Abstract
A requirement that an animal be able to feed to grow constrains how a cell can grow into an animal, and it forces an alternation between growth (increase in mass) and proliferation (increase in cell number). A growth-only phase that transforms a stem cell of ordinary proportions into a huge cell, the oocyte, requires dramatic adaptations to help a nucleus direct a 105-fold expansion of cytoplasmic volume. Proliferation without growth transforms the huge egg into an embryo while still accommodating an impotent nucleus overwhelmed by the voluminous cytoplasm. This growth program characterizes animals that deposit their eggs externally, but it is changed in mammals and in endoparasites. In these organisms, development in a nutritive environment releases the growth constraint, but growth of cells before gastrulation requires a new program to sustain pluripotency during this growth.
The cellular programs governing growth and proliferation during early embryogenesis depend on whether the egg is deposited externally (as in most animal species) or nurtured inside the mother (as in mammals).
The phrase “Nothing in biology makes sense except in the light of evolution,” originally introduced in an essay supporting the theory of evolution (Dobzhansky 1973), has been repurposed to chide biologists, who all too often ignore the origin and purpose of the biological processes and mechanisms they study. To look for how these processes “make sense,” one should look at the way they serve the life histories of the organisms in which they are used. Such an examination can suggest how a process might have been molded by evolution to achieve the benefit provided. I have been an advocate for such considerations, because I believe them to be the best guide to interesting and important questions for investigation. Here, I will synthesize observations made by examination of the natural histories of diverse organisms to describe four almost universal phases of growth and proliferation in animal biology. I will be focusing on the two earliest phases: the growth that produces a huge egg and the transformation of this single large cell into an embryo. The subsequent phases of growth were described in previous reviews (O’Farrell 2004, 2011; O’Farrell et al. 2004). But first, I will provide some context for the comparisons that are made, and a brief synopsis of the four growth phases that convert an ordinary-sized stem cell into a big metazoan adult.
MAMMALS CAME LATE
It is perhaps important to point out to the reader that I will first focus on animals that deposit their eggs externally, as this oviparous lifestyle is the most general and ancestral mode of animal development. Mammals, of course, are interesting to us, but if we wish to understand our connection to the rest of biology, we should first recognize the context within which mammals appeared. Mammals are a small clade of ∼4000 species out of more than two million animal species, and they are derived chordates that arose relatively late in animal evolution. Furthermore, mammalian evolution invested in a special program, which, by housing embryos in a nutritive environment, introduced an unusual fashion of dealing with the growth constraints faced by all embryos. Understanding the specializations of mammalian development in this context can be illuminating (O’Farrell et al. 2004). I will end with an effort to point out these mammalian specializations and deep connections to programs showed by their evolutionary predecessors. But we will begin with a few guiding generalizations that give perspective on the coordination of growth with animal development.
AN ORGANISM MUST DEVELOP FEEDING STRUCTURES BEFORE IT CAN INCREASE IN MASS
Although there are some exceptionally large single-celled organisms, large body plans are substantially the domain of multicellular organisms. However, the production of a large body from a single-cell zygote must deal with a fundamental problem. Any animal whose nutrition depends on a complex body plan with its specialized feeding structures must be able to develop these specialized structures before it can feed and grow independently. In these cases, a complex body plan needs to be produced during development before there is significant growth of the organism (O’Farrell 2004).
We will view life histories of organisms in the light of this problem, which appears to have acted as a constraint throughout the evolution of the animals. Despite great diversity, this vantage point reveals relationships that lead to the global concept of four phases of growth and proliferation in the life plans of animal species.
ALIGNING PROGRAMS OF DEVELOPMENT AT A CONSERVED STAGE
The strategy used when comparing distantly related protein sequences suggests a general approach when looking for distant homologies. Rather than just aligning sequences at the amino terminus, one first identifies the most conserved domains and uses these for alignment. Similarly, in comparing the programs of growth and proliferation of diverse species, I have chosen to align the life histories of different organisms at a particularly conserved point (O’Farrell 2004; O’Farrell et al. 2004).
The early developmental biologists, von Baer and Haeckel, were fascinated by the similarities of vertebrate embryos of very diverse organisms at a stage following establishment of the body axis and formation of the neural tube. Recognition of this conserved stage and description of the morphogenetic events that brought increasingly dramatic distinctions to the embryos of different species were taken by some as evidence for a claim that ontogeny recapitulates phylogeny—or that the developmental sequence passes through all of the evolutionary steps as if “more advanced” organisms achieve their distinctions by late additions to the developmental sequence (e.g., Graham and Richardson 2012). The attractive phrase, “ontogeny recapitulates phylogeny,” appears to be somewhat of an overstatement and the idea is often described as discredited. Nonetheless, the existence of a relatively similar embryonic morphology is strongly supported.
A common embryonic morphology is also found among embryos within different invertebrate phyla. This is particularly apparent in arthropods. Arthropod embryos of very diverse species show remarkably similar morphologies on the initial establishment of the body plan. These similar looking embryos also use similar molecules to guide the formation of embryonic patterns (Patel et al. 1989; Prud’homme et al. 2003). This point in development has been called the phylotypic stage to reflect the fact that diverse organisms within a phylum show a “typical morphology” at this stage.
It is notable that the “phylotypic” stages of different phyla occur at a similar point during embryogenesis, just after establishment of the basic body plan, and embryos of even different phyla have a very similar organization at this stage. These distant similarities suggested by morphologies have been supported by molecular analyses (Domazet-Lošo and Tautz 2010; Kalinka et al. 2010). Key mechanisms patterning the phylotypic embryo are conserved across phyla. For example, a gradient in a signaling molecule of the transforming growth factor (TGF)-β type guides dorsoventral polarity of the embryo, and local expression of different homeotic-type transcription factors subdivide the anteroposterior axis of the body plan (Shen 2007). These findings suggest mechanistic parallels and argue for evolution by descent. That is, even distantly related animals evolved from a common predecessor with a similar embryonic stage, which was patterned by mechanisms that continue to be used in extant animals. Consequently, the phylotypic stage can be taken as an especially conserved stage of development at which the embryos of different species can be compared.
The original view exemplified by the “ontogeny recapitulates phylogeny” phrase presumes that embryogenesis begins with a common ancestral morphology that adds species-specific specializations as embryogenesis progresses. However, the phylotypic stage occurs after important early steps in embryogenesis and the modern view considers development more like an hourglass (Raff 1996; Prud’homme and Gompel 2010). Eggs are diverse, and distinctive programs and morphologies are seen during very early embryogenesis, but diversity shrinks with progression to the phylotypic stage and then reexpands in later development. The bottleneck in morphological diversity at the phylotypic stage presumably is a reflection of the early evolution of a successful strategy for building a basic body plan, and the developmental reexpansion of diversity after the phylotypic stage is a likely an indication that modification and addition of specializations were major routes of later evolutionary diversification.
SUBDIVIDING ANIMAL GROWTH
Embryos at the phylotypic stage are not only similar in general morphology, but they are the same size, or relatively close (O’Farrell 2004). Taking a little license with the exact stage that is defined as the phylotypic stage, the anterior/posterior axis of the phylotypic embryo is roughly a millimeter. A few examples are a little smaller (e.g., Caenorhabditis elegans) or bigger (e.g., grasshopper), and some embryos continue to extend their length for some time after the initial elaboration of the body axis. Nonetheless, sizes are rather similar. This similarity in size applies to the embryos of fruit flies, frogs, and blue whales, organisms whose adult masses, 1 mg, 100 g, and 100 metric tons, range over 1011-fold. This near-constancy in the size of the embryo at the phylotypic stage allows us to divide our consideration of growth in animals into two. How do embryos of ∼10 µg (wet weight of cytoplasm) at the phylotypic stage grow to the adult sizes? How does oogenesis and early embryogenesis transform a single ordinarily sized stem cell into a phylotypic embryo, a miniorganism with a roughed-out body plan?
In this presentation, I consider growth as an exponential process. Thus, the seemingly massive growth of a 7-ton blue whale calf into a 150-ton adult is viewed as ∼20-fold and, hence, considerably less impressive than the ∼100,000-fold growth of a normally sized stem cell to produce a 10-µg Drosophila egg.
Examination of the life histories of numerous species reveals four dramatically different phases of growth in the life plans of many animals:
Sponsored growth: During oogenesis, a stem cell grows enormously in size without division. The mother sponsors this growth for the benefit of the progeny.
Subdivision: Following deposition of an egg in an external environment, the massive egg divides rapidly without any growth to produce the phylotypic embryo.
Expansion: Diverse but identifiable strategies are used in the growth of the phylotypic embryo to the adult.
Size control: Adult organisms usually constrain their growth, a process that relies on both quiescence and balanced replacement strategies.
The central features of growth in phases 3 and 4 were described in two previous reviews (O’Farrell 2004, 2011). Together with the considerations presented here, these reviews reveal a perspective gained from examination of the natural histories of organisms and their development and growth. It is strikingly different from the views we develop when focusing on some of our models of cell growth and proliferation. I suggest that the biological perspective gained in an exercise of considering how growth and proliferation are controlled in animal development will uncover new and important facets of the control of these processes.
PHASE 1—SPONSORED GROWTH: GROWING A HUGE CELL (OOGENESIS)
General Background
The development of an externally deposited egg into a feeding organism depends on supplies received from the mother. This maternal dowry has two parts: a large volume of cytoplasm, often in the range of 100,000 times the amount found in a normal cell, and a supply of nutrients, largely in the form of yolk. The first growth phase is maternally sponsored growth of a stem cell of ordinary proportions into a large oocyte that is often more than 105-fold bigger than an average cell. This phase represents growth without division and it involves highly specialized processes that are well studied in only a few systems.
The vast majority of the millions of animal species deposit their eggs in the external environment in which, without further nutritional input, they develop within protective shells or coating that largely isolate the embryo from its environment. The duration and extent of the development that occurs during this autonomous stage varies substantially in proportion to the nutritional supplies from the mother. From species to species, the amount of yolk included in an egg differs over many orders of magnitude. Avian eggs are at the upper end of this spectrum. For example, a chicken egg has a yolk vesicle of ∼17 g that is actually one extraordinarily large cell, the oocyte, before fertilization. Imagine a dinosaur oocyte! The chicken oocyte is more than a hundred-thousand-million-fold (1011) larger than a typical cell, which is ∼5 μm in diameter (or ∼10−10 cm3, here approximated as 10−10 g). On a geometric scale, the 1011-fold growth to produce this large cell in an ovary of the hen totally belittles the roughly 100-fold growth of the hatchling to an adult chicken. This growth of the single-celled oocyte is, of course, growth without proliferation. Although the growth of the yolk-rich avian oocyte is above average, and so exaggerates the contribution of oogenesis to overall growth, it is nonetheless true that, on a geometric scale, the growth of an oocyte makes huge contributions to the growth of organisms throughout animal phylogeny.
Yolk versus Cytoplasm
Before embarking on a consideration of the phases of growth, I would like to deconstruct the extravagant process that occurs in the hen. Besides familiarity, I use the chicken egg as an example because of the dramatic size of the oocyte. As mentioned, the size of the yolk supplies provided by the mother differs widely between species, and it is the extravagant supply of yolk that makes the avian oocyte a standout in size. But chicken oocytes also have a very large cytoplasm, and here eggs of even very diverse organisms are remarkably similar. In the case of the chicken oocyte, the nonyolky cytoplasm forms but a small whitish disc, the blastodisc, on the surface of the huge yolk. Although small on a relative scale, this small white area includes ∼10 μg of cytoplasm, a cytoplasmic volume of 100,000 times that of an ordinary cell. The growth that produces this volume of cytoplasm amounts to a remarkable achievement for one cell, and I consider this growth as the first of the four phases of growth.
After fertilization, this egg cytoplasm will divide rapidly to generate a cap of cells on top of the massive yolk vesicle, and these cells will undergo morphogenesis to produce the elementary body plan of the phylotypic embryo. This subdivision of cytoplasm and production of a phylotypic embryo represents phase 2 of the growth and proliferation program, the subdivision phase that is largely attributed to proliferation without growth. For the moment, what I want to get across is that yolk contributes little to this process. To a large extent, it is the nonyolky cytoplasm that undergoes this phase 2 program to produce an elemental phylotypic embryo, and later the yolk is called on to fund a third phase in the growth and proliferation program, the expansion phase (O’Farrell 2004, 2011; O’Farrell et al. 2004). Thus, we will ignore yolk in our considerations of phase 1 and phase 2 growths. We will instead focus on the cytoplasmic accumulation in oogenesis and its subdivision in early embryogenesis.
Transcriptional Capacity, a Constraint on Production of Large Cells
There are numerous examples of huge cells: the Drosophila salivary gland cells, the cells of Ascaris (a nematode that has a volume a billion-fold bigger than C. elegans but the same cellular anatomy), the giant cells of Aplasia, and ciliates of extraordinary size. But these cells have to either overcome or tolerate a biological constraint. The genome has a limited ability to support growth or rapid changes in a large cell. As pointed out by Woodland (1982), the rate at which a gene can produce transcripts is limited, and this limitation imposes practical limitations on the growth and function of large cells. Let us look at this limitation.
Rates of initiation of transcription cannot be increased indefinitely. There is a packing limit on the density of RNA polymerase molecules that can be loaded along a DNA molecule. Once initiation rates are high enough to load a sequence to this limit, no further increase in the rate of RNA synthesis can be achieved. Irrespective of the length of a gene, at the maximum packing density, polymerases are lined up one behind the other at spacing estimated to be 50–100 bp. A new transcript will be produced only when a polymerase advances to the end of the gene, and the rate at which each successive polymerase arrives at the end—corresponding to the rate of product synthesis—depends only on the rate at which the polymerase traverses the distance separating it from the previous polymerase. If we use 30 bp per sec as an approximation of transcriptional elongation rate and 60 bp as the maximal packing density, a new polymerase will advance to the end of the gene every 2 sec. Thus, at maximal loading, a single-copy gene can produce a new transcript once every 2 sec.
A normally sized animal cell has ∼100,000 messenger RNAs. The most abundantly expressed genes in most cells represent several percent of the total and would be present at levels between ∼1000 and 10,000 copies. At the maximal rate of gene expression, it would take between 2000 and 20,000 sec, or between 30 min and 5 h, to make this amount of transcript, and half this time in a diploid and half again in a G2 cell, which has four copies of the genome. A growing cell must be able to produce a new complement of all of its mRNAs every time it doubles. For many animal cells, this requirement is not a serious constraint because doubling times are usually considerably longer than the time required to produce the most abundant transcripts. However, as a cell enlarges, the amount of mRNA in the cell increases in rough proportion to the cytoplasmic volume. Many oocytes achieve a size ∼100,000 times that of a normal cell. It would take comparably longer to make the transcripts to supply this large cytoplasm. Above, we calculated that a normal cell would take 0.5 to 5 h to make its most abundant transcripts. If we increase the lower value 100,000-fold, we find that it would take ∼50,000 h or ∼2000 d. For a G2 cell, with its four copies of each gene, this reduces to 500 d. According to this, the growth of cells to a very large size should be an extremely slow process, and take roughly a year.
If an organism produces exceptionally large cells, it must do so very slowly or find a solution to the transcriptional rate constraint. In many cases, and in all of the examples of large cells given above other than oocytes, the genome is amplified to increase coding potential. In accord with the idea that transcriptional potential might limit growth, manipulations of the ploidy of Drosophila salivary gland cells have shown that the size attained depends on ploidy (Follette et al. 1998; Hayashi and Yamaguchi 1999). But an increase in ploidy does not appear to be an option for the germline, as this would disrupt the normal and faithful transmission of chromosomes from generation to generation. Ciliated protozoa have found a way to enjoy the benefits of polyploidy while retaining stable genetics. Here, a disposable polyploid macronucleus supports large cells, while a distinct diploid germ nucleus, the micronucleus, bears the responsibility for inheritance. How do oocytes in animals manage to grow despite the limited transcriptional potency of their nuclei?
Nurse Cell Support of Oocyte Growth
One effective way to speed the growth of a single large cell is to enlist the help of other cells. A special organization is required if other cells are to contribute transcripts to a growing oocyte. In many species of arthropods and annelids, the precursor of the oocyte divides, but only incompletely, leaving sister cells interconnected by cytoplasmic bridges that develop into specialized and stable intercellular conduits called ring canals. The interconnected group of sister cells is called a cyst when it first forms, and, in insects in which it is a stable unit in oogenesis, it is called an egg chamber once it is invested in a coating of independently derived follicle cells. Although one cell of the interconnected group becomes the oocyte, the remaining interconnected cells serve as nurse cells that supply the oocyte. The nurse cells amplify their genomes and their capacity to produce RNA in endoreduplication cycles consisting of distinct rounds of S phase without cell division. The resulting large polyploid cells efficiently fuel the growth of an immense egg cytoplasm. For example, in Drosophila, a precursor cell called a cystocyte divides four times to create an interconnected “cyst” of 16 cells that develops into an egg chamber in which 15 of the cells become highly polyploidy, and one cell, which will grow into the oocyte, arrests in meiotic prophase. Although the oocyte nucleus remains transcriptionally inactive, the nurse cells produce an abundance of RNA and cytoplasmic constituents that are transferred to the oocyte, which grows enormously over a few days at the expense of the nurse cells.
The nurse cell strategy is particularly effective in overcoming the limitation imposed by transcriptional capacity and it supports the rapid increase in cytoplasmic volume of the developing oocyte. The mother can also make other contributions to the development of the egg. Most notably, a different approach is generally used to accelerate the accumulation of the yolky nutrient supply of the oocyte. Other tissues of the mother, even distant tissues, such as liver, can contribute yolk protein, vitellogenin, lipids, and other nutrients that are taken up by the oocyte often with the assistance of surrounding follicle cells. The accumulation of yolk is of major importance to nutrition of the embryo and later growth. The mother also provides further contributions in the elaboration of protective coatings and shells. But here we focus on the roughly 100,000-fold growth of the cytoplasm, exclusive of yolk, as the germline stem cell expands to produce the oocyte. The numerous copies of the genome available in nurse cells, altogether a few thousand copies in the egg chamber of Drosophila melanogaster, provide abundant template for rapid accumulation of transcripts and growth of the oocyte. In organisms that use this strategy, a stem cell can mature into a large oocyte relatively quickly, ∼10 d in the case of Drosophila.
Autonomous Growth of Oocytes
Cytology and electron microscopy have been used to survey oogenesis in numerous organisms. These analyses define different oogenesis strategies. In many species, the precursors of oocytes either lack or lose connections with sister cells and the individual oocytes grow autonomously, often with an isolating coating of follicle cells that do not have detectable cytoplasmic continuity with the growing oocyte. In these organisms, the oocyte nucleus appears to support the growth in cytoplasmic volume, and the contributions of the maternal environment appear to be limited to provision of yolk and other nutrients and production of protective coatings.
Given the limitation imposed by the transcriptional potency of a single nucleus, how does autonomous growth succeed at producing oocytes? To a large degree, the answer is that oocytes using this program grow much more slowly, generally taking several months to more than a year to reach a mature size (Smiley 1990). But even this pace appears to rely on several striking specializations that appear designed to enhance provision of key transcripts.
As expected, when growth is autonomous, the oocyte nucleus is transcriptionally active (Gall et al. 2004). The growth phase still occurs during an arrested prophase of meiosis I, but meiosis is modified to incorporate a special transcriptional phase generally not seen in systems using nurse-cell-supported growth. Meiosis begins normally. The chromatids condense on the start of prophase, and progress through the meiotic events of homolog pairing and recombination before a diplotene arrest. At this arrest, large loops of chromatin unfurl from a still-visible chromosome axis. The axes outline the meiotic arrangement of paired chromosomes, suggesting a change in chromatin compaction without disruption of the meiotic chromosome configuration. The chromatin loops are packed with RNA polymerase and elongating transcripts giving a striking appearance noted by early microscopists. Because they resembled the long fuzzy brushes that were once used to clean the soot from the glass chimneys on oil-burning lamps, they are called lampbrush chromosomes. This stage often persists for months as the oocyte enlarges, and studies in Xenopus showed that oocyte transcripts accumulate during this period (Gall 2012; Kloc et al. 2014).
The lampbrush chromosome stage provides a long period of highly active transcription with a postreplicative nucleus that has four copies of the genome. Still, our rough calculations would suggest that a very long time would be required for the transcript accumulation needed for a 100,000-fold increase in cytoplasmic volume. It would not be surprising if there were additional adaptations that might reduce the severity of the transcriptional constraint so that oocytes could be produced more rapidly. Indeed, there is dramatic evidence for such adaptations.
Selective Amplification of Genes Encoding Ribosomal RNA
Unlike protein-coding RNAs whose impact is further amplified by translation, structural RNAs, such as ribosomal RNA, function directly, and there is no way to bypass or minimize the need for a high transcriptional capacity to accumulate a high level of functional product. Furthermore, there are a lot of ribosomes in even an ordinary cell, roughly several million (Blobel and Potter 1967; Wolf and Schlessinger 1977), a few orders of magnitude more than most abundant mRNAs. If there were only one gene encoding ribosomal RNA, the nucleus would take about a month to meet the transcriptional demand required to reproduce a normally sized cell. However, ribosomal RNA is encoded in repeating arrays of genes at loci often called nucleolar organizers. The repeats increase the transcriptional capacity, but phenotypes resulting from partial deletions of the recombinant DNA (rDNA) repeats in Drosophila suggest that, even with this repetition, there is little reserve capacity (Gersh 1968). In species in which oocytes grow autonomously, the coding capacity of the rDNA repeats would be severely taxed and should limit growth. However, a remarkable process takes care of this limitation.
As oocytes progress through meiotic prophase to their arrest in diplotene, they hugely amplify the ribosomal repeats (Brown and Dawid 1968; Gall 1968). The replication associated with this amplification has been detected by labeling, and the DNA copy increase was assessed by molecular and cytological methods. This amplification can be so extensive that it increases the total amount of DNA in the nucleus several-fold. The amplified sequences are produced as free circles that form hundreds of small nucleoli throughout the oocyte nucleus. Amplification occurs in diverse organisms that use oocyte-autonomous growth and is presumed to support the massive increase in ribosomes during growth.
We have seen three striking biological innovations that promote growth of oocytes: nurturing by nurse cells, transcription from lampbrush chromosomes, and amplification of rDNA. Apparently, the nurse-cell strategy reduces the need for other adaptations to increase transcriptional output, as oocytes supported by nurse cells appear to be transcriptionally silent and to lack lampbrush chromosomes (Ganot et al. 2008). These relatively widespread specializations modify three fundamental biological processes. First, cytokinesis is rendered incomplete and the arrested cleavage furrows are transformed into ring canals that interconnect nurse cells and oocytes. Second, although transcription is normally discontinued on mitotic or meiotic chromosomes, the lampbrush chromosome stage is created by reactivating transcription of condensed meiotic chromosomes. Finally, although DNA replication is ordinarily tightly regulated to ensure that all sequences are replicated once and only once, these controls are bypassed to massively amplify the rDNA repeats. These are not oddities of a peculiar branch of evolution—they are modifications making key contributions to a central step in the life history of nearly all animal species, the production of an oocyte.
Unbalanced Growth
The accepted truism about cell growth is that all of the contents of a cell need to double, at least on average, every time a cell divides. This applies to the much-studied case of exponentially growing cells in culture. However, growth to produce an organism is not simply a matter of increasing everything equally. Growth in vivo is often unbalanced, and, at a cellular level, this is particularly dramatic when one considers the huge growth of a stem cell into an oocyte. Extraordinary changes occur in the size of some structures, and the stoichiometry of some key cellular components. As I describe here, this places huge stress on growth itself as a result of disproportionate changes in pathways contributing to growth.
The nucleus of the oocyte is huge—so big that it has its own special name, the germinal vesicle. Despite having only the DNA content of a normal G2 cell, the nucleus expands in rough proportion to the entire oocyte, ∼100,000 times. The DNA occupies only a tiny volume of the germinal vesicle, and forms an easily stained compact structure called the karyosome within a sea of nucleoplasm. A variety of specialized structures have been described in the germinal vesicle (Gall et al. 2004; Gardner et al. 2012). One of these structures, the mininucleoli, which appear throughout the germinal vesicle, results from the amplification of ribosomal repeats and is clearly a response to the disproportionate demand for ribosomal RNA synthesis described above.
The growth/multiplication of mitochondria in the oocyte faces huge distortions in the balance of different mitochondria gene products, those encoded in the nucleus and those encoded by the mitochondrial DNA. The mitochondrial genome is replicated in proportion to the expansion of the cytoplasm. In Xenopus, 7 × 108 copies of mitochondrial DNA are produced during oogenesis (Chase and Dawid 1972), whereas the nuclear DNA is unchanged. This represents nearly a million-fold increase in mitochondrial genome copy number above that in a typical cell. Because nuclear and mitochondrial DNA–encoded gene products assemble stoichiometically to make the respiratory complexes, continued production of functional mitochondria must be managed in the face of a changing imbalance in the two contributions. We know nothing of the mechanisms that deal with this supply imbalance, one that is faced to a smaller degree in other enlarging cells, such as neurons.
The great bulk of the multiplication of mitochondria occurs in a specialized structure, the Balbiani body. This structure, which is found in the oocytes of a wide range of animals, is seen as a differentiated area of cytoplasm that can be detected early in oogenesis, just as precursor cells are committing to oogenesis (Kloc et al. 2014). It is usually adjacent to the nucleus. Often, it is a dominating structure obvious in the light microscope. In the electron microscope, it is seen as a congregation of mitochondria, endoplasmic reticulum, and an RNA-rich structure called nuage (French for “cloud”). Nuage appears to be universally associated with developing oocytes and more generally with the germline. Localization, genetic dissection and identification of molecular constituents have tied nuage not to one, but to several functions—storing mRNAs for the future embryo, specifying the vegetal or posterior pole of the egg, defining the germline, and protecting against infectious DNA elements by housing an immunity system based on a category of small RNAs, piRNAs (Marlow and Mullins 2008; Kloc et al. 2014). Obviously, there are many tasks to be performed in the oocyte, and rapid progress is being made in defining the contributions of nuage. But here, we focus on the mitochondria.
The cytoplasmic Balbiani body of Xenopus, which is densely packed with mitochondria, grows to be huge. Its increase in size is paralleled by an increase in mitochondria and mitochondrial DNA, and the structure only disperses months later when the oocyte turns its attention to yolk accumulation. Mitochondrial morphology and DNA labeling argue that Balbiani body mitochondria proliferate (Chase and Dawid 1972; Webb and Smith 1977; Kloc et al. 2014). Indeed, it appears that more than 16 doublings of the mitochondrial genome occur before dispersal of the Balbiani body in Xenopus.
The Balbiani body is not always a dominating morphological feature. In Drosophila, it is difficult to see by light microscopy without specialized tags, and while obvious by electron microscopy, it is rather transient in comparison to its persistence in autonomously growing oocytes (Cox and Spradling 2003). This is likely linked to delegation of some responsibility for production of various components to the multiple nurse cells. In earwigs, an insect wherein a single large nurse cell supports each oocyte, a Balbiani body is found in the nurse cell as well as the oocyte (Tworzydlo et al. 2009).
Nurse-cell support of oogenesis, as exemplified by Drosophila, reduces one type of stress. The increased nuclear transcription capacity provided by the polytene nurse cells reduces the imbalance in nuclear and mitochondrial copy number. However, the transcription of nuclear products occurs in a distinct, albeit interconnected, nurse cell compartment. Now, there is a need to manage the flow of gene product to the oocyte where mitochondria are actively replicating. Perhaps more impressive is that most of the 100,000-fold increase in the number of mitochondrial genomes occurs in a few days in this more rapid style of oogenesis.
One final feature of mitochondrial multiplication dramatically emphasizes the fact that growth in vivo is “unbalanced”—that not all cellular components double when cells double. In C. elegans, a supply of ∼100,000 mitochondrial genomes is accumulated during oogenesis, and no further increase occurs until nearly adult stages. Additionally, a mutant lacking the late increase in mitochondrial genomes produces mature adults (Tsang and Lemire 2002; Bratic et al. 2009). Thus, the supply of genomes made during oogenesis suffices for the life of the worm. It is likely that the behavior seen in C. elegans is broadly representative, although likely less extreme in other systems. After the huge increases in mitochondrial DNA copy number during oogenesis in Xenopus and in Drosophila, little or no further increase occurs during all of embryogenesis (Chase and Dawid 1972; H Ma and PH O’Farrell, unpubl.).
Although these descriptions of mitochondrial multiplication during oogenesis do not reveal mechanism, they give a very different view of what mechanisms we should be looking for. Mitochondrial genome replication is uncoupled from cellular growth and proliferation, but is strikingly regulated during development and shows a remarkable departure from balanced growth. To understand how the copy number of this genome is regulated, we need to look for developmental inputs, and we might also expect special regulatory circuits that handle the rapid multiplication in the face of a dramatically changing context during oocyte growth.
Constrained Change versus Conservation
The specialized features described above can be found in diverse organisms with representatives in more than one phylum. For example, lampbrush chromosomes have been described in organisms representing mollusks, arthropods, echinoderms, and chordates (Bedford 1966; Gruzova and Batalova 1979; Smiley 1990; Gall et al. 2004). This gives the impression of conservation, and conservation often comes with a connotation of stability. However, change is persistent, and it imposes a constant diversifying pressure that is resisted by selection. Constraints imposed by a requirement for function limit some change, but not all. Thus, like the cars produced over the past several decades, which continue to have four wheels (generally), a steering mechanism and, brakes, a lot of changes occur “under the hood.” Variations in the format of oogenesis and the distributions of the variants in phylogeny reveal changes, but the retained features suggest operation of powerful and lasting constraints that limited the range of successful formats.
Although our sampling of species is still relatively small, the phylogenetic distribution of nurse-cell-supported oogenesis versus autonomous oogenesis is of interest. Nurse-cell-supported oogenesis is seen in numerous insect species and has been particularly well studied in Drosophila; however, there are also many insects including members of one of the most diverse groups of animals on earth, the beetles, which show autonomous growth of their oocytes. Because the nurse-cell format of oogenesis is found, at least so far, in derived insect lineages, it is suspected that this program itself is derived. This interpretation is consistent with findings of autonomous oocyte growth in sampled species of various other animal phyla, including chordates, echinoderms, mollusks, and cnidarians, and a group of more primitive animals, such as jellyfish (Smiley 1990; Eckelbarger and Larson 1992; Eckelbarger and Young 1997; Marlow and Mullins 2008). However, the nurse-cell program is not limited to derived insect lineages. Nurse cells are also used in annelids, and a related nurse-cell type of oogenesis has been discovered in a group of chordate species whose lifestyle relies on rapid oogenesis (Ganot et al. 2007). Furthermore, a group of frogs that carry their eggs in a brooding pouch, marsupial frogs, does oogenesis differently than other frogs, and the part that is especially different has features like nurse-cell-supported oocyte growth. In these frogs, oogonia undergo multiple incomplete divisions to create a dramatically multinucleate oocyte: these nuclei develop lampbrush chromosomes, apparently all contributing transcripts for growth of the oocyte, but eventually all but one germ nucleus is eliminated (Del Pino and Humphries 1978; Macgregor and Del Pino 1982). Also, nematodes produce eggs from a syncytial gonad in which many nuclei contribute to a cytoplasm that is partitioned into the rapidly formed large egg (O’Farrell 2004). Thus, helper cells, or helper nuclei, promote the growth of large oocytes by means of shared cytoplasm in at least a few animal phyla. This might represent convergent evolution to this strategy.
An alternative to convergent evolution is divergence from an unrecognized common progenitor. Indeed, at least one observation might suggest a deep connection between nurse-cell-supported oogenesis and autonomous oogenesis. In numerous organisms, cytokinesis of the precursors to the oocyte is incomplete and produces specialized bridges interconnecting the sister cells (Pepling et al. 1999; Marlow and Mullins 2008). In organisms with nurse-cell programs, only one cell of the interconnected group becomes the oocyte, and the bridges persist so that the other cells, the nurse cells, can transmit their cytoplasmic endowment to the oocyte. However, in organisms showing autonomous oocyte growth, the relationship is transient, is lost before oocyte growth, and usually each of the originally connected cells makes an oocyte. Although a common progenitor to nurse-cell programs and autonomous oogenesis has not been recognized, we should not forget that sex appeared earlier in evolution than animals. Perhaps vestiges of early gametogenesis programs provided a foundation for the different strategies present in animal species today.
Regardless of the path taken in evolution, representative animals in every major branch of phylogeny grow large oocytes, and these varied organisms call on a limited set of specialized mechanisms to support this growth. I suggest that this reflects the persistent operation of constraints that guide selection. An operational necessity to produce an egg that can support development to an autonomous feeding stage might have been the driver of other constraints as follows. Development of a feeding organism without growth demanded a large egg. Production of a large egg required extensive growth of the oocyte. Production of very large cytoplasm harboring only a single nucleus distorted the usual biological relationships, and the resulting stresses produced selection for special adaptations.
PHASE 2—FROM EGG TO EMBRYO
Fertilization and egg deposition marks a major turnaround for growth. After growing to huge dimensions as a single cell, nutrients are no longer freely available as the egg sets out to make an embryo. In doing so, the huge egg is subdivided into many cells and is restructured, initially without growth. It is not a time to tarry. The egg is a defenseless immobile package of nutrients, and speed of development is one way that externally deposited eggs minimize predation.
We tend to think of development as a relatively protracted affair, perhaps because of our own gestation time. But the long periods of fetal growth in mammals are more about growth than development, as is the time of yolk-supported growth of birds. The relationship between developmental time and growth is perhaps most obvious in birds, in which the time to hatching in different species increases in proportion to the size of the egg and the hatchling it eventually produces (Rahn et al. 1974). Externally deposited eggs with little yolk often develop quickly. C. elegans develops from a fertilized egg into a functional organism in 13 h. Some clam species hatch as ciliated trochophore larvae within 24 h, a Drosophila larva crawls away from its eggshell 24 h after fertilization, and zebrafish swim after 3 d. Apparently, the extraordinary processes of morphogenesis and differentiation that build an organism do not need to take a long time, but growth does. To sidestep variations associated with growth, we will consider development from fertilization to the time of the phylotypic embryo as the second phase of “growth” and proliferation. I have called this phase “subdivision,” because it primarily involves subdivision of the mass of the oocyte, without growth (i.e., without a substantial increase in mass).
As we saw for phase 1 growth, the limited transcriptional potency of a nucleus also constrains early developmental events during phase 2. Slow accumulation of RNA was a chief limitation to the growth of a huge oocyte, and now the egg has this same cumbersomely large cytoplasm and a single nucleus. Slow accumulation of transcripts would stymie the progress of development (Woodland 1982). As a result, eggs initially avoid relying on transcription.
The egg immediately embarks on a program that will resolve the discordance between the cytoplasmic volume and the nucleus. Rapid cell cycles exponentially increase the number of nuclei, whereas the cytoplasmic volume remains constant and so reduces the cytoplasm for which each nucleus is responsible. As the nuclear to cytoplasmic ratio increases, transcription can, and does, begin to play an important role in regulation.
Importantly, the early cell cycles that ramp up the transcriptional potential of the embryo are themselves independent of transcription. Regulated gene expression normally plays a major role in the control of a whole host of biological processes, including regulation of the cell cycle, and, by forsaking transcriptional control, the embryo has to extensively restructure biological regulatory mechanisms. Consequently, the egg, at the starting block of development, uses unusual biological mechanisms. As I will discuss below, the unusual aspects of this ancestral program of fast, transcription-independent cell cycles has a pervasive impact on the biology of the early embryo and influences the groundwork on which subsequent development has been built. But first, I will very briefly review more studied aspects of this early embryonic program.
Rapid Cell Cycles and the Midblastula Transition (MBT)
The rapid early embryonic divisions have been an attractive model for cell-cycle studies, and the return to reliance on transcription has been an interesting area of developmental analysis. Fast, early cell cycles are widespread. The mitotic cycles of newly fertilized eggs of Drosophila, sea urchin, and Xenopus are 8.6, 30, and 30 min, respectively. These cycles lack gap phases, and occur even if transcription is prevented—indeed, in frog and sea urchin, they have been shown to occur even if the nucleus is eliminated (Harvey 1935). After several periodic divisions, the cycles begin to slow, at first gradually and then more abruptly, as the embryo approaches gastrulation.
In addition to its transcriptional independence, the early cell cycle is redesigned in several ways to achieve speed. It lacks the gap phases, G1 and G2, which usually separate S phase and mitosis. Shortening of S phase by as much as 100-fold further quickens these cycles (Farrell and O’Farrell 2014). These features of the early cell cycle are seen in diverse settings—the rapidly dividing teleoblasts of annelid embryos, the syncytial mitotic cycles of Drosophila, the early divisions of sea urchin, and those of Xenopus. Thus, these cycles, which are often viewed as peculiar and distinct from “normal,” are used during the initial steps of development of diverse animals. I suggest that the constancy of these features is the result of constraints demanding rapid expansion of transcriptional potential in the unwieldy large-egg cytoplasm.
One of the most fascinating questions regarding these early cycles is: What terminates them? The egg is primed by its maternal dowry to initiate relentless and rapid cycles that have no obvious outside input. But the rapid cycles have to end, and they slow in a stereotyped fashion just before gastrulation. Furthermore, the maternally deposited gene products need to relinquish control to transcription of the zygotic genome. This changeover, which involves destruction of maternally provided RNAs and proteins and activation of zygotic transcription, is referred to as the maternal-to-zygotic transition (MZT). In part, this is a progressive transition with additional zygotic functions coming into play as persisting maternal functions are successively eliminated (Wieschaus 1996; Follette and O’Farrell 1997). Nonetheless, during this process, there are abrupt transitions when the nearly silent nuclei activate expression of new panels of genes (Newport and Kirschner 1982a,b; Edgar and Schubiger 1986; Edgar et al. 1986). The slowing of the cell cycle, MZT, and onset of the cell-shape changes and movements that initiate morphogenesis occur over a relatively short time span that is called the midblastula transition (MBT) (Farrell and O’Farrell 2014).
Although it is not yet understood how the MBT is controlled, it seems natural to believe that the embryo is running out of something—perhaps a key component is used up, or a maternal supply becomes inadequate as nuclei increase. Experimental manipulation of the ratio of nuclei to cytoplasm suggests that its increase has an input into the MBT (Newport and Kirschner 1982a,b; Edgar and Schubiger 1986; Edgar et al. 1986; Pritchard and Schubiger 1996). Progress has been made in understanding how the cell cycle slows. For example, inhibitory phosphorylation leads to a decline in activity of the cell-cycle kinase, Cdk1, prolongs S phase, and introduces a G2 phase at the MBT in Drosophila (Edgar and O’Farrell 1990; Farrell and O’Farrell 2013). Increasing nuclear density appears to one of the signals triggering slowing of the cell cycles; however, the mechanism of this coupling remains unclear. It is also uncertain whether the nuclear to cytoplasmic ratio acts directly on other MBT events, or whether its effects are mediated by changes in the cell cycle.
Distinctive Regulation during Early Cell Cycles
Beyond suggesting that growth issues underlie a universal program of early cell cycles without growth or transcription, creation of this special proliferation stage has been associated with a major retooling of regulation so that embryonic development begins in a special context.
In addition to impressive diversity in nature, evolution creates distinctions between tissues and between stages in the life histories of an organism. Exemplifying the latter, evolution optimizes larval and adult forms for different lifestyles. Phase 2 of animal growth is present in the life histories of all animals, and evolution might have optimized this stage. As evidence for such an evolutionary trajectory, I present three aspects of regulatory biology that are widely conserved among eukaryotes, and yet are not followed by early embryos.
Cells coordinate the synthesis of the precursors of DNA, the deoxynucleotide triphosphates (dNTPs), with replication. By providing the dNTPs just as they are used, their concentration never rises very high, and hydroxyurea inhibition of ribonucleotide reductase, an enzyme key to producing dNTPs, quickly brings DNA replication to a standstill in organisms from bacteria to human. Things are different in the early embryo. Xenopus eggs have high levels of dNTPs (Woodland and Pestell 1972). Additionally, hydroxyurea does not block the early S phases of Xenopus or of Drosophila (Landström et al. 1975; Newport and Dasso 1989; Howe et al. 1995; K Yuan, AW Shermoen, PH O’Farrell, unpubl.). Sea urchin eggs initiate cleavage cycles without one of the subunits of ribonucleotide reductase and must similarly have a preformed pool of dNTPs (Evans et al. 1983). Thus, eggs in at least three different phyla deviate from an otherwise universal program of regulation and carry a pool of dNTPs that provides DNA precursors for all of the early cell cycles.
Histones assemble newly replicated DNA into chromatin, and, like the dNTPs, histones are usually made only during S phase. Again early embryos are different. The egg carries a stockpile of histone proteins. The egg also has a large capacity to store this depot in complex with chaperones, such as nucleoplasmin (Laskey et al. 1978). This deviation from an otherwise universal program regulating histone supply could have a major consequence on the chromatin assembled.
Two types of chromatin exist in eukaryotes from yeast to humans: heterochromatin and euchromatin. The especially compacted heterochromatin, originally defined by intense histologic staining, is now more commonly distinguished by molecular hallmarks (e.g., a histone modification, H3-K9 methylation, and binding of heterochromatin protein 1 [HP1]). These marks are considered repressive because heterochromatin is generally transcriptionally quiescent. Additionally, heterochromatic DNA replicates later than euchromatic sequences, and the sequential replication of different regions of the genome is the main reasons that S phase is normally long (Shermoen et al. 2010). But during the early cell cycles, chromatin is relatively decompacted, repressive marks are absent, and all sequences replicate at the same time (Shermoen et al. 2010; Li et al. 2014; Yuan et al. 2014). Thus, the specialization of chromatin into euchromatic and heterochromatic domains, a feature of regulatory biology that spans known eukaryotic biology, is abandoned in the early embryonic cell cycles.
It is dangerous to suggest why things are the way they are, but it seems likely that the dramatic distinctions in mechanisms that characterize the early embryo are secondary to the devoted attention to the pell-mell cell cycles. Cell-cycle events can disrupt other processes. The spindle usurps cytoskeletal components, and cytokinesis disrupts intercellular membranes. Furthermore, nascent transcripts are aborted during mitosis, interrupting ongoing transcription until new RNA polymerases transit the entire gene (Shermoen and O’Farrell 1991). In the early embryonic cycles, cells are in mitosis for a large fraction of the cycle, and many cell biological processes appear to be held in abeyance.
In summary, constraints forced an alternation between different styles of growth. Each style of growth elicited evolutionary adaptations that altered fundamental features of biological regulation. Consequently, the transitions from one growth phase to another are widely transformative.
Erasing the Slate for Development
Because animals evolved with alternating growth phases as a consistent context, developmental processes might be optimized to function in their phase-specific biological setting. In this way, the unusual biology of the proliferation-only phase might have become an integral part of programs governing early animal development. Here I consider pluripotency—the ability of cells to form multiple cell and tissue types.
Induction of pluripotent stem cells in mammals is highly topical as result of current excitement about stem cells and their therapeutic potential. However, the capacity of a cell to produce diverse cell types is broadly important. As development progresses and cells differentiate, their genomes and the chromatin that packages these genomes are modified to restrict gene expression. But all life forms must begin each generation anew with a totipotent progenitor.
Though the epigenetic marks that specify the fates of cells can be transmitted over many cell divisions, John Gurdon’s classical experiments showed that a nucleus of a differentiated cell, when transplanted into an enucleated frog egg, could support development of a viable and fertile frog (Gurdon et al. 1958). Thus, the egg, acting either directly or by driving the implanted nucleus through a rapid series of maternally programmed divisions, can expand the developmental potency of an otherwise inadequate nucleus.
The resetting of epigenetic marks is a complex topic as there are many types of marks and complex interactions influencing their production, inheritance, and reversal. It is now known that pluripotency can be established by experimental treatments, but the egg cytoplasm is special in that it is both capable of resetting epigenetic marks and is the natural environment in which such a resetting should occur. To get a view of how the egg might reprogram a nucleus, I consider its specialized ability to activate one of the most repressed and condensed nuclei known, that of the sperm.
Following fertilization, the highly condensed sperm nucleus swells and the chromatin decompacts, replicates, and forms the male pronucleus, which joins the female pronucleus to participate in the rapid early-division cycles. In the early stages of this process, sperm nuclear basic proteins (SNBPs), which include specialized histones and protamines, are replaced with more conventional histones. Xenopus egg extracts promote this decompaction of sperm nuclei. Immunodepletion showed that this activity depends on nucleoplasmin, the abundant histone chaperone (Philpott et al. 1991). Nucleoplasmin-like chaperones in flies, sea urchins, and frogs appear to be involved in sperm decompaction.
The octomeric nucleosome is built in a two-step process by deposition of H3/H4 tetramer followed by addition of a H2A/H2B tetramer. Nucleoplasmin interacts with H2A/H2B, and so appears specialized for the second step. A fly mutant, hira, was unable to decompact the sperm nucleus, which then failed to contribute to zygotic events (Loppin et al. 2005). Hira is a conserved member of a protein complex that acts as an H3/H4 chaperone to promote replication-independent deposition of nucleosomes (Orsi et al. 2013). The Hira homologs in fish and mouse are similarly required for decompaction (Zhao et al. 2011; Lin et al. 2014).
Decompaction is a general activity of egg extracts with extracts of one species being able to decompact the sperm of various species, despite differences in the SNBPs. Furthermore, these extracts will decompact the chromatin of other nuclei, such as the inactive nuclei of avian red blood cells. I suggest that the ability to exchange basic chromatin proteins, whether protamines or histones, with a pool of naïve histones contributes to the ability of the egg to reprogram the epigenetic state of a nucleus (see Lin et al. 2014).
In contrast to egg extracts, oocyte extracts are ineffective at sperm decompaction. Nucleoplasmin is extensively phosphorylated at the time of egg activation, and dephosphorylation of nucleoplasmin compromises its activity in decompaction (Philpott et al. 2000). Phosphorylation of nucleoplasmin persists until the MBT is dependent on high-cyclin-dependent kinase activity, a specialization of the proliferation-only phase. Additionally, the egg and early cleavage stage embryo are well positioned to reprogram epigenetic marks because they have especially high levels of the histone chaperones and a unique pool of naïve histones available for exchange.
The speed of the early embryonic cycles might also contribute to reprogramming by outpacing the mechanisms responsible for epigenetic inheritance. Histone methylation and DNA methylation marks are added after DNA replication and histone deposition, and, if cells progress immediately to mitosis and another round of replication, there may not be adequate time to modify the newly replicated DNA and newly deposited histones. Indeed, during the extremely rapid early cell cycle in Drosophila embryos, chromatin lacks hallmarks of heterochromatin, H3K9 methylation, and bound HP1 binding, and these only make their appearance as the cycles slow. When the features of heterochromatin do appear, they do so in a sequence as if heterochromatin formation is an integral part of the developmental program (Shermoen et al. 2010).
I suggest that erasure of the epigenetic marks in externally deposited eggs is coupled to the unusual context in the egg and early embryo, and that restoration of epigenetic marks awaits slowing of the cell cycle when chromatin becomes less dynamic and can again accumulate modifications. As discussed below, even though these mechanisms are set in a different context in mammalian development, an understanding of the ancestral mechanism will give us deeper insight into this process in mammals (Cañon et al. 2011; Sánchez-Sánchez et al. 2011; Marandel et al. 2012; Lee et al. 2013).
WHY ARE MAMMALS SO DIFFERENT?
Mammalian eggs are smaller than externally deposited eggs, and their early cleavages are slow. So, despite a long heritage of predecessors that adhered to the four phases of growth that I have described, the early growth phases of mammalian embryos are altered. Here, I explore the nature of the difference, and why the mammalian program differs.
The Alignment Problem
One of the most dramatic oddities of mammalian development is that it does not start at the same point as the development of other animals. A developmental module generating extra embryonic tissues is inserted at the beginning of mammalian development. This insertion obscures relationships even with close vertebrate relatives (O’Farrell et al. 2004). Certainly, when I was faced with descriptions that began with fertilization, I was hard pressed to see common features among of the developmental programs of key vertebrate models—frog, chicken, and mouse.
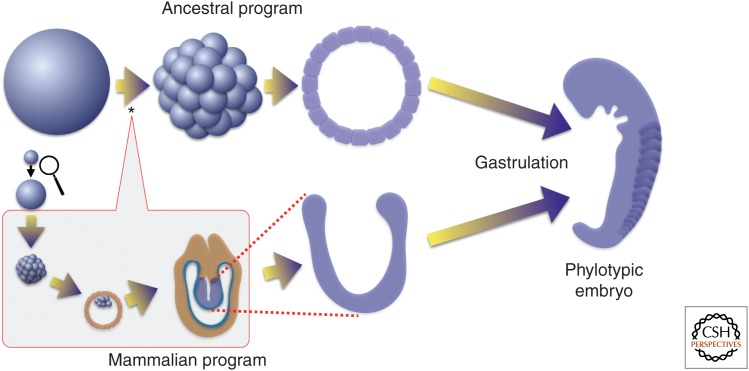
If, instead, we align development at the phylotypic stage (O’Farrell et al. 2004), one is struck by the close relationships among the embryos of these model vertebrate organisms, as was von Baer two centuries ago. Moving from this point of alignment to earlier stages reveals a point of divergence in the programs. In all three organisms, gastrulation precedes establishment of the phylotypic body plan and is regulated by related molecules and mechanisms, even if differences in topology of the movements obscure the parallel. In frog and chicken, as well as a vast number of organisms throughout phylogeny, the rapid cleavage cycles of the egg immediately precede gastrulation so that the blastomeres produced by cleavage make up the gastrula. In contrast, cleavage of the mouse egg begins 6.5 d before the start of gastrulation, and the inner cell mass (ICM) cells and trophoblast cells that result from these cleavages do not initiate gastrulation (Fig. 1), but instead begin a growth program discussed below. Recognition of the changes associated with the insertion of the mammalian specific module gives us new ways of thinking about the parallels between organisms.
Figure 1.
Insertion of a new module of development distinguishes early mammalian embryogenesis from that of its predecessors. Most animals deposit very large eggs in the external environment where they develop into a feeding organism without nutrition or growth. Cleavage cell cycles divide the egg into blastomeres that immediately initiate pattern formation and differentiation to produce an embryo, called phylotypic, because all of the species of a phylum look similar at this stage. Even the embryos of other phyla are similar in shape and size at this stage. Mammalian embryogenesis, as exemplified by the mouse, begins with a much smaller egg (magnified somewhat here) and a distinctly different early program of development. The cells resulting from the initial divisions do not gastrulate. First, extra embryonic tissues are produced as the embryo grows enormously and rapidly. Only then does it gastrulate and establish strong parallels to the ancestral program of embryogenesis. The mammalian module of development (boxed) is inserted early during development (*) so that the earliest divisions are dramatically changed and many of the attributes of the early events of the ancestral program are deferred along with gastrulation.
Insertion of Mass Increase into Phase 2 Growth
Protected and fed by the maternal environment, the mammalian embryo is released from growth constraint, and its altered development depends on the relaxation of the constraint. Mammalian eggs are small, and have ∼100–1000 times less cytoplasm than a typical externally deposited egg. Consequently, mammalian embryos need to grow to produce a phylotypic embryo of ordinary proportions. A growth period added to embryogenesis provides the chance to “catch up” to the embryos of externally deposited eggs.
The early mammalian egg begins development at a leisurely pace. One to two cell divisions per day are typical in early mammalian embryos. This slow beginning is a feature that appears in the derived mammalian lineage and is not seen in closely related vertebrates. For example, in the time it takes a mouse embryo to reach its second division, a chicken egg has generated a 60,000-cell blastodisc and is preparing to gastrulate. At 3 d, the still small mouse embryo has formed a blastocyst with 8 to 10 inner cell mass cells, which will contribute to the embryo proper. But these cells are far from ready to begin development at this point. Over the course of the next day and a half, the blastocyst hatches out of its protective shell (zona pellucida) and implants. On hatching, it begins to grow rapidly. During the next 3 d, the embryo grows in volume 500-fold (Snow 1981). There is also an impressive increase in cell number arriving at 15,000 cells at the time of gastrulation. Although this catch-up phase of growth makes a mammalian gastrula that is comparable in size and cell number to the gastrulae formed by externally deposited eggs, it also changes the foundation for gastrulation. It diminishes maternal input into embryogenesis and separates the cleavage cycles from the onset of patterning.
To understand the relationship of the mammalian program to the ancestral program, it is important to understand at what point the mammalian module has been inserted. I suggest that this module was inserted near the very beginning (see asterisk on the ancestral program in Fig. 1). Accordingly, the “cleavage” of the mammalian egg is part of the inserted program; hence, it is not surprising that its features bear little resemblance to the features of early divisions in externally deposited eggs. This arrangement suggests that the rapid cleavage cycles of the ancestral program have been displaced and are part of the growth program, a position that seems incongruous for “cleavage cycles,” but we will see that, although there are important modifications to the early division program, this alignment fits numerous observations.
Realignment of Early Development
According to the proposed displacement of early developmental events to a later stage in mammals, we should be able to identify parallels between the proliferative division that amplify cell number in the mammalian embryo and the cleavage cell cycle. One parallel is timing with respect to conserved features of embryogenesis—both types of divisions immediately precede gastrulation; another is speed.
The cell cycle just before gastrulation of the mammalian embryo are the fastest known in mammals. A 5.1-h cell-cycle time would be needed to explain the increasing cell number if growth were uniform. However, proliferation is not uniform, and, based on mitotic index, Snow estimated that cells just anterior to the streak, the morphological marker of gastrulation, double every 2–3 h (Snow 1977). Rat embryos show a similarly fast cell-cycle rate of ∼3–3.5 h near the streak (Mac Auley et al. 1993). Thus, these cycles, like the cleavage cycles of externally deposited eggs, are exceptionally fast. In addition to being fast, other similarities that I have previously reviewed suggest parallels between these pregastrulation cell cycles in mammals and the early cleavage cycles (O’Farrell et al. 2004). Here, I present the relationship between mammalian embryonic stem (ES) cells and the inserted growth phase, as well as describing apparent parallels of ES cell cycles to early embryonic cycles in externally deposited eggs.
ES cells were derived from the ICM of mouse blastocysts (Martin 1981). As Martin suggested in her report of the successful isolation of ES cells, perhaps the cells “represent a selected population of completely normal embryonic cells that are programmed to divide until they receive the appropriate signals for differentiation.” Indeed, the ICM cells divide and grow during the rapid-growth phase of the implanted mammalian embryo, and these cells are still growing at the egg cylinder stage, which also successfully yields pluripotent stem cells (Evans and Kaufman 1981). Thus, growing ES cells provide a model for the growth phase of the pregastrulation mammalian embryo.
ES cells also have a peculiar cell cycle that reverts to more a typical mode of regulation when these cells are induced to differentiate (Savatier et al. 1996). Before differentiation, ES cells divide quickly for a cultured cell, although not nearly as fast as the estimates for the embryonic cells. The ES cells also have remarkably short G1 and G2 phases, reminiscent of the absence of G1 and G2 in the cleavage cycles of externally deposited eggs (Coronado et al. 2013). Furthermore, the cycle is regulated unlike other cultured mammalian cells. During G1, the canonical regulators of progress to S phase, Rb, and cyclin E, are inappropriately phosphorylated and abundant, respectively (White et al. 2005). Furthermore, oscillations of numerous key cell-cycle regulators are greatly dampened, such that there is peculiarly high cyclin:Cdk activity in interphase (Savatier et al. 1996; Fujii-Yamamoto et al. 2005; Yang et al. 2011; van der Laan et al. 2013). Although these features of the ES cell cycle have puzzled investigators expecting an invariant mode of cell-cycle regulation, they are familiar to those of us exploring the changes in cell-cycle regulation during early development in other organisms. Notably, work in Drosophila has shown that the early rapid cycles are associated with very high interphase cyclin:Cdk activity, and near absence of oscillation in the abundance of key cell-cycle regulators (Edgar et al. 1994). Furthermore, there is not just one divergent mode of regulating the early cell cycle in Drosophila. As development progresses, the mode of regulating cell-cycle changes in several steps as S phase slows, a G2 is added and then a G1 is introduced. Rb and cyclin E come to play important regulatory roles only late in this process, consistent with their behavior in ES cells. I propose that cell-cycle regulation in ES cells, as well as the embryonic cells from which they are derived, is modified, and these modifications reflect their derivation from the early embryonic cycles of externally deposited eggs. It will be interesting to learn the extent of the parallels.
In summary, it appears that the mammalian module of development was inserted within the early rapid cell divisions with modifications to these divisions. This insertion displaced many developmental events to a later stage.
Pluripotency and the Transfer of Responsibility to the Zygotic Program
Perhaps the chief modification of the early embryonic programs is a switch from maternal programing to zygotic programming of the rapid divisions in mammals. The maternal programming of cleavage divisions used by externally deposited eggs is not practical in mammals because the rapid divisions that continue after extensive growth would have diluted maternal gene products. Furthermore, mammals have little need to rely on maternal programming. Given that their eggs are small and that the protected mammalian egg develops at a leisurely pace, even a single nucleus can modify transcript levels fast enough to regulate events. Indeed, mammalian embryos become dependent on zygotic gene expression within the first few divisions with kinetics that differ slightly between species. This early zygotic program faces requirements absent in externally deposited eggs. The zygotic program of mammalian embryos sustains a growing population of pluripotent cells and then provides these cells with a zygotic mimic of the ancestral maternally run cleavage program.
The addition of the zygotically supported embryonic growth program in mammalian development created a new need for a mechanism to sustain pluripotency. In externally deposited eggs, the early rapid divisions that produce the blastomeres appear to be part of the program that refreshes chromatin structures to create a naïve state for the beginning of development. Because these newly produced blastomeres immediately begin morphogenesis and differentiation at the time that they initiate zygotic gene expression, they have no need to maintain their pluripotency. But the mammalian embryo has to zygotically create pluripotency and maintain it during a growth phase that amplifies cell number >500-fold before initiating embryogenesis.
Tremendous advances in the study of stem cells have identified mammalian factors that can reprogram differentiated cells to pluripotency (Takahashi and Yamanaka 2006). These pluripotency factors are expressed in normal embryonic cells, and sustain pluripotency as the ICM cells of the blastocyst grow and proliferate before gastrulation (Marandel et al. 2012; Le Bin et al. 2014; Sun et al. 2014). If the pluripotent cells of the mammalian embryo are analogous to the cleavage stage blastomeres of externally deposited eggs, perhaps homologs of the pluripotency factors will be expressed in the blastomeres in the predecessors of mammals. Oct4, Sox2, and Nanog, three factors involved in maintenance of pluripotency in mammals, do have homologs in several nonmammalian vertebrates, and they are expressed in early pregastrulation embryos.
As a result of maternal supply and very early zygotic expression, the zebrafish homologs of Oct4, Sox2, and Nanog are present in the early embryo. They function jointly to promote early transcription. In their absence, transcription of many genes fails and the embryo does not gastrulate (Lee et al. 2013). These factors appear to act widely to direct transcription of genes necessary to stimulate the developmental potential of the blastomeres of the zebrafish embryo (Lee et al. 2013; Leichsenring et al. 2013). These findings suggest analogies between the pluripotency of mammalian embryonic cells and transcription factors directing the onset of the initial zygotic program of gene expression in fish.
Despite relatedness, Oct4, Sox2, and Nanog have somewhat different jobs in fish and mammals. In fish, these transcription factors act as a kick-starter to activate the first set of genes that are transcribed as the embryo transitions to zygotic expression. But the initial wave of transcription is transient as the embryo immediately advances to gastrulation and differentiation and new tiers of zygotic gene expression. In mammals, the role of pluripotency factors is less transient. These transcription factors are required to sustain developmental potency during growth and proliferation until the time of the deferred activation of gastrulation and differentiation. Presumably, vertebrates coopted a preexisting transcription program that drove early gene expression in vertebrate ancestors, and reconfigured the wiring of the program so that the expression of these early activators, their targets, and the early properties of the cells persist during the growth phase.
In addition to simple persistence, the pluripotency factors may have another job. In contrast to fish eggs, which are preloaded with maternal products to run the rapid embryonic cycles, the mammalian embryo has to support its dramatic growth by zygotic gene expression. Perhaps early embryonic transcription factors took on this growth-promoting role and now stimulate expression of cell-cycle and growth genes to drive the rampant pregastrulation growth in mammals.
The presence of the “pluripotency factors” in cleavage stage zebrafish embryos might suggest conservation of the program that erases epigenetic marks. However, relatively rapid divergence of these transcription factors outside of mammals shows that pluripotency in other species can be accomplished independent of these factors. I suggest that the extremely fast maternally run divisions in externally deposited eggs makes a major contribution to erasing epigenetic marks, and that the pluripotency factors took over this role in conjunction with the abandonment of the maternally driven division program in mammals.
Releasing the Growth Constraint Unleashes Evolutionary Change
I have argued that the simple requirement that an egg be able to produce a feeding organism gave rise to constraints on the growth program of animals. In particular, it forced the large-egg paradigm, and the requirement for a large egg generated new constraints that underlie the curious features of growth and proliferation in phases 1 and 2. A viviparous lifestyle opens up new possibilities. Development of the egg in a protected uterine environment in which nutrients can be provided frees evolution of these growth constraints. Once freed of constraints, change is possible and to some extent inevitable.
Several of the major changes seen in early mammalian development might be explained by the release of the constraint on growth. Clearly, growth of the embryo following hatching from the zona pellucida depends on the release of the growth constraint. It also seems likely that the small size of the mammalian egg and absence of yolk accumulation were allowable changes because of the release of the growth constraint. But because many coupled changes occurred during the evolution of mammals, it is hard to argue that release of the growth constraint was the driving event. Perhaps other examples of a release of the growth constraint can give us additional perspective on this.
It turns out that the constraint on growth of the embryo has been released numerous times in evolution, perhaps most frequently in organisms called endoparasites that deposit their eggs within the bodies of other organisms. This lifestyle has evolved independently numerous times (Grbić et al. 1998; Grbić 2000; Wiegmann et al. 2011), and a diversity of changes in early development are found among the species with this lifestyle (Grbić et al. 1998; Grbić 2000; Wiegmann et al. 2011). Although perhaps not an appealing analogy, the life histories of endoparasitic wasps provides a number of independent examples of branches of evolution in which eggs are released of their responsibility for providing for the development to a feeding adult stage. Additionally, the changes in development associated with these events can be analyzed in some detail because close relatives of the endoparasitic wasps are known and these adhere closely to a canonical program of early development described in detail in Drosophila.
The endoparasitic wasps lay their eggs inside the larvae of bigger insects where they develop in the nutrient-rich hemolymph (Grbić et al. 1998; Grbić 2000; Wiegmann et al. 2011). The eggs of such wasps are small, in the same size range as a mammalian egg, and lack yolk. The early development of these small eggs differs dramatically from the canonical insect program. Notably, a new developmental module is inserted at the beginning of development. The early mitotic cycles are greatly changed, and patterning events and gastrulation are delayed. During this period, the embryo grows dramatically reaching the same size as the externally developing embryos that began with vastly bigger eggs. The morphology of the gastrulating embryo differs from its close relatives; nonetheless, the endoparasitic wasps express classical insect-patterning genes in canonical patterns during this stage and generate a phylotypic embryo resembling that of its relatives. Although there is great diversity in the details, the overall features of the changes are similar in various lineages and are not unlike the reorganization of early embryogenesis in mammals. Notably, it is clear that development is appropriately aligned at the phylotypic stage, and that a direct comparison of the cleavage cycles between even closely related species would give the impression that there was no relationship.
Many endoparasitic wasps lay only one egg in an individual host larva, but, in a number of species, numerous progeny eventually emerge form this parasitized larvae. This is the result of a remarkable adaptation in which the embryo twins repeatedly in a process called polyembryony (Grbić et al. 1998; Grbić 2000; Wiegmann et al. 2011). This process, which is particularly dramatic and well studied in the wasp Copidosoma floridanum, takes advantage of the growth stage that was inserted into the early embryonic programs of the endoparasitic wasps. The pregastrulation embryo of this species forms a hollow ball of growing and proliferating cells. The shell of cells subdivides to produce additional balls as it grows. This process can produce in the range of 2000 embryos before patterning, gastrulation, and differentiation begin. This program, which is of obvious advantage in producing more wasps, is also an indication of the flexibility of the early developmental program once the growth constraint is removed.
Although the relatively close relationship of endoparasitic wasps with other wasps provides a particularly clear illustration of the consequences of the release of the growth constraint, other parasites provide additional examples of modification of the ancestral early embryonic program. Flukes, or trematodes, comprise a substantial class of parasitic flatworms with about four times as many species as there are species of mammals. Usually, the primary or definitive host is a vertebrate. The parasites, often hermaphroditic, produce small encysted eggs that are released from the infected primary host (Butcher et al. 2002). Typically, these eggs, if ingested by a mollusk, will develop into a primitive larva called a miracidium that takes in nourishment from the host mollusk and grows and develops germinative tissue that generates egg-like cells that enlarge and go through stages recognizable as cleavage, gastrulation, and establishment of a phylotypic body plan (Podvyaznaya and Galaktionov 2014; Skála et al. 2014). Notably, growth occurs throughout this embryogenesis. The miracidium nourishes many embryos in a uterus-like sac. On release, the progeny, now called cercariae, can infect the vertebrate host to begin a new cycle. Like the egg of the wasp, the egg of the fluke manages to produce many embryos—polyembryony. However, instead of simple serial twining of the embryo, a complex zygotic program of development in the molluskan host has been added to zygotically generate many egg-like cells that then initiate embryogenesis. This program suggests that, once the growth constraint is released, the earliest stage of development is evolutionarily plastic. In contrast to the dramatic changes in early zygotic development, events close to the phylotypic stage persist although in a context divorced from the usually close association with fertilization and initial division of the egg.
It is interesting to consider pluripotency in the context of polyembryony. This lifestyle requires that embryonic cells grow and proliferate but still retain pluripotency. The transcription factors responsible for zygotic maintenance of pluripotency in mammals are not conserved in insects, which appear to use different transcription factors (e.g., zelda in Drosophila) to promote early embryonic transcription. Perhaps the endoparasitic wasps will have independently evolved a means of supporting pluripotency during growth and proliferation. For example, persistent expression of a Zelda-like transcription factor might provide a wasp embryo with the equivalent of a pluripotency factor. We might expect that ES cell lines could be established from these organisms (see Jourdane and Theron 1980).
Overall, the rapid evolution of the early developmental programs in endoparasitic wasps supports the interpretation that the growth constraint played a major role in restraining such divergence. Despite occasional release of the constraint, we expect that many aspects of the regulatory programs that control growth and proliferation were molded by an unchanging constraint that influenced much of animal evolution.
SUMMARY
Consideration of oogenesis and early development across a wide swath of the animal kingdom suggests that the issue of growth has been an enduring constraint influencing much of evolution. It is suggested that this constraint emerges from a simple requirement that an externally deposited egg be able to produce a feeding animal to have a workable life cycle. This demand was met by production of large eggs. This then produced the widespread pattern of cell growth in phase 1 (oocyte production) and cell proliferation in phase 2 (early embryogenesis) of an overall four-phase animal program of growth and proliferation. This solution stressed the capacities of ordinary growth and division. The growth of an oocyte and its division as an egg shows numerous accommodations to these stresses. These accommodations are evident in representatives of all animal phyla, arguing that the biology of these stages has been molded by the pressures of this enduring constraint. Furthermore, embryonic development evolved with these imposed constraints, so the programs of development are interwoven with the programs for growth and proliferation. Finally, I argue that mammalian development ought to be viewed as a diverged branch of the mainline of animal evolution, which was dramatically influenced by the release of the growth constraint that was originally formative. Introduction of an early growth phase in mammalian embryogenesis required a novel feature, persistent pluripotency of the proliferating cells, which go on to ultimately support the deferred embryogenesis. Parasitic organisms, which also evade the growth constraint, show diverged early programs including small eggs, early embryonic growth, and acquisition of a proliferating pluripotent population of cells. These examples of the release of the growth constraint are an indication of the powerful influence that this constraint has exerted on animal evolution.
ACKNOWLEDGMENTS
I thank current and past members of the laboratory, especially Jeffrey Farrell, Jennifer Page, and Isaac Strong, as well as Danielle O’Farrell for help in preparing this article. Our efforts are supported by Grants R37GM037193 and R01ES020725 to P.H.O.
Footnotes
Editors: Rebecca Heald, Iswar K. Hariharan, and David B. Wake
Additional Perspectives on Size Control in Biology: From Organelles to Organisms available at www.cshperspectives.org
REFERENCES
- Bedford L. 1966. The electron microscopy and cytochemistry of oogenesis and the cytochemistry of embryonic development of the prosobranch gastropod Bembicium nanum L. J Embryol Exp Morphol 15: 15–37. [PubMed] [Google Scholar]
- Blobel G, Potter VR. 1967. Studies on free and membrane-bound ribosomes in rat liver. I: Distribution as related to total cellular RNA. J Mol Biol 26: 279–292. [DOI] [PubMed] [Google Scholar]
- Bratic I, Hench J, Henriksson J, Antebi A, Bürglin TR, Trifunovic A. 2009. Mitochondrial DNA level, but not active replicase, is essential for Caenorhabditis elegans development. Nucleic Acids Res 37: 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Dawid IB. 1968. Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science 160: 272–280. [DOI] [PubMed] [Google Scholar]
- Butcher AR, Brealey JK, Grove DI, Dymock RB. 2002. Brachylaima cribbi (Digenea: Brachylaimidae): Scanning electron microscopical observations of the life-cycle stages. J Helminthol 76: 207–215. [DOI] [PubMed] [Google Scholar]
- Cañon S, Fernandez-Tresguerres B, Manzanares M. 2011. Pluripotency and lineages in the mammalian blastocyst: An evolutionary view. Cell Cycle 10: 1731–1738. [DOI] [PubMed] [Google Scholar]
- Chase JW, Dawid IB. 1972. Biogenesis of mitochondria during Xenopus laevis development. Dev Biol 27: 504–518. [DOI] [PubMed] [Google Scholar]
- Coronado D, Godet M, Bourillot PY, Tapponnier Y, Bernat A, Petit M, Afanassieff M, Markossian S, Malashicheva A, Iacone R, et al. 2013. A short G1 phase is an intrinsic determinant of naïve embryonic stem cell pluripotency. Stem Cell Res 10: 118–131. [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC. 2003. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130: 1579–1590. [DOI] [PubMed] [Google Scholar]
- Del Pino EM, Humphries AA. 1978. Multiple nuclei during early oogenesis in Flectonotus pygmaeus and other marsupial frogs. Biol Bull 154: 198–212. [Google Scholar]
- Dobzhansky T. 1973. Nothing in biology makes sense except in the light of evolution. J Hered 68: 3–10. [DOI] [PubMed] [Google Scholar]
- Domazet-Lošo T, Tautz D. 2010. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468: 815–818. [DOI] [PubMed] [Google Scholar]
- Eckelbarger KJ, Larson R. 1992. Ultrastructure of the ovary and oogenesis in the jellyfish Linuche unguiculata and Stomolophus meleagris, with a review of ovarian structure in the Scyphozoa. Marine Biol 114: 633–643. [Google Scholar]
- Eckelbarger KJ, Young CM. 1997. Ultrastructure of the ovary and oogenesis in the methane-seep mollusc Bathynerita naticoidea (Gastropoda: Neritidae) from the Louisiana slope. Invertebr Biol 116: 299–312. [Google Scholar]
- Edgar BA, O’Farrell PH. 1990. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell 62: 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. 1986. Parameters controlling transcriptional activation during early Drosophila development. Cell 44: 871–877. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Kiehle CP, Schubiger G. 1986. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell 44: 365–372. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Sprenger F, Duronio RJ, Leopold P, O'Farrell PH. 1994. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev 8: 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156. [DOI] [PubMed] [Google Scholar]
- Evans T, Rosenthal ET, Youngblom J, Distel D, Hunt T. 1983. Cyclin: A protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33: 389–396. [DOI] [PubMed] [Google Scholar]
- Farrell JA, O’Farrell PH. 2013. Mechanism and regulation of Cdc25/twine protein destruction in embryonic cell-cycle remodeling. Curr Biol 23: 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JA, O’Farrell PH. 2014. From egg to gastrula: How the cell cycle is remodeled during the Drosophila mid-blastula transition. Annu Rev Genet 48: 269–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follette PJ, O’Farrell PH. 1997. Connecting cell behavior to patterning: Lessons from the cell cycle. Cell 88: 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follette PJ, Duronio RJ, O’Farrell PH. 1998. Fluctuations in cyclin E levels are required for multiple rounds of endocycle S phase in Drosophila. Curr Biol 8: 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii-Yamamoto H, Kim JM, Arai KI, Masai H. 2005. Cell cycle and developmental regulations of replication factors in mouse embryonic stem cells. J Biol Chem 280: 12976–12987. [DOI] [PubMed] [Google Scholar]
- Gall JG. 1968. Differential synthesis of the genes for ribosomal RNA during amphibian oögenesis. Proc Natl Acad Sci 60: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. 2012. Are lampbrush chromosomes unique to meiotic cells? Chromosome Res 20: 905–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Wu Z, Murphy C, Gao H. 2004. Structure in the amphibian germinal vesicle. Exp Cell Res 296: 28–34. [DOI] [PubMed] [Google Scholar]
- Ganot P, Bouquet JM, Kallesøe T, Thompson EM. 2007. The Oikopleura coenocyst, a unique chordate germ cell permitting rapid, extensive modulation of oocyte production. Dev Biol 302: 591–600. [DOI] [PubMed] [Google Scholar]
- Ganot P, Moosmann-Schulmeister A, Thompson EM. 2008. Oocyte selection is concurrent with meiosis resumption in the coenocystic oogenesis of Oikopleura. Dev Biol 324: 266–276. [DOI] [PubMed] [Google Scholar]
- Gardner EJ, Nizami ZF, Talbot CC, Gall JG. 2012. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev 26: 2550–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersh ES. 1968. Mutants at the bobbed locus in Drosophila melanogaster: Relation to ribosomal RNA synthesis? Science 162: 1139. [DOI] [PubMed] [Google Scholar]
- Graham A, Richardson J. 2012. Developmental and evolutionary origins of the pharyngeal apparatus. Evodevo 3: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić M. 2000. “Alien” wasps and evolution of development. Bioessays 22: 920–932. [DOI] [PubMed] [Google Scholar]
- Grbić M, Nagy LM, Strand MR. 1998. Development of polyembryonic insects: A major departure from typical insect embryogenesis. Dev Genes Evol 208: 69–81. [DOI] [PubMed] [Google Scholar]
- Gruzova MN, Batalova FM. 1979. Nuclear structures in the telotrophic ovarioles of nocturnal ground beetles (Tenebrionidae, Polyphaga). II: The oocyte nucleus of Blaps lethifera and Gnaptor spinimanus. Light optical data. Ontogenez 10: 323–331. [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M. 1958. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182: 64–65. [DOI] [PubMed] [Google Scholar]
- Harvey EB. 1935. Cleavage without nuclei. Science 82: 277. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Yamaguchi M. 1999. Kinase-independent activity of Cdc2/cyclin A prevents the S phase in the Drosophila cell cycle. Genes Cells 4: 111–122. [DOI] [PubMed] [Google Scholar]
- Howe JA, Howell M, Hunt T, Newport JW. 1995. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes Dev 9: 1164–1176. [DOI] [PubMed] [Google Scholar]
- Jourdane J, Theron A. 1980. Schistosoma mansoni: Cloning by microsurgical transplantation of sporocysts. Exp Parasitol 50: 349–357. [DOI] [PubMed] [Google Scholar]
- Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Ohler U, Bergman CM, Tomancak P. 2010. Gene expression divergence recapitulates the developmental hourglass model. Nature 468: 811–814. [DOI] [PubMed] [Google Scholar]
- Kloc M, Jedrzejowska I, Tworzydlo W, Bilinski SM. 2014. Balbiani body, nuage and sponge bodies—The germ plasm pathway players. Arthropod Struct Dev 43: 341–348. [DOI] [PubMed] [Google Scholar]
- Landström U, Løvtrup-Rein H, Løvtrup S. 1975. Control of cell division and cell differentiation by deoxynucleotides in the early embryo of Xenopus laevis. Cell Differ 4: 313–325. [DOI] [PubMed] [Google Scholar]
- Laskey RA, Honda BM, Mills AD, Finch JT. 1978. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275: 416–420. [DOI] [PubMed] [Google Scholar]
- Le Bin GC, Muñoz-Descalzo S, Kurowski A, Leitch H, Lou X, Mansfield W, Etienne-Dumeau C, Grabole N, Mulas C, Niwa H, et al. 2014. Oct4 is required for lineage priming in the developing inner cell mass of the mouse blastocyst. Development 141: 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. 2013. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503: 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mössner R, Driever W, Onichtchouk D. 2013. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 341: 1005–1009. [DOI] [PubMed] [Google Scholar]
- Li XY, Harrison MM, Villalta JE, Kaplan T, Eisen MB. 2014. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 10.7554/eLife.03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. 2014. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell 30: 268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B, Bonnefoy E, Anselme C, Laurençon A, Karr TL, Couble P. 2005. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437: 1386–1390. [DOI] [PubMed] [Google Scholar]
- Mac Auley A, Werb Z, Mirkes PE. 1993. Characterization of the unusually rapid cell cycles during rat gastrulation. Development 117: 873–883. [DOI] [PubMed] [Google Scholar]
- Macgregor HC, Del Pino EM. 1982. Ribosomal gene amplification in multinucleate oocytes of the egg brooding hylid frog Flectonotus pygmaeus. Chromosoma 85: 475–488. [DOI] [PubMed] [Google Scholar]
- Marandel L, Labbe C, Bobe J, Le Bail PY. 2012. nanog 5′-upstream sequence, DNA methylation, and expression in gametes and early embryo reveal striking differences between teleosts and mammals. Gene 492: 130–137. [DOI] [PubMed] [Google Scholar]
- Marlow FL, Mullins MC. 2008. Bucky ball functions in Balbiani body assembly and animal–vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev Biol 321: 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci 78: 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Dasso M. 1989. On the coupling between DNA replication and mitosis. J Cell Sci 12: 149–160. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. 1982a. A major developmental transition in early Xenopus embryos. I: Characterization and timing of cellular changes at the midblastula stage. Cell 30: 675–686. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. 1982b. A major developmental transition in early Xenopus embryos. II: Control of the onset of transcription. Cell 30: 687–696. [DOI] [PubMed] [Google Scholar]
- O’Farrell PH. 2004. How metazoans reach their full size: The natural history of bigness. In Cell growth: Control of cell size (ed. Hall M, et al. ). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- O’Farrell PH. 2011. Quiescence: Early evolutionary origins and universality do not imply uniformity. Phil Trans R Soc B 366: 3498–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell PH, Stumpff J, Su TT. 2004. Embryonic cleavage cycles: How is a mouse like a fly? Curr Biol 14: R35–R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi GA, Algazeery A, Meyer RE, Capri M, Sapey-Triomphe LM, Horard B, Gruffat H, Couble P, Aït-Ahmed O, Loppin B. 2013. Drosophila Yemanuclein and HIRA cooperate for de novo assembly of H3.3-containing nucleosomes in the male pronucleus. PLoS Genet 9: e1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH, Martin-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. 1989. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell 58: 955–968. [DOI] [PubMed] [Google Scholar]
- Pepling ME, de Cuevas M, Spradling AC. 1999. Germline cysts: A conserved phase of germ cell development? Trends Cell Biol 9: 257–262. [DOI] [PubMed] [Google Scholar]
- Philpott A, Leno GH, Laskey RA. 1991. Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell 65: 569–578. [DOI] [PubMed] [Google Scholar]
- Philpott A, Krude T, Laskey RA. 2000. Nuclear chaperones. Semin Cell Dev Biol 11: 7–14. [DOI] [PubMed] [Google Scholar]
- Podvyaznaya IM, Galaktionov KV. 2014. Trematode reproduction in the molluscan host: An ultrastructural study of the germinal mass in the rediae of Himasthla elongata (Mehlis, 1831) (Digenea: Echinostomatidae). Parasitol Res 113: 1215–1224. [DOI] [PubMed] [Google Scholar]
- Pritchard DK, Schubiger G. 1996. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev 10: 1131–1142. [DOI] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N. 2010. Evolutionary biology: Genomic hourglass. Nature 468: 768–769. [DOI] [PubMed] [Google Scholar]
- Prud’homme B, de Rosa R, Arendt D, Julien JF, Pajaziti R, Dorresteijn AWC, Adoutte A, Wittbrodt J, Balavoine G. 2003. Arthropod-like expression patterns of engrailed and wingless in the annelid Platynereis dumerilii suggest a role in segment formation. Curr Biol 13: 1876–1881. [DOI] [PubMed] [Google Scholar]
- Raff RA. 1996. The shape of life. University of Chicago Press, Chicago. [Google Scholar]
- Rahn H, Paganelli CV, Ar A. 1974. The avian egg: Air-cell gas tension, metabolism and incubation time. Respir Physiol 22: 297–309. [DOI] [PubMed] [Google Scholar]
- Sánchez-Sánchez AV, Camp E, Mullor JL. 2011. Fishing pluripotency mechanisms in vivo. Int J Biol Sci 7: 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB, Samarut J. 1996. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene 12: 309–322. [PubMed] [Google Scholar]
- Shen MM. 2007. Nodal signaling: Developmental roles and regulation. Development 134: 1023–1034. [DOI] [PubMed] [Google Scholar]
- Shermoen AW, O’Farrell PH. 1991. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell 67: 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, McCleland ML, O’Farrell PH. 2010. Developmental control of late replication and S phase length. Curr Biol 20: 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skála V, Bulantová J, Walker AJ, Horák P. 2014. Insights into the development of Notocotylus attenuatus (Digenea: Notocotylidae) in Lymnaea stagnalis: From mother sporocyst to cercariae. Parasitol Int 63: 94–99. [DOI] [PubMed] [Google Scholar]
- Smiley S. 1990. A review of echinoderm oogenesis. J Electron Microsc Tech 16: 93–114. [DOI] [PubMed] [Google Scholar]
- Snow MHL. 1977. Gastrulation in the mouse: Growth and regionalization of the epiblast. J Embryol Exp Morphol 42: 293–303. [Google Scholar]
- Snow MH. 1981. Growth and its control in early mammalian development. Br Med Bull 37: 221–226. [DOI] [PubMed] [Google Scholar]
- Sun LT, Yamaguchi S, Hirano K, Ichisaka T, Kuroda T, Tada T. 2014. Nanog co-regulated by Nodal/Smad2 and Oct4 is required for pluripotency in developing mouse epiblast. Dev Biol 392: 182–192. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. [DOI] [PubMed] [Google Scholar]
- Tsang WY, Lemire BD. 2002. Mitochondrial genome content is regulated during nematode development. Biochem Biophys Res Commun 291: 8–16. [DOI] [PubMed] [Google Scholar]
- Tworzydlo W, Kloc M, Bilinski SM. 2009. The Balbiani body in the female germline cells of an earwig, Opisthocosmia silvestris. Zool Sci 26: 754–757. [DOI] [PubMed] [Google Scholar]
- van der Laan S, Tsanov N, Crozet C, Maiorano D. 2013. High Dub3 expression in mouse ESCs couples the G1/S checkpoint to pluripotency. Mol Cell 52: 366–379. [DOI] [PubMed] [Google Scholar]
- Webb AC, Smith LD. 1977. Accumulation of mitochondrial DNA during oogenesis in Xenopus laevis. Dev Biol 56: 219–225. [DOI] [PubMed] [Google Scholar]
- White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. 2005. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell 16: 2018–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmann BM, Trautwein MD, Winkler IS, Barr NB, Kim JW, Lambkin C, Bertone MA, Cassel BK, Bayless KM, Heimberg AM, et al. 2011. Episodic radiations in the fly tree of life. Proc Natl Acad Sci 108: 5690–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E. 1996. Embryonic transcription and the control of developmental pathways. Genetics 142: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SF, Schlessinger D. 1977. Nuclear metabolism of ribosomal RNA in growing, methionine-limited, and ethionine-treated HeLa cells. Biochemistry 16: 2783–2791. [DOI] [PubMed] [Google Scholar]
- Woodland H. 1982. The translational control phase of early development. Biosci Rep 2: 471–491. [DOI] [PubMed] [Google Scholar]
- Woodland HR, Pestell RQ. 1972. Determination of the nucleoside triphosphate contents of eggs and oocytes of Xenopus laevis. Biochem J 127: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang VS, Carter SA, Hyland SJ, Tachibana-Konwalski K, Laskey RA, Gonzalez MA. 2011. Geminin escapes degradation in G1 of mouse pluripotent cells and mediates the expression of Oct4, Sox2, and Nanog. Curr Biol 21: 692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, Shermoen AW, O’Farrell PH. 2014. Illuminating DNA replication during Drosophila development using TALE-lights. Curr Biol 24: R144–R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZK, Li W, Wang MY, Zhou L, Wang JL, Wang YF. 2011. The role of HIRA and maternal histones in sperm nucleus decondensation in the gibel carp and color crucian carp. Mol Reprod Dev 78: 139–147. [DOI] [PubMed] [Google Scholar]