Abstract
Aire controls immunologic tolerance by inducing the ectopic thymic expression of many tissue-specific genes, acting broadly by removing stops on the transcriptional machinery. To better understand Aire’s specificity, we performed single-cell RNAseq and DNA methylation analysis in Aire-sufficient and -deficient medullary epithelial cells (mTECs). Each of Aire’s target genes was induced in only a minority of mTECs, independently of DNA methylation patterns, as small inter-chromosomal gene clusters activated in concert in a proportion of mTECs. These microclusters differed between individual mice, and thus suggest an organization of the DNA or of the epigenome that results from stochastic determinism, but is bookmarked and stable through mTEC divisions, ensuring more effective presentation of self-antigens, and favoring diversity of self-tolerance between individuals.
Aire is a fascinating transcription factor, with a unique function in promoting immunological tolerance of differentiating thymocytes1. First, it induces ectopic expression in medullary epithelial cells (mTECs) of a large set of genes whose products are typically associated with fully differentiated parenchymal cells (so-called peripheral-tissue antigens, PTAs)2. In addition, Aire controls factors that modulate the presentation of peptides derived from these PTAs by MHC molecules at the mTEC surface, or their cross-presentation by dendritic cells3. These peptides mold the T cell repertoire by inducing negative selection of self-reactive specificities4, 5 or by promoting positive selection of regulatory T cells6. The physiological consequences of Aire’s activity are profound, as humans and mice with loss-of-function mutations in AIRE/Aire loci develop multi-organ autoimmunity1.
Even if its structural domains are shared with conventional motif-specific transcription factors, Aire is a very unusual transcription factor. It impacts a large number of genes, generally allowing transcription of genes that would not be expected to be expressed in a given cell-type. Aire contains a SAND domain typically involved in DNA binding, but it does not have a clear DNA-binding motif, although it has been suggested to recognize methylated CpG residues in association with the meCpG-binding factor MBD17. Rather, its transcriptional activity seems to depend on the recognition of non-specific markers of low-activity chromatin, such as hypomethylated amino-terminal tail of histone H38, 9 or transcriptional start sites (TSS) with a surfeit of paused polymerases10. Aire also interacts with a variety of non-specific elements of the transcriptional and splicing machinery11, 12. Indeed, recent data derived from a variety of experimental approaches argue that Aire’s major modus operandi is to release promoter-proximal RNA polymerase-II (Pol-II) pausing10, 13, 14.
Aire’s action has an element of stochasticity. Single-cell PCR analysis suggested that individual mTECs, otherwise indistinguishable, express distinct patterns of PTAs15–18. Gene-expression profiling of mTECs from individual mice also suggested that inter-individual “noise” in gene expression between genetically identical mice was higher for Aire target genes than for the bulk of transcripts19. In spite of these clues, a coherent framework that explains Aire’s action in individual cells has remained elusive.
Single-cell RNA sequencing (scRNAseq) opens completely new vistas on the analysis of gene expression20 by combining the globality of genome-wide transcriptome profiling with the unique granularity brought by single-cell technologies like flow cytometry. It can reveal unrecognized subpopulation structure and avoid erroneous averaging, and can provide information on the fluctuations (“noise”) in gene expression21, 22 in an otherwise homogeneous population of cells23–25. Some of this noise can result from transcriptional bursting26, but it may also reveal coordinated activation of specific transcriptional programs that can be important in determining cellular differentiation or responses. Recent technical innovations make scRNAseq more performant and robust, with cell multiplexing, molecular barcoding and microfluidic devices27. scRNAseq data analysis remains challenging, however. First, with efficacies of molecular conversion of 20% at best, the low-expressed portion of the transcriptome is unreliably assessed in any one cell. Second, because real replicates are innately impossible with single-cell analysis, estimation of technical variance remains uncertain. Finally, the data must be interpreted in the context of sampling statistics, which makes analysis less intuitive than conventional profiling data, and necessitates complex statistical models24, 25, 28.
scRNAseq seemed to provide an attractive opportunity to explore the distribution of PTA expression in individual mTECs. This perspective, much broader than was achieved earlier by PCR15, 17, allowed us to ask how frequently individual Aire target genes are expressed in mTECs, and whether Aire changes the frequency of cells expressing particular transcripts or instead boosts the intensity of transcript expression in cells in which they are already present. Although Aire-induced gene expression proved to be very noisy, affecting genes with low frequency of expression, we uncovered unexpected order in this chaos detecting a number of Aire-induced transcripts whose expression clustered in small groups of mTECs, with no apparent logic, and varied between individual mice. These observations have direct implications for the efficiency of tolerance induction, and individual susceptibility to autoimmune deviation.
RESULTS
Range of Aire-induced gene expression
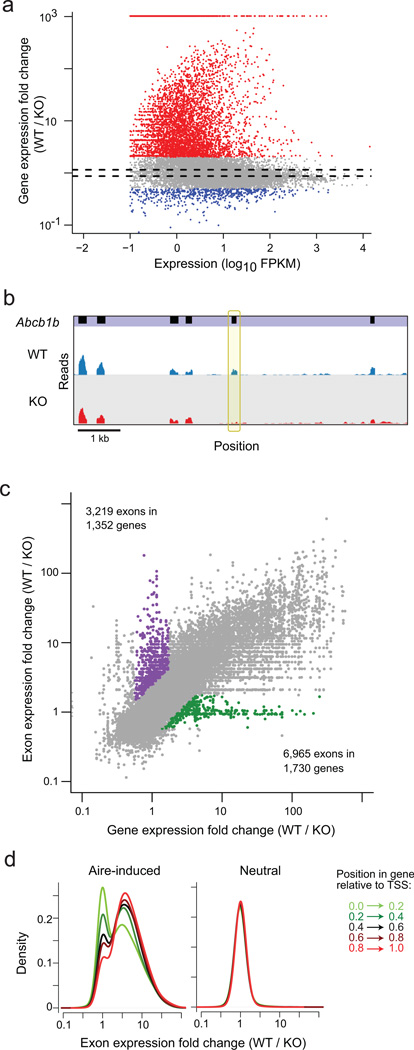
As a prelude to single-cell analysis, we performed standard RNA-seq on bulk-sorted mTECs. CD45−Ly51loMHCIIhiGFPhi cells were prepared in duplicate from Aire-gfp transgenic mice29, which were crossed with mice carrying the Aire-knockout mutation2 to generate wild-type (wildtype) and deficient (hereafter KO) littermates. In the TruSeq libraries generated (11.8 to 31.3 × 106 mapped reads per sample), we observed a biased and very deep impact of Aire: of the 19,772 genes expressed (at a threshold of 1 Fragments Per Kilobase of exon per Million; FPKM) in these datasets, 2995 were “Aire-induced” and 766 “Aire-repressed” genes (at an arbitrary FoldChange >2; (Fig. 1a). These results are consistent with prior microarray analyses19 and more recent RNA-seq data30 showing that Aire regulates a large fraction of the transcriptome. The sets of Aire-induced and -neutral genes defined here will be those tracked in the scRNAseq analyses below.
Fig. 1. Aire increases the repertoire and diversity of mTEC transcriptome.
a) Mean read counts (as FPKM) vs wild-type (wildtype) / Aire−/− (KO) FoldChange of gene expression for all gene in TrueSeq RNA-seq libraries generated from whole mTEC RNA from Aire-deficient mice or wildtype littermates (data pooled from 2 mice per group). Genes up- or down-regulated by 2-fold or more highlighted in red and blue, or between the dashed lines (FC<1.1) are designated as the Aire-induced, Aire-repressed and Aire-neutral set, respectively, in the following analyses. b) Example of Aire-induced exon in the Aire-neutral gene Abcb1b. Data from (a), read pileups are shown for Aire-wildtype and KO samples, below the exon positions (in black). The differentially spliced exon is bracketed in yellow. c)Aire-wildtype / KO FoldChange per gene (x-axis) or per exon (y-axis) in mTEC RNAseq data from (a).. Exons up-regulated > 2-fold at gene-level but less than 1.1-fold at the exon-level are highlighted in green, and vice versa in purple. d) Distribution of Aire-wildtype / KO exon FoldChange according to the relative position within the gene, for Aire-induced (left) and -neutral genes (right).
In addition to these consequences on entire transcripts, these RNAseq data showed that Aire further exerts more subtle effects on the usage of individual exons within genes. Several transcripts whose overall levels were little affected in mTECs by the absence of Aire showed Aire-dependent inclusion of particular exons (Fig. 1b). A more complete analysis of this phenomenon revealed 3,219 such exons with Aire-dependent expression, in contrast to the majority of exons whose representation correlated with that of the gene as a whole (175,216 exons, Fig. 1c). Alternative splicing has long been known to affect tolerance, as initially recognized for autoimmune responses to the Plp1 gene product31, 32. As also speculated by Keane et al33, Aire may help maximize exposure to genome-encoded peptides by enhancing exon inclusion, a property consistent with the splicing factors with which it interacts11. Conversely, this analysis also revealed the presence of a set of exons whose abundance remained invariant in the presence or absence of Aire, while the whole transcript was induced (Fig. 1c). These exons were particularly prevalent at the beginning of the transcripts (Fig. 1d), which is consistent with our earlier demonstration that the representation of the first exon shows comparatively little change in the absence of Aire10, reflecting polymerases that transcribe a short portion of the gene before stalling in Aire’s absence. The match between the degree of induction of genes and exons by Aire increased progressively along the transcript (Fig. 1d), suggesting that this effect may actually extend quite some distance from the TSS.
Overall diversity in transcriptomes of individual mTECs
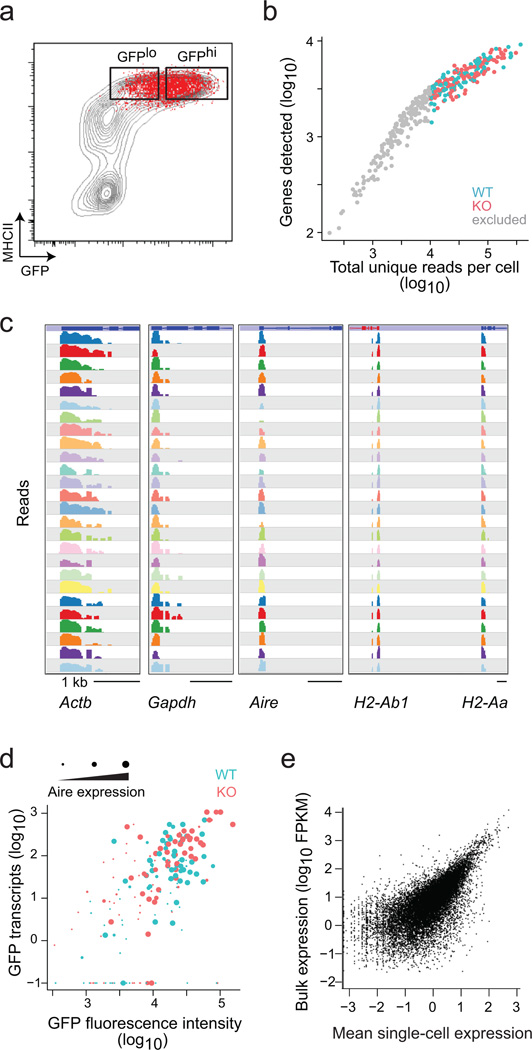
With these reference data in hand, we proceeded with analyzing Aire-controlled gene expression in individual mTECs through scRNAseq. Index sorting of single cells into wells of microtiter plates was used, such that we could relate the RNAseq profiles to the marker characteristics of the cells (Fig. 2a). We generated sequencing libraries from 360 single mTECs from two pairs of wildtype and KO mice, using a protocol modified from the original CEL-Seq technique34. This protocol includes oligo-dT priming with barcodes to allow attribution of each sequence read to its cell of origin, as well as Unique Molecular Identifiers for tagging each original molecule, in order to avoid artefacts from over-amplification of small numbers of initial molecules35. Although most single-cell libraries yielded high-quality data, for robustness we restricted our further analysis to 201 cells that generated at least 104 unique mappable reads per cell (Fig. 2b). Several findings validated these single-cell data. First, there was good representation of the MHC-II and housekeeping transcripts expected in mTECs (Fig. 2c). Second, the intensity of the Aire-GFP fluorescence as detected by flow cytometry matched the GFP and Aire transcript counts in each cell (Fig. 2d; note that the Aire-KO mutation abolishes function, but not the transcript). Third, the total number of reads per gene, obtained by aggregating all the scRNAseq datasets, recapitulated well the data at the population level (Fig. 2e, Pearson r=0.72).
Fig. 2. Single-cell RNAseq in mTECs.
a) Sorting of single mTECs (red) from wildtype mice for scRNAseq (representative of 2 wildtype and 2 KO mice). b) Number of unique mappable reads vs the number of genes detected, for each cell, in the scRNAseq datasets. Cells omitted from further analysis are in gray. c) Representative read pileups for 26 representative cells at 5 illustrative genes in the scRNAseq datasets. Only one exon shows because our scRNAseq technique only tags sequences next to the polyA. d) Correlation between the GFP fluorescence intensity during the sort vs the number of GFP mRNA reads observed in each cell (Pearson r=0.56). The size of the dot indicates the number of reads from the Aire transcript. e) Mean single-cell read counts per gene compared with bulk read counts of those same genes.
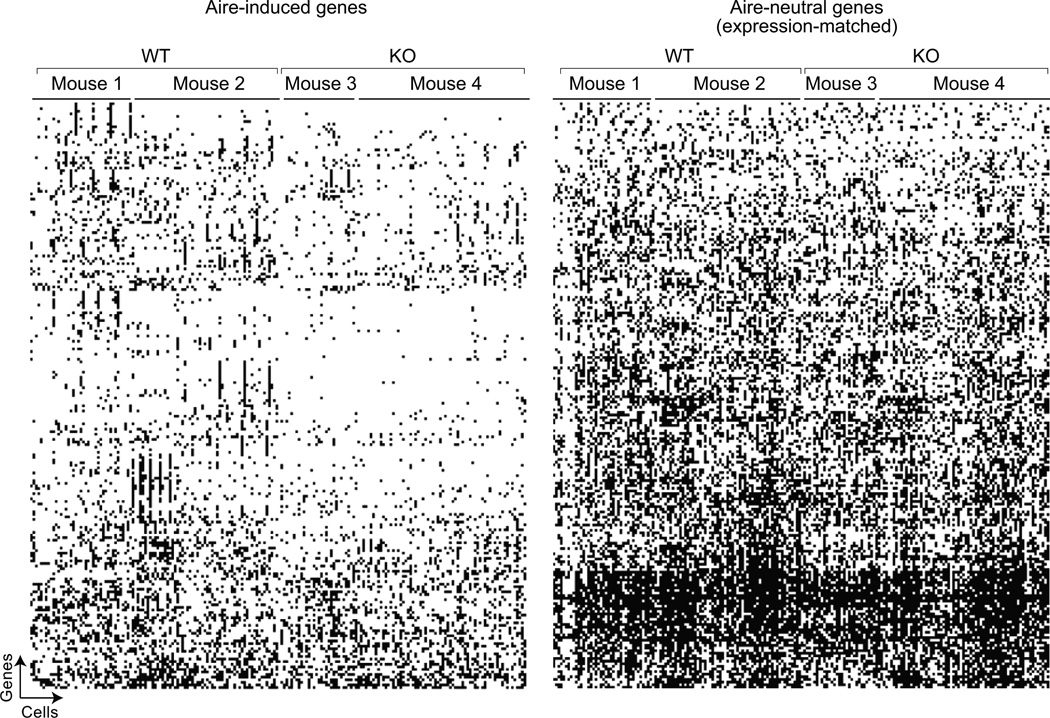
We then analyzed computationally the expression of Aire targets in these scRNAseq data. Examination of Fig. 3, which depicts the presence or absence of individual transcripts in each cell, reveals each of the points that will be substantiated and validated in Figs 4–6: (i) Aire mainly targets transcripts expressed at a low frequency (Aire-induced more sparse than Aire-neutral transcripts). (ii) Aire increases this frequency, higher in Aire-wildtype than in Aire-KO cells. (iii) Discrete clusters of Aire-induced genes show coordinated expression. (iv) Accordingly, there are groups of mTECs with comparable expression of small gene clusters. (v) mTEC clusters are different in individual mice.
Fig. 3. Graphic summary of the single-cell expression results.
Presence or absence of individual transcripts in Aire-wildtype and KO mTECs, for Aire-induced transcripts (left) and a set of Aire-neutral genes (matched for mean expression levels in positive cells) from the scRNAseq data of Fig. 2. Genes are arranged in rows by hierarchical clustering, cells in columns according to genotype and mouse. The weighted probability of expression of each transcript in each cell was computed, per the Bayesian approach of Kharchenko et al28; black squares denote the presence of the transcript (regardless of intensity) and white squares represent no expression (most at high confidence of not being dropouts by SCDE analysis, per Fig. 4).
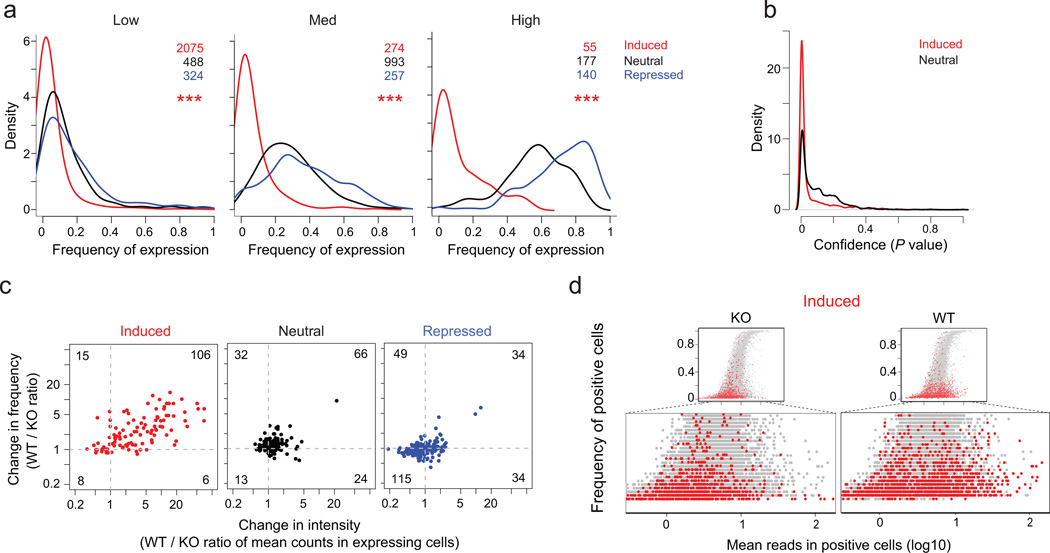
Fig. 4. Aire increases the intensity and frequency of otherwise rare transcripts.
a) Distribution of the frequency of expression in single-cells (scRNAseq data of Fig. 2, 2 KO mice) for genesets matched by expression levels in bulk RNAseq data (Low: 1–5 FPKM; Med: 10–25 FPKM; High: 50–100 FPKM)); ***: Wilcoxon p<10−15b) Bayesian28 confidence (p.value) that genes scored as unexpressed a given cell are not sampling dropouts, for expression-matched Aire-induced and –neutral genes, in the scRNAseq data of Fig. 2, 2 KO mice. c) Change in expression intensity (wildtype/KO ratio of mean count per gene in expressing cells) vs change in the frequency of expression (wildtype/KO ratio of frequencies of expressing cells), computed for expression-matched transcripts (window of 25–50 counts per gene) Aire-induced, -neutral or –repressed genes (scRNAseq data of Fig. 2, 2 wildtype and 2 KO mice). d) Mean counts in positive cells versus frequency of expression for Aire-induced genes (red) relative to genome-wide distribution (grey); the bottom section is expanded to focus on the shift in Aire-induced genes (scRNAseq data of Fig. 2, 2 wildtype and 2 KO mice).
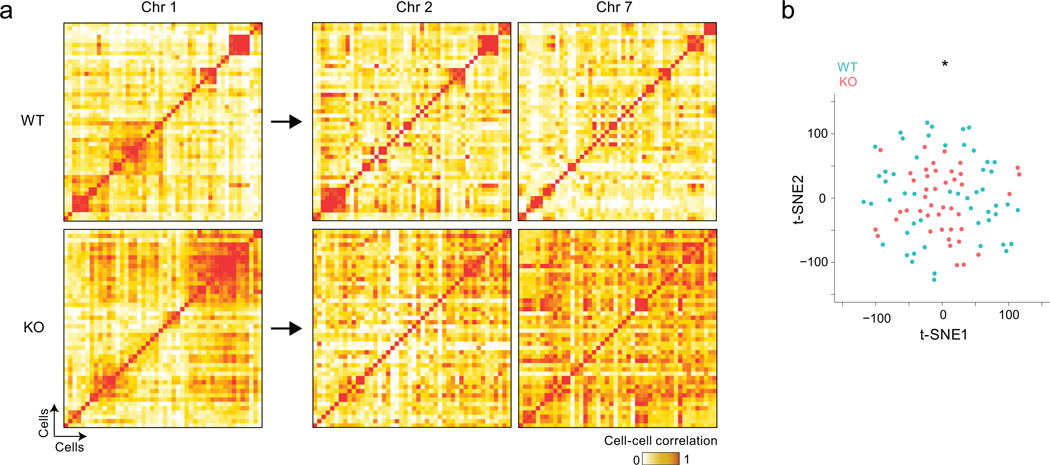
Fig. 6. Aire-dependent interchromosomal gene networks generate diverse and distinct mTEC subsets.
a) Cell-by-cell Pearson correlation between individual mTECs (scRNAseq data of Fig. 2, 2 wildtype and 2 KO mice per group) among wildtype (top) and KO mTECs (bottom), computed from Aire-induced genes from different chromosomes. Clustering was determined by affinity propagation for genes on chromosome 1 (left), and the same cell order was applied for correlations values computed with Aire-induced genes from chromosomes 2 or 7 (right). b) Relative distances computed by t-SNE reduction to 2D space of wildtype (blue) and KO (red) mTECs, based on the expression of Aire-induced genes (scRNAseq data of Fig. 2, 2 wildtype and 2 KO mice); Wilcoxon p<10−3.
Aire mainly targets transcripts expressed at low frequency
Density plots of the frequency of mTECs expressing individual genes were generated for Aire-induced, -neutral or -repressed mRNAs (transcripts matched for expression in bulk RNA-seq). This analysis showed that most Aire-induced genes were active in only 5–20% of the sampled Aire-KO mTECs (Fig. 4a). Aire-neutral and -repressed genes were more frequently expressed in these mTECs, and the difference was significant across the three expression levels (Fig. 4a). Two points argued that this low frequency of Aire-induced transcripts was not merely a consequence of statistical sampling, which can be a concern for scRNAseq. First the frequency of false-negatives (dropouts) from sampling is directly related to the intensity of expression, and these dropouts would be expected at the same frequency for expression-matched Aire-induced or -neutral transcripts, which is clearly not the case (Fig. 4a). Second, we plotted the probability that cells with no reads for a given gene were statistical dropouts. Most Aire-induced transcripts had very low probabilities of being false-negatives (68.1% with nominal p<0.05, Fig. 4b).
Aire mainly increases the level of target-gene expression
The increase observed in wild-type mTEC in comparison to KO mTECs for a given Aire-induced gene in bulk population profiling could result from increases in either the amount of transcript per cell, or in the proportion of cells expressing the transcript. When we compared the wildtype/KO changes in mean expression intensity in mTECs positive for a given transcript, as well as the changes in the frequency of cells expressing this transcript, we found that Aire expression appeared to increase both (Fig. 4c, chisq.test p < 10−15). Curiously, a limited but significant shift was also observed between KO and wildtype mTECs for transcripts in the Aire-neutral category (chisq.test p< 10−7), indicating that Aire subtly activates the majority of genes in the cell. A potential confounder of this analysis is that higher read number per cell lead to more frequent detection of positive cells, simply because higher intensities favor lower dropout rates. To test for such bias we plotted the frequency of cells expressing Aire-induced or –neutral genes vs the mean intensity of expression in mTECs that do express them, in wildtype and KO mTECs (Fig. 4d). This analysis showed that the presence of Aire resulted in a predominant shift in the distribution towards higher per-cell intensities, a shift that did not merely follow the main intensity-frequency relation. Indeed, we found that the shift in expression intensity in Aire’s presence led to less increase in the expression frequency of its targets than predicted from the dropout distribution of gene pairs randomly drawn from the Aire-neutral distribution (Supplementary Fig. 1). Thus, Aire target genes remained less frequently expressed than the genome-wide norm, even after transcriptional activation by Aire.
Discrete clusters of Aire-induced genes show coordinated expression
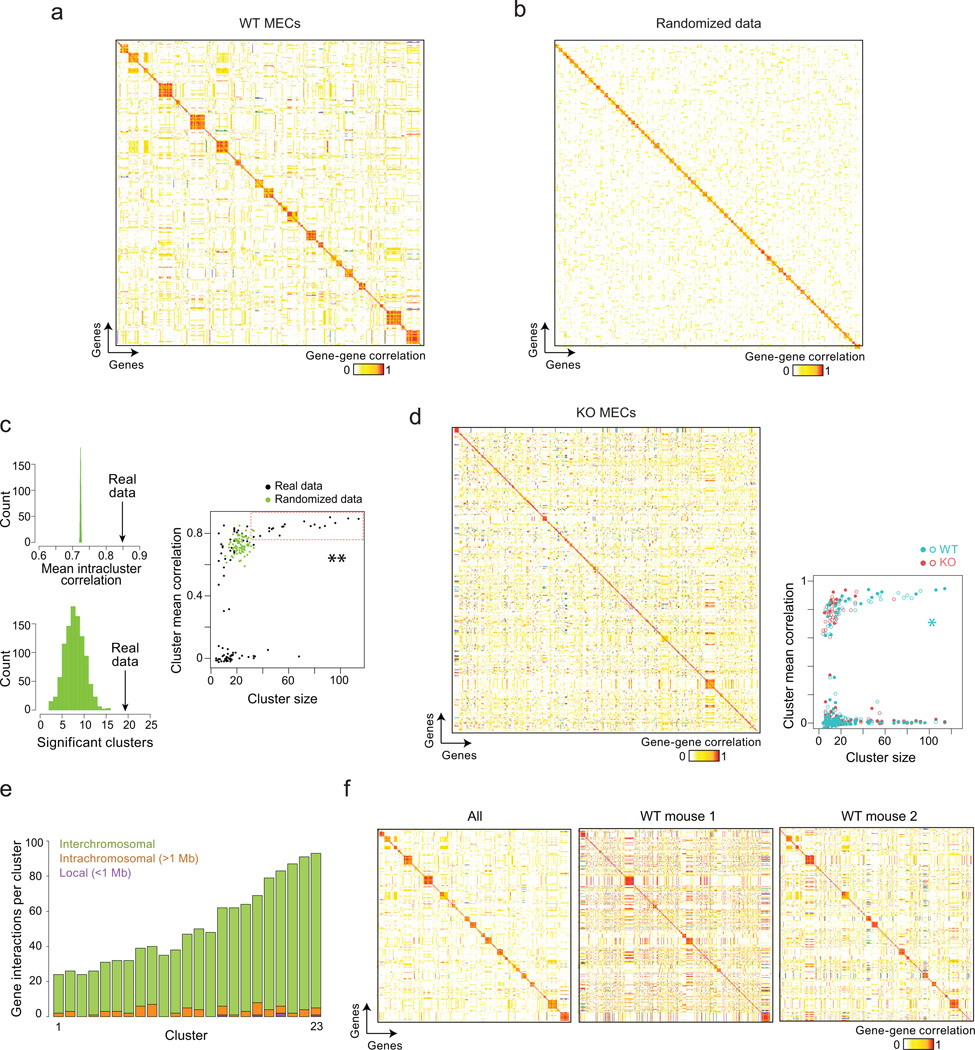
The short vertical streaks in the checkerboard plot of Fig. 3 suggested that subsets of genes are expressed in concert. To better evidence such structures in the data, we computed gene-by-gene correlations for all Aire-induced genes (on the basis of weighted expression matrix24), and performed a partition clustering using an affinity propagation (AP) algorithm36. We found a high degree of structure in the scRNAseq datasets from wild-type mTECs, as 51% of Aire-induced transcripts grouped into 19 clusters with an internal mean correlation >0.75 (Fig. 5a); these clusters were small (33 to 114 transcripts; median 57) and largely distinct from each other. The significance of these clusters was verified by permutation (randomly shuffling the expression levels per gene between cells), which did not reproduce the same degree of cluster structure (Fig. 5b); comparable cluster sizes and internal correlations were not achieved in 1000 random permutations (Fig. 5c, Wilcoxon p=0.001). Fewer such clusters were detected with the scRNAseq datasets from KO mTECs (Fig. 5d, Wilcoxon p=0.002), or when computed from expression-matched Aire-neutral genes (Supplementary Fig. 2, Wilcoxon p=8 × 10−5), further substantiating their significance, and indicating that Aire is required for the appearance of these co-expressed clusters.
Fig. 5. Aire coordinates discrete interchromosomal gene networks.
a) Gene-by-gene Pearson correlations computed from the weighted expression matrix for Aire-induced genes in all wildtype mTECs (scRNAseq data of Fig. 2, 2 wildtype mice). Genes are ordered according to Affinity Propagation clustering36, with no preset number of clusters. b) Same computation as in (a), but in a control data matrix generated by random permutation of gene expression values. c) Results of 1000 random permutations and Affinity Propagation clustering of the Aire-wildtype scRNAseq data, recording the mean within-cluster correlation and the number of significant correlations in each iteration (significant clusters being defined as clusters with more than 30 genes and mean correlation > 0.75); **: Wilcoxon p=0.001. d) As in (a), but correlations within KO mTECs, in the scRNAseq data of Fig. 2, pooled from 2 mice. The size and internal correlation of clusters in wildtype and KO mTECs are compared at right; *: Wilcoxon p=0.002. e) Quantification of significant gene-gene correlations that are local (purple, less than 1Mb distance on same chromosome), intrachromosomal (orange, more than 1Mb distance on same chromosome), and interchromosomal (green, different chromosome) in the top 23 clusters of the wildtype scRNAseq datasets from Fig. 2. f) Gene-gene correlations between Aire-induced transcripts computed as in (a) for all wildtype mTECs (left), and computed independently in mTECs from the two different wildtype mice (middle and right); (scRNAseq data of Fig. 2, 2 wildtype mice).
It was previously reported that Aire-induced PTAs tend to fall in local gene clusters37, 38. We observed such co-regulated activity of local segments, as illustrated for the Sprr and Mup loci (Supplementary Fig. 3). On the other hand, these localized coregulation events contributed little to the overall gene clusters, most of which rested on correlations across chromosomes (Fig. 5e). Therefore, mTECs co-express discrete, interchromosomal gene networks. We searched for commonalities between the transcripts that form these small clusters. GeneOntology or pathway analysis (MSigDB, PANTHER) failed to reveal any gene function or pathway shared by members of any of these clusters; nor did cluster members share specificity of expression when analyzed across the GNF compendium of gene expression39; nor did the promoter regions of cluster members show enrichment for binding motifs for a particular transcription factor (data not shown). Therefore, these co-expressed gene clusters seem unrelated in terms of genomic position, biological function, or transcriptional regulation.
Aire-induced gene networks define distinct mTEC subgroups
Given these small clusters of Aire-induced genes, we asked how their expression demarcates individual mTECs. Correlation analysis based on probability values for Aire-induced genes showed that wild-type mTECs partitioned into discrete groups (Fig. 6). These groupings were based on inter-chromosomal gene networks, because the cell-to-cell correlation maps computed on the basis of transcripts from one chromosome were reproduced, for the most part, when computed with transcripts from other chromosomes (Fig. 6a, right panels). For a broader perspective on mTEC heterogeneity in wildtype and KO thymi, we analyzed the cell-to-cell correlation matrix with t-SNE40, a dimensionality reduction algorithm that computes the probability for two cells to be neighbors, and displays the best fit in 2D space. Aire-deficient mTECs tended to group close together at the center (Fig. 6b). Wild-type mTECs, although distributed around the same center, radiated further (Wilcoxon p<10−3 in repeated runs), and were more distant from each other than were KO mTECs (p<10−60). Predictably, these small t-SNE groups coincided with the cell clusters of Fig. 6a. Thus, Aire diversifies gene expression, not in a completely random fashion, but with some degree of coordination between cells.
mTEC clusters are different in individual mice
It was already apparent from Fig. 3 that the small gene clusters expressed in mTECs were not shared between mice. Indeed, when gene-gene correlations were computed independently from scRNAseq mTEC datasets from each wildtype mouse, correlations within a cluster applied for mTECs of only one mouse, but not in the other (Fig. 5f). Thus, these gene networks are most likely established by stochastic events, and not by hardwired molecular cues.
DNA methylation in mTECs does not account for Aire specificity
Epigenetic regulatory mechanisms make attractive candidates to explain two prominent characteristics of Aire transcriptional specificity, the predilection to activate infrequently expressed genes, and the interchromosomal clusters that are coordinately expressed in small groups of mTECs. DNA methylation at CpG dinucleotides is one such candidate, as variable but heritable methylation patterns could be at root. Indeed, Waterfield et al. have proposed that Aire associates with the methyl-CpG binding protein MBD1 and uses this factor’s ability to preferentially recognize methylation at the TCGCA motif for preferential PTA targeting7. In addition, analysis of DNA methylation by reduced representation bisulfite sequencing (RRBS) is inherently a single-cell methodology that measures the frequency of DNA methylation marks at specific locations, and was thus a good complement to the single-cell analyses above.
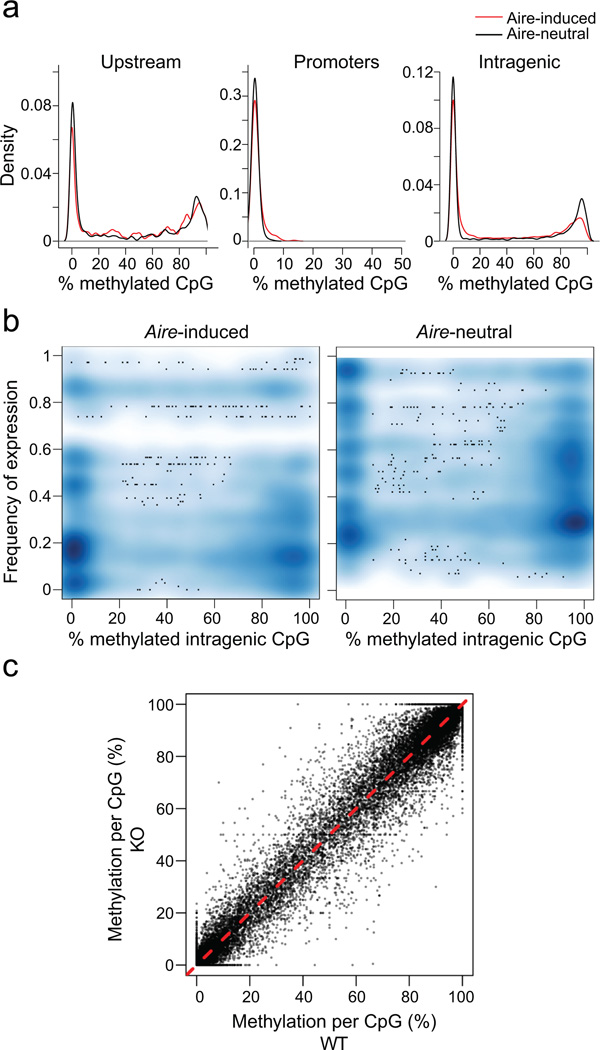
To determine their DNA methylation status, mTECs were sorted as described above, and their DNA was processed for RRBS41. The distribution of CpG methylation at different positions did not differ markedly between mTECs from wild-type and KO mice, for Aire-induced and -neutral loci (Fig. 7a). CpG positions in upstream enhancer elements were similarly represented in both sets of genes, and the region surrounding the TSS was uniformly unmethylated in both cases (Fig. 7a). This observation held for MBD1 sites in particular, which were uniformly unmethylated in all loci (Supplementary Fig. 4). In fact, the frequency of TCGCA sites in Aire target genes was the same in Aire-induced TSS and Aire-neutral TSS, and reanalysis of the published expression data7 showed that MBD1 has a limited transcriptional impact in mTECs, which overlaps very little with Aire’s transcriptional signature (Supplementary Fig. 4c), indicating that MBD1 plays little or no role in Aire-dependent PTA expression.
Fig. 7. Little or no difference in DNA CpG methylation in Aire-induced genes.
a) Proportion of methylated CpG residues in RRBS methylation libraries from mTECs of KO mice (2 mice pooled); frequencies computed in Upstream (−1 to −50 Kb from the TSS), Promoter (−100 to −1 Kb from the TSS), or Intragenic (25% or more of gene length beyond the TSS) regions of Aire-induced (red) or Aire-neutral (black) genes. b) Relationship between the mean methylation frequency at intragenic CpGs and the frequency of expression for Aire-induced genes, in KO mTECs. c) Comparison of methylation frequencies at each CpG position in DNA from wildtype and KO mTECs (data from (a), pooled from 2 mice/group).
CpG methylation increases in frequency at intragenic positions42, and this trend was slightly less pronounced for Aire-induced loci than for expression-matched neutral loci (Fig. 7a). We asked whether the intragenic CpG methylation frequency might relate to the frequency of expression of corresponding Aire-induced genes in wild-type mTECs. The majority of intragenic CpGs were either not methylated or highly methylated, and both of these methylation statuses were associated with a range of expression frequencies (Fig. 7b). Finally these methylation profiles, including the sites of variable methylation, were not Aire-dependent, as evidenced by the high correlation between wild-type and KO MECs (Fig. 7c). Therefore, Aire itself does not alter the DNA methylation profiles in mTECs, and methylation patterns do not provide any obvious clue as to the frequency distribution of Aire target genes.
Discussion
This study has revealed several novel aspects of Aire’s function as a transcription factor, which likely have direct consequences on its function in central tolerance induction.
First, Aire increased mTEC transcriptome diversity by inducing thousands of Aire-dependent transcripts, as well as Aire-dependent exons in otherwise Aire-neutral genes. This observation is consistent with recent predictions33 that ectopic PTA expression involves different splicing patterns relative to the tissues in which PTAs are “normally” expressed. Alternative splicing is known to have important consequences on autoimmune responses (e.g.31, 32), but Aire’s role in this process had not been recognized. It seems likely that Aire’s impact on differential exon inclusion is tied to its close interactions with splicing factors of the transcriptional machinery and its preferential effect on spliced transcripts in cultured cells11, 14. This broad effect on the mTEC transcriptome maximizes the representation of peptides presented to developing thymocytes.
Second, Aire seems to preferentially target genes that are expressed in a minority of cells, and increases the intensity of expression of its target genes. This is consistent with the notion that Aire recognizes generic features of gene and chromatin organization, such as unmethylated H3K4 or promoters with a surfeit of paused polymerases8, 10, 13, 43. Thus, it isn’t as if Aire had any particular specificity for PTAs, but rather that it keys on these generic features of poorly expressed genes. But, with regards to regulation of gene expression, what does an “infrequently expressed” gene really mean, for a rather homogenous primary cell population like the Aire+ mTECs analyzed here? It is a notion far removed from the deterministic gene expression programs usually envisaged for lineage differentiation, but rather related to notions of “noisy” gene expression. There are several sources of noise in gene expression that can be important in allowing progression through differentiation or cellular adaptation processes44. Infrequent gene expression can correspond to a “burst” of transcription, where any given gene is actively transcribed only a small fraction of the time26 and produces relatively short-lived transcripts. Then, a low frequency of positive cells can simply denote the odds of catching a cell during such a transcriptional burst. Alternatively, low-frequency expression can result from a particular organization of the DNA or epigenetic modifications, which are set stochastically in every cell, but are then stable for some period of time. Aire is predominantly found in tight nuclear “speckles” 45, thought to be sites of active transcription, and one might speculate that the set of genes ectopically expressed by a mTEC are those which have been threaded into these Aire-containing speckles.
Clues to the basis of low expression frequency of Aire-controlled genes may be found in the small clusters of co-regulated genes whose expression is shared by discrete groups of mTECs. The existence of these discrete microclusters of expression is not easily compatible with a burst model, because it is unlikely that genes would burst at the same time in different cells, but rather with a model where infrequent expression results from stable organization of the genome or epigenome. Because the genes within these expression clusters do not share discernable sequence motifs or chromosomal locations that might explain their coordinated transcription, their co-expression in a fraction of the mTECs is perhaps most easily interpreted in terms of clonal relationship. mTEC that share PTA clusters could plausibly be daughters of the same epithelial cell progenitor46, implying that Aire-target selection within a mTEC clone is “bookmarked” across cell division. Bookmarking (the recovery of gene expression programs after mitosis 47) can be explained for conventional transcription by the persistence of networks of specific transcription factors, but is puzzling for a mode of regulation that does not depend on the transcription factors that normally activate specific PTAs 15. Epigenetic cues such as DNA modifications, albeit probably not methylation, or remanent histone marking might be involved in bookmarking PTA expression. Of note, Brd4, which binds acetylated histones and promotes the release of PolII stalled at the promoter, is involved in trans-mitotic bookmarking47 and is an essential Aire cofactor (Yoshida et al, in press), and one might imagine that Brd4, together with Aire and other cofactors, forms trans-mitotically stable complexes with fixed DNA regions. Thus, one might propose that an inherently stochastic mechanism initially selects and marks groups of loci, whose coexpression is then bookmarked and transmissible. A parallel could be made to the stochastically determined repertoire of activating and inhibitory receptors in NK cells.
In terms of tolerance induction, the low expression frequency of Aire-target genes and the existence of expression microclusters implies that mTECs are “splitting the burden” of PTA expression, and that there is a higher local concentration of any gene product than if all PTAs were uniformly expressed in all mTECs. Since immature thymocytes scan the thymic medulla and negative selection is effective with small pockets of antigen-positive presenting cells 48, 49, negative selection should be more effective than with lower but widespread amounts of PTA expression in mTECs.
Importantly, these co-expressed gene clusters were not the same in the two genetically identical wild-type mice whose mTECs we analyzed by scRNAseq, which has important implications for the inter-individual variation in tolerance within a species. We had previously reported, on the basis of microarray profiling data, that Aire-induced transcripts show significantly greater inter-individual variability than do Aire-independent transcripts19. The present data now provide a cellular explanation for this observation. One caveat of the present study, however, is that we cannot formally know if these co-regulated clusters persist and reflect constant inter-individual differences, or fluctuate and represent the state of the mTEC pool at the time of cell preparation for scRNAseq. However, since the expressed clusters of Aire-induced genes were not shared at the time of the experiment, the two analyzed mice exposed their immature thymocytes to slightly different sets of self-peptides, thereby generating T cell repertoires with slightly different autoreactivities to peripheral tissues. Such diversity may be favorable at the level of the species in ensuring a diversity of potential responses to pathogens without uniform holes in the repertoire, albeit at the price of susceptibility to autoimmune diseases.
ONLINE METHODS
Mice
All mice were housed and bred under specific-pathogen–free conditions at the Harvard Medical School Center for Animal Resources and Comparative Medicine (Institutional Animal Care and Use Committee protocol 2954). Aire-driven Igrp-GFP (Adig) mice 29 were generously provided by Dr. Mark Anderson.
Thymic Epithelial Cell Isolation
Thymus tissue was dissociated in RPMI and digested with 0.5mg/mL collagenase/dispase (Roche) and 0.2mg/mL DNase (Sigma) in RPMI for 30 min with agitation every 10 minutes. Following staining with primary antibodies (MHC II(I-A/I-E)—APC; Ly51-PE; CD45-PE-Cy5), CD45+ cells were depleted by MACS separation with anti-PE beads (Miltenyi). DAPI−,CD45−,Ly51lo,MHCIIhi,GFPhi MECs (5–10 × 104 per mouse) were sorted on a MoFlo (Cytomation) into Trizol for RNA preparation (for TruSeq library preparation) or into RPMI medium (Gibco) for RRBS library preparation. For scRNAseq, similar gating, also including GFPlo MECs, was used for sorting at one cell per well of a 96-well plate on an Aria sorter (BD).
TruSeq library preparation and analysis
Bulk RNAseq libraries were prepared using TruSeq (Illumina) following the manufacturer’s protocol from 5–10 × 104 sorted MECs (one mouse) per sample. Sequencing (single-end, 50bp) was performed on a HiSeq2000 (Illumina), and reads were aligned to mm10 using Tophat2. Duplicated reads were filtered out from further analysis. Normalized counts per transcript (FPKM) were calculated using Cufflinks. Exon level expression was calculated using SeqMonk (Babraham).
RRBS library preparation and analysis
Reduced Representation Bisulfite Sequencing (RRBS) libraries were prepared as described 41 from 5–10 × 104 sorted MECs (one mouse) except that EZ DNA Methylation-Direct (Zymo) was used for bisulfite conversion. Sequencing (single-end, 50bp) was performed on a HiSeq2000. Prior to alignment, reads were trimmed to remove adapter sequences using the RRBS option in TrimGalore! (Babraham). Trimmed reads were aligned to mm10 using Bismark 50 (Babraham), and methylation calls per CpG were calculated using SeqMonk (Babraham). Only those CpG sites covered by at least 20 reads were considered for subsequent analysis. Relating CpG positions to the closest genes was determined in SeqMonk relative to the mm10 mouse genome release.
scRNAseq library construction
Single cell RNA sequencing libraries were performed using a modified CEL-Seq protocol 34. First single cells were index sorted using a BD FACSAria II in 96 well hard-shell PCR plates (BioRad #HSP9631) filled with 4.4uL of lysis buffer containing 0.125µL of RNAseOut (40 U/µL stock, Invitrogen 10777-019), 0.25µL of Reverse Transcription (RT) primer (25ng/µL stock) and 4µL of RNAse-free water (Ambion AM9932). Each of the RT primer contained a T7 promoter, the 5’ TruSeq Illumina adapter, unique molecular barcodes (4–9 bp), a single cell DNA barcode (8–16bp) and oligodT sequence (24bp) (see table). Three wells were filled with a different mix in order to process the carrier RNA and in which single cells were not sorted. These wells contained 0.5µL Hela total RNA (1µg/µL, Ambion AM7852), 0.25µL of a T7-oligo-dT primer that did not contain the Illumina adapter or barcodes (initially provided in the MessageAmpTM II aRNA Amplification Kit, AM1751), 0.25µL of RNAseOut and 3.5µL of RNAse-free water. Quickly after sorting, the plates were covered with an aluminum seal (AlumaSeal 96, Excel Scientific #F96100), vortexed for 10s, centrifuged for 1 min at maximum speed (>2250g at 4°C), frozen on dry ice and kept at −80°C for up to 3 weeks.
RNA denaturation was performed by incubating the plates for 3 min at 70°C (lid 80°C) in a thermocycler. 2µL of the First Strand Reverse Transcription mix was then added to each well (ArrayScript Reverse Transcriptase, Ambion AM2048) containing 1µL of dNTP mix (10mM each stock, Invitrogen 18427-013), 0.5µL of First Strand Buffer 10x, 0.25µL of ArrayScript (200 U/µL stock) and 0.25µL of RNAse Inhibitor (40 U/µL stock, Ambion AM2682). The plates were then incubated for 2 hours at 42°C (lid 50°C).
Second Strand Reverse Transcription was performed using the mRNA Second strand synthesis module (NEBNext #E6111L). 15µL of the Second Strand Reverse Transcription containing 12µL of RNAse-free water, 2 µL of 10X Second Strand Synthesis Reaction Buffer and 1 µL of the Second Strand Synthesis Enzyme Mix was added to each well of the plates. The plates were then incubated for 2.5 hours at 16°C (open lid). cDNA clean-up and size selection were then performed using the Agencourt RNAClean XP beads (Beckman Coulter A63987). First, single cell cDNA libraries containing different barcodes were pooled in one tube with a Hela carrier cDNA library. In our case, 30 single cell cDNA libraries were pooled together with a carrier Hela cDNA library (3 pools per plate). Each pool was mixed with 0.8x volume of Agencourt RNAClean XP beads, incubated 15min at room temperature, then placed 5 min on the magnet. The supernatant was carefully removed and the beads were washed twice while still on the magnet with fresh 70% ethanol. Beads were dried out 15 min before elution was carried out in 50µL of RNAse-free water. A second bead purification was performed similarly with 1x volume of RNA AMPure XP Beads and eluted in 6µL of RNase-free water.
In vitro transcription was then conducted using the MEGAshortscript T7 transcription kit (Ambion AM1354). 10.4 µL of a mix containing 1.6µL of ATP (75mM stock), 1.6µL of UTP (75mM stock), 1.6µL of GTP (75mM stock), 1.6µL of CTP (75mM stock), 1.6µL of T7 10X Reaction Buffer, 1.6µL of T7 Enzyme Mix and 0.8µL of RNAseOut was added to each 6µL cDNA pool, and incubated at 37°C lid 70°C for 14h. Illumina libraries were then constructed as follows. First the fragmentation of the amplified RNA (aRNA) was performed using the Magnesium RNA Fragmentation Module (NEBNext E6150S). 4µL of the fragmentation mix containing 2µL of RNAse-free water and 2µL of the RNA Fragmentation Buffer (10X) was added to 16µL of aRNA. Samples were immediately incubated at 94°C (lid 105°C) for 2 minutes, then immediately transferred onto ice and the reaction was stopped by adding quickly 2µL of 10X RNA Fragmentation Stop Solution. The fragmented aRNA was then cleaned up using the RNeasy MinElute Cleanup Kit (Qiagen 74204) following the manufacturer’s protocol and eluting in 10µL of RNAse-free water twice.
The size distribution and quantity of fragmented aRNA was then assessed by running 1µL of each sample in a BioAnalyzer using the Agilent RNA 6000 pico Kit (Agilent 5067-1513). The samples were then treated as follows. First the 5’ end of the aRNA was dephosphorylated by adding 4 µL of a mix containing 2µL of 10X Antarctic Phosphatase Reaction Buffer, 1µL of Antartic phosphatase (5U/µL stock, NEB M0289) and 1µL of RNAseOut to 16µL of each aRNA pool and incubating at 37°C for 30min and 65°C for 5min. Then the RNA was 3’ dephosphorylated and 5’ phosphorylated by adding 30µL of a mix containing 21.5µL of RNAse-free water, 5µL of 10X Antarctic Phosphatase Reaction Buffer (NEB M0289), 0.5µL of ATP (100mM stock, ATP Tris buffered Thermo Scientific #R1441), 1µL of RNAseOut and 2µL of T4 PolyNucleotide Kinase (10U/µL, NEB M0201S), and incubating at 37°C for 1 hour. The phosphatase and PNK treated RNA was then purified using RNeasy MinElute Cleanup Kit following the manufacturer’s protocol and eluted in 14µL of RNAse-free water. The samples were then dried down to 5µL using a vaccum concentrator (55°C, 5–7min). The 3’ Illumina adapter (RA3) (see table) was then ligated to the treated RNA using T4 RNA Ligase 2, truncated (Enzymatics L6070L). 3µL of a mix containing 1µL of 10x truncated T4 RNA Ligase 2 buffer, 1µL of DMSO (Sigma D9170) and 1µL of the 3’ adapter (10µM stock) was added to 5µL of the treated RNA. The samples were incubated at 70°C lid 80°C for 2min, placed immediatelly in ice and 2µL of a mix containing 0.5µL of RNAse Inhibitor (40U/µL, Enzymatics Y9240L) and 1.5µL of truncated T4 RNA Ligase 2 (5U/µL stock) was added. The ligation was performed at 22°C open lid for 1 hour. The ligated RNA was then reverse transcribed using SuperScript II (Invitrogen 18064-014). 8.5µL of a mix containing 2µL of RNA RT primer (RTP,10µM stock, see table) and 6.5µL of RNAse-free water was added to 10µL of the ligated RNA. The samples were then incubated at 70°C lid 80°C for 2min, placed immediatelly in ice and 10.5µL of a mix containing 4µL of 5X First Strand Buffer, 0.5µL of dNTP (25mM mix) 2µL of DTT (100mM stock), 2µL of RNAseOut and 2µL of SuperScript II (200U/µL stock) was added.
Reverse transcription was performed at 50°C lid 70°C for 1 hour and the library was then amplified using Kapa HotStart ReadyMix (Kapa KK2602). 71µL of the following mix was added to each reverse transcription reaction: RNAse-free water 17µL, 50µL of the Kapa HotStart ReadyMix 2X, and 4µL of the P5_Rd1_Primer_F (10µM stock, see table). To each reaction 4µl of a uniquely indexed P7_Rd2_Primer_idxN_R (10µM stock, see table) was added, and PCR cycles were performed as follows : 95°C 3 min ; 18 cycles of 20s at 98°C, 30s at 60°C, 30s at 72° C; 5min at 72°C. The PCR product was then cleaned up and size selected using two rounds of Agencourt AMPure XP Beads (A63880), as described above with the following modifications. The first purification used 1x volume of beads and elution was performed in 32µL of water ; the second purification used 1.2x volume and 12µL of elution water.
The size distribution and quantity of the library was assessed by running 1µL of each sample in a BioAnalyzer using the Agilent High Sensitivity DNA Kit (Agilent 5067-4626). Samples were pooled for sequencing on a MiSeq (nano kits) and HiSeq 2500 (rapid mode). Paired-end 50bp sequencing was performed using custom primers (100µM stock in water, see table) : 75bp for read 1 (custom_Read_1_seq), 7bp for the index sequencing (custom_i7_seq), 25bp for Read 2 (custom_Read_2_seq). Read 1 reads through the transcript sequence. Read 2 reads through the single cell barcode and unique molecular identifiers.
scRNAseq data processing
Raw data were processed using custom scripts. Read 1 contains the transcript sequence, and Read 2 contains the single cell barcode and unique molecular identifiers. Raw reads were first trimmed using the FASTX-Tollkit v0.0.13 (fastx_trimmer –Q 33). Read 2 was trimmed in order to extract the single cell barcode (8bp) and the UMI (4–8bp), and Read 1 was trimmed to 30bp get rid of a potential oligo-dT sequence. After merging the different parts (barcode, umi and transcript sequence), reads were filtered for quality (more than 80% of the sequence having a Sanger Phred+33 quality score > 33) using fastq_quality_filter -v -Q 33 -q 20 -p 80. Then the reads were assigned to each single cell by using the 8bp barcode and the fastx_barcode_splitter.pl tool script for a maximum of 2 mismatches. Reads assigned to each single cell were then trimmed again to retrieve the transcript sequence using fastx_trimmer.
Mapping was performed using Tophat2 to the mm10 mouse transcriptome and keeping the strand information with the following options : tophat -p 2 --library-type fr-firststrand --read-mismatches 5 --read-gap-length 5 --read-edit-dist 5 --no-coverage-search --segment-length 15 --transcriptome-index. Duplicated mapping reads were filtered out using the unique molecular barcodes as follows. First duplicated mapped reads were marked using picard-tools-1.79/MarkDuplicates.jar. Then the genomic position of the duplicated reads were extracted and for each of these positions, only reads having unique molecular identifiers were then kept. Reads that mapped to multiple positions were filtered out using samtools 0.1.19 flag 256. Finally reads were assigned to genes using htseq-count, biomart_mm10_gene.gff and the following options : -s yes -m intersection-nonempty. The script was modified in order to assign reads that overlapped in several genes to the one closest to a 3’ end.
Counts were normalized between cells by quantile normalization using the normalize.quantiles function in preprocessCore to account for differences in read depths between cells. However, there are inherent sampling biases that can occur when analyzing single cell RNAseq data which can cause some transcripts, particularly those expressed at low levels, to be undetected. These undetected events are known as ‘dropouts.’ Therefore, to account for these sampling biases, we calculated probabilities that a given transcript was unsampled (versus genuinely unexpressed) using the scde.failure.probability function in SCDE 28. “Confidence probabilities” were calculated as 1 – SCDE dropout probability. An event with a confidence p.value of less than 0.05 was considered a genuinely unexpressed event. Conversely, events with greater than 0.95 confidence values were considered significantly confident.
To determine how frequently individual genes were expressed in the MEC population, we calculated frequency of expression per gene as the number of MECs expressing a given gene (specifically, if >0 reads were detected for a given transcript in a given cell) divided by the total number of MECs; Aire WT and Aire KO frequencies were calculated independently. Similarly, we calculated mean counts from expressing cells to determine the transcriptional output of individual genes when that gene is expressed; therefore, we simply averaged non-zero counts per gene for Aire WT and Aire KO MECs, separately.
Gene set definitions
Aire-induced and -neutral gene lists used in many of our analyses were defined in Figure 1. Specifically, Aire-induced genes were those that were at least 2-fold higher in Aire WT versus Aire KO MECs at the population level. Aire-neutral genes were defined as those that did not differ more than 1.1-fold in Aire WT and Aire KO MECs.
To control for unrelated effects that could result simply from different levels of transcriptional output from different loci, we used expression-matched gene sets defined by scRNAseq data in many of our analyses. To this end, we selected genes that fell within indicated lower and upper bounds for second highest max read counts (that is, the number of counts per gene that was the second highest among all cells in that group) among our single cell data. We specifically did not use max read counts to avoid confounding, outlier events.
Simulation of intensity-frequency joined distributions
We aimed at testing the change, between Aire KO and WT MECs, in gene expression frequency versus the change in mean expression for Aire-induced genes,. To take into account the higher dropout probability observed for low expressed genes, we derived a null distribution for Aire-neutral genes of the changes in frequency that might result from changes in mean intensity in positive cells for Aire-induced gene: For each Aire-induced gene Gi, we randomly sampled 50 random Aire-neutral genes expressed at the same level as Gi in KO cells, and another 50 genes expressed at the same level as Gi in WT MECs. We then computed the average change in frequency for these 50 random pairs, and plotted it against their mean difference in expression (grey dots in Fig. S1).
Correlation and clustering analyses
We used a row standardized expression matrix weighted by the confidence of expression (1-dropout) as performed in earlier studies 24. Specifically, expression levels per gene were standardized among all cells using the scale function in R. For zero read events in the raw count data, the standardized expression value was multiplied by the expression confidence value (1-dropout) to correct for dropout biases.
Gene-gene and cell-cell Pearson correlations were performed with the cor function in R. To identify co-expressed gene networks and highly similar cell subsets, we clustered our expression data using affinity propagation based on Pearson correlations using the corSimMat function in apcluster 36). Affinity Popagation was useful in this case as it does not require a known number of clusters a priori.
To test the validity of the gene clusters observed in the Aire WT dataset, we shuffled our real data by randomly redistributing read counts per gene among Aire WT cells using the sample function in R per row of the data matrix. We shuffled the data and ran apcluster as before for 1000 permutations, storing cluster size and mean correlation per cluster for all permutations, using a custom script.
For cell clusters, affinity propagation using apcluster was used to determine cell groups based on the expression of Aire-induced genes located on chromosome 1. To determine whether the same cell groups were still highly similar based on the expression of Aire-induced genes from other chromosomes, we maintained the same order determined by our initial analysis using chromosome 1 genes and calculated cell-cell Pearson correlations based on Aire-induced genes from chromosomes 2 and 7.
We utilized t-SNE computation to visualize the cell-cell heterogeneity we observed in our Aire WT and KO MECs as a simple 2D representation. We calculated t-SNE components based on Pearson correlations of Aire-induced gene confidence probabilities using the tsne package 40.
Gene cluster chromosomal distances
To determine what genomic distances the components of the gene clusters spanned, we matched each gene per cluster with its most highly correlated partner (using cor function in R). Based on the TSS positions of the genes, each gene was designated as interchromosomal (located on different chromosomes), intrachromosomal (same chromosome, but >1Mb away), or local (same chromosome and <1Mb away) based on the distance to that gene’s most highly correlated partner.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs S. Mostafavi for key advice on the computational analysis, M. Anderson for the Aire-gfp line, K. Hattori, G. Buruzula, and K. Waraska for help with mice, sorting and sequencing. This work was supported by grant DK060027 from the NIH. MM was supported by an NIH Training Grant at Children’s Hospital in Pediatric Gastroenterology, DZ by a fellowship from the Boehringer Ingelheim Fonds. Data are available at NCBI repositories (SRR2038194-97, SRR2038206, SRR2038210, SRR2038212-13)
Footnotes
The authors have no conflicting financial interests.
REFERENCES
- 1.Peterson P, Org T, Rebane A. Transcriptional regulation by AIRE: molecular mechanisms of central tolerance. Nat. Rev. Immunol. 2008;8:948–957. doi: 10.1038/nri2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 3.Hubert FX, et al. Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood. 2011;118:2462–2472. doi: 10.1182/blood-2010-06-286393. [DOI] [PubMed] [Google Scholar]
- 4.Liston A, et al. Aire regulates negative selection of organ-specific T cells. Nat. Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 5.Anderson MS, et al. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23:227–239. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Malchow S, et al. Aire-dependent thymic development of tumor-associated regulatory T cells. Science. 2013;339:1219–1224. doi: 10.1126/science.1233913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waterfield M, et al. The transcriptional regulator Aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nat. Immunol. 2014;15:258–265. doi: 10.1038/ni.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Org T, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–376. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koh AS, et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl. Acad Sci U S. A. 2008;105:15878–15883. doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraud M, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A. 2012;109:535–540. doi: 10.1073/pnas.1119351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abramson J, Giraud M, Benoist C, Mathis D. Aire's partners in the molecular control of immunological tolerance. Cell. 2010;140:123–135. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 12.Gaetani M, et al. AIRE-PHD fingers are structural hubs to maintain the integrity of chromatin-associated interactome. Nucleic Acids Res. 2012;40:11756–11768. doi: 10.1093/nar/gks933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oven I, et al. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol. 2007;27:8815–8823. doi: 10.1128/MCB.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giraud M, et al. An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proc Natl Acad Sci U S A. 2014;111:1491–1496. doi: 10.1073/pnas.1323535111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villasenor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc Natl. Acad Sci U S. A. 2008;105:15854–15859. doi: 10.1073/pnas.0808069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taubert R, Schwendemann J, Kyewski B. Highly variable expression of tissue-restricted self-antigens in human thymus: implications for self-tolerance and autoimmunity. Eur. J Immunol. 2007;37:838–848. doi: 10.1002/eji.200636962. [DOI] [PubMed] [Google Scholar]
- 17.Derbinski J, et al. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl. Acad Sci U S. A. 2008;105:657–662. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto S, et al. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc Natl Acad Sci U S A. 2013;110:E3497–E3505. doi: 10.1073/pnas.1308311110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venanzi ES, Melamed R, Mathis D, Benoist C. The variable immunological self: genetic variation and nongenetic noise in Aire-regulated transcription. Proc Natl Acad Sci U S A. 2008;105:15860–15865. doi: 10.1073/pnas.0808070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 21.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 22.Ozbudak EM, et al. Regulation of noise in the expression of a single gene. Nat. Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 23.Kalmar T, et al. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS. Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shalek AK, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross IL, Browne CM, Hume DA. Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol. Cell Biol. 1994;72:177–185. doi: 10.1038/icb.1994.26. [DOI] [PubMed] [Google Scholar]
- 27.Wu AR, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat. Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner JM, et al. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science. 2008;321:843–847. doi: 10.1126/science.1159407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sansom SN, et al. Population and single cell genomics reveal the Aire-dependency, relief from Polycomb silencing and distribution of self-antigen expression in thymic epithelia. Genome Res. 2014 doi: 10.1101/gr.171645.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein L, et al. Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat. Med. 2000;6:56–61. doi: 10.1038/71540. [DOI] [PubMed] [Google Scholar]
- 32.Anderson AC, et al. High frequency of autoreactive myelin proteolipid protein-specific T cells in the periphery of naive mice: mechanisms of selection of the self-reactive repertoire. J Exp. Med. 2000;191:761–770. doi: 10.1084/jem.191.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keane P, Ceredig R, Seoighe C. Promiscuous mRNA splicing under the control of AIRE in medullary thymic epithelial cells. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu785. [DOI] [PubMed] [Google Scholar]
- 34.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Islam S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014;11:163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 36.Bodenhofer U, Kothmeier A, Hochreiter S. APCluster: an R package for affinity propagation clustering. Bioinformatics. 2011;27:2463–2464. doi: 10.1093/bioinformatics/btr406. [DOI] [PubMed] [Google Scholar]
- 37.Johnnidis JB, et al. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl. Acad Sci U S. A. 2005;102:7233–7238. doi: 10.1073/pnas.0502670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derbinski J, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl. Acad. Sci. U. S. A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Maaten L, Hinton G. Visualizing high-dimensional data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- 41.Gu H, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 42.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Org T, et al. AIRE activated tissue specific genes have histone modifications associated with inactive chromatin. Hum. Mol. Genet. 2009;18:4699–4710. doi: 10.1093/hmg/ddp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tao Y, et al. AIRE recruits multiple transcriptional components to specific genomic regions through tethering to nuclear matrix. Mol Immunol. 2006;43:335–345. doi: 10.1016/j.molimm.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 46.Gill J, Malin M, Hollander GA, Boyd R. Generation of a complete thymic microenvironment by MTS24(+) thymic epithelial cells. Nat. Immunol. 2002;3:635–642. doi: 10.1038/ni812. [DOI] [PubMed] [Google Scholar]
- 47.Zhao R, et al. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat Cell Biol. 2011;13:1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le BM, et al. The impact of negative selection on thymocyte migration in the medulla. Nat. Immunol. 2009;10:823–830. doi: 10.1038/ni.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merkenschlager M, Benoist C, Mathis D. Evidence for a single-niche model of positive selection. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11694–11698. doi: 10.1073/pnas.91.24.11694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.