Abstract
Recent successes of adeno-associated virus (AAV)–based gene therapy have created a demand for large-scale AAV vector manufacturing and purification techniques for use in clinical trials and beyond. During the development of purification protocols for rh.10, hu.37, AAV8, rh.64R1, AAV3B, and AAV9 vectors, based on a widely used affinity resin, AVB sepharose (GE), we found that, under the same conditions, different serotypes have different affinities to the resin, with AAV3B binding the best and AAV9 the poorest. Further analysis revealed a surface-exposed residue (amino acid number 665 in AAV8 VP1 numbering) differs between the high-affinity AAV serotypes (serine in AAV3B, rh.10, and hu.37) and the low-affinity ones (asparagine in AAV8, rh.64R1, and AAV9). The residue locates within a surface-exposed, variable epitope flanked by highly conserved residues. The substitution of the epitope in AAV8, rh.64R1, and AAV9 with the corresponding epitope of AAV3B (SPAKFA) resulted in greatly increased affinity to AVB sepharose with no reduction in the vectors’ in vitro potency. The presence of the newly identified AVB-binding epitope will be useful for affinity resin selection for the purification of novel AAV serotypes. It also suggests the possibility of vector engineering to yield a universal affinity chromatography purification method for multiple AAV serotypes.
Introduction
Adeno-associated virus (AAV) is a single-stranded DNA virus which is so far not linked to human disease. In recent years after decades of persistent effort, the field of AAV vector-based gene therapy has enjoyed several successes. Alipogene tiparvovec (trade name: Glybera), an AAV1-LDL vector treating lipoprotein lipase deficiency, was approved in Europe in 2012.1 In addition, good outcomes have been reported from clinical trials in the treatments of Leber’s congenital amaurosis2 and hemophilia B3 with AAV vectors. With more AAV vector–based therapeutic candidates entering the pipeline, and with the discovery of novel AAVs with versatile tropisms4,5 and their increasing applications in clinical trials, the need for large-scale GMP-grade AAV vector–manufacturing processes with the capability of producing and purifying a number of AAV serotypes is acute.
The purification phase of the AAV gene therapy vector–manufacturing process is critical for guaranteeing the safety, potency, and overall yield of the drug product. Various methods for the purification of AAV vectors have been explored, mainly based on buoyant density (ultra)centrifugation using cesium chloride6 and iodixanol gradients,7 as well as chromatography-based methods including size exclusion,8 ion exchange,9–11 and affinity-based methods and combinations thereof. Ceramic hydroxyapatite has also been used in the purification of AAV2 (ref. 12) and AAV9.13 Various ligands have been used for affinity chromatography including heparin,7,14 mucin,15 A20 monoclonal antibody,16 and AVB Sepharose High Performance.17,18 Other less conventional methods involving manipulation of the AAV capsid have also been proposed to purify AAV vectors by displaying certain well-studied binding epitopes on the surface of the AAV capsid and then capturing the vector with ligands to those epitopes. Examples include biotinylated AAV and hexa-histidine-tagged AAV captured with avidin and Ni-NTA columns, respectively.19,20
The ligand of the AVB resin is a single-chain Llama antibody produced from yeast which is conjugated to Sepharose beads.21,22 The resin is reported by the manufacturer to capture AAV serotypes 1, 2, 3, and 5 but has also been recently used to purify AAV1 through AAV8, AAVrh.10, and AAV12 from crude cell lysates of the baculovirus-insect cell AAV production platform.23 In the same study, the authors were unable to demonstrate binding of AAV9 and AAV11 serotypes.
During development of purification protocols using the AVB resin for the purification of hu.37, AAV8, rh.64R1, AAV3B, and AAV9 serotypes, we found that although the AVB resin was able to capture these serotypes, their respective affinities were quite different. In the course of investigating the molecular determinants of these differences using a combined approach of sequence alignment, structural analysis, and AVB-binding studies, we successfully identified an AAV capsid epitope specific for the AVB resin. The presence or absence of this epitope determines the differential binding of the different serotypes to the resin. The implications and potential uses of these techniques and findings are discussed.
Results
The affinity of AVB resin for AAV serotypes varied significantly
To test the affinity of AVB resin for AAV8, rh.64R1, and hu.37 serotypes, AAV vector preparations were mixed together, and the rh.10 serotype was added as an internal positive control. This mixing of preparations was performed in order to minimize variations during chromatography. Because of limited choices of real-time PCR probes, two types of vector mixes were made, AAV8 + hu.37 + rh.10 and rh.64R1 + rh.10, and run on the AVB affinity column. The rh.10 vector genome distribution among the different fractions collected was very similar between the two runs (data not shown), so the average of the two runs was used for reporting the rh.10 data. As shown in Figure 1, 84% of the loaded rh.10 vector genome was present in the elution fraction. The affinity of the hu.37 vector was similar to rh.10, with 82% in the elution fraction. On the contrary, both AAV8 and rh.64R1 vectors bound AVB resin poorly, with only 20 and 22% in the elution fraction, respectively. The affinity of AAV3B for AVB resin was remarkable, with 98% of vector genomes recovered in the elution fraction.
Figure 1.
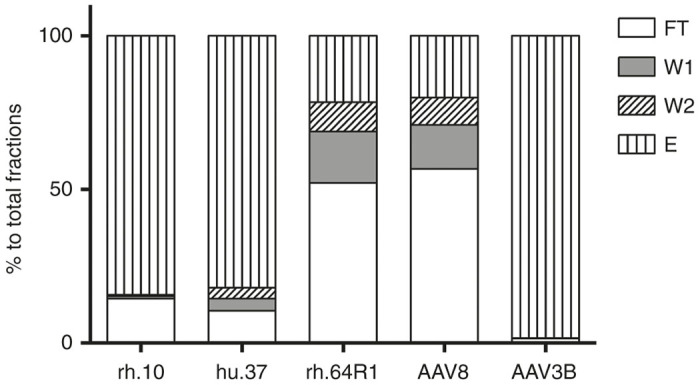
Vector genome distribution among the AVB column fractions. AAV vectors were diluted in binding buffer AVB.A (for AAV3B, culture supernatant was buffer-exchanged into the binding buffer) and then loaded onto the AVB column. Fractions from flow through (FT), AVB.A wash (W1), AVB.C wash (W2), and elution (AVB.B) (E) were collected. Vector genome copies were determined by real-time PCR.
Sequence alignment and structure analysis showed that the amino acid region 665–670 (AAV8 VP1 numbering) was the most diverse region on the capsid surface between the high AVB-affinity AAV serotypes, AAV3B, rh.10, and hu.37, and the low-affinity serotypes, AAV8 and rh.64R1. Among the residues exposed on the surface of the AAV8 capsid (PDB accession number: 2QA0 (ref. 24)), the following 26 residues are identical between rh.10 and hu.37 serotypes but different from AAV8 (numbering format: AAV8 residue-AAV8 VP1 numbering-rh.10/hu.37 residue): A269S, T453S, N459G, T462Q, G464L, T472N, A474S, N475A, T495L, G496S, A507G, N517D, I542V, N549G, A551G, A555V, D559S, E578Q, I581V, Q594I, I595V, N665S, S667A, N670A, S712N, V722T. Among the 26 residues, only residue 665 (AAV8 VP1 numbering) is identical among AAV1, 2, 3B, AAV5, rh.10, and hu.37. As shown in Figure 2a, all the poor-affinity AAV serotypes (AAV8, rh.64R1, and AAV9) have an Asn residue at this position while the high-affinity serotypes have Ser. The 665 residue is located in a small variable patch (665–670, AAV8 VP1 numbering) of the AAV capsid. The entire patch is exposed at the capsid surface, near the pore region (Figure 2b), and this whole epitope was therefore selected for swapping experiments. Because the affinity of the AAV3B serotype for AVB resin is very good, we chose the SPAKFA epitope from AAV3B to swap into the AAV8, rh.64R1, and AAV9 serotypes using site-specific mutagenesis. The resulting mutants were denoted as AAVx-SPAKFA. As a control, a reverse swap mutant was made where the corresponding epitope of AAV9 (NKDKLN) was swapped into the AAV3B capsid; the resulting mutant was named AAV3B-NKDKLN. The vector production yield of the SPAKFA epitope mutants was 81% (AAV8), 82% (rh.64R1), and 137% (AAV9) of their wild-type counterparts. The yield of AAV3B-NKDKLN was 28% of AAV3B.
Figure 2.
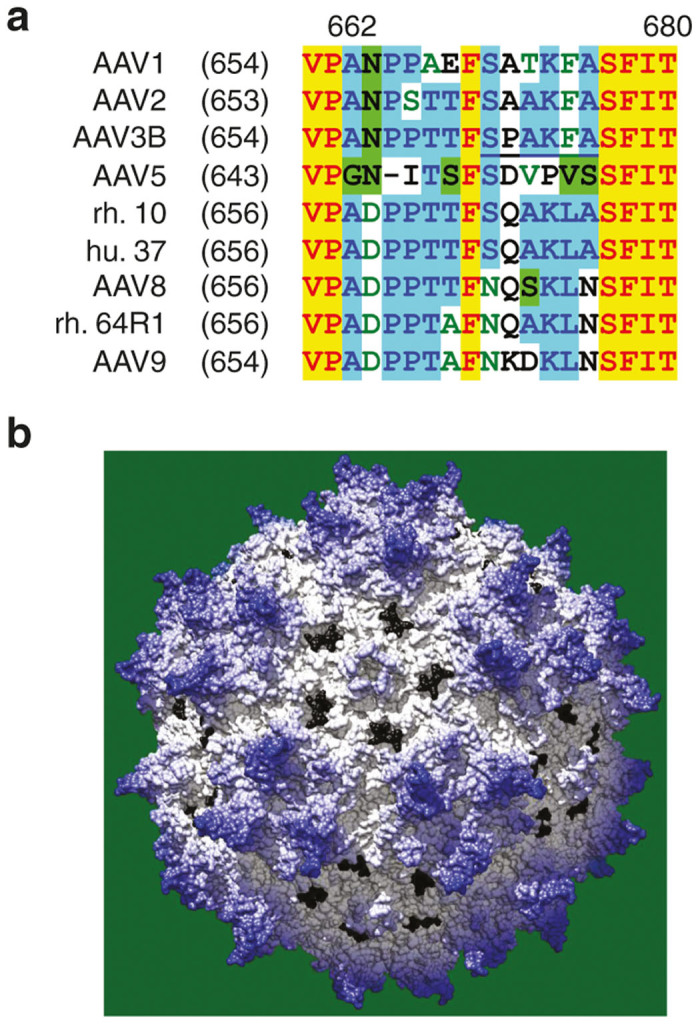
AAV serotype sequence alignment. (a) The alignment was performed with Vector NTI using ClustalW algorithm. The 665–670 region is shown with the SPAKFA epitope of AAV3B underlined. (b) The region corresponding to SPAKFA is shown in black on AAV8 capsid.
SPAKFA epitope exchange greatly improved the AVB affinity of AAV8, rh.64R1, and AAV9 serotypes
As shown in Figure 3, after SPAKFA substitution, clear improvement in the affinity of AAV8, rh.64R1, and AAV9 serotypes was shown, with the percentage recovery of loaded vector in the elution fraction rising from 30, 18, and 0.6% of total fractions to 93, 91, and 51%, respectively. In contrast, when the NKDKLN epitope of AAV9 was swapped into AAV3B, the elution fraction yield decreased from 98 to 87%, and the fractions of flow-through (FT), wash 1 (W1) and wash 2 (W2), rose from 1.3 to 8.2%, 0.0 to 0.1%, and 0.3 to 4.6%, respectively. The majority of AAV3B-NKDKLN was still in the elution fraction however, indicating the existence of other epitope(s) which are involved in binding to AVB resin.
Figure 3.
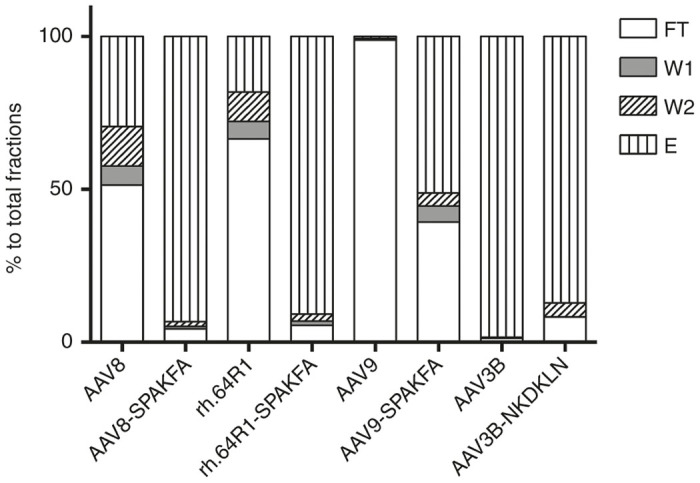
Substitution mutant vector genome distribution among the AVB column fractions. AAV vectors and their SPAKFA mutants were loaded onto an AVB column. Fractions for flow through (FT), DPBS wash (W1), AVB.C wash (W2), and elution (E) were collected for real-time PCR titration and represented as percent genome copies of the total. Each AAV and its mutant were compared head-to-head from production to titration. For AAV8, AAV9, and rh.64R1, mutants were made by substituting the corresponding region to SPAKFA based on sequence alignments shown in Figure 2a. For the AAV3B mutant, the SPAKFA epitope was replaced by NKDKLN.
SPAKFA epitope exchange did not lower the in vitro infectivity of AAV8, rh.64R1, and AAV9 serotypes
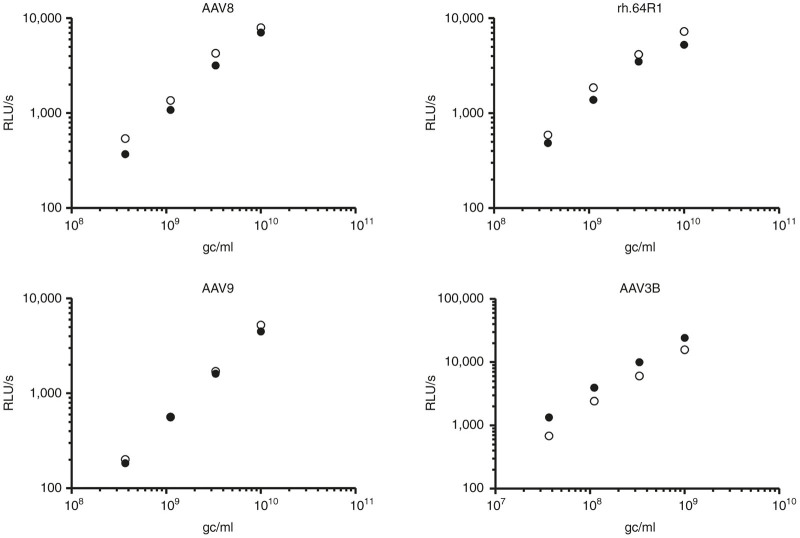
One key question was whether the epitope swapping performed impaired the potency of the recipient vector. To address this question, an in vitro infectivity assay was performed with the epitope substitution mutants in Huh7 cells. A range of vector concentrations were used for infection in order to avoid the possible saturation of transduction pathways at high multiplicity of infection. For AAV3B and AAV3B-NKDKLN, the vector concentration used for infection was 1 log lower than that for the other AAV vectors due to the very high Huh7 cell infectivity of the AAV3B serotype (data not shown) and its sibling, AAV3.25–28 As shown in Figure 4, the infectivity of the AAV8-, rh.64R1-, and AAV9-SPAKFA mutants was 130, 128, and 108% relative to the corresponding wild-type AAV vectors. Conversely, the infectivity of AAV3B-NKDKLN was only 59% of AAV3B.
Figure 4.
Huh7 cell transduction of AAVs and their SPAKFA mutants. The transgene cassette was CB7.CI.ffluciferase. Huh7 cells were infected with AAV vectors (filled circles) and their SPAKFA mutants (empty circles) at various concentrations (x-axis). The substitution mutant for AAV3B was AAV3B-NKDKLN. Luciferase expression was read 3 days after infection and denoted as RLU/s. gc, vector genome copies; RLU, relative luminescence unit.
Discussion
A simple, efficient, generic, and easily scalable purification protocol that can be used for all AAV serotypes is highly desirable. Affinity resins such as AVB will likely play an important role in enabling such a process as recently demonstrated in a study by Mietzsch et al.,23 in which 10 serotypes (AAV1-8, rh.10, and AAV12) were purified in a single step from clarified crude lysate using the AVB resin. However, our present study shows that although AAV8, rh.64R1, hu.37, rh.10, and AAV3B can be captured by AVB resin, the affinity of the resin for these different serotypes is very different, with AAV3B having a strong affinity and AAV8 and rh.64R1 binding more poorly. While further optimization of buffers and flow rate can improve binding of AAV8 in our hands (data not shown), conditions and the resulting resin capacity are still not optimal for process scale-up.
The variation in AVB affinity for AAV serotypes rh.10, AAV8, hu.37, and rh.64R1 was intriguing since they all belong to Clade E and display a high degree of sequence similarity. By contrast, another serotype, AAV5, binds well to AVB but is distantly related to Clade E members. These observations led us to speculate that some subtle sequence differences may play a role in the different binding affinities of these serotypes to AVB. Sequence alignment and structure analysis of the VP3 capsid proteins of these serotypes led us to narrow in on residue 665. At this position, AAV8 and rh.64R1 are asparagine, while rh.10, hu.37, and AAV5 are serine. Because the sequence patch around residue 665 is a small variable region, it was decided to swap the whole patch of AAV8, rh.64R1, and AAV9 with the patch (SPAKFA) from AAV3B. The clear improvement in affinity observed following these substitutions indicates that the SPAKFA sequence patch is an epitope of the AVB resin. Importantly, the substitutions did not impair capsid fitness, in terms of lowering yield or in vitro infectivity.
Another interesting observation was made when the corresponding sequence patch from the AAV9 serotype, NKDKLN, was substituted in place of the SPAKFA epitope in the AAV3B capsid. While the affinity of the AAV3B-NKDKLN vector was apparently weakened, as evidenced by the appearance of the vector in the flow-through fraction, the majority still bound to the column. This result, in conjunction with the fact that substitution of the SPAKFA epitope into AAV9 did not produce the affinity observed with AAV3B, suggests that there are other epitopes besides SPAKFA in the AAV3B VP3 amino acid sequence which contribute to AVB binding. One epitope candidate is the region containing residues 328–333 (Figure 5). This region is at the outside surface of the pore wall and is spatially close to the region containing residues 665–670. Residue 333 is especially close in spatial terms to the region containing residues 665–670 and for weak AVB binders such as AAV8, rh.64R1, and AAV9, this residue is Lysine, while in stronger binding serotypes such as AAV3B, it is threonine. The hypothesis suggested by these observations is that the regions containing residues 665–670 and 328–333 both contribute to AVB binding, although residues 665–670 make the major contribution. The AVB-binding data generated in this study, in addition to the AAV3B-NKDLN data described above, support this hypothesis. Serotypes with high SPAKFA homology in the 665–670 region and a threonine residue at position 333 bind best to AVB (AAV3B, AAV1, AAV2, and AAV5). Serotypes with low SPAKFA homology and a lysine residue at position 333 bind poorly (AAV8, rh64R1, and AAV9). Intermediate cases such as serotypes rh10, hu37, and epitope-substituted mutants which contain SPAKFA but have lysine rather than threonine at position 333 (AAV8-SPAKFA and rh64R1-SPAKFA) do bind to AVB resin but less well than serotypes such as AAV3B. Further mutagenic analysis of the 328–333 region and confirmation of its role in AVB binding is complicated because it overlaps the coding sequences for the assembly-activation protein (AAP) in another reading frame.29 However, with careful mutagenesis design, this problem might be overcome and would further enhance our understanding of the necessary interactions.
Figure 5.
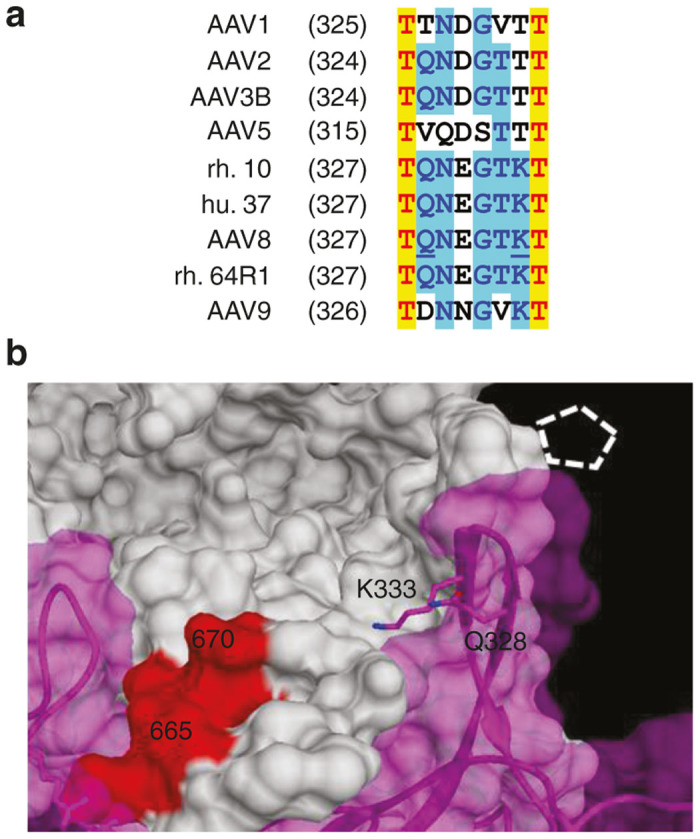
The 328–333 region. The sequence alignment of the 328–338 region of AAV VP1 is shown in (a) with the 328 and 333 residues of AAV8 underlined. Panel (b) demonstrates the two residues on AAV8 crystal structure. Two neighboring monomers of AAV8 capsid are shown (white and magenta). The white, dashed pentagon indicates the pore. The red region is the 665–670 region of the white monomer.
The discovery of the SPAKFA epitope might be useful in predicting whether AVB is a suitable resin for purification of some of the less commonly used AAV serotypes. For example, among the clade E members, rh.8, rh.43, and rh.46 serotypes have sequences very similar to AAV8 at residues 665–670 and so their affinity for AVB will probably be low. On the other hand, rh.39, rh.20, rh.25, AAV10, bb.1, bb.2, and pi.2 serotypes are likely to bind well because their sequences in this region are identical (or very similar) to rh.10. Similarly, for many clade D members, the 665–670 amino acid sequence is TPAKFA, and thus, these serotypes are likely to display high affinity to AVB, while the rh.69 serotype is likely to bind poorly since the 665–667 amino acid sequence is NQAKLN.
An intriguing possibility suggested by the studies presented here is that substitution of the SPAKFA epitope into the capsids of poor-affinity AAV serotypes such as AAV9 would permit for the use of AVB as a universal affinity chromatography resin for all AAV serotypes. While such an approach would provide process uniformity and simplicity, it is important to establish that the epitope substitutions do not affect vector yields, infectivity, and tropism. In the studies presented here, yields and infectivity of vectors receiving the SPAKFA sequence were not adversely affected; however, the impact on tropism was not investigated since it was beyond the scope of this work. However, there are reports which show that the tropism of AAV8 vectors relates mainly to hyper-variable region VII (AAV8 549–564) and IX (AAV8 708–720),30 and/or the subloop 1 (AAV8 435–482) and subloop 4 (AAV8 574–643)31 of the AAV8 capsid. Neutralizing epitope mapping data also supports the notion that the pore structure of AAV capsids and its nearby regions, which are responsible for binding to AVB resin, are not involved in cell transduction and therefore tropism. Neutralizing epitopes identified so far mainly locate around the threefold protrusion of the AAV capsid.32–36 Indeed, for AAV2, switching the tip (RGNR) of the threefold protrusion resulted in dramatic changes in the tropism of the vector.37 Another relevant antibody study was performed with monoclonal mouse antibody 3C5 raised against AAV5. This antibody is not neutralizing,38 and one of its epitopes locates in the 665–670 region.36 This observation therefore suggests that antibody binding in this region does not affect cell transduction and by extension, tropism.
In summary, the work presented here increases our understanding of the AAV capsid sequences necessary for binding to the commercially available affinity resin AVB sepharose. The use of AAV serotypes other than those commonly vectored for gene therapy applications might be a useful approach to attain specifically desired tropisms and may be valuable in avoiding immune responses in cases where readministration of a vector transgene is required. One factor in the selection of an appropriate AAV serotype for such gene therapy applications may be the ease by which it can be purified. In this regard, the ability to screen for AVB resin binding based upon the primary amino acid sequence as suggested by the data here, would greatly facilitate the selection process. For those serotypes where AVB resin binding is predicted to be poor, the substitution of the SPAKFA epitope may present a viable solution and enable the institution of a universal purification process for multiple serotypes.
Materials and Methods
Plasmids
Constructs pAAV2/8, pAAV2/rh.64R1, pAAV2/9, and pAAV2/3B expressing the AAV8, rh.64R1, AAV9, and AAV3B capsid protein respectively were used. Mutagenesis of these plasmids was carried out with QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, CA), following the manual’s instructions. The primers for the mutagenesis were: 5′- ATCCTCCGACCACCTTCAGCCCTGCCAAGTTTGCTTCTTTCATCACGCAATA -3′ and 5′- TATTGCGTGATGAAAGAAGCAAACTTGGCAGGGCTGAAGGTGGTCGGAGGAT -3′ for pAAV2/8 (NQSKLN→SPAKFA), 5′-ATCCTCCAACAGCGTTCAGCCCTGCCAAGTTTGCTTCTTTCATCACGCAGTA -3′ and 5′- TACTGCGTGATGAAAGAAGCAAACTTGGCAGGGCTGAACGCTGTTGGAGGAT -3′ for pAAV2/rh.64R1 (NQAKLN→SPAKFA), 5′- ATCCTCCAACGGCCTTCAGCCCTGCCAAGTTTGCTTCTTTCATCACCCAGTA -3′ and 5′- TACTGGGTGATGAAAGAAGCAAACTTGGCAGGGCTGAAGGCCGTTGGAGGAT -3′ for pAAV2/9 (NKDKLN→SPAKFA), 5′-ATCCTCCGACGACTTTCAACAAGGACAAGCTGAACTCATTTATCACTCAGTA-3′ and 5′-TACTGAGTGATAAATGAGTTCAGCTTGTCCTTGTTGAAAGTCGTCGGAGGAT-3′ for pAAV2/3B (SPAKFA→NKDKLN).
Vectors
Purified vector preparations AAV2/8.CMV.ffluciferase.SV40 AAV2/rh.64R1.CMV.PI.EGFP.WPRE.bGH, AAV2/hu.37.TBG.EGFP.bGH, and AAV2/rh.10.CMV.PI.Cre.RBG were produced and titrated by Penn Vector Core as previously described.39 Briefly, one cell stack (Corning, NY) of HEK293 cells was transfected with triple-plasmid cocktail by polyethylenimine when the cell confluency reached around 85%. Culture supernatant was harvested 5 days posttransfection and digested with turbonuclease (Accelagen, CA). NaCl was added to 0.5 mol/l, and the treated supernatant was then concentrated with tangential flow filtration. Concentration was followed by iodixanol density gradient ultracentrifugation and final formulation by buffer-exchange through Amicon Ultra-15 (EMD Millipore, MA) into DPBS (Dulbecco’s phosphate-buffered saline without calcium and magnesium, 1×; Mediatech, VA) with 35 mmol/l NaCl. Glycerol was added to 5% (v/v), and the vectors were stored at −80 °C until use. For titration, real-time PCR with Taqman reagents (Applied Biosystems, Life Technologies, CA) was performed targeting RBG, bGH, and SV40 polyadenylation sequences. AAV2/3B.CB7.CI.ffluciferase.RBG was made the same way, except that at the tangential flow filtration step, AVB.A buffer (Tris pH 7.5, 20 mmol/l, NaCl 0.4 mol/l) was used for buffer exchange. The retentate was then stored at 4 °C and 0.22 µm filtered before application to the AVB column.
For the vectors used for the wild-type–SPAKFA mutant comparison, each wild-type capsid and its mutants were made in parallel from one 15-cm plate, using a version of the protocol described above but scaled down proportionally according to the culture area of the plate. Culture supernatant was treated with turbonuclease and then stored at −20 °C. Before application to the AVB column, the supernatant was clarified at 47,360g and 4 °C for 30 minutes followed by 0.22-µm filtration. The transgene cassette for these vectors was CB7.CI.ffluciferase.RBG.
Chromatography
An AKTAFPLC system (GE Healthcare Life Sciences, NJ) was used for all binding studies. The HiTrap column (1 ml) used was prepacked with AVB Sepharose High Performance resin (GE Healthcare Life Sciences). AAV vectors were reconstituted in AVB.A buffer and loaded onto a column equilibrated in the same buffer. The column was washed with 6 ml of AVB.A buffer and 5 ml of AVB.C buffer (Tirs pH 7.5, 1 mol/l NaCl) and then eluted with 3 ml of AVB.B buffer (20 mmol/l sodium citrate, pH 2.5, 0.4 mol/l NaCl). The eluted peak fractions were immediately neutralized with 1/10 × volume of BTP buffer (0.2 mol/l Bis tris-propane, pH 10). The flow rate was 0.7 ml/min (109 cm/hour).
For testing the affinity of AAV vectors for the AVB resin, equal genome copy numbers (GC) of purified AAV8, rh.10, and hu.37 vectors were mixed together before loading. For rh.64R1, the load consisted of an equal amount (GC) of rh.10 and rh.64R1 in AVB.A buffer. For AAV3B, the clarified AAV3B product was loaded directly onto the AVB column.
For the comparison of AAV vectors and their SPAKFA mutants, 9.5 ml of the clarified product was loaded onto the AVB column at 0.7 ml/min, followed by washing with 8 ml of DPBS and 5 ml of AVB.C at 1 ml/min, and then eluted with 4 ml of AVB.B at 0.25 ml/min. The eluate was immediately neutralized as above.
In vitro infectivity assay
Huh7 cells were seeded in 96-well plates at a density of 5e4 cells/well. The cells were then infected with AAV vectors carrying the CB7.CI.ffluciferase.RBG transgene cassette 48 hours after seeding. Three days postinfection, luciferase activity was measured using a clarity luminometer (BioTek, VT).
Sequence alignment and structure analysis
Sequence alignments were done with the ClustalW algorithm by the AlignX component of Vector NTI Advance 11.0 (Invitrogen, CA). The protein sequences were: AAV1 (accession:NP_049542), AAV2 (accession:YP_680426), AAV3 (accession:NP_043941), AAV3B (accession:AAB95452), AAV5 (accession:YP_068409), rh.10 (accession:AAO88201), hu.37 (accession:AAS99285), AAV8 (accession:YP_077180), rh.64R1 (accession:ACB55316), AAV9 (accession:AAS99264). Structure analysis was performed with the Chimera program40,41 and the AAV8 capsid structure (PDB: 2QA0 (ref. 24)).
Acknowledgments
We thank Julie Johnson, Arbans Sandhu, Shu-Jen Chen, and Zeling Niu of the University of Pennsylvania Vector Core (Philadelphia, PA, USA) for providing vectors and other materials used in the study. The study was funded by a grant from Dimension Therapeutics.
Footnotes
M.L. and M.R.A. are inventors on patents of AAVs licensed to various biopharmaceutical companies, including ReGenX. J.M.W. is an advisor to REGENXBIO, Dimension Therapeutics, Solid Gene Therapy, and Alexion and is a founder of, holds equity in, and has a sponsored research agreement with REGENXBIO and Dimension Therapeutics. In addition, he is a consultant to several biopharmaceutical companies and is an inventor on patents licensed to various biopharmaceutical companies. Q.W., A.J.P., and B.P. have nothing to disclose.
References
- Kastelein, JJ, Ross, CJ and Hayden, MR (2013). From mutation identification to therapy: discovery and origins of the first approved gene therapy in the Western world. Hum Gene Ther 24: 472–478. [DOI] [PubMed] [Google Scholar]
- Maguire, AM, Simonelli, F, Pierce, EA, Pugh, EN Jr, Mingozzi, F, Bennicelli, J et al. (2008). Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N Engl J Med 358: 2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani, AC, Tuddenham, EG, Rangarajan, S, Rosales, C, McIntosh, J, Linch, DC et al. (2011). Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med 365: 2357–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, G, Vandenberghe, LH and Wilson, JM (2005). New recombinant serotypes of AAV vectors. Curr Gene Ther 5: 285–297. [DOI] [PubMed] [Google Scholar]
- Vandenberghe, LH, Wilson, JM and Gao, G (2009). Tailoring the AAV vector capsid for gene therapy. Gene Ther 16: 311–319. [DOI] [PubMed] [Google Scholar]
- Xiao, X, Li, J and Samulski, RJ (1996). Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol 70: 8098–8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin, S, Byrne, BJ, Mason, E, Zolotukhin, I, Potter, M, Chesnut, K et al. (1999). Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther 6: 973–985. [DOI] [PubMed] [Google Scholar]
- Smith, RH, Ding, C and Kotin, RM (2003). Serum-free production and column purification of adeno-associated virus type 5. J Virol Methods 114: 115–124. [DOI] [PubMed] [Google Scholar]
- Gao, G, Qu, G, Burnham, MS, Huang, J, Chirmule, N, Joshi, B et al. (2000). Purification of recombinant adeno-associated virus vectors by column chromatography and its performance in vivo. Hum Gene Ther 11: 2079–2091. [DOI] [PubMed] [Google Scholar]
- Davidoff, AM, Ng, CY, Sleep, S, Gray, J, Azam, S, Zhao, Y et al. (2004). Purification of recombinant adeno-associated virus type 8 vectors by ion exchange chromatography generates clinical grade vector stock. J Virol Methods 121: 209–215. [DOI] [PubMed] [Google Scholar]
- Lock, M, Alvira, MR and Wilson, JM (2012). Analysis of particle content of recombinant adeno-associated virus serotype 8 vectors by ion-exchange chromatography. Hum Gene Ther Methods 23: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan, CR, Lachapelle, AL, Vincent, KA and Wadsworth, SC (2000). Scaleable chromatographic purification process for recombinant adeno-associated virus (rAAV). J Gene Med 2: 444–454. [DOI] [PubMed] [Google Scholar]
- Zhou, J, Yang, X, Wright, JF, High, KA, Couto, L and Qu, G (2011). PEG-modulated column chromatography for purification of recombinant adeno-associated virus serotype 9. J Virol Methods 173: 99–107. [DOI] [PubMed] [Google Scholar]
- Zolotukhin, S, Potter, M, Zolotukhin, I, Sakai, Y, Loiler, S, Fraites, TJ Jr et al. (2002). Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods 28: 158–167. [DOI] [PubMed] [Google Scholar]
- Auricchio, A, O’Connor, E, Hildinger, M and Wilson, JM (2001). A single-step affinity column for purification of serotype-5 based adeno-associated viral vectors. Mol Ther 4: 372–374. [DOI] [PubMed] [Google Scholar]
- Grimm, D, Kern, A, Rittner, K and Kleinschmidt, JA (1998). Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum Gene Ther 9: 2745–2760. [DOI] [PubMed] [Google Scholar]
- Smith, RH, Levy, JR and Kotin, RM (2009). A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol Ther 17: 1888–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L, Veres, G, and Knop, D (2009). A rapid, scalable, two-step column chromatography purification of rAAV vectors. Mol Ther 17: S273. [Google Scholar]
- Arnold, GS, Sasser, AK, Stachler, MD and Bartlett, JS (2006). Metabolic biotinylation provides a unique platform for the purification and targeting of multiple AAV vector serotypes. Mol Ther 14: 97–106. [DOI] [PubMed] [Google Scholar]
- Koerber, JT, Jang, JH, Yu, JH, Kane, RS and Schaffer, DV (2007). Engineering adeno-associated virus for one-step purification via immobilized metal affinity chromatography. Hum Gene Ther 18: 367–378. [DOI] [PubMed] [Google Scholar]
- Oranje, PPA, Verheesen, P, Verbart, D, Mijnsbergen, Y, Haard, HJW, Hermans, P et al. (2004). 423. isolation of an adeno-associated virus (AAV)-specific camelid-derived single chain antibody fragment: a novel tool for purification of AAV vectors of different serotypes. Mol Ther 9: S162–S162. [Google Scholar]
- Ayuso, E, Mingozzi, F and Bosch, F (2010). Production, purification and characterization of adeno-associated vectors. Curr Gene Ther 10: 423–436. [DOI] [PubMed] [Google Scholar]
- Mietzsch, M, Grasse, S, Zurawski, C, Weger, S, Bennett, A, Agbandje-McKenna, M et al. (2014). OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1-12 vectors for gene therapy. Hum Gene Ther 25: 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, HJ, Lane, MD, Padron, E, Gurda, B, McKenna, R, Kohlbrenner, E et al. (2007). Structure of adeno-associated virus serotype 8, a gene therapy vector. J Virol 81: 12260–12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glushakova, LG, Lisankie, MJ, Eruslanov, EB, Ojano-Dirain, C, Zolotukhin, I, Liu, C et al. (2009). AAV3-mediated transfer and expression of the pyruvate dehydrogenase E1 alpha subunit gene causes metabolic remodeling and apoptosis of human liver cancer cells. Mol Genet Metab 98: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, C, Lu, Y, Kalsi, JK, Jayandharan, GR, Li, B, Ma, W et al. (2010). Human hepatocyte growth factor receptor is a cellular coreceptor for adeno-associated virus serotype 3. Hum Gene Ther 21: 1741–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, B, Ling, C, Dai, Y, Lu, Y, Glushakova, LG, Gee, SW et al. (2012). Development of optimized AAV3 serotype vectors: mechanism of high-efficiency transduction of human liver cancer cells. Gene Ther 19: 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, C, Wang, Y, Zhang, Y, Ejjigani, A, Yin, Z, Lu, Y et al. (2014). Selective in vivo targeting of human liver tumors by optimized AAV3 vectors in a murine xenograft model. Hum Gene Ther 25: 1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag, F, Schmidt, K and Kleinschmidt, JA (2010). A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc Natl Acad Sci U S A 107: 10220–10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney, RM, Bell, CL and Wilson, JM (2014). AAV8 capsid variable regions at the two-fold symmetry axis contribute to high liver transduction by mediating nuclear entry and capsid uncoating. Virology 454–455: 227–236. [DOI] [PubMed] [Google Scholar]
- Shen, X, Storm, T and Kay, MA (2007). Characterization of the relationship of AAV capsid domain swapping to liver transduction efficiency. Mol Ther 15: 1955–1962. [DOI] [PubMed] [Google Scholar]
- Gurda, BL, Raupp, C, Popa-Wagner, R, Naumer, M, Olson, NH, Ng, R et al. (2012). Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J Virol 86: 7739–7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi, K, Enoki, T, Kawano, Y, Veraz, M and Nakai, H (2014). Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat Commun 5: 3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko, M, Chen, L, van Roey, M, Donahue, BA, Snyder, RO, McArthur, JG et al. (2000). Epitope mapping of human anti-adeno-associated virus type 2 neutralizing antibodies: implications for gene therapy and virus structure. J Virol 74: 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus, CE, Hügle-Dörr, B, Girod, A, Petersen, G, Hallek, M and Kleinschmidt, JA (2000). Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J Virol 74: 9281–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurda, BL, DiMattia, MA, Miller, EB, Bennett, A, McKenna, R, Weichert, WS et al. (2013). Capsid antibodies to different adeno-associated virus serotypes bind common regions. J Virol 87: 9111–9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan, A, Conway, JC, Phillips, JL, Li, C, Hegge, J, Sinnott, R et al. (2010). Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat Biotechnol 28: 79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison, CE, Weichert, WS, Gurda, BL, Chiorini, JA, Agbandje-McKenna, M and Parrish, CR (2012). Examining the cross-reactivity and neutralization mechanisms of a panel of mAbs against adeno-associated virus serotypes 1 and 5. J Gen Virol 93(Pt 2): 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock, M, Alvira, M, Vandenberghe, LH, Samanta, A, Toelen, J, Debyser, Z et al. (2010). Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum Gene Ther 21: 1259–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, EF, Goddard, TD, Huang, CC, Couch, GS, Greenblatt, DM, Meng, EC et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Sanner, MF, Olson, AJ and Spehner, JC (1996). Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38: 305–320. [DOI] [PubMed] [Google Scholar]