Abstract
Bacterial cells divide by targeting a transmembrane protein machine to the division site and regulating its assembly and disassembly so that cytokinesis occurs at the correct time in the cell cycle. The structure and dynamics of this machine (divisome) in bacterial model systems are coming more clearly into focus, thanks to incisive cell biology methods in combination with biochemical and genetic approaches. The main conserved structural element of the machine is the tubulin homologue FtsZ, which assembles into a circumferential ring at the division site that is stabilized and anchored to the inner surface of the cytoplasmic membrane by FtsZ-binding proteins. Once this ring is in place, it recruits a series of transmembrane proteins that ultimately trigger cytokinesis. This review will survey the methods used to characterize the structure of the bacterial divisome, focusing mainly on the Escherichia coli model system, as well as the challenges that remain. These methods include recent super-resolution microscopy, cryo-electron tomography and synthetic reconstitution.
Keywords: FtsZ, FtsA, cytokinesis, bacteria, septum
1. First sightings
Cell-division genes of Escherichia coli were originally designated fts genes, because thermosensitive mutants of these genes conferred a filamentous temperature-sensitive phenotype. At the non-permissive temperature (usually 42°C), fts mutant cells continue to elongate without dividing, forming filaments that can be longer than 150 µm in rich growth medium. As newborn E. coli cells are approximately 3 µm long by 1 µm wide, this represents at least 50 mass doublings, all the while continuing to extend the cell wall and membrane continuously, as well as replicate and segregate their nucleoids as visualized with DAPI staining. These multinucleate filaments indicated that the fts genes were involved specifically in cytokinesis, but electron microscopy of thin sections of E. coli (and many other bacteria) could never reveal any type of structure in normal dividing cells visible by negative staining. The isolation of min mutants of E. coli that made anucleate minicells at the cell pole suggested that the divisome-centring mechanism could be disrupted, but again no specific structures within min mutant cells could be discerned.
The first breakthrough came in 1991 when Erfei Bi and Joe Lutkenhaus used immunogold labelling to identify FtsZ, a product of the final fts gene in a highly conserved segment of cell wall and cell-division genes called the dcw (division and cell wall) cluster. Their work showed that gold particles clustered exclusively at the site of division at midcell in thin sections of E. coli, indicating that FtsZ was a structural component of the divisome and formed what is now called the Z ring [1]. Soon thereafter, new methods adapted for bacterial cell biology would set the stage for direct visualization of divisome structural proteins in intact cells.
The first of these methods, immunofluorescence microscopy (IFM), had been used for many years in eukaryotic cells but was first adapted for Bacillus subtilis cells by Liz Harry and Kit Pogliano in Rich Losick's laboratory. In addition to cell fixation and incubation with a primary antibody followed by a fluorescent secondary antibody, the key step involved cell permeabilization by limited lysozyme treatment, allowing the antibodies to enter the bacterial cells [2]. Using IFM, Arigoni et al. [3] found that the sporulation phosphatase SpoIIE localized to the asymmetric septum that separates the B. subtilis mother cell from the developing spore. IFM was quickly adapted for use in E. coli and other species, and was used to confirm that FtsZ strongly localized to the divisome at midcell, between segregated daughter nucleoids [4,5]. A series of papers from several groups then used IFM to demonstrate that other known products of fts genes, including FtsA, FtsQ, FtsW and FtsI, also localized sharply to division sites, provided that FtsZ was there [6–9]. Using a combination of the fts mutants and IFM, a new fts gene called ftsN was found to localize only when the other fts genes were intact, indicating that it needed preassembled ring components for recruitment and probably acted late in cell division [10]. This use of both cytology and genetics enabled the first rough understanding of a recruitment dependency, which in turn suggested a temporal hierarchy. This would have been very difficult to dissect with genetics alone.
At nearly the exact same time that IFM for bacteria was developed, green fluorescent protein (GFP) was rediscovered as a genetically encodable fluorescent tag [11]. As with IFM, eukaryotic cells were the first application of this exciting new technology, but the Losick laboratory soon adapted GFP for use in bacteria and used it to localize proteins in specific cell compartments during B. subtilis sporulation [12]. Shortly thereafter, our laboratory used FtsZ–GFP fusions to visualize FtsZ and FtsA for the first time in living cells [13]. With help from David Ehrhardt, who applied a computationally intensive method called deconvolution, or wide-field optical sectioning, originally developed by John Sedat's group [14], we reported the first three-dimensional view of the Z ring. GFP tagging now enabled the localization of any protein, without the need for specific antibodies or for cell fixation. This technology ushered in further breakthroughs that would be impossible with IFM alone, as described in §2.
However, tagging with fluorescent proteins comes with risks, including perturbation of the target protein by the tag [15,16]. Indeed, E. coli FtsZ tagged with GFP is not fully functional and as with other GFP-tagged proteins, artefacts can result, including dominant negative effects and spurious aggregation [13]. Fortunately, when FtsZ–GFP is produced at low levels in cells containing native FtsZ, it seems to behave consistently with IFM data. Moreover, other divisome proteins that bind directly to FtsZ, such as ZipA and ZapA in E. coli, and EzrA in B. subtilis, among others, can mimic FtsZ localization quite faithfully without perturbing the system much and thus serve as proxies for FtsZ [17–19].
2. Casting call: towards a low-resolution structure of the divisome
The use of IFM and fluorescent protein tags, ideally in combination, soon resulted in a fairly comprehensive understanding of the low-resolution structure of the divisome in several model organisms, including E. coli, B. subtilis and Caulobacter crescentus, and enabled researchers to demonstrate localization of similar homologues in other diverse bacterial species. As a result, we learned that many diverse bacteria, euryarchaea, plant plastids and even a few primitive mitochondria harbour Z rings, and that perturbing localization of the rings results in cytokinesis defects. Moreover, fluorescence microscopy has been crucial in screening for Z ring binding proteins [20–23], particularly as many of these proteins would be very hard to identify by genetic or biochemical methods because of transient interactions and/or modest phenotypes when inactivated.
To understand more about divisome structure, several avenues have been taken. One successful approach identified which divisome proteins could still properly localize after removal of other divisome proteins. As knockouts of divisome protein genes are generally lethal, these types of experiments have mostly been done in model systems that have sophisticated genetic tools such as thermosensitive mutants, regulatable promoters or suicide plasmids that can rapidly induce the depletion of a specific protein from the cell. Because filamentous cells of many rod-shaped bacteria such as E. coli can remain viable for several generations, IFM or fluorescent protein-tagged divisome proteins were then used to determine whether a given divisome protein depended on another. For example, in E. coli, the divisome proteins FtsQ, FtsL and FtsI fail to localize to the divisome in the absence of FtsZ, FtsA or FtsK but can localize in the absence of FtsN [24]. This demonstrates that FtsQ, FtsL and FtsI depend on FtsZ, FtsA and FtsK but not on FtsN, placing them in the middle of the recruitment order. This dependency order was roughly consistent with the actual timing of their detectable accumulation at the Z ring, with the more dependent proteins arriving later [25].
Another approach towards a divisome structure has been to define the binary protein–protein interactions within the divisome (figure 1). The most powerful genetic approach has been bacterial two-hybrid assays based on two fragments of the Bordetella pertussis adenylate cyclase [26]. These fragments normally do not interact efficiently, but if each is attached to a protein that interacts with another protein in vivo, they bring the two adenylate cyclase fragments together, and cAMP is synthesized. In a cya− cell incapable of synthesizing cAMP endogenously, the production of cAMP results in the induction of the lac operon, which in turn can be measured by colorimetric substrates such as X-gal or MacConkey dye (figure 1a). In addition, although adenylate cyclase is cytoplasmic, the enzyme itself is anchored to the membrane and thus provides a membrane-bound environment similar to most divisome proteins. The studies using this assay have identified numerous binary interactions between divisome proteins [27–29]. However, bacterial two-hybrid analysis, like any two-hybrid assay, is prone to false positives and negatives and should be used mainly as a genetic screen and to generate hypotheses that can be tested more rigorously using other methods (see below).
Figure 1.
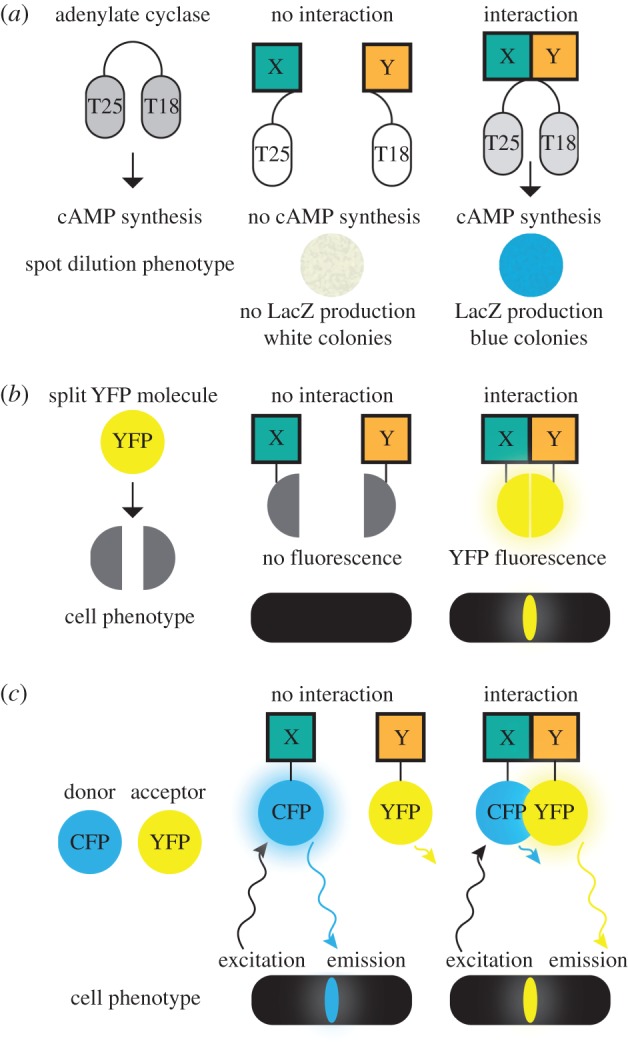
Schematic of protein–protein interaction methods. Bacterial two-hybrid assay: domains of adenylate cyclase are fused to two divisome proteins, X and Y (a). Interaction results in cAMP synthesis and production of β-glalctosidase (LacZ) in colonies. Bimolecular fluorescence complementation: two parts of a YFP molecule are fused to two divisome proteins, X and Y (b). Interaction results in YFP fluorescence at the divisome. Förster resonance energy transfer: CFP and YFP are fused to two divisome proteins, X and Y (c). Interaction results in an increase in fluorescence of the acceptor fluorophore, YFP.
In contrast to the sequential recruitment order of divisome proteins suggested by genetic experiments, the bacterial two-hybrid studies hint at a more integrated network of overlapping interactions. In addition to the known interactions among FtsZ and FtsA, ZipA and the Zap proteins in E. coli, some additional interactions among later recruits to the divisome have been confirmed using purified proteins. For example, it has been known for some time that FtsQ, FtsL and FtsB form a defined subcomplex [30], and the most attractive model to date is that they are in a 2 : 2 : 2 stoichiometry. As each of these proteins is present at approximately 200–400 molecules per cell [31], there may be approximately 100–200 FtsQLB complexes per divisome. More detailed interaction data for FtsQLB comes from photo-cross-linking of these proteins using unnatural amino acids placed in strategic positions [32]. The advantage of this method is that it can be done in growing cells in the natural membrane environment [33]. The cross-linking study pinpointed several regions within the periplasmic domain of FtsQ that are important for interacting with FtsB and/or FtsL. Another well-established interaction in the divisome is between FtsA and FtsN [34]. This was at first surprising, as FtsA is an early recruit to the Z ring, while FtsN is mainly a late recruit [10]. However, it was found that a small amount of FtsN molecules are recruited early by FtsA [35], which initiates a positive feedback loop that involves septal wall synthesis [36], consistent with FtsN's ability to contact peptidoglycan directly [37]. The result is a robust localization of FtsN to the divisome at late stages.
Two other methods have been used to measure protein–protein interactions and the location of those interactions in E. coli cells. One of these, called bimolecular fluorescence complementation or BIFC, relies on expressing the N-terminal and C-terminal fragments of GFP or derivatives such as yellow fluorescent protein (YFP). Normally, these fragments are non-fluorescent. However, if they are brought in close proximity via a strong protein–protein interaction, they will anneal to reconstitute the fluorescent protein and thus display a fluorescent pattern that mimics the interaction [38] (figure 1b). BIFC was used to confirm interactions between FtsZ and ZipA, as well as ZapB with itself [39]. In addition, BIFC identified new putative ZipA–ZapA and ZipA–ZapB interactions. BIFC has several advantages. It can pinpoint a close interaction in a live cell, and is sufficiently stable that even transient interactions can be marked. It can be used with standard microscope set-ups and requires no complex image processing. However, the disadvantage is that the reconstitution is relatively inefficient, so false negatives will be high. Moreover, the reconstitution of intact GFP/YFP is irreversible, making the BIFC fluorescence signal also irreversible; therefore, BIFC cannot be used to monitor off rates.
The other in situ method is Förster resonance energy transfer (FRET). FRET is the transfer of energy from a donor fluorophore, whose emission wavelength overlaps the excitation wavelength of a higher-wavelength receptor fluorophore. This transfer only occurs efficiently when the donor and acceptor fluorophores are in close contact, for example, between 1 and 8 nm apart. In this range, the FRET signal rises sharply with proximity. FRET can be measured as an increase in donor emission upon bleaching of the acceptor, as the donor retains the energy normally transferred to the acceptor when donor and acceptor are close together (figure 1c). Alternatively, a decrease in donor emission and/or an increase in acceptor emission can be detected. The power of microscopic FRET is that like BIFC, it can measure as well as pinpoint protein–protein interactions in living cells. As a result, it has been used for several applications in bacteria. For example, protein–protein interactions and dynamics within chemoreceptor clusters have been demonstrated by FRET, using protein fusions to the donor cyan fluorescent protein (CFP) and acceptor YFP [40,41].
In the first use of FRET to investigate protein–protein interactions in the bacterial divisome, Tanneke den Blaauwen's group measured interactions among FtsZ, ZapA, FtsQ, FtsW, FtsI and FtsN of E. coli [42]. Using the mKO fluorescent protein as donor and mCherry as acceptor fusion tags, they detected strong FRET between FtsZ and FtsZ, ZapA and ZapA, and between FtsZ and ZapA, as expected. They also found that the later divisome proteins FtsW, FtsI and FtsN interacted, and FtsN interacted with itself. More surprisingly, they measured significant FRET for ZapA–FtsN and ZapA–FtsI. These results, along with direct FtsA–FtsN interactions demonstrated biochemically [34] indicate that early and late divisome proteins, while temporally separated, are not necessarily spatially segregated; moreover, as mentioned above, a small proportion of late divisome protein FtsN arrives early [35]. Further biochemical studies will need to be done to confirm these FRET results, as it is always possible that FRET can lead to false positives if fluorescent proteins are in proximity but their attached divisome proteins do not directly interact. Likewise, false negatives can arise if two proteins interact but their attached fluorescent proteins are sterically hindered and/or not properly oriented for optimum FRET.
A powerful combination of FRET and fluorescence lifetime imaging, or FLIM, was used to assess the interactions between FtsZ and two positive spatial regulators of FtsZ positioning, SsgA and SsgB, in Streptomyces [43]. The advantage of FRET–FLIM over FRET alone is that measuring changes in the short lifetime of the fluorophores upon close interactions overcome some of the limitations of FRET, including background (non-FRET) donor fluorescence in the acceptor channel and vice versa.
3. Visualizing rapid dynamics of cell-division proteins and regulators in living cells
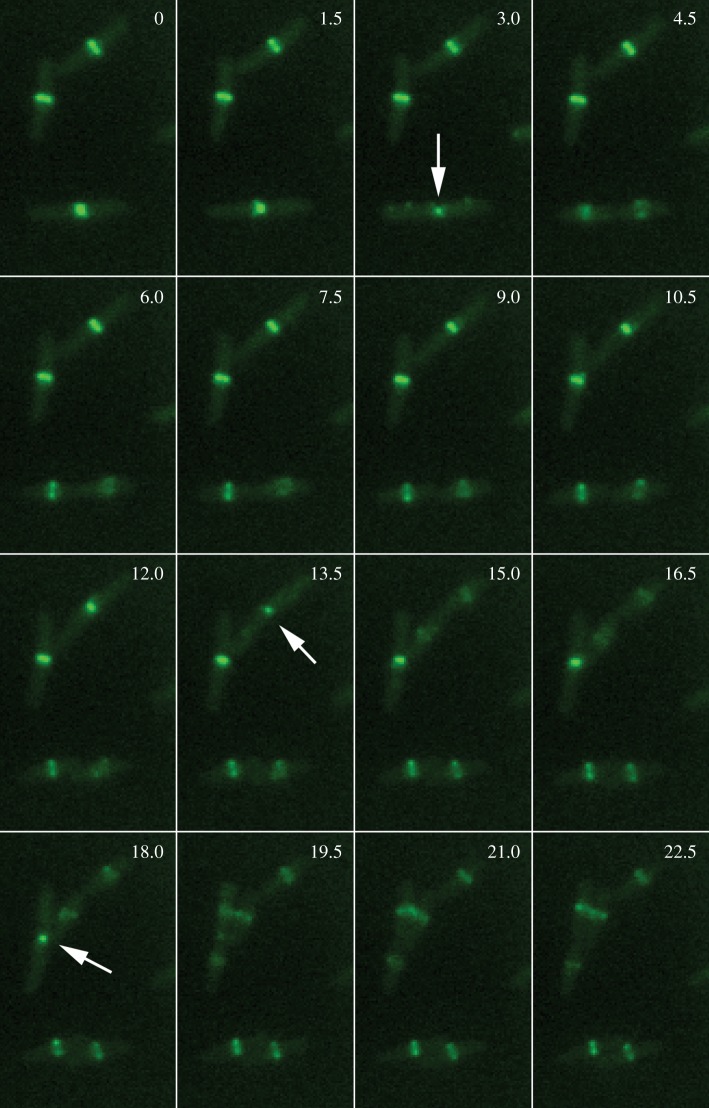
As mentioned in §2, the real power of tagging with fluorescent proteins lay in being able to follow protein dynamics in live cells over time. The first example of this was the demonstration that FtsZ–GFP in E. coli assembles into Z rings with some discontinuities but a clear lumen, which then constricted down to a diffraction-limited spot before disappearing [44] (figure 2). We now know that most of the FtsZ leaves the constriction zone prior to other Fts proteins such as FtsK and before complete septum formation [45,46]. During constriction, a portion of FtsZ–GFP can be seen to spiral out of the closing ring, and non-ring FtsZ (as FtsZ–GFP) seems to move rapidly in spiral patterns [47,48]. Such FtsZ spirals, also visible by IFM, have also been observed in B. subtilis cells during the transition from medial to asymmetric septation as part of the sporulation pathway, as well as during vegetative septation [49–51]. However, in other species, FtsZ often forms mobile foci prior to forming the Z ring [52,53].
Figure 2.
Dynamics of FtsZ in dividing E. coli cells. Low levels of FtsZ–GFP were produced in otherwise wild-type E. coli cells to monitor the localization of FtsZ during growth in time lapse on an agar pad under the microscope. Elapsed time is shown in minutes. Arrows point to constricting Z rings just prior to septum completion and cell separation for the three cells in the original field. Note that FtsZ–GFP fluorescence rapidly relocalized to future division sites.
Advanced microscopy techniques have also been used to measure how quickly FtsZ subunits turn over in what appears to be a fairly static Z ring structure. The Erickson laboratory used fluorescence recovery after photobleaching (FRAP) to monitor the time it took for a bleached section of a Z ring decorated with FtsZ–GFP molecules to become fluorescent again. As photobleaching with a laser permanently ablates the fluorescence of an FtsZ–GFP molecule in the ring, the only way that the bleached part of the ring can recover fluorescence is if unbleached FtsZ–GFP molecules from elsewhere in the cell (ring or non-ring FtsZ) replace the bleached molecules. If this recovery did not occur, it would indicate that the FtsZ–GFP molecules in the Z ring are static, with little turnover. However, recovery within the Z ring reproducibly occurs within approximately 10 s, demonstrating that there is rapid interchange between FtsZ molecules in the Z ring and elsewhere in the cell [54]. This turnover is slower in cells carrying a mutant of FtsZ that has lower GTPase activity, consistent with the connection between GTP hydrolysis and FtsZ assembly [55]. Similar rapid turnover dynamics have been observed in active Z rings of B. subtilis and Streptomyces, indicating that fast subunit exchange is a widely conserved property of FtsZ [43,54]. GFP-tagged FtsA had similar dynamics as FtsZ [56], consistent with their proposed coassembly at the membrane.
Time-lapse analyses of GFP fusions were essential for uncovering the mechanism of Z ring centring in E. coli. It has been known for some time that MinC and MinD were cell-division inhibitors and MinE somehow antagonized them to centre the Z ring [57]. But the breakthrough came when David Raskin in Piet de Boer's laboratory discovered that the bulk of GFP–MinD oscillated from pole to pole, forming large, transient arrays at the membrane [58]. GFP–MinC followed essentially the same pattern, whereas MinE–GFP formed a mobile ring at the edge of the MinD array [58–61]. These observations, in combination with biochemical characterization of Min protein activities, led to a plethora of mathematical models, still being refined today, that accurately simulate the oscillations and Z ring centring [62]. None of these models could have been developed or tested without the ability to monitor movement of Min proteins in cells in real time.
More evidence in support of Min protein oscillations and their effects on the Z ring came from more recent studies of GFP-tagged divisome protein dynamics. Although FtsZ by IFM or FtsZ–GFP in live cells forms a sharp band at midcell, a considerable amount of FtsZ, accounting for at least half of the total cellular FtsZ, is also present outside the ring [63]. It follows that other FtsZ-binding proteins such as ZipA, ZapA, ZapC and ZapD in E. coli will also be in significant numbers outside the ring. Interestingly, bulk FtsZ, ZipA and ZapA proteins, as visualized with fluorescent protein tags, all oscillate in response to Min oscillations [48,64,65]; when MinCD is at one pole, most of the divisome proteins are at the opposite pole because of the transient initiation of FtsZ assembly away from MinCD.
4. Super-resolution of the divisome: is the Z-ring a patchy toroid?
As the bacterial divisome is essentially invisible by electron microscopy, the advent of methods to resolve fluorescent tags at resolutions lower than the diffraction limit of light (approx. 200–300 nm) has led to significant insights into our understanding of its fine structure. Several new super-resolution technologies have paved the way (figure 3), although more refinements will surely follow [73]. One of these, three-dimensional structured illumination microscopy (3D-SIM), spatially patterns the excitation laser light to resolve two fluorescent point sources that are as little as 100 nm apart in the plane of the cover glass (XY), although the resolution is lower in the Z plane [74,75]. Although it requires a sophisticated instrument and software, the advantage of 3D-SIM is that it does not need to detect single molecules and thus can be used with fluorescent protein tags in multiple colours. The speed of this method allows the use of time-lapse imaging of live cells, although laser illumination during acquisition of multiple images needed to make the reconstruction can result in significant photobleaching, which can cause reconstruction artefacts. Another method that does not require single molecule detection but can achieve high resolution is stimulated emission depletion (STED), which also requires a specialized microscope/laser system [76]. STED can theoretically provide much higher resolution than 3D-SIM, because the technique is based on physically masking the out of focus light from a point source. However, this is technically challenging at present, and image acquisition is slow.
Figure 3.
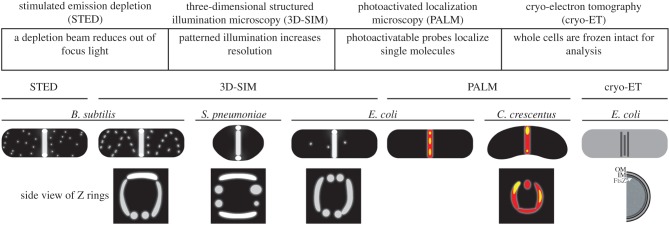
Examples of Z ring localization using different methods of super-resolution microscopy. The main principle of each method is listed in the table. The side views of Z rings represent a rotated image of the cell above them. For the PALM images, yellow areas represent areas of higher signal intensity. Cartoons from left to right were drawn based on Z-ring localization reported in [66–72].
The first super-resolution images of the bacterial divisome came from IFM of B. subtilis FtsZ (STED), B. subtilis cells producing FtsZ-GFP as the only copy of FtsZ in the cell (3D-SIM), and Staphylococcus aureus cells producing FtsZ-GFP as a label with native FtsZ present (3D-SIM) [66,68,76]. These studies revealed that FtsZ formed a patchy, discontinuous-appearing structure, with clear areas of low fluorescence around the ring circumference. Such discontinuities confirmed a pattern suggested previously by lower-resolution deconvolution studies [44], but the sharp 3D-SIM images removed any doubt. Moreover, the rapid image acquisition permitted by 3D-SIM showed that B. subtilis FtsZ–GFP patches in the ring moved around, suggesting that these assemblages move along the cytoplasmic membrane [68]. The unevenly distributed Z ring detected by this method suggested a model for how increased FtsZ density might contribute to force generation during constriction [68,77]. In E. coli, FtsA and ZipA also formed patches that often, but not always, colocalized [67]. This supports the idea that FtsZ and its membrane anchors assemble together in clusters. In agreement with previous conventional microscopic studies, the super-resolution images showed that FtsZ often formed a spiral structure at the midpoint of cells that were not yet actively constricting. 3D-SIM was also used to show the existence of double rings of a cell-division protein in B. subtilis. Unlike the Min system of E. coli, the B. subtilis Min system lacks MinE and instead MinCD are recruited, via another protein called MinJ, to rings that lie to either side of the Z ring. These rings, which are difficult to discern by conventional fluorescence microscopy but visualized distinctly by 3D-SIM, are formed by a protein called DivIVA, and recruitment of MinCD to these rings keeps the MinCD inhibitor away from the active Z ring while preventing spurious Z rings from assembling nearby [78].
Another class of super-resolution methods has been used that can locate single molecules to a very small region. Instead of patterning the excitation light to increase the resolution of a large number of fluorescent molecules, these methods, called PALM (photoactivated localization microscopy) or STORM (stochastic optical reconstruction microscopy) rely on fluorescent tags that are in the off (often dark) state until turned on by a specific excitation light. PALM and STORM were originally named for separate applications but describe essentially the same single molecule technology. The position of a single molecule can then be measured by calculating the centre of the diffraction-limited fluorescent spot that is spread among multiple pixels. Controlled photoactivation of hundreds of individual tagged molecules and calculation of their positions within the cell results in nanoscale resolution, better than anything currently achievable by 3D-SIM or STED. Using photoswitchable fluorescent proteins such as mEos fused to FtsZ for PALM of live cells, Jie Xiao's laboratory has found that the E. coli Z ring is approximately 110 nm wide and, in agreement with the other super-resolution studies, is a patchy assemblage of FtsZ filaments [79]. A 3D-PALM study of the Caulobacter Z ring using FtsZ-Dendra fusions supports these results, concluding that the Z ring in normal cells is patchy [72]. Interestingly, the PALM studies of E. coli indicate that FtsZ polymers within the Z ring are not all aligned circumferentially, but are often positioned at odd angles, sometimes parallel with the cell's long axis [69]. A study using fluorescence polarization found a similar lack of circumferential FtsZ polymer orientation in E. coli and B. subtilis, although the polymers were more aligned circumferentially in Caulobacter [80]. It is likely that there are significant species-specific differences in Z ring organization and ultrastructure, depending on the mode of cytokinesis and other factors. For example, most Gram-positive bacteria and many Gram-negative bacteria including E. coli form a clear septum, whereas some alpha-proteobacteria such as C. crescentus divide solely by constriction [81] and not by a combination of septation and constriction like E. coli. These differences are reflected not only in the intrinsic behaviour of FtsZ polymers, but also in the FtsZ-associated factors that modify FtsZ polymer shape, length and turnover.
In addition to refining what we know about FtsZ polymer density and orientation, super-resolution microscopy has also helped visualize how far the Z ring extends into the cell. In E. coli, the divisome consists of many periplasmic proteins such as the Tol/Pal complex, which extends all the way to the outer membrane and helps coordinate outer membrane invagination with Z ring constriction and formation of the division septum [82]. Combining molecular biology methods with PALM, the Xiao laboratory recently provided evidence that FtsZ, ZapA, ZapB and a protein called MatP form a protein network that extends from the cytoplasmic membrane inward to the chromosomal replication terminus, with which the DNA-binding protein MatP interacts directly. Deletion of the matP gene speeds the rate of Z ring constriction, indicating that the entire network is important for proper regulation of the divisome and normally helps to keep it in check [83]. A different type of super-resolution method using an astigmatic point spread function in combination with wide-field imaging found that the Z ring in Caulobacter is quite thick. Taken together, these results suggest that the Z ring is actually a patchy toroid instead of a ring [84,85].
Finally, super-resolution methods have been recently used to define divisome complexes in smaller bacterial cells that previously stretched the limits of conventional fluorescence microscopy. As mentioned above, 3D-SIM was used to visualize a patchy Z ring in S. aureus. In addition, 3D-SIM was used to localize divisome components of Streptococcus pneumoniae, which grow and divide as small ovococci. Using 3D-SIM for IFM, Malcolm Winkler's group found that penicillin-binding proteins Pbp1A and Pbp2x localize to division septa like FtsZ, but form rings clearly outside the Z ring and remain at maturing division septa even after FtsZ has migrated to future division sites [70]. Moreover, at a certain point in the S. pneumoniae division cycle, the Pbp1a ring is detectably outside the Pbp2x ring, suggesting that these proteins form concentric layers. Although it was possible to detect ZapA and FtsZ outside an inner ZapB ring in dividing E. coli cells by conventional resolution [86], clearly the new methods are poised to refine our views of divisome ultrastructure in intact cells. Moreover, 3D-SIM was recently used to distinguish the effects of MapZ, an FtsZ positioning regulator, on Z ring structure in S. pneumoniae [87].
5. Viewing native divisome structures by cryo-electron tomography
Yet another relatively new and powerful microscopic method has made inroads in understanding nanoscale protein assembly structure in bacteria: cryo-electron tomography (cryo-ET) (figure 3). Unlike conventional negative-stain transmission EM, for which cells are fixed, often sliced into thin sections, and then stained with uranyl acetate to reveal protein structures, specimens for whole-cell cryo-ET are plunge-frozen without further staining. The resulting images are low, by contrast, but can be computationally enhanced, and do not suffer from fixation, labelling or staining artefacts. Although the small size of bacteria is a disadvantage for most light microscopy, thin cells are actually required for sufficient image contrast. As a result, species that generate thin cells, such as Rhodobacter sphaeroides and C. crescentus, make it possible to visualize protein complexes if they are sufficiently concentrated. E. coli and other larger rod-shaped cells are difficult to image because they are too thick, although tiny minicells of E. coli and other species have recently been exploited to visualize surface protein assemblies [88–90].
Caulobacter crescentus is particularly suitable for cryo-ET analysis of the divisome as it divides solely by constriction, thus making a very thin bridge between daughter cells that is optimal for this technique. An initial cryo-ET study of the C. crescentus Z ring visualized long polymers of presumably FtsZ, which were periodically anchored to the cytoplasmic membrane [91]. Such an arrangement is consistent with known interactions between FtsZ and its membrane anchor proteins such as FtsA (ZipA is absent in C. crescentus), which are lower in abundance than FtsZ and should periodically tether FtsZ polymers to the membrane. A more recent cryo-ET study also visualized long polymers, and more controls were done that suggested these polymers were FtsZ itself [71]. A major conclusion of this study was that the Z ring is comprised at least partially of continuous long FtsZ polymers, which seems to be at odds with the patchy nature of Z rings visualized by 3D-SIM and PALM. It is possible that some long FtsZ polymers or polymer bundles in the Z ring might be difficult to detect by 3D-SIM if they are sparse (although they would be easier to detect with PALM). As mentioned in §1, the lack of complete function of FtsZ fusions to fluorescent proteins could result in these fusions not assembling into continuous polymers in vivo; however, cells producing native FtsZ also displayed patchy Z rings by IFM, ruling this out as a significant problem [67,79]. Future studies with different high resolution methods, including correlated light and cryo-EM that can match a structure with a specific protein or proteins [92], will be needed to clarify these results.
6. Reconstitution of the divisome on membranes in vitro
Perhaps the most striking advance in the field of bacterial cell division has been the recent successes in reconstituting parts of the divisome on membranes. In this ‘bottom-up’ approach, individual proteins can be manipulated, along with cofactors, to understand mechanistic details that are too difficult to parse in a whole-cell system. There are three notable examples of reconstitution with a cell-division phenotype. The first is the interaction between FtsZ and liposomes, investigated by Harold Erickson's group. Initially using a version of FtsZ fused to an artificial membrane anchor, they found that FtsZ polymers could assemble into rings inside liposomes and pinch them in the presence of GTP [93]. More recently, they found that purified FtsZ together with FtsA*, a variant of FtsA that can divide an E. coli cell in the absence of ZipA, could form a ring and divide a small proportion of liposomes to apparent completion in the presence of GTP for FtsZ and ATP for FtsA* [94]. These experiments suggested that FtsZ and FtsA were sufficient to exert a significant inward pinching force on a membrane and could be doing so in the cell. Cryo-EM of inside-out Z ring complexes on liposomes displayed long parallel bundled filaments on the membrane [71,95], supporting the in vivo cryo-ET data.
The second type of reconstitution comes from Martin Loose and Tim Mitchison, who investigated the interplay between purified FtsA and FtsZ on supported lipid bilayers (SLBs) [96]. By placing FtsA on top of an SLB and then adding a fluorescently tagged FtsZ, they found that these two proteins self-organized into dynamic polymer networks upon addition of GTP and ATP [97]. These networks often organized into ring-like structures on the flat surface of the membranes, indicating that curved polymers of both FtsZ and FtsA are favoured. The nature of the dynamics suggested that FtsZ polymers were moving by treadmilling, which can be described as subunits adding onto the front of a polarized polymer while simultaneously sloughing off the rear. This type of dynamics would be analogous to the treadmilling observed in eukaryotic actin filaments and microtubules.
The third notable example of reconstitution, from Petra Schwille's group, is the use of SLBs to mimic the size and shape of bacterial cell membranes. Addition of purified MinD and MinE labelled with fluorescent tags resulted in the formation of migrating waves on flat SLBs, with MinE lagging behind MinD just like in the cell [98]. More recently, SLBs have been fashioned in bathtub-like containers. When MinE and MinD are added to these, they oscillate from one end to the other [99]. Most impressively, when MinC and FtsZ are added to the mix, FtsZ ends up being positioned at midcell. Although there are clearly other regulatory factors involved in Z ring positioning, these reductionist experiments show that the basic patterns of cytokinesis can be reconstituted in vitro from a minimal set of components. Maxicells, which are E. coli cells devoid of chromosomes, also contain an oscillating Min system and a centred Z ring, supporting the idea that cell membrane geometry can position a Z ring without the need of the bacterial nucleoid [100,101].
7. Concluding remarks
The recent advent of super-resolution imaging methods has made a significant impact on the field of bacteriology by increasing the precision with which protein localization can be viewed in the smallest cells. Combining the power of super-resolution microscopy methods with genetic and biochemical approaches will further our understanding of how the divisome functions to divide bacterial cells and shed light on the protein–protein interactions that regulate this process.
Authors' contributions
W.M. and V.W.R. wrote the paper.
Competing interests
We have no competing interests.
Funding
W.M. and V.W.R. were funded by grant R01-GM61074 from the National Institutes of Health, and V.W.R. was additionally funded by the University of Texas Graduate School of Biomedical Sciences.
References
- 1.Bi E, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164. ( 10.1038/354161a0) [DOI] [PubMed] [Google Scholar]
- 2.Harry EJ, Pogliano K, Losick R. 1995. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 177, 3386–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arigoni F, Pogliano K, Webb CD, Stragier P, Losick R. 1995. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science 270, 637–640. ( 10.1126/science.270.5236.637) [DOI] [PubMed] [Google Scholar]
- 4.Levin P, Losick R. 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 10, 478–488. ( 10.1101/gad.10.4.478) [DOI] [PubMed] [Google Scholar]
- 5.Addinall SG, Bi E, Lutkenhaus J. 1996. FtsZ ring formation in fts mutants. J. Bacteriol. 178, 3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss DS, Pogliano K, Carson M, Guzman L-M, Fraipont C, Nguyen-Distéche M, Losick R, Beckwith J. 1997. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol. Microbiol. 25, 671–681. ( 10.1046/j.1365-2958.1997.5041869.x) [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Khattar MK, Donachie WD, Lutkenhaus J. 1998. FtsI and FtsW are localized to the septum in Escherichia coli. J. Bacteriol. 180, 2810–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buddelmeijer N, Aarsman ME, Kolk AH, Vicente M, Nanninga N. 1998. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J. Bacteriol. 180, 6107–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Addinall SG, Lutkenhaus J. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178, 7167–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Addinall SG, Cao C, Lutkenhaus J. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25, 303–309. ( 10.1046/j.1365-2958.1997.4641833.x) [DOI] [PubMed] [Google Scholar]
- 11.Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher DC. 1994. Green fluorescent protein as a marker for gene expression. Science 263, 802–805. ( 10.1126/science.8303295) [DOI] [PubMed] [Google Scholar]
- 12.Webb CD, Decatur A, Teleman A, Losick R. 1995. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J. Bacteriol. 177, 5906–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma X, Ehrhardt DW, Margolin W. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc. Natl Acad. Sci. USA 93, 12 998–13 003. ( 10.1073/pnas.93.23.12998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agard DA, Hiraoka Y, Shaw P, Sedat JW. 1989. Fluorescence microscopy in three dimensions. Methods Cell Biol. 30, 353–377. ( 10.1016/S0091-679X(08)60986-3) [DOI] [PubMed] [Google Scholar]
- 15.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. 2012. Segregation of molecules at cell division reveals native protein localization. Nat. Methods 9, 480–482. ( 10.1038/nmeth.1955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margolin W. 2012. The price of tags in protein localization studies. J. Bacteriol. 194, 6369–6371. ( 10.1128/JB.01640-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556. ( 10.1101/gad.1014102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haeusser DP, Schwartz RL, Smith AM, Oates ME, Levin PA. 2004. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol. Microbiol. 52, 801–814. ( 10.1111/j.1365-2958.2004.04016.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hale CA, de Boer PA. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185. ( 10.1016/S0092-8674(00)81838-3) [DOI] [PubMed] [Google Scholar]
- 20.Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. 2012. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol. 194, 3189–3198. ( 10.1128/JB.00176-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. 2011. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J. Bacteriol. 193, 1405–1413. ( 10.1128/JB.01258-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goley ED, Dye NA, Werner JN, Gitai Z, Shapiro L. 2010. Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol. Cell 39, 975–987. ( 10.1016/j.molcel.2010.08.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hale CA, Shiomi D, Liu B, Bernhardt TG, Margolin W, Niki H, de Boer PA. 2011. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J. Bacteriol. 193, 1393–1404. ( 10.1128/JB.01245-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goehring NW, Beckwith J. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15, 514–526. ( 10.1016/j.cub.2005.06.038) [DOI] [PubMed] [Google Scholar]
- 25.Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55, 1631–1645. ( 10.1111/j.1365-2958.2005.04502.x) [DOI] [PubMed] [Google Scholar]
- 26.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl Acad. Sci. USA 95, 5752–5756. ( 10.1073/pnas.95.10.5752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187, 2233–2243. ( 10.1128/JB.187.7.2233-2243.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Lallo G, Fagioli M, Barionovi D, Ghelardini P, Paolozzi L. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149, 3353–3359. ( 10.1099/mic.0.26580-0) [DOI] [PubMed] [Google Scholar]
- 29.Maggi S, Massidda O, Luzi G, Fadda D, Paolozzi L, Ghelardini P. 2008. Division protein interaction web: identification of a phylogenetically conserved common interactome between Streptococcus pneumoniae and Escherichia coli. Microbiology 154, 3042–3052. ( 10.1099/mic.0.2008/018697-0) [DOI] [PubMed] [Google Scholar]
- 30.Buddelmeijer N, Beckwith J. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52, 1315–1327. ( 10.1111/j.1365-2958.2004.04044.x) [DOI] [PubMed] [Google Scholar]
- 31.Li G-W, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635. ( 10.1016/j.cell.2014.02.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van den Berg van Saparoea HB, Glas M, Vernooij IGWH, Bitter W, den Blaauwen T, Luirink J. 2013. Fine-mapping the contact sites of the Escherichia coli cell division proteins FtsB and FtsL on the FtsQ protein. J. Biol. Chem. 288, 24 340–24 350. ( 10.1074/jbc.M113.485888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin JW, Martin AB, King DS, Wang L, Schultz PG. 2002. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl Acad. Sci. USA 99, 11 020–11 024. ( 10.1073/pnas.172226299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Busiek KK, Eraso JM, Wang Y, Margolin W. 2012. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J. Bacteriol. 194, 1989–2000. ( 10.1128/JB.06683-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busiek KK, Margolin W. 2014. A role for FtsA in SPOR-independent localization of the essential Escherichia coli cell division protein FtsN. Mol. Microbiol. 92, 1212–1226. ( 10.1111/mmi.12623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerding MA, Liu B, Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. 2009. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J. Bacteriol. 191, 7383–7401. ( 10.1128/JB.00811-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ursinus A, van den Ent F, Brechtel S, de Pedro M, Holtje JV, Löwe J, Vollmer W. 2004. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J. Bacteriol. 186, 6728–6737. ( 10.1128/JB.186.20.6728-6737.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu CD, Chinenov Y, Kerppola TK. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798. ( 10.1016/S1097-2765(02)00496-3) [DOI] [PubMed] [Google Scholar]
- 39.Pazos M, Natale P, Margolin W, Vicente M. 2013. Interactions among the early Escherichia coli divisome proteins revealed by bimolecular fluorescence complementation. Environ. Microbiol. 15, 3282–3291. ( 10.1111/1462-2920.12225) [DOI] [PubMed] [Google Scholar]
- 40.Sourjik V, Berg HC. 2002. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA 99, 12 669–12 674. ( 10.1073/pnas.192463199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sourjik V, Vaknin A, Shimizu TS, Berg HC. 2007. In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol. 423, 365–391. ( 10.1016/S0076-6879(07)23017-4) [DOI] [PubMed] [Google Scholar]
- 42.Alexeeva S, Gadella JTW, Verheul J, Verhoeven GS, den Blaauwen T. 2010. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol. Microbiol. 77, 384–398. ( 10.1111/j.1365-2958.2010.07211.x) [DOI] [PubMed] [Google Scholar]
- 43.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 25, 89–99. ( 10.1101/gad.600211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Q, Margolin W. 1998. FtsZ dynamics during the cell division cycle of live Escherichia coli. J. Bacteriol. 180, 2050–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Söderström B, Skoog K, Blom H, Weiss DS, von Heijne G, Daley DO. 2014. Disassembly of the divisome in Escherichia coli: evidence that FtsZ dissociates before compartmentalization. Mol. Microbiol. 92, 1–9. ( 10.1111/mmi.12534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Possoz C, Sherratt DJ. 2005. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 19, 2367–2377. ( 10.1101/gad.345305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu L, Yu J. 2008. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys. J. 95, 2009–2016. ( 10.1529/biophysj.108.128751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thanedar S, Margolin W. 2004. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol. 14, 1167–1173. ( 10.1016/j.cub.2004.06.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peters PC, Migocki MD, Thoni C, Harry EJ. 2007. A new assembly pathway for the cytokinetic Z ring from a dynamic helical structure in vegetatively growing cells of Bacillus subtilis. Mol. Microbiol. 64, 487–499. ( 10.1111/j.1365-2958.2007.05673.x) [DOI] [PubMed] [Google Scholar]
- 50.Michie KA, Monahan LG, Beech PL, Harry EJ. 2006. Trapping of a spiral-like intermediate of the bacterial cytokinetic protein FtsZ. J. Bacteriol. 188, 1680–1690. ( 10.1128/JB.188.5.1680-1690.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ben-Yehuda S, Losick R. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109, 257–266. ( 10.1016/S0092-8674(02)00698-0) [DOI] [PubMed] [Google Scholar]
- 52.Chiu SW, Roberts MA, Leake MC, Armitage JP. 2013. Positioning of chemosensory proteins and FtsZ through the Rhodobacter sphaeroides cell cycle. Mol. Microbiol. 90, 322–337. [DOI] [PubMed] [Google Scholar]
- 53.Quardokus E, Din N, Brun YV. 1996. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc. Natl Acad. Sci. USA 93, 6314–6319. ( 10.1073/pnas.93.13.6314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson DE, Gueiros-Filho FJ, Erickson HP. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186, 5775–5781. ( 10.1128/JB.186.17.5775-5781.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukherjee A, Lutkenhaus J. 1998. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17, 462–469. ( 10.1093/emboj/17.2.462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geissler B, Shiomi D, Margolin W. 2007. The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology 153, 814–825. ( 10.1099/mic.0.2006/001834-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Boer PAJ, Crossley RE, Rothfield LI. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine the proper placement of the division site in Escherichia coli. Cell 56, 641–649. ( 10.1016/0092-8674(89)90586-2) [DOI] [PubMed] [Google Scholar]
- 58.Raskin DM, de Boer PA. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181, 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu X, Shih YL, Zhang Y, Rothfield LI. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl Acad. Sci. USA 98, 980–985. ( 10.1073/pnas.98.3.980) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu Z, Lutkenhaus J. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34, 82–90. ( 10.1046/j.1365-2958.1999.01575.x) [DOI] [PubMed] [Google Scholar]
- 61.Raskin DM, de Boer PA. 1997. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91, 685–694. ( 10.1016/S0092-8674(00)80455-9) [DOI] [PubMed] [Google Scholar]
- 62.Bonny M, Fischer-Friedrich E, Loose M, Schwille P, Kruse K. 2013. Membrane binding of MinE allows for a comprehensive description of Min-protein pattern formation. PLoS Comput. Biol. 9, e100347 ( 10.1371/journal.pcbi.1003347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stricker J, Maddox P, Salmon ED, Erickson HP. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl Acad. Sci. USA 99, 3171–3175. ( 10.1073/pnas.052595099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bisicchia P, Arumugam S, Schwille P, Sherratt D. 2013. MinC, MinD, and MinE drive counter-oscillation of early-cell-division proteins prior to Escherichia coli septum formation. mBio 4, e00856-13. ( 10.1128/mBio.00856-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tonthat NK, Milam SL, Chinnam N, Whitfill T, Margolin W, Schumacher MA. 2013. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc. Natl Acad. Sci. USA 110, 10 586–10 591. ( 10.1073/pnas.1221036110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jennings PC, Cox GC, Monahan LG, Harry EJ. 2011. Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron 42, 336–341. ( 10.1016/j.micron.2010.09.003) [DOI] [PubMed] [Google Scholar]
- 67.Rowlett VW, Margolin W. 2014. 3D-SIM super-resolution of FtsZ and Its membrane tethers in Escherichia coli cells. Biophys. J. 107, L17–L20. ( 10.1016/j.bpj.2014.08.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 2012. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 10, e1001389 ( 10.1371/journal.pbio.1001389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. 2010. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS ONE 5, e12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsui H-CT, et al. 2014. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol. Microbiol. 94, 21–40. ( 10.1111/mmi.12745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szwedziak P, Wang Q, Bharat TAM, Tsim M, Löwe J. 2014. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3, e04601 ( 10.7554/eLife.04601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holden SJ, Pengo T, Meibom KL, Fernandez Fernandez C, Collier J, Manley S. 2014. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc. Natl Acad. Sci. USA 111, 4566–4571. ( 10.1073/pnas.1313368111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.MacDonald L, Baldini G, Storrie B. 2015. Does super-resolution fluorescence microscopy obsolete previous microscopic approaches to protein co-localization? Methods Mol. Biol. 1270, 255–275. ( 10.1007/978-1-4939-2309-0_19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turnbull L, Strauss MP, Liew ATF, Monahan LG, Whitchurch CB, Harry EJ. 2014. Super-resolution imaging of the cytokinetic Z ring in live bacteria using fast 3D structured illumination microcopy (f3D-SIM). J. Vis. Exp. 91, e51469 ( 10.3791/51469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shao L, Kner P, Rego EH, Gustafsson MGL. 2011. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat. Methods 8, 1044–1046. ( 10.1038/nmeth.1734) [DOI] [PubMed] [Google Scholar]
- 76.Muller T, Schumann C, Kraegeloh A. 2012. STED microscopy and its applications: new insights into cellular processes on the nanoscale. Chemphyschem 13, 1986–2000. ( 10.1002/cphc.201100986) [DOI] [PubMed] [Google Scholar]
- 77.Lan G, Daniels BR, Dobrowsky TM, Wirtz D, Sun SX. 2009. Condensation of FtsZ filaments can drive bacterial cell division. Proc. Natl Acad. Sci. USA 106, 121–126. ( 10.1073/pnas.0807963106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eswaramoorthy P, Erb ML, Gregory JA, Silverman J, Pogliano K, Pogliano J, Ramamurthi KS. 2011. Cellular architecture mediates DivIVA ultrastructure and regulates Min activity in Bacillus subtilis. MBio 2, pe00257-11. ( 10.1128/mBio.00257-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buss J, Coltharp C, Huang T, Pohlmeyer C, Wang SC, Hatem C, Xiao J. 2013. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol. Microbiol. 89, 1099–1120. ( 10.1111/mmi.12331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Si F, Busiek K, Margolin W, Sun SX. 2013. Organization of FtsZ filaments in the bacterial division ring measured from polarized fluorescence microscopy. Biophys. J. 105, 1976–1986. ( 10.1016/j.bpj.2013.09.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH. 2005. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J. Bacteriol. 187, 6874–6882. ( 10.1128/JB.187.20.6874-6882.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. 2007. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 63, 1008–1025. ( 10.1111/j.1365-2958.2006.05571.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buss J, Coltharp C, Shtengel G, Yang X, Hess H, Xiao J. 2015. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 11, e1005128 ( 10.1371/journal.pgen.1005128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biteen JS, Goley ED, Shapiro L, Moerner WE. 2010. Three-dimensional super-resolution imaging of the midplane protein FtsZ in live Caulobacter crescentus cells using astigmatism. Chemphyschem 13, 1007–1012. ( 10.1002/cphc.201100686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Piro O, Carmon G, Feingold M, Fishov I. 2013. Three-dimensional structure of the Z-ring as a random network of FtsZ filaments. Environ. Microbiol. 15, 3252–3258. ( 10.1111/1462-2920.12197) [DOI] [PubMed] [Google Scholar]
- 86.Galli E, Gerdes K. 2010. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol. Microbiol. 76, 1514–1526. ( 10.1111/j.1365-2958.2010.07183.x) [DOI] [PubMed] [Google Scholar]
- 87.Fleurie A, et al. 2014. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516, 259–262. ( 10.1038/nature13966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. 2012. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc. Natl Acad. Sci. USA 109, E1481–E1488. ( 10.1073/pnas.1200781109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu B, Morado DR, Margolin W, Rohde JR, Arizmendi O, Picking WL, Picking WD, Liu J. 2015. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc. Natl Acad. Sci. USA 112, 1047–1052. ( 10.1073/pnas.1411610112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu B, Margolin W, Molineux IJ, Liu J. 2013. The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science 339, 576–579. ( 10.1126/science.1231887) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Z, Trimble MJ, Brun YV, Jensen GJ. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26, 4694–4708. ( 10.1038/sj.emboj.7601895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang Y-W, Chen S, Tocheva EI, Treuner-Lange A, Lobach S, Sogaard-Andersen L, Jensen GJ. 2014. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat. Methods 11, 737–739. ( 10.1038/nmeth.2961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Osawa M, Anderson DE, Erickson HP. 2008. Reconstitution of contractile FtsZ rings in liposomes. Science 320, 792–794. ( 10.1126/science.1154520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Osawa M, Erickson HP. 2013. Liposome division by a simple bacterial division machinery. Proc. Natl Acad. Sci. USA 110, 11 000–11 004. ( 10.1073/pnas.1222254110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Milam SL, Osawa M, Erickson HP. 2012. Negative-stain electron microscopy of inside-out FtsZ rings reconstituted on artificial membrane tubules show ribbons of protofilaments. Biophys. J. 103, 59–68. ( 10.1016/j.bpj.2012.05.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Loose M, Schwille P. 2009. Biomimetic membrane systems to study cellular organization. J. Struct. Biol. 168, 143–151. ( 10.1016/j.jsb.2009.03.016) [DOI] [PubMed] [Google Scholar]
- 97.Loose M, Mitchison TJ. 2014. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 16, 38–46. ( 10.1038/ncb2885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. 2008. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320, 789–792. ( 10.1126/science.1154413) [DOI] [PubMed] [Google Scholar]
- 99.Zieske K, Schwille P. 2014. Reconstitution of self-organizing protein gradients as spatial cues in cell-free systems. eLife 3, e03949 ( 10.7554/eLife.03949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pazos M, Casanova M, Palacios P, Margolin W, Natale P, Vicente M. 2014. FtsZ placement in nucleoid-free bacteria. PLoS ONE 9, e91984 ( 10.1371/journal.pone.0091984) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun Q, Yu X-C, Margolin W. 1998. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol. Microbiol. 29, 491–504. ( 10.1046/j.1365-2958.1998.00942.x) [DOI] [PubMed] [Google Scholar]