Abstract
For some time, cancer has not been thought of as a disease, but as a multifaceted, heterogeneous complex of genotypic and phenotypic manifestations leading to tumorigenesis. Due to recent technological progress, the outcome of cancer patients can be greatly improved by introducing in clinical practice the advantages brought about by the development of next generation sequencing techniques. Biomedical suppliers have come up with various applications which medical researchers can use to characterize a patient’s disease from molecular and genetic point of view in order to provide caregivers with rapid and relevant information to guide them in choosing the most appropriate course of treatment, with maximum efficiency and minimal side effects. Breast cancer, whose incidence has risen dramatically, is a good candidate for these novel diagnosis and therapeutic approaches, particularly when referring to specific sequencing panels which are designed to detect germline or somatic mutations in genes that are involved in breast cancer tumorigenesis and progression. Benchtop next generation sequencing machines are becoming a more common presence in the clinical setting, empowering physicians to better treat their patients, by offering early diagnosis alternatives, targeted remedies, and bringing medicine a step closer to achieving its ultimate goal, personalized therapy.
Keywords: breast cancer, benchtop sequencers, genome, next generation sequencing (NGS), personalized therapy
Background and aims
Science alone could never answer the great questions of humanity, nor could technology lead to improvement on its own. But when the two meet, progress can take place, and such is the case of discovering the molecular profile of phenotypic manifestations of various illnesses, which can eventually lead to the discovery of suitable and effective targeted therapies. Nowadays, molecular diagnosis for cancer patients prior to establishing the course of treatment is becoming a common presence in clinical practice, since nucleic acid sequencing has developed into a useful tool in the hands of medical researchers.
During the past years, the attention of healthcare practitioners and clinical researchers has begun to shift from treating patients already affected by various diseases to what is known as “predictive, preventive, and personalized medicine” (PPPM), and nowhere is this trend better illustrated than in the field of oncology [1]. For a while now, cancer has not been thought of as a disease, but a multifaceted, heterogeneous complex of genotypic and phenotypic manifestations that lead to tumorigenesis. Among tumors, breast cancer is the most frequent type of cancer diagnosed in women across the world, with an incidence of 22.9% according to GLOBOCAN-2008 [2], and the second most common type of cancer for both sexes combined, after lung cancer; at the same time, it is the principal cause of cancer death in women [2].
In the pursuit for the best response to therapy for each patient, caregivers are beginning to rely on more than the histopathological grading and staging of the disease and the general health of the patient. Due to recent advances in molecular and genetic research, clinicians now consider more specific information when deciding on therapeutic approaches: gene and molecular expression of tumors, mutation status of genes involved in a particular disease, polymorphisms or copy number variations. As an example, it is worth mentioning the increased capabilities of oncologists to predict the response to the recombinant monoclonal antibody trastuzumab (Herceptin) in HER2 positive breast cancer patients [3], or the response to cetuximab and panitumumab in mutated BRAF and/or KRAS colorectal adenocarcinoma patients [4]. The ability to foresee if a patient will respond to a therapeutic scheme based on their molecular profile does not only translate into potentially improving their outcome, but it also means an important step towards achieving the goal of personalized medicine.
As Bateman and Quackenbush stated in a 2009 editorial from the Bioinformatics Journal [5], the most outstanding outcome of sequencing and publishing the first reference human genome was not only knowing the genome itself, but all the technological advances that contributed to it, and the accelerated scientific progress derived from the Human Genome Project (HGP) [6]. Among the recent scientific and technical advances that impact medical research is next generation sequencing, which has the potential to improve clinicians’ approach on diagnosis and targeted therapies [7,8].
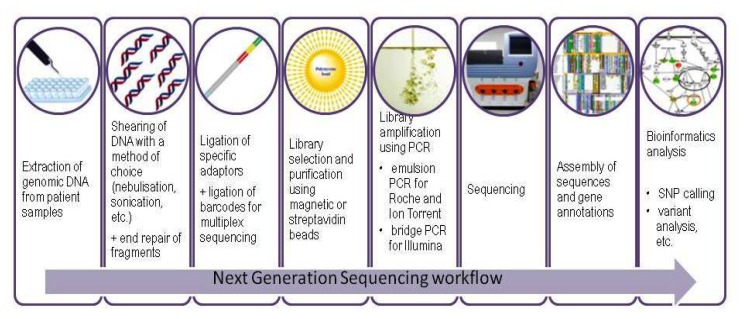
Leading suppliers of biomedical equipment have used these recent technological discoveries to produce various sequencing platforms based on the most recent advances in the field of next generation sequencing. Or, to be more precise, in the field of second generation sequencing, to better differentiate between Sanger sequencers and the more recent SMRT sequencing techniques (Single Molecule Real Time) developed by Pacific Biosciences [9], which are not the object of this paper. Many second generation sequencing platforms are available on the market, displaying a wide range of performances, reading capabilities, prices and possible applications, as seen in Table I [8,10–16] but they all need certain crucial steps which are summarized in Figure 1. These machines, which are becoming a familiar presence in molecular biology and clinical research laboratories, are produced and developed by the key players in the field, Roche (454 GS FLX, GS Junior), Illumina/Solexa (GAII, MiSeq, HiSeq) and Life Technologies (SOLiD, Ion Torrent, Ion Proton). The purpose of this paper is to provide a brief overview of some of the applications for breast cancer research developed for these platforms, as well as an outline of the sequencing workflow, focusing on the benchtop instruments which are more likely to be found in clinical research laboratories (GS Junior from Roche, MiSeq from Illumina, Ion Torrent from Life Technologies).
Table I.
Brief characterization of some Second Generation Sequencing platforms.
| Producer | Roche | Illumina/Solexa | Life Technologies | |||||
|---|---|---|---|---|---|---|---|---|
| Platform | 454 GS FLX | 454 GS Junior | GAII | HISeq models | MiSeq | SOLiD | Ion Torrent | Ion Proton |
| Seq method | SBS Pyro | SBS Pyro | SBS RDT | SBS RDT | SBS RDT | SBS | SBS H+ | SBS H+ |
| Amplification | emPCR | emPCR | Bridge PCR | Bridge PCR | Bridge PCR | emPCR | emPCR | emPCR |
| Read length | 400–700 bp* | 400 bp | 36–151 bp | up to 2×150 bp** | 36–151 bp | 35–75 bp | > 100 bp***** | up to 200 bp |
| Type of read | SE, PE | SE, PE | SE, PE | SE, PE | SE, PE | SE, PE | SE, PE | SE, PE |
| Output Gb/run | 0.45 – 0.7 | 0.35 | ≤ 95 | up to 600*** | > 1 | 77–155**** | > 1***** | up to 10 |
| Run time | up to 24 h | 10 h | 2–14 days | 27h to 11 days | 4–27 h | 2–7 days | 2 h | 2–4 h |
| Error rate (%) | 1 | 1 | ≥0.1 | ≥0.1 | >0.1 | >0.01 | ~1 | |
| Error type | Indels | Indels | Substitutions | Substitutions | Substitutions | Substitutions | Indels | |
| Cost of machine | up to $ 500k | Approx $115k | approx $ 540k | up to $740k | approx $128k | $525k | approx $80k | approx $150k |
| Pros | Longer reads, faster | Good read length, fast | Widely used, mature platform | Ultra high output, ease of use | Fully automated workflow | Ultra high output, scalable runs; can use 96 bar codes | Cheap, past, label free chemistry, highly scalable | WGS applications, 384 barcodes supported by the Torrent Suite Software |
| Cons | High cost per Mb, errors with homo polymer repeats | Like the FLX, plus lowest output among small sequencing instruments | High cost per Mb, low multiplexing capabilities | Less scalability, high overall costs | Insufficiently used | Shorter read length, long time for clonal preparation | Homopolymer errors, laborious (yet semi-automated) library preparation | Possibly high cost per Mb and errors |
SBS = Sequencing by synthesis; SBL = Sequencing by ligation; RDT = Reverse dye terminator; SE = Single-ended read; PE = Paired-ended read
at the lower limit of the range for the Titanium
PE for the 1500 and 2500 on rapid run
for the 2000 and 2500 on high output
155 Gb for the XL model
for the 318 chip
Figure 1.
Overview of the main steps in Next Generation Sequencing workflow.
Methods
454/Roche technology and workflow
454 Life Sciences, a biotechnology company from Connecticut, launched the first massively parallel DNA sequencer, the 454, probably the most widely used platform today [13,15]. After the first 454 machine became available on the market, it underwent several improvements regarding its performance and output, leading up to the benchtop version, the GS Junior. The technology employed by all 454 systems is sequencing by synthesis, and the actual sequencing is performed by pyrosequencing, in which a complex of enzymes and optical devices discriminate each pyrophosphate that is released during the attachment of nucleotides by the polymerase, but only if there are no extended homopolymer repeats [17–19]. The workflow is comparable to the one used when sequencing on the Ion Torrent. The input samples can be genomic DNA from BACs, plastids, bacteria, plants or animals for de novo sequencing, amplicons generated for targeted sequencing or re-sequencing, or cDNA.
The samples are first quantified and the DNA is randomly fragmented usually by nebulisation. After performing some end-repair procedures, adaptors are ligated to the double stranded DNA fragments. Two types of adaptors are used, A and B, the latter carrying a biotin group which contributes to the final separation of fragments that can be used for pyrosequencing. After ligation and denaturation, the mixture of fragments is exposed to streptavidin beads for library purification. Since the homozygous A-A fragments do not contain any biotin marker, they are flushed away. The homozygous B-B fragments are also eliminated, being constrained by the double biotin-streptavidin bond. What remains are the heterozygous fragments that constitute the DNA library which, prior to sequencing, needs to undergo an amplification step via emulsion PCR (emPCR) [7]. This is performed by using DNA capture beads on which each fragment is attached via the A adaptor at a one-to-one ratio by using precise and limiting dilutions. The water-in-oil emulsion is designed to trap each bead with a single DNA fragment attached, creating a high number of micro-reactors equipped with all the necessary PCR ingredients, where massive parallel amplification takes place. In this manner, hundreds of thousands of clonally amplified DNA fragments are generated on the surface of each capture bead [20]. After the water-in-oil emulsion is broken, the amplified library is placed on a fiber optic “MicroTiterPlate” containing millions of wells sized in such a way that each well is occupied by only one capture bead [21]. Enzyme beads are also added to each well, containing DNA polymerase, luciferase and ATP sulfurylase, necessary for recording each pyrophosphate released upon nucleotide incorporation.
During the following step, the sequencing per se, the plate is flushed with separate solutions containing each of the four nucleotides (dATP, dTTP, dGTP, dCTP) in a sequential manner and a precise, pre-defined order. As each nucleotide is incorporated into the chain based on complementarity, the pyrophosphate that is displaced during phosphodiester bond formation is converted by sulfurylase into ATP. During a subsequent reaction, luciferase oxidizes luciferin in the presence of ATP and oxygen, leading to the formation of a series of compounds and a quantum of light which is recorded by the CCD camera of the sequencer. If the template strand contains a homopolymer stretch, the intensity of the emitted light is directly proportional to the number of identical bases which were added to the new, complementary strand, but this may be a source of errors, since the light signal that is recorded is not always conclusive for the exact number of incorporated bases [22]. Other errors are prevented by flushing the PicoTiter plate with the enzyme apyrase between each run of nucleotides added to the reaction.
The raw data is processed and recorded in the form of a flowgram, and then the primary reads are assembled by overlapping multiple reads into longer strands to obtain consensus reads. The sequences are further mapped and compared against the known information from various databases.
Illumina/Solexa technology and workflow
Illumina uses a different approach for sequencing, the dye terminator technique, a method originally developed by the researchers from Manteia Predictive Medicine and from Solexa, a company bought by Illumina in 2007. Another particularity consists in the fact that the sequencing uses DNA libraries amplified through bridge-PCR. For this, the genomic DNA is randomly fragmented by sonication, the ends are repaired, and poly-A overhangs are added to the 3′ ends of each fragment by Klenow DNA polymerase exo(−) [9]. The poly-adenylated tails are used to add partially complementary specific adaptors to both ends of the double stranded fragments. The library is then size selected and placed on the flow cell for bridge-PCR amplification [7,15]. The flow cell used by Illumina is a solid support with the inner surface functionalized with two distinct oligonucleotide primers, forward and reverse, which are complementary to the two adaptors ligated on the DNA library [18]. The primers are attached with their 5′ end to the surface of the flow cell by a flexible linker, so that molecules that are amplified from a particular DNA fragment from the original library remain clustered together in groups of about 1,000 clonal amplicons. Prior to the actual sequencing, the amplicons are linearized and sequencing primers are hybridized to the 3′ ends. Then, a series of reagents is added to the flow cell for the “sequencing by synthesis” step.
Each round of interrogation is based on the elongation of the newly synthesized strand with only one nucleotide. The nucleotides used by Illumina platforms are modified in two ways. First, each of the four bases (A, T, C, G) is attached to a specific fluorochrome, so they do not need to be flushed over the flow cell separately [23]. The second modification consists in the fact that the bases resemble the dideoxynucleotides used in Sanger sequencing, but they are “reversible terminators”, since the 3′-OH is blocked by a cleavable fraction that prevents the incorporation of more than one base to the sequencing strand [8]. Therefore, each query is recorded independently, and the sequencing cannot continue until the terminator tag is released from the nucleotide to expose the hydroxyl group. This method overcomes the limitation of the 454 platforms regarding the biased output when incorporating homopolymer repeats. It also offers real-time feedback after each synthesis cycle, and provides faster results [17]. Nonetheless, sequencing with Illumina platforms is prone to some weaknesses, mainly concerning read length, and the reasons may be connected to incomplete and uneven removal of the terminating group. Still, the accuracy of reads makes Illumina the platform of choice in many laboratories, especially after the release of its bench top model, MiSeq, suitable for small genomes, exomes or targeted sequencing [11].
Ion Torrent technology and workflow
Ion Torrent, the “Personal Genome Machine” (PGMTM) released by Life Technologies in 2011, brings a new approach to second generation sequencing. Although the technology is still based on sequencing by synthesis, it does not require fluorescently labeled or chemiluminescent dNTPs, or an image-based detection of incorporated nucleotides. Instead, it uses a disposable chip built on the semiconductor technology to detect not the pyrophosphate released during nucleotide incorporation, but the proton that is also discharged during this process. In other words, the Ion Torrent chip acts as a pH-meter, detecting subtle pH shifts which occur when a phosphodiester bond is formed during elongation of the sequencing strand [23]. This novel technology, which does not require any optical devices and allows faster runs, together with its low price and automated possibilities for library preparation, make the Ion Torrent a suitable instrument for targeted sequencing or small genome analyses.
The workflow starts with the construction of DNA libraries. Genomic or cDNA is first fragmented into 200–400 bp strands, the ends are repaired, and the library is amplified and purified; then specific adaptors are attached to the ends of the fragments, and the gaps are filled in. When performing multiplex sequencing, such as sequencing DNA for more than one patient, specific barcodes are added to the fragments. The library is then size-selected and quality-controlled, either by using an Agilent Bioanalyzer or traditional quantitative PCR (qPCR). Next step is amplification via emulsion PCR [7]. After amplification, which resembles what we previously described for the 454 platform, the amplicon library undergoes template bead enrichment. For the PCR amplification and library enrichment, Ion Torrent developed the “Ion OneTouch” system, which automates the process and reduces hands-on time a great deal [12]. After this step, the DNA library is loaded on the chip and centrifuged, to ensure that each well of the chip is occupied by a template bead, and the sequencing process is initiated [15].
As previously mentioned, Ion Torrent technology is based on sequencing by synthesis, which consists in flooding the chip with individual, native nucleotides in a defined order. When complementary nucleotides are added to the sequencing strand by the DNA polymerase, the proton that is released causes a pH modification that is measured by sensors located at the bottom of each well. Since there are no optical devices involved, the sequencing reactions can proceed at a reasonably high rate [10,24]. Still, aside from the improvements brought by the Ion Torrent PGMTM, the problem of the homopolymer repeats still remains unsolved for the pyrosequencing based platforms, regardless of the way they detect nucleotide incorporation – as fluorescent signal or as a pH shift [14,25]. However, the release of the new Ion Proton and the rapid advancements in this field might lead to a solution to this limitation.
Results and discussion
NGS applications in breast cancer research
Second generation sequencing covers a wide range of applications, from whole genome sequencing (WGS) for individual patients or for other groups of organisms to exome sequencing, targeted re-sequencing and functional studies focused on the DNA – protein interactions (chromatin immunoprecipitation ChIP), which all have the potential to bring improvement in the quest for personalized medicine [26]. Human genetic variation applications developed for NGS include the evaluation of Single Nucleotide Polymorphisms (SNPs), Structural Variations (SVs) and Indels. The knowledge derived from these investigations can provide grounds for assessing possible links between genetic variations and different diseases, with high applicability in the field of oncology. Among these studies, the most common are genome-wide association studies (GWAS) and functional characterization of SNPs [16,27]. Many of these single nucleotide polymorphisms are connected to various pathological conditions either by abnormal gene expression or by altered protein structure and/or function, so they hold the potential to be used as genetic markers in the clinic to predict the risk of developing a certain disease, prognostic or response to therapy [28,29].
Breast cancer epidemic has reached alarming levels, so considerable efforts are made to learn more about this disease and the genetic alterations that lead to tumorigenesis, including cell growth, migration, angiogenesis and metastasis. Another aspect which requires attention is the lack of consistent and predictable response to main-stream chemotherapy used for mammary tumors classified according to traditional histopathological or other commonly used grading systems (TNM, Nottingham, etc). The reason for this is breast cancer heterogeneity – both inter-tumor and intra-tumor, which causes tumor-cell populations to behave differently and unpredictably [30–32].
The extensive knowledge unraveled at the end of the Human Genome Project enabled biochemical companies to produce commercial kits designed for the study of breast cancer, and cancer in general, with the help of NGS. Today, the market offers a wide range of cancer panels which can be used to discover the degree of tumor-specific genomic alterations by targeted re-sequencing of cancer genomes [13]. Some of the kits are produced by the manufacturers of next generation sequencers, to be used on their own machines – such as the IonAmpliseq Cancer Panel from Life Technologies, and the TruSeq Cancer Panel from Illumina, while others are generic and can be performed on many instruments. The cancer panels differ also with regard to the number of genes they contain and to the specificity of the results they produce. For instance, BRCA MASTR™ Dx from Multiplicom was designed in order to identify mutations in two tumor suppressor genes responsible for inherited breast and ovarian cancer, namely BRCA1 and BRCA2. On the other hand, the IonAmpliseq Comprehensive Cancer panel from Life Technologies can pinpoint mutations in 409 genes that are linked to the process of tumor formation and progression. Some information regarding the most frequently used commercial kits for NGS based studies in cancer research is presented in Table II [33–39].
Table II.
Some commercially available cancer panel kits.
| Company | Seq panel | Genes included | Targeted disease | Preferred seq machine |
|---|---|---|---|---|
| Ambry Genetics | BreastNext | ATM, BARD1, BRIP1, CDH1, CHEK2, MRE11A, MUTYH, NBN, PALB2, PTEN, RAD50, RAD51C, STK11, TP53 | Mainly hereditary breast and/or ovarian cancer; some genes associated with other cancers | Any NGS platform |
| Ambry Genetics | OvaNext | ATM, BARD1, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MRE11A, MSH2,SH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD50, RAD51C, STK11, TP53 | Hereditary gynecological cancers (breast, ovarian, uterus) | Any NGS platform |
| Ambry Genetics | CancerNext | APC,ATM, BARD1, BRIP1, BMPR1A, CDH1, CHEK2, EPCAM, MLH, MRE11A, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD50, RAD51C,SMAD4, STK11, TP53 | Breast, colon, ovarian, uterine, other | Any NGS platform |
| Multiplicom | BRCA MASTR™ Dx | BRCA1, BRCA2 | Hereditary breast and/or ovarian cancer | Any NGS platform |
| Roche NimbleGen | NimbleGen Sequence Capture arrays | User-designed panels, according to the research aim | Any type of cancer, known to be related to a particular gene/group of genes | Roche 454/GS Junior |
| Ion Torrent/ Life Technologies | Ion AmpliSeq Cancer Hotspot Panel | ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, VHL, EZH2, IDH2 | It targets the hotspot regions of 50 oncogenes and tumor suppressor genes which have been related to various types of cancer | Ion Torrent PGM |
| Ion Torrent/ Life Technologies | Ion AmpliSeq Comprehensive Cancer Panel | 409 oncogenes and tumor suppressor genes (including the 50 genes covered by the Cancer Hotspot Panel) | It targets genes frequently cited in tumorigenesis, as well as tumor suppressors | Ion Torrent PGM |
| Illumina | TruSeq Amplicon – Cancer Panel | ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAQ, GNAS, HNF1A, HRAS, IDH1, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, VHL | Able to identify somatic mutations in a high number of mutational hotspots in cancer genomes from various samples | Illumina, including MiSeq |
| University of Washington | BROCA Cancer Risk Panel | APC, ATM, ATR, BABAM1, BAP1, BARD1, BMPR1A, BRCC36, BRIP1, CDH1, CDK4, CDKN2A, CHEK1, CHEK2, EPCAM, FAM175A (Abraxas), MLH1, MRE11A, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PRSS1, PTEN, RAD50, RAD51, RAD51B, RAD51C, RAD51D, RBBP8, RET, SMAD4, STK11, TP53, TP53BP1, UIMC1, VHL, XRCC2, and XRCC3 | Mainly hereditary breast and/or ovarian cancer; some genes associated with increased risk of other cancers | Any NGS platform |
Five to ten percent of all mammary tumors are hereditary, and are connected to germline mutations in the genes BRCA1 and BRCA2 [40,41]. Still, only about 25% of familial breast cancers are associated with mutations in the two tumor suppressor genes [35,42]. Although scientists are far from pinpointing all genomic alterations that lead to this multifactorial, heterogeneous disease, some mutated genes were observed to take an active part in tumorigenesis, such as tumor suppressors (APC, CHEK 2, TP53, PTEN), DNA repair genes (NBN, MUTYH, MLH1, BRIP1), members of important signaling pathways implicated in cell division, proliferation and survival (ABL1, ATM, BRAF, FGFR1, JAK2, KRAS) or genes that encode for cell adhesion molecules (CDH1, CTNNB1, EPCAM) (See Supplementary Table I) [43]. These genes and others can be targeted and analyzed with the commercial cancer panels designed for next generation sequencing.
Conclusions
For the time being, the main purpose of these targeted re-sequencing studies is to assess risk or prognosis, but, with NGS platforms and reagents becoming more affordable and approaching the $1,000/genome threshold, sequencing can achieve more clinical applications [20]. Automation made benchtop NGS machines affordable also from the time perspective, since the results for targeted sequencing can be obtained within a work day, so they have the potential of becoming familiar presences in clinics. Thus, geneticists can provide caregivers rapid and essential information on the molecular profile of tumors, which can help in diagnosis, assessment of disease, and in planning the most appropriate treatment scheme, with maximum efficiency and minimal side effects [24,28]. Once this is accomplished, medicine will be one step closer to achieving its ultimate goal, personalized therapy. And when this aim is reached, chances are that next generation sequencing will have played a crucial role in this endeavor.
Supplementary materials
Supplementary Table I.
Some of the genes involved in tumorigenesis, which are targeted in commercially available sequencing kits.
| Gene | Location | Full name | UniGene | Function | |
|---|---|---|---|---|---|
| Native | When mutated | ||||
| ABL1 | 9q34.1 | c-abl oncogene 1, non-receptor tyrosine kinase | Hs.431048 | Cell growth, survival, adhesion, differentiation, DNA damage response, apoptosis | Chronic myeloid leukemia (CML), when translocated with BCR - also found in AML and ALL |
| APC | 5q21-q22 | adenomatous polyposis coli | Hs.158932 | Tumor suppressor, controls cell division | Colorectal cancer, Familial adenomatous polyposis |
| ATM | 11q22.3 | ataxia telangiectasia mutated | Hs.367437 | Cell division, DNA repair, normal development of nervous and immune system | When homozygous, ataxia-telangiectasia. When heterozygous, breast cancer and others |
| BARD1 | 2q34-q35 | BRCA1 associated RING domain 1 | Hs.591642 | Cell growth and division, together with BRCA1 | Uncertain risk in breast cancer and neuroblastoma |
| BRAF | 7q34 | v-raf murine sarcoma viral oncogene homolog B1 | Hs.550061 | In RAS/MAPK pathway, role in differentiation, migration, apoptosis | Cardiofaciocutaneous and multiple lentigines syndromes. Oncogene, somatic mutations associated with many cancers |
| BRCA1 | 17q21 | breast cancer 1, early onset | Hs.194143 | Tumor suppressor, gene regulation | Breast cancer, ovarian cancer, pancreatic cancer |
| BRCA2 | 13q12.3 | breast cancer 2, early onset | Hs.34012 | Tumor suppressor, gene regulation | Breast cancer, ovarian, pancreatic, prostate. Also Fanconi anemia type D1 when homozygous |
| BRIP1 | 17q22.2 | BRCA1 interacting protein C-terminal helicase 1 | Hs.128903 | DNA damage repair | Breast cancer when inherited heterozygous. Fanconi anemia when homozygous |
| CDH1 | 16q22.1 | cadherin 1, type 1, E-cadherin (epithelial) | Hs.461086 | Cell adhesion, cell signaling | Breast cancer, hereditary diffuse gastric cancer |
| CHEK2 | 22q12.1 | checkpoint kinase 2 | Hs.291363 | Tumor suppressor, detection of DNA damage and strand breaks | Breast cancer, Li-Fraumeni syndrome, other cancers |
| CTNNB1 | 3p21 | catenin (cadherin-associated protein), beta 1 | Hs.476018 | Cell adhesion, cell signaling | Pilomatricoma; colorectal, liver, ovarian cancer, medulloblastoma; desmoid fibromatosis |
| EPCAM | 2p21 | epithelial cell adhesion molecule | Hs.542050 | Calcium independent cell adhesion molecule in normal epithelium and gastrointestinal carcinoma | Hereditary nonpolyposis colorectal cancer, congenital tufting enteropathy |
| ERBB2 | 17q12 | Her-2/neu, v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 | Hs.446352 | Encodes for growth factor receptors | Amplification found breast cancer, overexpression in receptors leads to aggressive forms of cancer |
| FGFR1 | 8p12 | fibroblast growth factor receptor 1 | Hs.264887 | Cell division, regulation of cell growth and maturation | Alterations found in cancers via proliferation, migration, angiogenesis |
| HRAS | 11p15.5 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Hs.37003 | Regulates cell division through signal transduction | Oncogenes; involved in bladder, thyroid, kidney cancer; Costello syndrome |
| JAK2 | 9p24 | Janus kinase 2 | Hs.656213 | Cell growth and proliferation, role in hematopoiesis | Leukemia, essential thrombocythemia, primary myelofibrosis |
| KRAS | 12p12.1 | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | Hs.505033 | GTPase involved in cell division and differentiation | Oncogenes; involved in pancreatic, lung, and colorectal cancers; cardiofaciocutaneous and Noonan syndromes |
| MLH1 | 3p21.3 | mutL homolog 1, colon cancer, nonpolyposis type 2 | Hs.195364 | DNA damage repair | Lynch syndrome; when homozygous, also causes leukemia and neurofibromatosis |
| MRE11A | 11q21 | MRE11 meiotic recombination 11 homolog A | Hs.192649 | DNA double-strand break repair, telomere length maintenance, homologous recombination via exo- and endonuclease activity | Ataxia telangiectasia-like disorder, blocks meiotic recombination |
| MSH2 | 2p21 | mutS homolog 2, colon cancer, nonpolyposis type 1 | Hs.597656 | DNA damage repair | Lynch syndrome; endometrium, stomach, intenstine, liver cancer, etc. Homozygous mutations can also cause leukemia or lymphoma |
| MSH3 | 5q11-q12 | mutS homolog 3 | Hs.280987 | Heterodimer with MSH2, involved in post-replicative DNA mismatch repair system | Endometrial cancer |
| MSH6 | 2p16 | mutS homolog 6 | Hs.445052 | Post-replicative DNA damage repair | Lynch syndrome; endometrium, stomach, intenstine, liver cancer, etc. Homozygous mutations can also cause leukemia or lymphoma |
| MUTYH | 1p34.1 | mutY homolog | Hs.271353 | DNA damage repair by MYH glycosylase | Familial adenomatous polyposis |
| NRAS | 1p13.2 | neuroblastoma RAS viral (v-ras) oncogene homolog | Hs.486502 | Cell division and differentiation | Noonan syndrome, melanoma, other types |
| NBN | 8q21 | nibrin | Hs.492208 | DNA damage repair | Breast, ovarian, prostate cancer, melanoma, leukemia, Nijmegen breakage syndrome |
| PALB2 | 16p12.2 | partner and localizer of BRCA2 | Hs.444664 | Interacts with BRCA2; tumor suppressor | Breast cancer when inherited heterozygous. Fanconi anemia when homozygous |
| PIK3CA | 3q26.3 | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha | Hs.553498 | Role in signaling cascades involved in cell growth, survival, proliferation, motility and morphology | Oncogene, in breast, colorectal, ovarian, liver, gastric, lung cancers |
| PTEN | 10q23.3 | phosphatase and tensin homolog | Hs.500466 | Tumor suppressor, involved in apoptosis | Somatic mutations in prostate and endometrial cancer, glioblastoma, astrocytoma, melanoma |
| RAD50 | 5q31 | RAD50 homolog | Hs.633509 | DNA damage repair by holding the broken ends together during process | Suggested to contribute to breast cancer |
| RAD51C | 17q25.1 | RAD51 homolog C | Hs.412587 | DNA damage repair and meiotic homologous recombination | Hereditary breast and ovarian cancer, Fanconi anemia |
| SMAD4 | 18q21.1 | SMAD family member 4 | Hs.75862 | Controls gene activity and regulates cell proliferation | Colon, pancreas cancer; hereditary hemorrhagic telangiectasia, juvenile polyposis syndrome |
| SRC | 20q12-q13 | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog | Hs.195659 | Proto-oncogene with role in regulation of embryonic development and cell growth | Increased activity in colon carcinoma cells |
| STK11 | 19p13.3 | serine/threonine kinase 11 | Hs.515005 | Tumor suppressor, role in tissue polarization and apoptosis | Breast cancer, non-small cell lung carcinoma, melanoma, pancreatic cancer; Peutz-Jeghers syndrome |
| TP53 | 17p13.1 | tumor protein p53 | Hs.437460 | Tumor suppressor, “guardian of the genome” (Lane, 1992) | Breast, bladder, colorectal cancer, osteosarcoma, Li-Fraumeni syndrome |
Acknowledgment
Roxana Cojocneanu Petric is a fellow of POSDRU grant no. 159/1.5/S/138776 with title ”Model colaborativ institutional pentru translatarea cercetarii stiintifice biomedicale in practica clinica - TRANSCENT” [Institutional collaborative model for the translation of biomedical research into medical practice].
Laura Pop is a fellow of POSDRU grant no. 159/1.5/S/138776 with title ”Model colaborativ institutional pentru translatarea cercetarii stiintifice biomedicale in practica clinica - TRANSCENT” [Institutional collaborative model for the translation of biomedical research into medical practice].
References
- 1.Bodrova TA, Kostyushev DS, Antonova EN, Slavin S, Gnatenko DA, Bocharova MO, et al. Introduction into PPPM as a new paradigm of public health service: an integrative view. The EPMA journal. 2012;3(1):16. doi: 10.1186/1878-5085-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer Journal international du cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Davoli A, Hocevar BA, Brown TL. Progression and treatment of HER2-positive breast cancer. Cancer chemotherapy and pharmacology. 2010;65(4):611–623. doi: 10.1007/s00280-009-1208-1. [DOI] [PubMed] [Google Scholar]
- 4.Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut. 2011;60(1):116–129. doi: 10.1136/gut.2009.206250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman A, Quackenbush J. Bioinformatics for next generation sequencing. Bioinformatics. 2009;25(4):429. doi: 10.1093/bioinformatics/btp037. [DOI] [PubMed] [Google Scholar]
- 6.Marcus-Kalish MaHM. Healthcare Overview New Perspectives: Advances in Predictive, Preventive and Personalised Medicine. 2012 ed. Springer; Simultaneous Systematic Approach to Enable Predictive, Preventive and Personalized Medicine – Women Healthcare as a Case Study. [Google Scholar]
- 7.Casey G, Conti D, Haile R, Duggan D. Next generation sequencing and a new era of medicine. Gut. 2013;62(6):920–932. doi: 10.1136/gutjnl-2011-301935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metzker ML. Sequencing technologies - the next generation. Nature reviews Genetics. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 9.Sam LT, Lipson D, Raz T, Cao X, Thompson J, Milos PM, et al. A comparison of single molecule and amplification based sequencing of cancer transcriptomes. PloS one. 2011;6(3):e17305. doi: 10.1371/journal.pone.0017305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glenn TC. Field guide to next-generation DNA sequencers. Molecular ecology resources. 2011;11(5):759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Li Y, Li S, Hu N, He Y, Pong R, et al. Comparison of next-generation sequencing systems. Journal of biomedicine & biotechnology. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nature biotechnology. 2012;30(5):434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 13.Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: a practical perspective. The Clinical biochemist Reviews / Australian Association of Clinical Biochemists. 2011;32(4):177–195. [PMC free article] [PubMed] [Google Scholar]
- 14.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stranneheim H, Lundeberg J. Stepping stones in DNA sequencing. Biotechnology journal. 2012;7(9):1063–1073. doi: 10.1002/biot.201200153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X, Ren L, Meng Q, Li Y, Yu Y, Yu J. The next-generation sequencing technology and application. Protein & cell. 2010;1(6):520–536. doi: 10.1007/s13238-010-0065-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92(5):255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Shendure J, Ji H. Next-generation DNA sequencing. Nature biotechnology. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 19.Su Z, Ning B, Fang H, Hong H, Perkins R, Tong W, et al. Next-generation sequencing and its applications in molecular diagnostics. Expert review of molecular diagnostics. 2011;11(3):333–343. doi: 10.1586/erm.11.3. [DOI] [PubMed] [Google Scholar]
- 20.Natrajan R, Reis-Filho JS. Next-generation sequencing applied to molecular diagnostics. Expert review of molecular diagnostics. 2011;11(4):425–444. doi: 10.1586/erm.11.18. [DOI] [PubMed] [Google Scholar]
- 21.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith AM, Heisler LE, St Onge RP, Farias-Hesson E, Wallace IM, Bodeau J, et al. Highly-multiplexed barcode sequencing: an efficient method for parallel analysis of pooled samples. Nucleic acids research. 2010;38(13):e142. doi: 10.1093/nar/gkq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niedringhaus TP, Milanova D, Kerby MB, Snyder MP, Barron AE. Landscape of next-generation sequencing technologies. Analytical chemistry. 2011;83(12):4327–4341. doi: 10.1021/ac2010857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross JS, Cronin M. Whole cancer genome sequencing by next-generation methods. American journal of clinical pathology. 2011;136(4):527–539. doi: 10.1309/AJCPR1SVT1VHUGXW. [DOI] [PubMed] [Google Scholar]
- 25.Hui P. Next generation sequencing: chemistry, technology and applications. Topics in current chemistry. 2014;336:1–18. doi: 10.1007/128_2012_329. [DOI] [PubMed] [Google Scholar]
- 26.Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome biology. 2009;10(3):R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Chiodini R, Badr A, Zhang G. The impact of next-generation sequencing on genomics. Journal of genetics and genomics = Yi chuan xue bao. 2011;38(3):95–109. doi: 10.1016/j.jgg.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz-Cano SJ. Tumor heterogeneity: mechanisms and bases for a reliable application of molecular marker design. International journal of molecular sciences. 2012;13(2):1951–2011. doi: 10.3390/ijms13021951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weigelt B, Pusztai L, Ashworth A, Reis-Filho JS. Challenges translating breast cancer gene signatures into the clinic. Nature reviews Clinical oncology. 2012;9(1):58–64. doi: 10.1038/nrclinonc.2011.125. [DOI] [PubMed] [Google Scholar]
- 30.Almendro V, Fuster G. Heterogeneity of breast cancer: etiology and clinical relevance. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2011;13(11):767–773. doi: 10.1007/s12094-011-0731-9. [DOI] [PubMed] [Google Scholar]
- 31.Saunders NA, Simpson F, Thompson EW, Hill MM, Endo-Munoz L, Leggatt G, et al. Role of intratumoural heterogeneity in cancer drug resistance: molecular and clinical perspectives. EMBO molecular medicine. 2012;4(8):675–684. doi: 10.1002/emmm.201101131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanton C, Burrell RA, Futreal PA. Breast cancer genome heterogeneity: a challenge to personalised medicine? Breast cancer research : BCR. 2011;13(1):104. doi: 10.1186/bcr2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ion AmpliSeq Cancer Hotspot Panel v2 (tools.lifetechnologies.com).
- 34.Ion AmpliSeq Comprehensive Cancer Panel (tools.invitrogen.com).
- 35.Hereditary Cancer Panels from Ambry Genetics (ambrygen.com).
- 36.BRCA Mastr DX kit from Multiplicom (multiplicom.com).
- 37.TruSeq Amplicon Cancer Panel (illumina.com).
- 38.BROCA - Cancer Risk Panel (web.labmed.washington.edu).
- 39.The Case for Clinical Adoption of Hereditary Breast Cancer Panel Testing (ambrygen.com).
- 40.Petrucelli N, Daly MB, Feldman GL. Hereditary breast and ovarian cancer due to mutations in BRCA1 and BRCA2. Genetics in medicine : official journal of the American College of Medical Genetics. 2010;12(5):245–259. doi: 10.1097/GIM.0b013e3181d38f2f. [DOI] [PubMed] [Google Scholar]
- 41.Vaca-Paniagua F, Alvarez-Gomez RM, Fragoso-Ontiveros V, Vidal-Millan S, Herrera LA, Cantu D, et al. Full-exon pyrosequencing screening of BRCA germline mutations in Mexican women with inherited breast and ovarian cancer. PloS one. 2012;7(5):e37432. doi: 10.1371/journal.pone.0037432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meindl A, Ditsch N, Kast K, Rhiem K, Schmutzler RK. Hereditary breast and ovarian cancer: new genes, new treatments, new concepts. Deutsches Arzteblatt international. 2011;108(19):323–330. doi: 10.3238/arztebl.2011.0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Genetics Home Reference Database (ghr.nlm.nih.gov/BrowseGenes).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table I.
Some of the genes involved in tumorigenesis, which are targeted in commercially available sequencing kits.
| Gene | Location | Full name | UniGene | Function | |
|---|---|---|---|---|---|
| Native | When mutated | ||||
| ABL1 | 9q34.1 | c-abl oncogene 1, non-receptor tyrosine kinase | Hs.431048 | Cell growth, survival, adhesion, differentiation, DNA damage response, apoptosis | Chronic myeloid leukemia (CML), when translocated with BCR - also found in AML and ALL |
| APC | 5q21-q22 | adenomatous polyposis coli | Hs.158932 | Tumor suppressor, controls cell division | Colorectal cancer, Familial adenomatous polyposis |
| ATM | 11q22.3 | ataxia telangiectasia mutated | Hs.367437 | Cell division, DNA repair, normal development of nervous and immune system | When homozygous, ataxia-telangiectasia. When heterozygous, breast cancer and others |
| BARD1 | 2q34-q35 | BRCA1 associated RING domain 1 | Hs.591642 | Cell growth and division, together with BRCA1 | Uncertain risk in breast cancer and neuroblastoma |
| BRAF | 7q34 | v-raf murine sarcoma viral oncogene homolog B1 | Hs.550061 | In RAS/MAPK pathway, role in differentiation, migration, apoptosis | Cardiofaciocutaneous and multiple lentigines syndromes. Oncogene, somatic mutations associated with many cancers |
| BRCA1 | 17q21 | breast cancer 1, early onset | Hs.194143 | Tumor suppressor, gene regulation | Breast cancer, ovarian cancer, pancreatic cancer |
| BRCA2 | 13q12.3 | breast cancer 2, early onset | Hs.34012 | Tumor suppressor, gene regulation | Breast cancer, ovarian, pancreatic, prostate. Also Fanconi anemia type D1 when homozygous |
| BRIP1 | 17q22.2 | BRCA1 interacting protein C-terminal helicase 1 | Hs.128903 | DNA damage repair | Breast cancer when inherited heterozygous. Fanconi anemia when homozygous |
| CDH1 | 16q22.1 | cadherin 1, type 1, E-cadherin (epithelial) | Hs.461086 | Cell adhesion, cell signaling | Breast cancer, hereditary diffuse gastric cancer |
| CHEK2 | 22q12.1 | checkpoint kinase 2 | Hs.291363 | Tumor suppressor, detection of DNA damage and strand breaks | Breast cancer, Li-Fraumeni syndrome, other cancers |
| CTNNB1 | 3p21 | catenin (cadherin-associated protein), beta 1 | Hs.476018 | Cell adhesion, cell signaling | Pilomatricoma; colorectal, liver, ovarian cancer, medulloblastoma; desmoid fibromatosis |
| EPCAM | 2p21 | epithelial cell adhesion molecule | Hs.542050 | Calcium independent cell adhesion molecule in normal epithelium and gastrointestinal carcinoma | Hereditary nonpolyposis colorectal cancer, congenital tufting enteropathy |
| ERBB2 | 17q12 | Her-2/neu, v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 | Hs.446352 | Encodes for growth factor receptors | Amplification found breast cancer, overexpression in receptors leads to aggressive forms of cancer |
| FGFR1 | 8p12 | fibroblast growth factor receptor 1 | Hs.264887 | Cell division, regulation of cell growth and maturation | Alterations found in cancers via proliferation, migration, angiogenesis |
| HRAS | 11p15.5 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Hs.37003 | Regulates cell division through signal transduction | Oncogenes; involved in bladder, thyroid, kidney cancer; Costello syndrome |
| JAK2 | 9p24 | Janus kinase 2 | Hs.656213 | Cell growth and proliferation, role in hematopoiesis | Leukemia, essential thrombocythemia, primary myelofibrosis |
| KRAS | 12p12.1 | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | Hs.505033 | GTPase involved in cell division and differentiation | Oncogenes; involved in pancreatic, lung, and colorectal cancers; cardiofaciocutaneous and Noonan syndromes |
| MLH1 | 3p21.3 | mutL homolog 1, colon cancer, nonpolyposis type 2 | Hs.195364 | DNA damage repair | Lynch syndrome; when homozygous, also causes leukemia and neurofibromatosis |
| MRE11A | 11q21 | MRE11 meiotic recombination 11 homolog A | Hs.192649 | DNA double-strand break repair, telomere length maintenance, homologous recombination via exo- and endonuclease activity | Ataxia telangiectasia-like disorder, blocks meiotic recombination |
| MSH2 | 2p21 | mutS homolog 2, colon cancer, nonpolyposis type 1 | Hs.597656 | DNA damage repair | Lynch syndrome; endometrium, stomach, intenstine, liver cancer, etc. Homozygous mutations can also cause leukemia or lymphoma |
| MSH3 | 5q11-q12 | mutS homolog 3 | Hs.280987 | Heterodimer with MSH2, involved in post-replicative DNA mismatch repair system | Endometrial cancer |
| MSH6 | 2p16 | mutS homolog 6 | Hs.445052 | Post-replicative DNA damage repair | Lynch syndrome; endometrium, stomach, intenstine, liver cancer, etc. Homozygous mutations can also cause leukemia or lymphoma |
| MUTYH | 1p34.1 | mutY homolog | Hs.271353 | DNA damage repair by MYH glycosylase | Familial adenomatous polyposis |
| NRAS | 1p13.2 | neuroblastoma RAS viral (v-ras) oncogene homolog | Hs.486502 | Cell division and differentiation | Noonan syndrome, melanoma, other types |
| NBN | 8q21 | nibrin | Hs.492208 | DNA damage repair | Breast, ovarian, prostate cancer, melanoma, leukemia, Nijmegen breakage syndrome |
| PALB2 | 16p12.2 | partner and localizer of BRCA2 | Hs.444664 | Interacts with BRCA2; tumor suppressor | Breast cancer when inherited heterozygous. Fanconi anemia when homozygous |
| PIK3CA | 3q26.3 | phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha | Hs.553498 | Role in signaling cascades involved in cell growth, survival, proliferation, motility and morphology | Oncogene, in breast, colorectal, ovarian, liver, gastric, lung cancers |
| PTEN | 10q23.3 | phosphatase and tensin homolog | Hs.500466 | Tumor suppressor, involved in apoptosis | Somatic mutations in prostate and endometrial cancer, glioblastoma, astrocytoma, melanoma |
| RAD50 | 5q31 | RAD50 homolog | Hs.633509 | DNA damage repair by holding the broken ends together during process | Suggested to contribute to breast cancer |
| RAD51C | 17q25.1 | RAD51 homolog C | Hs.412587 | DNA damage repair and meiotic homologous recombination | Hereditary breast and ovarian cancer, Fanconi anemia |
| SMAD4 | 18q21.1 | SMAD family member 4 | Hs.75862 | Controls gene activity and regulates cell proliferation | Colon, pancreas cancer; hereditary hemorrhagic telangiectasia, juvenile polyposis syndrome |
| SRC | 20q12-q13 | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog | Hs.195659 | Proto-oncogene with role in regulation of embryonic development and cell growth | Increased activity in colon carcinoma cells |
| STK11 | 19p13.3 | serine/threonine kinase 11 | Hs.515005 | Tumor suppressor, role in tissue polarization and apoptosis | Breast cancer, non-small cell lung carcinoma, melanoma, pancreatic cancer; Peutz-Jeghers syndrome |
| TP53 | 17p13.1 | tumor protein p53 | Hs.437460 | Tumor suppressor, “guardian of the genome” (Lane, 1992) | Breast, bladder, colorectal cancer, osteosarcoma, Li-Fraumeni syndrome |