Abstract
Nasopharyngeal carcinoma (NPC) is a malignant epithelial tumor with tremendous invasion and metastasis capacities, and it has a high incidence in southeast Asia and southern China. Previous studies identified that far upstream element-binding protein 1 (FBP1), a transcriptional regulator of c-Myc that is one of the most frequently aberrantly expressed oncogenes in various human cancers, including NPC, is an important biomarker for many cancers. Our study aimed to investigate the expression and function of FBP1 in human NPC. Quantitative real-time RT-PCR (qRT-PCR), western blot and immunohistochemical staining (IHC) were performed in NPC cells and biopsies. Furthermore, the effect of FBP1 knockdown on cell proliferation, colony formation, side population tests and tumorigenesis in nude mice were measured by MTT, clonogenicity analysis, flow cytometry and a xenograft model, respectively. The results showed that the mRNA and protein levels of FBP1, which are positively correlated with c-Myc expression, were substantially higher in NPC than that in nasopharyngeal epithelial cells. IHC revealed that the patients with high FBP1 expression had a significantly poorer prognosis compared with the patients with low expression (P=0.020). In univariate analysis, high FBP1 and c-Myc expression predicted poorer overall survival (OS) and poorer progression-free survival. Multivariate analysis indicated that high FBP1 and c-Myc expression were independent prognostic markers. Knockdown of FBP1 reduced cell proliferation, clonogenicity and the ratio of side populations, as well as tumorigenesis in nude mice. These data indicate that FBP1 expression, which is closely correlated with c-Myc expression, is an independent prognostic factor and promotes NPC progression. Our results suggest that FBP1 can not only serve as a useful prognostic biomarker for NPC but also as a potential therapeutic target for NPC patients.
Nasopharyngeal carcinoma (NPC) presents a remarkable geographic pattern and has a high incidence in Southern China, Hong Kong, Taiwan and Southeast Asia.1, 2 NPC is associated with Epstein–Barr virus (EBV) infection.3, 4, 5, 6 Serum EBV antibodies and EBV-DNA serves as a useful tools for the diagnosis in NPC.7, 8, 9 Although there have been improvements in diagnosis techniques, irradiation and chemo-radiotherapy, local recurrence or distant metastasis are the main reasons for treatment failure.10 Therefore, it is particularly important to look for diagnostic markers and therapeutic targets for NPC. c-Myc is one of the most frequently dysregulated oncogenes in various human cancers, including NPC11 and its expression is induced by EBV-encoded genes,12 such as LMP1 (ref. 13) and LMP2A.12, 14, 15 The aberration of c-Myc in cancers could occur at different levels, such as genetic amplification, upregulation of transcription, post-transcriptional regulation and post-translational regulation.16, 17, 18, 19 It has been shown that gain of chromosome 8, where c-Myc is localized, was observed on 30% of primary NPCs. However, overexpression of the c-Myc mRNA or protein has been observed in ~80–90% of NPC samples.20 Thus, transcriptional or post-transcriptional regulation of c-myc may be the major mechanism for its aberrant expression in NPC. The far upstream element (FUSE)-binding proteins (FBPs) are a family of three regulatory proteins, termed FBP1, FBP2 and FBP3.21 FBP1 was first identified as a DNA-binding protein that regulates the expression of c-Myc by binding to a single-stranded FUSE located upstream of the c-Myc promoter22, 23 FBP1 is also an RNA-binding protein that binds to the Hepatitis C and Enterovirus 71 RNAs and mediates their replication during retroviral infection.24, 25 FBP1 modulates viral replication by interacting with the untranslated regions of the Japanese encephalitis virus RNA.26 In apoptotic cells, FBP1 may translocate from the nucleus to the cytoplasm. This location is associated with the fact that FBP1 may be cleaved by caspase-3 and -7.27 Recently, FBP1 has been indentified as a potential biomarker for oligodendrogliomas,28 non-small cell lung cancer,29 breast cancer,30, 31 liver cancer,32, 33 colon cancer34, 35 and clear cell renal cancer.36 However, little is known about the prognostic significance and function of FBP1 and its role in the regulation of c-Myc in NPC.
In this study, we investigated FBP1 expression in NPC tissues using immunohistochemistry (IHC) and analyzed the correlation between FBP1, c-Myc and the clinicopathological characteristics. In addition, we also explored the function of the FBP1 protein in NPC cells using two independent small interference RNAs (siRNAs) to knock down the expression of FBP1, and found that silencing FBP1 suppresses cell proliferation, colony-formation abilities, side populations (SPs) and markedly reduces tumorigenesis in nude mice. Taken together, these results indicate that FBP1 was overexpressed in NPC and had an important role in the development of NPC.
Results
Expression of FBP1 positively correlates with c-Myc in NPC cells and tissues
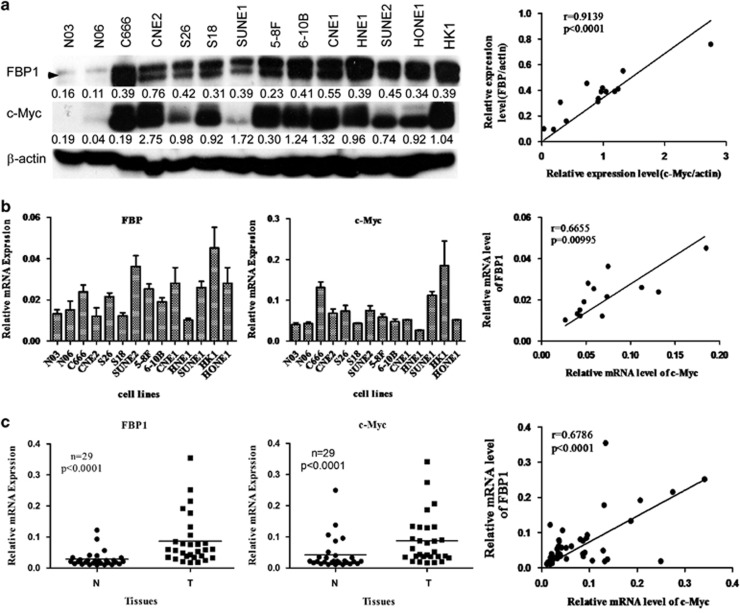
To explore the expression of FBP1 in NPC cells and tissues, we first analyzed the levels of the FBP1 protein and mRNA in two non-cancerous primary human nasopharyngeal epithelial cells and 12 NPC cell lines by western blot and quantitative real-time PCR (qRT-PCR). As shown in Figures 1a and b, the levels of the FBP1 protein and transcript were much higher in NPC cells than in normal cells. It has been reported that FBP1 is the major transcriptional regulator for c-Myc, and there is a correlation between FBP1 and c-Myc expression in many cancers.37 Therefore, we also analyzed c-Myc expression in these samples. As expected, the expression of c-Myc was consistent with FBP1. Furthermore, we used the Spearman's rank correlation analysis to examine the relationship between FBP1 and c-Myc expression and found that FBP1 expression was positively associated with c-Myc at the protein (Spearman's r=0.9137, P<0.0001; Figure 1a, right panel) and mRNA levels (Spearman's r=0.6655, P=0.00095; Figure 1b, right panel).
Figure 1.
The expression of FBP1 directly correlates with c-Myc in nasopharyngeal carcinoma. (a) Western blots of normal human nasopharyngeal epithelial cell samples (N03 and N06) and samples of nasopharyngeal carcinoma cell lines using antibodies against FBP1 and c-Myc (left panel). The relationship between FBP1 and c-Myc is shown in the right panel. β-Actin was used as a control for protein loading and integrity. (b) FBP1 and c-Myc mRNA levels in a series of NPC cell lines compared with the normal human nasopharyngeal epithelial cells. mRNA levels are presented as the means±S.D. and are normalized to the housekeeping gene β-actin using qRT-PCR. (c) The relative expression of FBP1 and c-Myc in 29 pairs of matched NPC and non-tumor tissues at the transcript level (left and middle panels). The right panel shows the association between FBP1 and c-Myc
To further determine the expression of FBP1 and c-Myc in NPC tissue samples, we analyzed the transcript levels of FBP1 and c-Myc in 29 NPC tissues and 29 adjacent non-cancerous tissues by qRT-PCR. We observed that the expression level of FBP1 and c-Myc in NPC tissues was significantly higher than in non-cancerous tissues (Figure 1c), and also found that the expression of FBP1 correlated with c-Myc (Spearman's r=0.6786, P<0.0001; Figure 1c, right panel). Taken together, these data demonstrated that FBP1 was frequently upregulated and its expression was positively associated with c-Myc in both NPC cells and tissues.
Inhibition of FBP1 expression suppresses NPC cell proliferation and clonogenicity
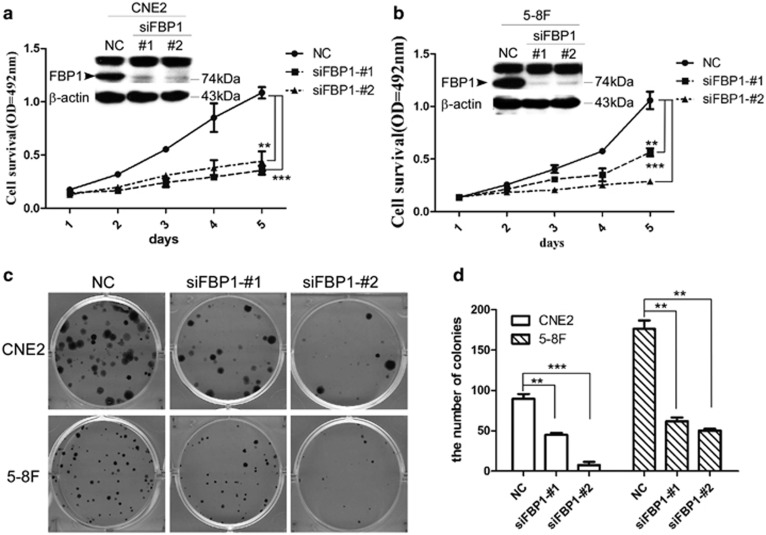
To explore the role of the FBP1 gene in NPC, we used two independent siRNAs to knock down FBP1 expression in CNE2 and 5-8F cells. A significant reduction was observed at the transcript level (data not shown) and protein level (Figures 2a and b). We next performed the MTT assay to test whether knocking down the expression of FBP1 suppressed cell proliferation. The results showed that FBP1 knockdown reduced the tumor cell viability (Figures 2a and b, CNE2-NC versus siFBP1-1#, P<0.01; CNE2-NC versus siFBP1-2#, P<0.0001; 5-8F-NC versus siFBP1-1#, p<0.01; 5-8F-NC versus siFBP1-2#, P<0.001). In addition, we also observed that inhibition of FBP1 in CNE2 or 5-8F cells resulted in significantly fewer and smaller colonies compared with the control (Figures 2c and d, CNE2-NC versus siFBP1-1#, P<0.01; CNE2-NC versus siFBP1-2#, P<0.001; 5-8F NC versus siFBP1-1# and siFBP1-2#, P<0.01). All of these results suggested that the FBP1 protein could exhibit oncogenic functions in NPC.
Figure 2.
FBP1 silencing suppressed the proliferation and clone formation of NPC cells. (a, b) Western blot samples from CNE2 (a) and 5-8F (b) cells were transfected with NC- or FBP-targeting (KD) siRNAs. β-Actin was used as an internal control (upper panel). The MTT assay measured the viability of CNE2 and 5-8F cells with NC- or FBP1-targeting (KD) siRNAs. The data represent the means±S.E.M. (lower panel; **P<0.01, ***P<0.001 by the paired t-test). (c, d) The representative pictures (c) and the quantitative analysis (d) of colony-formation assay of CNE2 and 5-8F cells transfected with NC- or FBP1-targeting (KD) siRNAs. The assay was performed in triplicate. The data represent the means± S.E.M. (**P<0.01, ***P<0.001 by the paired t-test)
Knockdown of FBP1 inhibits the side population in NPC cell
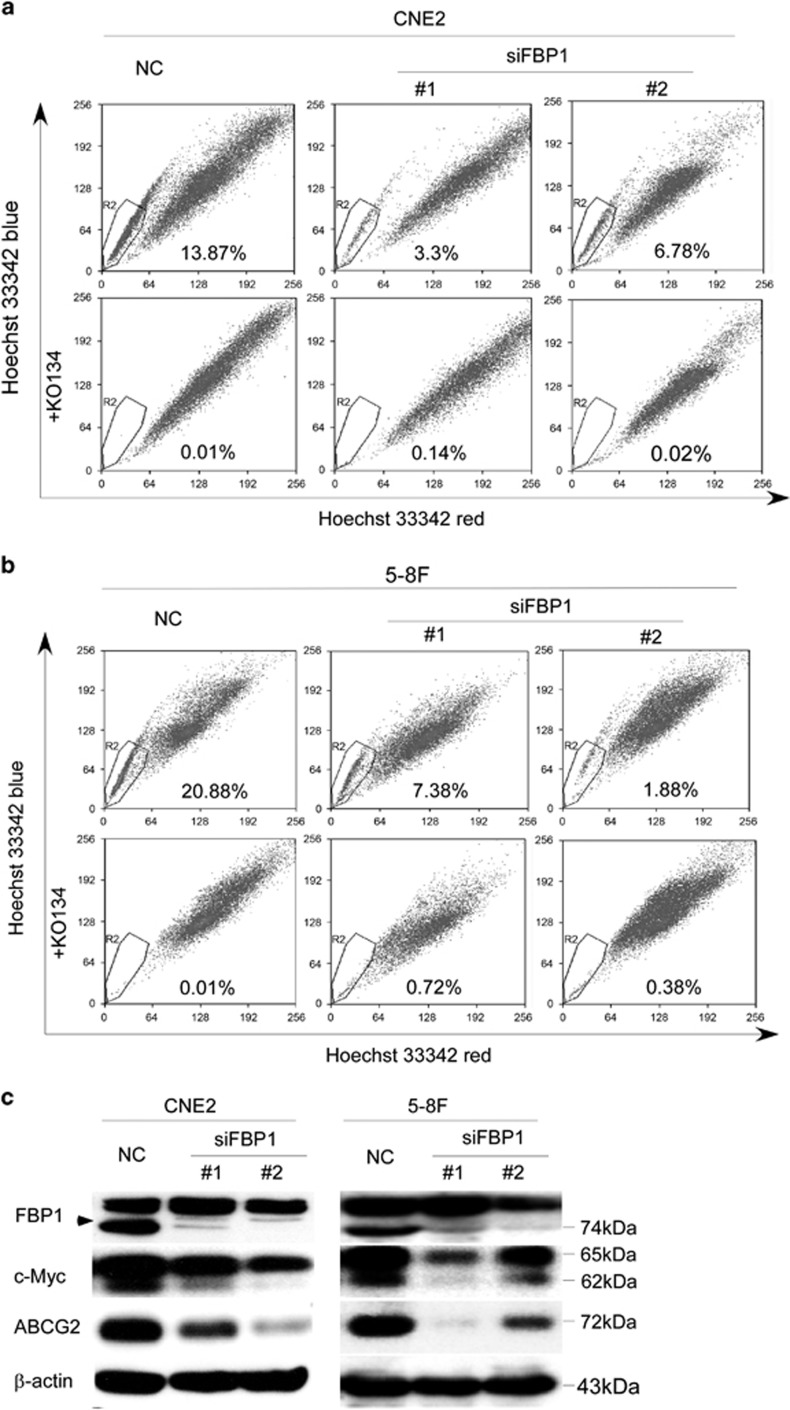
It has been known that FBP1 is involved in carcinogenesis via c-Myc-independent or -dependent pathways.32, 36, 38 Several studies demonstrated that the ABC family is a direct transcriptional target of the Myc family.39, 40, 41 To further explore whether the FBP/MYC/ABCG2 pathway contributed to the induction of SP cells in NPC, we performed Hoechst 33342 flow cytometry and found an apparent decrease of the number of SP when FBP1 was knocked down (Figures 3a and b), whereas Ko143 (a specific inhibitor of ABCG2) obviously inhibited the efflux of Hoechst 33342. Moreover, an obvious reduction of the c-Myc and ABCG2 proteins was present in FBP silenced cells (Figure 3c). These results indicated that FBP1 may activate the c-Myc/ABCG2 pathway and induce an SP phenotype in NPC cells.
Figure 3.
Knockdown of FBP1 decreased the side population in NPC cells. (a) Flow cytometry analysis of Hoechst 33342 staining was performed in CNE2 cells transfected with NC- or FBP1-targeting (KD) siRNAs (upper panel) and treated with ko143 (final concentration: 5 μm) before Hoechst 33342 staining (lower panel). The percentage of SP cells is indicated. (b) Flow cytometry analysis of Hoechst 33342 staining was performed and 5-8F cells transfected with NC- or FBP1-targeting (KD) siRNAs (upper panel) and treated with ko143 (final concentration: 5 μm) before Hoechst 33342 staining (lower panel). The percentage of SP cells is indicated. (c) Representative western blots of FBP1, c-Myc, ABCG2 and β-actin in CNE2 and 5-8F cells transfected with NC- or FBP1-targeting (KD) siRNAs. β-Actin served as a loading control
FBP1 silencing reduces the chemosensitvity and radiosensitvity of NPC cells
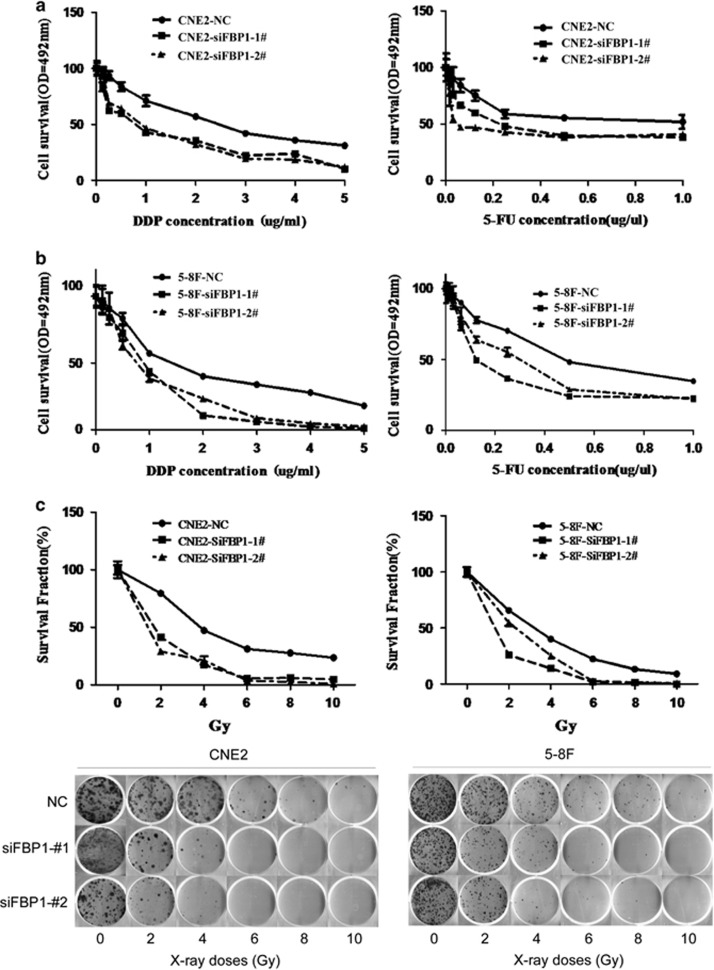
The SP cells that exhibited cancer stem cell characteristics have been reported in NPC.42, 43 As we showed that inhibition of FBP1 expression could reduce the ratio of CSC-like SP, we explored whether FBP1 also had a role in tumor resistance to chemotherapy and radiotherapy. Two common chemotherapeutic drugs (cisplatin (DDP); 5-flurouracil (5-FU)) for NPC were used to examine the cellular response after suppressing FBP1 expression. The results showed that suppressing FBP1 expression increased the tumors' sensitivity to both DDP and 5-FU (Figures 4a and b). Radiotherapy is a main treatment for NPC. Thus, we treated the cells that were transfected with the siRNA duplexes with different doses and observed that knocking down of FBP1 decreased the radioresistance in NPC cells. (Figure 4c; CNE2-NC versus CNE2-siFBP1-1#1, P<0.05; CNE2-NC versus CNE2-siFBP1-2#, P<0.01; 5-8F-NC versus 5-8F-siFBP1-1#, P<0.0264; 5-8F-NC versus 5-8F-siFBP1-2#, P<0.01). Together, targeting FBP1 can reduce both chemotherapy and radiotherapy resistance.
Figure 4.
Results of radiation and chemotherapy sensitivity. (a) The growth curves of CNE2 cells transfected with NC- or FBP1-targeting (KD) siRNAs at 48 h post treatment with the indicated doses of DDP (left panel) and 5-FU (right panel). (CNE2-NC versus CNE2-siFBP1-1#1 and siFBP1-2#, P<0.001 by the paired t-test). (b) The growth curves of 5-8F cells transfected with NC- or FBP1-targeting (KD) siRNAs at 48 h post treatment with the indicated doses of DDP (left panel) and 5-FU (right panel). (5-8F-NC versus 5-8F-siFBP1-1# and 5-8F-siFBP1-2#, P<0.05, by the paired t-test). (c) The survival fraction (SF) curves (upper panel) and the representative pictures of CNE2 and 5-8F cells transfected with NC- or FBP1-targeting (KD) siRNAs at 10 days after X-ray irradiation with the indicated Gy doses (lower panel). (NC versus siFBP1-1#1 in CNE2 and 5-8F, P<0.05; NC versus siFBP1-2# in CNE2 and 5-8F, P<0.01 by the paired t-test)
Immunohistochemical analysis of FBP1 or c-Myc expression and their correlations with clinicopathological characteristics
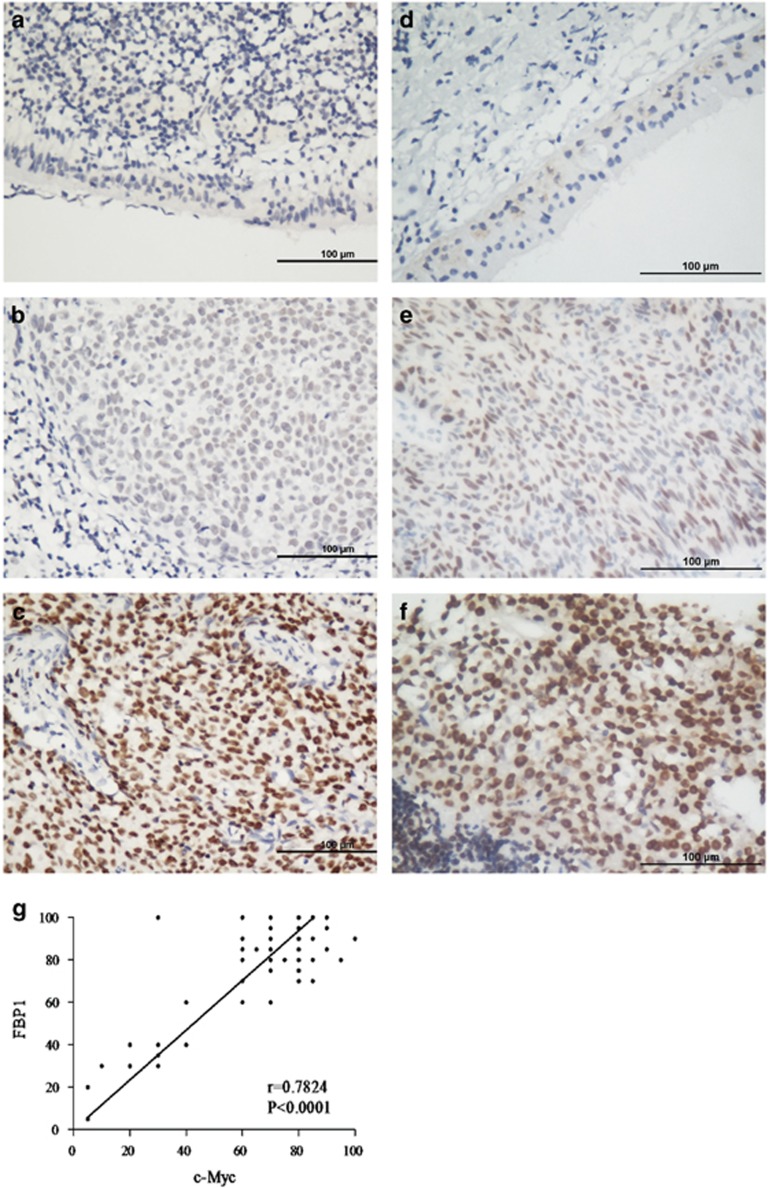
To reveal a potential role for FBP1 in NPC, 83 NPC samples from patients were stained with the human FBP1 antibody and c-Myc antibody using IHC. We found that both FBP1 and c-Myc were predominantly located in the nuclei of tumor cells in the NPC samples (Figure 5). Representative images of FBP1 and c-Myc IHC staining in NPC tissues are shown in Figures 5a–f. The normal nasopharyngeal epithelial cells has no or very low staining intensity of FBP1 (Figure 5a) and c-Myc (Figure 5d). The non-stained area is constituted by stromal cells and lymphocyte, suggesting that the IHC staining of FBP1 (Figure 5b and c) and c-Myc (Figure 5e and f) are specific to NPC cells. In addition, we also observed that FBP1 expression is positively associated with c-Myc44 (r=0.7824, P<0.001, Figure 5g). We further analyzed the correlation between the FBP1 and c-Myc expression and clinicopathological characteristics. As presented in Table 1, both FBP1 and c-Myc expression correlated with the clinical stage (P=0.010). In addition, c-Myc was also associated with N classification (P=0.040), whereas FBP1 was associated with locoregional failure (P=0.049) and distant metastasis (P=0.038). However, there were no significant correlations with other clinical features, including age, gender, T classification, EBV-DNA, EBV-VCA-IgA, EBV-EA-IgA, tumor progression and death.
Figure 5.
The expression of FBP1 in nasopharyngeal carcinoma (NPC) by immunohistochemical (IHC) staining Positive expression of FBP1 was primarily detected in the nucleus of the NPC cells. (a) Negative expression of FBP1 in normal nasopharyngeal tissues. (SP x400, left upper panel). (b) Moderate expression of FBP1 in NPC. (SP x400, right upper panel). (c) High expression of FBP1 in NPC. (SP x400, right lower panel). (d) Negative expression of c-Myc in normal nasopharyngeal tissues. (SP x400, left upper panel). (e) Moderate expression of c-Myc in NPC. (SP x400, right upper panel). (f) High expression of c-Myc in NPC. (SP x400, right lower panel). (g) The correlation of FBP1 and c-Myc expression by IHC
Table 1. Characteristics of the patients.
| Characteristics | No. of patients |
Expression of FBP1 |
P-value |
Expression of c-Myc |
P-value | ||
|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||
| Patients | 83 | 41 | 42 | 41 | 42 | ||
| Age | |||||||
| Median | 46 | ||||||
| Range | 24~77 | ||||||
| ≤46 | 43 | 24 | 19 | 0.225 | 9 | 34 | 0.305 |
| >46 | 40 | 17 | 23 | 5 | 35 | ||
| Gender | |||||||
| Female | 19 | 2 | 17 | 0.506 | 2 | 17 | 0.506 |
| Male | 64 | 12 | 52 | 12 | 52 | ||
| T classification | |||||||
| T1–T2 | 21 | 11 | 10 | 0.752 | 3 | 18 | 1.000 |
| T3–T4 | 62 | 30 | 32 | 11 | 51 | ||
| N classification | |||||||
| N0–N1 | 33 | 19 | 14 | 0.226 | 9 | 24 | 0.040 |
| N2–N3 | 50 | 22 | 28 | 5 | 45 | ||
| Clinical stage | |||||||
| I–II | 8 | 4 | 4 | <0.001 | 2 | 6 | 0.022 |
| III–IV | 75 | 37 | 38 | 12 | 63 | ||
| EBV-DNA | |||||||
| <3200 | 44 | 18 | 44 | 0.931 | 7 | 37 | 0.804 |
| ≥3200 | 39 | 15 | 24 | 7 | 32 | ||
| EBV-VCA-IgA | |||||||
| <1:160 | 19 | 7 | 12 | 0.440 | 4 | 15 | 0.439 |
| ≥1:160 | 64 | 30 | 64 | 19 | 54 | ||
| EBV-EAIA2-IgA | |||||||
| <1 : 20 | 33 | 15 | 18 | 0.559 | 6 | 27 | 0.795 |
| ≥1 : 20 | 50 | 26 | 24 | 8 | 42 | ||
| Locoregional failure | |||||||
| Yes | 2 | 0 | 2 | 0.049 | 0 | 2 | 1.000 |
| No | 81 | 41 | 40 | 14 | 67 | ||
| Distant metastasis | |||||||
| Yes | 18 | 5 | 13 | 0.038 | 1 | 17 | 0.284 |
| No | 65 | 36 | 29 | 13 | 52 | ||
| Tumor progression | |||||||
| Yes | 19 | 1 | 18 | 0.172 | 13 | 51 | 0.172 |
| No | 64 | 13 | 51 | 1 | 18 | ||
| Death | |||||||
| Yes | 12 | 3 | 9 | 0.068 | 1 | 11 | 0.681 |
| No | 71 | 38 | 33 | 13 | 58 | ||
Abbreviations: EBV, Epstein–Barr virus; FBP1, far upstream element-binding protein 1
Relationship between FBP1 or c-Myc and patient survival
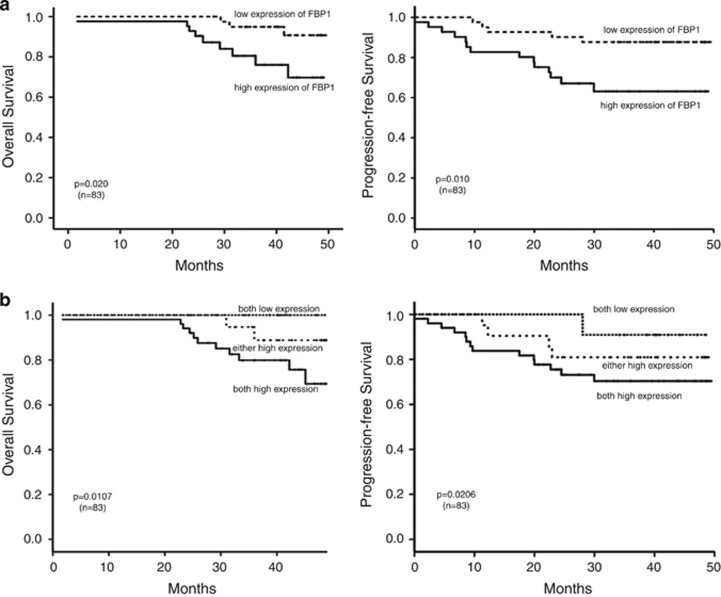
We carried out the Kaplan–Meier test to analyze the association between the expression of FBP1 and patient survival. Patients with low FBP1 expression have longer OS (P=0.020; Figure 6a, left panel). Consistently, progression-free survival (PFS) was shorter in the FBP1 high expression group than in those with low FBP1 expression (P=0.010, Figure 6a, right panel). We further analyzed the combined the phenotypes of FBP1 and c-Myc and found that patients with low expression of both FBP1 and c-Myc have longer OS and PFS than those with high levels of expression of both FBP1 and c-Myc (Figure 6b). The clinicopathological variables that were relevant to the OS are presented in Supplementary Figure S1.
Figure 6.
Relationship between FBP1 and patient survival. (a) Overall survival curves and progression-free survival curves of patients with low and high FBP1 expression. (b) Overall survival curves and progression-free survival curves of patients with high expression of both FBP1/c-Myc, low expression of both FBP1/c-Myc and high expression of either FBP1 or c-Myc
Next, we performed univariate and multivariate analyses using the COX proportional hazards model to investigate whether the gender, age, T stage, N stage, clinical stage, FBP1 or c-Myc expression were correlated with OS and PFS. As shown in Table 2, FBP1 was a prognostic factor for OS and PFS. The T stage and clinical stage were prognostic factors for OS, whereas gender was a prognostic factor for PFS. From the multivariate analyses, we found that both the clinical stage and FBP1 expression were independent indicators for both OS and PFS (Table 3).
Table 2. Univariate analysis with the COX proportional hazards for predictor of OS, and PFS of NPC patients.
| Prognostic factors | P-value | HR |
95% CI for HR |
|
|---|---|---|---|---|
| Lower | Upper | |||
| OS | ||||
| Age (years; >46 versus ≤46) | 0.194 | 1.759 | 0.667 | 7.384 |
| Gender (female versus male) | 0.799 | 1.185 | 0.321 | 4.380 |
| T stage (T3–4 versus T1–2) | 0.047 | 2.220 | 0.835 | 8.035 |
| N stage (N2–3 versus N0–1) | 0.297 | 2.010 | 0.540 | 7.459 |
| Clinical stage (III–IV versus I–II) | 0.015 | 2.332 | 0.965 | 10.632 |
| c-Myc (high versus low) | 0.400 | 0.415 | 0.053 | 3.218 |
| FBP1 (high versus low) | 0.033 | 4.184 | 1.126 | 15.552 |
| PFS | ||||
| Age (years; >46 versus ≤46) | 0.365 | 1.019 | 0.978 | 1.036 |
| Gender (female versus male) | 0.048 | 2.654 | 1.008 | 6.989 |
| T stage (T3–4 versus T1–2) | 0.308 | 1.900 | 0.553 | 6.521 |
| N stage (N2–3 versus N0–1) | 0.310 | 1.651 | 0.627 | 4.346 |
| Clinical stage (III–IV versus I–II) | 0.493 | 2.002 | 0.270 | 15.149 |
| c-Myc (high versus low) | 0.141 | 0.220 | 0.029 | 1.654 |
| FBP1 (high versus low) | 0.015 | 3.555 | 1.275 | 9.099 |
Abbreviations: CI, confidence interval; FBP1, far upstream element-binding protein 1; HR, hazard ratio; OS, overall survival; PFS, progression-free survival
Table 3. Multivariate analysis with the COX proportional hazards for the predictor of OS and PFS of the NPC.
| Prognostic factors | P-value | HR |
95% CI for HR |
|
|---|---|---|---|---|
| Lower | Upper | |||
| OS | ||||
| Age (years; >46 versus ≤46) | 0.170 | 0.420 | 0.122 | 1.450 |
| Gender (female versus male) | 0.982 | 1.105 | 0.267 | 3.864 |
| T stage (T3–4 versus T1–2) | 0.866 | 0.872 | 0.178 | 4.276 |
| N stage (N2–3 versus N0–1) | 0.401 | 0.553 | 0.139 | 2.220 |
| Clinical stage (III–IV versus I–II) | 0.048 | 1.900 | 0.984 | 6.399 |
| c-Myc (high versus low) | 0.031 | 0.512 | 0.063 | 4.151 |
| FBP1 (high versus low) | 0.039 | 1.405 | 0.680 | 4.268 |
| PFS | ||||
| Age (years; >46 versus ≤46) | 0.055 | 0.376 | 0.138 | 1.021 |
| Gender (female versus male) | 0.340 | 1.623 | 0.600 | 4.386 |
| T stage (T3–4 versus T1–2) | 0.453 | 0.558 | 0.122 | 2.559 |
| N stage (N2–3 versus N0–1) | 0.186 | 0.480 | 0.161 | 1.424 |
| Clinical stage (III–IV versus I–II) | 0.823 | 1.352 | 0.097 | 18.832 |
| c-Myc (high versus low) | 0.240 | 0.384 | 0.123 | 1.194 |
| FBP1 (high versus low) | 0.014 | 0.958 | 1.330 | 6.037 |
Abbreviations: CI, confidence interval; FBP1, far upstream element-binding protein 1; HR, hazard ratio; OS, overall survival; PFS, progression-free survival
Repression of FBP1 inhibits tumorigenesis in nude mice
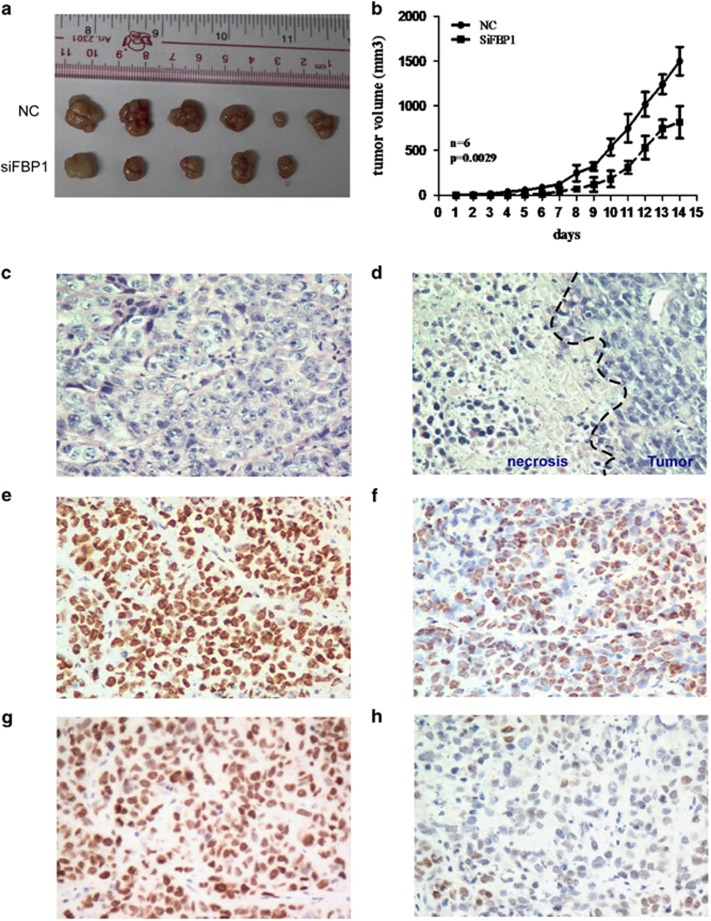
To further analyze the role of FBP1 in the progress of NPC, CNE2 cells, which were treated with the siRNA targeting FBP1, were injected into the subcutaneous area of 5-week-old female BALB/c nude mice. Tumor formation was assessed after 2 weeks. The repression of FBP1 reduced the volume and weight of the xenografts (Figures 7a and b, NC versus siFBP1, P=0.0029) The tumors obtained from injection of CNE2-NC/SiFBP1-1# were performed to assess the tumor pathology by HE staining (Figures 7c and d), FBP1 IHC staining (Figures 7e and f) and c-Myc IHC staining (Figure 7g and h), and then analyzed by a pathologist. We found that there were lymphocyte infiltrate and necrosis in the xenografts of siFBP group (Figure 7d), whereas the intensity of FBP1 and c-Myc was weak in the xenografts of siFBP group compared with that in NC group (Figures 7f and h). In summary, our data suggested that knockdown of FBP1 repressed the tumor progression in nude mice, probably attributable to the downregulation of c-Myc expression.
Figure 7.
FBP1 silencing reduced the tumorigenicity of NPC cells. (a) Images of the tumors formed by CNE2 cells transfected with NC or siFBP1. (b) The growth curve of tumors formed by CNE2 cells transfected with NC or siFBP1. The data are presented as the means±S.D. (n=6 mice in each group). (c–d) HE staining pictures of mice treated with NC (c) or with siFBP1 (d) (SP x400). (e–f) IHC staining pictures of FBP1 expression in the primary xenografts from cells treated with NC (e) or with siFBP1 (f) (SP x400). (g–h) IHC staining pictures of c-Myc expression in the primary xenografts from cells treated with NC (g) or with siFBP1 (h) (SP x400)
Discussion
In the present study, it was the first time to show that the expression of FBP1 was higher in NPC cell lines and NPC tissues than that in primary NPECs and normal nasopharyngeal tissues (Figure 1). In addition, the patients with high levels of FBP1 have a lower OS time than patients with low FBP1 expression, and there is a strong correlation between FBP and c-Myc expression.
c-Myc is located at 8q24.21, and encodes a transcription factor that has essential roles in cell proliferation, cell growth, differentiation and apoptosis.45 It has been reported that c-Myc is an oncogene and transcription factor involved in the tumorigenesis of multiple cancers.46, 47, 48, 49, 50, 51 Fan, CS et al52 indicated that c-Myc overexpression is a frequent genetic abnormality in NPC. Several studies showed that c-Myc induces the proliferation, migration and invasion of NPC cells.53, 54, 55 In addition, c-Myc was reported to promote radioresistance in a stem cell-like population of NPC cells.19 Mounting evidence indicates that c-Myc expression in the tumor may be an important strategy for designing appropriate treatment, using lower drug dosages and overcoming drug resistance and drug-related toxicity.56, 57, 58, 59 Hence, many investigators and international pharmaceutical companies have turned to identifying c-Myc inhibitors to cure cancers. However, the development of c-Myc inhibitors for clinical applications has not been very successful to date. Thus, a potential strategy may be to target the major protein that regulates c-Myc expression rather than only targeting c-Myc itself.
The FUSE-binding protein 1 (FBP1) was first discovered as transcriptional regulator of the proto-oncogene c-Myc. FBP1 regulated the expression of c-Myc by binding to the FUSE in the c-Myc promoter region at 1.5-kb upstream of the transcription start site.22 A number of reports demonstrate that FBP1 is involved in the regulation of cellular processes, including gene expression, differentiation and apoptosis.22, 34, 38, 60, 61 Many studies suggest that FBP1 is a proto-oncogene, because there is a close relationship between FBP1 expression and tumor development. In our study, we showed that the expression of FBP1 is positively correlated to the progress of NPC. Moreover, we also found that patients with high levels of the FBP1 protein had poorer OS and PFS (Figure 6a). In addition, higher expression of both the FBP1 protein and the c-Myc protein were remarkably correlated with patients' shorter survival time (Figure 6b). This result is consistent with a previous study in renal carcinomas.36
Previous study showed that FBP1 binds to c-Myc promoter and regulates its expression and effects its downstream targets.22, 23, 62 Downregulation of c-Myc reduced the cyclin-dependent kinase (Cdk) 4/6 activity and proliferating cell nuclear antigen (PCNA), Cdc2 and Rb-binding protein,63 causing cell cycle arrest at the G1 phase.64 In this study, we explored the function of FBP1 in cell proliferation and clonality using siRNA in CNE2 and 5-8F cells, and found that knockdown of FBP1 decreased the expression of c-Myc, then significantly inhibited the cell proliferation and clone formation by MTT assay and clonogenicity analysis, suggesting that FBP1 promotes NPC cell proliferation and colony formation ability, probably through c-Myc (Figure 2).
Several studies demonstrated that Myc can directly regulate the ATP-binding cassette drug transporters, which shuttle hydrophobic lipophilic compounds across the membranes in an ATP-dependent manner.40, 65, 66 However, there has been no evidence showing that FBP1 induces tumorigenesis through the enhancement of cancer stem-like cell (CSC). Here we first reported that the ratio of the SP in NPC was reduced after knocking down FBP1 expression by siRNA (Figures 3a and b). Western blot analysis suggested that FBP1 may induce the CSC-like characteristics by activating the FBP/C-Myc/ABCG2 pathway. It is known that CSC contributes to the resistance to irradiation and chemotherapy.43 Knockdown of FBP1 consistently reduced the chemoresistant and radioresistant characteristics of NPC cells (Figure 4).
In summary, our study demonstrated that FBP1 is a novel biomarker and can serve as a potential predictor for tumor progression in NPC. In addition, FBP is strongly correlated with c-Myc expression, and knocking down FBP1 expression could suppress NPC progression in vitro and in vivo, which suggested that FBP1 could be a potential therapeutic target in reducing chemoresistance and radioresistance as well as reducing tumor progression in NPC.
Materials and Methods
Patients and tissue specimens
Twenty-nine NPC biopsy samples and an equal number of non-cancerous nasopharyngeal tissues were acquired from Sun Yat-sen University Cancer Center (SYSUCC), Guangzhou, China and used for qRT-PCR. For IHC analysis, the paraffin-embedded NPC specimens were acquired from 83 patients (median age: 46 years,range 24–77 years) diagnosed with NPC in 2009–2013 at Cancer Center, Sun Yat-sen University, Guangzhou, China. This study was approved by the Institutional Research Ethics Committee at the Cancer Center. The clinical characteristics of the NPC patients were described in Table 1.
Cell culture
Normal primary NPECs67, 68, 69(NPEC03 and NPEC06) were cultured in Keratinocyte serum-free medium (Invitrogen, Carlsbad, CA, USA). The NPC cell lines70, 71 (C666, CNE2, S18, S26, SUNE2, 5-8F, 6-10B, SUNE1, CNE1, HNE1, HK1 and HONE1) were cultured in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) in a humidified 5% CO2 incubator at 37 °C.
siRNA transfection
The siRNAs targeting the mRNA of human FBP1 (Gene ID: 8880 NM_001303433.1) were denoted as siFBP1-#1 (GGUCAAGGCAACUGGAACA) and siFBP1-#2 (GGACAGGUUGAUUAUACCA). The negative control (NC)72, 73, 74 RNA duplex for the siRNA was indicated as NC and was not homologous to any human genome sequences. The CNE2 and 5-8F cells were plated in 6-well plates for 18 h, and then transfected with the 20 nM of the RNA duplex and 5 μl Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer's instructions. After 48 h, the cells were harvested for further experiments, including qRT-PCR, western blotting, MTT, colony formation assay and nude mice xenograft assay.
RNA isolation and reverse transcriptase PCR analysis
Total RNA was extracted from the tissue specimens and NPC cell lines using the TRIzol reagent (Invitrogen), according to the manufacturer's instructions. The reverse transcriptase kit (Promega, Madison, Wisconsin, USA,) was used to synthesize the complementary DNA from 2 μg of the total RNA. qRT-PCR was performed using the Power SYBR Green qPCR SuperMix-UDG (Invitrogen) to detect the mRNA level of the target genes using a LightCycler 480 II (Roche, Basel, Swiss). β-Actin was used as an internal control. The relative expression of TACC3 was normalized to the expression of β-actin, which yielded a 2-▵ct value. All reactions were performed in triplicate in three independent experiments. The sequences of the real-time PCR primers were as follows:
FBP1 sense: 5′-GGAGGTGATGCAGGGACATC-3′
FBP1 antisense: 5′-CTGCTGATGCATCGGTGGTA-3′
β-actin sense: 5′-CGCGAGAAGATGACCCAGAT-3′
β-actin antisense: 5′-GGGCATACCCCTCGTAGATG-3′.
Western blotting analysis
Cells were collected and lysed in SDS sample buffer (62.5 mM Tris-HCl (pH 6.8), 3% SDS, 10% glycerol, 50 mM DL-dithiothreitol and 0.1% bromophenol blue) with protease inhibitors (Roche, Indianapolis, IN, USA). The protein concentrations were determined by the BCA method (Pierce, Thermo Fisher Scientific, Rockford, IL, USA). The proteins (20 μg) were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Bovine serum albumin (BSA; 5%) in TBS-T (1 mol/l Tris-HCl (pH 7.5), 0.8% NaCl and 0.1% Tween 20) was used to block the membrane. Then, the membrane was incubated with anti-FBP1 (Santa-Cruz, sc-374342, Dallas, TX, USA), anti-c-Myc (Cell Signaling, #9402, Danvers, MA, USA), anti-ABCG2 (Abcam, ab3380, Cambridge, MA, USA), and anti-β-actin (Sigma-Aldrich, A5441, St Louis, MO, USA) antibodies at 4 °C overnight. The blots were then treated with an HRP-conjugated secondary antibody (Pierce).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide reduction (MTT) assay
The MTT assay was used to measure the viability of the NPC cells. Cells transfected with specific siRNA were seeded onto a 96-well plate at a density of 1000 cells per well. The cells were incubated with 0.2% MTT for 4 h at 37 °C, then 200 μl dimethyl sulfoxide (DMSO) per well was added to the culture cells to dissolve the crystals, and the cells were counted daily by reading the absorbance at 490 nm. The concentration at which DDP or 5-FU produced 50% growth inhibition (IC50) was calculated by the relative survival curve. Cells transfected with specific siRNA were seeded onto a 96-well plate at a density of 1000 cells per well. After 8 h for adherence, cells treated with chemotherapy drugs such as DDP and 5-FU at the increasing concentrations for 48 h. Finally, about 4 h before test, MTT (5 mg/ml) was added into the medium and 200 μl DMSO was added to dissolve the purple crystals. The 96-well plates were read at the wavelength of 490 nm.
Colony formation assay
After transfection with the specific siRNAs, cells (200 cells per well) were seeded in a 6-well plate and cultured for 14 days in a humidified 5%CO2 incubator at 37 °C.
Colonies were fixed in methanol for 10 min, and then stained with 0.5% crystal violet for 15 min. All visible colonies were quantified. The radiation dose that produced 50% growth inhibition was calculated by the relative survival fraction. At least three independent experiments were performed for each assay.
Detection of the side population of NPC cell lines
Forty-eight hours after transfection with specific siRNAs, CNE2 and 5-8F cells were trypsinized and resuspended at density of 1 × 106 cells per ml. The DNA-binding dye, Hoechst 33342 (Sigma-Aldrich), was then added at a final concentration of 7.5 μg/ml and incubated for 90 min in the dark with periodic mixing. After washing twice with PBS, the cells were placed at 4 °C in the dark before flow cytometry (EPICS ALTRA Flow Cytosorter, Beckman Coulter, Indianapolis, IN, USA) using dual-wavelength analysis. However, a fraction of the cell preparation was incubated with 5 μM Ko143 (Ko143, a specific inhibitor of ABCG2, Sigma-Aldrich) for 10 min at 37 °C before adding Hoechst 33342 to determine whether Ko143 would block the fluorescent efflux of SP cell.
Immunohistochemistry
The 4 μm paraffin-embedded NPC sections were deparaffinized in xylene and an alcohol gradient to rehydrate the sections. Next, the sections were treated with a Citrate Antigen Retrieval Solution (pH=8.0) in a pressure cooker for 5 min. Subsequently, 5% BSA in PBS (25 mM Tris, 0.8% NaCl, 2.68 mM KCl (pH 7.4)) was added to block non-specific binding, and the sections were then incubated with a mouse monoclonal anti-FBP1 antibody (1 : 100, Santa-Cruz Biotechnology, sc-374342, Dallas, TX, USA) or anti-c-Myc antibody (1 : 200, Cell Signaling, #9402, Danvers, MA, USA) in a moist chamber overnight at 4 °C. The secondary antibodies were incubated for 30 min at 37 °C on the next day.
Finally, the sections were incubated in 3, 3-diaminobenzidine for 2 min and counterstained with 10% Mayer's hematoxylin before being dehydrated and mounted. As a negative control, the primary antibodies were replaced with normal mouse and rabbit serum.
Two independent pathologists who were blinded to the clinical status of the patients scored the stained sections under a microscope. Semi-quantitative analysis was used to score the staining results. Compared with the control, the intensity was scored as follows: 0, negative staining; 1, weak staining; 2, moderate staining; and 3, strong staining. According to the percentages of the positive-stained areas, extent of staining was scored as follows: 0, <1% positive tumor cells; 1, 1–15% 2, 15–30% 3, 30–50% and 4, >51%. The final staining scores (0–7) for FBP1 or c-Myc were the sum of the scores for percentage and the intensity. Samples with the score of ≤4 were considered to have low expression, whereas the samples having a score of >4 were considered to have high expression.75
Nude mice xenograft assay and histopathology
To analyze tumor formation, the nude mice were injected with cells that had repressed FBP1. Female BALB/c nude mice were purchased from Shanghai Slac Laboratory Animal, (Shanghai, China) and maintained in microisolator cages. The animals were maintained under protocols approved by the China Care Committee Institute and this method was routinely performed as previously described.76, 77, 78, 79 All 12 mice were randomly assigned to two groups and underwent subcutaneous injection of 100 μl of a viable cell suspension mixture (2 × 106) containing 75% CNE2-NC or CNE2-siFBP1 cell suspension and 25% matrigel. When the tumors could be touched, the size of the tumor was measured by calipers daily. The tumor volume was calculated using a simplified formula: L × W 2 × 0.5, where L is the largest dimension and W is the perpendicular diameter. All mice were killed on the second week after injection, and the individual tumors were weighed. All mice were killed on the second week after injection. Individual tumors were weighted and embedded in 10% paraffin. Each tissue was subjected to analyze the expression of markers (FBP1 and c-Myc) by IHC, as described previously.
Statistical analysis
All data were analyzed using SPSS standard version 16.0 (SPSS, Chicago, USA) and GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA, USA). The χ2-test or Fisher's exact test was used to assess the correlation between the clinical features and FBP1 expression. The nonparametric Spearman's rank correlation coefficient was used to evaluate the correlation between FBP1 and c-Myc expression. The Kaplan–Meier method and the log-rank test were used to determine the differences in the actual survival rates. The univariate and multivariate Cox proportional hazards models were performed to test the relative risks of FBP1 expression and other predictive variables. Data were presented as the mean±S.E.M. obtained from three independent experiments. A P-value of <0.05 was considered to be statistically significant.
Acknowledgments
This study was supported by the grants from the Ministry of Science and Technology of China (2013BAI01B07, 2012CB967003 and 2015AA020931) and the National Natural Science Foundation of China (81230045, 91440106 and 81202137).
Glossary
- NPC
nasopharyngeal carcinoma
- FBP1
FUSE-binding protein 1
- qRT-PCR
quantitative real-time PCR
- siRNA
small interference RNA
- MTT
3-(4,5-dimethylthiazol 2-yl)-2,5-diphenylterazoliumbromide
- DMSO
dimethyl sulfoxide
- SP
side population
- DDP
cisplatin;5-FU, 5-flurouracil
- IHC
immuohistochemistry
- OS
overall survival
- PFS
progression-free survival
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by A Stephanou
Supplementary Material
References
- 1Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev 2006; 15: 1765–1777. [DOI] [PubMed] [Google Scholar]
- 2Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer 2004; 4: 757–768. [DOI] [PubMed] [Google Scholar]
- 3Kamran SC, Riaz N, Lee N. Nasopharyngeal carcinoma. Surg Oncol Clin N Am 2015; 24: 547–561. [DOI] [PubMed] [Google Scholar]
- 4Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet 2005; 365: 2041–2054. [DOI] [PubMed] [Google Scholar]
- 5Tsang CM, Deng W, Yip YL, Zeng MS, Lo KW, Tsao SW. Epstein-Barr virus infection and persistence in nasopharyngeal epithelial cells. Chin J Cancer 2014; 33: 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Lun SW, Cheung ST, Lo KW. Cancer stem-like cells in Epstein-Barr virus-associated nasopharyngeal carcinoma. Chin J Cancer 2014; 33: 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Lin X, Chen S, Xue X, Lu L, Zhu S, Li W et al. Chimerically fused antigen rich of overlapped epitopes from latent membrane protein 2 (LMP2) of Epstein-Barr virus as a potential vaccine and diagnostic agent. Cell Mol Immunol 2015; e-pub ahead of print 13 April 2015; doi:10.1038/cmi.2015.29. [DOI] [PMC free article] [PubMed]
- 8Stoker SD, Novalic Z, Wildeman MA, Huitema AD, Verkuijlen SA, Juwana H et al. Epstein-Barr virus-targeted therapy in nasopharyngeal carcinoma. J Cancer Res Clin Oncol 2015; 141: 1845–1857. [DOI] [PubMed] [Google Scholar]
- 9Tsang CM, Tsao SW. The role of Epstein-Barr virus infection in the pathogenesis of nasopharyngeal carcinoma. Virol Sin 2015; 30: 107–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Sun X, Su S, Chen C, Han F, Zhao C, Xiao W et al. Long-term outcomes of intensity-modulated radiotherapy for 868 patients with nasopharyngeal carcinoma: an analysis of survival and treatment toxicities. Radiother Oncol 2014; 110: 398–403. [DOI] [PubMed] [Google Scholar]
- 11Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv Cancer Res 2002; 84: 81–154. [DOI] [PubMed] [Google Scholar]
- 12Allday MJ. How does Epstein-Barr virus (EBV) complement the activation of Myc in the pathogenesis of Burkitt's lymphoma? Semin Cancer Biol 2009; 19: 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Yang J, Deng X, Deng L, Gu H, Fan W, Cao Y. Telomerase activation by Epstein-Barr virus latent membrane protein 1 is associated with c-Myc expression in human nasopharyngeal epithelial cells. J Exp Clin Cancer Res 2004; 23: 495–506. [PubMed] [Google Scholar]
- 14Fish K, Chen J, Longnecker R. Epstein-Barr virus latent membrane protein 2A enhances MYC-driven cell cycle progression in a mouse model of B lymphoma. Blood 2014; 123: 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Bieging KT, Amick AC, Longnecker R. Epstein-Barr virus LMP2A bypasses p53 inactivation in a MYC model of lymphomagenesis. Proc Natl Acad Sci USA 2009; 106: 17945–17950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 2014; 511: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Selmi A, de Saint-Jean M, Jallas AC, Garin E, Hogarty MD, Benard J et al. TWIST1 is a direct transcriptional target of MYCN and MYC in neuroblastoma. Cancer Lett 2015; 357: 412–418. [DOI] [PubMed] [Google Scholar]
- 18Wang HB, Liu GH, Zhang H, Xing S, Hu LJ, Zhao WF et al. Sp1 and c-Myc regulate transcription of BMI1 in nasopharyngeal carcinoma. FEBS J 2013; 280: 2929–2944. [DOI] [PubMed] [Google Scholar]
- 19Wang WJ, Wu SP, Liu JB, Shi YS, Huang X, Zhang QB et al. MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res 2013; 73: 1219–1231. [DOI] [PubMed] [Google Scholar]
- 20Luo J, Xiao J, Tao Z, Li X. Detection of c-myc gene expression in nasopharyngeal carcinoma by nonradioactive in situ hybridization and immunohistochemistry. Chin Med J 1997; 110: 229–232. [PubMed] [Google Scholar]
- 21Rydziel S, Delany AM, Canalis E. AU-rich elements in the collagenase 3 mRNA mediate stabilization of the transcript by cortisol in osteoblasts. J Biol Chem 2004; 279: 5397–5404. [DOI] [PubMed] [Google Scholar]
- 22Avigan MI, Strober B, Levens D. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J Biol Chem 1990; 265: 18538–18545. [PubMed] [Google Scholar]
- 23Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M et al. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev 1994; 8: 465–480. [DOI] [PubMed] [Google Scholar]
- 24Huang PN, Lin JY, Locker N, Kung YA, Hung CT, Lin JY et al. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic Acids Res 2011; 39: 9633–9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Zheng Y, Miskimins WK. Far upstream element binding protein 1 activates translation of p27Kip1 mRNA through its internal ribosomal entry site. Int J Biochem Cell Biol 2011; 43: 1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Chien HL, Liao CL, Lin YL. FUSE binding protein 1 interacts with untranslated regions of Japanese encephalitis virus RNA and negatively regulates viral replication. J Virol 2011; 85: 4698–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Jang M, Park BC, Kang S, Chi SW, Cho S, Chung SJ et al. Far upstream element-binding protein-1, a novel caspase substrate, acts as a cross-talker between apoptosis and the c-myc oncogene. Oncogene 2009; 28: 1529–1536. [DOI] [PubMed] [Google Scholar]
- 28Chan AK, Pang JC, Chung NY, Li KK, Poon WS, Chan DT et al. Loss of CIC and FUBP1 expressions are potential markers of shorter time to recurrence in oligodendroglial tumors. Mod Pathol 2014; 27: 332–342. [DOI] [PubMed] [Google Scholar]
- 29Singer S, Malz M, Herpel E, Warth A, Bissinger M, Keith M et al. Coordinated expression of stathmin family members by far upstream sequence element-binding protein-1 increases motility in non-small cell lung cancer. Cancer Res 2009; 69: 2234–2243. [DOI] [PubMed] [Google Scholar]
- 30Lasserre JP, Fack F, Revets D, Planchon S, Renaut J, Hoffmann L et al. Effects of the endocrine disruptors atrazine and PCB 153 on the protein expression of MCF-7 human cells. J Proteome Res 2009; 8: 5485–5496. [DOI] [PubMed] [Google Scholar]
- 31Xu SG, Yan PJ, Shao ZM. Differential proteomic analysis of a highly metastatic variant of human breast cancer cells using two-dimensional differential gel electrophoresis. J Cancer Res Clin Oncol 2010; 136: 1545–1556. [DOI] [PubMed] [Google Scholar]
- 32Malz M, Weber A, Singer S, Riehmer V, Bissinger M, Riener MO et al. Overexpression of far upstream element binding proteins: a mechanism regulating proliferation and migration in liver cancer cells. Hepatology 2009; 50: 1130–1139. [DOI] [PubMed] [Google Scholar]
- 33Zubaidah RM, Tan GS, Tan SB, Lim SG, Lin Q, Chung MC. 2-D DIGE profiling of hepatocellular carcinoma tissues identified isoforms of far upstream binding protein (FUBP) as novel candidates in liver carcinogenesis. Proteomics 2008; 8: 5086–5096. [DOI] [PubMed] [Google Scholar]
- 34Matsushita K, Tomonaga T, Kajiwara T, Shimada H, Itoga S, Hiwasa T et al. c-myc suppressor FBP-interacting repressor for cancer diagnosis and therapy. Front Biosci 2009; 14: 3401–3408. [DOI] [PubMed] [Google Scholar]
- 35Matsushita K, Tomonaga T, Shimada H, Shioya A, Higashi M, Matsubara H et al. An essential role of alternative splicing of c-myc suppressor FUSE-binding protein-interacting repressor in carcinogenesis. Cancer Res 2006; 66: 1409–1417. [DOI] [PubMed] [Google Scholar]
- 36Weber A, Kristiansen I, Johannsen M, Oelrich B, Scholmann K, Gunia S et al. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. BMC Cancer 2008; 8: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Zhang J, Chen QM. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene 2013; 32: 2907–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Rabenhorst U, Beinoraviciute-Kellner R, Brezniceanu ML, Joos S, Devens F, Lichter P et al. Overexpression of the far upstream element binding protein 1 in hepatocellular carcinoma is required for tumor growth. Hepatology 2009; 50: 1121–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Huynh T, Norris MD, Haber M, Henderson MJ. ABCC4/MRP4: a MYCN-regulated transporter and potential therapeutic target in neuroblastoma. Front Oncol 2012; 2: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Porro A, Haber M, Diolaiti D, Iraci N, Henderson M, Gherardi S et al. Direct and coordinate regulation of ATP-binding cassette transporter genes by Myc factors generates specific transcription signatures that significantly affect the chemoresistance phenotype of cancer cells. J Biol Chem 2010; 285: 19532–19543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Wang Z, Xu Y, Meng X, Watari F, Liu H, Chen X. Suppression of c-Myc is involved in multi-walled carbon nanotubes' down-regulation of ATP-binding cassette transporters in human colon adenocarcinoma cells. Toxicol Appl Pharmacol 2015; 282: 42–51. [DOI] [PubMed] [Google Scholar]
- 42Hoe SL, Tan LP, Jamal J, Peh SC, Ng CC, Zhang WC et al. Evaluation of stem-like side population cells in a recurrent nasopharyngeal carcinoma cell line. Cancer Cell Int 2014; 14: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Wang J, Guo LP, Chen LZ, Zeng YX, Lu SH. Identification of cancer stem cell-like side population cells in human nasopharyngeal carcinoma cell line. Cancer Res 2007; 67: 3716–3724. [DOI] [PubMed] [Google Scholar]
- 44Johnson PC, McAulay KA, Montgomery D, Lake A, Shield L, Gallagher A et al. Modeling HLA associations with EBV-positive and -negative Hodgkin lymphoma suggests distinct mechanisms in disease pathogenesis. Int J Cancer 2015; 137: 1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Henriksson M, Luscher B. Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res 1996; 68: 109–182. [DOI] [PubMed] [Google Scholar]
- 46Abba MC, Laguens RM, Dulout FN, Golijow CD. The c-myc activation in cervical carcinomas and HPV 16 infections. Mutat Res 2004; 557: 151–158. [DOI] [PubMed] [Google Scholar]
- 47Avet-Loiseau H, Gerson F, Magrangeas F, Minvielle S, Harousseau JL, Bataille R et al. Rearrangements of the c-myc oncogene are present in 15% of primary human multiple myeloma tumors. Blood 2001; 98: 3082–3086. [DOI] [PubMed] [Google Scholar]
- 48Baker VV, Borst MP, Dixon D, Hatch KD, Shingleton HM, Miller D. c-myc amplification in ovarian cancer. Gynecol Oncol 1990; 38: 340–342. [DOI] [PubMed] [Google Scholar]
- 49Herms JW, von Loewenich FD, Behnke J, Markakis E, Kretzschmar HA. c-myc oncogene family expression in glioblastoma and survival. Surg Neurol 1999; 51: 536–542. [DOI] [PubMed] [Google Scholar]
- 50Miranda Peralta EI, Valles Ayoub Y, Hernandez Mendoza L, Rangel Ramirez LM, Castrejon Rojas A, Collazo-Jaloma J et al. [MYC protein and proteins antigenically related with MYC in acute lymphoblastic leukemia]. Rev Invest Clin 1991; 43: 139–145. [PubMed] [Google Scholar]
- 51Treszl A, Adany R, Rakosy Z, Kardos L, Begany A, Gilde K et al. Extra copies of c-myc are more pronounced in nodular melanomas than in superficial spreading melanomas as revealed by fluorescence in situ hybridisation. Cytometry B Clin Cytom 2004; 60: 37–46. [DOI] [PubMed] [Google Scholar]
- 52Fan CS, Wong N, Leung SF, To KF, Lo KW, Lee SW et al. Frequent c-myc and Int-2 overrepresentations in nasopharyngeal carcinoma. Hum Pathol 2000; 31: 169–178. [DOI] [PubMed] [Google Scholar]
- 53Solares G, Blanco E, Pulgar S, Diago C, Ramos F. Carcinoid syndrome and intravenous cyproheptadine. Anaesthesia 1987; 42: 989–992. [DOI] [PubMed] [Google Scholar]
- 54Zhao Y, Pang TY, Wang Y, Wang S, Kang HX, Ding WB et al. LMP1 stimulates the transcription of eIF4E to promote the proliferation, migration and invasion of human nasopharyngeal carcinoma. FEBS J 2014; 281: 3004–3018. [DOI] [PubMed] [Google Scholar]
- 55Zhou W, Feng X, Ren C, Jiang X, Liu W, Huang W et al. Over-expression of BCAT1, a c-Myc target gene, induces cell proliferation, migration and invasion in nasopharyngeal carcinoma. Mol Cancer 2013; 12: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56Albihn A, Loven J, Ohlsson J, Osorio LM, Henriksson M. c-Myc-dependent etoposide-induced apoptosis involves activation of Bax and caspases, and PKCdelta signaling. J Cell Biochem 2006; 98: 1597–1614. [DOI] [PubMed] [Google Scholar]
- 57Bidwell GL 3rd, Raucher D. Enhancing the antiproliferative effect of topoisomerase II inhibitors using a polypeptide inhibitor of c-Myc. Biochem Pharmacol 2006; 71: 248–256. [DOI] [PubMed] [Google Scholar]
- 58Grassilli E, Ballabeni A, Maellaro E, Del Bello B, Helin K. Loss of MYC confers resistance to doxorubicin-induced apoptosis by preventing the activation of multiple serine protease- and caspase-mediated pathways. J Biol Chem 2004; 279: 21318–21326. [DOI] [PubMed] [Google Scholar]
- 59Penn LJ, Laufer EM, Land H. C-MYC: evidence for multiple regulatory functions. Semin Cancer Biol 1990; 1: 69–80. [PubMed] [Google Scholar]
- 60He L, Liu J, Collins I, Sanford S, O'Connell B, Benham CJ et al. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J 2000; 19: 1034–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61Kim MJ, Park BJ, Kang YS, Kim HJ, Park JH, Kang JW et al. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat Genet 2003; 34: 330–336. [DOI] [PubMed] [Google Scholar]
- 62Liu J, He L, Collins I, Ge H, Libutti D, Li J et al. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell 2000; 5: 331–341. [DOI] [PubMed] [Google Scholar]
- 63Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol 2006; 16: 253–264. [DOI] [PubMed] [Google Scholar]
- 64Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol 1999; 19: 4672–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65Dean M, Hamon Y, Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res 2001; 42: 1007–1017. [PubMed] [Google Scholar]
- 66Higgins CF. ABC transporters: from microorganisms to man. Ann Rev Cell Biol 1992; 8: 67–113. [DOI] [PubMed] [Google Scholar]
- 67Liao WT, Wang HM, Li MZ, Song LB, Zhang L, Mai HQ et al. [Establishment of three-dimensional culture models related to different stages of nasopharyngeal carcinogenesis]. Ai Zheng 2005; 24: 1317–1321. [PubMed] [Google Scholar]
- 68Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest 2009; 119: 3626–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69Song LB, Zeng MS, Liao WT, Zhang L, Mo HY, Liu WL et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res 2006; 66: 6225–6232. [DOI] [PubMed] [Google Scholar]
- 70Peterson BR, Nelson BL. Nonkeratinizing undifferentiated nasopharyngeal carcinoma. Head Neck Pathol 2013; 7: 73–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71Thompson L. World Health Organization classification of tumours: pathology and genetics of head and neck tumours. Ear Nose Throat J 2006; 85: 74. [PubMed] [Google Scholar]
- 72Li BS, Zuo QF, Zhao YL, Xiao B, Zhuang Y, Mao XH et al. MicroRNA-25 promotes gastric cancer migration, invasion and proliferation by directly targeting transducer of ERBB2, 1 and correlates with poor survival. Oncogene 2015; 34: 2556–2565. [DOI] [PubMed] [Google Scholar]
- 73Li Y, Zeng X, Wang S, Sun Y, Wang Z, Fan J et al. Inhibition of autophagy protects against PAMAM dendrimers-induced hepatotoxicity. Nanotoxicology 2015; 9: 344–355. [DOI] [PubMed] [Google Scholar]
- 74Zhang JP, Zhang H, Wang HB, Li YX, Liu GH, Xing S et al. Down-regulation of Sp1 suppresses cell proliferation, clonogenicity and the expressions of stem cell markers in nasopharyngeal carcinoma. J Transl Med 2014; 12: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75Xie F, Liu H, Zhu YH, Qin YR, Dai Y, Zeng T et al. Overexpression of GPR39 contributes to malignant development of human esophageal squamous cell carcinoma. BMC Cancer 2011; 11: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76Shirasaki T, Maruya S, Mizukami H, Kakehata S, Kurotaki H, Yagihashi S et al. Effects of small interfering RNA targeting thymidylate synthase on survival of ACC3 cells from salivary adenoid cystic carcinoma. BMC Cancer 2008; 8: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77Mehta G, Scheinman RI, Holers VM, Banda NK. A new approach for the treatment of arthritis in mice with a novel conjugate of an anti-C5aR1 antibody and C5 small interfering rNA. J Immunol 2015; 194: 5446–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78Jung HS, Rajasekaran N, Song SY, Kim YD, Hong S, Choi HJ et al. Human papillomavirus E6/E7-specific sirna potentiates the effect of radiotherapy for cervical cancer in vitro and in vivo. Int J Mol Sci 2015; 16: 12243–12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79Chen S, Liu X, Gong W, Yang H, Luo D, Zuo X et al. Combination therapy with VEGFR2 and EGFR siRNA enhances the antitumor effect of cisplatin in non-small cell lung cancer xenografts. Oncol Rep 2013; 29: 260–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.