Abstract
Gram-negative bacteria contain a double membrane which serves for both protection and for providing nutrients for viability. The outermost of these membranes is called the outer membrane (OM), and it contains a host of fully integrated membrane proteins which serve essential functions for the cell, including nutrient uptake, cell adhesion, cell signalling and waste export. For pathogenic strains, many of these outer membrane proteins (OMPs) also serve as virulence factors for nutrient scavenging and evasion of host defence mechanisms. OMPs are unique membrane proteins in that they have a β-barrel fold and can range in size from 8 to 26 strands, yet can still serve many different functions for the cell. Despite their essential roles in cell survival and virulence, the exact mechanism for the biogenesis of these OMPs into the OM has remained largely unknown. However, the past decade has witnessed significant progress towards unravelling the pathways and mechanisms necessary for moulding a nascent polypeptide into a functional OMP within the OM. Here, we will review some of these recent discoveries that have advanced our understanding of the biogenesis of OMPs in Gram-negative bacteria, starting with synthesis in the cytoplasm to folding and insertion into the OM.
Keywords: β-barrel membrane protein, outer membrane, protein folding, β-barrel assembly machinery complex, insertase, lateral gate
1. Introduction
Gram-negative bacteria are surrounded by both an inner membrane (IM) and outer membrane (OM). The OM is unique in its composition and asymmetrical distribution of lipids, with the inner leaflet containing phospholipids, whereas the outer leaflet is composed of lipopolysaccharide (LPS), a highly negatively charged molecule that protrudes into the bacterial environment [1]. This unusual membrane environment is home to lipoproteins and integral membrane proteins called outer membrane proteins (OMPs). As much as 3% of the Gram-negative bacterial genome may code for OMPs [2]. Every OMP is synthesized in the cytoplasm, a location and environment distinct from the OM. The bacterium must transport OMP precursors from their point of synthesis to their destination in the OM, and do so while preventing their aggregation and maintaining them in a competent state for membrane insertion. This is a daunting task. Not only do nascent OMPs need to traverse the hydrophobic IM, but also the aqueous and crowded periplasmic space [3]. Once OMPs arrive at the OM, they must be correctly inserted into this membrane and fold into their final functional state. The bacterium achieves these vital tasks by the efforts of soluble chaperones and membrane-embedded machines. The function of these machines is made all the more extraordinary given the lack of chemical energy in the periplasmic compartment and at the OM.
OMPs are integral membrane proteins which adopt a β-barrel architecture in the membrane with short loops between strands on the periplasmic side and large, extended loops on the extracellular side. To date, almost all structures of these type of proteins show an even number of β-strands arranged in an antiparallel pattern [4]. This structural feature contributes to their high stability in this membrane, helping them withstand the sometimes harsh and variable environment. OMPs are exposed to the outside of the bacterial cell and are the first line of contact between the bacterium and its surroundings. Given their key location, OMPs have many diverse roles, acting as adhesion factors in virulence, channels for the uptake of nutrients, siderophore receptors and enzymes such as proteases and lipases, to name but a few.
Despite the wide variety of functions that OMPs perform, only two OMPs have so far been found to be essential in Escherichia coli. The first is LptD, a 26-stranded β-barrel which forms a complex with the lipoprotein LptE and together they are responsible for the fundamental process of insertion of LPS into the outer leaflet of the OM, maintaining the asymmetry of the bilayer [5]. The second essential OMP in E. coli is BamA [6].
BamA is a component of the β-barrel assembly machinery (BAM) complex, a complex that acts to insert β-barrel proteins into the OM. BamA is a 16-stranded β-barrel with five polypeptide transport-associated (POTRA) domains that sit in the periplasm. The full-length structure has been solved by X-ray crystallography, encompassing the β-barrel domain and all five POTRAs from Neisseria gonorrhoeae [7], as well as N-terminal (NT) truncated versions from Haemophilus ducreyi [7] and E. coli [8,9]. BamA is the largest and most highly conserved component of the BAM complex, which, in E. coli, consists of five lipoproteins including components BamB–E, which will be discussed in §§2 and 4. Only BamA and BamD are essential in E. coli [10–13], but the presence of all five is needed for the most efficient insertion of OMP precursors. It is thought the BAM complex recognizes the C-terminal (CT) strand of OMPs (β-signal or motif) as the signal for the insertion of these precursors into the OM [14].
2. Outer membrane protein trafficking into the periplasm across the inner membrane
OMPs are synthesized, like all bacterial proteins, by ribosomes in the cytoplasm. They traverse the IM barrier in an unfolded state by passage through a small pore in the centre of the SecYEG translocon [15]. Precursors are delivered to the Sec translocase by one of two well-documented mechanisms: either post-translationally by the use of the SecAB chaperones which maintain the protein in an unfolded conformation [16], or co-translationally during their synthesis on the ribosome [17]. Proteins are targeted to the Sec translocon by an NT signal peptide, a tripartite motif which can be cleaved by periplasmic peptidases upon delivery to this compartment [18].
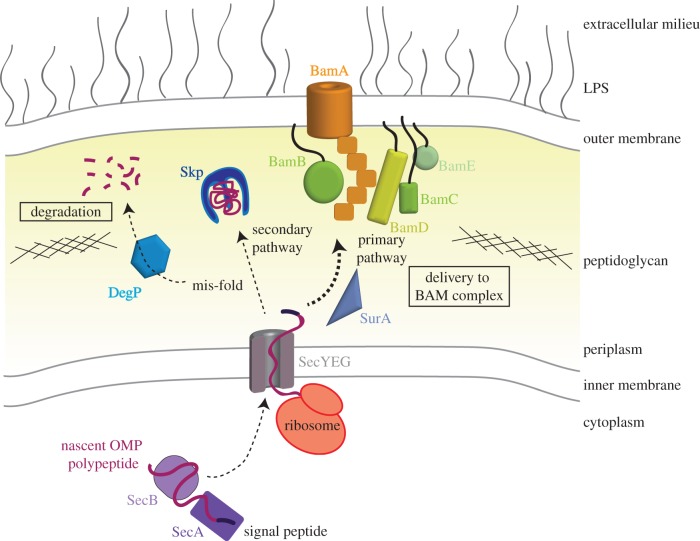
Upon emerging from the Sec translocon, the unfolded OMP encounters the periplasm, an aqueous environment in which they are prone to aggregation. To prevent their misfolding, the OMP precursor polypeptide chains interact with periplasmic chaperones. These soluble proteins then escort the nascent OMPs through the periplasmic space and deliver them to the BAM complex (figure 1). The BAM complex inserts the nascent polypeptide chains into the OM, a mechanism which is now being defined at the atomic level due to recent structural and functional studies on the membrane-embedded BamA component [7,19]. However, the shuttling and quality control pathways in the periplasm are much less well understood.
Figure 1.
The OMP journey to the outer membrane. Nascent OMPs (maroon) are synthesized in the bacterial cytoplasm and delivered by the SecA/SecB chaperones (purple) or by the ribosome (red) to the Sec translocase (grey) for passage through the IM. Once in the periplasm, nascent OMPs can be delivered to the BAM complex by the periplasmic chaperones SurA (slate) or Skp (blue). Any OMPs which misfold during their journey will be degraded by the protease DegP (light blue) to prevent aggregation.
Quality control for the biogenesis of OMPs involves not only chaperones that promote the correct folding and prevent misfolding, but also proteases which degrade any OMPs which fall off the folding pathway, so preventing their accumulation in the periplasm. Chaperones are generally defined as proteins which bind to unfolded, but not folded, precursor proteins. There are three main classes of periplasmic chaperones that have so far been identified. Firstly, there are disulfide bond catalysts [20], which are involved in the formation of disulfide bonds in the oxidizing environment of the periplasm. Secondly, peptidyl-prolyl cis–trans isomerases (PPIases), which catalyse the cis–trans isomerization of peptide bonds involving a proline residue. Finally, proteins with general chaperone activity were identified. Proteins from this third class are thought to function in the biogenesis of OMPs [21]. The three most thoroughly characterized and important periplasmic chaperones present in E. coli are Survival protein A (SurA), Seventeen kilodalton protein (Skp) and DegP.
Survival protein A (SurA) was first identified for its essential role in cell survival during stationary phase. It has since been shown to have general chaperone activity in the biogenesis of OMPs [22] and has been shown to contact BamA directly in vivo [23]. Seventeen kilodalton protein (Skp) has been shown to bind very tightly to unfolded, but not mature, OMPs [24] with dissociation constants in the nanomolar range. A trimer of Skp was shown to bind and form stoichiometric complexes with various OMPs [25], the formation of which prevents their aggregation [26], indicating a chaperone role in the periplasm.
DegP has homologues in almost all species of bacteria, as well as in eukaryotic chloroplasts and mitochondria [27]. Its presence in organelles which possess a functionally analogous double membrane and intermembrane space suggests DegP plays an important role in biogenesis of OMPs. Indeed, DegP is essential in Gram-negative bacteria under heat shock conditions [28]. It has been previously believed that DegP had both protease and chaperone activity, and the ability to switch between these two roles during a transition from low to high temperatures [29]. However, recent data suggest that DegP functions primarily as a protease to degrade misfolded and aggregated OMPs and has very little chaperone activity at both high and low temperatures [30]. The precise role performed by DegP is therefore still unclear.
Defining the specific roles and relative importance of the periplasmic chaperones is key in understanding the pathway OMPs take to the OM. Do subsets of the chaperones act in parallel, partially overlapping pathways, or do they work in a sequential manner, handing over the precursor proteins from one chaperone to the next? The relationships between each of the three major chaperones, SurA, Skp and DegP have been investigated by the use of mutant and depletion strains and the assessment of OMP assembly in the cell membrane.
Strains in which SurA has been mutated have previously shown a decrease in OMP density and the misfolding of many of the OMPs [31]. These mutant strains display typical phenotypes associated with OM defects, such as an increased sensitivity to detergents and antibiotics [31,32]. By contrast, mutation of DegP or Skp has minimal effects on OM composition [23], and skp strains are only marginally more sensitive to detergents and antibiotics than wt strains [23,33]. Double mutants of surA skp and surA degP are synthetically lethal, whereas skp degP cells are viable [34]. Taken together, these results led to a model in which SurA plays the major role in OMP biogenesis, and Skp/DegP act together in a separate, secondary pathway. However, complications arise when assessing the effects of mutation of SurA. Mutation of SurA induces the σE stress response, a response triggered by the presence of a large number of unfolded OMPs or heat shock [31]. This leads to downregulation of OMP expression levels [35] and an increase in the synthesis of molecular chaperones to assist with folding within the periplasm. A decrease in OMP synthesis will contribute to the observed decrease in the level of OMPs present in the surA strain [36,37] and will also lessen the burden on the Skp/DegP pathway by reducing the number of substrates for this pathway to handle.
In order to rigorously assess the relationships between these three chaperones on the global level, a differential proteomic approach has been used to assess the overall levels of every OMP present in chaperone depletion strains in E. coli. This is a powerful approach as it allows the level of every OMP to be assessed holistically, not just a select few as in previous studies. Alongside the proteomics, the levels of mRNAs of certain OMPs were measured in order to address whether the σE stress response is responsible in part for their depletion at the cell's surface. The proteomic experiments showed that SurA depletion does lead to a decrease in levels of OMPs, two of which (LptD and FhuA) were independent of the σE response [38]. These two OMPs may be specifically dependent on SurA for their biogenesis. By contrast, there were no OMPs whose levels were affected by the deletion of Skp, yet depleting SurA in a skp strain led to a significant decrease in the abundance of 14 β-barrel proteins [39]. These results lend significant support to the model in which most OMPs prefer the SurA pathway, yet can use the Skp/DegP pathway when SurA is absent.
There is, however, evidence that SurA does not have a primary role in the biogenesis of all OMPs. A greater dependence on other chaperones in the case of some specific OMPs in E. coli and in other bacterial species has been observed. In Shigella, Skp is necessary for biogenesis of the autotransporter IcsA, with the NT passenger domain being responsible for the Skp requirement [40,41]. It has been demonstrated that deletion of Skp in N. meningitidis decreases the levels of the OMPs PorA and PorB, an effect not seen in a surA mutant [41,42]. Additionally, despite the importance of the three major E. coli chaperones mentioned above, it is also clear that there are other chaperones that assist in the periplasm. FkpA, BepA and Spy have been shown to act in the OMP pathway. FkpA has been identified as a multicopy suppressor of the lethal phenotype seen in a surA skp strain, and may be functionally redundant to SurA under heat shock conditions [43]. In E. coli, it has been reported that Skp is involved in the assembly of LptD, but that the role performed by Skp may also be carried out by the chaperone FkpA or BamB [44], which mask the role of Skp when these two proteins remain present in skp deletion strains. BepA has been implicated in the biogenesis and degradation of the essential OMPs LptD and BamA, indicating a chaperone-like role [45]. Finally, overexpression of the periplasmic chaperone Spy has been shown to increase LptD assembly in a skp fkpA mutant [44].
Given the abundance of OMPs and periplasmic chaperones, bacteria may have evolved numerous back-up mechanisms to perform the essential tasks of OMP delivery to the OM. This may lead to the apparent redundancy of pathways, and species differences.
3. Outer membrane protein interactions with periplasmic chaperones
Cytoplasmic chaperones use the ATP present within the cell to drive cycles of their conformational change. These cycles are paired with precursor binding and release to help drive the precursor to its native fold [46]. The periplasm is devoid of energy in the form of electrochemical gradients or ATP. The periplasmic chaperones and the BAM complex insertase must therefore perform their roles independently of ATP-driven conformation changes. Several studies have been undertaken to try to understand the molecular nature of how each chaperone specifically interacts with the unfolded OMP precursors, and how they might prevent their misfolding and aggregation.
(a). Survival protein A
The crystal structure of SurA shows a four domain architecture, an NT domain, two PPIase domains and a CT domain (figure 2a). The NT and CT domains are structured around the first PPIase domain leaving the second PPIase domain as a satellite domain, distinct from the rest of the structure by a linker of approximately 30 Å [47]. The PPIase activity [48], shown to be present in only the second, distal domain [22], is not required for SurA function in vivo [22,31]. Whether the PPIase activity can be supplemented by other periplasmic chaperones is still unknown. Phage display experiments demonstrated that SurA has micromolar affinity for peptides that contain aromatic residues arranged in the repeating pattern Ar-X-Ar (with Ar as an aromatic residue and X being any residue) [49]. This pattern is seen in the CT sequence motif of OMP proteins, the so-called β-signal sequence which is thought to target them to the BAM complex for insertion into the OM [50]. The ability of SurA to bind these peptides indicates a potential mechanism for recognition of the precursor OMPs as they emerge into the periplasmic space.
Figure 2.
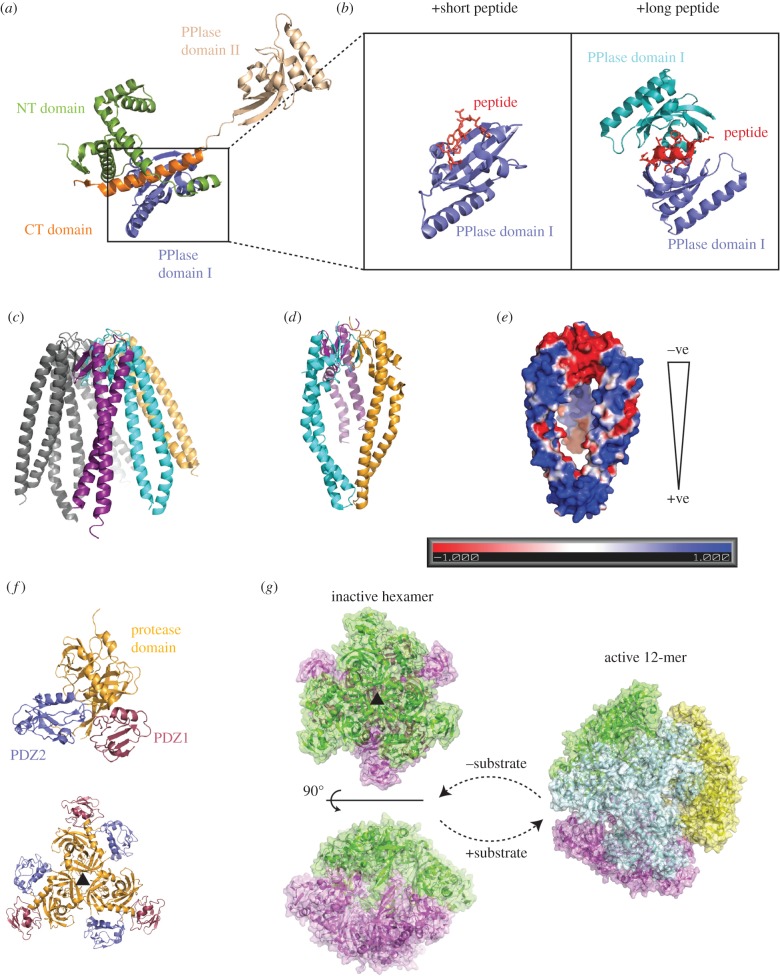
Structures of the major periplasmic chaperones involved in OMP biogenesis. (a) The overall structure of full-length SurA alone and in (b), the PPIase domain bound to two different length peptides. (c) The structure of prefoldin with three of the six monomers coloured. The structure of Skp with each monomer coloured as for prefoldin (d) and the electrostatic surface potential calculated by ABPS (e). (f) The trimeric building block of DegP binds to another trimer to form an inactive hexamer (g, left) in the absence of substrate. Upon substrate addition, the trimers rearrange and form a larger cage-like structures, such as a 12-mer (g, right).
Peptide selection and isothermal titration calorimetry suggested that the P1 domain is responsible for binding to OMP-like peptides [51]. The P1 domain is, however, not essential to SurA function, since a fusion of the NT and CT domains of SurA shows chaperone-like activity in vitro and is able to significantly complement a SurA deletion strain in vivo [22]. This may suggest the interactions mediated by the PPIase domain with OMP-like peptides allow SurA to interact specifically with OMP precursors, whereas chaperone activity lies in the NT and CT domains. Crystallographic analysis of constructs of SurA bound to two types of artificial peptides of differing lengths [51] indicated SurA interacts preferentially with aromatic residues independently of whether the peptide binds in an extended or a helical arrangement. A change in SurA conformation coupled with dimerization of the P1 domains was observed when SurA was bound to the longer peptide, suggesting SurA is able to bind a variety of aromatic sequences within OMPs by altering its overall domain structure (figure 2b).
Beyond binding to OMP precursors, what role does SurA perform? In contrast to Skp (discussed in §3b) spectroscopic and SDS-PAGE folding assays into liposomes found SurA does not directly facilitate OMP folding into lipids, indicating that in vitro SurA acts by another mechanism [52]. SurA is the only soluble chaperone that has been cross-linked in vivo to BamA [23], and addition of SurA increases the rate of OmpT folding in an in vitro reconstituted BAM system [53]. These data may indicate that SurA acts to deliver the OMPs across the periplasm specifically to the BAM complex, in an insertion-competent state which may increase their rate of insertion into the OM.
(b). Seventeen kilodalton protein
Arguably the most well-investigated OMP chaperone is Skp. Various crystallographic, NMR and spectroscopic studies have been used in the attempt to understand how this chaperone performs its role. The X-ray crystal structure of Skp shows a jellyfish-like umbrella architecture, with three pronged arms that form a central cavity below the base that comprises the oligomerization domain of the protein [26]. Skp has a strikingly similar shape to the hexameric eukaryotic cytoplasmic chaperone, prefoldin [54] (figure 2c). Prefoldin belongs to the holdase category of chaperones, a family which do not facilitate folding of the substrates themselves; instead they protect partially folded proteins from aggregation by binding substrates in their central cavity before passing them onto a downstream folding chaperone, such as CCT in the case of prefoldin [55].
Precursor OMPs, rich in aromatic and hydrophobic residues, can bind to the internal cavity of Skp, and so be shielded from the aqueous surroundings of the periplasm. However, the size of the cavity inside the Skp trimer (figure 2d) is not large enough to accommodate OMPs that have significant periplasmic or extracellular domains, such as OmpA. A site-directed fluorescence study and a subsequent combination of cross-linking and NMR relaxation methods determined that the β-barrel domain of OmpA is bound deep within the Skp cavity in an unfolded state, whereas the periplasmic domain residing outside the cavity was fully folded and able to move relatively independently of the Skp : β-barrel complex [56,57]. These data imply that Skp is not only selective for unfolded OMPs, but also for their hydrophobic β-barrel domain specifically. High-resolution NMR of Skp when bound to OmpX and the β-barrel domain of OmpA [58] showed the OMP is highly dynamic inside the Skp cage, and the high affinity interaction is caused by many transient and non-sequence interactions. The authors hypothesized that downstream in the folding pathway the sequence specific, and thus higher affinity, interaction between the β-signal and the BAM complex would allow the motif to be fished out of the conformational ensemble mediating the handover from Skp.
It appears that electrostatic interactions may play a role in the function of Skp. Firstly, OMPs and Skp no longer form stoichiometric complexes at pIs at which they are both no longer charged in vitro [25]. Secondly, the folding of OmpA into various phospholipid bilayers in the presence of Skp exhibits a strong dependence on the charge of the lipids, with Skp catalysing OMP folding into negatively charged membranes yet impeding folding into positively charged membranes [59]. The electrostatic potential of Skp exhibits a large dipole (figure 2e), with a net positive charge at the ‘head’ of the molecule. The data above suggest that the positively charged head region of Skp could target the precursor OMP to a negatively charged membrane. There are also large patches of negative charge present on the POTRA domains of BamA [60,61], which could be involved in attracting Skp. However, the importance of these observations to the mechanism in vivo is still to be investigated.
(c). Periplasmic serine endoprotease DegP
The protease DegP is responsible for the clearance of misfolded and aggregated OMPs from the periplasm. DegP is able to cleave peptides between pairs of hydrophobic residues, an activity that is induced by the presence of peptides containing these sequences [62].
The structure of DegP is composed of a protease domain and two PDZ domains, PDZ1 and PDZ2 [63]. DegP does not exist in a monomeric form, instead it forms a variety of higher order structures, hexamers, 12-mers and 24-mers, all composed of a common trimeric building block (figure 2f). The hexameric form is built from two trimers that are arranged face-to-face (figure 2g, left). In this structure, a regulatory loop from one molecule is inserted into the active site of a neighbouring protease domain, altering both the conformation and accessibility of the protease active site, rendering the protease inactive.
Binding misfolded substrates triggers higher order oligomerization of DegP. The 12-mers and 24-mers are formed by rearrangement of trimers into large and impressive cage-like structures [64,65] (figure 2g, right). The architecture of the cages suggests a method for DegP to adapt the size of the central cavity to the shape of the aggregated substrate. Access to the fully formed chamber is only possible through small holes, which may allow DegP to filter for unfolded substrates. It has also been speculated that this cavity could serve a dual purpose and act as a protective folding chamber for OMPs en route to the OM, and so would reconcile with a potential chaperone role of DegP [29].
The rearrangement of the trimer subunits upon forming the higher order oligomers also removes the regulatory loop from the protease active site, so switching on the protease activity. This switch suggests substrate binding followed by trimer rearrangement may be a reversible means of protease regulation. However, recent data suggest that cage assembly may not be a strict requirement of DegP protease regulation as activation of the core trimeric unit can be uncoupled from the higher order oligomerization. Mutants which are trapped in one oligomeric state in the presence or the absence of substrate still display proteolytic activity, and cells expressing these mutants can sustain high-temperature growth similar to wild-type [66].
4. Outer membrane protein delivery to the β-barrel assembly machinery complex for folding and insertion
Upon delivery of the nascent OMPs to the OM by the periplasmic chaperones SurA and Skp, a conserved multicomponent complex called the BAM complex then folds and inserts the OMPs into the OM [67–69]. Exactly how the BAM complex performs its insertase function remains unknown. However, the past decade has provided significant advances to our current understanding of how the BAM complex may function. The BAM complex was first identified in Neisseria meningitidis and then E. coli and was later determined to be composed of multiple components [10,12]. In E. coli, the BAM complex consists of BamA, an OMP itself, and four lipoproteins called BamB, BamC, BamD and BamE, which are all anchored to the inner leaflet of the OM via an NT post-translational lipid modification (figure 1) [67,68]. BamB and BamD interact directly with BamA, with BamC and BamE interacting directly with BamD [11,61,70]. BamA and BamD form the core of the complex and are the essential components required for cell viability [10]. The BAM complex can be isolated as two separate modules, a BamA/B module and a BamC/D/E module [53,71]. The composition of the BAM complex varies with bacterial species. For example, N. meningitidis appear to lack BamB [72]. Representative structures of all the components of the BAM complex have now been solved and have been instrumental in guiding ongoing research [7–9,60,73–82] (figure 3).
Figure 3.
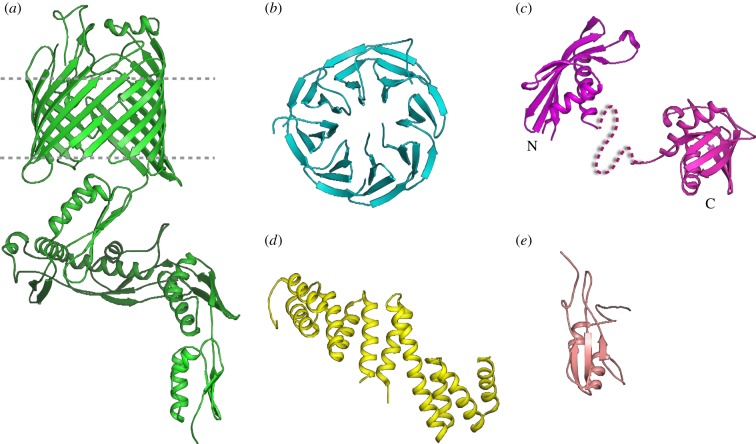
Structures of the components of the BAM complex. (a) The crystal structure of full-length BamA from Neisseria gonorrhoeae. (b) The crystal structure of BamB from E. coli. (c) The crystal structure of the two domains of BamC from E. coli. (d) The crystal structure of BamD from E. coli. (e) The NMR structure of BamE from E. coli.
BamA is an OMP itself and is the central and essential component of the BAM complex [10,67,68]. BamA contains a large periplasmic domain consisting of five POTRA repeat domains. These POTRA domains form the scaffold that mediates the interaction with BamB and BamD and are also thought to play a role in the initial recognition of substrate [61,83]. Studies have suggested that SurA may directly hand off nascent OMPs to the POTRA domains of BamA and that the POTRA domains may somehow assist in the folding or threading of the OMPs into the OM by a process known as β-augmentation [60,61,82]. The POTRA domains have been shown to be flexible, assuming bent and extended conformations, and sample open and closed conformations relative to the barrel domain [60,82]. Exactly what role the other components of the BAM complex serve in substrate recognition or interactions is not well understood, yet it is clear that they are important in the overall mechanism of OMP biogenesis. In particular, BamB has been suggested to either directly participate in the handoff of substrate to BamA or possibly serve as a scaffold, since knocking out either SurA or BamB was found to have very similar phenotypes on OMP assembly [83,84]. All OMPs are thought to contain something called a β-signal, which is typically the last strand of the β-barrel domain and pairs with the first strand in an antiparallel fashion to close the barrel domain. While the mechanism is still unknown, the β-signal is important for the proper recognition and folding of an OMP, and studies have indicated it may also play a role in species-specific recognition of nascent OMPs [50,85].
BamA is an OMP itself and contains a 16-stranded β-barrel domain which sits in the OM. With crystal structures now available, it is clear the barrel domain of BamA is unique in its shape and properties [7–9]. For instance, the first and last strands are only weakly associated, and molecular dynamics simulations have indicated that they can relatively easily be separated to form a direct portal into the OM for OMP insertion [7]. This, along with cross-linking studies which locked the barrel in a closed state rendering BamA inactive, has been the foundation for the idea that the barrel domain of BamA may undergo lateral opening to allow the newly formed OMPs to be inserted into the membrane [19]. The lateral opening would presumably mediate the direct insertion of the newly formed β-strands. An additional feature of the barrel domain was the identification of a putative substrate exit pore that sits above the site of lateral opening [9,19]. The exit pore has been suggested to allow the formation of extracellular loops, some of which can be nearly 100 residues large. Exactly how these features of the BamA structure contribute to the biogenesis of OMPs remains unknown; however, two proposed mechanisms will be discussed in §5.
Studies have shown that OMPs can spontaneously insert into liposomes much more efficiently in thinner membranes rather than thicker ones, indicating that the kinetic barrier is lessened [86–88]. Molecular dynamics simulations with BamA have shown that the barrel domain can mimic this behaviour on the membrane by acting as a catalyst to significantly thin and destabilize the local membrane in proximity of the lateral opening site [7]. This has been further supported by studies which have used electron microscopy to directly observe depressions or localized thinning in BamA proteoliposomes and by in vitro folding assays which show that OMP folding and insertion can be accelerated by preloading liposomes with BamA [87,89]. While these studies have focused on the role of BamA, it is clear from other studies that the other components of the BAM complex, particularly BamB and BamD, play an important role in vivo in the biogenesis of the OMPs. However, exactly what roles these other components play remains to be determined.
5. Folding and insertion into the outer membrane by the β-barrel assembly machinery-mediated complex
Despite having crystal structures of all the components of the BAM complex now available, including complex structures of BamAPOTRA/B and BamC/D, the exact mechanism for how OMPs are folded and inserted into the OM remains elusive. However, recent structural and function studies on BamA, which contains the only fully integrated membrane domain of the BAM complex, has assisted in narrowing down to two proposed mechanisms which have existed for a number of years, which we are now referring to the BamA-assisted and the BamA-budding models [69].
The BamA-assisted model is based on the observation and long-standing idea that some OMPs can spontaneously fold into membranes very efficiently in the absence of folding machinery. Folding and insertion are further increased in thinned or destabilized membranes, indicating that the changes in the membrane are yielding a reduction in the kinetic barrier for folding [86–88]. Molecular dynamics simulations have shown that the barrel domain of BamA is unique in that it can significantly affect the localized membrane in proximity to the lateral opening site, by both thinning the membrane and destabilizing the local lipids [7]. Therefore, the BamA-assisted model proposes that chaperones only need to escort the nascent OMPs to the BAM complex and in proximity to the locally ‘primed’ membrane where they can spontaneously insert into the membrane. Here, the BAM complex probably serves two functions, as a trafficking complex to localize the nascent OMPs to the OM, and as a catalyst to ‘prime’ the local membrane for insertion.
The BamA-budding model is based on the observation that some larger OMPs do not spontaneously fold into membranes efficiently and are inherently more complex by having more strands, large soluble domains and/or large extracellular loops. Here, chaperones would escort the nascent OMPs to the BAM complex where the POTRA domains of BamA, possibly along with other Bam components, would thread the new OMPs through the barrel domain of BamA, likely starting with a β-hairpin since the N- and C-termini of all OMPs in Gram-negative bacteria are found within the periplasm. The threading mechanism is supported by the fact that another member of this family, FhaC, is alone able to mediate the secretion of a substrate across a membrane [90]. But instead of secreting across a membrane as with FhaC, BamA ‘secretes’ substrate directly into the membrane. This threading, and/or possibly interaction with the β-signal of the new OMP, would then trigger the lateral opening of BamA, exposing unpaired β-strands which then serve as a template for forming new strands by β-augmentation with the new OMP, thereby forming a BamA : OMP intermediate. Initiation of folding would begin when the β-signal of the new OMP pairs with the first strand of BamA. Extracellular loops would be formed by the substrate exit pore and β-strands would continue to grow off the initial strands of the BamA : OMP intermediate, forming a ‘super-barrel’. To prevent this ‘super-barrel’ from growing too large and creating a ‘super-pore’ in the membrane, the newly forming OMP would eventually begin to ‘bud’ away from the BamA barrel, extending laterally into the membrane. Folding and insertion would then be terminated once the first strand of the new OMP arrives at the membrane, which would lead to strand exchange of the β-signal of the new OMP from BamA to its own first strand to close the barrel domain, thereby allowing diffusion away from BamA into the membrane. Like the BamA-assisted model, the BamA-budding model also relies on the BAM complex, via BamA, to ‘prime’ the membrane for new OMP insertion, allowing the energy from folding to be sufficient to fuel biogenesis into the OM. It should be noted that while evidence has been reported in support of the BamA-assisted model, little to no evidence has been reported yet directly addressing the BamA-budding model. However, the BamA-budding model satisfies the requirements for systematically inserting the amphipathic β-strands into the hydrophobic environment of the OM.
6. Summary and future directions
OMPs can only be found in the OMs of Gram-negative bacteria, mitochondria and chloroplasts. While the pathway for the biogenesis of these OMPs varies depending on the cell or organelle, the core of the machinery remains conserved. Here we have reviewed the pathways that are taken for the biogenesis of OMPs in Gram-negative bacteria starting with biosynthesis in the cytoplasm, transport across the IM and the periplasm, and ending with folding and insertion into the OM by the BAM complex. The overall picture for how OMPs are inserted and the routes they take are becoming more and more clear. However, the finer details for how they are folded and inserted and the role of the various components of the BAM complex remains to be elucidated. Exactly how the BAM complex recognizes substrates for insertion is still not known. Many in the field have expressed the hypothesis that folding of OMPs by the BAM complex may be initiated immediately after first being exported across the IM by the Sec translocon which is an attractive idea; however, this would directly conflict with the models for insertion since the β-signal has been proposed to play a role in substrate trafficking in the BamA-assisted model and a role in initiating substrate folding in the BamA-budding model. And what exactly is the role of the β-signal itself? This is not fully clear either. What about chaperones? How do they interact with substrate and does this offer any additional clues about whether substrates might be preformed to some degree within the periplasm? Lastly, while the structural studies have greatly advanced our understanding of the biogenesis of OMPs, more structures are needed, particularly of chaperone : OMP complexes and of the fully assembled BAM complex, both with and without substrate. The vast amount of work done over the past decade has laid a solid foundation for the next wave of discoveries, making this is an exciting time in the field since the structures of all the components have now been reported. The task at hand now is to determine how these pieces all come together to function as a finely tuned machine.
Acknowledgements
All figures were prepared using PyMOL (Schrodinger), Adobe Photoshop and Adobe Illustrator.
Authors' contributions
All authors contributed significantly to the design, research and writing of the manuscript.
Competing interests
We have no competing interests.
Funding
M.A.S. and N.N. are supported by the Department of Biological Sciences at Purdue University and by the National Institute of Allergy and Infectious Diseases (1K22AI113078-01). S.E.R. and S.K.B. are supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases. S.E.R. is also supported by a Sir Henry Wellcome Post-Doctoral Fellowship (103040/Z/13/Z).
References
- 1.Gan L, Chen S, Jensen GJ. 2008. Molecular organization of Gram-negative peptidoglycan. Proc. Natl Acad. Sci. USA 105, 18 953–18 957. ( 10.1073/pnas.0808035105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wimley WC. 2003. The versatile β-barrel membrane protein. Curr. Opin. Struct. Biol. 13, 404–411. ( 10.1016/S0959-440X(03)00099-X) [DOI] [PubMed] [Google Scholar]
- 3.Weiner JH, Li L. 2008. Proteome of the Escherichia coli envelope and technological challenges in membrane proteome analysis. Biochim. Biophys. Acta Biomembranes 1778, 1698–1713. ( 10.1016/j.bbamem.2007.07.020) [DOI] [PubMed] [Google Scholar]
- 4.Fairman JW, Noinaj N, Buchanan SK. 2011. The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr. Opin. Struct. Biol. 21, 523–531. ( 10.1016/j.sbi.2011.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu T, McCandlish AC, Gronenberg LS, Chng S-S, Silhavy TJ, Kahne D. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 103, 11 754–11 759. ( 10.1073/pnas.0604744103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genevrois S, Steeghs L, Roholl P, Letesson JJ, van der Ley P. 2003. The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane. EMBO J. 22, 1780–1789. ( 10.1093/emboj/cdg174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK. 2013. Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390. ( 10.1038/nature12521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albrecht R, Schutz M, Oberhettinger P, Faulstich M, Bermejo I, Rudel T, Diederichs K, Zeth K. 2014. Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr. Sect. D 70, 1779–1789. ( 10.1107/S1399004714007482) [DOI] [PubMed] [Google Scholar]
- 9.Ni D, et al. 2014. Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J. 28, 2677–2685. ( 10.1096/fj.13-248450) [DOI] [PubMed] [Google Scholar]
- 10.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245. ( 10.1016/j.cell.2005.02.015) [DOI] [PubMed] [Google Scholar]
- 11.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl Acad. Sci. USA 104, 6400–6405. ( 10.1073/pnas.0701579104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299, 262–265. ( 10.1126/science.1078973) [DOI] [PubMed] [Google Scholar]
- 13.Onufryk C, Crouch M-L, Fang FC, Gross CA. 2005. Characterization of six lipoproteins in the σE regulon. J. Bacteriol. 187, 4552–4561. ( 10.1128/jb.187.13.4552-4561.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Struyvé M, Moons M, Tommassen J. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218, 141–148. ( 10.1016/0022-2836(91)90880-F) [DOI] [PubMed] [Google Scholar]
- 15.Berg Bvd, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. 2004. X-ray structure of a protein-conducting channel. Nature 427, 36–44. ( 10.1038/nature02218) [DOI] [PubMed] [Google Scholar]
- 16.Economou A, Wickner W. 1994. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78, 835–843. ( 10.1016/S0092-8674(94)90582-7) [DOI] [PubMed] [Google Scholar]
- 17.Matlack KES, Mothes W, Rapoport TA. 1998. Protein translocation: tunnel vision. Cell 92, 381–390. ( 10.1016/S0092-8674(00)80930-7) [DOI] [PubMed] [Google Scholar]
- 18.Hegde RS, Bernstein HD. 2006. The surprising complexity of signal sequences. Trends Biochem. Sci. 31, 563–571. ( 10.1016/j.tibs.2006.08.004) [DOI] [PubMed] [Google Scholar]
- 19.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. 2014. Lateral opening and exit pore formation are required for BamA function. Structure 22, 1055–1062. ( 10.1016/j.str.2014.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamoto H, Bardwell JCA. 2004. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim. Biophys. Acta Mol. Cell Res. 1694, 111–119. ( 10.1016/j.bbamcr.2004.02.012) [DOI] [PubMed] [Google Scholar]
- 21.Duguay AR, Silhavy TJ. 2004. Quality control in the bacterial periplasm. Biochim. Biophys. Acta Mol. Cell Res. 1694, 121–134. ( 10.1016/j.bbamcr.2004.04.012) [DOI] [PubMed] [Google Scholar]
- 22.Behrens S, Maier R, de Cock H, Schmid FX, Gross CA. 2001. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 20, 285–294. ( 10.1093/emboj/20.1.285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sklar JG, Wu T, Kahne D, Silhavy TJ. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21, 2473–2484. ( 10.1101/gad.1581007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen R, Henning U. 1996. Aperiplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol. Microbiol. 19, 1287–1294. ( 10.1111/j.1365-2958.1996.tb02473.x) [DOI] [PubMed] [Google Scholar]
- 25.Qu J, Mayer C, Behrens S, Holst O, Kleinschmidt JH. 2007. The trimeric periplasmic chaperone Skp of Escherichia coli forms 1:1 complexes with outer membrane proteins via hydrophobic and electrostatic interactions. J. Mol. Biol. 374, 91–105. ( 10.1016/j.jmb.2007.09.020) [DOI] [PubMed] [Google Scholar]
- 26.Walton TA, Sousa MC. 2004. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol. Cell 15, 367–374. ( 10.1016/j.molcel.2004.07.023) [DOI] [PubMed] [Google Scholar]
- 27.Pallen MJ, Wren BW. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26, 209–221. ( 10.1046/j.1365-2958.1997.5601928.x) [DOI] [PubMed] [Google Scholar]
- 28.Strauch KL, Johnson K, Beckwith J. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171, 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spiess C, Beil A, Ehrmann M. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97, 339–347. ( 10.1016/S0092-8674(00)80743-6) [DOI] [PubMed] [Google Scholar]
- 30.Ge X, Wang R, Ma J, Liu Y, Ezemaduka AN, Chen PR, Fu X, Chang Z. 2014. DegP primarily functions as a protease for the biogenesis of beta-barrel outer membrane proteins in the Gram-negative bacterium Escherichia coli. FEBS J. 281, 1226–1240. ( 10.1111/febs.12701) [DOI] [PubMed] [Google Scholar]
- 31.Rouvière PE, Gross CA. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 10, 3170–3182. ( 10.1101/gad.10.24.3170) [DOI] [PubMed] [Google Scholar]
- 32.Lazar SW, Kolter R. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 178, 1770–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schäfer U, Beck K, Müller M. 1999. Skp, a molecular chaperone of Gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274, 24 567–24 574. ( 10.1074/jbc.274.35.24567) [DOI] [PubMed] [Google Scholar]
- 34.Rizzitello AE, Harper JR, Silhavy TJ. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183, 6794–6800. ( 10.1128/jb.183.23.6794-6800.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. 2005. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4, e2 ( 10.1371/journal.pbio.0040002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. 1993. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7, 2618–2628. ( 10.1101/gad.7.12b.2618) [DOI] [PubMed] [Google Scholar]
- 37.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113, 61–71. ( 10.1016/S0092-8674(03)00203-4) [DOI] [PubMed] [Google Scholar]
- 38.Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet J-F. 2009. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. PROTEOMICS 9, 2432–2443. ( 10.1002/pmic.200800794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denoncin K, Schwalm J, Vertommen D, Silhavy TJ, Collet J-F. 2012. Dissecting the Escherichia coli periplasmic chaperone network using differential proteomics. PROTEOMICS 12, 1391–1401. ( 10.1002/pmic.201100633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purdy GE, Fisher CR, Payne SM. 2007. IcsA surface presentation in Shigella flexneri requires the periplasmic chaperones DegP, Skp, and SurA. J. Bacteriol. 189, 5566–5573. ( 10.1128/jb.00483-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner JK, Heindl JE, Gray AN, Jain S, Goldberg MB. 2009. Contribution of the periplasmic chaperone Skp to efficient presentation of the autotransporter IcsA on the surface of Shigella flexneri. J. Bacteriol. 191, 815–821. ( 10.1128/jb.00989-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volokhina EB, Grijpstra J, Stork M, Schilders I, Tommassen J, Bos MP. 2011. Role of the periplasmic chaperones Skp, SurA, and DegQ in outer membrane protein biogenesis in Neisseria meningitidis. J. Bacteriol. 193, 1612–1621. ( 10.1128/jb.00532-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge X, Lyu Z-X, Liu Y, Wang R, Zhao XS, Fu X, Chang Z. 2014. Identification of FkpA as a key quality control factor for the biogenesis of outer membrane proteins under heat shock conditions. J. Bacteriol. 196, 672–680. ( 10.1128/jb.01069-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwalm J, Mahoney TF, Soltes GR, Silhavy TJ. 2013. Role for Skp in LptD assembly in Escherichia coli. J. Bacteriol. 195, 3734–3742. ( 10.1128/jb.00431-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narita S-i, Masui C, Suzuki T, Dohmae N, Akiyama Y. 2013. Protease homolog BepA (YfgC) promotes assembly and degradation of β-barrel membrane proteins in Escherichia coli. Proc. Natl Acad. Sci. USA 110, E3612–E3621. ( 10.1073/pnas.1312012110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horwich AL, Fenton WA. 2009. Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys. 42, 83–116. ( 10.1017/S0033583509004764) [DOI] [PubMed] [Google Scholar]
- 47.Bitto E, McKay DB. 2002. Crystallographic structure of SurA, a molecular chaperone that facilitates folding of outer membrane porins. Structure 10, 1489–1498. ( 10.1016/S0969-2126(02)00877-8) [DOI] [PubMed] [Google Scholar]
- 48.Dartigalongue C, Raina S. 1998. A new heat-shock gene, ppiD, encodes a peptidyl–prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 17, 3968–3980. ( 10.1093/emboj/17.14.3968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bitto E, McKay DB. 2003. The periplasmic molecular chaperone protein SurA binds a peptide motif that is characteristic of integral outer membrane proteins. J. Biol. Chem. 278, 49 316–49 322. ( 10.1074/jbc.M308853200) [DOI] [PubMed] [Google Scholar]
- 50.Robert V, Volokhina EB, Senf F, Bos MP, Gelder PV, Tommassen J. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4, e377 ( 10.1371/journal.pbio.0040377) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu X, Wang S, Hu Y-X, McKay DB. 2007. The periplasmic bacterial molecular chaperone SurA adapts its structure to bind peptides in different conformations to assert a sequence preference for aromatic residues. J. Mol. Biol. 373, 367–381. ( 10.1016/j.jmb.2007.07.069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMorran LM, Bartlett AI, Huysmans GHM, Radford SE, Brockwell DJ. 2013. Dissecting the effects of periplasmic chaperones on the in vitro folding of the outer membrane protein PagP. J. Mol. Biol. 425, 3178–3191. ( 10.1016/j.jmb.2013.06.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagan CL, Kim S, Kahne D. 2010. Reconstitution of outer membrane protein assembly from purified components. Science 328, 890–892. ( 10.1126/science.1188919) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korndorfer IP, Dommel MK, Skerra A. 2004. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat. Struct. Mol. Biol. 11, 1015–1020. ( 10.1038/nsmb828) [DOI] [PubMed] [Google Scholar]
- 55.Martín-Benito J, Boskovic J, Gómez-Puertas P, Carrascosa JL, Simons CT, Lewis SA, Bartolini F, Cowan NJ, Valpuesta JM. 2002. Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J. 21, 6377–6386. ( 10.1093/emboj/cdf640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qu J, Behrens-Kneip S, Holst O, Kleinschmidt JH. 2009. Binding regions of outer membrane protein A in complexes with the periplasmic chaperone Skp. A site-directed fluorescence study. Biochemistry 48, 4926–4936. ( 10.1021/bi9004039) [DOI] [PubMed] [Google Scholar]
- 57.Walton TA, Sandoval CM, Fowler CA, Pardi A, Sousa MC. 2009. The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc. Natl Acad. Sci. USA 106, 1772–1777. ( 10.1073/pnas.0809275106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burmann BM, Wang C, Hiller S. 2013. Conformation and dynamics of the periplasmic membrane-protein–chaperone complexes OmpX–Skp and tOmpA–Skp. Nat. Struct. Mol. Biol. 20, 1265–1272. ( 10.1038/nsmb.2677) [DOI] [PubMed] [Google Scholar]
- 59.Patel GJ, Behrens-Kneip S, Holst O, Kleinschmidt JH. 2009. The periplasmic chaperone Skp facilitates targeting, insertion, and folding of OmpA into lipid membranes with a negative membrane surface potential. Biochemistry 48, 10 235–10 245. ( 10.1021/bi901403c) [DOI] [PubMed] [Google Scholar]
- 60.Gatzeva-Topalova PZ, Walton TA, Sousa MC. 2008. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure 16, 1873–1881. ( 10.1016/j.str.2008.09.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964. ( 10.1126/science.1143993) [DOI] [PubMed] [Google Scholar]
- 62.Jones CH, Dexter P, Evans AK, Liu C, Hultgren SJ, Hruby DE. 2002. Escherichia coli DegP protease cleaves between paired hydrophobic residues in a natural substrate: the PapA pilin. J. Bacteriol. 184, 5762–5771. ( 10.1128/jb.184.20.5762-5771.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416, 455–459. ( 10.1038/416455a) [DOI] [PubMed] [Google Scholar]
- 64.Krojer T, Sawa J, Schafer E, Saibil HR, Ehrmann M, Clausen T. 2008. Structural basis for the regulated protease and chaperone function of DegP. Nature 453, 885–890. ( 10.1038/nature07004) [DOI] [PubMed] [Google Scholar]
- 65.Jiang J, Zhang X, Chen Y, Wu Y, Zhou ZH, Chang Z, Sui S-F. 2008. Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc. Natl Acad. Sci. USA 105, 11 939–11 944. ( 10.1073/pnas.0805464105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim S, Sauer RT. 2012. Cage assembly of DegP protease is not required for substrate-dependent regulation of proteolytic activity or high-temperature cell survival. Proc. Natl Acad. Sci. USA 109, 7263–7268. ( 10.1073/pnas.1204791109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ricci DP, Silhavy TJ. 2012. The Bam machine: a molecular cooper. Biochim. Biophys. Acta 1818, 1067–1084. ( 10.1016/j.bbamem.2011.08.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hagan CL, Silhavy TJ, Kahne D. 2011. beta-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80, 189–210. ( 10.1146/annurev-biochem-061408-144611) [DOI] [PubMed] [Google Scholar]
- 69.Noinaj N, Rollauer SE, Buchanan SK. 2015. The beta-barrel membrane protein insertase machinery from Gram-negative bacteria. Curr. Opin. Struct. Biol. 31, 35–42. ( 10.1016/j.sbi.2015.02.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61, 151–164. ( 10.1111/j.1365-2958.2006.05211.x) [DOI] [PubMed] [Google Scholar]
- 71.Anwari K, et al. 2010. A modular BAM complex in the outer membrane of the alpha-proteobacterium Caulobacter crescentus. PLoS ONE 5, e8619 ( 10.1371/journal.pone.0008619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Volokhina EB, Beckers F, Tommassen J, Bos MP. 2009. The beta-barrel outer membrane protein assembly complex of Neisseria meningitidis. J. Bacteriol. 191, 7074–7085. ( 10.1128/JB.00737-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noinaj N, Fairman JW, Buchanan SK. 2011. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. J. Mol. Biol. 407, 248–260. ( 10.1016/j.jmb.2011.01.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heuck A, Schleiffer A, Clausen T. 2011. Augmenting beta-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J. Mol. Biol. 406, 659–666. ( 10.1016/j.jmb.2011.01.002) [DOI] [PubMed] [Google Scholar]
- 75.Kim KH, Paetzel M. 2011. Crystal structure of Escherichia coli BamB, a lipoprotein component of the beta-barrel assembly machinery complex. J. Mol. Biol. 406, 667–678. ( 10.1016/j.jmb.2010.12.020) [DOI] [PubMed] [Google Scholar]
- 76.Kim KH, Aulakh S, Paetzel M. 2011. Crystal structure of beta-barrel assembly machinery BamCD protein complex. J. Biol. Chem. 286, 39 116–39 121. ( 10.1074/jbc.M111.298166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim KH, Aulakh S, Tan W, Paetzel M. 2011. Crystallographic analysis of the C-terminal domain of the Escherichia coli lipoprotein BamC. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1350–1358. ( 10.1107/S174430911103363X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dong C, Hou HF, Yang X, Shen YQ, Dong YH. 2012. Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly. Acta Crystallogr. D Biol. Crystallogr. 68, 95–101. ( 10.1107/S0907444911051031) [DOI] [PubMed] [Google Scholar]
- 79.Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. 2011. Crystal structure of BamD: an essential component of the beta-barrel assembly machinery of Gram-negative bacteria. J. Mol. Biol. 409, 348–357. ( 10.1016/j.jmb.2011.03.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim KH, Kang HS, Okon M, Escobar-Cabrera E, McIntosh LP, Paetzel M. 2011. Structural characterization of Escherichia coli BamE, a lipoprotein component of the beta-barrel assembly machinery complex. Biochemistry 50, 1081–1090. ( 10.1021/bi101659u) [DOI] [PubMed] [Google Scholar]
- 81.Knowles TJ, et al. 2011. Structure and function of BamE within the outer membrane and the beta-barrel assembly machine. EMBO Rep. 12, 123–128. ( 10.1038/embor.2010.202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. 2010. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure 18, 1492–1501. ( 10.1016/j.str.2010.08.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vuong P, Bennion D, Mantei J, Frost D, Misra R. 2008. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J. Bacteriol. 190, 1507–1517. ( 10.1128/JB.01477-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. 2007. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J. Bacteriol. 189, 446–454. ( 10.1128/JB.01103-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paramasivam N, Habeck M, Linke D. 2012. Is the C-terminal insertional signal in Gram-negative bacterial outer membrane proteins species-specific or not? BMC Genomics 13, 510 ( 10.1186/1471-2164-13-510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burgess NK, Dao TP, Stanley AM, Fleming KG. 2008. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J. Biol. Chem. 283, 26 748–26 758. ( 10.1074/jbc.M802754200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gessmann D, Chung YH, Danoff EJ, Plummer AM, Sandlin CW, Zaccai NR, Fleming KG. 2014. Outer membrane beta-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc. Natl Acad. Sci. USA 111, 5878–5883. ( 10.1073/pnas.1322473111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stanley AM, Fleming KG. 2008. The process of folding proteins into membranes: challenges and progress. Arch. Biochem. Biophys. 469, 46–66. ( 10.1016/j.abb.2007.09.024) [DOI] [PubMed] [Google Scholar]
- 89.Sinnige T, Weingarth M, Renault M, Baker L, Tommassen J, Baldus M. 2014. Solid-state NMR studies of full-length BamA in lipid bilayers suggest limited overall POTRA mobility. J. Mol. Biol. 426, 2009–2021. ( 10.1016/j.jmb.2014.02.007) [DOI] [PubMed] [Google Scholar]
- 90.Fan E, Fiedler S, Jacob-Dubuisson F, Muller M. 2012. Two-partner secretion of Gram-negative bacteria: a single beta-barrel protein enables transport across the outer membrane. J. Biol. Chem. 287, 2591–2599. ( 10.1074/jbc.M111.293068) [DOI] [PMC free article] [PubMed] [Google Scholar]