Abstract
B lymphoma Mo-MLV insertion region 1 (Bmi1) is a polycomb-family transcriptional factor critical for self-renewal in many adult stem cells and human neoplasia. We sought to identify microRNAs regulated by Bmi1 that could play a role in multipotent cardiac progenitor cell (CPC) decisions. We found that miR-300, a poorly characterized microRNA mapping in the Dlk1-Dio3 microRNA cluster, was positively regulated by Bmi1 in CPCs. Forced expression of miR-300 in CPCs promoted an improved stemness signature with a significant increase in Oct4 levels, a reduction in senescence progression and an enhanced proliferative status via p19 activation and inhibition of p16 accumulation. Endothelial and cardiogenic differentiation were clearly compromised by sustained miR-300 expression. Additionally, RNA and protein analysis revealed a significant reduction in key cardiac transcription factors, including Nkx2.5 and Tbx5. Collectively, these results suggest that some functions attributed to Bmi1 are due to induction of miR-300, which decreases the cardiogenic differentiation potential of multipotent CPCs in vitro and promotes self-renewal.
Bmi1 (B lymphoma Mo-MLV insertion region 1), a member of polycomb repressive complex 1 (PCR1), is a transcription factor involved in multiple biological processes, including embryonic development, organ formation, tumorogenesis, and stem cell self-renewal and differentiation.1 Deficiency in Bmi1 results in progressive postnatal growth retardation and neurological defects, confirming that Bmi1 is required for the self-renewal of stem cells in the peripheral and central nervous systems, but not for their survival or differentiation.2 In the absence of Bmi1, the cyclin-dependent kinase inhibitor p16Ink4a is generally upregulated,2, 3 reducing the rate of cellular proliferation. BMI1 is downregulated by replicative senescence in human fibroblasts and overexpression of BMI1 extends replicative life span, correlating with INK4A/ARF deregulation.4 Bmi1 has been reported to play crucial roles during the self-renewal and maintenance of hematopoietic, intestinal, bronchioalveolar, pancreatic, prostate, lung, and epithelial stem cells2, 3, 4, 5, 6 and more recently the tongue and rodent incisors.7 Thus, Bmi1 is possibly the most universal adult stem cell marker and a remarkable player in many cancer stem cells models.8, 9 In all models tested, Bmi1 dependence seems to distinguish stem cell self-renewal from committed progenitor proliferation.2
Multiple functions have been ascribed to Bmi1, underscoring its broad importance in numerous processes. Bmi1 facilitates the repression of Hox genes to prevent inappropriate differentiation and induces downregulation of Dickopf (DKK) proteins, resulting in upregulation of the Wnt target c-Myc, which in turn leads to transcriptional autoactivation of Bmi1.7 Reinforcement of this positive feedback loop regulating Bmi1 expression seems relevant in promoting cancer and maintaining the stem cell phenotype.9, 10 In addition, Bmi1 is transcriptionally controlled by phosphorylated Nanog,11, 12 and is inhibited by a plethora of miRNAs that are significantly downregulated in cancer models.12
Extensive studies on primed differentiation of murine embryonic stem (ES) and adult stem cells have established that efficient stem cell maintenance requires a highly concerted regulation of gene expression,13, 14 involving both coding genes and noncoding RNAs (ncRNAs). Among the regulatory elements involved in stem cell function, Oct4 protein, encoded by the Pou5f1 gene, is essential for the stemness properties of ES cells and is a key regulator of pluripotency in mammalian development. The Oct4 transcription factor is also indispensible for somatic cell reprogramming. Besides transcription factors, microRNAs (miRNAs) also play important roles in stem cell homeostasis. miRNAs are an abundant class of small ncRNAs that regulate the translation, stability, and localization of target messenger RNAs. Functional studies in ES cells have demonstrated critical roles for miRNAs, especially in regulating the balance between self-renewal and differentiation.14, 15 Less information is available about the role(s) of specific miRNAs in the regulation of adult stem cell systems, but some studies have explored possible collaborations; for example, miR-142-3p controls specification of definitive hemangioblasts during ontogeny.16, 17 Also, miR-21 strongly inhibits SOX2 expression in human mesenchymal stem cells (MSCs), resulting in reduced clonogenic and proliferative potential and cell-cycle arrest.18 Finally, Lin28a, a highly conserved RNA-binding protein expressed during embryogenesis, plays important roles in development, pluripotency, and metabolism (reviewed in Shyh-Chang et al.19), and its expression in several adult injury models improves repair processes in a manner mediated by its inhibition of let-7 miRNA biogenesis.19 Regarding the involvement of miRNAs in Bmi1 regulation, there are multiple descriptions of miRNAs targeting Bmi1 and many of them impact the corresponding stem or cancer stem cell function (Supplementary Table S1).
Adult resident cardiac stem/progenitor cells (CSCs/CPCs) are implicated in homeostatic turnover of the adult heart (reviewed in Malliaras et al.20). Among the different populations of putative CSCs isolated to date (reviewed in Martin-Puig et al.21 Urbanek et al.22 and Li et al.23), those expressing surface markers such as c-kit, SCA1, ATP-binding cassette Abcg2, or PDGFRα are the principal candidates. Indeed, resident CPCs with genetic elimination of Sca1 do not properly respond to pathological damage in vivo and show impaired growth and survival in vitro.24 In another study, Ellison et al.25 have proposed that cKIT+ CSCs are necessary and sufficient for adult heart turnover, although a recent lineage-tracing experiment questioned the relevance of cKIT+ CSCs to in vivo cardiomyocyte turnover.26 These authors claimed that, in all probability, diversity in CSC populations could be related to some heterogeneity within the CSC compartment.25 Nevertheless, two clinical evaluations have been initiated with promising results in early phases.27, 28
Here, using murine CPCs, we attempted to identify miRNAs that could be directly regulated by Bmi1. We hypothesized that those miRNAs upregulated in parallel with Bmi1 could be involved in the maintenance of the undifferentiated state. Conversely, miRNAs downregulated with Bmi1 might be involved in early commitment/differentiation decisions. We show that miR-300 is positively regulated in parallel with Bmi1 expression in CPCs, while miR-188 and miR-362 are inversely regulated. Furthermore, we confirm that miR-300 is a target of Bmi1 and show that forced expression of miR-300 favors the undifferentiated state and impedes differentiation of CPCs. Indeed, CPCs engineered to overexpress miR-300 exhibit compromised spontaneous endothelial differentiation and show a poor response to cardiomyogenic stimuli, an outcome that is possibly mediated by severe inhibition of Nkx2.5 expression.
Results
Bmi1 regulates miR-300 expression in CPCs
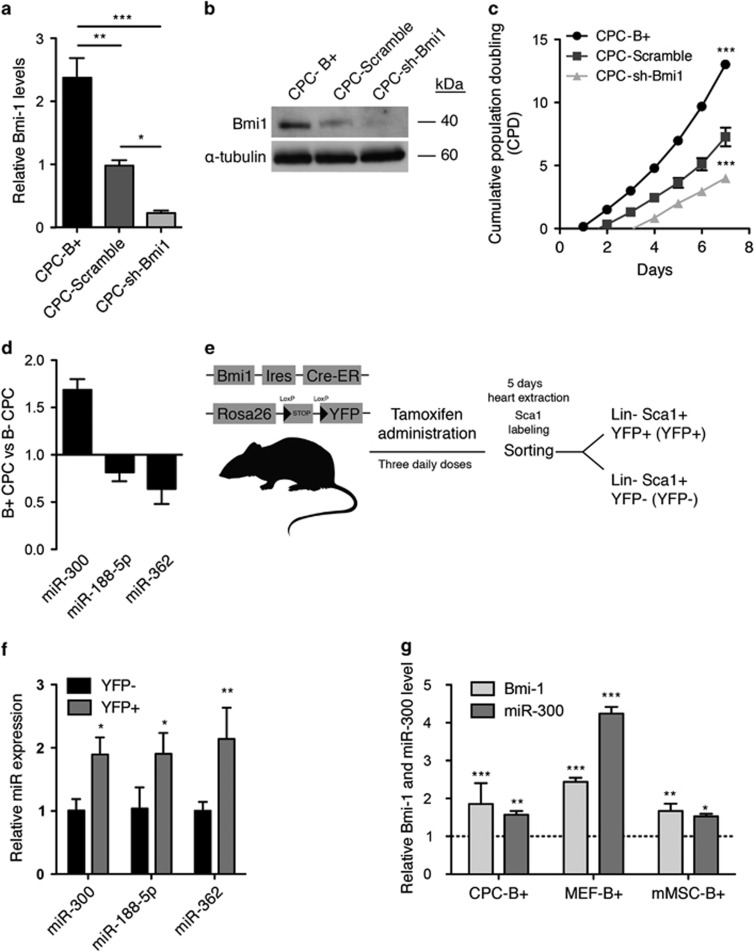
Freshly isolated Lin-SCA1+ CPCs (SCA1-CPCs) from heart are defined as Cd45 negative and express a panel of multipotency-related genes, including Tbx5, Klf4, c-Myc, Bmi1, and Meg3, and a distinctive profile of endothelial, fibroblast, and cardiac markers (Supplementary Figure S1). To assess the potential involvement of Bmi1 in the control of miRNA expression in SCA1-CPCs (hereafter named CPCs), we transduced freshly isolated CPCs with lentiviral vectors (Supplementary Figure S2a) engineered to overexpress or silence Bmi1 (B+ and sh-Bmi1, respectively). We first confirmed Bmi1 up- or downregulation in transduced cells by RT-qPCR and western blotting (Figures 1a and b). As expected, CPC-B+ cells grew significantly faster than equivalent cells expressing an shRNA against Bmi1 or a scrambled shRNA control (Figure 1c). We then compared the respective miRNA expression profiles (Microarray v1.0) of B+ and sh-Bmi1 CPCs as described,29 and considered miRNAs that showed a significant upregulation in B+ CPCs and a reduction in sh-Bmi1 CPCs. From these results (GSE66813), we compiled a short list of candidate miRNAs showing Bmi1-dependent changes in expression (Supplementary Figure S3a). The expression profile of three miRNAs, miR-300, miR-188, and miR-362, was validated by RT-qPCR, but miR-346 was not confirmed (Figure 1d). The expression profile of the three confirmed miRNAs in CPCs corresponded to miR-362>miR-188>miR-300 (Supplementary Figure S3b). We next evaluated endogenous expression of these miRNAs in CPCs with moderate-high expression of Bmi1. To obtain this population, we marked Bmi1-expressing cells in the mouse and traced their descendants using an inducible Bmi1 CreERT2 strain30 crossed to a Rosa26-floxed yellow fluorescent protein (YFP) reporter line. Induction of Cre recombination by tamoxifen results in permanent expression of YFP in Bmi1-expressing cells and their progeny (Figure 1e). Subsequently, we compared CPC-B+ cells labeled by YFP (YFP+) with the remaining CPC compartment (YFP−). Results confirmed an upregulation of miR-300 in the YFP+ (CPC-B+) population, but not miR-188 or miR-362, which were not overrepresented in the YFP− population (Figure 1f). We validated this result in other cell lines. Accordingly, overexpression of Bmi1 in mouse embryonic fibroblasts (MEFs) and adipose tissue-derived MSCs induced the upregulation of miR-300 relative to control cells (GFP), although the relative increase was cell-type dependent (Figure 1g). Specifically, Bmi1-induced upregulation of miR-300 in CPCs (1.35-fold) was found to be similar to MSCs (1.5-fold), but lower than that observed in MEFs (4.3-fold). Taken together, these results suggest that Bmi1 might directly or indirectly regulate miR-300 expression.
Figure 1.
miR-300 expression is increased in Bmi1-overexpressing cells. CPCs were transduced with lentiviral vectors expressing Bmi1, sh-Scramble (encoding a negative control shRNA), and sh-Bmi1. Transduced cells were purified by FACS. Bmi1 expression was assessed in purified cells by (a) RT-qPCR and (b) western blot. (c) Cumulative population doubling (CPD) was estimated in CPC-B+ (closed circles), CPC-Scramble (gray squares), and CPC-sh-Bmi1 (triangles) cells. (d) RT-qPCR validation of the array results for miR-300, miR-188, and miR-362; miR-346 was not validated. (e) Isolation of an identified primary subpopulation of Sca1-CPCs characterized by moderate-high expression of Bmi1 using conditional Bmi1-YFP mice. (f) Comparison of miRNA expression in Sca1 compartments (YFP+ versus YFP−). (g) Relative expression of miR-300 in correlation with Bmi1 expression in cardiac progenitor cells (CPCs), mouse embryonic fibroblasts (MEFs), and mesenchymal stem cells (MSCs). ***P<0.001, **P<0.01, *P<0.05 (two-way ANOVA followed by Bonferroni post-test; means±S.E.M., n=5)
miR-300 counteracts CPC replicative senescence
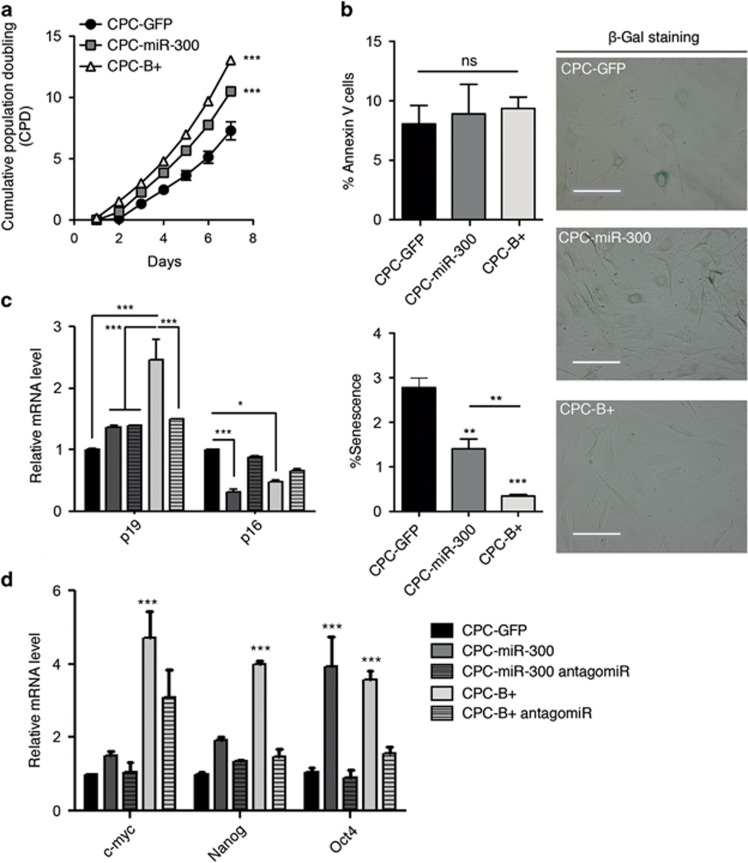
Because Bmi1 upregulation increased cellular proliferation, we questioned whether this was mediated by miR-300. We therefore overexpressed miR-300 in CPCs using a non-viral piggyBac vector (Supplementary Figure S2b). CPC-miR-300 cells exhibited a ≈5-fold increase in miR-300 compared with control (CPC-GFP) cells, whereas CPC-B+ cells had a ≈1.7-fold increase (Supplementary Figure S3c). Growth rates of CPC-miR-300 and CPC-B+ cells, at all points analyzed, were significantly higher than CPC-GFP cells (Figure 2a). This increase in growth was not the result of changes in apoptosis since annexin V staining was equivalent in the three groups (Figure 2b, top left); however, significant differences were found in the proportion of senescent cells measured by β-galactosidase activity (Figure 2b, right panels). Accordingly, the number of β-Gal+ cells was reduced 2.2-fold in CPC-miR-300 and 6-fold in CPC-B+ populations compared with control (Figure 2b, bottom left), suggesting that miR-300 overexpression contributes to reduce and/or delay senescence. Consistent with a modification in senescence development, differences were found in cell-cycle distribution in miR-300-expressing cells (Supplementary Figure S4). Gene expression analysis of cell-cycle senescence-associated and stemness markers in CPC-miR-300 and CPC-B+ cells demonstrated a significant upregulation in p19, c-myc, Nanog, and Oct4 levels and a clear reduction in p16 levels (Figures 2c and d). To further examine whether miR-300 is a relevant downstream effector of Bmi1, we knocked down miR-300 expression in CPC-miR-300 and CPC-B+ cells using anti-miR-300 oligonucleotides. We titrated the anti-miR concentration required to achieve at least a 50% reduction of miR-300 expression in both cell lines (Supplementary Figure S5). Results showed that anti-miR-300 (50 pmol) suppressed the Bmi1-mediated effect on several target genes (Figures 2c and d). In particular, BMI1-induced Oct4 expression was fully dependent on miR-300. Thus, knockdown of miR-300 in CPC-B+ and CPC-miR-300 cells reduced Oct4 expression to basal levels compared with control cells. Furthermore, miR-300 overexpression induced an upregulation of Oct4 mRNA to levels similar to those in CPC-B+ cells (Figures 2c and d). Collectively, these results suggest that miR-300 is necessary for Bmi1-dependent expression of genes involved in cell-cycle dynamics, which might preserve CPCs in a more undifferentiated state. Indeed, genes related to multipotency and stemness were found to be overrepresented in CPC-B+ and CPC-miR-300 cells (Figures 2c and d), and recovered to normal levels when miR-300 was downregulated. In summary, miR-300 enhances the expression of a panel of multipotent genes, indicating a plausible association with the maintenance of CPCs in a more undifferentiated state.
Figure 2.
miR-300 delays CPC senescence. (a) Cumulative population doubling over several culture passages of CPC-GFP (closed circles), CPC-miR-300 (gray squares), and CPC-Bmi1(B+ triangles). (b) Apoptosis and senescence determined by annexin V staining (left upper panel) and β-gal staining (left lower panel), respectively; the right panels show representative β-gal staining images. Bars, 100 μm. (c, d) Panels of relative expression levels of p19 and p16 (c) and multipotency-related genes (d) in CPC-manipulated cells; where indicated, cells were transfected with mirVana miR-300 inhibitor. ***P<0.001; *P<0.05; ns, not significant (two-way ANOVA followed by Bonferroni post-test; means±S.E.M., n=5)
miR-300 in the Dlk1-Dio3 context
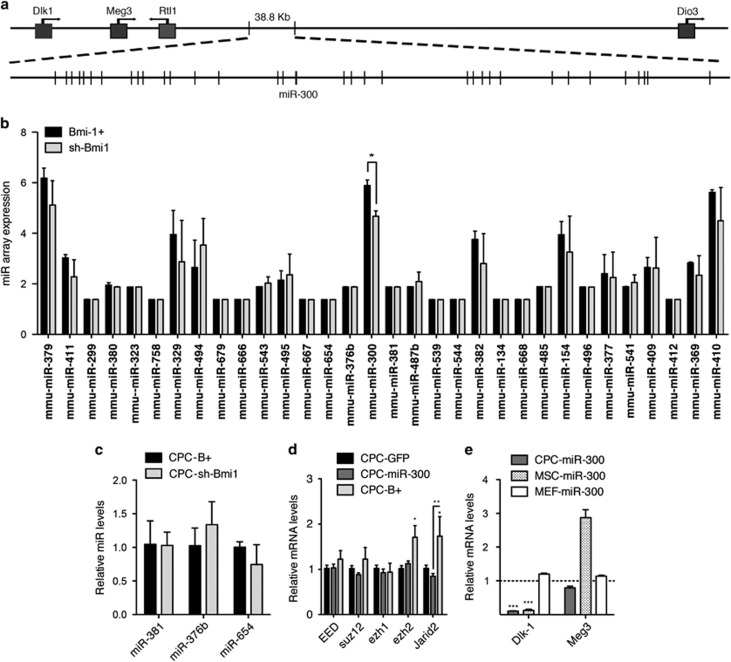
In humans, miR-300 localizes to a dense miRNA cluster residing in chromosome 14, region 14q32.31 (DLK1-MEG3; Figure 3a). In the mouse, this region corresponds to the Dlk1/Gtl2 domain located on chromosome 12qF1.31 Detailed miRNA array expression analysis of the whole miRNA-encoding locus in CPC-B+ and CPC-sh-Bmi1 cells showed that miR-300 was the only miRNA significantly modulated by overexpression of Bmi1 (Figure 3b). This was confirmed by RT-qPCR analysis of neighboring miRNAs (Figure 3c). Because DLK1-MEG3 regulation is dependent on polycomb-containing complexes,28 we assessed whether miR-300 could modulate members of the polycomb complexes PRC1 and PRC2. We found a modest decrease in the expression of Suz12, Ezh1, and Jarid2 in CPC-miR-300 cells relative to CPC-GFP cultures (Figure 3d). In contrast, CPC-B+ cells displayed a different expression pattern, with a moderate increase in Eed and Suz12 expression and a significant upregulation of Ezh2 and Jarid2 (Figure 3d). Additionally, we found that modulation of miR-300 expression also affected other elements of the DLK1-MEG3 domain in different cell types (Figure 3e), which was contingent on Bmi1 regulation (Supplementary Figure S6). Accordingly, miR-300 overexpression provoked a decrease in Dlk1 mRNA levels in CPCs and MSCs, but not in MEFs (Figure 3e), suggesting a context-dependent regulation. This effect was also observed for Meg3, which was reduced by miR-300 overexpression in CPCs, but was increased in MSCs (Figure 3e). Collectively, these data illustrate the great complexity of the regulatory mechanisms underlying gene expression at this locus (reviewed in Royo and Cavaille32), but also highlight the apparent selectivity of Bmi1 for miR-300 expression and the consequent impact on some elements of the DLK1-MEG3 domain.
Figure 3.
miR-300 belongs to the imprinted Dlk1-Dio3 cluster. (a) Simplified map (5′→ 3′) of the mouse DlK-1-Dio3 cluster (chromosome 12qF1); miR-300 is shown. (b) miR array results at the DlK-1-Dio3 cluster. (c) Direct validation by RT-qPCR of miR-300 neighbors, miR-381, miR-376b, and miR-654, confirmed the specificity of association of Bmi1 and increased expression of miR-300. (d) RT-qPCR analysis of different polycomb repressor complex 1/2 members and recruiters in CPC-GFP, CPC-B+, and CPC-miR-300 cells. (e) RT-qPCR analysis of different Dlk1-Dio3 cluster inductors in CPC-miR-300, MEF-miR-300, and MSC-miR-300 cells relative to their respective control (−GFP) cultures. ***P<0.001, **P<0.01, *P<0.05 (two-way ANOVA followed by Bonferroni post-test; means±S.E.M., n=5)
Forced expression of miR-300 impairs CPC differentiation
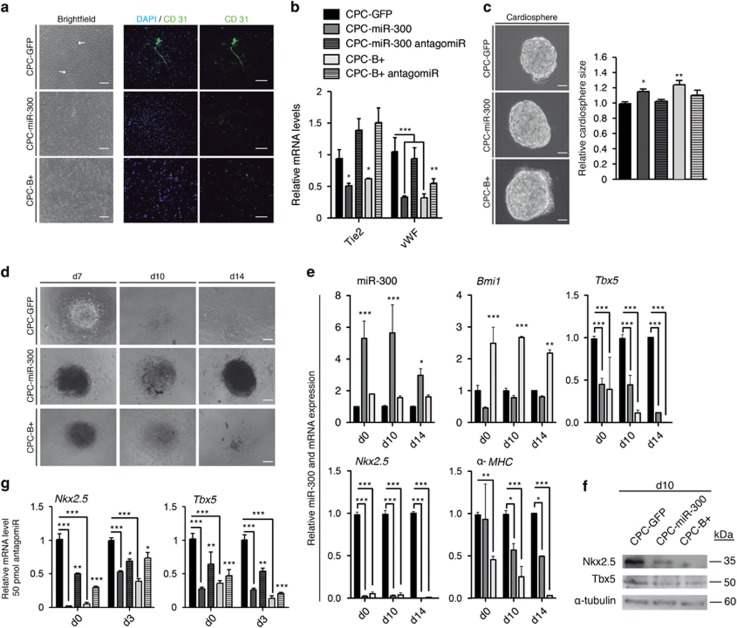
To test the role of miR-300 in CPC differentiation, cells were first allowed to spontaneously differentiate in culture and endothelial commitment was measured. CPC-GFP cells, but not CPC-miR-300 or CPC-B+ cells, formed reticular structures after 10–12 days (Figure 4a, brightfield), which were positive for CD31 (Figure 4a). Moreover, mRNA analysis of endothelial markers revealed that Tie2 and vWf expression was significantly reduced in CPC-miR-300 and CPC-B+ cells compared with control CPC-GFP cells (Figure 4b). These data suggest a functional impairment of endothelial cell differentiation in miR-300 and Bmi1-expressing cells. To test the generality of this response, we used the same cell populations and induced cardiac differentiation by the hanging drop method. After 6 days in culture, cardiospheres from CPC-miR-300 and CPC-B+ cells were significantly larger than those formed from CPC-GFP cells (Figure 4c), corroborating the finding of increased proliferation potential (Figure 2a). Additionally, knockdown of miR-300 in CPC-miR-300 cells led to a significant reduction in cardiosphere size (Figure 4c, right panel). Following retinoic acid treatment to promote cardiac differentiation, plated CPC-B+ cardiospheres presented an apparently slower differentiation kinetic as observed by a reduction in cell spreading compared with control cells. Also, CPC-miR-300 cardiospheres continued to proliferate with minimal differentiation compared with CPC-GFP control cells, which showed greater expansion (Figure 4d). Time course analysis of cardiac marker gene expression revealed that, compared with control cells, levels of α-MHC, Nkx2.5, and Tbx5 were significantly reduced in CPC-B+ and CPC-miR-300 cells throughout the differentiation period (Figure 4e). Indeed, at d14 of differentiation, α-MHC gene expression was almost abolished in CPC-B+. Of note, Nkx2.5 gene expression was scarcely detected at any period in CPC-B+ and CPC-miR-300 (Figure 4e), and this was confirmed by western blot analysis (Figure 4f). Because Nkx2.5 is a critical cardiogenic transcription factor,30 its evident absence in these cells likely contributes to the impaired cardiogenic differentiation observed. Finally, to confirm that miR-300 is a downstream effector of Bmi1 during the early steps of CPC cardiogenic differentiation, we analyzed Nkx2.5 and Tbx5 expression early after anti-miR-300 transfection. Exposure to anti-miR-300 for as little as 3 days resulted in a partial rescue of Nkx2.5 and Tbx5 expression in CPC-B+ and CPC-miR-300 cells compared with control cells (Figure 4g).
Figure 4.
miR-300 diminishes CPC differentiation potential in cardiospheres. (a) Representative images of spontaneous endothelial differentiation in transduced cells after 10–12 days of differentiation. Arrows indicate tubular structures in brightfield, which were positive for CD31. Bars, 200 μm. (b) RT-qPCR analysis of endothelial genes (Tie2 and vWF) in CPC-GFP (black bars), CPC-miR-300 (gray bars), and CPC-B+ (white bars) cells. (c) Representative cardiosphere images at day 6 of differentiation. Bars, 100 μm (left). Relative cardiosphere size (right). (d) Cardiac differentiation induced by retinoic acid; representative images of transfected CPCs at day (d)7, d10, and d14 of the differentiation protocol. Bars, 200 μm. (e) Selected cardiac genes were measured by RT-qPCR in transfected cells during differentiation (d0, d10, and d14). (f) Western blotting of total protein extracts was performed at d10. (g) RT-qPCR analysis of selected cardiac genes in CPCs transfected with mirVana miR-300 inhibitor. ***P<0.001, **P<0.01, *P<0.05 (two-way ANOVA followed by Bonferroni post-test; means±S.E.M., n=6)
miR-300/Bmi1 interaction network in CPC
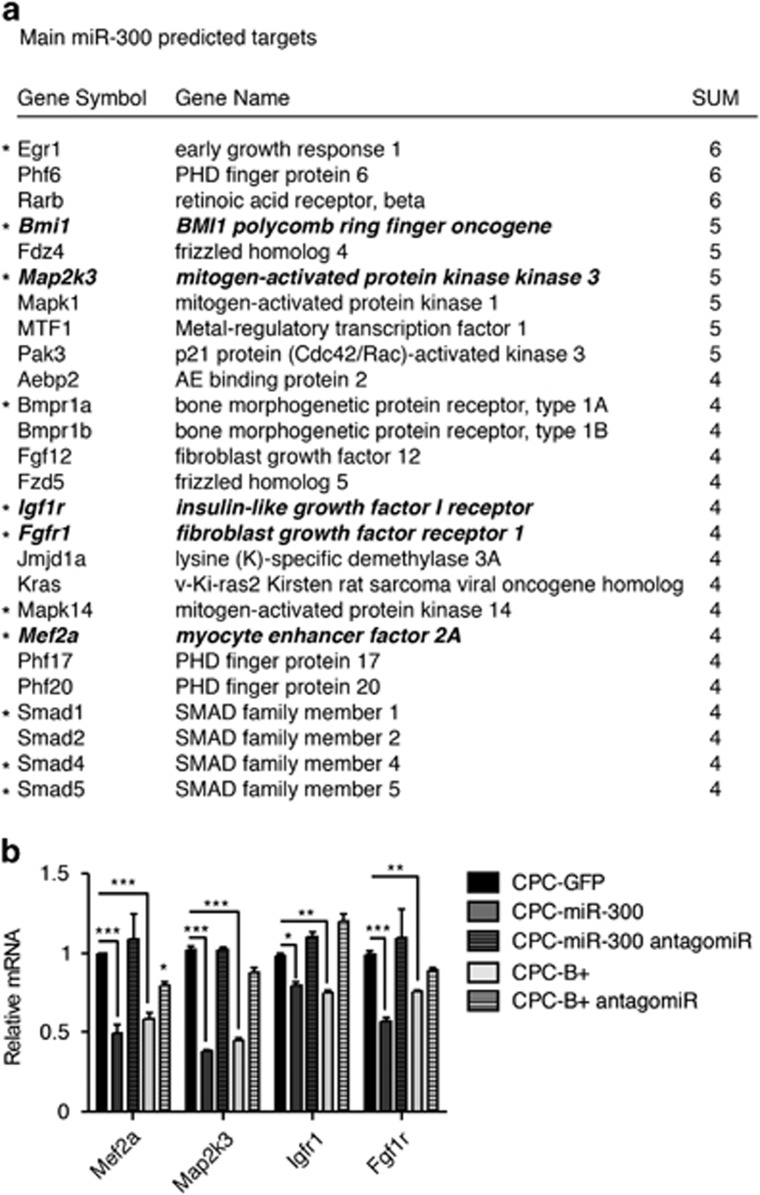
Little is known about the physiological roles of miR-300. We compared all potential miR-300 target genes predicted by specialized databases and Ingenuity Pathway Analysis (IPA) was used to link the direct targets. The most significant targets found by IPA are shown in Figure 5. These included genes in the Tgf-β signaling pathways (Map2k3) and factors involved in vertebrate cardiogenesis (Smad2, Mapk14, Bmpr1a) (Figure 5a). The Tgf-β pathway is associated with the regulation of cardiac differentiation.33 Another target, Map2k3, of the Fgf signaling pathway has been also described to be important incardiac differentiation34 and affects mainly the p38 Mapk pathway.35, 36 Eleven putative targets were selected for validation (Figure 5b and Supplementary Figure S6). RT-qPCR analysis revealed that Mef2a, Map2k3, Igfr1, and Fgf1r expression was dependent on miR-300 expression, and this decrease in expression could be reversed by transfection of an anti-miR-300 (five shown in bold in Figure 5a). A similar modulation was found in CPC-B+ cells (Figure 5b). Expression of the remaining putative miR-300 targets tested was not influenced by miR-300, although all were inhibited by Bmi1 overexpression (Supplementary Figure S6). The finding of Bmi1 as a highly probable target of miR-300 suggests that it could directly regulate Bmi1.
Figure 5.
miR-300 regulatory potential. (a) Main plausible targets of miR-300 were identified by comparing all predicted miRNA target sites in miRWalk database produced by Diana, Pictar4, miRanda, Pita, TargetScan, and RNA22 databases, containing all the putative targets. Main miR-300 predicted targets with at least four hits in these databases (indicated in the table) were selected. Asterisks indicate tested targets (11 in total) and in bold-italic those validated targets. (b) RT-qPCR analysis of CPC; where indicated, cells were transfected with mirVana miR-300 inhibitor. ***P<0.001, **P<0.01, *P<0.05 (two-way ANOVA followed by Bonferroni post-test; means±S.E.M., n=5)
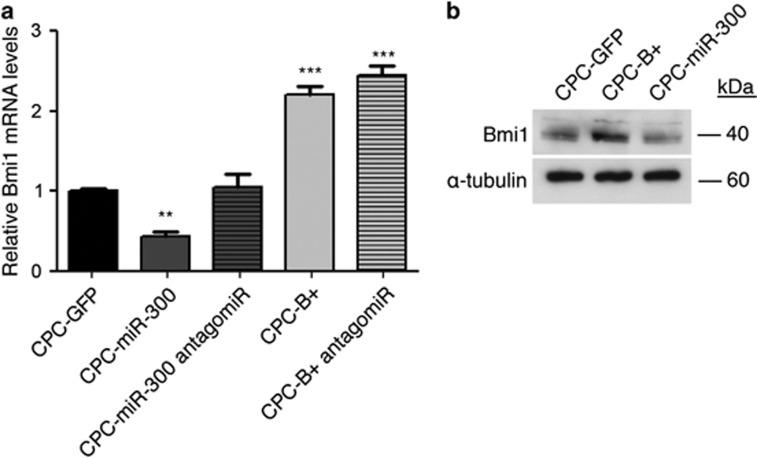
Because activation of Bmi1 in CPCs results in increased levels of miR-300 (Figure 1), we tested whether miR-300 activation affected the levels of Bmi1 mRNA and protein. We examined Bmi1 expression in CPC-B+ and CPC-miR-300 cells before and after transfection with anti-miR-300. Both cell lines exhibited an increase in Bmi1 expression following suppression of miR-300 expression (Figures 6a and b). These data suggest that Bmi1 positively regulates miR-300 and, reciprocally, miR-300 negatively regulates Bmi1 expression in a negative feedback loop.
Figure 6.
Bmi1/miR-300 negative feedback loop. (a) RT-qPCR analysis of Bmi1 expression in CPC-GFP, CPC-miR-300, and CPC-B+ cells, before and after treatment with the antagomiR. (b) Bmi1 protein levels measured by western blot; α-tubulin was used as a loading control. ***P<0.001, **P<0.01 (two-way ANOVA followed by Bonferroni post-test; means±S.E.M., n=5)
Taken together, our study provides evidence that miR-300 could play an important role in maintaining multipotent cell status, favouring Oct4 transcription factor expression, and negatively regulating differentiation. Since none of the confirmed genes modulated by overexpression of miR-300 or Bmi1 (Oct4, Nkx2.5, Tbx5, and vWF) are predicted as direct targets (Figure 5), our results suggest a non-direct effect.
Discussion
Bmi1 is required for the self-renewal of adult stem cells in many tissues.3, 6, 30, 37, 38, 39 Because Bmi1 knockdown reduces proliferation of mouse CPCs (Figure 1), we attempted to identify regulatory elements controlled by Bmi1. We used lentiviral vectors to overexpress (B+) or inhibit (sh) Bmi1, to study its function in CPCs. CPC-B+ cells had an enhanced proliferation capacity relative to CPC-sh-Bmi1 cells, suggesting that Bmi1 is important for the maintenance of some CPC primitive properties. Because miRNAs can fine tune cell homeostasis,16, 17 we screened for miRNAs regulated by Bmi1. To the best of our knowledge, all miRNAs identified in association with Bmi1 expression are inhibitors (Supplementary Table S1). Therefore, we searched for miRNAs that could be regulated by Bmi1 in CPCs, as plausible modulators of stem cell function.
We confirmed that miR-300 is upregulated when Bmi1 levels are high. Also, miR-300 is highly expressed in a primary subpopulation of CPCs characterized by medium-high expression of Bmi1. Presumably, because of the comparatively low basal expression of miR-300 (Supplementary Figure S3b), it was not included in a recently published study on the miRNA repertoire of adult mouse CPCs.40 Expression of Bmi1 in other murine cell lineages (MEFs and MSCs) consistently activated miR-300 expression, although the levels of overexpression were variable. All these results suggest that Bmi1 regulates miR-300. BMI1 is known as a transcriptional repressor.7 To upregulate c-myc mRNA, Bmi1 activates the Wnt pathway by repressing the Dickkopf (Dkk) family of Wnt inhibitors. Repression of Dkk genes by Bmi1, in particular Dkk1, drives the upregulation of Wnt and finally the Wnt target gene, c-myc.11 We hypothesize that a similar mechanism of suppression of a transcriptional repressor may operate to activate miR-300 when Bmi1 is overrepresented.
Bmi1 plays a central role in senescence and aging, regulating the expression of relevant genes involved in aging and cancer.41, 42, 43 Overexpression of Bmi1 results in repression of the tumor suppressor p16INK4a, which has emerged as a major regulator of aging and age-associated pathologies.44 We show that in CPCs, Bmi1, and miR-300 oppositely regulate expression of p16Ink4a and p19ARF. Expression of p16Ink4a is decreased in CPCs upon Bmi1 and miR-300 upregulation (Figure 2c); in contrast, p19ARF levels are upregulated. These two genes are localized in the same locus, sharing the common exons 2 and 3, but differing in their first exons and their respective promoters. As neither p16Ink4a or p19ARF are miR-300 targets, the present evidence suggests that the observed changes to p16INK4a and p19ARF expression are due mainly to changes in the transcriptional activity of their respective promoters. This effect on the INK4a/ARF locus has been also observed in cells treated with histone deacetylase inhibitors, with consequences for chromatin remodeling.45 It is possible that variations in BMI1 protein levels could affect PRC1 complex activity and chromatin structure; such changes seem to alter INK4a/ARF transcription in a way similar to histone deacetylase inhibitors. We hypothesize that the DLK1-MEG3 cluster, where miR-300 is localized, could be regulated in a similar manner.
It has previously been demonstrated that p16Ink4a accumulates in conjunction with p38 activation in ras-induced senescent cells, and that this accumulation is essential for senescence.35 Although prevention of senescence in CPC-miR-300 cells is less effective than in CPC-B+, p16 levels are significantly reduced in CPC-miR-300 cells. This could be related to the different levels of overexpression of miR-300 in the cell populations (Supplementary Figure S3c). Thus, it is possible that miR-300 upregulation may at least partially account for the downregulation of p16Ink4a and the p38 pathway, thereby bypassing cell senescence. Since the most important known senescence-relevant target of Bmi1 is p16Ink4a, reduction of senescence in CPCs by miR-300 is almost certainly associated with reduction of p16Ink4a expression. In agreement with this, CPC-miR-300 cells increase the expression levels of Oct4. Additionally, it has been recently shown that the Oct4/p16Ink4a ratio directly correlates with culture life span. This result suggests that low p16Ink4a and high Oct4 levels govern the senescent state of MSCs and therefore may be a good predictor of in vitro viability.45 We speculate that the same principle operates in CPCs.
Forced expression of miR-300 reduced spontaneous endothelial differentiation of CPCs, and a similar effect was obtained after Bmi1 overexpression. Cardiac differentiation induced by retinoic acid was also reduced in CPC-miR-300 cells. These cells generated similar cardiosphere-type bodies to control cells (d0–d8), but did not differentiate further. This blockage seemed to be even stronger than that observed in CPC-B+ cells. From a panel of critical cardiogenic genes analyzed, the significant reduction found in the expression of Nkx2.5 would be sufficient to explain the phenotype observed.46 Because Nkx2.5 is not a plausible direct target for miR-300 (Figure 5), we infer that this effect is indirect.
In conclusion, miR-300 is a new member of the family of genes that control stem cell function regulated by Bmi1 in multipotent CSCs. miR-300 favors maintenance of the undifferentiated state, inhibits differentiation, and establishes a negative feedback loop to control levels of Bmi1.
Materials and Methods
Animals
C57BL/6 mice (8–12 weeks old) were provided by Charles River Laboratories (Wilmington, MA, USA). Where indicated, Bmi1CreER/+; Rosa26YFP/+ (Bmi1-YFP) mice were generated by crossing Bmi1CreER/+ mice27 with Rosa26YFP/+ reporter mice (Valiente-Alendí et al. in press). Bmi1CreER/+; Rosa26YFP/+ double heterozygous male and female mice were injected intraperitoneally with tamoxifen (TM; Sigma; 9 mg per 40 g of body weight), every 24 h on three consecutive days between postnatal days 30 (P30) and P60. Animals were maintained and handled according to the recommendations of the CNIC Institutional Ethics Committee.
Lentiviral construction and non-viral vectors
Lentivirus production and titration were carried out as described.47 Viral supernatant stocks contained 1 × 107 TU/ml, with a 1:100 TU/physical particles ratio as measured by qPCR. Cells were transduced with cleared supernatants at a multiplicity of infection of 10, in 3 ml of culture medium and polybrene (Sigma) at 8 μg/ml, for 8 h. Then, inoculum was removed and cell cultures were refreshed with medium before assay.
Cellular transfection
Non-viral piggy-bac vectors (1–2 μg) were co-transfected with mouse transposase-expression vector (0.5 μg) in SCA1+ cells, MEFs, and MSCs, using Lipofectamine 2000 Transfection Reagent (11668-027, Sigma). MirVana miRNA inhibitor (Applied Biosystems, Carlsbad, CA, USA) was used to knockdown mir-300 expression in a separate set of experiments. Transfection studies, in parallel with negative control miRNA inhibitor experiments, were carried out with lipofectamine RNAiMax Transfection Reagent (13778030-Sigma).
Isolation of SCA1+ and Bmi1+ CPC cells from adult heart and cell culture
Sca1+ CPCs were isolated essentially as previously described.48 Enrichment for SCA1+ cells was achieved by incubating cells with a SCA1-biotinylated rat antibody (ab25196; Abcam, Cambridge, UK) and anti-rat kappa microbeads (Miltenyi Biotec, Madrid, Spain; 130-047-401), followed by purification with magnetic selection. Purified cells were plated in culture plates precoated with 0.1% gelatin and were maintained at 37 °C, 3% oxygen with the corresponding medium (IMDM, 10% FCS, 1% penicillin and streptomycin, and 1% l-glutamine, supplemented with 10 ng/ml EGF (E9644, Sigma), 20 ng/ml FGF (450-33, Pretech, London, UK), and LIF (103 U; ESG1107, Millipore, Madrid, Spain). Medium was refreshed every 2 days. Cells were passaged at 70–80% confluency by trypsinization.
Bmi1+ CPC cells were obtained from hearts of Bmi1-YFP mice 5 days after TM induction. Hearts were perfused and processed as above. The resulting single-cell suspension was passed through a 40 μm filter to remove debris. YFP+ cells were separated from the total heart mass with a BD FacsAria II Special Order System cell sorter fitted with a 488 nm laser to excite YFP (collected in the 525/550 channel). To discriminate YFP+ cells from autoflorescent cells, a 488 nm laser was used to excite cells, followed by collection in the 585 channel (phycoerythrin). Purified YFP+ cells were cultured at 37 oC, 3% O2 and 5% CO2 in the same expansion medium used for Sca1+ cells.
MicroRNA microarray
After SCA1+CPC transduction of the indicated lentiviral vectors (Supplementary Figure S2a), positively transduced cells were sorted, briefly expanded, and total RNA was isolated using the mirVana kit (Ambion, Madrid, Spain). RNA was quantified and integrity was checked by agarose gel electrophoresis. The total RNA required for analysis (500 ng) was prepared in 10 μl RNase-free water and analyzed using Agilent miRNA microarrays (Microarray v1.0). After normalization, only those probes present in at least one sample and with average expression over the 20th percentile of all average expressions were considered for further analysis (197 miRNAs). We then used lineal models as implemented in the limma Bioconductor package.26 We considered the experimenter as a random variable. The analysis was performed at the CNIC Bioinformatics Unit.
Endothelial and retinoic acid-induced cardiac differentiation
For endothelial differentiation, SCA1+ cells were cultured for 10 days in the above-indicated expansion conditions until spontaneous tubular structures appeared. For cardiac differentiation, SCA1+ cells were trypsinized and diluted to 50 000 cells/ml. Cardiospheres were prepared using hanging drop culture (20 μl approx 1000 cells) for 3 days in DMEM GlutaMax supplemented with 20% FBS, 1% non-essential amino acids, and β-mercaptoethanol. Cardiospheres were harvested, washed, and cultured for three additional days on nonadherent Petri culture dishes (10 cm) in fresh medium. Cardiospheres were then subsequently treated with all-trans retinoic acid (10 nM) in DMEM GlutaMax supplemented with 15% FBS, 1% non-essential amino acids, and β-mercaptoetanol (50 μM). The medium was replenished every 2 days.
Apoptosis and senescence assays
To determine programmed cell death of SCA1+ cells, cells were harvested and examined for annexin V staining (APC-conjugated; 550474, BD Pharmingen, Madrid, Spain), using an apoptosis detection kit (556570 BD Pharmingen). For detection of senescence detection, cells were cultured to a density of 20 000 cells/cm2 in β-galactosidase staining buffer for 24 h (Cell Signaling Technology, Danvers, MA, USA). Cells were then fixed and stained.
Immunofluorescence
For immunostaining, cells were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature. After washing three times in PBS – 0.1% BSA for 5 min, cells were permeabilized with 0.2% Triton (Sigma, T9284) in PBS for 20 min, and washed in PBS – 0.1% BSA. Primary antibody against CD31 (MAB1398Z, Millipore) was diluted in PBS – 0.1% BSA to 1/200 and incubated overnight at 4 °C. After washing, cells were incubated with a FITC-conjugated goat anti-hamster secondary antibody (127-095-099, Jackson, Bar Harbor, ME, USA) used at a 1/1000 dilution in the presence of DAPI for 45 min (1 h at room temperature). Vectashield mounting medium without DAPI (Vectors Lab, Burlingame, CA, USA; H-1000) was applied to all slides. Fluorescent images were taken using a Nikon A1-R inverted confocal microscope.
Gene expression analysis and western blotting
Total RNA was isolated from cultured cells using the Direct-zol RNA Miniprep Kit (Zymo, Irvine, CA, USA) and reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) for mRNA and with the TaqMan MicroRNA Reverse Transcription Kit (4366596, Applied Biosystems, Madrid, Spain) for miRNAs. Complementary DNAs were analyzed by real-time PCR using the Power SYBR Green PCR Master Mix (Applied Biosystems) for mRNA and No AmpErase TaqMan 2X Universal PCR Master Mix (S08590, Applied Biosystems) for miRNA. Amplification, detection, and data analysis were carried out with an ABI PRISM 7900HT Sequence Detection System. The crossing threshold values for individual mRNAs were normalized to GusB expression for mRNAs and U6 for miRNAs. Changes in mRNA expression were denoted as the fold change relative to the control (see supplementary Table S2 for primers used).
For western blotting, total proteins were isolated using RIPA buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS) and quantified by the DC Protein assay (Bio-Rad). Approximately 50 μg of protein was resolved on each lane on 10% SDS-PAGE gels, electrotransferred onto nitrocellulose membrane, and probed with specific antibodies (see Supplementary Table S3 for antibodies used).
Statistics
Data were analyzed by two-way ANOVA and Student's t-test. Error bars represent S.E.M. In all corresponding figures, ∗P<0.05, ∗∗P<0.01, ∗∗∗P<0.001, and ns P>0.05.
Acknowledgments
This study was supported by grants from the Ministry of Science and Innovation (SAF2012-34327, PLE2009-0147, and PSE-010000-2009-3), the Comunidad Autónoma de Madrid (S2010/BMD-2420), the Instituto de Salud Carlos III (RETICS.TERCEL), and the European Commission (Proposal 242038) to AB, (BFU2012-35258 and RYC-2009-04341) to JAB, and fellowship FPU-AP2010-5951 to FMC. We thank JL Torán and S Mendez-Ferrer for critical discussions of the manuscript; RM Carmona for help with the animal procedure; Fátima Sánchez Cabo for statistic and bioinformatic assistance, and K. McCreath for editorial support. The CNB-CSIC and CNIC are supported by the Spanish Ministry of Economy and Competitiveness. The CNIC is additionally supported by the Pro-CNIC Foundation. The authors declare no competing financial interests.
Glossary
- BMI1
B lymphoma Mo-MLV insertion region 1
- CSCs/CPCs
cardiac stem/progenitor cells
- GFP
green fluorescent protein
- MEFs
mouse embryonic fibroblast
- miR-300
microRNA miR-300
- MSCs
mesenchymal stem cells
- Nkx2.5
NK2 homeobox 5
- Oct4
POU class 5 homeobox 1
- Tbx5
T-box 5
- YFP
yellow fluorescent protein
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Raschellà
Supplementary Material
References
- 1van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev 1994; 8: 757–769. [DOI] [PubMed] [Google Scholar]
- 2Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 2003; 425: 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 2003; 423: 302–305. [DOI] [PubMed] [Google Scholar]
- 4Itahana K, Zou Y, Itahana Y, Martinez JL, Beausejour C, Jacobs JJ et al. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol 2003; 23: 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Tanaka T, Komai Y, Tokuyama Y, Yanai H, Ohe S, Okazaki K et al. Identification of stem cells that maintain and regenerate lingual keratinized epithelial cells. Nat Cell Biol 2013; 15: 511–518. [DOI] [PubMed] [Google Scholar]
- 6Zacharek SJ, Fillmore CM, Lau AN, Gludish DW, Chou A, Ho JW et al. Lung stem cell self-renewal relies on BMI1-dependent control of expression at imprinted loci. Cell Stem Cell 2011; 9: 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Biehs B, Hu JK, Strauli NB, Sangiorgi E, Jung H, Heber RP et al. BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat Cell Biol 2013; 15: 846–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Allegra E, Trapasso S, Pisani D, Puzzo L. The role of BMI1 as a biomarker of cancer stem cells in head and neck cancer: a review. Oncology 2014; 86: 199–205. [DOI] [PubMed] [Google Scholar]
- 9Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells 2012; 30: 372–378. [DOI] [PubMed] [Google Scholar]
- 10Cho JH, Dimri M, Dimri GP. A positive feedback loop regulates the expression of polycomb group protein BMI1 via WNT signaling pathway. J Biol Chem 2013; 288: 3406–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Xie X, Piao L, Cavey GS, Old M, Teknos TN, Mapp AK et al. Phosphorylation of Nanog is essential to regulate Bmi1 and promote tumorigenesis. Oncogene 2014; 33: 2040–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009; 138: 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 2008; 134: 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C et al. Dissecting self-renewal in stem cells with RNA interference. Nature 2006; 442: 533–538. [DOI] [PubMed] [Google Scholar]
- 15Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009; 10: 126–139. [DOI] [PubMed] [Google Scholar]
- 16Schoeftner S, Scarola M, Comisso E, Schneider C, Benetti R. An Oct4-pRb axis, controlled by MiR-335, integrates stem cell self-renewal and cell cycle control. Stem Cells 2013; 31: 717–728. [DOI] [PubMed] [Google Scholar]
- 17Nimmo R, Ciau-Uitz A, Ruiz-Herguido C, Soneji S, Bigas A, Patient R et al. MiR-142-3p controls the specification of definitive hemangioblasts during ontogeny. Dev Cell 2013; 26: 237–249. [DOI] [PubMed] [Google Scholar]
- 18Trohatou O, Zagoura D, Bitsika V, Pappa KI, Antsaklis A, Anagnou NP et al. Sox2 suppression by miR-21 governs human mesenchymal stem cell properties. Stem Cells Transl Med 2014; 3: 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM et al. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 2013; 155: 778–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Mallliaras K, Ibrahim A, Tseliou E, Liu W, Sun B, Middleton RC et al. Stimulation of endogenous cardioblasts by exogenous cell therapy after myocardial infarction. EMBO Mol Med 2014; 6: 760–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell 2008; 2: 320–331. [DOI] [PubMed] [Google Scholar]
- 22Urbanek K, Cesselli D, Rota M, Nascimbene A, De Angelis A, Hosoda T et al. Stem cell niches in the adult mouse heart. Proc Natl Acad Sci USA 2006; 103: 9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Li Y, Fukuda N, Yokoyama S, Kusumi Y, Hagikura K, Kawano T et al. Effects of G-CSF on cardiac remodeling and arterial hyperplasia in rats. Eur J Pharmacol 2006; 549: 98–106. [DOI] [PubMed] [Google Scholar]
- 24Bailey B, Fransioli J, Gude NA, Alvarez R Jr, Zhang X, Gustafsson ÅB et al. Sca-1 knockout impairs myocardial and cardiac progenitor cell function. Circ Res 2012; 111: 750–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Ellison GM, Vicinanza C, Smith AJ, Aquila I, Leone A, Waring CD et al. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell 2013; 154: 827–842. [DOI] [PubMed] [Google Scholar]
- 26van Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, Lin SC et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 2014; 509: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Bolli R, Chugh AR, D'Amario D, Loughran JH, Stoddard MF, Ikram S et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet 2011; 378: 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, Berman D et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet 2012; 379: 895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: Article3. [DOI] [PubMed] [Google Scholar]
- 30Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 2008; 40: 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Benetatos L, Hatzimichael E, Londin E, Vartholomatos G, Loher P, Rigoutsos I et al. The microRNAs within the DLK1-DIO3 genomic region: involvement in disease pathogenesis. Cell Mol Life Sci 2013; 70: 795–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Royo H, Cavaille J. Non-coding RNAs in imprinted gene clusters. Biol Cell 2008; 100: 149–166. [DOI] [PubMed] [Google Scholar]
- 33Behfar A, Zingman LV, Hodgson DM, Rauzier JM, Kane GC, Terzic A et al. Stem cell differentiation requires a paracrine pathway in the heart. FASEB J 2002; 16: 1558–1566. [DOI] [PubMed] [Google Scholar]
- 34Rosenblatt-Velin N, Lepore MG, Cartoni C, Beermann F, Pedrazzini T. FGF-2 controls the differentiation of resident cardiac precursors into functional cardiomyocytes. J Clin Invest 2005; 115: 1724–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Fu XB, Xing F, Yang YH, Sun TZ, Guo BC. Activation of phosphorylating-p38 mitogen-activated protein kinase and its relationship with localization of intestinal stem cells in rats after ischemia-reperfusion injury. World J Gastroenterol 2003; 9: 2036–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Williamson D, Gallagher P, Harber M, Hollon C, Trappe S. Mitogen-activated protein kinase (MAPK) pathway activation: effects of age and acute exercise on human skeletal muscle. J Physiol 2003; 547: 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Sangiorgi E, Capecchi MR. Bmi1 lineage tracing identifies a self-renewing pancreatic acinar cell subpopulation capable of maintaining pancreatic organ homeostasis. Proc Natl Acad Sci USA 2009; 106: 7101–7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Chatoo W, Abdouh M, David J, Champagne MP, Ferreira J, Rodier F et al. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J Neurosci 2009; 29: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Robson LG, Di Foggia V, Radunovic A, Bird K, Zhang X, Marino S. Bmi1 is expressed in postnatal myogenic satellite cells, controls their maintenance and plays an essential role in repeated muscle regeneration. PloS One 2011; 6: e27116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Bras-Rosario L, Matsuda A, Pinheiro AI, Gardner R, Lopes T, Amaral A et al. Expression profile of microRNAs regulating proliferation and differentiation in mouse adult cardiac stem cells. PloS One 2013; 8: e63041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 2011; 479: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Matheu A, Klatt P, Serrano M. Regulation of the INK4a/ARF locus by histone deacetylase inhibitors. J Biol Chem 2005; 280: 42433–42441. [DOI] [PubMed] [Google Scholar]
- 43Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S et al. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res 2009; 69: 9090–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 1999; 397: 164–168. [DOI] [PubMed] [Google Scholar]
- 45Piccinato CA, Sertie AL, Torres N, Ferretti M, Antonioli E. High OCT4 and Low p16(INK4A) Expressions determine in vitro lifespan of mesenchymal stem cells. Stem Cells Int 2015; 2015: 369828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Pashmforoush M, Lu JT, Chen H, Amand TS, Kondo R, Pradervand S et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 2004; 117: 373–386. [DOI] [PubMed] [Google Scholar]
- 47Torres R, Garcia A, Paya M, Ramirez JC. Non-integrative lentivirus drives high-frequency cre-mediated cassette exchange in human cells. PloS One 2011; 6: e19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Izarra A, Moscoso I, Levent E, Cañón S, Cerrada I. Díez-Juan et al. miR-133a enhances the protective capacity of cardiac progenitors cells after myocardial infarction. Stem Cell Rep 2014; 3: 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.