Abstract
LMO2 was first discovered through proximity to frequently occurring chromosomal translocations in T cell acute lymphoblastic leukaemia (T-ALL). Subsequent studies on its role in tumours and in normal settings have highlighted LMO2 as an archetypical chromosomal translocation oncogene, activated by association with antigen receptor gene loci and a paradigm for translocation gene activation in T-ALL. The normal function of LMO2 in haematopoietic cell fate and angiogenesis suggests it is a master gene regulator exerting a dysfunctional control on differentiation following chromosomal translocations. Its importance in T cell neoplasia has been further emphasized by the recurrent findings of interstitial deletions of chromosome 11 near LMO2 and of LMO2 as a target of retroviral insertion gene activation during gene therapy trials for X chromosome-linked severe combined immuno-deficiency syndrome, both types of event leading to similar T cell leukaemia. The discovery of LMO2 in some B cell neoplasias and in some epithelial cancers suggests a more ubiquitous function as an oncogenic protein, and that the current development of novel inhibitors will be of great value in future cancer treatment. Further, the role of LMO2 in angiogenesis and in haematopoietic stem cells (HSCs) bodes well for targeting LMO2 in angiogenic disorders and in generating autologous induced HSCs for application in various clinical indications.
Keywords: LMO2, haematopoiesis, angiogenesis, chromosomal translocations, leukaemia, X chromosome-linked severe combined immuno-deficiency syndrome
1. The discovery of LMO2
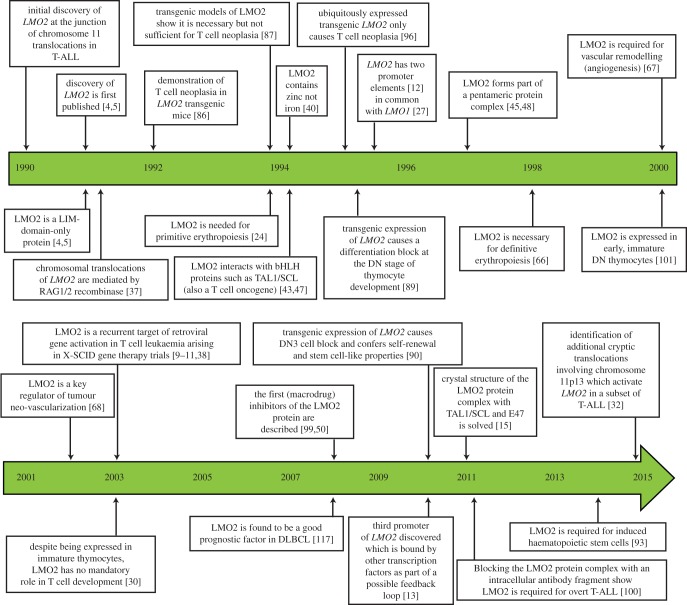
Developments in molecular biology in the 1980s led to the finding that recurrent, cancer-associated chromosomal translocations result in either perturbed oncogene control resulting from joining with antibody or T cell receptor (TCR) genes or creation of novel fusion protein with chimaeric functions (reviewed in [1,2]). Studies of TCR gene rearrangement and gene location suggested that T cell cancer-associated chromosomal translocations would have oncogene activations [3]. This proved correct, and 25 years ago LMO2 was discovered, and published the following year [4,5], as a recurrent chromosomal translocation partner of TCR loci in a subset of patients with T cell acute lymphoblastic leukaemia (T-ALL). Since then, research into this remarkable protein has shown that LMO2 is highly conserved among evolutionary orthologues (http://www.imm.ox.ac.uk/the-lmo-genes-and-proteins) and that it is capable of eliciting a multitude of cellular effects, ranging from a proto-oncogenic role in T cells to an essential role in haematopoiesis and vascular remodelling, as well as a major function in stem cell biology. This review outlines the developing understanding of LMO2 cancer and normal biology, illustrating how LMO2 acts as a paradigm for genes activated in acute forms of cancer. Figure 1 indicates the chronological milestones in this process.
Figure 1.
Milestones in LMO2 research: timeline indicating the major steps in LMO2 research from the gene discovery in 1990 to present.
2. LMO2 belongs to the LIM-domain-only family of proteins
The progenitor gene in the family to which LMO2 belongs was LMO1 (formerly known as RBTN1, Rhombotin-1 or Ttg-1). LMO1 was one of the first T-ALL translocation proto-oncogenes to be isolated, located on chromosome 11 and involved in translocations t(11;14)(p15;q11) [6–8]. It was suggested that other similar proto-oncogenes may exist within the genome and subsequently LMO2 (formerly known as RBTN2, Rhom-2 or Ttg-2) was first discovered through filter hybridization experiments aimed at uncovering additional genes homologous to LMO1 [4] and by direct cloning from the t(11;14)(p13;q11) T-ALL translocation breakpoints [5]. Thus, LMO2, like LMO1, is located on the short arm of human chromosome 11 but at band 11p13 rather than band 11p15 (LMO1). Leukaemias carrying translocations involving the 11p13 cluster are found more frequently in T-ALL patient samples than the 11p15 translocations [4] (figure 2).
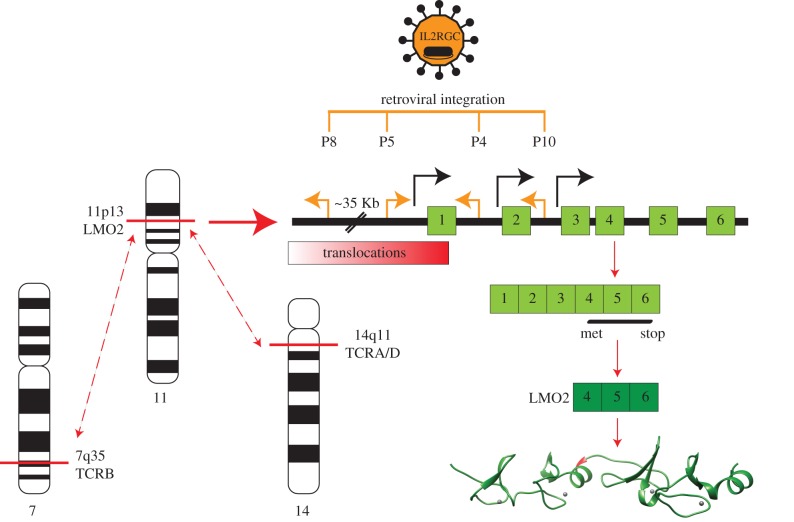
Figure 2.
Chromosomal translocations and X-SCID retroviral insertions associated with LMO2 gene activation. Diagram of the chromosomal bands of TCRA/D and TCRB and LMO2 involved in T cell ALL translocations resulting in LMO2 activation. Also indicated are the retroviral insertions found in the X-SCID gene therapy trial leukaemias (orange lines, with orientation of insertion indicated by orange arrows) [9–11]. The distal, proximal [12] and intermediate [13] gene promoters are shown (black arrows). LMO2 comprises six exons (light green boxes, numbered) of which exons 4–6 (dark green boxes, numbered) are protein coding (green ribbon structure) drawn in USCF Chimaera [14] from PDB file 2XJY [15]. The coding region of LMO2 is unaltered after either the chromosomal translocations or the retroviral insertions. (Adapted from [16].)
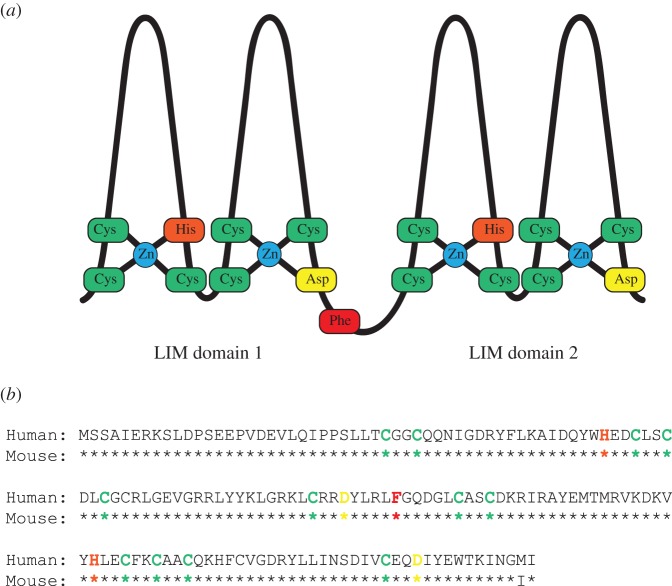
The LMO family of proteins (so-called because these proteins comprise the LIM-domain-only proteins; see below) is now known to contain four genes (table 1), LMO1, 2, 3 and 4 (formerly RBTN1, 2, 3, 4) [17–20]. The name of this family comes from their tertiary structure that is composed of two tandemly arranged regions called LIM-domains (viz. LIM1 and LIM2). In turn, the LIM-domain was named after the transcription factors Lin-11, Isl-1 and Mec-3 [21–23]. It is a cysteine-rich motif, characterized by the consensus amino acid sequence C-X2-C-X17−19-H-C-X2-C-X2-C-X16−20-C-X2-C-X/C/D (figure 3).
Table 1.
Chromosomal location of the LMO gene family in human and mouse genomes. Chromosomal translocations known to occur in proximity to the genes are listed, with associated malignancies and knock-out mouse phenotypic defects indicated. For references, see detail in relevant text sub-sections.
| chromosome band |
||||||
|---|---|---|---|---|---|---|
| protein | human | mouse | knock-out phenotypic defects | chromosomal translocation | T-ALL | other cancers |
| LMO1 | 11p15 | 7E3 | CNS | t(11;14)(p15;q11) | ✓ | neuroblastoma |
| LMO2 | 11p13 | 2E2 | haematopoiesis | t(11;14)(p13;q11) t(7;11)(q35;p13) |
✓ | B cell lymphoma B-ALL |
| LMO3 | 12p12 | 6G1 | CNS | t(7;12)(q35;p12) | ✓ | neuroblastoma |
| LMO4 | 1p22.3 | 3H2 | neural tube development | not known | not known | breast cancer neuroblastoma |
Figure 3.
Diagrammatic structure of the LIM-Only proteins and LMO2 amino acid sequence comparing human with mouse. (a) Schematic diagram of the LMO proteins showing the tandem arrangement of the two LIM-domains. Each domain comprises two zinc-finger-like structures, which coordinate a zinc atom between four residues. The two fingers of each domain are linked by two amino acid residues which are conserved between species and confer specificity of subsequent PPI. (b) An alignment of the human and mouse LMO2 proteins to illustrate species conservation and the homology and differences between them. Residues are highlighted to correlate with their position in the schematic structure shown in (a); green denotes cysteine, orange indicates histidine, yellow is aspartate while the key hinge region residue, phenylalanine 88, is highlighted in red [15].
LMO1 and LMO3 share the highest degree of sequence conservation, being 98% identical. LMO1, 2 and 3 are all involved in T-ALL by chromosomal translocations (table 1), but LMO4 (uncovered through two hybrid screening [18,20]) is the most divergent member of the family and currently has no known leukaemia translocations. Gene targeting has been implemented to discover the gene functions in mice. The homozygous phenotypes of the four genes in knock-out mice show their essential role in cell fate decisions (table 1), and there is phenotypic synergy when homozygous loss of Lmo1 and Lmo3 occurs in mice [24,25]. These phenotypic properties in part led to the proposal of the ‘master gene’ hypothesis [1] based on highly conserved, developmentally important, transcriptional activators [1,26]. LMO2 is an archetypal example of such a master transcription regulator, but also of a chromosomal translocation oncogene, which is discussed further in the following sections.
3. Chromosomal translocations of LMO2 and the involvement of RAG recombinase
Human LMO2 has six exons, of which the last three encode the protein comprising 158 amino acids and has two major transcription promoters [12,27,28] and a recently described third ‘intermediate’ promoter [13] (indicated in figure 2). Gene expression analyses have shown LMO2 is expressed in a range of tissues during development [28,29] but not in normal mature thymocytes other than tumourigenic T cells [5,17,24,30]. This suggests that LMO2 expression in T cells is reliant upon activation through mechanisms such as chromosomal translocations.
Chromosomal aberrations, including translocations, deletions and insertions are frequent in childhood T-ALL, often resulting in the temporally and spatially incorrect activation and expression of developmental regulatory genes [2]. Aberrant expression of LMO2 resulting from such gross genetic abnormalities is documented in approximately 9% of childhood T-ALL cases [31,32] but expression has been reported in as much as 45% of T-ALL [33].
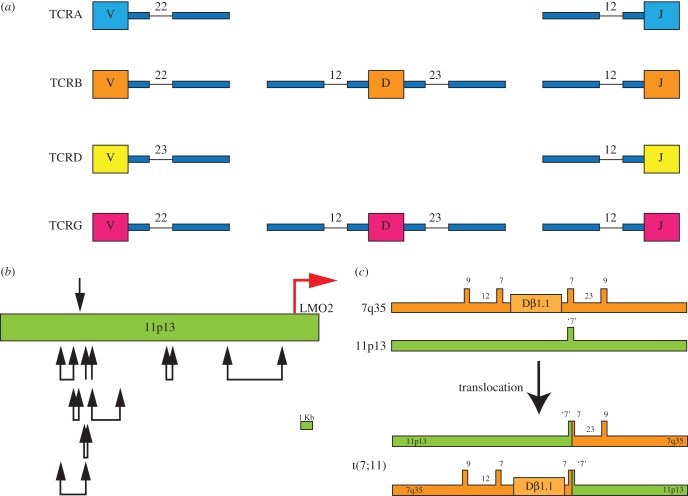
LMO2 occurs at the junction of common T-ALL-associated translocations, namely the translocation t(11;14)(p13;q11) involving the TCRD/A from 14q11 or the t(7;11)(q35;p13) translocation with the TCRB locus from 7q35 (figure 2). These translocations encompass approximately 5% of primary paediatric T-ALL cases investigated by karyotypic analysis [32]. The mediator of these translocations, and of other similar leukaemic TCR-locus translocations such as involving TAL1/SCL, herein referred to as TAL1, [34,35], appears to be the RAG1/2 recombinase complex that is normally responsible for intra-chromosomal antigen receptor gene rearrangement in thymic T cell maturation (reviewed in [36]), but that makes occasional errors of inter-chromosomal translocation [37]. In many cases, this error is sequence-specific as the chromosomal translocation breakpoint sequences on chromosome 11p13 (i.e. near LMO2) can have heptamers typical of recombination signal sequences (RSS) that could be the normal recognition for RAG recombinase, albeit generally lacking the nonamer stretch found at genuine RSS locations. Figure 4a illustrates normal TCR heptamer and nonamer RSS for variable (V), diversity (D) and joining (J) for TCRD and TCRB, and V-J RSS for TCRA and TCDG. When the breakpoint cluster region near LMO2 on chromosome 11 was sequenced, heptamer-like RSS were found near translocation breakpoints (figure 4b) and in three independent tumours, the break occurs at the same heptamer-like RSS on chromosome 11 [37].
Figure 4.
Site-specific mechanism of LMO2 chromosomal translocations mediated by RAG recombinase. (a) TCR gene segments (V, D and J) are linked to RSS. Each segment has heptamer and nonamer sequences as indicated (and at the spacings in TCR V, D and J segments shown). (b) Heptamer sequences are found near LMO2 at chromosome 11p13 (green), specifically around translocation breakpoint regions denoted by black arrows [37]. Although not all were found to possess joining via the heptamer sequences, hallmark sequences of Rag recombinase were noted. The location of the proximal LMO2 promoter is shown by a red arrow. (c) Detailed analysis of the reciprocal translocation breakpoints of LMO2 has shown the involvement of the VDJ RAG recombinase in the translocations. An example of the organization of reciprocal t(7;11)(q35;p13) translocation regions is shown (lower part of the panel) compared with germline organization. Note the presence of a heptamer-like sequence at the junction of the 11p13 breakpoint [37].
The precision with which the chromosomal translocations subvert TCR rearrangement is illustrated in figure 4c, which shows the molecular architecture of the one t(7;11) case [37]. Although the TCR-associated translocations are sequence-specific, their rarity doubtless reflects a restriction imposed by chromosomal territory in the T cell nuclei and less efficient processing when heptamer, but not nonamer, RSS are present.
LMO2 expression may also be activated by the cryptic deletion, del(11)(p12p13) in approximately 4% paediatric T-ALL patients [31], and by a plethora of cryptic and newly discovered translocations [32]. It is intriguing that the del(11) can involve the juxtaposition of RAG2 with LMO2 (and, apparently, deletion of RAG1). The deleted region, just upstream of LMO2, includes negative regulatory sequences, whose removal could permit LMO2 expression via its proximal promoter. These same sequences are also removed from LMO2 by the chromosomal translocations. However, the existence of translocation-negative, LMO2-positive clonal T cell tumours in up to 45% of other T-ALL [33] suggests that LMO2 may be activated through mechanisms other than gross genetic changes at the locus. This may arise through TAL1, LYL1 and/or ERG cooperatively engaging in a feedback loop at their respective regulatory elements to activate or increase expression [13] (see §5).
4. LMO2 is a preferred oncogene in X chromosome-linked severe combined immuno-deficiency syndrome gene therapy associated leukaemia
Aberrant expression of LMO2, following activation by retroviral insertion, has also led to a T-ALL-like leukaemia arising in four of the children treated in X chromosome-linked severe combined immuno-deficiency syndrome (X-SCID) gene therapy trials [9,10,38]. X-SCID is caused by a defect in the Interleukin-2 receptor gamma common chain (IL2RGC), which is necessary for high-affinity signalling of several interleukin (IL) receptors and is typified by a lack of mature T cells and natural killer cells. Gene correction was achieved by transduction of autologous CD34+ bone marrow stem cells with retrovirus expressing IL2RGC [39]. While several patients eventually had an immune cell repertoire that matched those of non-SCID children, some of the children involved in the trial developed T cell leukaemia [10,11,38,39]. Four cases have activated LMO2 caused by retroviral insertion into, or just upstream of, the LMO2 locus that consequently activates expression of LMO2 through the strong constitutive expression of the retroviral promoter/enhancer systems. Figure 2 shows the location of the insertion sites and that insertional mutagenesis occurs outside the LMO2 coding region in all four cases. We have shown with a mouse transgenic model that concurrent expression of LMO2 and IL2RGC in thymocytes accelerates the formation of clonal T cell neoplasias compared with those arising in transgenic aberrantly expressing only LMO2, while aberrant expression of IL2RGC alone has no discernable oncogenic effect in thymocytes (K. Ruggero & THR, unpublished data). This demonstrates that LMO2 and IL2RGC expression synergizes in thymus cells to cause T cell neoplasias in mice and, by inference, describes the T cell adverse effects that occur in the X-SCID gene therapy patients. A model for LMO2-associated X-SCID T-ALL is discussed in §9.
5. LMO2 protein structure and the multimeric DNA-binding complex
LMO2 is a LIM-domain-only protein. LIM-domains were originally thought to carry Fe-S centres but the demonstration of zinc in the proteins [40] led to the discovery that LIM-domain folds to form two LIM-fingers through the coordination of a zinc atom by the side-chains of three cysteine residues and a histidine residue (finger 1), or three cysteine residues and an aspartic acid residue (finger 2) (figure 3). The intervening peptide chain is forced outward to form the finger-like projections, arranged as two perpendicular anti-parallel β-sheets [41]. The LIM-fingers are distinct from the zinc-fingers of transcription factors such as those of the GATA-family, as no evidence exists of LIM-domain-only proteins directly binding DNA. Rather, the LIM-fingers provide a mechanism of protein : protein interaction (PPI) with a variety of binding partners [42–45]. Each finger is separated by two amino acid residues, the spacing and sequence of which has been shown to be vital for subsequent protein interactions in other LIM-domain-containing proteins [46].
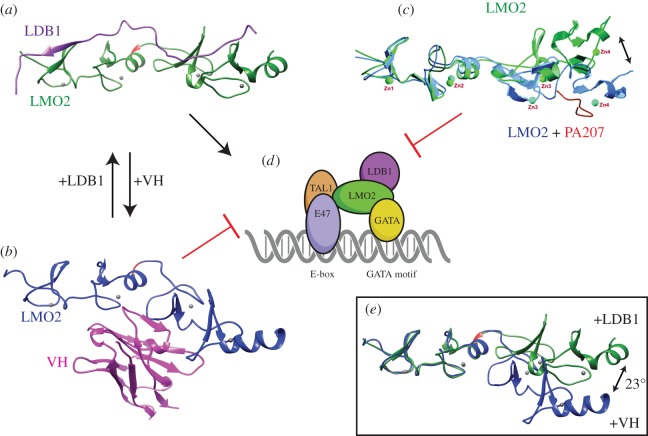
LMO2 does not bind DNA directly but instead acts by forming part of a bridge for DNA-binding proteins to create a bipartite multi-protein DNA-binding complex (illustrated in figure 5d). Newly synthesized LMO2 protein does not have a stable conformation, requiring interaction with other proteins to stabilize its inherently unstructured nature and also prevent proteasomal degradation, as in the case of TAL1 binding [51]. LMO2 was first shown in complex with TAL1, E47 and GATA-1 in erythroid cells [43], and subsequently shown to interact with several other basic-helix-loop-helix (bHLH) transcription factor proteins including LYL1 and TAL2 [47]. LMO2 binds directly to the bHLH protein TAL1 and it is intriguing that this partner is also a T cell oncogene [34,35,52–54]. TAL1 is a haematopoietic tissue-specific member of the class II bHLH transcription factor family, which also includes LYL1 and TAL2, both of which interact with LMO2 [47]. LMO2 can bind GATA-1, GATA-2 or GATA-3 [45,55,56]. The LMO2-complex contains an additional protein called the LIM-domain-binding protein 1 (LDB1) [45,57]. The LMO2-complex thus comprises the bipartite DNA-binding complex recognizing an E-box and GATA site, separated by one turn of the DNA helix (figure 5d). This complex was shown capable of substituting the GATA-factor for another TAL1/E47 heterodimer in transgenic LMO2-dependent T cell tumours [48] and also at the Ckit promoter in erythroid cells [58]. Similarly, a GATA-E-box-GATA motif has been shown at the erythroid Krüppel-like factor locus [59]. Taken together, these findings suggest that LMO2 has a flexible multi-protein complex nucleation function in different cell settings.
Figure 5.
Representation of the conformational flexibility of LMO2 in complex with LDB1 or inhibitors of LMO2. The pentameric LMO2-complex commonly found in erythroid lineage cells binds to a bipartite E-box-GATA motif [45] and is permitted or inhibited depending upon the stabilizing protein partner of LMO2 (a,d). LMO2 has been shown to nucleate variants of this complex, including the substitution of the GATA-factor and binding other bHLH transcription factor proteins [47], with resulting altered DNA recognition sequence binding specificity [48]. The structure and conformational stabilization of LMO2 (green) when bound with LDB1-LID (purple) [15] is indicated in (a). When LMO2 (shown in blue in panel (b)) is in complex with the anti-LMO2 VH (magenta), there is a conformational distortion, preventing nucleation of the LMO2-complex to the complex illustrated in (d) [49]. Superimposition of the normal LDB1- and VH-bound structures highlights the 23° contortion of the molecule when the anti-LMO2 VH is bound (e). The hinge region residue Phe88 (red), zinc atoms coordinated by the LIM-fingers (grey spheres) are indicated in both structures, drawn using UCSF Chimaera [14] from PDB accession codes 2XJY [15] and 4KFZ [49]. An in silico model of the structural effects on LMO2 induced by the peptide aptamer PA-207 is shown in (c). (Adapted from [50].)
As it is a naturally disordered protein, containing very little secondary structure, LMO2 presents an unstable conformation, making it inherently insoluble when made in recombinant form, and hence difficult to purify for structural studies. This has been partially overcome by expressing both LIM-domains individually or full-length LMO2 as a fusion protein with the LIM-interaction domain (LID) of LDB1 [15,60,61]. NMR [60,62] and crystallographic data [15,63] of the LMO2–LID fusion show that LID binds across the LIM1 and LIM2 domains. There is a flexible hinge region located between the two LIM-domains and the LDB1 LID interaction, leaving the LIM-finger-side of both domains unoccupied for other protein interactions [15]. Thus, the structural instability of LMO2 is resolved on binding LDB1 (as illustrated in figure 5a), allowing the nucleation of the additional members into a multimeric protein complex, such as the erythroid complex including TAL1/E47 and GATA1 binding to the bipartite E-box-GATA motif [45]. When bound to the N-terminal LIM-domain of LMO2, the TAL1/E47 heterodimer undergoes small conformational changes [64] that may be a feature allowing fine-tuning of the LMO2-complex to repress or activate specific genes in a temporally controlled fashion. Conformational change in LMO2 has been induced by binding to an antibody fragment [49] that accounts for an inhibitory effect on the LMO2 protein complex (figure 5b,e) (discussed further in §10).
6. LMO2 haematopoietic master regulator function portends its oncogenicity
After the identification of LMO2 as a proto-oncogene in T-ALL, the natural function of the protein was investigated in mouse and human cells to delineate its normal biochemistry and to instruct future work on development of anti-LMO2 therapies. A principle finding was the expression of LMO2 in haematopoietic cells and a pivotal function in specification of haematopoietic lineage. This suggested the means by which LMO2 function lies behind its role as a cancer inducer.
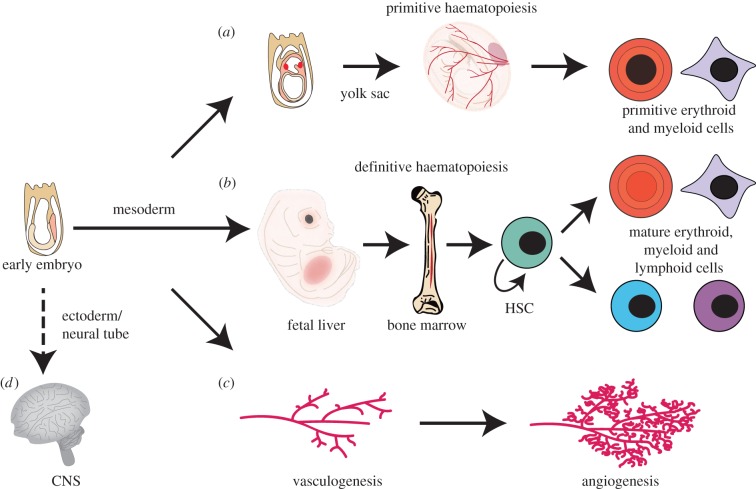
The development of gene targeting in mouse embryonic stem cells (ES) and generation of mice from these mutant pluripotent cells (technology reviewed in [65]) allowed LMO2 gene targeting to be implemented to discover the functions of LMO2 in mouse development [24,66]. Insertion of the β-galactosidase gene as reporter for Lmo2 expression showed mainly expression in haematopoiesis and in developing brain [67,68]. Figure 6 outlines LMO2 functions in development, determined mainly by gene targeting. In the haematopoietic hierarchy, there is an essential role for LMO2 in red cell development in the embryo yolk sac (primitive erythropoiesis) (figure 6a) as homozygous Lmo2 null mice died in utero at embryonic stage E9–E10 [24]. While Lmo2 null embryos displayed normal blood islands of the yolk sac, they were devoid of red cells. Intriguingly, it was later shown that the consequences of the homozygous Lmo2 null mutation was similar to those encountered in Tal1 [69,70], Gata-1 [71,72] and Gata-2-null embryos [73], presaging the finding that the LMO2 protein is part of a pentameric complex that includes TAL1 and GATA proteins [45,47,55].
Figure 6.
Role of LMO2 in mouse development. Gene knock-out of mouse LMO2 has revealed its involvement in several aspects of haemaotopoiesis and blood vessel formation. LMO2 has a necessary role in primitive [24] (a) and definitive haematopoiesis [66] (b), and it is required for remodelling of the vascular networks as part of angiogenesis [67] (c). LMO2 has been shown to be expressed in the developing central nervous system [17] and tissue staining using lacZ knock-in mice showed specific regions of LMO2 expression [66] although no discernable knock-out brain phenotype has been noted to date.
As Lmo2 null embryos do not survive to term, a role of LMO2 in adult haematopoiesis (definitive haematopoiesis) required a different strategy for analysis. Definitive haematopoiesis initiates in the mid-gestation embryo in the aorta–gonad–mesonephros region (reviewed in [74,75]), switches to the fetal liver and relocates to the bone marrow shortly before birth, where it persists throughout adult life. Accordingly, ES cells with homozygous Lmo2 null mutation were used to create chimaeric mice that have contribution of cells from both the recipient blastocyst and the donor homozygous Lmo2 null ES cells. These studies showed that homozygous Lmo2 null ES donor cells do not contribute to haematopoiesis in adult mice [24,66], suggesting that LMO2 function is crucial in the multi-potent haematopoietic stem cells (HSCs) (figure 6b). It was also shown that overexpression of LMO2 (and LDB1) in erythroblast cell lines resulted in immature, undifferentiated precursor erythroid cells, which established that LMO2 could act as a negative regulator of differentiation [76]. Furthermore, LMO2 mRNA expression has been demonstrated during both primitive [77] and definitive [78] haematopoiesis through in situ hybridization of mouse embryos.
Blood vessel endothelial cell formation (de novo vasculogenesis) also arises from mesoderm and vascular remodelling (angiogenesis) is growth of new blood vessels from existing vasculature through sprouting, migration and adhesion. The processes of blood cell and blood vessel specification are tightly linked (reviewed in [79]). By examining the contribution of Lmo2 null-LacZ ES cells in chimaeric mice, it was found that Lmo2 is not required for the de novo vessel formation during early embryogenesis but is needed for angiogenic remodelling of vascular networks (figure 6c) [67].
LMO2 also has a role in normal endothelial and lymphatic endothelial cells. It is found in lymphatic endothelial cells, which form new lymphatic vessels by lymphangiogenesis, a process similar to angiogenesis. Angiogenesis can be induced by various growth factors, including VEGF that increases endothelial cell and lymphatic endothelial cell levels of LMO2 and GATA2 [80]. Moreover, through an in silico screen [81], the E-box-GATA motif of the LMO2-transcriptional complex was found to occur in the promoter regions of many pro- and anti-angiogenic-related genes such as Angiopoietin-2.
LMO2 has long been implicated in developing CNS tissue (figure 6d) [17] and is expressed in the adult mouse brain, alongside LMO1 and LMO3 [82], particularly after epileptic seizures, possibly as a repair mechanism. Intriguingly, LMO2 can nucleate an altered transcriptional complex with LDB1, the bHLH E-box-binding transcription factor NSLC2 and BEX2 in fetal brain tissue [83] to regulate transcription. There remains no clear correlation between this CNS expression of LMO2 and function but it may be a link to the recent implication that LMO2 plays a role in glioma [84] (see §11).
7. LMO2, haemtopoietic stem cells, iPS and cancer-initiating cells
LMO2 has a role in stem cells and/or confers stem cell-like functions. As discussed in §6, LMO2 is needed in induction and maintenance of HSCs that can differentiate into all blood cell types (HSCs are described as pluripotent in the context of blood cell development to distinguish them from multi-potent progenitors). The gene is expressed in mouse lineage-negative, ScaI-positive, cKit-positive (LSK) haematopoietic progenitor cells in mice [85], and knock-out of Lmo2 prevents primitive and definitive haematopoiesis [24,66] (see also §6).
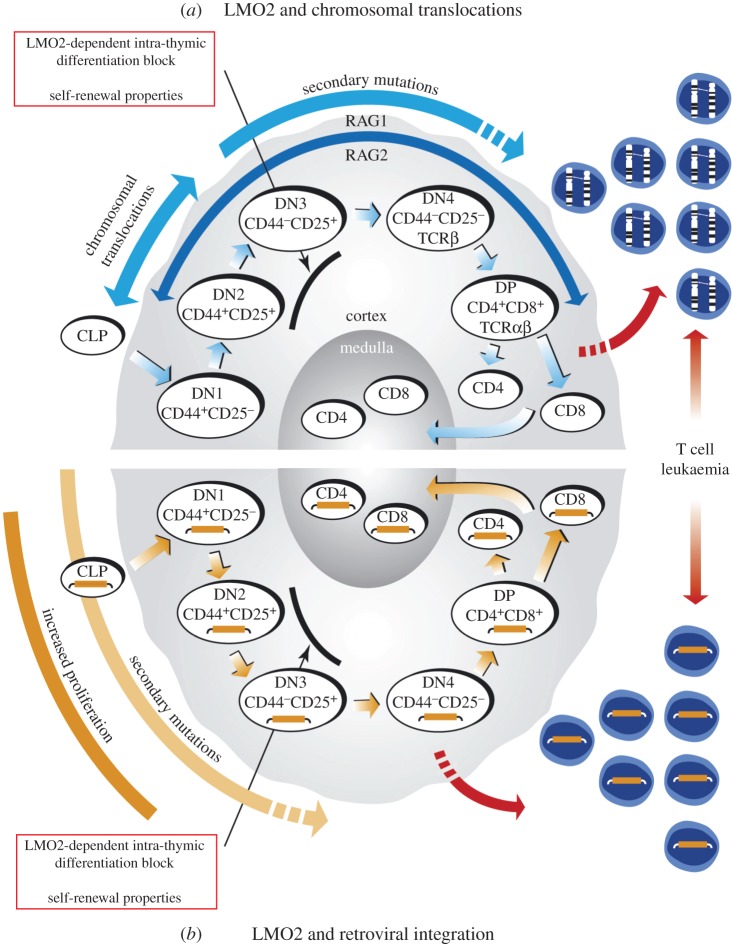
One of the key functions of LMO2 in T cell neoplasia is interference with intra-thymic T cell differentiation as the immature double negative (DN; not expressing CD4 or CD8) thymocytes accumulate in Lmo2 transgenic mice [86,87] (illustrated in figure 7a). This is in part achieved in DN3 cells, where LMO2 elicits self-renewing properties to these cancer precursors [90]. A critical function of LMO2 in T cell tumourigenesis is thus to form leukaemia-initiating cells (LICs) as a pool for further oncogenic mutations (discussed in §9).
Figure 7.
Model of LMO2-mediated leukaemogenesis. Schematic diagram to represent the current understanding of the effect of LMO2 in LMO2-translocation positive T-ALL (a) and in the X-SCID gene therapy trial leukaemias (b) where LMO2 expression was activated through retroviral integration. (a) Fully mature CD4 or CD8 single positive (SP) T cells develop in the thymus through DN CD4/CD8 precursor populations, DN1–4, which are characterized by their progressive changing expression of CD25 and CD44 surface markers. The RAG recombinase genes are expressed from the DN2 stage onwards and promote TCR gene recombination events to produce functioning receptor molecules during the normal maturation cycle of the early thymoctyes. Although some cells progress through to mature CD4/CD8 double positive or SP T cells, thymus-specific transgenic Lmo2 mice exhibit a block in T cell differentiation with an accumulation of DN2–DN3 stage immature thymocytes (indicated), which after a long asymptomatic phase develop clonal T cell neoplasias [87–89] after the acquisition of additional mutations [53]. This differentiation block coincides with the timing of RAG expression, directly mimicking the temporal occurrence, and effect of, chromosomal translocations involving the TCR genes and LMO2 loci [37]. Furthermore, the enforced expression of Lmo2 in the transgenics results in immature cells with acquiring self-renewal capacity, demonstrated by serial transplantation of the DN3 thymocytes [90]. (b) Following viral therapy in the X-SCID gene replacement trials, leukaemias were observed in which retroviral insertion had occurred into or upstream of the LMO2 gene and activated expression in thymocytes [9–11]. Expanding from the LMO2 tumourigenesis model in (a), it would appear that leukaemogenesis in the X-SCID patients could arise from lymphoid progenitor cells with activated LMO2 expression migrating to the thymus, as part of the normal maturation process, and resulting in LMO2 conferring the DN2/3 differentiation block that was observed in the transgenic model, promoting self-renewal properties, accumulation of secondary mutations and finally the severe clinical adverse effects of leukaemias seen. (Adapted from [16].)
An exciting consequence of LMO2 stem-cell properties is the recent demonstration of LMO2 as a factor in the generation of induced haematopoietic stem cells (iHSCs). This follows from work demonstrating the creation of pluripotent stem cells (iPSCs) through reprogramming of terminally differentiated adult cells by the expression of key transcription factors [91]. Three studies have defined the requirement for LMO2 together with other factors to support creation of stem cell properties in differentiated cells, to generate induced haematopoietic cells. One study shows that expandable haemangioblasts (capable of forming endothelial cells, multi-lineage haematopoietic cells and smooth muscle) can be made in vitro from ES cells, fetal liver cells and fibroblasts [92]. LMO2 with its partners TAL1 and GATA2 together with SOX17, PITX2 and MYCN are sufficient. iHSCs capable of multi-lineage development can be formed from fibroblasts by expressing LMO2 with its partners TAL1 and GATA2 together with ERG, RUNX1 [93], and it is possible to reprogram committed cells (myeloid and lymphoid committed cells) using a cocktail of factors in addition to LMO2 including RUN1T1, HLF, PRDM5, PBX1 and ZFP37 [94]. In the latter, it is possible that the LMO2 partners (GATA and TAL1) are provided by the cellular environment in the committed cells used for making the iHSC. Autologous iHSCs that are capable of multi-lineage differentiation and of long-term reconstitution have enormous potential clinical benefit. There may be a risk involving the oncogenic potential of LMO2 in reprogrammed cells [95], particularly given the appearance of leukaemias in the X-SCID trials following retroviral insertional activation of LMO2 [9,10,38] and the evidence of neoplasia in models of enforced lmo2 expression [87–89,96]. Stringent controls and safety measures may be necessary for further evaluation of these technologies, possibly through using exogenous protein rather than genetic manipulation, to avoid these problems [97].
8. LMO2 and the chromosomal translocation master gene model
The chromosomal translocation master gene model [1] was proposed based on the normal role of the genes found at the junctions of acute leukaemia chromosomal translocations, such as the two LIM-only genes LMO1 and LMO2, and HOX11. In particular, LMO1 was shown to have specific expression during hindbrain rhombomere development [98]. The essential features of the model were based on the findings that these genes play a role in lineage determination and/or tissue specification and the suggestion that the molecular mechanisms underlying these functions are subverted, after the chromosomal translocation gene activation, to create the ‘chromosomal translocation master gene’. The proteins are involved in PPI and DNA binding (indirectly like LMO2 or directly like TAL1 or HOX11) and thus control gene expression. Therefore, the cell that gains a chromosomal translocation (or other chromosome abnormality leading to aberrant gene expression) is committed to a transcriptional programme that the chromosomal translocation master gene dictates.
The subsequent work on LMO2 shows that it fulfils all the components of the master gene model; (i) the protein is active in controlling PPI by forming multimeric protein complexes that bind to DNA through two parts of the complex (such as bHLH and GATA components), (ii) it confers a differentiation block on T cell development (intra-thymic), and (iii) it also confers the stem-cell-like property of self-propagation on these cells. Inhibition of the LMO2 protein complex in mouse models of T cell neoplasia [50,99,100] show that LMO2 is necessary, but not sufficient [87], for the cancers, but that it is required at the stage of overt disease.
9. The LMO2 paradigm of chromosomal translocation genes in T cell acute lymphoblastic leukaemia
The combined studies of transgenic T cell neoplasias resulting from thymic expression of Lmo2 and the study of human T-ALL carrying LMO2 translocations led to a model of LMO2-mediated T-ALL (figure 7a) that is a paradigm for chromosomal translocation-associated acute cancers.
Transgenic expression of LMO2 from CD2 [87–89], Metallothionine [96] or Lck promoters (K. Ruggero & THR, unpublished data) results in long-latency, clonal T cell neoplasia. While not expressed in normal mature T cells, gene expression analyses have shown that LMO2 occurs in early, immature CD4/CD8 DN thymocytes (thymocytes that do not express TCRs), at the DN1 stage of their development alongside TAL1 [101], a member of the LMO2 multi-protein transcription factor complex. Normally, downregulation of LMO2 and TAL1 is concordant with upregulation of additional transcription factors that permit thymocyte maturation through the DN3, DN4 and double-positive CD4/CD8 stages, resulting in fully mature single CD4-positive or CD8-positive T cells. Despite evidence of LMO2 being expressed in the DN1 early thymocytes, lymphoid lineage-specific Lmo2 knock-out showed no abnormal development or irregular thymic cellularity [30].
The detailed study of tumourigenesis caused by transgenic LMO2 from the CD2 [87–89] or from the Lck promoter (K. Ruggero & THR, unpublished data) demonstrates that LMO2 exerts its oncogenic effect on immature T cells in the thymus, prior to TCR-based positive and negative selection (figure 7). The transgenic mice exhibit two phases in the appearance of overt T cell neoplasias, the first being a long asymptomatic phase during which there is an accumulation of immature thymocytes within the thymus [53,87,89] at the DN2/DN3 stages (defined, respectively, by CD25+/CD44+ or CD25+/CD44−). Later, overt T cell neoplasias develop with clonal TCR rearrangements. Thus, transgenic LMO2 is necessary in this model but not sufficient, and transition to frank haematopoietic cancer requires secondary mutations. One type of mutation that is found frequently in the transgenic mouse tumours are Notch1 mutations (K. Ruggero & THR, unpublished data), which parallels mutations seen in about 50% of human T-ALL [102].
The data suggest that, in transgenic models, T cell progenitors enter the thymus as DN1 cells (CD25−/CD44−) and begin to mature through DN2 and DN3 but are held up at the DN3 stage (figure 7). This is a stochastic block as some cells progress to make mature T cells expressing CD4+/CD8+ intermediates or CD4+ or CD8+ T cells. Serial transplantation studies show that the DN3 thymocytes of the CD2–LMO2 transgenic mice acquire stem cell-like properties as these cells have self-renewal properties in recipient mice [90]. We propose that this sub-population of self-renewing DN3 cells acts as a pool of LICs. The high penetrance of transgenic overt T cell neoplasia is a consequence of there being a pool of LICs in which secondary mutational events can arise. Of course, in the case of the human LMO2 chromosomal translocation T-ALL, cancers arise from single cells due to the rarity of the relevant translocation. Interestingly, transgenic mice expressing LMO2 with its partner protein TAL1 exhibit clonal T cell neoplasias at a faster rate than in LMO2-only transgenics [53,87], whereas TAL1-only mice did not show any neoplasias [103]. This latter finding may reflect the lack of endogenous LMO2 in the critical thymocyte (i.e. DN3) to partner the TAL1 transgenic protein, whereas the LMO2 transgenic protein can partner other endogenous bHLH proteins (e.g. LYL1) in the absence of TAL1 for tumourigenesis.
We discussed in §3 how the LMO2 chromosomal translocations are frequently mediated through mistakes made by RAG1/2 recombinase causing the inter-chromosomal events. As RAG1 and RAG2 begin expression at the early stages of the DN thymocyte differentiation, and are active in DN2/DN3 cells, it is likely that LMO2 chromosomal translocations occur before the DN3 stage and have a pathogenic consequence as the single intra-thymic translocation event will result in a LIC. By analogy with the transgenic mouse data, this cell will acquire self-renewal properties due to LMO2 expression and propagate to gradually produce a pool of cells within which the secondary mutations (such as NOTCH1) occur to give rise to frank leukaemia (figure 7a). An interesting possibility is that RAG-mediated translocations could occur throughout thymocyte differentiation but only manifest tumourigenic consequences if before or at the DN3 stage. In addition, it seems significant that LMO2 translocations do not appear in B cell neoplasias, despite B cells having RAG1 and RAG2 expression.
This chromosomal translocation model provides an intriguing back-drop to the incidence of LMO2-associated gene activation in the X-SCID gene therapy trials. Parallels with the effect of LMO2 chromosomal translocations are remarkable (figure 7b; reviewed in [16,104,105]). Autologous donor CD34+ bone marrow HSCs from X-SCID patients were infected with retrovirus expressing IL2RGC and in some recipients leukaemias developed with LMO2 activation. In the model (illustrated in figure 7b), it is envisaged that accessibility for retroviral insertion into LMO2 in the pluripotent CD34+ bone marrow cells occurs as LMO2 is expressed therein and thus the chromatin is ‘open’. But it is also predicted that this has no oncogenic consequence until the progenitors enter the thymus as all models show that LMO2 is only oncogenic during haematopoiesis in thymocytes. The cell(s) with the retrovirally activated LMO2, by analogy to chromosomal translocation-activated LMO2, can progress normally into the DN3 stage, where the presence of LMO2 will cause inhibition of T cell differentiation and the LIC phenotype. This clone will eventually acquire secondary mutation(s) that in turn lead to overt leukaemia. The cooperating property of the LIC(s) in the gene therapy patients, which influences the adverse outcome, is that the IL2RGC gene is the common chain of several IL receptors, including the IL2 receptor, which is expressed in DN3 cells (marked by CD25 surface protein which is IL2RA).
Recent clinical studies have identified a highly aggressive form of T-ALL, designated early T cell precursor-like ALL (ETP-ALL) [106], which is characterized by immature DN1-like immature thymocytes and which is often highly treatment-resistant. ETP-ALLs have been found to have high levels of LMO2 and LYL1 expression [107–109], and LYL1 appears to be the necessary bHLH transcription factor partner of LMO2 rather than TAL1 [108]. Together, the LMO2–LYL1 complex seems to correlate with enhanced rates of transition to full neoplasia, perhaps an effect from withdrawal of competitor cells within the early DN populations, promoting existing thymocyte self-renewal and thus the acquisition of further mutations [110].
10. Pre-clinical macrodrugs targeting the LMO2 protein complex
The transgenic models of LMO2-associated T cell neoplasia show that LMO2 is necessary but not sufficient for T cell neoplasia. Because of this, it is possible that LMO2 is only needed in the asymptomatic phase within the LIC and does not need to persist in overt tumours. Such a hit and run mechanism has been ruled out in the transgenic tumours by inhibiting the LMO2 protein with intracellular macrodrugs (macromolecular drugs) exemplified by a peptide aptamer [50], by an antibody single-chain Fv fragment [99] and by a single VH domain antibody fragment [49,100].
Each of these macrodrugs binds to LMO2 and interferes with LMO2 PPI by different mechanisms. The peptide aptamer confers a growth inhibition on LMO2 leukaemic cells in transplantation assays [50], and modelling suggested that it disrupts the LMO2 LIM finger 4 by competing for interaction with the zinc atom and thus destroying the ability of LMO2 to interact with components of the multimeric complex (figure 5c). While recent data imply broad binding of the aptamer across other zinc-finger-containing proteins [111], in vivo binding shows specificity for LMO2 and mutagenesis suggests predominance of binding to LIM finger 4 [50]. The intracellular antibody fragments also abrogate the neoplastic effect of LMO2 in transgenic LMO2 cell transplantation assays [99,100] mediated by the disruption of the LMO2-complex by preventing LDB1 interaction [100]. The crystal structure of the LMO2 : VH complex [49], compared with that of LMO2-LDB1-LID fusion [15], demonstrates the mechanism. When the VH binds to LMO2, the latter adopts a distorted conformation around its hinge region (figure 5b,e). This bending and twisting distortion of a normal LMO2 fold induced by the natural partner LDB1 LID, interferes with the interaction with natural partners of LMO2. Thus, the intracellular single-domain antibody fragment makes its binding partners unavailable for the normal protein complex formation and function.
11. Association of LMO2 with non-T cell tumours
The chromosomal abnormalities involving LMO2 appear to be restricted to T cell tumours, but LMO2 expression studies have implicated LMO2 in a range of other cancers. It is noteworthy nevertheless that widespread transgenic expression of LMO2 from a Metallothionine promoter only resulted in T cell tumour phenotype [96], implying that the tumourigenic effect in mice is restricted to the T cell lineage.
LMO2 expression has been observed in diffuse large B cell lymphoma (DLBCL) [112] and in some B cell acute leukaemias [113–115]. It is expressed in normal germinal centre (GC) B cells and in the GC-derived DLBCL [112,116] where the presence of LMO2 is a good prognostic factor [117]. This may reflect that LMO2 expression is a passenger effect of pre-existing expression in GC cells or it may be a consequence of there being different LMO2-binding partners in DLBCL, where it has been shown to bind to ELK1 (ETS-like gene 1), NFATc1 (nuclear factor of activated T cells 1) and LEF1 (lymphoid enhancer-binding factor 1) [116]. Moreover, when in complex with LMO2, the NFATc1 and LEF1 proteins exhibited increased and decreased transcriptional activity, respectively. LMO2 expression has also been reported in some B cell acute leukaemias (B-ALL) with differing prognostic correlations, where there is good prognostic association in some [115,118], but in others, such as B-ALL expressing the E2A–HLF fusion from t(17;19) (q22;p13), it is a poor prognostic feature [113,114].
Controversy surrounds the significance of LMO2 in epithelial cancers. One study reported LMO2 in 60% of pancreatic cancer samples and in 80–90% of high grade neoplasias tested, while being absent from normal pancreatic ductal epithelium [119]. However, tissue staining of a smaller set of pancreatic tumours did not find LMO2 expression except in the surrounding tumour vasculature [120], where it is known to be associated with tumour vascular remodelling [68]. The significance of LMO2 expression in pancreatic and other epithelial cancers requires more analysis. An intriguing observation has recently been published showing LMO2 exerting a stem cell phenotype in glioblastoma [84]. LMO2 could induce stem cell characteristics in mouse premalignant astrocytes and anti-LMO2 siRNA affected growth of human patient-derived glioma stem cells. Further studies are eagerly awaited.
12. Future of LMO2 therapy and LMO2 biology
While improved treatments have led to an increased survival rate, around one-fifth of paediatric T-ALL patients succumb to the disease, either through treatment resistance or relapse, while the figure for adult T-ALL is much higher [121,122]. LMO2 suffers chromosomal translocation or activation by interstitial deletion in 5% of T-ALL and is aberrantly expressed in approximately 45% of T-ALL not exhibiting defects at the LMO2 locus [33]. LMO2 is also expressed in the vasculature of many types of tumour [68,120], fulfilling its normal angiogenic role [67] in a disadvantageous setting. Increased angiogenesis is implicated in a number of clinical conditions, including eye diseases such as diabetic proliferative retinopathy, and also found to occur due to the inflammatory response in diseases such as rheumatoid arthritis and bowel disease (reviewed in [123]). Therefore, inhibition of LMO2 could be of clinical benefit for many cancers, but also for other diseases that have angiogenic processes.
In view of the role of LMO2 in multimeric protein complexes by PPI and its importance in several areas of haematopoiesis, development of LMO2 inhibitors is important but also very challenging, requiring new approaches and categories of molecule that will be efficacious and safe for use in the clinic. Relatively few small molecules that inhibit PPI have been developed, although the options are increasing (reviewed in [124]). At present, there are no small molecule inhibitors of LMO2. The protein is not an enzyme with an active site but rather works as part of the protein interaction network. The development of anti-LMO2 macrodrugs is under investigation as an alternative that will target the LMO2 PPI or the LMO2 mRNA. The latter could be targeted for destruction through the use of small interfering (si)RNA molecules. In a manner analogous to micro (mi)RNA miR-223 [125], anti-LMO2 siRNA molecules could potentially prevent translation of LMO2 mRNA. However, these small RNAs may produce off-target effects and are prone to rapid turnover within the cell. Targeting therapies to disrupt the function of the LMO2 protein may be more practicable. A peptide aptamer was developed that conferred a significant growth inhibition of transgenic LMO2 leukaemic cells in transplantation assays [50]. An alternative is the use of intracellular antibody fragments, in the form of a single-chain variable fragment and a single variable heavy chain (VH) that can readily be isolated using molecular biology techniques such as intracellular antibody capture (IAC; reviewed in [126]). The anti-LMO2 single VH domain developed in our laboratory through the IAC technology [100,127–130] binds to LMO2 with high affinity and induces a structural distortion, rendering LMO2 unable to efficiently nucleate the LMO2-complex [49] (figure 5b,e). Indeed, an exciting possibility is that these macrodrugs may be used to inform rational design of small molecule drug inhibitors of LMO2 with similar mechanistic properties.
Major problems with macrodrugs, as opposed to small molecule drugs, are cell penetration, immunogenicity, stoichiometry and target location inside the cell. Macrodrugs are generally too large to simply cross the cell membrane, and are at risk of degradation, natural clearing through the liver and possible immune response. Intracellular expression of the antibody fragment from nucleic acid would, however, ensure high levels of anti-LMO2 protein in target cells. As LMO2 has roles in the HSCs, erythropoiesis and angiogenesis, specific tumour targeting may be required. This could be achieved through immunoliposome or immune-nanoparticle delivery, using particles coated with antibody having recognition of a target cell surface antigen, for specific uptake. Another option is using therapeutic viruses engineered for specificity. Furthermore, enhancement of the anti-leukaemic effects could be achieved through linking effector domains to the intracellular antibody fragments, to accelerate LMO2 protein degradation or to initiate apoptosis of the tumour cells following LMO2 binding [131]. Ongoing work in our laboratory is being undertaken to answer these questions and evaluate ways to produce effective therapy for LMO2-dependent disease.
In the 25 years since its discovery, much has been learned about LMO2 and the other LIM-domain-only family members (reviewed in [19]). In addition to LMO2 in diseases like cancer, LMO2 has potential clinical benefit in the development of therapeutic stem cells, due to its ability to be involved in reprogramming differentiated cells to confer a stem cell-like phenotype [92,94] and in making autologous iHSC [93]. In all, LMO2 has proved to be a remarkable protein with diverse effects. Research into LMO2 and its functions continues, and we hope this will soon translate into better management of relevant human diseases.
Competing interests
We declare we have no competing interests.
Funding
The authors are funded by the Medical Research Council (J.C. and T.H.R.), the Leukaemia and Lymphoma Research (T.H.R.), the Wellcome Trust (T.H.R.) and the University of Oxford (J.C. and T.H.R.).
References
- 1.Rabbitts TH. 1991. Translocations, master genes, and differences between the origins of acute and chronic leukemias. Cell 67, 641–644. (doi:10.1016/0092-8674(91)90057-6) [DOI] [PubMed] [Google Scholar]
- 2.Rabbitts TH, Boehm T. 1991. Structural and functional chimerism results from chromosomal translocation in lymphoid tumors. Adv. Immunol. 50, 119–146. (doi:10.1016/s0065-2776(08)60824-x) [DOI] [PubMed] [Google Scholar]
- 3.Lefranc MP, Rabbitts TH. 1985. Two tandemly organized human genes encoding the T-cell gamma constant-region sequences show multiple rearrangement in different T-cell types. Nature 316, 464–466. (doi:10.1038/316464a0) [DOI] [PubMed] [Google Scholar]
- 4.Boehm T, Foroni L, Kaneko Y, Perutz MF, Rabbitts TH. 1991. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc. Natl Acad. Sci. USA 88, 4367–4371. (doi:10.1073/pnas.88.10.4367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Royer-Pokora B, Loos U, Ludwig WD. 1991. TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11). Oncogene 6, 1887–1893. [PubMed] [Google Scholar]
- 6.Boehm T, Foroni L, Kennedy M, Rabbitts TH. 1990. The rhombotin gene belongs to a class of transcriptional regulators with a potential novel protein dimerisation motif. Oncogene 5, 1103–1105. [PubMed] [Google Scholar]
- 7.Boehm T, Buluwela L, Williams D, White L, Rabbitts TH. 1988. A cluster of chromosome 11p13 translocations found via distinct D-D and D-D-J rearrangements of the human T cell receptor delta chain gene. EMBO J. 7, 2011–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGuire EA, Hockett RD, Pollock KM, Bartholdi MF, O'Brien SJ, Korsmeyer SJ. 1989. The t(11;14)(p15;q11) in a T-cell acute lymphoblastic leukemia cell line activates multiple transcripts, including Ttg-1, a gene encoding a potential zinc finger protein. Mol. Cell. Biol. 9, 2124–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hacein-Bey-Abina S, et al. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302, 415–419. (doi:10.1126/science.1088547) [DOI] [PubMed] [Google Scholar]
- 10.Howe SJ, et al. 2008. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 118, 3143–3150. (doi:10.1172/JCI35798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacein-Bey-Abina S, et al. 2008. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 118, 3132–3142. (doi:10.1172/JCI35700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Royer-Pokora B, Rogers M, Zhu TH, Schneider S, Loos U, Bolitz U. 1995. The TTG-2/RBTN2 T cell oncogene encodes two alternative transcripts from two promoters: the distal promoter is removed by most 11p13 translocations in acute T cell leukaemia's (T-ALL). Oncogene 10, 1353–1360. [PubMed] [Google Scholar]
- 13.Oram SH, et al. 2010. A previously unrecognized promoter of LMO2 forms part of a transcriptional regulatory circuit mediating LMO2 expression in a subset of T-acute lymphoblastic leukaemia patients. Oncogene 29, 5796–5808. (doi:10.1038/onc.2010.320) [DOI] [PubMed] [Google Scholar]
- 14.Pettersen EF, et al. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. (doi:10.1002/jcc.20084) [DOI] [PubMed] [Google Scholar]
- 15.El Omari K, Hoosdally SJ, Tuladhar K, Karia D, Vyas P, Patient R, Porcher C, Mancini EJ. 2011. Structure of the leukemia oncogene LMO2: implications for the assembly of a hematopoietic transcription factor complex. Blood 117, 2146–2156. (doi:10.1182/blood-2010-07-293357) [DOI] [PubMed] [Google Scholar]
- 16.McCormack MP, Rabbitts TH. 2004. Activation of the T-cell oncogene LMO2 after gene therapy for X-linked severe combined immunodeficiency. N. Eng. J. Med. 350, 913–922. (doi:10.1056/NEJMra032207) [DOI] [PubMed] [Google Scholar]
- 17.Foroni L, et al. 1992. The rhombotin gene family encode related LIM-domain proteins whose differing expression suggests multiple roles in mouse development. J. Mol. Biol. 226, 747–761. (doi:10.1016/0022-2836(92)90630-3) [DOI] [PubMed] [Google Scholar]
- 18.Grutz G, Forster A, Rabbitts TH. 1998. Identification of the LMO4 gene encoding an interaction partner of the LIM-binding protein LDB1/NLI1: a candidate for displacement by LMO proteins in T cell acute leukaemia. Oncogene 17, 2799–2803. (doi:10.1038/sj.onc.1202502) [DOI] [PubMed] [Google Scholar]
- 19.Matthews JM, Lester K, Joseph S, Curtis DJ. 2013. LIM-domain-only proteins in cancer. Nat. Rev. Cancer 13, 111–122. (doi:10.1038/nrc3418) [DOI] [PubMed] [Google Scholar]
- 20.Kenny DA, Jurata LW, Saga Y, Gill GN. 1998. Identification and characterization of LMO4, an LMO gene with a novel pattern of expression during embryogenesis. Proc. Natl Acad. Sci. USA 95, 11 257–11 262. (doi:10.1073/pnas.95.19.11257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Way JC, Chalfie M. 1988. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell 54, 5–16. (doi:10.1016/0092-8674(88)90174-2) [DOI] [PubMed] [Google Scholar]
- 22.Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. 1990. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature 344, 879–882. (doi:10.1038/344879a0) [DOI] [PubMed] [Google Scholar]
- 23.Freyd G, Kim SK, Horvitz HR. 1990. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature 344, 876–879. (doi:10.1038/344876a0) [DOI] [PubMed] [Google Scholar]
- 24.Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. 1994. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell 78, 45–57. (doi:10.1016/0092-8674(94)90571-1) [DOI] [PubMed] [Google Scholar]
- 25.Tse E, et al. 2004. Null mutation of the Lmo4 gene or a combined null mutation of the Lmo1/Lmo3 genes causes perinatal lethality, and Lmo4 controls neural tube development in mice. Mol. Cell. Biol. 24, 2063–2073. (doi:10.1128/MCB.24.5.2063-2073.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleary ML. 1991. Oncogenic conversion of transcription factors by chromosomal translocations. Cell 66, 619–622. (doi:10.1016/0092-8674(91)90105-8) [DOI] [PubMed] [Google Scholar]
- 27.Boehm T, Greenberg JM, Buluwela L, Lavenir I, Forster A, Rabbitts TH. 1990. An unusual structure of a putative T cell oncogene which allows production of similar proteins from distinct mRNAs. EMBO J. 9, 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boehm T, Spillantini MG, Sofroniew MV, Surani MA, Rabbitts TH. 1991. Developmentally regulated and tissue specific expression of mRNAs encoding the two alternative forms of the LIM domain oncogene rhombotin: evidence for thymus expression. Oncogene 6, 695–703. [PubMed] [Google Scholar]
- 29.Neale GA, Mao S, Parham DM, Murti KG, Goorha RM. 1995. Expression of the proto-oncogene rhombotin-2 is identical to the acute phase response protein metallothionein, suggesting multiple functions. Cell Growth Diff. 6, 587–596. [PubMed] [Google Scholar]
- 30.McCormack MP, Forster A, Drynan L, Pannell R, Rabbitts TH. 2003. The LMO2 T-cell oncogene is activated via chromosomal translocations or retroviral insertion during gene therapy but has no mandatory role in normal T-cell development. Mol. Cell. Biol. 23, 9003–9013. (doi:10.1128/MCB.23.24.9003-9013.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Vlierberghe P, et al. 2006. The cryptic chromosomal deletion del(11)(p12p13) as a new activation mechanism of LMO2 in pediatric T-cell acute lymphoblastic leukemia. Blood 108, 3520–3529. (doi:10.1182/blood-2006-04-019927) [DOI] [PubMed] [Google Scholar]
- 32.Wu L, et al. 2015. High frequency of cryptic chromosomal rearrangements involving the LMO2 gene in T-cell acute lymphoblastic leukemia. Haematologica 100, e233–e236. (doi:10.3324/haematol.2014.120089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrando AA, et al. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 1, 75–87. (doi:10.1016/S1535-6108(02)00018-1) [DOI] [PubMed] [Google Scholar]
- 34.Begley CG, et al. 1989. Chromosomal translocation in a human leukemic stem-cell line disrupts the T-cell antigen receptor delta-chain diversity region and results in a previously unreported fusion transcript. Proc. Natl Acad. Sci. USA 86, 2031–2035. (doi:10.1073/pnas.86.6.2031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finger LR, Kagan J, Christopher G, Kurtzberg J, Hershfield MS, Nowell PC, Croce CM. 1989. Involvement of the TCL5 gene on human chromosome 1 in T-cell leukemia and melanoma. Proc. Natl Acad. Sci. USA 86, 5039–5043. (doi:10.1073/pnas.86.13.5039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schatz DG, Ji Y. 2011. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11, 251–263. (doi:10.1038/nri2941) [DOI] [PubMed] [Google Scholar]
- 37.Garcia IS, et al. 1991. A study of chromosome 11p13 translocations involving TCR beta and TCR delta in human T cell leukaemia. Oncogene 6, 577–582. [PubMed] [Google Scholar]
- 38.Hacein-Bey-Abina S, et al. 2003. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Eng. J. Med. 348, 255–256. (doi:10.1056/NEJM200301163480314) [DOI] [PubMed] [Google Scholar]
- 39.Hacein-Bey-Abina S, et al. 2002. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N. Eng. J. Med. 346, 1185–1193. (doi:10.1056/NEJMoa012616) [DOI] [PubMed] [Google Scholar]
- 40.Archer VE, Breton J, Sanchez-Garcia I, Osada H, Forster A, Thomson AJ, Rabbitts TH. 1994. Cysteine-rich LIM domains of LIM-homeodomain and LIM-only proteins contain zinc but not iron. Proc. Natl Acad. Sci. USA 91, 316–320. (doi:10.1073/pnas.91.1.316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Alvarado GC, Kosa JL, Louis HA, Beckerle MC, Winge DR, Summers MF. 1996. Structure of the cysteine-rich intestinal protein, CRIP. J. Mol. Biol. 257, 153–174. (doi:10.1006/jmbi.1996.0153) [DOI] [PubMed] [Google Scholar]
- 42.Sadler I, Crawford AW, Michelsen JW, Beckerle MC. 1992. Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J. Cell Biol. 119, 1573–1587. (doi:10.1083/jcb.119.6.1573) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valge-Archer VE, Osada H, Warren AJ, Forster A, Li J, Baer R, Rabbitts TH. 1994. The LIM protein RBTN2 and the basic helix-loop-helix protein TAL1 are present in a complex in erythroid cells. Proc. Natl Acad. Sci. USA 91, 8617–8621. (doi:10.1073/pnas.91.18.8617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmeichel KL, Beckerle MC. 1994. The LIM domain is a modular protein-binding interface. Cell 79, 211–219. (doi:10.1016/0092-8674(94)90191-0) [DOI] [PubMed] [Google Scholar]
- 45.Wadman IA, et al. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16, 3145–3157. (doi:10.1093/emboj/16.11.3145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmeichel KL, Beckerle MC. 1997. Molecular dissection of a LIM domain. Mol. Biol. Cell 8, 219–230. (doi:10.1091/mbc.8.2.219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wadman I, et al. 1994. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 13, 4831–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grutz GG, Bucher K, Lavenir I, Larson T, Larson R, Rabbitts TH. 1998. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 17, 4594–4605. (doi:10.1093/emboj/17.16.4594) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sewell H, Tanaka T, El Omari K, Mancini EJ, Cruz A, Fernandez-Fuentes N, Chambers J, Rabbitts TH. 2014. Conformational flexibility of the oncogenic protein LMO2 primes the formation of the multi-protein transcription complex. Sci. Rep. 4, 3643 (doi:10.1038/srep03643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Appert A, et al. 2009. Targeting LMO2 with a peptide aptamer establishes a necessary function in overt T-cell neoplasia. Cancer Res. 69, 4784–4790. (doi:10.1158/0008-5472.CAN-08-4774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lecuyer E, Lariviere S, Sincennes MC, Haman A, Lahlil R, Todorova M, Tremblay M, Wilkes BC, Hoang T. 2007. Protein stability and transcription factor complex assembly determined by the SCL-LMO2 interaction. J. Biol. Chem. 282, 33 649–33 658. (doi:10.1074/jbc.M703939200) [DOI] [PubMed] [Google Scholar]
- 52.Baer R. 1993. TAL1, TAL2 and LYL1: a family of basic helix-loop-helix proteins implicated in T cell acute leukaemia. Semin. Cancer Biol. 4, 341–347. [PubMed] [Google Scholar]
- 53.Larson RC, et al. 1996. Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. EMBO J. 15, 1021–1027. [PMC free article] [PubMed] [Google Scholar]
- 54.Ono Y, Fukuhara N, Yoshie O. 1997. Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or -2 and induces TALLA1, a highly specific tumor marker of T-ALL. J. Biol. Chem. 272, 4576–4581. (doi:10.1074/jbc.272.7.4576) [DOI] [PubMed] [Google Scholar]
- 55.Osada H, Grutz G, Axelson H, Forster A, Rabbitts TH. 1995. Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc-finger protein GATA1. Proc. Natl Acad. Sci. USA 92, 9585–9589. (doi:10.1073/pnas.92.21.9585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ono Y, Fukuhara N, Yoshie O. 1998. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol. Cell. Biol. 18, 6939–6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. 1996. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384, 270–272. (doi:10.1038/384270a0) [DOI] [PubMed] [Google Scholar]
- 58.Vitelli L, Condorelli G, Lulli V, Hoang T, Luchetti L, Croce CM, Peschle C. 2000. A pentamer transcriptional complex including tal-1 and retinoblastoma protein downmodulates c-kit expression in normal erythroblasts. Mol. Cell. Biol. 20, 5330–5342. (doi:10.1128/MCB.20.14.5330-5342.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson KP, Crable SC, Lingrel JB. 1998. Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (eKLF) gene. J. Biol. Chem. 273, 14 347–14 354. (doi:10.1074/jbc.273.23.14347) [DOI] [PubMed] [Google Scholar]
- 60.Deane JE, Sum E, Mackay JP, Lindeman GJ, Visvader JE, Matthews JM. 2001. Design, production and characterization of FLIN2 and FLIN4: the engineering of intramolecular ldb1:LMO complexes. Protein Eng. 14, 493–499. (doi:10.1093/protein/14.7.493) [DOI] [PubMed] [Google Scholar]
- 61.Ryan DP, et al. 2006. Identification of the key LMO2-binding determinants on LDB1. J. Mol. Biol. 359, 66–75. (doi:10.1016/j.jmb.2006.02.074) [DOI] [PubMed] [Google Scholar]
- 62.Matthews JM, Visvader JE, Mackay JP. 2001. 1H, 15N and 13C assignments of FLIN2, an intramolecular LMO2:LDB1 complex. J. Biomol. NMR 21, 385–386. (doi:10.1023/A:1013373203772) [DOI] [PubMed] [Google Scholar]
- 63.El Omari K, Porcher C, Mancini EJ. 2010. Purification, crystallization and preliminary X-ray analysis of a fusion of the LIM domains of LMO2 and the LID domain of LDB1. Acta Crystallogr. Sec. F Struct. Biol. Crystal. Commun. 66, 1466–1469. (doi:10.1107/S1744309110032872) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.El Omari K, et al. 2013. Structural basis for LMO2-driven recruitment of the SCL:E47bHLH heterodimer to hematopoietic-specific transcriptional targets. Cell Rep. 4, 135–147. (doi:10.1016/j.celrep.2013.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rossant J, Bernelot-Moens C, Nagy A. 1993. Genome manipulation in embryonic stem cells. Phil. Trans. R. Soc. Lond. B 339, 207–215. (doi:10.1098/rstb.1993.0018) [DOI] [PubMed] [Google Scholar]
- 66.Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. 1998. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl Acad. Sci. USA 95, 3890–3895. (doi:10.1073/pnas.95.7.3890) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamada Y, Pannell R, Forster A, Rabbitts TH. 2000. The oncogenic LIM-only transcription factor Lmo2 regulates angiogenesis but not vasculogenesis in mice. Proc. Natl Acad. Sci. USA 97, 320–324. (doi:10.1073/pnas.97.1.320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamada Y, Pannell R, Forster A, Rabbitts TH. 2002. The LIM-domain protein Lmo2 is a key regulator of tumour angiogenesis: a new anti-angiogenesis drug target. Oncogene 21, 1309–1315. (doi:10.1038/sj.onc.1205285) [DOI] [PubMed] [Google Scholar]
- 69.Shivdasani RA, Mayer EL, Orkin SH. 1995. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373, 432–434. (doi:10.1038/373432a0) [DOI] [PubMed] [Google Scholar]
- 70.Robb L, Lyons I, Li R, Hartley L, Kontgen F, Harvey RP, Metcalf D, Begley CG. 1995. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc. Natl Acad. Sci. USA 92, 7075–7079. (doi:10.1073/pnas.92.15.7075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D'Agati V, Orkin SH, Costantini F. 1991. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349, 257–260. (doi:10.1038/349257a0) [DOI] [PubMed] [Google Scholar]
- 72.Simon MC, Pevny L, Wiles MV, Keller G, Costantini F, Orkin SH. 1992. Rescue of erythroid development in gene targeted GATA-1- mouse embryonic stem cells. Nat. Genet. 1, 92–98. (doi:10.1038/ng0592-92) [DOI] [PubMed] [Google Scholar]
- 73.Tsai FY, et al. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–226. (doi:10.1038/371221a0) [DOI] [PubMed] [Google Scholar]
- 74.Durand C, Dzierzak E. 2005. Embryonic beginnings of adult hematopoietic stem cells. Haematologica 90, 100–108. [PubMed] [Google Scholar]
- 75.Medvinsky A, Rybtsov S, Taoudi S. 2011. Embryonic origin of the adult hematopoietic system: advances and questions. Development 138, 1017–1031. (doi:10.1242/dev.040998) [DOI] [PubMed] [Google Scholar]
- 76.Visvader JE, Mao X, Fujiwara Y, Hahm K, Orkin SH. 1997. The LIM-domain binding protein LDB1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc. Natl Acad. Sci. USA 94, 13 707–13 712. (doi:10.1073/pnas.94.25.13707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silver L, Palis J. 1997. Initiation of murine embryonic erythropoiesis: a spatial analysis. Blood 89, 1154–1164. [PubMed] [Google Scholar]
- 78.Manaia A, et al. 2000. Lmo2 and GATA-3 associated expression in intraembryonic hemogenic sites. Development 127, 643–653. [DOI] [PubMed] [Google Scholar]
- 79.Lacaud G, Keller G, Kouskoff V. 2004. Tracking mesoderm formation and specification to the hemangioblast in vitro. Trends Cardiovasc. Med. 14, 314–317. (doi:10.1016/j.tcm.2004.09.004) [DOI] [PubMed] [Google Scholar]
- 80.Coma S, Allard-Ratick M, Akino T, van Meeteren LA, Mammoto A, Klagsbrun M. 2013. GATA2 and Lmo2 control angiogenesis and lymphangiogenesis via direct transcriptional regulation of neuropilin-2. Angiogenesis 16, 939–952. (doi:10.1007/s10456-013-9370-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee SK, et al. 2003. Identification of novel anti-angiogenic factors by in silico functional gene screening method. J. Biotechnol. 105, 51–60. (doi:10.1016/S0168-1656(03)00183-4) [DOI] [PubMed] [Google Scholar]
- 82.Hinks GL, et al. 1997. Expression of LIM protein genes Lmo1, Lmo2, and Lmo3 in adult mouse hippocampus and other forebrain regions: differential regulation by seizure activity. J. Neurosci. 17, 5549–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han C, et al. 2005. Human Bex2 interacts with LMO2 and regulates the transcriptional activity of a novel DNA-binding complex. Nucleic Acids Res. 33, 6555–6565. (doi:10.1093/nar/gki964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim SH, et al. In press. The LIM-only transcription factor LMO2 determines tumorigenic and angiogenic traits in glioma stem cells. Cell Death Diff. (doi:10.1038/cdd.2015.7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akashi K, et al. 2003. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood 101, 383–389. (doi:10.1182/blood-2002-06-1780) [DOI] [PubMed] [Google Scholar]
- 86.Fisch P, et al. 1992. T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene 7, 2389–2397. [PubMed] [Google Scholar]
- 87.Larson RC, et al. 1994T. cell tumours of disparate phenotype in mice transgenic for Rbtn-2. Oncogene 9, 3675–3681. [PubMed] [Google Scholar]
- 88.Cleveland SM, et al. 2013. Lmo2 induces hematopoietic stem cell-like features in T-cell progenitor cells prior to leukemia. Stem Cells 31, 882–894. (doi:10.1002/stem.1345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Larson RC, Osada H, Larson TA, Lavenir I, Rabbitts TH. 1995. The oncogenic LIM protein Rbtn2 causes thymic developmental aberrations that precede malignancy in transgenic mice. Oncogene 11, 853–862. [PubMed] [Google Scholar]
- 90.McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, Jane SM, Curtis DJ. 2010. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science 327, 879–883. (doi:10.1126/science.1182378) [DOI] [PubMed] [Google Scholar]
- 91.Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. (doi:10.1016/j.cell.2006.07.024) [DOI] [PubMed] [Google Scholar]
- 92.Vereide DT, Vickerman V, Swanson SA, Chu LF, McIntosh BE, Thomson JA. 2014. An expandable, inducible hemangioblast state regulated by fibroblast growth factor. Stem Cell Rep. 3, 1043–1057. (doi:10.1016/j.stemcr.2014.10.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Batta K, Florkowska M, Kouskoff V, Lacaud G. 2014. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep. 9, 1871–1884. (doi:10.1016/j.celrep.2014.11.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Riddell J, et al. 2014. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 157, 549–564. (doi:10.1016/j.cell.2014.04.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamanaka S. 2009. A fresh look at iPS cells. Cell 137, 13–17. (doi:10.1016/j.cell.2009.03.034) [DOI] [PubMed] [Google Scholar]
- 96.Neale GA, Rehg JE, Goorha RM. 1995. Ectopic expression of rhombotin-2 causes selective expansion of CD4-CD8- lymphocytes in the thymus and T-cell tumors in transgenic mice. Blood 86, 3060–3071. [PubMed] [Google Scholar]
- 97.Zhou H, et al. 2009. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4, 381–384. (doi:10.1016/j.stem.2009.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Greenberg JM, et al. 1990. Segmental and developmental regulation of a presumptive T-cell oncogene in the central nervous system. Nature 344, 158–160. (doi:10.1038/344158a0) [DOI] [PubMed] [Google Scholar]
- 99.Nam CH, Lobato MN, Appert A, Drynan LF, Tanaka T, Rabbitts TH. 2008. An antibody inhibitor of the LMO2-protein complex blocks its normal and tumorigenic functions. Oncogene 27, 4962–4968. (doi:10.1038/onc.2008.130) [DOI] [PubMed] [Google Scholar]
- 100.Tanaka T, Sewell H, Waters S, Phillips SE, Rabbitts TH. 2011. Single domain intracellular antibodies from diverse libraries: emphasizing dual functions of LMO2 protein interactions using a single VH domain. J. Biol. Chem. 286, 3707–3716. (doi:10.1074/jbc.M110.188193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T. 2000. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat. Immunol. 1, 138–144. (doi:10.1038/77819) [DOI] [PubMed] [Google Scholar]
- 102.Gutierrez A, Look AT. 2007. NOTCH and PI3K-AKT pathways intertwined. Cancer Cell 12, 411–413. (doi:10.1016/j.ccr.2007.10.027) [DOI] [PubMed] [Google Scholar]
- 103.Robb L, Rasko JE, Bath ML, Strasser A, Begley CG. 1995. scl, a gene frequently activated in human T cell leukaemia, does not induce lymphomas in transgenic mice. Oncogene 10, 205–209. [PubMed] [Google Scholar]
- 104.Pike-Overzet K, van der Burg M, Wagemaker G, van Dongen JJ, Staal FJ. 2007. New insights and unresolved issues regarding insertional mutagenesis in X-linked SCID gene therapy. Mol. Therapy 15, 1910–1916. (doi:10.1038/sj.mt.6300297) [DOI] [PubMed] [Google Scholar]
- 105.Staal FJ, Pike-Overzet K, Ng YY, van Dongen JJ. 2008. Sola dosis facit venenum. Leukemia in gene therapy trials: a question of vectors, inserts and dosage? Leukemia 22, 1849–1852. (doi:10.1038/leu.2008.219) [DOI] [PubMed] [Google Scholar]
- 106.Coustan-Smith E, et al. 2009. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 10, 147–156. (doi:10.1016/S1470-2045(08)70314-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang J, et al. 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163. (doi:10.1038/nature10725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McCormack MP, et al. 2013. Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL. Blood 122, 2093–2103. (doi:10.1182/blood-2012-09-458570) [DOI] [PubMed] [Google Scholar]
- 109.Shields BJ, Alserihi R, Nasa C, Bogue C, Alexander WS, McCormack MP. 2014. Hhex regulates Kit to promote radioresistance of self-renewing thymocytes in Lmo2-transgenic mice. Leukemia 29, 927–938. (doi:10.1038/leu.2014.292) [DOI] [PubMed] [Google Scholar]
- 110.Martins VC, et al. 2014. Cell competition is a tumour suppressor mechanism in the thymus. Nature 509, 465–470. (doi:10.1038/nature13317) [DOI] [PubMed] [Google Scholar]
- 111.Wilkinson-White L, Matthews JM. 2014. The PA207 peptide inhibitor of LIM-only protein 2 (Lmo2) targets Zinc Finger domains in a non-specific manner. Protein Pept. Lett. 21, 132–139. (doi:10.2174/09298665113206660116) [DOI] [PubMed] [Google Scholar]
- 112.Natkunam Y, et al. 2007. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B-cell lymphomas. Blood 109, 1636–1642. (doi:10.1182/blood-2006-08-039024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Deucher AM, Qi Z, Yu J, George TI, Etzell JE. 2015. BCL6 expression correlates with the t(1;19) translocation in B-lymphoblastic leukemia. Am. J. Clin. Pathol. 143, 547–557. (doi:10.1309/AJCPO4U4VYAAOTEL) [DOI] [PubMed] [Google Scholar]
- 114.de Boer J, Yeung J, Ellu J, Ramanujachar R, Bornhauser B, Solarska O, Hubank M, Williams O, Brady HJM. 2011. The E2A-HLF oncogenic fusion protein acts through Lmo2 and Bcl-2 to immortalize hematopoietic progenitors. Leukemia 25, 321–330. (doi:10.1038/leu.2010.253) [DOI] [PubMed] [Google Scholar]
- 115.Malumbres R, et al. 2011. LMO2 expression reflects the different stages of blast maturation and genetic features in B-cell acute lymphoblastic leukemia and predicts clinical outcome. Haematologica 96, 980–986. (doi:10.3324/haematol.2011.040568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cubedo E, et al. 2012. Identification of LMO2 transcriptome and interactome in diffuse large B-cell lymphoma. Blood 119, 5478–5491. (doi:10.1182/blood-2012-01-403154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Natkunam Y, et al. 2008. LMO2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. J. Clin. Oncol. 26, 447–454. (doi:10.1200/JCO.2007.13.0690) [DOI] [PubMed] [Google Scholar]
- 118.Lossos IS, et al. 2004. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N. Eng. J. Med. 350, 1828–1837. (doi:10.1056/NEJMoa032520) [DOI] [PubMed] [Google Scholar]
- 119.Nakata K, et al. 2009. LMO2 is a novel predictive marker for a better prognosis in pancreatic cancer. Neoplasia 11, 712–719. (doi:10.1593/neo.09418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gratzinger D, et al. 2009. The transcription factor LMO2 is a robust marker of vascular endothelium and vascular neoplasms and selected other entities. Am. J. Clin. Pathol. 131, 264–278. (doi:10.1309/AJCP5FP3NAXAXRJE) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pui CH, Relling MV, Downing JR. 2004. Acute lymphoblastic leukemia. N. Eng. J. Med. 350, 1535–1548. (doi:10.1056/NEJMra023001) [DOI] [PubMed] [Google Scholar]
- 122.Pui CH, Evans WE. 1998. Acute lymphoblastic leukemia. N. Eng. J. Med. 339, 605–615. (doi:10.1056/NEJM199808273390907) [DOI] [PubMed] [Google Scholar]
- 123.Carmeliet P. 2003. Angiogenesis in health and disease. Nat. Med. 9, 653–660. (doi:10.1038/nm0603-653) [DOI] [PubMed] [Google Scholar]
- 124.Wells JA, McClendon CL. 2007. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450, 1001–1009. (doi:10.1038/nature06526) [DOI] [PubMed] [Google Scholar]
- 125.Yuan JY, Wang F, Yu J, Yang GH, Liu XL, Zhang JW. 2009. MicroRNA-223 reversibly regulates erythroid and megakaryocytic differentiation of K562 cells. J. Cell. Mol. Med. 13, 4551–4559. (doi:10.1111/j.1582-4934.2008.00585.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang J, Rabbitts TH. 2014. Intracellular antibody capture: a molecular biology approach to inhibitors of protein–protein interactions. Biochim. Biophys. Acta 1844, 1970–1976. (doi:10.1016/j.bbapap.2014.05.009) [DOI] [PubMed] [Google Scholar]
- 127.Tse E, Lobato MN, Forster A, Tanaka T, Chung GT, Rabbitts TH. 2002. Intracellular antibody capture technology: application to selection of intracellular antibodies recognising the BCR-ABL oncogenic protein. J. Mol. Biol. 317, 85–94. (doi:10.1006/jmbi.2002.5403) [DOI] [PubMed] [Google Scholar]
- 128.Tanaka T, Rabbitts TH. 2012. Intracellular antibody capture (IAC) methods for single domain antibodies. Methods Mol. Biol. 911, 151–173. (doi:10.1007/978-1-61779-968-6_10) [DOI] [PubMed] [Google Scholar]
- 129.Tanaka T, Rabbitts TH. 2010. Protocol for the selection of single-domain antibody fragments by third generation intracellular antibody capture. Nat. Protoc. 5, 67–92. (doi:10.1038/nprot.2009.199) [DOI] [PubMed] [Google Scholar]
- 130.Visintin M, Tse E, Axelson H, Rabbitts TH, Cattaneo A. 1999. Selection of antibodies for intracellular function using a two-hybrid in vivo system. Proc. Natl Acad. Sci. USA 96, 11 723–11 728. (doi:10.1073/pnas.96.21.11723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tse E, Rabbitts TH. 2000. Intracellular antibody-caspase-mediated cell killing: an approach for application in cancer therapy. Proc. Natl Acad. Sci. USA 97, 12 266–12 271. (doi:10.1073/pnas.97.22.12266) [DOI] [PMC free article] [PubMed] [Google Scholar]