Abstract
Horseradish peroxidase (HRP), conjugated to antibodies and lectins, is widely used in medical diagnostics. Since recombinant production of the enzyme is difficult, HRP isolated from plant is used for these applications. Production in the yeast Pichia pastoris (P. pastoris), the most promising recombinant production platform to date, causes hyperglycosylation of HRP, which in turn complicates conjugation to antibodies and lectins. In this study we combined protein and strain engineering to obtain an active and stable HRP variant with reduced surface glycosylation. We combined four mutations, each being beneficial for either catalytic activity or thermal stability, and expressed this enzyme variant as well as the unmutated wildtype enzyme in both a P. pastoris benchmark strain and a strain where the native α-1,6-mannosyltransferase (OCH1) was knocked out. Considering productivity in the bioreactor as well as enzyme activity and thermal stability, the mutated HRP variant produced in the P. pastoris benchmark strain turned out to be interesting for medical diagnostics. This variant shows considerable catalytic activity and thermal stability and is less glycosylated, which might allow more controlled and efficient conjugation to antibodies and lectins.
Keywords: horseradish peroxidase, glyco-engineering, strain engineering, bioreactor cultivation, OCH1, site-directed mutagenesis
1. Introduction
The methylotrophic yeast Pichia pastoris is a widely used host organism for recombinant protein production. It can grow on cheap and defined media to high cell densities, is robust against stressful conditions and able to perform post-translational modifications, like glycosylation [1,2,3,4]. However, glycosylation in Pichia pastoris (P. pastoris) is characterized by a severe drawback: native glycosyltransferases recognize the amino acid motif Asn-X-Ser/Thr and link many glycan moieties to the asparagine residues of recombinant proteins yielding a hyperglycosylated product (e.g., [5,6,7]). Hyperglycosylation causes many problems as it alters the physicochemical properties of the product, hampers downstream processing [5,8] and prevents medical applications. One of the main disadvantages of yeast-derived glycosylation lies in its heterogeneous nature making subsequent conjugation to antibodies and lectins, a prerequisite for applications in medical diagnostics, extremely difficult. Two different approaches can be applied to reduce yeast-derived hyperglycosylation, namely (1) protein engineering and (2) strain engineering. The yeast P. pastoris is a prominent example for such efforts. In several studies detailed strain engineering approaches describing humanization of this yeast were reported (Table 1; e.g., [7,8,9,10,11,12,13,14]).
Table 1.
Selected studies focusing on the humanization of N-glycosylation in P. pastoris.
| Goal of the Study | Citation |
|---|---|
| Introduction of α-1,2-Mns and GntI, OCH1 inactivation via a knock-in plasmid | [9] |
| Introduction of an UDP-GlcNAc transporter, α-1,2-MnsIA, MnsII, GntI, GntII in a Δoch1::URA3 strain | [10] |
| Introduction of sialic acid biosynthesis pathway to produce sialylated glycoproteins | [11] |
| OCH1 knock-out and introduction of glycosidases and glycosyltransferases to produce terminally galactosylated glycoproteins | [14] |
Mns, mannosidase; Gnt, β-N-acetylglucosaminyltransferase; UDP-GlcNAc, uridine diphosphate-N-acetylglucosamine; OCH1, outer chain elongation gene.
We used P. pastoris for the recombinant production of the heme-containing plant enzyme horseradish peroxidase (HRP), an enzyme widely used in medical diagnostics (e.g., [5,15]). We chose this yeast as expression host because Morawski et al. [16,17] had shown P. pastoris to produce recombinant proteins with significant shorter surface glycans than Saccharomyces cerevisiae (S. cerevisiae). We investigated various aspects of the expression of HRP in P. pastoris: different HRP isoenzymes were produced [18,19], production strategies were developed and optimized [20,21,22,23], media supplementation and strain engineering for increased heme-incorporation were analyzed [24] and the methanol utilization pathway of P. pastoris was manipulated for higher productivity [25].
In more recent studies we applied both protein and strain engineering with the goal of producing more homogenously glycosylated HRP variants in P. pastoris. However, we did not use fully humanized yeast strains, which are proprietary, but produced HRP in a P. pastoris strain with a deleted och1 gene (Δoch1 strain; [26]). This gene codes for an α-1,6-mannosyltransferase responsible for triggering the uncontrolled addition of mannose residues to the recombinant protein. Although HRP produced in the Δoch1 strain was more homogeneously glycosylated with more than 70% of the enzyme being of the Man8 glycosylation type, the Δoch1 strain showed a growth impaired phenotype and was hard to cultivate [26]. In a subsequent study, we analyzed the effect of the process parameters temperature, pH and dissolved oxygen concentration on strain physiology and productivity [6]. We found that the space-time-yield (STY) of the glyco-engineered strain was eight-fold lower compared to an unmodified P. pastoris benchmark strain, making HRP production in the Δoch1 strain unattractive.
Thus, we glyco-engineered the enzyme HRP by mutating each of the eight N-glycosylation sites to produce in a benchmark strain but still get HRP with reduced surface glycosylation [27]. We found different effects of the single mutations on enzyme activity and stability and demonstrated that mutation of the respective N-glycosylation site in fact resulted in the absence of glycans there [27]. Although the combination of all eight mutations gave a non-glycosylated enzyme, both catalytic activity and thermal stability were dramatically reduced leading to the hypothesis that some of the N-glycosylation sites must not be mutated to obtain active and stable HRP.
In the present study we combined both (1) protein engineering and (2) strain engineering to obtain a stable and active HRP variant with reduced and more homogeneous surface glycosylation. We combined the four mutations we had identified as being beneficial for either catalytic activity or stability in a previous study (Table 2; [27]).
Table 2.
Biochemical characteristics of the unmodified horseradish peroxidase (HRP) wildtype enzyme (wt) and four different HRP variants [27].
| Enzyme | ABTS | H2O2 | Thermal Half-Life Time | ||||
|---|---|---|---|---|---|---|---|
| Variant | Km (mM) | vmax (U/mg) | vmax/Km (U/mg/mM) | Km (mM) | vmax (U/mg) | vmax/Km (U/mg/mM) | τ½ (min) |
| wt | 1.60 | 44.2 | 27.7 | 0.003 | 16.3 | 5433 | 20.6 |
| N13D | 2.90 | 47.2 | 16.3 | 0.005 | 14.7 | 3066 | 28.9 |
| N57S | 2.98 | 113 | 38.1 | 0.004 | 23.7 | 5378 | 38.5 |
| N255D | 1.72 | 51.5 | 29.9 | 0.005 | 21.6 | 4506 | 11.6 |
| N268D | 1.89 | 32.5 | 17.2 | 0.003 | 10.6 | 3642 | 61.9 |
ABTS, 2-2-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid); H2O2, hydrogen peroxide.
We produced the enzyme variant, called 4/8 HRP, in both an unmodified P. pastoris benchmark strain and a Δoch1 strain. Summarizing, enzymes produced in the Δoch1 strain were characterized by lower catalytic activity and stability. However, 4/8 HRP produced in the P. pastoris benchmark strain (enzyme wt4/8 HRP) showed considerable catalytic activity and increased thermal stability. In combination with its significantly reduced surface glycosylation, which might allow more controlled conjugation to antibodies and lectins, this variant might be useful for applications in medical diagnostics in the future.
2. Results and Discussion
2.1. Strain Physiology
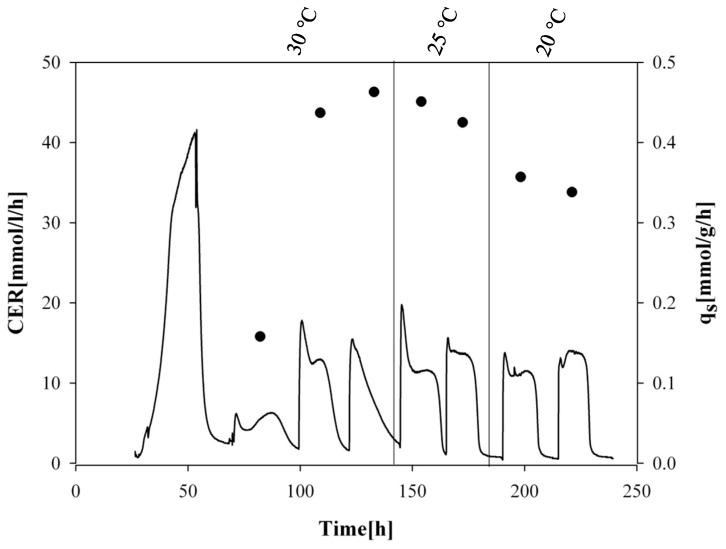
We cultivated the four recombinant P. pastoris strains in dynamic batch cultivations with methanol pulses at different temperatures and analyzed the effect of temperature on strain physiology. A typical cultivation is exemplarily shown for strain wt4/8 HRP in Figure 1.
Figure 1.
Schematic overview of the dynamic batch cultivation of strain wt4/8 HRP with methanol pulses at different temperatures. Black continuous line, carbon dioxide evolution rate (CER); black dots, specific methanol uptake rate (qs MeOH).
The most important strain physiological parameters are summarized in Table 3. Closing C-balances for the benchmark strains underline data validity. Similar to our previous studies, we observed that the Δoch1 strains lost metabolic activity over time and were affected by cell lysis [26]. Thus, C-balances did not close.
Table 3.
Physiological parameters of strains wtwt HRP, benchmark strain expressing the unmutated HRP enzyme; wt4/8 HRP, benchmark strain expressing the mutated 4/8 HRP variant; OCH1wt HRP, deleted OCH1 gene strain expressing the unmutated HRP enzyme and OCH14/8 HRP delete OCH1 gene strain expressing the mutated 4/8 HRP variant were determined in dynamic batch cultivations.
| Strain | μmax gly (h−1) | Δtime adapt (h) | qs adapt (mmol/g/h) | qs MeOH 20 °C (mmol/g/h) | qs MeOH 25 °C (mmol/g/h) | qs MeOH 30 °C (mmol/g/h) | C-Balance |
|---|---|---|---|---|---|---|---|
| wtwt HRP | 0.271 | 11.1 | 0.272 | 0.931 | 1.190 | 1.32 | 0.96 |
| wt4/8 HRP | 0.200 | 16.0 | 0.158 | 0.354 | 0.438 | 0.450 | 0.97 |
| OCH1wt HRP | 0.199 | 4.5 | 0.370 | 0.891 | 0.780 | 0.632 | const. Decreasing |
| OCH14/8 HRP | 0.182 | 3.8 | 0.400 | 1.02 | 0.955 | 0.800 | const. Decreasing |
When comparing the recombinant benchmark strains, great differences in specific methanol uptake rates (qs MeOH) were identified. While for both strains qs MeOH increased with increasing temperature, a correlation we also had observed before [6], the strain producing the 4/8 HRP variant showed a three-fold lower qs MeOH compared to the strain producing the unmutated enzyme at the respective temperature. Apparently, production of the glyco-engineered 4/8 HRP caused a physiological burden for the yeast, decelerating metabolism and thus methanol uptake.
When comparing the recombinant Δoch1 strains, we observed that qs MeOH for both strains decreased with increasing temperature, a phenomenon we also had described before [6]. However, qs MeOH of both Δoch1 strains were comparable at each temperature indicating that the mutated product did not cause any physiological burden. We speculate that the glyco-engineered 4/8 HRP variant might cause problems in the P. pastoris benchmark strain during secretion as it might get stuck in the cell wall. On the contrary, the same enzyme variant can be secreted without any problems in the Δoch1 strain, which has a completely altered cell wall structure [24]. However, this remains to be elucidated in detail.
2.2. Protein Purification
After harvest, we purified the different enzyme variants by a 1-step hydrophobic charge interaction chromatography (HCIC) purification strategy [8,19]. As shown in Table 4, the majority of each HRP variant was found in the respective flow-through fraction, where purification factors (PF) between 1.34 and 1.53 were achieved. However, enzyme OCH14/8 HRP showed a different result, as nearly 20% of the enzyme was retained on the resin. The eluate fraction showed a higher PF compared to the flow-through fraction (Table 4). We ascribe this phenomenon to the fact that this HRP variant was the least glycosylated one, as the four remaining glycosylation sites mainly carried Man8 instead of longer Man chains (respective detailed analyses had been performed in our previous study [26]). Apparently, the reduced glycosylation of enzyme OCH14/8 HRP allows physico-chemical interactions of the enzyme variant and the resin. One might speculate that the amount of enzyme variant able to interact with the resin should be even higher. We think that the rather stressful manner of cultivation with pulses and temperature shifts might have caused very heterogeneous glycosylation on the four remaining N-glycosylation sites and thus resulted in the still rather low fraction of only around 20% enzyme interacting with the resin. Detailed analysis of surface glycosylation by mass spectrometry, as we have done previously [24], could shed light on this speculation. However, we did not perform this analysis in this study, since variant OCH14/8 HRP did not turn out to be interesting for further applications due to low catalytic activity (see below).
Table 4.
Results of the hydrophobic charged interaction chromatography (HCIC) purification and Reinheitszahl (RZ) measurements for the four different HRP enzyme variants.
| HCIC | Concentrated Fraction | ||||
|---|---|---|---|---|---|
| Enzyme Variant | R% Total | R% FT | PF FT | Specific Activity (U/mg) | RZ (A404/A280) |
| wtwt HRP | 87.2 | 87.2 | 1.49 | 273.3 | 0.50 |
| wt4/8 HRP | 97.4 | 95.6 | 1.34 | 36.4 | 0.51 |
| OCH1wt HRP | 99.8 | 98.7 | 1.53 | 59.8 | 0.63 |
| OCH14/8 HRP | 93.1 | 76.1 | 1.47 | 11.5 | 0.19 |
| Variant | R% Total | R% Eluate | PF Eluate | Specific Activity (U/mg) | RZ (A404/A280) |
| OCH14/8 HRP | 93.1 | 17.5 | 2.43 | 19.1 | 0.31 |
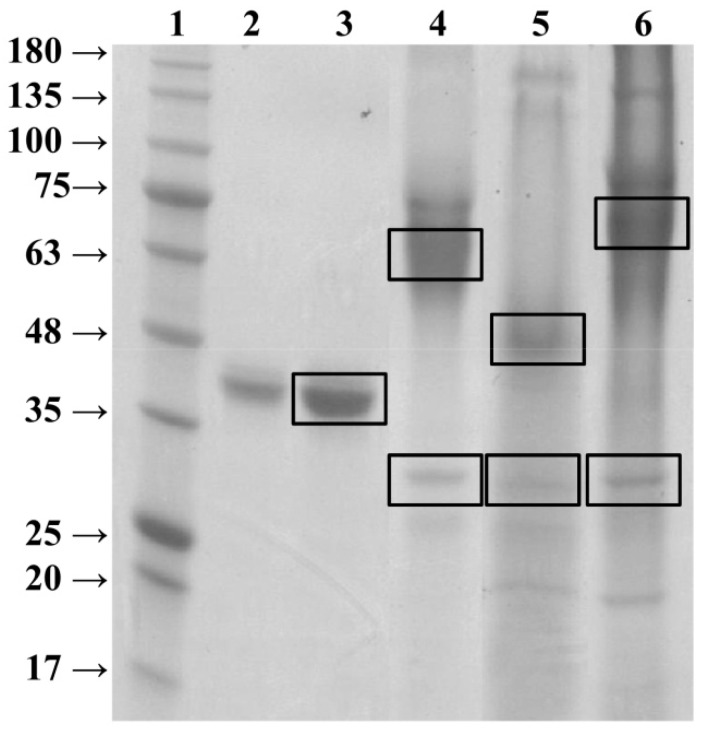
After HCIC, flow-through fractions showing highest PFs were pooled and concentrated to around 1.5 mL. To assess purity of the different enzyme preparations Reinheitszahl values (RZ; A404/A280) were determined (Table 4). Highly pure HRP preparations are known to have RZ values of more than 3.0 [16]. Although we did not get pure enzyme preparations by the one-step HCIC purification in this study, the RZ values of the different enzyme variants were in the same range. We also analyzed the different enzyme variants on SDS-PAGE gels, to identify potential differences in apparent size (Figure 2). Furthermore, interesting protein bands were excised and analyzed by mass spectrometry (respective bands are indicated in black boxes in Figure 2).
Figure 2.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of different horseradish peroxidase (HRP) variants. Black boxes indicate protein bands analyzed by mass spectrometry. Lane 1, BLUeye Prestained Protein Ladder; lanes 2 and 3, HRP from plant in two different concentrations (4 μg protein and 8 μg protein, respectively); lane 4, wtwt HRP, unmutated HRP enzyme expressed in the benchmark strain; lane 5, wt4/8 HRP, mutated 4/8 HRP variant expressed in the benchmark strain; lane 6, OCH14/8 HRP, mutated 4/8 HRP variant expressed in the deleted OCH1 gene strain.
The enzyme preparation from plant showed a rather distinct band at an apparent size of around 40 kDa on the SDS gel (lanes 2 and 3 in Figure 2), whereas the recombinant enzyme wtwt HRP showed a smear at an apparent size of around 65 kDa (lane 4 in Figure 2). Both proteins were identified as HRP by mass spectrometry (Table 5). The apparent size difference of around 20 kDa and the smeary appearance of wtwt HRP result from the heterogeneous yeast-derived glycosylation [17,20,21]. The second prominent protein band in lane 4 at an apparent size of around 30 kDa was identified as a glucosidase from P. pastoris (Table 5).
Table 5.
Identification of prominent protein bands by mass spectrometry.
| Lane | Apparent Size [kDa] | Rank | Peptides | Scores | Protein | Accession |
|---|---|---|---|---|---|---|
| 3 | 45 | 1 | 11 | 608.9 | Peroxidase C1A Organism species (OS)=Armoracia rusticana | PER1A_ARMRU |
| 4 | 65 | 1 | 6 | 386.9 | 1,3-β-glucanosyltransferase OS=Komagataella pastoris | Q0QCW1_PICPA |
| 2 | 9 | 368.0 | Peroxidase C1A OS=Armoracia rusticana | PER1A_ARMRU | ||
| 30 | 1 | 9 | 411.5 | Glucan 1,3-β-glucosidase OS=Komagataella pastoris | F2QPL8_PICP7 | |
| 5 | 50 | 1 | 9 | 454.5 | Alpha-1-antichymotrypsin 2 OS=Sus scrofa | Q9GMA6_PIG |
| 2 | 9 | 386.6 | Keratin, type II cytoskeletal 1 OS=Homo sapiens | K2C1_HUMAN | ||
| 3 | 9 | 317.4 | Peroxidase C1A OS=Armoracia rusticana | PER1A_ARMRU | ||
| 30 | 1 | 9 | 411.5 | Glucan 1,3-β-glucosidase OS=Komagataella pastoris | F2QPL8_PICP7 | |
| 6 | 70 | 1 | 20 | 1136.6 | Primary-amine oxidase OS=Komagataella pastoris | F2QTE6_PICP7 |
| 2 | 10 | 685.6 | 1,3-β-glucanosyltransferase OS=Komagataella pastoris | Q0QCW1_PICPA | ||
| 3 | 9 | 548.8 | 1,3-β-glucanosyltransferase OS=Komagataella pastoris | F2QQJ2_PICP7 | ||
| 30 | 1 | 9 | 411.5 | Glucan 1,3-β-glucosidase OS=Komagataella pastoris | F2QPL8_PICP7 |
As shown in Figure 2, the preparation of enzyme wt4/8 HRP showed a different protein pattern on the SDS gel, as the band at an apparent size of 65 kDa disappeared, whereas a prominent band at an apparent size of around 50 kDa appeared. In fact, this protein was identified as HRP (Table 5). Apparently, the mutation of four of the eight N-glycosylation sites resulted in the absence of glycans there and thus a size reduction of around 15 kDa. This nicely underlines the feasibility of reducing the vast and heterogeneous glycosylation of recombinant proteins from yeast by protein design. The second prominent band at an apparent size of around 30 kDa was again identified as a glucosidase (Table 5).
Lane 6 in Figure 2 shows the protein bands of enzyme preparation OCH14/8 HRP. Again, we observed a different protein pattern. The prominent band at an apparent size of around 70 kDa was identified as an oxidase and the band at an apparent size of around 30 kDa again as a glucosidase (Table 5). We expected to see a protein band at an apparent size of around 45 kDa representing enzyme OCH14/8 HRP. However, no respective band was visible on the SDS gel. We ascribe this absence to the extremely low protein production in the Δoch1 strain [6,26]. Furthermore, cell lysis during bioreactor cultivation resulted in a rather high impurity pattern, which is also demonstrated by the low RZ value for this enzyme variant (Table 4). However, we still measured enzymatic activity for OCH14/8 HRP and thus included this enzyme variant in the comparative biochemical characterization.
2.3. Biochemical Enzyme Characterisation
After purification, we biochemically characterized the different HRP variants. The kinetic parameters for ABTS and H2O2 are summarized in Table 6. We also included plant HRP for comparison. Introducing the four mutations N13D, N57S, N255D and N268D into HRP did not affect the affinity towards ABTS and H2O2 as Km values of enzymes wtwt HRP and wt4/8 HRP were comparable. However, the catalytic efficiency was reduced seven-fold for ABTS and six-fold for H2O2, respectively. Apparently, the introduced mutations did not only reduce surface glycosylation (Figure 2) but also affected the active site and reduced catalytic activity.
Table 6.
Kinetic constants of four different HRP enzyme variants and plant HRP.
| Enzyme | ABTS | H2O2 | ||||
|---|---|---|---|---|---|---|
| Variant | Km (mM) | vmax (U/mg) | vmax/Km (U/mg/mM) | Km (mM) | vmax (U/mg) | vmax/Km (U/mg/mM) |
| wtwt HRP | 1.50 | 152.9 | 101.9 | 0.009 | 55.2 | 6133 |
| wt4/8 HRP | 0.99 | 13.6 | 13.7 | 0.015 | 15.5 | 1030 |
| OCH1wt HRP | 1.56 | 26.8 | 17.2 | 0.008 | 10.4 | 1300 |
| OCH14/8 HRP | 1.34 | 1.28 | 0.96 | 0.016 | 1.29 | 80.7 |
| plant HRP | 1.75 | 567.2 | 324.8 | 0.033 | 377.8 | 11,589 |
ABTS, 2-2-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid); H2O2, hydrogen peroxide.
When we produced the HRP variants in the Δoch1 strain, we found that reduced surface glycosylation, caused by the absence of the native enzyme OCH1 [26], did not alter substrate affinity either. In fact, Km values of all four HRP variants for both ABTS and H2O2 were comparable. However, reducing surface glycosylation affected catalytic efficiency. Enzyme OCH1wt HRP showed a nearly six-fold reduced vmax/Km compared to enzyme wtwt HRP. Apparently, the benefit of having a more homogeneously glycosylated enzyme produced in the Δoch1 strain comes at cost of catalytic activity. Finally, we characterized the mutated 4/8 HRP produced in the Δoch1 strain (enzyme OCH14/8 HRP). This enzyme is the least glycosylated one in this study, only providing four out of eight N-glycosylation sites, which are mainly occupied by Man8-glycan structures [26]. However, compared to enzyme wtwt HRP the catalytic efficiency for ABTS was reduced 119-fold and for H2O2 76-fold, respectively. Although, enzyme OCH14/8 HRP is basically fit for medical applications, as it misses the heterogeneous yeast-derived glycan structures, the highly reduced catalytic activity as well as low productivity in the bioreactor will most likely prevent future applications.
Compared to the commercially available HRP preparation from plant, all recombinant HRP variants showed comparable Km values but reduced catalytic activity. This might be a result from incomplete heme incorporation or different surface glycosylation. However, the plant preparation contains a mixture of different HRP isoenzymes with varying surface glycosylation, which has to be isolated from its natural source in a rather cumbersome way. Furthermore, seasonal variation in the isoenzyme content and thus the variable production scenario describe only some of the disadvantages of plant HRP. Thus, even though recombinant HRP variants show lower catalytic activity, they are still interesting for industry, as they only describe a single isoenzyme that can be produced in a predictable manner in the controlled environment of a bioreactor. Furthermore, less glycosylated enzymes might allow more controlled and efficient conjugation to antibodies and lectins, which outweighs reduced catalytic activity.
2.4. Thermal Stability
Since we had determined mutations N13D, N57S and N268D to positively affect thermal stability of HRP before (Table 2; [27]), we also analyzed thermal half-life times of the four HRP variants at 60 °C. We again included plant HRP as standard and normalized all enzyme preparations to a protein concentration of 0.1 mg/mL before incubation to guarantee comparability. In Table 7 the respective results are summarized.
Table 7.
Thermal half-life times of four different HRP variants and the HRP plant preparation at 60 °C. All enzymes were normalized to a protein concentration of 0.1 mg/mL before incubation.
| Enzyme Variant | Protein Concentration (mg/mL) | τ1/2 60 °C (min) |
|---|---|---|
| wtwt HRP | 0.1 | 31.5 |
| wt4/8 HRP | 173.2 | |
| OCH1wt HRP | 3.3 | |
| OCH14/8 HRP | 19.3 | |
| plant HRP | 53.3 |
As shown in Table 7, the introduction of the four mutations into HRP caused a significant increase in thermal stability. Enzyme wt4/8 HRP had a 5.5-fold higher τ1/2 60 °C than enzyme wtwt HRP and was even 3.3-fold more stable than the enzyme preparation from plant. The same HRP variants produced in the Δoch1 strain showed a 10-fold reduced thermal stability compared to their respective counterparts from the benchmark strain. Reducing glycosylation to Man8 structures apparently affected stability to a greater extent than completely removing four N-glycosylation sites. In fact, mutating the four N-glycosylation sites significantly increased stability instead of decreasing it. It is remarkable that enzyme OCH14/8 HRP actually showed a similar thermal half-life time as enzyme wtwt HRP (Table 7).
Considering both, kinetic parameters and thermal stability, enzyme wt4/8 HRP might be interesting for future applications in medical diagnostics. This enzyme variant can be efficiently produced in bioreactor cultivations, is less glycosylated, which might allow more controlled and efficient conjugation to antibodies and lectins, still shows considerable catalytic activity and a 5.5-fold higher thermal stability compared to enzyme wtwt HRP. In fact, higher stability and reduced glycosylation could compensate for reduced catalytic activity.
3. Experimental Section
3.1. Chemicals
2,2′-Azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) diammonium salt (ABTS), D(+)-biotin and hemin were purchased from Sigma-Aldrich. Difco™ yeast nitrogen base w/o amino acids (YNB), Bacto™ tryptone and Bacto™ yeast extract were obtained from Becton Dickinson (Franklin Lakes, NJ, USA). Zeocin™ was obtained from InvivoGen (San Diego, CA, USA). Other chemicals were obtained from Carl Roth (Karlsruhe, Germany).
3.2. Strain Generation
All strains in this study are based on the P. pastoris wildtype strain CBS7435. Generation of the Δoch1 strain and the single HRP variants are described in detail in our previous studies [26,27]. In short, the four Asn, representing glycosylation sites of HRP, were mutated by site directed mutagenesis and subsequent digestion with DpnI. The mutagenic PCR was performed as: 98 °C for 30 s; then 10 cycles of 98 °C for 10 s, 57 °C for 20 s, 72 °C for 1 min-10 cycles of 98 °C for 10 s, 60 °C for 20 s, 72 °C for 1 min-10 cycles of 98 °C for 10 s, 63 °C for 20 s, 72 °C for 1 min; with a final incubation at 72 °C for 10 min. Each reaction contained 1× HF buffer (Fermentas, Waltham, MA, USA), 0.01 μg of plasmid DNA, 2.5 U Phusion DNA polymerase (Fermentas), 10 μM of each dNTP and 5 pmol of each primer in a total volume of 50 μL. All primers are listed in Supplementary Table S1 and were purchased from Microsynth (Balgach, Sweden).
After PCR, the methylated template DNA was degraded by digestion with 10 U of DpnI at 37 °C for at least three hours. The remaining PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and 5 μL of each purified PCR product were transformed into electro-competent E. coli TOP10 F′ cells. The successful introduction of the desired mutation and the absence of further mutations were confirmed by DNA sequencing (Microsynth). Transformation of approximately 2 μg SwaI-linearized pPpT4_S plasmid DNA harbouring the respective mutated HRP gene into the P. pastoris benchmark and the Δoch1 strain was done by electroporation. Stable transformants were generated via homologous recombination between the linearized plasmid DNA and genomic yeast DNA. Selection of successfully transformed clones was performed on Yeast Extract Peptone Dextrose medium (YPD; 10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose, 20 g/L agar) supplemented with 100 mg/L Zeocin. In total, we generated and compared four recombinant P. pastoris strains producing four different HRP enzyme variants (Table 8).
Table 8.
Recombinant P. pastoris strains and HRP enzyme variants in this study.
| P. pastoris Chassis Strain | HRP Variant | Name of Recombinant P. pastoris Strain and Enzyme |
|---|---|---|
| benchmark strain | wt HRP | wtwt HRP |
| 4/8 HRP | wt4/8 HRP | |
| Δoch1 strain | wt HRP | OCH1wt HRP |
| 4/8 HRP | OCH14/8 HRP |
3.3. Bioreactor Cultivations
3.3.1. Preculture
Precultures were grown in 100 mL YNB_Zeo medium (0.1 M potassium phosphate buffer, pH 6.0; 3.4 g/L YNB w/o amino acids and ammonium sulfate, 10 g/L (NH4)2SO4, 400 mg/L biotin, 20 g/L glucose, 100 mg/L Zeocin) in 1000 mL baffled shake flasks at 30 °C and 230 rpm for 24 h.
3.3.2. Dynamic Batch Cultivation
For dynamic batch cultivations, 3582.6 mL 2-fold concentrated basal salt medium (BSM; 26.7 mL/L 85% phosphoric acid, 1.17 g/L CaSO4·2H2O, 18.2 g/L K2SO4, 14.9 g/L MgSO4·7H2O, 4.13 g/L KOH, 0.3 mL/L Antifoam Struktol J650, 60 g/L glycerol) were sterilized in a 5 L working volume glass bioreactor (Infors, Molndal, Sweden). After sterilization, 4.35 mL PTM1 per litre medium were added (i.e., 17.4 mL for 4 litres) and pH was set to 5.0 with concentrated ammonia solution. Pre-cultures were aseptically transferred to the respective vessel (10% inoculation volume) and the batch phase on glycerol was carried out at 30 °C with the stirrer fixed at 1300 rpm. Aeration with compressed dry air was set to 1 volume per volume per minute (vvm). Dissolved oxygen (dO2) was measured with a sterilizable VisiFerm DO 225 probe (Hamilton, ON, Canada). The pH was measured with a sterilizable electrode (Mettler Toledo, Greifensee, Switzerland) and maintained constant at pH 5.0. Reactor weight was continuously recorded by a precision balance (Sartorius, Göttingen, Germany). Batch and methanol adaptation were performed at 30 °C. After the complete consumption of glycerol, indicated by an increase of dO2 and a drop in off-gas activity, the first methanol pulse at a final concentration of 0.5% volume per volume (v/v) was conducted with methanol supplemented with 12 mL/L PTM1. Following pulses were performed with 1% methanol/PTM1 (v/v) at different temperatures. At 30, 25 and 20 °C at least two pulses were applied, respectively. For each pulse, two samples were taken to determine the concentrations of substrate and product, as well as dry cell weight to calculate specific rates and yields.
3.3.3. Analysis of Growth and Expression Parameters
Dry cell weight (DCW) was determined by centrifugation of 2 × 5 mL fermentation broth (4800 rpm, 10 min, 4 °C), washing the pellet with 5 mL water and subsequent drying to a constant weight at 105 °C. Optical density at 600 nm (OD600) was determined in a spectrophotometer (Thermo Scientific, Waltham, MA, USA). Activity of HRP in the cell-free supernatant was determined using a previously described assay in a CuBiAn-XC enzymatic robot (Innovatis, Bielefeld, Germany) [27]. Protein concentration was determined using a Bradford protein assay kit (Thermo Scientific). All growth and protein expression parameters were determined in duplicates.
3.3.4. Analysis of Substrate Concentration
Concentrations of methanol were determined in cell free samples by HPLC (Agilent Technologies, Santa Clara, CA, USA) equipped with an ion-exchange column (Supelcogel C-610H, Sigma-Aldrich, St. Louis, MO, USA) and a refractive index detector (Agilent Technologies). The mobile phase was 0.1% H3PO4 with a constant flow rate of 0.5 mL/min and the system was run isocratic at 30 °C. Calibration was done by measuring standard points in the range from 0.1 to 10 g/L methanol. Measurements of biomass, product and substrate concentration were executed in duplicates.
3.3.5. Calculation of Strain Physiological Parameters
The relevant parameters to physiologically characterize the recombinant yeast strains were: carbon dioxide evolution rate (CER; mmol/L/h), adaptation time to methanol (h), specific methanol uptake rate (qMeOH; mmol/g/h), biomass yield (YX/MeOH; C-mol/C-mol), carbon dioxide yield (YCO2/MeOH; C-mol/C-mol) and C-balance. Details concerning the calculation of these parameters have been published before [20,21,27].
3.4. Protein Purification
Cell-free cultivation broth was diafiltrated with a Centramate 500S TFF system (Pall) using a 10 kDa MWCO membrane. The buffer was HCIC-A (20 mM NaOAc, 1.0 M NaCl, pH 6.0) and the protein solution was concentrated to a final volume of 100–120 mL. The HCIC resin MEP HyperCel™ was obtained from Pall (Port Washington, NY, USA) and HCIC was performed in flow-through mode [8,19]: a column containing 25 mL of MEP HyperCel™ resin was equilibrated with at least four column volumes (CV) buffer HCIC-A. The HRP solution in HCIC-A was loaded onto the column that was then washed with at least five CV of HCIC-A at a flow rate of 20 cm/h. Then a step elution to 100% buffer HCIC-B (50 mM Tris, pH 8.0) was performed. After elution, the column was washed with five CV 0.8 M NaOH before it was stored in EtOH 20%, 1.0 M NaCl. During all the steps, fractions were collected and analyzed for protein content and catalytic activity. Fractions showing HRP activity were pooled and concentrated to around 1.5 mL using Amicon Ultra-15 Centrifugal Filter Units with 10 kDa MWCO (Merck Millipore, Darmstadt, Germany).
The efficiency of the purification was evaluated by determining the purification factor (PF; Equation (1)) and the recovery yield of HRP activity in percentage (R%; Equation (2)). The suffixes “pre” and “post” indicate the respective values before and after the HCIC step.
| (1) |
| (2) |
Additionally, Reinheitszahl values (RZ; A404/A280) of the concentrated enzyme preparations were measured. Absorbance at 280 and 404 nm were determined in a quartz cuvette in a spectrophotometer (UV-1601; Shimadzu, Long Beach, CA, USA).
3.5. SDS-PAGE
Apparent sizes and purities of produced HRP variants were followed by SDS-PAGE according to the Laemmli protocol [28]. Electrophoresis was done using an Amersham ECL Gel 8%–16% gel (GE Healthcare, Buckinghamshire, UK) in 1× Tris-glycine buffer. Before loading, the gel had to be pre-run at 160 V for 12 min. Protein separation was performed at 140 V for about 2 h. BLUeye Prestained Protein Ladder (GeneDirex, Taoyuan County, Taiwan) was used as protein mass standard. Gels were stained with Coomassie Blue sensitive stain.
3.6. Protein Identification and Peptide Analysis Using LC-ESI-MS
The relevant protein bands were cut out from the SDS gel and digested in gel. S-alkylation with iodoacetamide and digestion with sequencing grade modified trypsin (Promega, Madison, WI, USA) were performed. The peptide mixture was analysed using a Dionex Ultimate 3000 system directly linked to an ion trap instrument (amaZon speed ETD, Bruker, Billerica, NA, USA) equipped with the standard ESI source in the positive ion, DDA mode (=switching to MSMS mode for eluting peaks). MS-scans were recorded (range: 400–1600 m/z; icc target: 100,000; max. accu time: 200 ms) and the 12 highest peaks were selected for fragmentation. Instrument calibration was performed using ESIcalibration mixture (Agilent). For separation of the peptides a Thermo BioBasic C18 separation column (5 μm particle size, 150 × 0.360 mm) was used. A gradient from 97% solvent A and 3% solvent B (Solvent A: 65 mM ammonium formiate buffer, B: 100% ACCN) to 32% B in 45 min was applied, followed by a 15 min gradient from 32% B to 75% B, at a flow rate of 6 μL/min. The analysis files were converted using Data Analysis 4.0 (Bruker) to XML files, which are suitable to perform MS/MS ion searches with MASCOT (embedded in ProteinScape 3.0, Bruker) for protein identification. Only proteins identified with at least 2 peptides with a protein score higher than 80 were accepted. For the searches the SwissProt database was used. Peptide MS/MS data were evaluated against the target sequence using X! Tandem (www.thegpm.org/tandem/) with the following settings: reversed sequences no; check parent ions for charges 1, 2 and 3 yes; models found with peptide loge lower −1 and proteins loge lower −1; residue modifications: oxidation M, W and deamidation N, Q; isotope error was considered; fragment type was set to monoisotopic; refinement was used with standard parameters; fragment mass error of 0.3 Da and ± 50 ppm parent mass error; fragment types b and y ions; maximum parent ion charge of 3; missed cleavage sites allowed was set to 1; semi-cleavage yes.
3.7. Biochemical Enzyme Characterization
We determined the basic kinetic parameters Km and vmax for the substrates ABTS and H2O2 for the different HRP variants in a spectrophotometer UV-1601 (Shimadzu). The reaction mixture with a final volume of 1.0 mL contained 20 μL of HRP variant, 50 mM potassium phosphate buffer, pH 6.5, and either varying concentrations of ABTS (0.01–10 mM) and a saturating concentration of H2O2 of 1.0 mM or varying concentrations of H2O2 (0.001–1.0 mM) and a saturating concentration of ABTS of 10.0 mM, respectively. The increase in absorption was followed at 420 nm at 30 °C for 180 s. Absorption curves were recorded with a software program (UVPC Optional Kinetics software; Shimadzu). The maximum reaction rate (vmax) and the Michaelis constant (Km) were calculated with the software Sigma Plot (Systat Software Inc., Chicago, IL, USA).
The thermal stability of individual HRP variants was tested at 60 °C. The residual activity with ABTS was measured after 1, 5, 10, 15, 30, 45, 60, 90 and 120 min of incubation at 60 °C in a PCR thermoblock. Protein concentrations were normalized to 0.1 mg/mL in Bis-Tris buffer (50 mM Bis-Tris, pH 6.5) to guarantee comparability. Residual activities were plotted versus the incubation time and the half-life times of thermal inactivation at 60 °C (τ½) were calculated using Equation (3):
| (3) |
kin: rate of inactivation (slope of the logarithmic residual activity).
4. Conclusions
In this study we combined protein and strain engineering to obtain an active and stable recombinant HRP variant with reduced surface glycosylation. We combined four mutations, which individually had shown beneficial effects on either catalytic activity or thermal stability before, and expressed this enzyme variant as well as the unmutated wildtype enzyme in both an unmodified P. pastoris benchmark strain and a Δoch1 strain. Our results can be summarized as:
Production of the 4/8 HRP variant caused a physiological burden for the P. pastoris benchmark strain slowing down its metabolism. In contrast, production of the same enzyme variant did not affect physiology of the Δoch1 strain. While qs MeOH increased with increasing temperature for the benchmark strain, the opposite was true for the Δoch1 strain. Based on strain physiological parameters identified in dynamic batch experiments, fed-batch strategies can be easily designed for future production of these enzyme variants.
Reduced and missing surface glycosylation did not affect substrate affinity of HRP, but significantly reduced catalytic activity.
Introducing the four mutations into HRP significantly boosted thermal stability. In fact, the 4/8 HRP variant produced in the P. pastoris benchmark strain even showed a 3.3-fold increased thermal half-life time compared to the HRP preparation from plant.
Considering both, enzyme activity and thermal stability, the 4/8 HRP variant produced in the P. pastoris benchmark strain might be interesting for future applications in medical diagnostics. This enzyme variant can be efficiently produced in the bioreactor, shows considerable catalytic activity and high thermal stability and is less glycosylated, which might allow more controlled and efficient conjugation to antibodies and lectins.
In this study we show that enzymes can be modified not only by mutation, but that a combination of both strain and protein engineering is a useful way to obtain enzyme variants tailored to specific needs.
Acknowledgments
The authors are grateful to the Austrian Science Fund FWF: project P24861-B19 for financial support. We further thank Alexander Pekarsky for assistance in the laboratory.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/10/23127/s1.
Author Contributions
Simona Capone, Lejla Ćorajević, Günther Bonifert and Patrick Murth performed experiments. Oliver Spadiut was responsible for experimental design. Daniel Maresch and Friedrich Altmann conducted mass spectrometry analyses. Oliver Spadiut and Christoph Herwig initiated, planned and supervised the study. Simona Capone and Oliver Spadiut wrote the manuscript. All authors read and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Mattanovich D., Sauer M., Gasser B. Yeast biotechnology: Teaching the old dog new tricks. Microb. Cell Factories. 2014;13:34. doi: 10.1186/1475-2859-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cregg J.M., Cereghino J.L., Shi J.Y., Higgins D.R. Recombinant protein expression in Pichia pastoris. Mol. Biotechnol. 2000;16:23–52. doi: 10.1385/MB:16:1:23. [DOI] [PubMed] [Google Scholar]
- 3.Cregg J.M. Pichia protocols, second edition. In: Cregg J.M., editor. Methods in Molecular Biology. Humana Press; Totowa, NJ, USA: 2007. p. 268. [Google Scholar]
- 4.Cregg J.M., Tolstorukov I., Kusari A., Sunga J., Madden K., Chappell T. Expression in the yeast Pichia pastoris. Methods Enzymol. 2009;463:169–189. doi: 10.1016/S0076-6879(09)63013-5. [DOI] [PubMed] [Google Scholar]
- 5.Spadiut O., Herwig C. Production and purification of the multifunctional enzyme horseradish peroxidase: A review. Pharm. Bioproc. 2013;1:283–295. doi: 10.4155/pbp.13.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gmeiner C., Saadati A., Maresch D., Krasteva S., Frank M., Altmann F., Herwig C., Spadiut O. Development of a fed-batch process for a recombinant Pichia pastoris Δoch1 strain expressing a plant peroxidase. Microb. Cell Factories. 2015;14:1. doi: 10.1186/s12934-014-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton S.R., Gerngross T.U. Glycosylation engineering in yeast: The advent of fully humanized yeast. Curr. Opin. Biotechnol. 2007;18:387–392. doi: 10.1016/j.copbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Spadiut O., Rossetti L., Dietzsch C., Herwig C. Purification of a recombinant plant peroxidase produced in Pichia pastoris by a simple 2-step strategy. Protein Expr. Purif. 2012;86:89–97. doi: 10.1016/j.pep.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Vervecken W., Kaigorodov V., Callewaert N., Geysens S., de Vusser K., Contreras R. In vivo synthesis of mammalian-like, hybrid-type N-glycans in Pichia pastoris. Appl. Environ. Microbiol. 2004;70:2639–2646. doi: 10.1128/AEM.70.5.2639-2646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton S.R., Bobrowicz P., Bobrowicz B., Davidson R.C., Li H.J., Mitchell T., Nett J.H., Rausch S., Stadheim T.A., Wischnewski H., et al. Production of complex human glycoproteins in yeast. Science. 2003;301:1244–1246. doi: 10.1126/science.1088166. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton S.R., Davidson R.C., Sethuraman N., Nett J.H., Jiang Y., Rios S., Bobrowicz P., Stadheim T.A., Li H., Choi B.K., et al. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science. 2006;313:1441–1443. doi: 10.1126/science.1130256. [DOI] [PubMed] [Google Scholar]
- 12.Li H., Miele R.G., Mitchell T.I., Gerngross T.U. N-linked glycan characterization of heterologous proteins. Methods Mol. Biol. 2007;389:139–150. doi: 10.1007/978-1-59745-456-8_10. [DOI] [PubMed] [Google Scholar]
- 13.Li H., Sethuraman N., Stadheim T.A., Zha D., Prinz B., Ballew N., Bobrowicz P., Choi B.K., Cook W.J., Cukan M., et al. Optimization of humanized IgGs in glycoengineered Pichia pastoris. Nat. Biotechnol. 2006;24:210–215. doi: 10.1038/nbt1178. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs P.P., Geysens S., Vervecken W., Contreras R., Callewaert N. Engineering complex-type N-glycosylation in Pichia pastoris using glycoswitch technology. Nat. Protoc. 2009;4:58–70. doi: 10.1038/nprot.2008.213. [DOI] [PubMed] [Google Scholar]
- 15.Krainer F.W., Glieder A. An updated view on horseradish peroxidases: Recombinant production and biotechnological applications. Appl. Microbiol. Biotechnol. 2015;99:1611–1625. doi: 10.1007/s00253-014-6346-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morawski B., Lin Z.L., Cirino P.C., Joo H., Bandara G., Arnold F.H. Functional expression of horseradish peroxidase in Saccharomyces cerevisiae and Pichia pastoris. Protein Eng. 2000;13:377–384. doi: 10.1093/protein/13.5.377. [DOI] [PubMed] [Google Scholar]
- 17.Morawski B., Lin Z.L., Bandara G., Arnold F.H. Expression of horseradish peroxidase (HRP) in S. cerevisiae and P. pastoris. Abstr. Pap. Am. Chem. Soc. 2000;219:U165–U165. doi: 10.1093/protein/13.5.377. [DOI] [PubMed] [Google Scholar]
- 18.Naatsaari L., Krainer F.W., Schubert M., Glieder A., Thallinger G.G. Peroxidase gene discovery from the horseradish transcriptome. BMC Genom. 2014;15:227. doi: 10.1186/1471-2164-15-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krainer F.W., Pletzenauer R., Rossetti L., Herwig C., Glieder A., Spadiut O. Purification and basic biochemical characterization of 19 recombinant plant peroxidase isoenzymes produced in Pichia pastoris. Protein Expr. Purif. 2013;95C:104–112. doi: 10.1016/j.pep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dietzsch C., Spadiut O., Herwig C. A fast approach to determine a fed batch feeding profile for recombinant Pichia pastoris strains. Microb. Cell Factories. 2011;10:85. doi: 10.1186/1475-2859-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dietzsch C., Spadiut O., Herwig C. A dynamic method based on the specific substrate uptake rate to set up a feeding strategy for Pichia pastoris. Microb. Cell Factories. 2011;10:14. doi: 10.1186/1475-2859-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spadiut O., Dietzsch C., Herwig C. Determination of a dynamic feeding strategy for recombinant Pichia pastoris strains. Methods Mol. Biol. 2014;1152:185–194. doi: 10.1007/978-1-4939-0563-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spadiut O., Zalai D., Dietzsch C., Herwig C. Quantitative comparison of dynamic physiological feeding profiles for recombinant protein production with Pichia pastoris. Bioprocess Biosyst. Eng. 2013;37:1163–1172. doi: 10.1007/s00449-013-1087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krainer F.W., Capone S., Jager M., Vogl T., Gerstmann M., Glieder A., Herwig C., Spadiut O. Optimizing cofactor availability for the production of recombinant heme peroxidase in Pichia pastoris. Microb. Cell Factories. 2015;14:4. doi: 10.1186/s12934-014-0187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krainer F.W., Dietzsch C., Hajek T., Herwig C., Spadiut O., Glieder A. Recombinant protein expression in Pichia pastoris strains with an engineered methanol utilization pathway. Microb. Cell Factories. 2012;11:22. doi: 10.1186/1475-2859-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krainer F.W., Gmeiner C., Neutsch L., Windwarder M., Pletzenauer R., Herwig C., Altmann F., Glieder A., Spadiut O. Knockout of an endogenous mannosyltransferase increases the homogeneity of glycoproteins produced in Pichia pastoris. Sci. Rep. 2013;3:3279. doi: 10.1038/srep03279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capone S., Pletzenauer R., Maresch D., Metzger K., Altmann F., Herwig C., Spadiut O. Glyco-variant library of the versatile enzyme horseradish peroxidase. Glycobiology. 2014;24:852–863. doi: 10.1093/glycob/cwu047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.