4E), which featured larger size on average (Figure 4F), and formed stress granules (Figure 4G).
Knock-down of C9ORF72 does not affect neuronal survival in vitro
Studies of familial C9-patients have reported an approximately 50% reduction in C9ORF72 mRNA levels (DeJesus-Hernandez et al., 2011; Renton et al., 2011), hypermethylation of these repeats and the neighboring CpG islands (Xi et al., 2013), suggesting haploinsufficiency as the possible initiating mechanism of neurodegeneration.
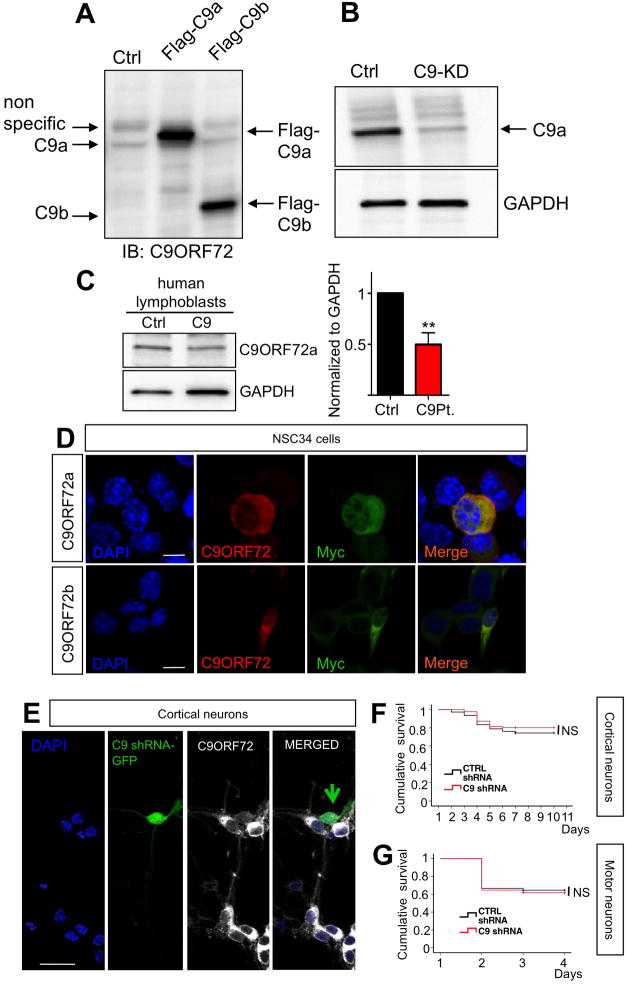
To test whether C9ORF72 is critical for neuronal survival, we characterized an shRNA-GFP construct to efficiently knock-down C9ORF72 in cortical and motor neurons. This approach also allowed the optimization of a commercially available anti-C9ORF72 antibody. The antibody detected C9ORF72 isoform a and b (Woollacott and Mead, 2014) when their respective Flag-C9ORF72 cDNAs were transfected in HEK-293 cells (Figure 5A). The antibody also detected endogenous C9ORF72a but not b isoform (Figure 5A). This antibody was then used to confirm specific knock down of C9ORF72a with the shRNA construct (Figure 5B). Because the shRNA sequence targets both C9ORF72 isoforms, the successful knock down of the endogenous isoform a (≥70%) can imply a similar outcome for isoform b. The fact that the antibody could not detect endogenous C9ORF72b, despite the antibody was recognizing both over-expressed isoforms with similar affinity, suggests that C9ORF72a is the dominant isoform (Figure 5A). When we used this antibody to probe the expression levels of C9ORF72 in human lymphoblast lines derived from C9-ALS/FTD patients, we found that expression levels of C9ORF72a was decreased by ~50% compared to controls (Figure 5C).
Figure 5. C9ORF72 knock-down in primary cortical and motor neurons does not cause cell death.
(A) Characterization of C9orf72 antibody and cytosolic localization of C9ORF72. Immunoblotting shows that the C9ORF72 antibody recognizes Flag tagged C9orf72 isoform a and b transiently expressed in NSC34 cells; for schematics of C9orf72 isoform a and b see (Woollacott and Mead, 2014). (B) C9orf72 isoform a is efficiently knocked down in NSC34 cells. GAPDH levels demonstrated equal protein loading in the control and C9orf72 knock down (C9-KD) lane. (C) C9ORF72 protein levels are reduced in lymphoblasts of C9ORF72 ALS patients. **p≤0.01; 2-tailed T-test. (D) C9orf72a and b isoforms, Myc tagged, were transfected in NCS34 cells. Double-label immunofluorescence shows cytosolic localization of transfected C9orf72a and b. Blue signals represent DAPI, red signals represent C9ORF72 (antibody Sigma cat.#HPA023873), which recognized both isoforms, and green represents Myc staining. Calibration bar is 10 μm. (E) Immunocytochemistry of C9ORF72 protein in cortical neurons transfected with C9orf72 shRNA construct GFP tagged. Nuclei are stained in blue (DAPI), green signals represent GFP which is co-expressed with C9orf72 shRNA, and white signals represent C9ORF72 protein. Calibration bar is 20 μm. (F) Kaplan-Meier survival analysis of cortical neurons co-transfected with C9orf72 shRNA or scrambled shRNA and Td-tomato constructs (4:1 ratio) shows that knockdown of C9orf72 does not directly cause cell death in primary cortical neurons. p=0.312. (G) Similarly, motor neuron survival was not affected by C9ORF72 knock down. P=0.444.
The sequence targeted by our shRNA construct is conserved amongst human, mouse and rat, so we could efficiently knock down C9ORF72 in primary cortical neurons as well. Immunocytochemistry analysis of NSC34 cells transfected with C9ORF72 cDNA tagged with Flag and Myc sequences showed prevalent cytosolic staining of C9ORF72 (Figure 5D). The cytosolic localization of C9ORF72 was further confirmed by immunocytochemistry analysis of cortical neurons transfected with C9ORF72-shRNA construct (Figure 5E).
To address whether down-regulation of C9ORF72 expression would lead to neuronal death, we transfected primary rat cortical neurons with C9ORF72 shRNA-EGFP or scrambled shRNA constructs as control. Cortical neurons were co-transfected with the synapsin promoter-driven Td-Tomato reporter construct for live-cell imaging. Survival curves of cortical neurons treated with shRNA or control scrambled shRNA were not statistically different (Figure 5F). Similarly, C9ORF72 knock down did not affect survival of rat primary motor neurons (Figure 5G), indicating C9ORF72 loss-of-function is not a primary cause of neurotoxicity in C9-ALS/FTD.
Ectopic expression of C9RAN translated PR50 dipeptide leads to neurodegeneration in Drosophila melanogaster
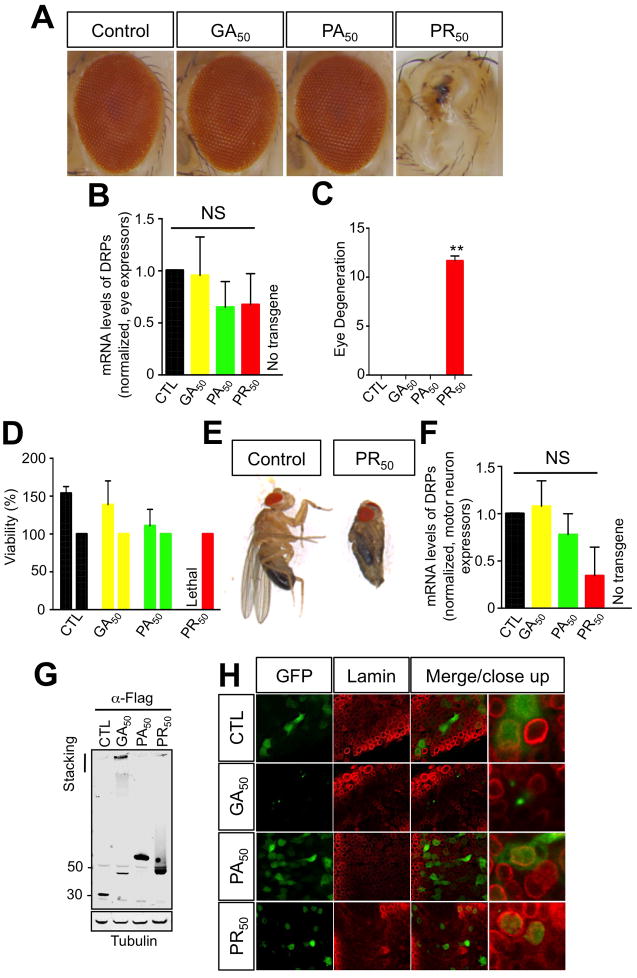
An in vivo model was recently developed to investigate the toxicity of expanded hexanucleotide transcripts. These transcripts caused progressive neurodegeneration in the eye and motor neurons of Drosophila (Xu et al., 2013). We wanted to investigate whether C9RAN PR dipeptides could also lead to a neurodegenerative phenotype in vivo and, therefore, generated transgenic flies over-expressing flag-EGFP tagged PR50. We also generated transgenic flies expressing GA50 and PA50 because, despite these DRPs lack in vitro toxicity, they displayed quite distinctive localization patterns, which could possibly result in a neurotoxic profile in vivo. These transgenes were inserted by PhiC31 integration at the exact same genomic locus and are therefore transcribed equally.
We found that targeted expression of PR50 caused severe neurodegeneration in Drosophila eyes (Figure 6A). PR50 expression was not detectable by western blot analysis of the eye in PR50 flies presumably due to massive loss of the eye neurons. Expression of mRNA of the DRP transgenes was verified in the pre-pupae stage and found to be statistically not different amongst the different fly lines, suggesting that the toxicity was not to be ascribed to difference in DRP expression levels (Figure 6B). On quantitation of the eye phenotype using our previously published criteria (Lanson et al., 2011; Pandey et al., 2007), the ommatidia appeared to be completely degenerated in flies ectopically expressing PR50, with loss of mechanosensory bristles and loss of pigmentation, whereas expression of GA50, PA50, or flag-EGFP control resulted in no obvious external eye degeneration (Figure 6A, C).
Figure 6. PR50 expression is neurotoxic in Drosophila melanogaster.
(A) Expression of C9ORF72 RAN proteins in Drosophila eyes. PR50 expression causes complete degeneration of the eyes. (B) mRNA of the DRPs was measured by in the eyes of the different fly lines by qPCR analysis (ANOVA; Kruskal-Wallis test; P=0.4373). A wild type, non-transgenic fly strain (w1118) was crossed with the GMR-GAL4 strain to rule out non-specific qPCR amplification reactions of the different DRP50 mRNAs. (C) Quantification of eye phenotypes. PR50 expression produces a significant toxic effect relative to the other RAN products and the control (**P<0.001; one-way ANOVA). (D) PR50 expressors do not develop successfully to adulthood. Viability of F1 flies that express RAN proteins is measured relative to viability of F1 flies that carry the TM6b balancer chromosome, which are produced in the same cross. (E) Flies that express PR50 in motor neurons are morphologically normal despite being trapped in the pupal case. The pupal case of the trapped PR50 expressor has been removed for this picture. The folded wings and legs are characteristic of the pupal state. (F) Expression of C9RAN dipeptide mRNAs is equally expressed in OK371-GAL4 motor neuron expressor Drosophila larvae (ANOVA; Kruskal-Wallis test; P=0.1251). (G) Western blot showing expression of C9ORF72 RAN proteins in Drosophila muscle tissue. (H) Subcellular localization of GFP tagged C9ORF72 RAN proteins in motor neurons of Drosophila larvae. Lamin staining delineates the nuclear envelope.
In order to understand the consequences of PR50 in the motor system, we targeted the expression of these poly-peptides to motor neurons using the OK371-GAL4 driver. Contrary to the other genotypes or control, PR50 expressors did not develop to adulthood (Figure 6D). They fully developed to the pupal stage and appeared morphologically normal, but we found that they were unable to escape out of the pupal case whereas flies expressing GA50, PA50 and control construct were able to escape normally. These PR50 expressing escaper flies had no movement and didn’t survive. Dissecting the PR50-expressing flies from their pupal case, we found that these flies had no obvious defects other than what could be expected from being paralyzed with folded wings and legs, wet appearance, and no movement (Figure 6E). Similarly to the eye expressor line, PR50 could not be detected by western blot analysis presumably due to massive loss of motor neurons. We, therefore, verified expression of the DRP transgenes by qPCR at the larvae stage of the transgenic flies and found that there was statistically no difference in the DRP mRNAs in the different lines (Figure 6F). Furthermore, we drove expression in muscles using MHC-GAL4 driver and confirmed effective expression and similar expression levels of the different C9RAN peptides by western blot analysis, with GA50 dipeptides often found in the stacking gel (Figure 6G). To confirm the subcellular localization pattern of the DRPs, we used anti-lamin antibody to stain the nuclear envelope in the Drosophila larvae neural tissue. Thus, staining inside the lamin ring is nuclear and outside the ring is cytoplasmic (Daigle et al., 2013; Lanson et al., 2011). We found that GA50 concentrated in perinuclear or cytosolic aggregates, while PA50 and GFP control displayed diffuse cytosolic localization. Similarly to cultured neurons, PR50 was predominantly aggregated in nuclei (Figure 6H).
PR aggregates are toxic to human iPS-derived neurons and are present in C9ORF72 ALS patient derived human induced motor neurons and postmortem spinal cord tissue
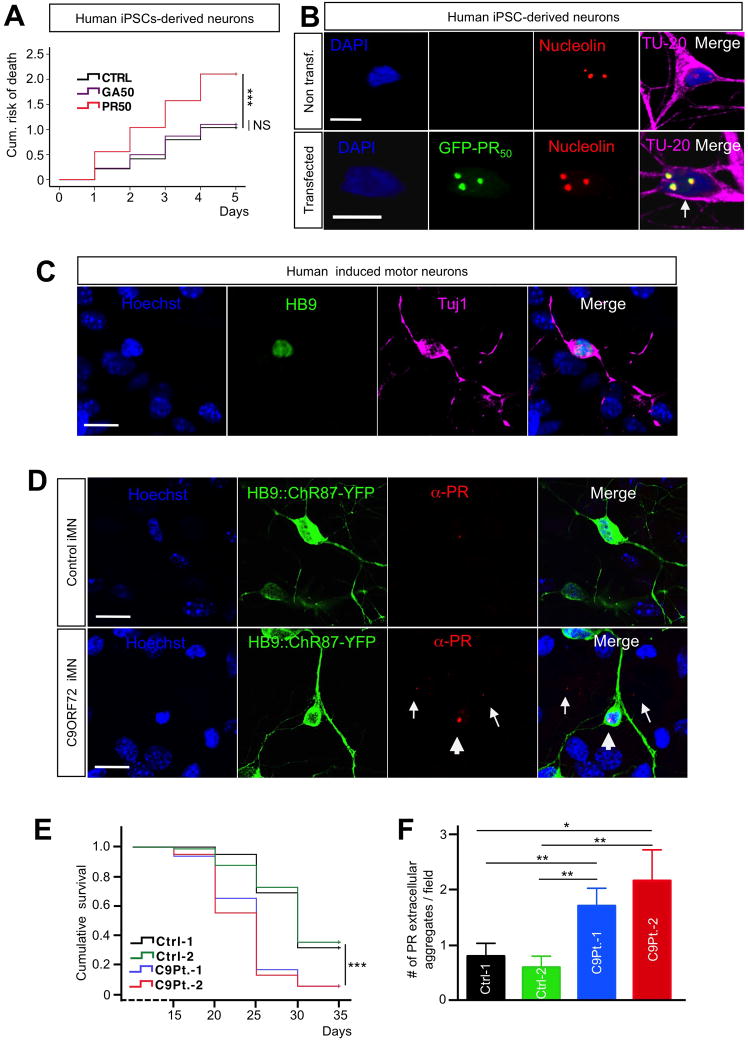
To establish whether expression of PR was also toxic to human neurons, we transfected human iPSCs-derived generic neurons with PR50, GA50 or control constructs. Transfection efficiency of the constructs in these neurons was somewhat lower that the one we achieved for rat primary neurons. Nevertheless, we were able to build Kaplan-Meier survival curves and hazard analysis for each experimental group and found that PR50 was potently toxic, whereas neurons expressing GA50 at similar levels had a cumulative risk of death non significantly different from the control transfected group (Figure 7A). Similarly to what reported for rat primary neurons, we found PR50 aggregates co-localized with nucleolin in nucleoli (Figure 7B). To determine whether PR aggregates are relevant to the human disease state, we determined if PR nuclear aggregates were also endogenously expressed by induced motor neurons from C9ORF72 ALS patients. We used transcription factor-mediated lineage conversion (Son et al., 2011) to generate induced motor neurons (iMNs) from iPSCs of C9ORF72 ALS patient and unaffected control. In both patient and control cultures, we observed cells with mature neuronal morphologies that expressed Hb9 and TuJ1, confirming their motor neuron identity (Figure 7C). The iMNs were labeled with an Hb9::Channelrhodopsin-YFP reporter lentivirus (Marchetto et al., 2008). In control iMN cultures, we rarely observed intranuclear PR+ aggregates, suggesting a low level of PR production in these cells. Similarly, we observed intranuclear PR+ aggregates in C9-ALS iMNs (Figure 7D). However, in contrast to control cultures, C9-ALS cultures contained a high number of extracellular PR+ aggregates, and significantly fewer iMNs survived in C9-ALS cultures after extended periods, indicating that they undergo accelerated neurodegeneration relative to control iMNs (Figure 7E, F). Interestingly, we also detected extracellular PR aggregates in primary cortical and motor neuron cultures transfected with PR-expressing constructs. Live-cell imaging showed that PR aggregates remained attached on the bottom of the dish once the PR-expressing neurons (either motor or cortical neurons) lost membrane integrity and died. (Figures 1C, S4A, S7). Overall, these results demonstrate that human C9-ALS derived motor neurons produce elevated levels of PR. Moreover, the presence of large numbers of extracellular PR+ aggregates and reduced numbers of iMNs in C9-ALS patient cultures agree with results in rodent cultures.
Figure 7. PR aggregates are toxic to human iPSCs-derived neurons and form in iPSC-derived induced motor neurons from C9ORF72 ALS patients.
(A) Cox propotional hazards analysis of iPSCs-derived neurons transfected with GA50, PR50 or control plasmid. Expression of PR50 significantly increases the risk of death compared to that of neurons expressing either GA50 or EGFP control. At least 45 neurons were followed/group; n=4 experiments; ***P<0.001. (B) Immunofluorescence analysis shows that PR nuclear aggregates co-localize with nucleolin in iPSCs-derived neurons transfected with PR50 (bottom pannel). DAPI: blue; PR50: green; Nucleolin: red; TU-20: magenta. Calibration bar is 10 μm. (C) Characterization of iMNs. iMNs express HB9 and Tuj1. Hoechst: blue; HB9: green; Tuj1: magenta. Calibration bar is 10 μm. (D) Immunofluorescence of PR aggregates in control iMNs (top) and C9ORF72 ALS iMNs (bottom). iMNs are labeled with Hb9::ChR87-YFP. Arrowheads indicate intranuclear aggregates, and arrows indicate extracellular aggregates. Hoechst: blue; Hb9::ChR87-YFP: green; PR: red. Calibration bar is 10 μm. (E) Kaplan-Meier survival analysis of iMNs generated from 2 C9-ALS patients compared to 2 control lines showed a significant iMN loss in C9-ALS patient lines. ***P<0.001. (F) Significantly more extracellular PR aggregates were observed in C9-ALS iMNs compared to control iMNs. ***P<0.001. 2 control lines and 2 patient lines were generated. 20 random fields were analyzed per group.
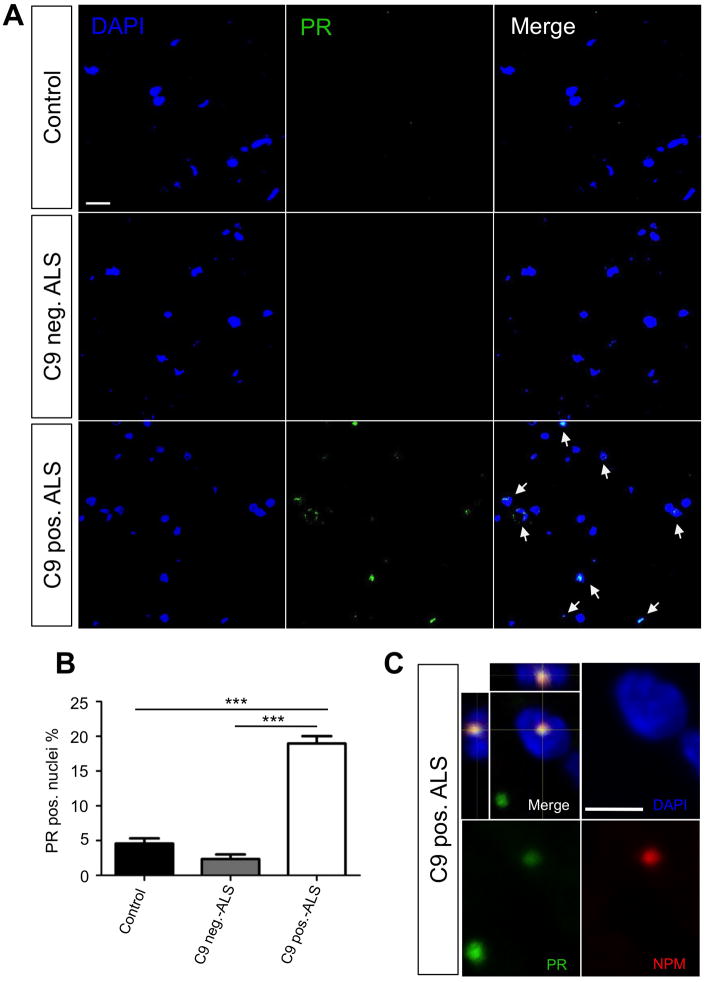
Confocal microscopy analysis of spinal cord tissues from C9-ALS and C9-ALS/FTD patients revealed the presence of PR+ nuclear as well as extranuclear inclusions (Figure 8A). Notably, PR+ aggregates were rarely detected in control non-diseased spinal cords and, even less, in non-C9 spinal cords (Figure 8B, S8C). In PR+ nuclei, poly-PR either aggregated in a relatively large nuclear area or they formed punctate aggregates that co-localized with nucleophosmin in nucleoli (Figure 8C, S8D). Nuclei-containing PR aggregates often appeared smaller and shaped irregularly compared to the well-rounded nuclei of the control tissue, suggesting nuclear stress (Figure 8A).
Figure 8. Nuclear PR inclusions are found in C9ORF72 ALS/FTD patient spinal cord tissues.
(A) Representative images of a 15 slice z-series performed on age-matched human lumbar spinal cord slices from various patient types (control, C9ORF72 mutation negative ALS, and C9ORF72 mutation positive ALS). PR dipeptides (green), DAPI (blue), and merged images show absent to low intensity staining in control and C9ORF72 mutation negative ALS tissues. C9ORF72 mutation positive tissues shows higher intensity staining as well as nuclear localization of the PR dipeptides. (B) Quantification of PR positive nuclei was performed on 20 15-slice z-series images for each patient type with ~400 nuclei quantified. C9ORF72 mutation positive tissues showed a significantly higher (P< 0.0001) percent of PR positive nuclei (18.96 ± 1.07) than control (4.464 ± 0.88) and C9ORF72 mutation negative ALS (2.44 ± 0.67). (C) Immunofluorescence analysis shows that PR aggregates co-localize with nucleophosmin (NPM) in the nucleus from C9ORF72 mutation positive patient spinal cord tissues. DAPI: blue; PR: green; NPM: red. Calibration bar is 5 μm.
DISCUSSION
Pathological findings in cells and tissues of C9ORF72-linked ALS/FTD patients suggest three possible pathogenic mechanisms of disease. The presence of RNA foci and accumulation of RAN translated proteins suggests an RNA-gain of function or an RAN translation-induced toxicity. The evidence that cells derived from C9ORF72 patients have decreased levels of C9ORF72 transcripts suggests instead a loss of function mechanism. Increasing lines of evidence point at RNA gain-of-function and RAN translation as the most likely mechanisms (Donnelly et al., 2013; Gendron et al., 2013b; Haeusler et al., 2014; Zu et al., 2013). However, whether one mechanism predominates over the other or whether they both contribute to neurodegeneration is not clear. Similarly, it remains unknown whether there are neuronal-specific RAN toxic species. Recent studies showed that arginine-rich RAN dipeptides were cytotoxic by impairing the biogenesis of ribosomal RNA when non-neuronal cells (ie. astrocytes and immortalized cell lines) were incubated with the synthetic peptides (Kwon et al., 2014). Toxicity of these arginine-rich peptides was also shown in vivo, in Drosophila models (Mizielinska and Isaacs, 2014). Another study showed, instead, that a different RAN translated species, poly-GA, caused toxicity measured by increased release of LDH, caspase-3 activation and endoplasmic reticulum stress when expressed in cultured cells and primary neurons (Zhang et al., 2014).
Here, we extended these findings by employing a systematic approach to determine the specific contribution to neurotoxicity of each of the three C9ORF72-related pathological instances using disease-relevant human and rat neuronal cultures coupled to longitudinal live-cell tracking imaging, as well as Drosophila models. We provided clear evidence that: (1) Amongst all the possible C9RAN translated dipeptides, the arginine-rich antisense transcript encoded PR, and the sense transcript encoded GR are the neurotoxic species, with GR, but not PR, showing a repeat length-dependent toxicity, whereas PR full toxicity was already manifested at the lowest repeat length tested; (2) arginine-rich C9RAN dipeptides toxicity follows their nuclear aggregation; (3) the arginine-rich dipeptides aggregate in nucleoli, causing a stress-response in cells, with a reduction in the number of processing bodies and formation of stress granules; (4) intronic G4C2 transcripts, but not loss of C9ORF72, are also toxic to motor and cortical neurons; (5) G4C2-neurotoxicity is independent from the appearance of RAN pathology, although the neurotoxic effects of intronic G4C2 transcripts synergize with that of PR aggregates. Finally, we demonstrated that the equal sensitivity of cortical and motor neurons to C9ORF72 is mutation specific.
The initiation of RAN translation is thought to depend on RNA hairpin structures that utilize GC pairing (Zu et al., 2011). A recent study demonstrated that an RNA sequence of 38-G4C2 repeats is sufficient to initiate this unconventional translation in a cell line (Mori et al., 2013). In the time frame of our experiments, we did not detect C9RAN DRPs in neurons expressing R42 transcripts, albeit the 42-G4C2 hexanucleotide repeats were properly expressed. This lack of RAN translation and DRP pathology suggested that RAN translation might require selective, still unknown, stressors to occur and provided us a suitable in vitro model to test the neurotoxic profile of G4C2 transcripts and DRPs independently from one another.
Intronic G4C2 transcripts and PR dipeptides are both sufficient to induce death of both cortical and motor neurons in a dose-dependent manner. However, sub-toxic or threshold toxic concentrations of R42 transcripts and PR dipeptides exhibit neurotoxic synergy when co-expressed in the same neurons. This opens up the possibility that C9ORF72 products, being G-quartets formed from sense C9-transcripts (Haeusler et al., 2014) or RAN translated PR dipeptides formed from antisense C9-transcripts could converge towards the same mechanism(s). More studies are needed to tease out the many possible toxic combinations that could originate from C9 transcripts, and more importantly, to determine what triggers them. Of the five possible C9RAN dipeptides tested, only the ariginine-rich PR and GR were neurotoxic. They were also the only two DRPs with a nuclear localization profile. In particular, the risk of death of neurons expressing PR considerably increased if nuclear aggregates were present. PR nuclear aggregation was confirmed both in human induced motor neurons derived from iPS cells and in postmortem spinal cord tissue of C9-ALS patients. Morever, aggressive toxicity and nuclear aggregation phenotype were also seen in human iPSCs-derived neurons from non-diseased individual transfected with PR50 and in vivo in a Drosophila model. Thus, our data suggested that the arginine-rich poly-peptides are the key toxic C9RAN products in C9-ALS/FTD. Live-cell imaging analysis showed that PR aggregates are stable and persist in the dish long after the neuron has died. Interestingly, we observed that in induced motor neuron cultures derived from C9 patients, there was an increased number of extracellular PR aggregates in the dish, which paralleled increased cell loss relative to control cultures, suggesting that PR-bearing C9 patient neurons did not survive in culture and the cells underwent a selection process. In line with recently reported histopathological analysis of DRP formation in postmortem CNS tissues of C9-ALS/FTD patients, our risk of death analysis underscores PR strong neurotoxic potential and predicts that only evidence of non-toxic DRP formation may be preponderant in these tissue autopsies. It is, therefore, not surprising that PR aggregates have been seldom found in post-mortem disease-affected tissue of C9-patients. On the other hand, GA, GR, PA, and GP inclusions, which are mild or not neurotoxic in our assays, have been found throughout different CNS areas, including those not affected by disease (Gendron et al., 2013a; Gendron et al., 2013b; Zu et al., 2013). Nonetheless, we provided here clear evidence for PR nuclear aggregates, including instances of nucleolar localization, in spinal cord tissues of C9-ALS patients. Similarly, Pickering-Brown and colleagues found evidence of PR proteins that displayed strong immunoreactivity of chromatin in pyramidal cells of the hippocampus, suggesting nuclear localization of the PR polypeptides (Mann et al., 2013). Whether PR aggregation is cell type specific in the human disease context remains to be further examined.
Our studies differentiated the possible role of RAN dipeptides in disease. While GA, GR, PA, and GP could represent pathological hallmarks detectable in surviving neurons, PR is the pathogenic candidate whose presence predisposes neurons to quickly degenerate and die. For disease-relevant mechanisms, the predictive nature of our culture assay is also validated by the cell- and mutation-specificity of our findings. Hippocampal neurons, which are seemingly less affected in C9-FTD and C9-ALS/FTD, are also significantly less sensitive to PR toxicity in cultures. Resilience to PR and/or R42 toxicity in cells not affected by disease and the mutant-specific vulnerability of different neuronal types implied the occurrence of molecular pathways capable of defining disease phenotypes. The message that different cells respond differently to same toxic insults and that pathological markers do not necessarily correlate with disease pathogenesis underscores the importance of dissecting disease mechanisms, even in relatively simple systems. The knowledge could be critical when identifying relevant and specific therapeutic targets.
But the most pressing question for the development of therapies aimed at stopping or even preventing neurotoxicity is how PR, or more in general, arginine-rich expanded poly-dipeptides give rise to neurodegeneration. Different groups have reported evidence for alterations in translational control of gene expression, characterized by alterations in processing bodies (P-bodies) and appearance of stress granules (SGs) in cell models of neurodegenerative disease (Nonhoff et al., 2007; Savas et al., 2008). In this respect, an interesting parallel can be drawn with the neurotoxicity of PR dipeptides. In neurons with nuclear PR aggregates, we found a significant reduction in the number of P-bodies concomitant to increased size average and the appearance of SGs. These RNA granules are silencing foci as they typically harbor transcripts circumstantially excluded from the translationally active pool (Thomas et al., 2011). P-bodies are constitutive, but respond to stimuli that affect mRNA translation and decay. SGs are specifically induced upon cellular stress, which triggers a global translational silencing of several pathways. Various changes in cell physiology are able to affect these RNA granules in size and numbers. At this point of our investigation, however, it is not possible to establish a firm temporal sequence of events leading to neurodegeneration as there is not enough functional data to support dysregulation of RNA granules as the leading mechanism of PR aggregates toxicity.
Interestingly, P-bodies dysregulation may result from nucleolar stress (Kedersha et al., 2008). We found that PR aggregates aberrantly accumulate in the nucleolus, which appears enlarged. Nucleoli control ribosome biogenesis and protein trafficking under cellular stress (Nalabothula et al., 2010). Thus, in the case of PR-containing neurons, where the nucleolus is enlarged and the number of P-bodies reduced, it is tempting to hypothesize PR-induced nucleolar stress as the mechanism that marks neurons destined to degenerate. Similarly to what we found here for PR-mediated neurotoxicity, Wang and colleagues have recently proposed nucleolar stress followed by alterations in P-bodies as possible mechanism(s) for G4C2 hexanucleotide repeat expansions toxicity (Haeusler et al., 2014). Additionally, the synergistic neurotoxic effect between R42 transcripts and PR dipeptides points at nucleolar stress as a converging mechanism between these two toxic insults.
Overall, the data presented here support a gain-of-function mechanism of C9ORF72-ALS/FTD toxicity and identify relevant toxic species in this gain-of-function pathway(s). Loss of C9ORF72 function achieved by knockdown approach did not impact survival of both, cortical and motor neurons, suggesting that loss of C9ORF72 function may not be one mechanism contributing to neurodegeneration. However, we do not know what could be the impact on neurons of knocking down C9ORF72 over the time scale of the human lifetime. Therefore, we cannot exclude for sure the loss-of-function hypothesis as one mechanism contributing to neurodegeneration in C9-ALS/FTD.
The identification of C9ORF72-linked toxic species could allow for the design of targeted therapeutics that are specifically tailored towards disrupting their formation rather than affecting the downstream cascade of events. In particular, this would reduce the risk for unwanted side effects. Since DRP toxicity seems dependent on PR nuclear accumulation, perhaps an approach aimed selectively at preventing or breaking these nuclear aggregates would be more likely to succeed.
EXPERIMENTAL PROCEDURES
Primary neuronal cultures and transfections
Primary cortical/hippocampal neurons were prepared from E19 rat embryos as previously described (Kayser et al., 2006). Cortical neurons were plated at a density of 90,000/well on poly-D-lysine and laminin coated 24-well plates or 12 mm coverslips. Hippocampal neurons were plated at a density of 25,000/well on poly-D-lysine and laminin coated 24-well plates or 12 mm coverslips. 5 μM Ara-C was added 3 days after plating to inhibit proliferation of dividing cells. Purified primary motor neurons were prepared from E14.5 rat embryos as previously described (Magrane et al., 2012). Motor neurons were plated at a density of 15,000/well on poly-D,L-ornithine and laminin coated 24-well plates or coverslips. Cortical/hippocampal neurons were transfected using Lipofectamine 2000 at DIV 10 (1 μg of total DNA/well), while motor neurons at DIV 5–7 (0.5 μg of total DNA/well). Synapsin-driven Td-Tomato reporter construct was co-transfected in cortical/hippocampal neurons and motor neurons with C9-related cDNA constructs at a ratio of 1:4.
Immunocytochemistry and confocal microscopy
Primary neuronal cultures were fixed in 3.6% PFA in PBS for 10 minutes at room temperature. Neurons were then permealized in 0.25% PBST for 15 minutes at room temperature and blocked in 1% BSA in PBST for 1 h at room temperature, incubated with primary antibodies at 4 °C overnight, secondary antibodies at room temperature for 1 h and mounted with anti-DAPI prolong-gold anti-fade mounting media (Invitrogen). Neurons were washed with PBS three times between each step. The following primary antibodies were used: α-C9ORF72 (Sigma#HPA023873, 1:200), α-Myc (Cell Signaling#2276, 1:1,000), α-TIAR (BD Transduction Laboratories#610352, 1:250), α-DCP1A (Sigma#D5444, 1:500), α-Nucleolin (ProteinTech#10556-1-AP, 1:2,000), α-SMI32 (Covance#SMI-32R, 1:1,000), α-MAP2 (Millipore#AB5622, 1:500), α-Fibrillarin (SantaCruz#sc-25397, 1:200) α-TU20 (Abcam#ab7751 1:500), α-HB9 (DSHB#81.5C10, 1:5), α-Tuj1 (Millipore#AB9354, 1:1,000), α-PR (ProteinTech#23979-1-AP, 1:1,000). Cells were imaged using confocal microscopy (Olympus FV1000). 6–10 images at a 0.3 μm step-size were acquired and projected onto one single image.
Longitudinal live-cell imaging analysis
For longitudinal live-cell imaging and neuronal survival analysis, we used an automated imaging system consisting of an inverted Nikon Eclipse Ti microscope equipped with PerfectFocus, a Tokai Hit stage top incubator with gas and temperature controller and a CoolSNAP ES2 High-performance CCD camera.
Quantification of nucleoli and Processing bodies (P-bodies)
The number and size of P-bodies and nucleoli size were quantified with ImageJ software (NIH) using a manual and an automatic method. The manual method is based on the Cell Counter plugin to mark individual P-bodies by hand. P-bodies were counted based on size of fluorescence. The automatic method relied on an unbiased approach using a computational algorithm built in. Confocal images showing fluorescence of the soma and P-bodies were converted to 16-bit grayscale and the soma was traced to create boundaries for the analysis. Threshold of the image was then adjusted to highlight P-bodies and the background was subtracted to split overlapping P-bodies. The P-bodies were then analyzed using the Analyze Particles plugin with a minimum pixel size of 2. The average size of the P-bodies was also retrieved through this method. Overall, there was no significant difference between the manual and automatic methods. Nucleoli size was quantified by mean of the nucleolin fluorescence staining using the automatic method in the ImageJ software. The nucleus for each cell was traced for the boundaries and the area of the nucleus analyzed using the measure function. Using the same boundaries, the threshold of the image was then adjusted to highlight nucleolin fluorescence area and the background was subtracted. Nucleolin area was then analyzed using the Analyze Particles plugin with a minimum pixel size of 2. The measured nucleolin area was compared as a percentage of the nucleus area.
Fly transgenesis and experiments
Transgenic flies that carry GAL4-activatable genes encoding RAN proteins were generated by the BestGene Inc. Eye expression, motor neuron expression and muscle expression were induced by crossing C9RAN protein flies with the GMR GAL4 strain (Bloomington stock Center), the OK371 GAL4 strain (Mahr and Aberle, 2006) and the MHC GAL4 strain at 25 °C respectively. Eye phenotype was scored and differences between the genotypes were assessed using one-way ANOVA and Scheffe’s post-hoc test. For the viability assay C9RAN protein flies containing the TM6b balancer were crossed with OK371 GAL4 driver flies. Further details are provided in supplemental material.
Statistics
For survival analysis, death was defined by the time at which a neuron was last imaged with intact neurites and no evidence of swollen soma or cell blebbing. Kaplan-Meier survival and Cox proportional hazards analysis were carried out using SPSS 19.0 software, while other statistical analysis was carried out using GraphPad Prism 6. Differences in Kaplan-Meier curves were assessed with the log-rank test. Comparisons between experimental groups in bar graphs were done by one-way ANOVA or t-test as indicated. Data are expressed as average ± sem. P<0.05 was considered statistically significant.
Supplementary Material
Figure S1. Randomized codon sequences encoding the different DRPs. Randomized codons sequences encoding the indicated DRPs. Sequences represent individual modules that are repeated in the construct to achieve the desired encoded DRP length. The cloning strategy is described in Supplementary Experimental Procedures.
Figure S2. Validation of DRP constructs and cellular localization of C9RAN DRPs. (A) Validation of DRP constructs by immunoblotting using transiently transfected NSC34 cells. DRP constructs (50 repeats long) in pcDNA3.1 vectors were transfected in NSC34 cells (1 μg DNA/well). Cells homogenates were prepared 48 hours post-transfection and analyzed on Tricine 10–20% gradient gels. (B) Analysis of DRPs expression levels was done by probing western blot of NSC34 cell homogenates with anti-GFP antibody. Among different DRPs, the arginine–rich dipeptides PR and GR showed consistently lower expression levels. (C) Immunofluorescence performed on NSC34 cells transfected with different DRPs. Confocal microscopy on cells stained with anti-GFP antibody at 48 hours post-transfection. Calibration bar is 20 μm. (D) Confocal microscopy imaging of cortical neurons co-transfected at DIV10 with the indicated DRP and synapsin-driven Td-tomato construct to aid visualization of neurons. Cortical neurons were imaged 48 hours post-transfection. Expression levels of Td-tomato reporter protein were noticed to be consistently lower in PR-expressing neurons. Exposure settings were uniform throughout confocal analysis. Arrows point at aggregates within the soma and neurites (GA50) and nucleus (GR50 and PR50). Calibration bar is 20 μm. (E) DRP constructs (50 repeats long) in pcDNA3.1 vectors were transfected in primary rat motor neurons at DIV5 (0.5 μg DNA/well). Neurons were imaged by confocal microscopy 24 hours post-transfection. Calibration bar is 20 μm. Arrows point at aggregates within neurites (GA50) and nucleus (GR50 and PR50). Calibration bar is 20 μm.
Figure S3. Time-lapse imaging of individual neurons in culture. (A) Schematic of live-cell longitudinal tracking experiments. Primary cortical neurons are transfected at DIV 10 and transfected neurons (Td-tomato+) in the same optical field are imaged at a 24h interval for up to 9 days post-transfection. To monitor individual neurons, a synapsin promoter driven Td-Tomato construct was co-transfected as a sensitive reporter of survival. (B) Representative images of cortical neurons co-transfected with Td-tomato reporter construct driven by synapsin promoter and GFP-control construct. Calibration bar is 100 μm. (C) Cortical neurons were imaged at transfection day 0 in bright field, which corresponds to DIV10. Image shows full maturation of the cortical neurons in vitro. The same neurons in the optical field were then imaged for 8 consecutive days post-transfection at 24-hour intervals using Td-tomato as fluorescent reporter. Arrows point at 4 neurons that were successfully transfected in this field. Time-lapse images show the increased expression of the Td-tomato reporter construct in those neurons that allows visualization of the in vitro neurites’ network. Calibration bar is 100 μm.
Figure S4. Survival analysis of different DRPs transfected in motor and hippocampal neurons. (A) Representative live-cell images of motor neurons co-transfected with Td-Tomato (0.1 μg/well; red fluorescence signal shown in top panels) and PR50 cDNA plasmids (0.4 μg/well; green fluorescence in bottom panels). Motor neuron with aggregates died, while motor neuron with diffused PR50 expression (orange arrow and inlet) did not undergo neurodegeneration. Calibration bar is 20 μm. (B) Kaplan-Meier survival analysis suggested that both PR50 and GR50 were toxic to primary motor neurons compared to control (***P<0.001). Although a trend was observed for GA50 expressing motor neurons difference with control did not reach significance. At least 40 neurons were followed/group; n=3 independent experiments. (C) Kaplan-Meier survival analysis of hippocampal neurons transfected with different DRP constructs showed that PR50 was toxic to hippocampal neurons. At least 80 neurons were followed/group; n=3 independent experiments. (D) However, hippocampal neurons were less vulnerable to PR50 compared to cortical neurons. *P<0.05, ***P<0.001. (E) Expression levels of GFP were quantified 72 hours post-transfection by confocal microscopy measuring immunofluorescence intensity in each neurons. At least 20 cortical and hippocampal neurons were imaged and quantified (Image J). Camera acquisition parameters were set the same between the two groups (unpaired t-test; P=0.0824). (F) Expression of untagged PR50 causes cortical neuron death, which is not significantly different from that caused by GFP-PR50 as shown by Kaplan-Meier survival analysis. At least 40 neurons/group; n = 4–6 experiments; ***P<0.001. (G) Immunoflurescence analysis using α-PR antibody shows that untagged PR50 forms nuclear aggregates. Td-Tomato signal indicates a neuron transfected with untagged PR50. (H) Western blot using lysates of NSC34 transfected with untagged PR50 shows that untagged PR50 is recognized by α-PR antibody and runs at predicted molecular weight. (I, J) Survival analysis of primary cortical and motor neurons transfected with human wild type SOD1 or the ALS-linked SOD1-G93A mutant. Expression of ALS-linked mutant SOD1-G93A did not affect survival of cortical neurons (I), while it was neurotoxic to motor neurons (J). At least 80 cortical and 40 motor neurons are followed/group; at least 3 independent experiments. ***P<0.001.
Figure S5. Generation of GFP plasmids containing intronic G4C2 repeats expansion. (A) Schematic of GFP construct harboring intronic GGGGCC repeat expansions. The insert has been subcloned into the GFP splicing reporter plasmid, pGint (plasmid 24217; Addgene; pEGFP-N1 vector backbone) (B) Restriction analysis showing the correct calculated size for the R0, R21 and R42 inserts. Lanes indicate individual bacterial colonies from which the DNA has been extracted. Confirmation of the presence or absence of GGGGCC repeat sequences in R0, R21 and R42 constructs was obtained by sequencing. Analysis of the R21 and R42 sequence was only partially successful due to their high GC content. (C) Representative images of co-transfection strategy. Cortical neurons were co-transfected with 0.2 μg of Td-tomato cDNA and 0.8 μg of R42 cDNA constructs/well at DIV10 and live imaged over time. Each neuron expressing Td-tomato fluorescent protein also expressed GFP from the R42 construct. Calibration bar is 100 μm. (D) Immunoblotting analysis of NSC34 cells transfected with R0–42 constructs showed that the presence of G4C2 repeats in the intronic sequence of the construct did not affect GFP expression in NSC34 cells, suggesting correct splicing. (E) Fluorescent in situ hybridization (FISH) analysis shows RNA nuclear inclusions detected in NSC34 cells transfected with R21–42 constructs. R0 construct served as control did not produce nuclear inclusions. DAPI was used to stain the nucleus. Cells were fixed and in situ hybridized with Cy3-linked (CCCCGG)4 probe. Red staining represents positive hybridization of the probe. Calibration bar is 5 μm.
Figure S6. Nuclear localization of PR50 and GR50 and association with nucleolin. (A) Immunofluorescence staining of PR50 transfected motor neuron with SMI32 and nucleolin showed that PR nuclear aggregates co-localized with nucleolin. DAPI: blue; Nucleolin: red; GFP-CTRL or GFP-PR50: green; SMI32: magenta. (B) 3D reconstructions of cortical neurons transfected with PR50 and stained for nucleolin. Nuclear aggregates formed by PR appeared enclosed by nucleolin. DAPI: blue; Nucleolin: red; GFP-PR50: green. (C, D) PR nucleolar staining was confirmed by its co-localization with fibrillarin. Orthogonal view showing co-localization of PR nuclear aggregates and fibrillarin. DAPI: blue; GFP-PR50: green; Fibrillarin: magenta. Calibration bars are 10 μm. (E) 3D reconstruction of cortical neurons transfected with GR50 and stained for nucleolin. DAPI: blue; Nucleolin: red; GFP-GR50: green. Calibration bars are 10 μm.
Figure S7. PR aggregates persist in the dish after the neuron has degenerated and lost membrane integrity. Motor neurons were co-transfected at DIV 5 with Td-Tomato and GFP-PR50 and monitored over time. At 24 hours post-transfection, distinct nuclear aggregates were detected. At 28 hours post-transfection, nuclear aggregates became more prominent and the soma swelled. At 36 hours post-transfection, the motor neuron lost its membrane integrity and died, while prominent PR aggregates remained. Calibration bar is 100 μm.
Figure S8. Nuclear localization of PR aggregates in spinal cord tissues from C9 ALS and C9 ALS/FTD patients. (A, B) Characterization of the α-PR antibody from ProteinTech. HEK-293 cells were transfected with DRP constructs and the control construct, and cell lysates were subjected to immunoblotting. The α-PR antibody from ProteinTech recognized poly-PR at predicted molecular weight with decent specificity. Actin was used as a loading control. Immunofluorescence analysis suggests that the α-PR antibody from ProteinTech specifically recognizes poly-PR. DAPI: blue; GFP-PR50: green; α-PR: red. Calibration bar is 10 μm. (C) Intensity of nuclear and extranuclear PR staining in human tissues was quantified subjectively by 3 blinded investigators. 10 representative 15-slice z-series images from each patient (a total of 80 images) were randomized for each investigator. The investigators ranked the PR fluorescent levels as absent, light, moderate, or heavy. The rankings were added up and taken as a percent of images quantified. − = 0%; +/− = 1 – 25%; + = 26 – 50%; ++ = 51 – 75%; +++ = 76 – 100%. (D) Orthogonal view of nuclear PR inclusion in C9-ALS/FTD patient spinal cord section. DAPI: blue; PR: red. Calibration bar is 10 μm.
Highlights.
Knock down of C9ORF72 is not toxic to cortical and motor neurons;
Poly-PR nuclear aggregates are potently neurotoxic in vitro and in vivo;
Cell stress responses mark poly-PR mediated neurotoxicity;
Intronic G4C2 transcripts and poly-PR proteins synergize to cause neurotoxicity.
Acknowledgments
We thank Dr. Petrucelli for providing antibodies against poly-DRPs, Drs. Hanamura and Dalva for providing embryonic rat cortices for cortical neuron preparations and Td-tomato construct, and Louise Menendez for technical help. We also thank members of the Weinberg Unit for ALS research for suggestions and helpful discussion. Human post-mortem tissues were obtained from the Alzheimer’s Disease Research Center Neuropathology core(NIA P50 AG05142), and Target ALS at Columbia. This work was supported by the Farber Family Foundation (to D.T. and P.P.), the National Institutes of Health (NIH) grants RO1-NS44292 (to D.T.), RO1-NS051488 (to P.P.), RO1-NS081303, the Robert Packard Center for ALS research, the ALS Association (to U.B.P.), R00NS07743 and the Donald E. and Delia B. Baxter Foundation (to J.K.I.).
Footnotes
AUTHOR CONTRIBUTIONS
X.W. was involved in the design and analysis of all the experiments of this study, performed live-cell imaging, confocal microscopy, western blotting, and optimized and prepared neuronal cultures. W.T. generated constructs for cell transfections and optimized and prepared neuronal cultures, and performed FISH analysis. T.W. optimized human tissue staining and performed imaging analysis and iPSC-derived neuron survival analysis. J.M. generated constructs for fly transgenesis. J.M. and U.B.P. were involved in the analysis of the phenotype of transgenic flies. K.K., S.S., P.P. were involved in human tissue and related imaging analysis. N.A.S. provided human tissue autopsies and contributed to tissue analysis. Y.S., S.L. and J.K.I. generated human-induced motor neurons and performed immunofluorescence analysis on human-induced motor neurons. D.T. wrote the manuscript, oversaw project development, experimental design and data interpretation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne S, Elamin M, Bede P, Shatunov A, Walsh C, Corr B, Heverin M, Jordan N, Kenna K, Lynch C, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012;11:232–240. doi: 10.1016/S1474-4422(12)70014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, Brice A, Kabashi E. Loss of function of C9orf72 causes motor deficits in a zebrafish model of Amyotrophic Lateral Sclerosis. Ann Neurol. 2013 doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- Daigle JG, Lanson NA, Jr, Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22:1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elamin M, Bede P, Byrne S, Jordan N, Gallagher L, Wynne B, O’Brien C, Phukan J, Lynch C, Pender N, Hardiman O. Cognitive changes predict functional decline in ALS: a population-based longitudinal study. Neurology. 2013;80:1590–1597. doi: 10.1212/WNL.0b013e31828f18ac. [DOI] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013a;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Cosio DM, Petrucelli L. c9RAN translation: a potential therapeutic target for the treatment of amyotrophic lateral sclerosis and frontotemporal dementia. Expert Opin Ther Targets. 2013b;17:991–995. doi: 10.1517/14728222.2013.818659. [DOI] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014 doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, McClelland AC, Hughes EG, Dalva MB. Intracellular and trans-synaptic regulation of glutamatergic synaptogenesis by EphB receptors. J Neurosci. 2006;26:12152–12164. doi: 10.1523/JNEUROSCI.3072-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Tisdale S, Hickman T, Anderson P. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods in enzymology. 2008;448:521–552. doi: 10.1016/S0076-6879(08)02626-8. [DOI] [PubMed] [Google Scholar]
- Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, Burrell JR, Zoing MC. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanson NA, Jr, Maltare A, King H, Smith R, Kim JH, Taylor JP, Lloyd TE, Pandey UB. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet. 2011;20:2510–2523. doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, et al. Hexanucleotide Repeats in ALS/FTD Form Length-Dependent RNA Foci, Sequester RNA Binding Proteins, and Are Neurotoxic. Cell reports. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuisse C, Martin LJ. Long-term culture of mouse cortical neurons as a model for neuronal development, aging, and death. J Neurobiol. 2002;51:9–23. doi: 10.1002/neu.10037. [DOI] [PubMed] [Google Scholar]
- Magrane J, Sahawneh MA, Przedborski S, Estevez AG, Manfredi G. Mitochondrial dynamics and bioenergetic dysfunction is associated with synaptic alterations in mutant SOD1 motor neurons. J Neurosci. 2012;32:229–242. doi: 10.1523/JNEUROSCI.1233-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene expression patterns: GEP. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simon-Sanchez J, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M, et al. Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta neuropathologica communications. 2013;1:68. doi: 10.1186/2051-5960-1-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Muotri AR, Mu Y, Smith AM, Cezar GG, Gage FH. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Isaacs AM. C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia: gain or loss of function? Curr Opin Neurol. 2014;27:515–523. doi: 10.1097/WCO.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Nalabothula N, Indig FE, Carrier F. The Nucleolus Takes Control of Protein Trafficking Under Cellular Stress. Molecular and cellular pharmacology. 2010;2:203–212. [PMC free article] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Molecular biology of the cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biology of the cell/under the auspices of the European Cell Biology Organization. 1985;54:123–133. doi: 10.1111/j.1768-322x.1985.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Phukan J, Elamin M, Bede P, Jordan N, Gallagher L, Byrne S, Lynch C, Pender N, Hardiman O. The syndrome of cognitive impairment in amyotrophic lateral sclerosis: a population-based study. J Neurol Neurosurg Psychiatry. 2012;83:102–108. doi: 10.1136/jnnp-2011-300188. [DOI] [PubMed] [Google Scholar]
- Phukan J, Pender NP, Hardiman O. Cognitive impairment in amyotrophic lateral sclerosis. Lancet Neurol. 2007;6:994–1003. doi: 10.1016/S1474-4422(07)70265-X. [DOI] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SY, Macgregor RB, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9ORF72 gene forms tract length-dependent uni- and multi-molecular RNA G-quadruplex structures. J Biol Chem. 2013 doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Heckman MG, Dejesus-Hernandez M, Baker MC, Soto-Ortolaza AI, Rayaprolu S, Stewart H, Finger E, Volkening K, Seeley WW, et al. Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype. Neurobiology of aging. 2012;33:2950 e2955–2957. doi: 10.1016/j.neurobiolaging.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Makusky A, Ottosen S, Baillat D, Then F, Krainc D, Shiekhattar R, Markey SP, Tanese N. Huntington’s disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci U S A. 2008;105:10820–10825. doi: 10.1073/pnas.0800658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cellular signalling. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren JD, Rohrer JD, Rossor MN. Clinical review. Frontotemporal dementia. BMJ. 2013;347:f4827. doi: 10.1136/bmj.f4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollacott IO, Mead S. The C9ORF72 expansion mutation: gene structure, phenotypic and diagnostic issues. Acta Neuropathol. 2014;127:319–332. doi: 10.1007/s00401-014-1253-7. [DOI] [PubMed] [Google Scholar]
- Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C, Zheng Y, Ghani M, Dib S, Keith J, et al. Hypermethylation of the CpG Island Near the G4C2 Repeat in ALS with a C9orf72 Expansion. Am J Hum Genet. 2013;92:981–989. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, Jin P. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014;128:505–524. doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Randomized codon sequences encoding the different DRPs. Randomized codons sequences encoding the indicated DRPs. Sequences represent individual modules that are repeated in the construct to achieve the desired encoded DRP length. The cloning strategy is described in Supplementary Experimental Procedures.
Figure S2. Validation of DRP constructs and cellular localization of C9RAN DRPs. (A) Validation of DRP constructs by immunoblotting using transiently transfected NSC34 cells. DRP constructs (50 repeats long) in pcDNA3.1 vectors were transfected in NSC34 cells (1 μg DNA/well). Cells homogenates were prepared 48 hours post-transfection and analyzed on Tricine 10–20% gradient gels. (B) Analysis of DRPs expression levels was done by probing western blot of NSC34 cell homogenates with anti-GFP antibody. Among different DRPs, the arginine–rich dipeptides PR and GR showed consistently lower expression levels. (C) Immunofluorescence performed on NSC34 cells transfected with different DRPs. Confocal microscopy on cells stained with anti-GFP antibody at 48 hours post-transfection. Calibration bar is 20 μm. (D) Confocal microscopy imaging of cortical neurons co-transfected at DIV10 with the indicated DRP and synapsin-driven Td-tomato construct to aid visualization of neurons. Cortical neurons were imaged 48 hours post-transfection. Expression levels of Td-tomato reporter protein were noticed to be consistently lower in PR-expressing neurons. Exposure settings were uniform throughout confocal analysis. Arrows point at aggregates within the soma and neurites (GA50) and nucleus (GR50 and PR50). Calibration bar is 20 μm. (E) DRP constructs (50 repeats long) in pcDNA3.1 vectors were transfected in primary rat motor neurons at DIV5 (0.5 μg DNA/well). Neurons were imaged by confocal microscopy 24 hours post-transfection. Calibration bar is 20 μm. Arrows point at aggregates within neurites (GA50) and nucleus (GR50 and PR50). Calibration bar is 20 μm.
Figure S3. Time-lapse imaging of individual neurons in culture. (A) Schematic of live-cell longitudinal tracking experiments. Primary cortical neurons are transfected at DIV 10 and transfected neurons (Td-tomato+) in the same optical field are imaged at a 24h interval for up to 9 days post-transfection. To monitor individual neurons, a synapsin promoter driven Td-Tomato construct was co-transfected as a sensitive reporter of survival. (B) Representative images of cortical neurons co-transfected with Td-tomato reporter construct driven by synapsin promoter and GFP-control construct. Calibration bar is 100 μm. (C) Cortical neurons were imaged at transfection day 0 in bright field, which corresponds to DIV10. Image shows full maturation of the cortical neurons in vitro. The same neurons in the optical field were then imaged for 8 consecutive days post-transfection at 24-hour intervals using Td-tomato as fluorescent reporter. Arrows point at 4 neurons that were successfully transfected in this field. Time-lapse images show the increased expression of the Td-tomato reporter construct in those neurons that allows visualization of the in vitro neurites’ network. Calibration bar is 100 μm.
Figure S4. Survival analysis of different DRPs transfected in motor and hippocampal neurons. (A) Representative live-cell images of motor neurons co-transfected with Td-Tomato (0.1 μg/well; red fluorescence signal shown in top panels) and PR50 cDNA plasmids (0.4 μg/well; green fluorescence in bottom panels). Motor neuron with aggregates died, while motor neuron with diffused PR50 expression (orange arrow and inlet) did not undergo neurodegeneration. Calibration bar is 20 μm. (B) Kaplan-Meier survival analysis suggested that both PR50 and GR50 were toxic to primary motor neurons compared to control (***P<0.001). Although a trend was observed for GA50 expressing motor neurons difference with control did not reach significance. At least 40 neurons were followed/group; n=3 independent experiments. (C) Kaplan-Meier survival analysis of hippocampal neurons transfected with different DRP constructs showed that PR50 was toxic to hippocampal neurons. At least 80 neurons were followed/group; n=3 independent experiments. (D) However, hippocampal neurons were less vulnerable to PR50 compared to cortical neurons. *P<0.05, ***P<0.001. (E) Expression levels of GFP were quantified 72 hours post-transfection by confocal microscopy measuring immunofluorescence intensity in each neurons. At least 20 cortical and hippocampal neurons were imaged and quantified (Image J). Camera acquisition parameters were set the same between the two groups (unpaired t-test; P=0.0824). (F) Expression of untagged PR50 causes cortical neuron death, which is not significantly different from that caused by GFP-PR50 as shown by Kaplan-Meier survival analysis. At least 40 neurons/group; n = 4–6 experiments; ***P<0.001. (G) Immunoflurescence analysis using α-PR antibody shows that untagged PR50 forms nuclear aggregates. Td-Tomato signal indicates a neuron transfected with untagged PR50. (H) Western blot using lysates of NSC34 transfected with untagged PR50 shows that untagged PR50 is recognized by α-PR antibody and runs at predicted molecular weight. (I, J) Survival analysis of primary cortical and motor neurons transfected with human wild type SOD1 or the ALS-linked SOD1-G93A mutant. Expression of ALS-linked mutant SOD1-G93A did not affect survival of cortical neurons (I), while it was neurotoxic to motor neurons (J). At least 80 cortical and 40 motor neurons are followed/group; at least 3 independent experiments. ***P<0.001.
Figure S5. Generation of GFP plasmids containing intronic G4C2 repeats expansion. (A) Schematic of GFP construct harboring intronic GGGGCC repeat expansions. The insert has been subcloned into the GFP splicing reporter plasmid, pGint (plasmid 24217; Addgene; pEGFP-N1 vector backbone) (B) Restriction analysis showing the correct calculated size for the R0, R21 and R42 inserts. Lanes indicate individual bacterial colonies from which the DNA has been extracted. Confirmation of the presence or absence of GGGGCC repeat sequences in R0, R21 and R42 constructs was obtained by sequencing. Analysis of the R21 and R42 sequence was only partially successful due to their high GC content. (C) Representative images of co-transfection strategy. Cortical neurons were co-transfected with 0.2 μg of Td-tomato cDNA and 0.8 μg of R42 cDNA constructs/well at DIV10 and live imaged over time. Each neuron expressing Td-tomato fluorescent protein also expressed GFP from the R42 construct. Calibration bar is 100 μm. (D) Immunoblotting analysis of NSC34 cells transfected with R0–42 constructs showed that the presence of G4C2 repeats in the intronic sequence of the construct did not affect GFP expression in NSC34 cells, suggesting correct splicing. (E) Fluorescent in situ hybridization (FISH) analysis shows RNA nuclear inclusions detected in NSC34 cells transfected with R21–42 constructs. R0 construct served as control did not produce nuclear inclusions. DAPI was used to stain the nucleus. Cells were fixed and in situ hybridized with Cy3-linked (CCCCGG)4 probe. Red staining represents positive hybridization of the probe. Calibration bar is 5 μm.
Figure S6. Nuclear localization of PR50 and GR50 and association with nucleolin. (A) Immunofluorescence staining of PR50 transfected motor neuron with SMI32 and nucleolin showed that PR nuclear aggregates co-localized with nucleolin. DAPI: blue; Nucleolin: red; GFP-CTRL or GFP-PR50: green; SMI32: magenta. (B) 3D reconstructions of cortical neurons transfected with PR50 and stained for nucleolin. Nuclear aggregates formed by PR appeared enclosed by nucleolin. DAPI: blue; Nucleolin: red; GFP-PR50: green. (C, D) PR nucleolar staining was confirmed by its co-localization with fibrillarin. Orthogonal view showing co-localization of PR nuclear aggregates and fibrillarin. DAPI: blue; GFP-PR50: green; Fibrillarin: magenta. Calibration bars are 10 μm. (E) 3D reconstruction of cortical neurons transfected with GR50 and stained for nucleolin. DAPI: blue; Nucleolin: red; GFP-GR50: green. Calibration bars are 10 μm.
Figure S7. PR aggregates persist in the dish after the neuron has degenerated and lost membrane integrity. Motor neurons were co-transfected at DIV 5 with Td-Tomato and GFP-PR50 and monitored over time. At 24 hours post-transfection, distinct nuclear aggregates were detected. At 28 hours post-transfection, nuclear aggregates became more prominent and the soma swelled. At 36 hours post-transfection, the motor neuron lost its membrane integrity and died, while prominent PR aggregates remained. Calibration bar is 100 μm.
Figure S8. Nuclear localization of PR aggregates in spinal cord tissues from C9 ALS and C9 ALS/FTD patients. (A, B) Characterization of the α-PR antibody from ProteinTech. HEK-293 cells were transfected with DRP constructs and the control construct, and cell lysates were subjected to immunoblotting. The α-PR antibody from ProteinTech recognized poly-PR at predicted molecular weight with decent specificity. Actin was used as a loading control. Immunofluorescence analysis suggests that the α-PR antibody from ProteinTech specifically recognizes poly-PR. DAPI: blue; GFP-PR50: green; α-PR: red. Calibration bar is 10 μm. (C) Intensity of nuclear and extranuclear PR staining in human tissues was quantified subjectively by 3 blinded investigators. 10 representative 15-slice z-series images from each patient (a total of 80 images) were randomized for each investigator. The investigators ranked the PR fluorescent levels as absent, light, moderate, or heavy. The rankings were added up and taken as a percent of images quantified. − = 0%; +/− = 1 – 25%; + = 26 – 50%; ++ = 51 – 75%; +++ = 76 – 100%. (D) Orthogonal view of nuclear PR inclusion in C9-ALS/FTD patient spinal cord section. DAPI: blue; PR: red. Calibration bar is 10 μm.