Abstract
Social insects such as ants have evolved collective rather than individual immune defence strategies against diseases and parasites at the level of their societies (colonies), known as social immunity. Ants frequently host other arthropods, so-called myrmecophiles, in their nests. Here, we tested the hypothesis that myrmecophily may partly arise from selection for exploiting the ants’ social immunity. We used larvae of the wax moth Galleria mellonella as ‘model myrmecophiles’ (baits) to test this hypothesis. We found significantly reduced abundance of entomopathogens in ant nests compared with the surrounding environment. Specific entomopathogen groups (Isaria fumosorosea and nematodes) were also found to be significantly less abundant inside than outside ant nests, whereas one entomopathogen (Beauveria brongniartii) was significantly more abundant inside nests. We therefore hypothesize that immunological benefits of entering ant nests may provide us a new explanation of why natural selection acts in favour of such a life-history strategy.
Keywords: symbiosis, immunocompetence, hygiene
1. Introduction
Group life has many advantages compared with a solitary lifestyle, and some of the ecologically most dominant organisms live in groups [1]. However, social life often comes at the cost of increased risk of infectious disease, as the frequent interactions between social organisms and the high densities in which they normally occur facilitate transmission of pathogens and parasites [2–4]. Some group-living animals have therefore evolved collective immune defence against diseases, known as social immunity [5]. Social insects have developed behavioural and chemical countermeasures against diseases in order to avoid infection and transmission [5]. A widespread behavioural strategy against diseases is hygienic behaviour such as allogrooming [6] and removal of dead corpses from the nest [7].
Ants frequently host other arthropod species that have evolved to live closely with them or even inside ants’ nests (inquilines) [8]. Little is known as to how inquilinism in distantly related invertebrates evolves. In myrmecophilous butterflies that do not live in ant nests as inquilines, pupation can still take place inside ant nests [9]. This could explain how inquilines arise, when selection favours the penetration of ant nests. Once inquilinism is established, benefits that the host ants provide to these myrmecophiles seem obvious: shelter, protection from natural enemies and often food. In consequence, like traditional parasites, inquilines may lose vital functions of their free-living ancestors, because selection for their maintenance is lost. For example, inquilines could lose their immunocompetence as they free-ride on the social immunity provided by the ants.
To examine the basis of this hypothesis, we studied the abundance of entomopathogens in nests of Myrmica rubra (Linnaeus) and Myrmica ruginodis (Nylander), both host ants of various myrmecophiles [10], using larvae of the wax moth Galleria mellonella (Linnaeus) as ‘model myrmecophiles’ (baits).
2. Material and methods
2.1. Collection of soil samples and soil baiting
A total of 166 soil samples from 13 sites (55 samples from M. rubra nests, 28 samples from M. ruginodis nests and 83 matched controls) were collected in August 2012 from sites in the area of northeastern Zealand (Denmark; table 1). Each sample was collected as a cylindrical core of approximately 10 cm depth and 5 cm diameter from the uppermost soil layer (including the soil surface) using a bulb planter (model: Gardena 3412). The bulb planter was rinsed in 70% ethanol and air-dried between samples. Samples were taken from nest cores of M. rubra group ants (M. rubra or M. ruginodis) as well as from a control point in the surrounding of the nest, together forming a sample pair. The ants were identified in the field by use of a 10× magnification hand lens. The control point was always chosen within a radius of 2 m from the ant nest, from the ant free point with the visually most similar type of soil compared with that of the ant nest. The soil was collected in plastic freezer bags and transferred to a 5°C climate chamber on the day of collection, where it was stored until analysis. All samples were processed by removing roots, stones and large pieces of wood, and homogenized by crushing soil clumps manually, and thoroughly mixing the sample, in plastic bags. Ants were removed using soft forceps sterilized in 70% ethanol, which were subsequently rinsed with distilled water between samples. All ants were kept in 96% ethanol as voucher specimens. Soil baiting was carried out as suggested in Meyling [11]. Moist soil from each individual sample point was distributed in even volumes into two plastic cups per sample, and each cup received 10 second- or third-instar (ca 10–15 mm) larvae of G. mellonella. The larvae came from a continuous culture kept at the University of Copenhagen and were heat-treated as described in Meyling & Eilenberg [12] prior to baiting in order to destroy the silk glands, so increasing their exposure to entomopathogens present in the soil by reducing the ability of the larvae to encapsulate inside webs. Soil samples were checked weekly during four weeks, dead larvae were rinsed with distilled water twice and isolated individually into 30 ml medicine cups containing a ca 2×2 cm piece of moist filter paper to maintain high humidity. Fungi emerging from dead G. mellonella larvae were classified to morphospecies level based on spore morphology using an Olympus BH-2 microscope at 100–400× magnification according to the key of Humber [13]. Spores from each detected morphospecies of entomopathogenic fungus per soil sample were transferred to standard selective media plates [11] until a clean culture could be established. Nematodes were identified as entomopathogenic when occurring in very large numbers, approximately replacing the biomass of the dead G. mellonella larva or when they showed ‘ambush behaviour’ [14]. If isolated nematodes were observed on cadavers, they were classified as ‘soil nematodes’ and not included. Nematodes were stored in 96% ethanol.
Table 1.
Distribution and diversity of entomopathogens in G. mellonella baited soil collected from Myrmica ant nests and control samples per locality.
| site | latitude (°N) | longitude (°E) | altitude (m) | no. samples (ant nest/control) | ant species | pathogens in soil from Myrmica ant nests | pathogens in surrounding soils (controls) |
|---|---|---|---|---|---|---|---|
| Vaserne | 55.82 | 12.45 | 23 | 10/10 | M. rubra | M. brunneum | M. brunneum |
| I. fumosorosea | nematodes | ||||||
| Allerød Sø | 55.87 | 12.36 | 45 | 9/9 | M. rubra | M. brunneum | M. brunneum |
| B. bassiana | B. bassiana | ||||||
| B. brongniartii | I. fumosorosea | ||||||
| I. fumosorosea | nematodes | ||||||
| Orehøjvej | 55.80 | 12.32 | 39 | 1/1 | M. ruginodis | none | none |
| Nymøllevej | 55.82 | 12.30 | 28 | 10/10 | M. rubra | M. brunneum | M. brunneum |
| I. fumosorosea | |||||||
| nematodes | |||||||
| Gadevang | 55.97 | 12.27 | 41 | 5/5 | M. rubra | none | M. brunneum |
| nematodes | |||||||
| Klampenborgvej | 55.77 | 12.56 | 11 | 4/4 | M. rubra | B. bassiana | M. brunneum |
| nematodes | B. bassiana | ||||||
| I. fumosorosea | |||||||
| Dyrehavn | 55.78 | 12.58 | 16 | 5/5 | M. rubra | M. brunneum | M. brunneum |
| nematodes | |||||||
| Dyrehavn | 55.78 | 12.58 | 15 | 2/2 | M. ruginodis | M. brunneum | M. brunneum |
| I. fumosorosea | I. fumosorosea | ||||||
| nematodes | nematodes | ||||||
| Birkerød | 55.83 | 12.45 | 37 | 2/2 | M. ruginodis | M. brunneum | M. brunneum |
| B. brongniartii | B. bassiana | ||||||
| nematodes | |||||||
| Brødeskov | 55.89 | 12.32 | 39 | 7/7 | M. ruginodis | M. brunneum | M. brunneum |
| B. bassiana | I. fumosorosea | ||||||
| nematodes | nematodes | ||||||
| Allerød forest | 55.87 | 12.37 | 54 | 9/9 | M. ruginodis | M. brunneum | M. brunneum |
| I. farinosa | B. pseudobassiana | ||||||
| nematodes | I. fumosorosea | ||||||
| nematodes | |||||||
| Rudegårds alle | 55.82 | 12.45 | 25 | 3/3 | M. rubra | none | M. brunneum |
| B. caledonica | |||||||
| nematodes | |||||||
| Hestetangsvej | 55.81 | 12.32 | 31 | 9/9 | M. rubra | M. brunneum | M. brunneum |
| M. flavoviride | M. flavoviride | ||||||
| B. bassiana | B. pseudobassiana | ||||||
| nematodes | nematodes | ||||||
| Logsø | 55.84 | 12.47 | 46 | 7/7 | M. ruginodis | nematodes | nematodes |
2.2. Molecular identification of entomopathogenic soil fungi
The selected fungal isolates were individually inoculated into sterile flasks containing liquid medium (2% peptone, 3% sucrose and 0.2% yeast extract) and incubated on a shaker (170 r.p.m.) at room temperature for 3 days. The resulting fungal material was filtered under suction and lyophilized overnight. The DNA extraction from dried fungal tissue was carried out using DNeasy blood and tissue kits (Qiagen). Entomopathogenic fungi of the genus Isaria were sequenced at the internally transcribed spacer region of the 18S nuclear ribosomal DNA, using the primer pair ITS1 (forward) and ITS4 (reverse) [15,16]. PCRs were carried out in 25 μl volumes. Each reaction contained 1×PCR gold buffer, 4 mM MgCl2, 200 μM of each nucleotide, 1 μM of each primer, 1.25 U AmpliTaq Goldr DNA polymerase (Invitrogen), 1 μl template DNA and ddH2O added to the total volume. The PCR programme consisted of 5 min initial denaturation at 95°C, followed by 35 cycles of 15 s denaturation at 95°C, 15 s annealing at 55°C and 90 s elongation at 72°C. The reaction ended with a final elongation at 72°C for 7 min and a hold temperature of 4°C.
Beauveria and Metarhizium spp. were identified by sequencing the 5′ end of the translation elongation factor 1-α gene (5′-TEF) using the primers EF2F (forward) and EFjR (reverse) [17]. The PCRs were set up in 50 μl volumes. Each reaction comprised 10 μl Phusion HF buffer (1.5 mM MgCl2), 200 μM of each nucleotide, 1 μM of each primer and 0.5 U Phusion high fidelity polymerase (Finzymes), 1 μl template DNA and sterile Milli-Q H2O added to the total volume. The PCRs were as follows: 30 s initial denaturation at 98°C, followed by 10 cycles of a touchdown programme with denaturation at 98°C for 10 s and annealing/extension during 90 s. The annealing/extension temperature started at 70°C and was lowered by 1°C per cycle until it had reached 60°C. Thereafter, the reaction continued with 38 cycles of denaturation at 98°C for 10 s, annealing at 60°C for 30 s and elongation at 72°C during 30 s, followed by a final elongation at 72°C for 10 min. All PCR products were quantified in 2% agarose gels, run at 150 V and 100 A for 35 min before sequencing. PCR products were purified using ExoSAP-ITr (Affymetrix) and kit purification GFX DNA/gel band 100 RXN. After purification, the samples were sent to Beijing Genomics Institute or Eurofins MWG for cycle sequencing. The returning chromatograms were edited and assembled in Geneious v. 6.1.6 (Biomatters; http://www.geneious.com/).
In order to identify the entomopathogenic fungi, we compared their 5′-TEF sequences with a set of TEF reference sequences of identified specimens stored at the USDA Agricultural Research Service Collection of Entomopathogenic Fungi (ARSEF). To identify Beauveria species, we chose a selection of ARSEF reference sequences (TEF) for the genus Beauveria from Rehner et al. [18] and a selection of Beauveria strains found and described in Meyling et al. [17]. Reference sequences for Metarhizium sp. were obtained by searching for the single sequence found in BLAST. We then chose three TEF sequences from the ARSEF collection with the highest pairwise identity as references. All sequences were aligned by use of ClustalW alignment [19] in Geneious v. 6.1.6. A phylogram of TEF sequences including the reference sequences was created using Bayesian inference as implemented in MrBayes v. 3.2.2x86 [20,21]. The HKY+G model of sequence evolution [22] was chosen after calculating the likelihood scores for the alignment in the software jModelTest [23] and the corrected Akaike information criterion [24,25]. Two parallel runs of MrBayes were carried out with one cold and three heated chains (chain heats: 1=cold, 0.91, 0.83, 0.77) for five million generations with every 1000 trees sampled. Convergence was assessed by examination of the minimal effective sample size of TL (ESS=1233) and the potential scale reduction factor (=1.000) [26].
2.3. Statistical analysis
The statistical comparisons of the total number of pathogen-infected G. mellonella larvae per sample in ant nests versus controls and the total number of G. mellonella larvae infected with entomopathogenic fungi per sample in ant nests versus control were carried out using generalized linear mixed models (GLMMs) with zero-inflated Poisson errors in the package glmmADMB [27] in R v. 3.0.1 [28]. The presence/absence of ants was treated as a fixed effect and ‘sample pair’ (the nest sample and its control) as a random effect to correct for variation in pathogen abundance between sample pairs. An analogous model was repeated to test for the effect of treatment (control, M. rubra and M. ruginodis) again including ‘sample pair’ as a random effect. Abundance of single pathogen groups in ant nest samples was compared with that in controls using a binomial GLMM (‘logit’ link) in the R package lme4 [29] with 1/0 coding for ant presence/absence. This analysis was carried out at the level of the single G. mellonella larvae. ‘Pathogen’ was included as a fixed effect (missing if no pathogen emerged) and ‘sample pair’ as a random effect.
3. Results
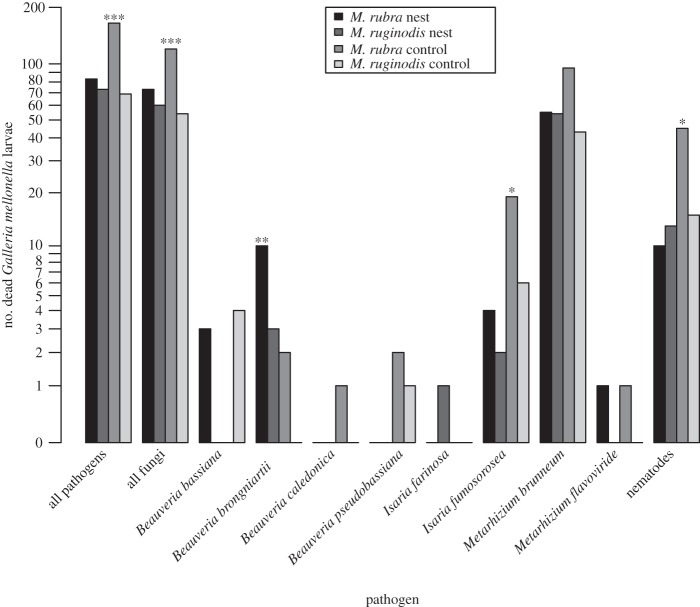
An overview of all isolated entomopathogens, their identification and abundance is given in figures 1 and 2 and tables 1 and 2. The number of G. mellonella larvae per sample found dead with a pathogen was lower in soil from ant nests compared with soil from control points (GLMM, nests versus controls: z=−4.15, p<0.0001). The same was found for entomopathogenic fungi excluding nematodes (z=−2.80, p=0.005, figure 1). More specifically, M. rubra nests contained significantly fewer cadavers with pathogens than the controls (M. rubra versus controls: z=−4.93, p<0.0001, n=55 nests), whereas this was not the case for M. ruginodis (M. ruginodis versus controls: z=+0.99, p=0.323, n=29 nests; figure 1). Several single pathogen species or groups differed significantly in abundance in ant nests compared with controls. Abundance in ant nests was reduced in the fungus Isaria fumosorosea (Wize; z=−2.51, p=0.012) and in entomopathogenic nematodes (z=−2.02, p=0.044). More surprisingly, the fungus Beauveria brongniartii (Saccardo) was more abundant inside than outside ant nests (z=+2.89, p=0.004), but this was much less abundant than the species that were reduced in ant nests (figure 1), and did not change the overall pattern of decreased G. mellonella mortality when exposed to ant nest soil. In all cases, death of a G. mellonella larva could be associated with the presence of either a single pathogen or none (i.e. multiple infections were not found).
Figure 1.
Bar graph shows the total number of G. mellonella larvae found per pathogen species or group comparing M. rubra and M. ruginodis nest samples with their respective controls. Significant differences are marked by asterisks above each set of bars (*p<0.05, **p<0.01, ***p<0.001). The identification of entomopathogenic fungi was carried out by sequencing at either the ITS or 5′-TEF region. For M. brunneum, one representative was sequenced per sampling locality.
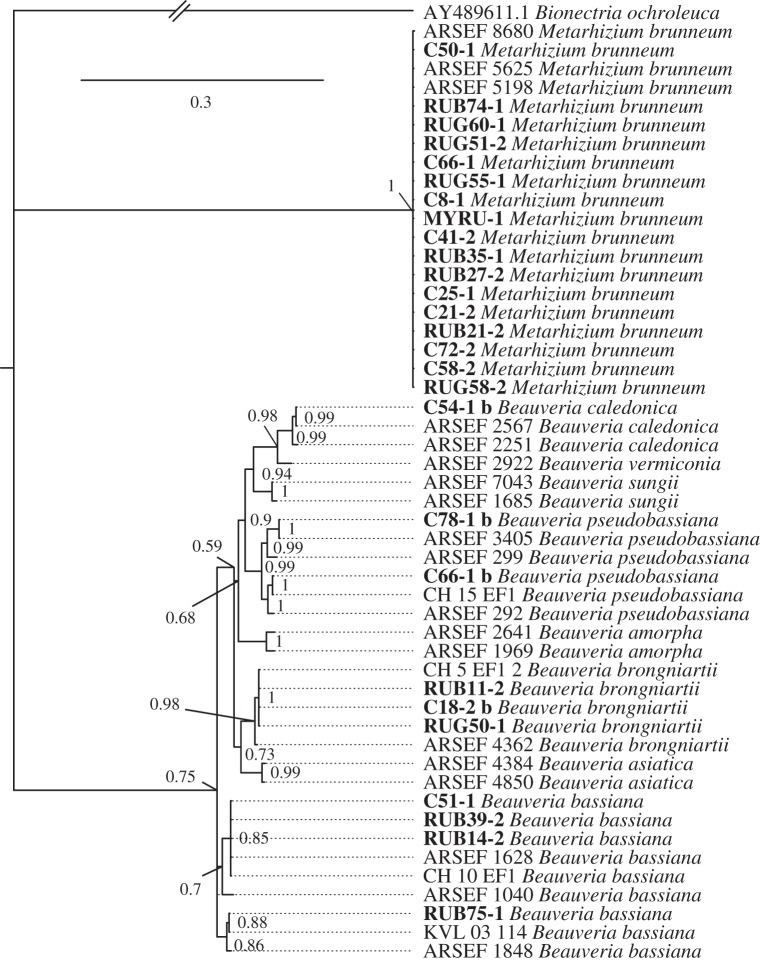
Figure 2.
A Bayesian inference phylogram of the 5′-TEF gene of the entomopathogenic fungi sequenced and a set of verified TEF reference sequences from GenBank, representing identified species stored at ARSEF. The phylogram is the consensus tree of 5000 trees obtained from two converged runs in the software MrBayes v. 3.2.2. Node labels show node support (posterior probability ≥0.5). Voucher IDs in bold correspond to samples from this study, those in regular font were used as references obtained from GenBank.
Table 2.
Entomopathogenic fungi identified by sequencing: site of collection, treatment (from ant nest or control soil), primers used for amplification, the fungal species, length of the sequenced fragment and GenBank accession number. All sequenced isolates of Isaria fumosorosea and Metarhizium brunneum had identical sequences.
| voucher ID | site | treatment (rug=M. ruginodis, rub=M. rubra) | primer pair | fungal species | length of sequence (bp) | GenBank accession no. |
|---|---|---|---|---|---|---|
| RUB14-2_b | Allerød Sø | Myrmica rubra | EF2F/EFjR | Beauveria bassiana (Bals. Criv. Vuill.) | 732 | KJ908271 |
| C51-1_b | Birkerød | control (rug) | EF2F/EFjR | Beauveria bassiana | 732 | KJ908272 |
| RUB39-2_b | Klampenborgvej | Myrmica rubra | EF2F/EFjR | Beauveria bassiana | 732 | KJ908273 |
| RUB75-1_b | Hestetangsvej | Myrmica rubra | EF2F/EFjR | Beauveria bassiana | 732 | KJ908274 |
| RUB11-2_b | Allerød Sø | Myrmica rubra | EF2F/EFjR | Beauveria brongniartii | 740 | KJ908275 |
| C18-2_b | Allerød Sø | control (rub) | EF2F/EFjR | Beauveria brongniartii | 740 | KJ908277 |
| RUG50-1_b | Birkerød | Myrmica ruginodis | EF2F/EFjR | Beauveria brongniartii | 740 | KJ908276 |
| C54-1_b | Rudegards alle | control (rub) | EF2F/EFjR | Beauveria caledonica [30] | 753 | KJ908270 |
| C66-1_b | Allerød forest | control (rug) | EF2F/EFjR | Beauveria pseudobassiana[18] | 734 | KJ908279 |
| C78-1_b | Hestetangsvej | control (rub) | EF2F/EFjR | Beauveria pseudobassiana | 736 | KJ908278 |
| RUG62_i | Allerød forest | Myrmica ruginodis | ITS1&4 | Isaria farinosa(Holmsk.) | 495 | KJ908284 |
| C12_i | Allerød Sø | control (rub) | ITS1&4 | Isaria fumosorosea | 488 | KJ908283 |
| C16_i | Allerød Sø | control (rub) | ITS1&4 | Isaria fumosorosea | 488 | KJ908283 |
| C19_i | Allerød Sø | control (rub) | ITS1&4 | Isaria fumosorosea | 488 | KJ908283 |
| C20_i | Allerød Sø | control (rub) | ITS1&4 | Isaria fumosorosea | 488 | KJ908283 |
| RUB42_i | Dyrehavn | Myrmica rubra | ITS1&4 | Isaria fumosorosea | 488 | KJ908283 |
| C38_i | Klampenborgvej | control (rub) | ITS1&4 | Isaria fumosorosea | 488 | KJ908283 |
| C39_i | Klampenborgvej | control (rub) | ITS5&4 | Isaria fumosorosea | 488 | KJ908283 |
| C66-1_m | Allerød forest | control (rug) | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| C21-2_m | Allerød Sø | control (rub) | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| RUB21-2_m | Allerød Sø | Myrmica rubra | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| RUG51-2_m | Birkerød | Myrmica ruginodis | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| C50-1_m | Birkerød | control (rug) | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| RUB35-1_m | Gadevang | Myrmica rubra | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| C72-2_m | Hestetangsvej | control (rub) | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| RUB74-1_m | Hestetangsvej | Myrmica rubra | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| C41-2_m | Klampenborgvej | control (rub) | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| C25-1_m | Nymøllevej | control (rub) | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| RUB27-2_m | Nymøllevej | Myrmica rubra | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| RUG55-1_m | Brødeskov | Myrmica ruginodis | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| RUG58-2_m | Brødeskov | Myrmica ruginodis | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| C58-2_m | Brødeskov | control (rug) | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| RUG60-1_m | Brødeskov | Myrmica ruginodis | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| C8-1_m | Vaserne | control (rub) | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
| MYRU-1_m | Vaserne | Myrmica rubra | EF2F/EFjR | Metarhizium brunneum | 682 | KJ908282 |
4. Discussion
We found evidence for reduced entomopathogen abundance in nests of M. rubra group ants. Especially, M. rubra (n=55 nests) seems either to reduce the abundance of pathogens inside their nest, or is able to avoid infected nest sites. We cannot distinguish between the two here. Previous studies have shown that some ants do not avoid pathogen-rich nesting sites [31,32], and hygienic behaviour, as observed frequently in ant species including M. rubra, such as removal of dead nest-mates to midden piles outside the nest [33], is likely to reduce pathogen abundance. Allogrooming and antimicrobial secretions of the metapleural glands are further thought to prevent pathogens from becoming abundant inside ant nests [34]. It has previously been shown that non-sterile soils from nests of the red imported fire ant Solenopsis invicta mitigated effects of B. bassiana compared with sterilized soil, suggesting an antagonistic effect of micro-organisms present in the soils of ant nests [35,36]. Similarly, increased microorganismal activity in Myrmica ant nests could eventually explain the reduced abundance of entomopathogenic fungi compared with control soil. In nests of M. ruginodis (n=28), no evidence for a reduction of entomopathogens was found: the number of G. mellonella larvae with pathogens did not differ significantly between soil samples from nests and controls, with slightly more entomopathogens inside nests. This may suggest extraordinary tolerance of M. ruginodis to entomopathogens, leading to a competitive advantage over other ant species (e.g. M. rubra) for M. ruginodis in pathogen-rich sites. For myrmecophiles, this could mean that M. ruginodis is a less suitable host species than M. rubra. Indeed, the myrmecophilous butterfly Maculinea alcon has been suggested to use M. rubra as a main host and M. ruginodis as a secondary host in Denmark [37].
We conclude that this study supports the idea of an immunological benefit of myrmecophily and suggest to test this theory further using facultatively and obligatorily myrmecophilous species.
Supplementary Material
Acknowledgements
We thank Malin Dörre for technical assistance, Anders A. Illum for feeding caterpillars, Bernhardt M. Steinwender for help with the classification of fungi, Ibtissem Ben Fekih for help in the laboratory and two anonymous reviewers for comments on the manuscript.
Data accessibility
The data used in this study are accessible as electronic supplementary material. DNA sequences are accessible through GenBank.
Authors' contributions
S.S. carried out fieldwork, experiments, molecular laboratory work, statistical analysis and sequence alignments, participated in the design of the study and drafted the manuscript; L.L. participated in experiments, carried out molecular laboratory work and helped draft the manuscript; N.M. participated in design of the study and data analysis, provided the equipment and environment for the laboratory work, helped carrying out experiments and molecular laboratory work and helped draft the manuscript; D.N. conceived the study, participated in design of the study and statistical analysis and helped draft the manuscript. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This study was supported by the Danish National Research Foundation via a grant to the Centre for Social Evolution, Department of Biology, University of Copenhagen (DNRF57).
References
- 1.Wilson EO. 1975. Sociobiology. The new synthesis. Cambridge, MA: Harvard University Press. [Google Scholar]
- 2.Schmid-Hempel P. 1998. Parasites in social insects. Princeton, NJ: Princeton University Press. [Google Scholar]
- 3.Kermack WO, McKendrick AG. 1927. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. A. 115, 700–721. (doi:10.1098/rspa.1927.0118) [Google Scholar]
- 4.Hochachka WM, Dhondt AA. 2000. Density-dependent decline of host abundance resulting from a new infectious disease. Proc. Natl Acad. Sci. USA 97, 5303–5306. (doi:10.1073/pnas.080551197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cremer S, Armitage SAO, Schmid-Hempel P. 2007. Social immunity. Curr. Biol. 17, R693–R702. (doi:10.1016/j.cub.2007.06.008) [DOI] [PubMed] [Google Scholar]
- 6.Hölldobler B, Wilson EO. 1990. The ants. Cambridge, MA: Harvard University Press. [Google Scholar]
- 7.Rothenbuhler WC. 1964. Behaviour genetics of nest cleaning in honey bees. I. Responses of four inbred lines to disease-killed brood. Anim. Behav. 12, 578–583. (doi:10.1016/0003-3472(64)90082-X) [DOI] [PubMed] [Google Scholar]
- 8.Kronauer DJC, Pierce NE. 2011. Myrmecophiles. Curr. Biol. 21, R208–R209. (doi:10.1016/j.cub.2011.01.050) [DOI] [PubMed] [Google Scholar]
- 9.Pontin AJ. 1988[90] Plebejus argus L. (Lep., Lycaenidae) pupae in nests of Lasius niger (L.) (Hym., Formicidae). Entomol. Mon. Mag. 126, 74. [Google Scholar]
- 10.Witek M, Barbero F, Markó B. 2014. Myrmica ants host highly diverse parasitic communities: from social parasites to microbes. Insect. Soc. 61, 307–323. (doi:10.1007/s00040-014-0362-6) [Google Scholar]
- 11.Meyling NV. 2007. Methods for isolation of entomopathogenic fungi from the soil environment. See http://orgprints.org/11200/1/11200.pdf.
- 12.Meyling NV, Eilenberg J. 2006. Occurrence and distribution of soil borne entomopathogenic fungi within a single organic agroecosystem. Agric. Ecosyst. Environ. 113, 336–341. (doi:10.1016/j.agee.2005.10.011) [Google Scholar]
- 13.Humber RA. 2012. Chapter VI - Identification of entomopathogenic fungi. In Manual of techniques in invertebrate pathology, 2nd edn (ed Lacey L.), pp. 151–187. San Diego, CA: Academic Press. [Google Scholar]
- 14.Adams BJ, Nguyen KB. 2002. Taxonomy and systematics. In Entomopathogenic nematology (ed. Gaugler R.), pp. 1–34 Wallingford, UK: CABI Publishing. [Google Scholar]
- 15.White TJT, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR protocols: a guide to methods and applications (eds Innis MA, Gelfand DH, Sninsky JJ, White TJ), pp. 315–322. New York, NY: Academic Press. [Google Scholar]
- 16.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. (doi:10.1111/j.1365-294X.1993.tb00005.x) [DOI] [PubMed] [Google Scholar]
- 17.Meyling NV, Pilz C, Keller S, Widmer F, Enkerli J. 2012. Diversity of Beauveria spp. isolates from pollen beetles Meligethes aeneus in Switzerland. J. Invertebr. Pathol. 109, 76–82. (doi:10.1016/j.jip.2011.10.001) [DOI] [PubMed] [Google Scholar]
- 18.Rehner SA, Minnis AM, Sung G-H, Luangsa-ard JJ, Devotto L, Humber RA. 2011. Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia 103, 1055–1073. (doi:10.3852/10-302) [DOI] [PubMed] [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. (doi:10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 21.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa M, Kishino H, Yano T. 1985. Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174. (doi:10.1007/BF02101694) [DOI] [PubMed] [Google Scholar]
- 23.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256. (doi:10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 24.Sugiura N. 1978. Further analysis of the data by Akaike’s information criterion and the finite corrections. Commun. Stat. A Theor. 7, 13–26. (doi:10.1080/03610927808827599) [Google Scholar]
- 25.Hurvich CM, Tsai C-L. 1993. A corrected Akaike information criterion for vector autoregressive model selection. J. Time Series Anal. 14, 271–279. (doi:10.1111/j.1467-9892.1993.tb00144.x) [Google Scholar]
- 26.Gelman A, Rubin DB. 1992. Inference from iterative simulation using multiple sequences. Stat. Sci. 457–472. (doi:10.1214/ss/1177011136) [Google Scholar]
- 27.Fournier DA, Skaug HJ, Ancheta J, Ianelli J, Magnusson A, Maunder M, Nielsen A, Sibert J. 2012. AD model builder: using automatic differentiation for statistical inference of highly parameterized complex nonlinear models. Optim. Method Softw. 27, 233–249. (doi:10.1080/10556788.2011.597854) [Google Scholar]
- 28.R Development Core Team. 2011. R: a language and environment for statistical computing. See http://www.R-project.org/. [Google Scholar]
- 29.Bates D, Maechler M, Bolker B, Walker S. 2014. lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1–7. See http://CRAN.R-project.org/package=lme4.
- 30.Bissett J, Widden P. 1988. A new species of Beauveria isolated from Scottish moorland soil. Can. J. Bot. 66, 361–362. (doi:10.1139/b88-057) [Google Scholar]
- 31.Brütsch T, Felden A, Reber A, Chapuisat M. 2014. Ant queens (Hymenoptera: Formicidae) are attracted to fungal pathogens during the initial stage of colony founding. Myrmecol. News 20, 71–76. [Google Scholar]
- 32.Pontieri L, Vojvodic S, Graham R, Pedersen JS, Linksvayer TA. 2014. Ant colonies prefer infected over uninfected nest sites. PLoS ONE 9, e111961 (doi:10.1371/journal.pone.0111961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diez L, Lejeune P, Detrain C. 2010. Ants’ survival and waste management in Myrmica rubra nests. In Abstracts for the XVI Congress of the Int. Union for the Study of Social Insects, IUSSI, Copenhagen, Denmark, 8–13 August 2010, p. 376.
- 34.Yek SZ, Mueller UG. 2011. The metapleural gland of ants. Biol. Rev. 86, 774–791. (doi:10.1111/j.1469-185X.2010.00170.x) [DOI] [PubMed] [Google Scholar]
- 35.Pereira RM, Alves SB, Stimac JL. 1993. Growth of Beauveria bassiana in fire ant nest soil with amendments. J. Invertebr. Pathol. 62, 9–14. (doi:10.1006/jipa.1993.1067) [Google Scholar]
- 36.Pereira RM, Stimac JL, Alves SB. 1993. Soil antagonism affecting the dose–response of workers of the red imported fire ant, Solenopsis invicta, to Beauveria bassiana conidia. J. Invertebr. Pathol. 61, 156–161. (doi:10.1006/jipa.1993.1028) [Google Scholar]
- 37.Nash DR, Als TD, Maile R, Jones GR, Boomsma JJ. 2008. A mosaic of chemical coevolution in a large blue butterfly. Science 319, 88–90. (doi:10.1126/science.1149180) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this study are accessible as electronic supplementary material. DNA sequences are accessible through GenBank.