Abstract
The design of scaffolds which mimic the stiffness, nanofiber structure, and biochemistry of the native extra-cellular matrix (ECM) has been a major objective for the tissue engineering field. Furthermore, mimicking the innate three dimensional (3D) environment of the ECM has been shown to significantly alter cellular response compared to traditional two dimensional (2D) culture. We report the development of a self-assembling, fibronectin-mimetic, peptide-amphiphile nanofiber scaffold for 3D cell culture. To form such a scaffold, 5 mol% of a bioactive PR_g fibronectin-mimetic peptide-amphiphile was mixed with 95 mol% of a diluent peptide-amphiphile (E2) whose purpose was to neutralize electrostatic interactions, increase the gelation kinetics and promote cell survival. Atomic force microscopy verified the fibrilar structure of the gels and the mechanical properties were characterized for various weight percent (wt%) formulations of the 5 mol% PR_g - 95 mol% E2 peptide-amphiphile mixture. The 0.5 wt% formulations had an elastic modulus of 429.0 ± 21.3 Pa while the 1.0 wt% peptide-amphiphile hydrogels had an elastic modulus of 808.6 ± 38.1 Pa. The presence of entrapped cells in the gels decreased the elastic modulus and the decrease was a function of the cell loading. While both formulations supported cell proliferation, the 0.5 wt% gels supported significantly greater NIH3T3/GFP fibroblast cell proliferation throughout the gels than the 1.0 wt% gels. However, compared to the 0.5 wt% formulations, the 1.0 wt% hydrogels promoted greater increase in mRNA expression and production of fibronectin and type IV collagen ECM proteins. This study suggests that this fibronectin-mimetic scaffold holds great promise in the advance of 3D culture applications and cell therapies.
INTRODUCTION
The importance of the mechanical, structural, and biochemical properties of the native ECM has been recognized and has led to the development of new techniques and chemistries to form and functionalize scaffolds for cell culture. In addition to designing scaffolds which mimic the nanofibrous structure, stiffness and biochemical signals of the ECM, mimicking the innate 3D environment of the ECM have been shown to be critically important for tissue engineering applications.1–4 Traditional 2D cell culture introduces an artificial polarity between cells’ upper and lower surfaces resulting in non-native cell morphologies and receptor and protein expression profiles. 5 Additionally, significant differences in the expression of integrins and other cell surface receptors between 2D and 3D environments have been reported.6,7
Encapsulation of cells within a 3D scaffold is most effectively accomplished by introducing cells during gelation in order to ensure uniform cell distribution. Many of the common hydrogel systems currently used for 3D cell entrapment require low pH, as with PuraMatrix;8 low temperature, as with collagen and Matrigel;9 or exposure to UV light to initiate gelation, as with polyethylene glycol (PEG),10,11 which can damage cells and lead to cell death. Peptide-amphiphiles are an attractive material for the design of cell scaffolds which mimic the ECM due to their ability to self-assemble into nanofibrous hydrogels and incorporate relevant bioactive, biomimetic peptide sequences.12 Peptide-amphiphiles have been shown to form hydrogels in cell culture media and have been used as 3D scaffolds for a variety of applications, including the culture and differentiation of dental stem cells,13,14 mesenchymal stem cells,15–17 neural progenitor cells,18–20 cartilage, 21 and pancreatic islets.22,23 It has been demonstrated that RGD-containing peptide-amphiphile scaffolds support enhanced proliferation and osteogenic differentiation of mesenchymal stem cells compared to peptide-amphiphile scaffolds without the RGD peptide.16 Peptide-amphiphiles containing the laminin mimetic peptide, IKVAV, have been shown to support neuron differentiation of neural progenitor cells.18 Peptide-amphiphiles gels have also been shown to be biocompatible in vivo, with significant degradation within the first 30 days of implantation, followed by complete degradation after 60 days with no signs of acute or chronic inflammation.24 Other work has shown that entrapped cells internalize the peptide- amphiphile nanofibers and possibly utilize peptide-amphiphiles in their metabolic pathways.25
We have previously developed a fibronectin-mimetic peptide-amphiphile, called PR_g, which self-assembles in water to form nanofibrous hydrogels.26 The PR_g peptide contains both the primary cell binding site, RGD, as well as the PHSRN synergy site. These two sites are separated by a linker which accurately mimics the distance and the overall hydrophobicity/hydrophilicity between these binding domains in fibronectin.27,28 We have previously demonstrated that the PR_g peptide specifically binds the α5β1 integrin27 with a dissociation constant of 76.3 ± 6.3 nM.29 PR_g peptide-amphiphile hydrogels have been shown to support increased cell adhesion, proliferation, and ECM secretion as 2D substrates compared to PEG hydrogels functionalized with fibronectin and PuraMatrix hydrogels.30
Electrolytes or diluent peptide-amphiphiles of opposite charge have been used before in the literature to screen charges and enable faster gelation.25,31 In the absence of any electrolytes or other amphiphiles, PR_g peptide-amphiphiles were prepared in Milli-Q water and aged for 48 h at room temperature before use for our 2D studies.30 In order to translate our PR_g hydrogels from 2D to 3D, a diluent peptide-amphiphile, E2 (C16-GGGSSSESE), was designed to screen charges on the PR_g peptide-amphiphile (C16-GGGSSSPHSRN(SG)5RGDSP) and facilitate faster gelation (30 min to 1 h at room temperature or 37 °C in Milli-Q water with CaCl2 or in cell culture media).
The goal of this study is to investigate cell responses to changes in weight percentages (wt%) of a hydrogel composed of a mixture of PR_g and E2 peptide-amphiphiles. Given the amount of evidence in the literature which shows that 5 mol% of an RGD-containing peptide is optimal, or at least sufficient, to facilitate cell adhesion and survival,32–38 a hydrogel formulation containing 5 mol% PR_g peptide-amphiphile and 95 mol% E2 peptide-amphiphile (PR_g/E2) was chosen for this study. Here we characterize the hydrogel microstructure and mechanical properties of 0.5 and 1.0 wt% PR_g/E2 peptide-amphiphile hydrogels in the presence or absence of entrapped cells, as well as their ability to support cell survival, proliferation and ECM production in a 3D environment.
EXPERIMENTAL
Materials
All chemicals were purchased from Thermo Fisher Scientific except otherwise stated. Atomic force microscopy contact mode rectangular Si cantilevers with an Al-coated backside (NSC36/Al BS) were acquired from MikroMasch (Lady’s Island, SC). Ruby mica sheets, V2 quality, were purchased from S&J Trading Inc. (Glen Oaks, NY). Reagents for histology and immunohistochemistry were purchased from Biocare Medical (Concord, CA) unless stated otherwise. Water, purified to a resistivity of 18.2 MΩ/cm, was obtained from a Milli-Q water system (EMD Millipore, Billerica, MA) and sterilized by autoclaving before using it to prepare hydrogels. NIH3T3/GFP fibroblasts were purchased from Cell Biolabs Inc. (San Diego, CA) and Dulbecco’s modified Eagle medium (DMEM) from Life Technologies (Grand Island, NY).
Synthesis of peptide-amphiphiles
The protected PR_g peptide (GGGSSSPHSRN(SG)5RGDSP) and E2 diluent peptide (GGGSSESE) were synthesized by the Oligo-nucleotide and Peptide Synthesis Facility at the University of Minnesota using standard Fmoc solid phase synthesis on peptide amide linker resin. The Fmoc protecting group was removed using a solution of 20% piperidine in dimethylformamide (DMF). A palmitic acid, C16, tail was coupled to the N-terminus of the peptide with 4.5 molar equivalents of N,N,N′,N′-tetramethyl-O-(1H-benzotriazol -1-yl)uronium hexafluorophosphate (HBTU), 4.5 molar equivalents of palmitic acid, and 6 molar equivalents of diisopropylethylamine in DMF for 4 h. The Kaiser test was used to verify complete coupling of the tails to the peptide amide. The resin beads were washed twice in DMF, dichloromethane, and methanol and dried overnight under vacuum. Peptide-amphiphiles were cleaved from resin in a cleavage cocktail of 90% trifluoroacetic acid (TFA), 5% thioanisole, 3% 1,2-ethandithiol, and 2% anisole for 2 h. The solution was collected and precipitated in 20X excess cold isopropyl ether. The precipitate was collected by centrifugation (10 min at 7,000 RCF), redissolved in Milli- Q water and purified using reversed phase high performance liquid chromatography (HPLC) on an Agilent 1100 Series system with a Waters Xterra Prep MS C18 column, using a water/methanol gradient with 0.1% TFA for PR_g or 0.1% ammonium hydroxide for E2. The pure peptide-amphiphile product was analyzed with the Bruker Bio- TOF II to verify that the product had the expected mass. Peptide-amphiphiles were stored as lyophilized powder at −20 °C. Stock solutions of peptide-amphiphiles were prepared in Milli-Q water from the lyophilized peptide-amphiphiles and aliquots were stored at −20 °C to avoid freeze-thaw. All peptide-amphiphile gels in this study contained 5 mol% PR_g and 95 mol% E2. Specifically, gels contained 0.24 mM PR_g and 4.4 mM E2 or 0.48 mM PR_g and 8.8 mM E2 for the 0.5 wt% and 1.0 wt% gels respectively.
Atomic force microscopy (AFM)
2X concentrations of peptide-amphiphile solutions containing 5 mol% PR_g and 95 mol% E2 were mixed with an equal volume of DMEM media supplemented with 10 mM CaCl2 on 15 mm mica discs. Gels were allowed to dry for 5 days at room temperature in a laminar flow hood or at 37 °C. AFM imaging of the dehydrated samples was performed with a Nanoscope V Multimode 8 SPM (Bruker, Santa Barbara, CA) in contact mode in air using rectangular Si cantilevers with a typical probe tip radius of 8 nm.
Rheology
The mechanical properties of peptide-amphiphile hydrogels with and without cells were characterized using an AR-G2 rheometer from TA Instruments. 2X concentrations of peptide-amphiphile solutions containing 5 mol% PR_g and 95 mol% E2 were loaded onto the peltier plate and mixed with an equal volume of DMEM media supplemented with 10 mM CaCl2. For the gel characterization with the entrapped cells, 2X concentrations of peptide-amphiphile solutions containing 5 mol% PR_g and 95 mol% E2 were loaded onto the peltier plate and mixed with an equal volume of DMEM media supplemented with 10 mM CaCl2 containing 2.7 x 106 or 1.35 x 106 NIH3T3/GFP cells/mL. Therefore, the final concentration of the entrapped cells in the gel was 1.35 x 106 or 6.75 x 105 cells/mL respectively. Immediately after mixing, the 8 mm parallel plate was lowered to a gap of 500 μm for the gels without the cells, or to a gap of 1,000 μm for the gels with the cells, and the peltier plate was heated to 37 °C. The gels were allowed to mature in a hydrated chamber for 2 h before frequency sweep measurements were made from 0.1 to 10 rad/s at 1% strain. Strain sweeps performed prior to frequency sweeps showed that 1% strain fell within the linear viscoelastic regime. For time sweep experiments, gels were prepared with and without cells as described above and coated with low viscosity oil (dimethylpolysiloxane) to prevent dehydration.
Cell culture
NIH3T3 fibroblasts expressing green fluorescent protein (GFP), NIH3T3/GFP, were cultured in DMEM supplemented with 0.1 mM non-essential amino acids, 100 units/mL penicillin, 100 μg/mL streptomycin, 10 μg/mL blasticidin (Life Technologies, Grand Island, NY), and 10% fetal bovine serum (FBS) (Atlas Biologicals, Fort Collins, CO). NIH3T3 fibroblasts were cultured in DMEM supplemented with 100 units/mL penicillin, 100 μg/mL streptomycin and 10% FBS. Media was changed every 2–3 days until cells reached confluency and were passaged. Cells were used between passage 3 and 7.
Cell viability and proliferation assays
0.5 wt% and 1.0 wt% gels were prepared from peptide-amphiphile stock solutions in Milli-Q water. 2X concentrated peptide- amphiphile solutions were mixed with an equal volume of cell (NIH3T3/GFP or NIH3T3) suspension containing 2.7 x 106 cells/mL in complete DMEM media supplemented with 10 mM CaCl2. The final concentration of the entrapped cells in the gel was 1.35 x 106 cells/mL. The peptide- amphiphile gels were allowed to mature at 37 °C for 1 h before DMEM media without CaCl2 was added to the top of the gels. Media was replenished every 2 days. For the Live/Dead assay, viability was examined at 24 h and 5 day time points using the Live/DeadR Viability/Cytoxicity Kit (Invitrogen, Grand Island, NY) following the manufacturer’s instructions. Imaging was done on a Ziess Cell Observer SD Spinning Disk Confocal at the University Imaging Center at the University of Minnesota and cells were maintained at 37 °C with 5% CO2. GFP cell fluorescence was quantified using a Biotek Synergy H1 plate reader (Winooski, VT) with a 485/528 filter. Background from gels without cells was subtracted from all measurements and fluorescence was normalized to the 3 h time point. The persistence of GFP fluorescence in dead cells was examined using fluorescence microscopy. NIH3T3/GFP fibroblasts were plated in 96-well plates at a density of 250 cells/mm2 and were allowed to adhere overnight. Attached fibroblasts were imaged using transmitted light and the GFP cube (Ex: 470 nm, Em: 525 nm) on an EVOS FL microscope (Life Technologies, Grand Island, NY). 30% ethanol was added to wells and cells were imaged again after 5 min.
Real-time reverse transcription polymerase chain reaction (RT-PCR) for ECM mRNA
RNA was extracted from the 1.35 x 106 NIH3T3/GFP cells/mL, 3 h after gel encapsulation on day 0 and on day 5 of culture using the E.Z.N.A.R Total RNA Kit I (Omega Biotek, Norcross, GA). The reverse transcription to cDNA was done with qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD) following the manufacturer’s instructions. Primers for fibronectin (FN1), collagen IV (Col4α1, Col4α2, Col4α3), and housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were designed using the NCBI primer blast tool (Table 1). Real-time RT-PCR was done in 25 μL reactions following manufacturer’s instructions using PerfeCTa SYBR Green, Low ROX (Quanta Biosciences, Gaithersburg, MD) and a Stratagene Mx3000P system.
Table 1.
Forward (F) and reverse (R) primer sequences for fibronectin (FN1), collagen IV (Col4α1, Col4α2, Col4α3), and housekeeping gene GAPDH.
| Gene | Primer | Sequence |
|---|---|---|
| FN1 | F | GCCACCATTACTGGTCTGGA |
| R | GGTTGGTGATGAAGGGGGTC | |
| Col4α1 | F | TTAGCAGGTGTGCGGTTTGT |
| R | GGCGAGCCAAAAGCTGTAAG | |
| Col4α2 | F | CGGCGTAATCTCAAAAGGCG |
| R | GGCCTCTGCTTCCTTTCTGT | |
| Col4α3 | F | GAGAACCATCCGTAGGCAGG |
| R | CTTCGACCTGGTTGCCCTTT | |
| GAPDH | F | GAAACCTGCCAAGTATGATGACA |
| R | CTTGCTCAGTGTCCTTGCTG |
Histology and immunohistochemistry
1.35 x 106 NIH3T3/GFP cells/mL entrapped for 5 days in 0.5 wt% and 1.0 wt% gels were fixed for 15 min in 4% paraformaldehyde in tris-buffered saline (TBS), and then washed 3 times in TBS. Fixed gels were submitted to the histology and immunohistochemistry lab at the University of Minnesota in TBS. HistoGel (Richard-Allan Scientific, Kalamazoo, MI) was used according to the manufacturer’s protocol to protect the gel samples during processing. Unstained cell block sections (4 μm) were de-paraffinized and rehydrated using standard methods. For antigen retrieval, slides were incubated in Reveal Decloaker reagent, pH 6.0, in a steamer at 95–98°C for 30 min, and then cooled for 20 min. Slides were rinsed in tap water and immersed in TBS/0.1% triton (TBST). Slides were placed in the Nemesis 7200 (Biocare Medical, Concord, CA) and endogenous peroxidase activity was quenched by immersing slides in Peroxidazed 1, a 3% hydrogen peroxide solution, for 10 min followed by a TBST rinse. Sections were blocked for 30 min in Background Sniper, a serum-free blocking solution, before application of diluted primary antibodies in 10% blocking solution/90% TBST for 1 h at room temperature. Anti-fibronectin (1:200, Abcam, Cambridge, MA) or anti-collagen IV (1:200, Millipore, Temecula, CA) stained slides were rinsed in TBST and Novolink post primary (Leica Microsystems, Buffalo Grove, IL) was applied for 30 min at room temperature. Slides were rinsed in TBST followed by Novolink polymer (Leica Microsystems, Buffalo Grove, IL) for 30 min at room temperature, rinsed again in TBST, and incubated in CAT Hematoxylin for 5 min. Slides were rinsed in Milli-Q water, de- hydrated using standard methods, cover slipped, and imaged using a Leica DME light microscope.
RESULTS AND DISCUSSION
Hydrogel microstructure
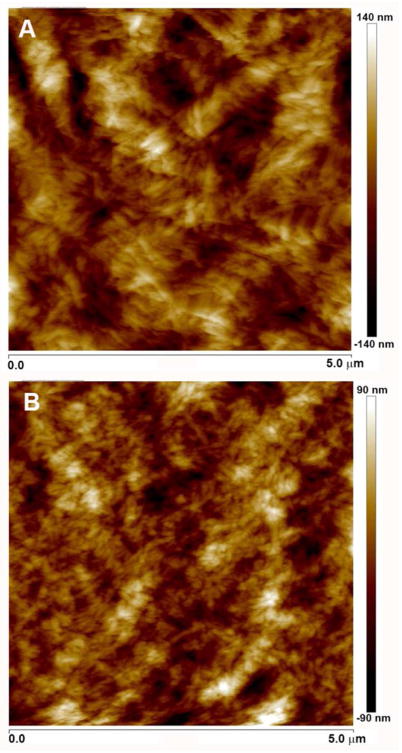
Pure PR_g hydrogels result from the self-assembly of the amphiphiles in water to form nanofibers and further aggregation of the nanofibers into a network of bundles as shown by cryogenic transmission electron microscopy (cryo-TEM) evaluation of PR_g samples.24 The nanoscale surface morphology of the pure PR_g peptide-amphiphile hydrogels was investigated with scanning electron microscopy (SEM) and cryo-SEM and verified the nanofibrous network structure of the PR_g hydrogels and the presence of fiber bundles.28 In this work AFM was used to evaluate the surface morphology of PR_g/E2 gels for the 0.5 wt% and 1.0 wt% hydrogels (Figure 1). Gels were left to dry at room temperature or at 37 °C and AFM images were collected in air. Results showed that the method of dehydration had no effect on the surface topography. Figure 1 verified the nanofibrous structure of the PR_g/E2 gels and the presence of bundles of nanofibers that varied in diameter between 50–130 nm for both wt%, in agreement with our previous PR_g cryo- TEM images.24
Figure 1.
AFM images (5 μm x 5 μm) in air of 5 mol% PR_g – 95 mol% E2 peptide-amphiphile hydrogels at (A) 0.5 wt% and (B) 1.0 wt%.
Rheology
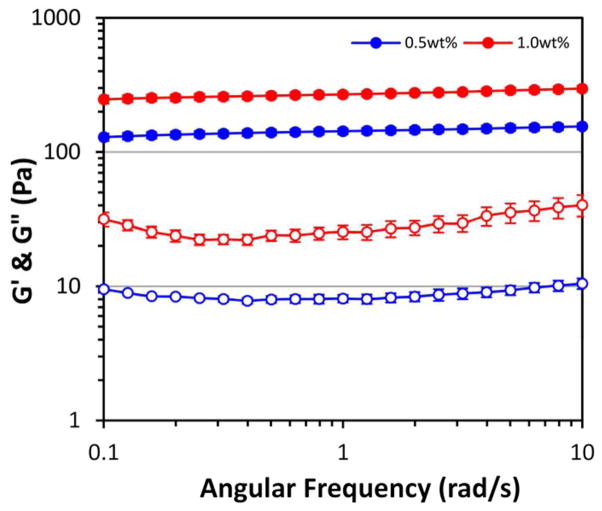
The mechanical properties of 0.5 wt% and 1.0 wt% PR_g/E2 hydrogels were measured using oscillatory rheology (Figure 2). Both gel formulations have a storage modulus (G’) greater than the loss modulus (G”), indicating that they do in fact form gels. Frequency sweeps in Figure 2 show that both hydrogels have minimal frequency dependence, indicating that there is a high degree of physical crosslinking. Because the measurement of storage and loss moduli were obtained in the linear viscoelastic regime, the elastic modulus, E, can be calculated by the following equation:
Figure 2.
Rheology of 5 mol% PR_g - 95 mol% E2 peptide-amphiphile hydrogels. Data are shown as the mean ± standard error from 3 independent experiments (n=3). Filled symbols represent the storage modulus (G’) and open symbols represent the loss modulus (G”).
The elastic modulus of the 0.5 wt% gels was 429.0 ± 21.3 Pa and the elastic modulus of the 1.0 wt% gels was almost twice the modulus of 0.5 wt% gels, at 808.6 ± 38.1 Pa.
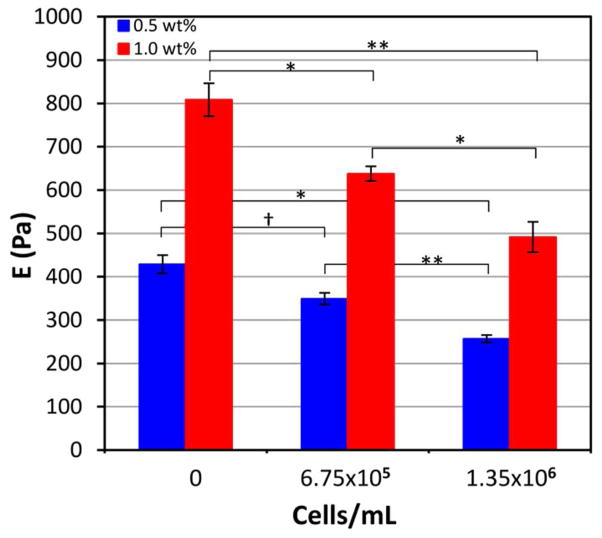
Rheological characterization of peptide-amphiphile hydrogels with entrapped cells (Figure S1, Supporting Information) revealed that the presence of cells decreased the elastic modulus, and the decrease was a function of the cell loading (Figure 3). As Figure 3 shows, the presence of 1.35 x 106 NIH3T3/GFP cells/mL entrapped in the gels decreased the elastic modulus by approximately 40%, to 256.8 ± 8.7 Pa and 491.9 ± 35.2 Pa for the 0.5 wt% and 1.0 wt% hydrogels respectively. The modulus of adherent cells has been reported to be between 30 Pa and 150 Pa when measured with magnetic twisting cytometry and optical tweezers.39,40 Much higher cell moduli have been reported from AFM measurements, although there is evidence to suggest that the substrate on which the cells are adhered and the state of adherence can significantly alter these measurements.41 The observed decrease in hydrogel modulus after cell encapsulation may be a result of encapsulating cells that are softer than the gel, however it is more likely due to the disruption of physical interactions between nanofibers and nanofiber bundles. This is the first evidence in the peptide-amphiphile literature showing the drastic changes in mechanical properties of the gel as a result of cell entrapment. While others have reported a dependence of the modulus on the concentration of the hydrogel solution for physical hydrogels used for cell culture,42–44 these measurements were typically made in the absence of cells. Our results indicate that the effect of cell encapsulation on the elastic modulus of a physical hydrogel is more significant than previously thought.
Figure 3.
Elastic modulus of 5 mol% PR_g - 95 mol% E2 peptide-amphiphile hydrogels as a function of NIH3T3/GFP cell loading. Data are shown as the mean ± standard error from 3 independent experiments (n=3). ANOVA analysis was performed and statistical significance noted for the bracketed data (* p < 0.05, ** p < 0.005, † p > 0.05).
Time sweep experiments were performed to determine how the elastic modulus of the hydrogels changes over time in the presence and absence of cells. Technical challenges, including sample dehydration (even though the samples were coated with low viscosity oil to prevent dehydration) and loss of contact between the hydrogel and parallel plate, prevented measurement for 5 days. For the 42 to 65 h for which data could be collected, no changes in hydrogel modulus were observed (Figure S2, Supporting Information). These results indicate that the hydrogels do not lose mechanical integrity during the time examined.
Both the 0.5 wt% and 1.0 wt% gels, with and without cells, have appropriate moduli to be used as scaffolds for tissue engineering applications, falling within the range of elastic moduli of soft tissue, 100 and 10,000 Pa.45,46 In particular, hepatic and pancreatic applications would be appropriate for our hydrogel system, with moduli between 400-600 Pa47 and between 640–1160 Pa48 respectively.
Cell viability and proliferation within peptide-amphiphile hydrogels
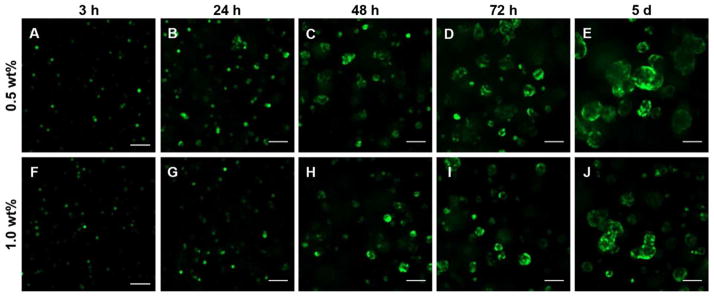
NIH3T3/GFP fibroblasts were selected in order to easily visualize cells within our hydrogel constructs and to avoid other methods of measuring cell viability and proliferation which can be complicated by limited diffusion through the gels and high background signals from the gels themselves. Live cells expressing GFP could be readily observed within the PR_g/E2 gels using confocal microscopy, and z-stacks were collected at 3, 24, 48, 72 h and 5 days. Movies showing rotating 3D representations of sequential z-stacked confocal images from the samples shown in Figure 4 are provided in Supporting Information (Movie S1–S10). After 3 and 24 h cells appeared to be rounded with little spreading apparent in both the 0.5 wt% and the 1.0 wt% PR_g/E2 gels (Figure 4 and Movie S1–S2, S6–S7, Supporting Information). This observation differs from previously reported behavior of cell attachment and spreading on 2D PR_g hydrogels, where cells were able to attach and spread within the first 24 h.30 In a 3D environment, cells appear to require more time to attach and spread. This is consistent with reports that cells embedded in a 3D matrix must first remodel their ECM before spreading, migrating, or depositing their own ECM.49–52 After 48 h an increase in cell number was observed (Figure 5) as well as cell clusters or aggregates, with larger clusters observed in the 0.5 wt% versus the 1.0 wt% gel (Figure 4 and Movie S3 and S8, Supporting Information). We hypothesize that the clustering is likely a result of cell proliferation. At both 72 h and 5 days cell number continues to increase (Figure 5) along with the size of the cell clusters (Figure 4 and Movie S4–S5 and S9–S10, Supporting Information), with more and larger cell clusters observed in the 0.5 wt% gels. Higher magnification images of cells at 24 h and 5 days in the 0.5 wt% and the 1.0 wt% PR_g/E2 gels are shown in Figure S3, Supporting Information.
Figure 4.
Representative confocal images of NIH3T3/GFP fibroblasts in 5 mol% PR_g - 95 mol% E2 peptide-amphiphile hydrogels over time. Images shown are 100 μm above the bottom of the well and 900 μm below the surface of (A-E) 0.5 wt% gels and (F-J) 1.0 wt% gels. NIH3T3/GFP cells are shown in green. The scale bar is 100 μm for all images.
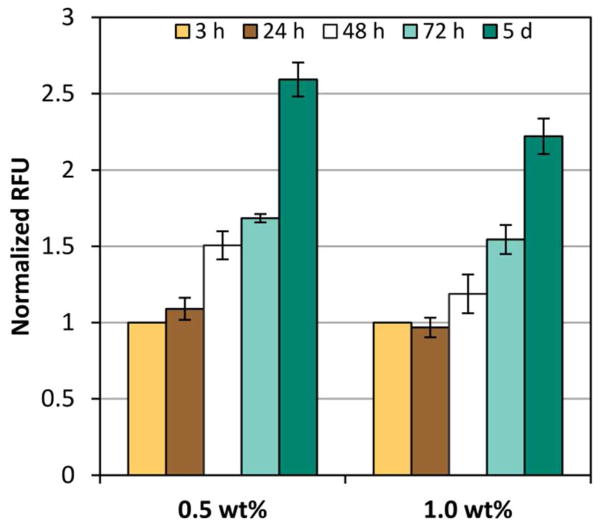
Figure 5.
Fluorescence quantification from NIH3T3/GFP fibroblasts entrapped in 5 mol% PR_g - 95 mol% E2 gels over time. Data described are the relative fluorescence units (RFU) detected from the GFP-expressing cells normalized to the 3 h time point. Data shown as mean ± standard error from four independent experiments (n=4) done in triplicates. ANOVA analysis was performed and p-values are reported in Tables S1–S2, Supporting Information.
The quantification of GFP fluorescence in Figure 5 from the 0.5 wt% gels was statistically significantly higher than the fluorescence from the 1.0 wt% gels at 48 h, 72 h and 5 days (p-values from ANOVA analysis of all the fluorescent data can be found in Table S1–S2, Supporting Information). Confocal images combined with the fluorescence quantification indicate that cells within the 0.5 wt% gels proliferate more compared to cells within the 1.0 wt% gels. The observed increase in proliferation in the lower wt% gel formulation is consistent with observations of fibroblasts encapsulated in protein 3D scaffolds and polymer hydrogels.53,54 For example, lower protein wt% resulted in bigger pore sizes and porosities due to lower protein mass per unit volume in the preparation solution, and bigger pore sizes led to better cell proliferation.53 Increased porosity and interconnectivity within scaffolds has also been associated with cell migration within porous 3D scaffolds, allowing more cells to proliferate thus reducing cell death due to overcrowding.53 These results are also consistent with the suggestion that highly cross-linked hydrogels and gels with higher elastic modulus make it more difficult for cells to spread and proliferate in 3D.54,55 In particular, it has been shown that the proliferation and spreading of fibroblasts encapsulated in PEG hydrogels functionalized with RGD-containing peptides increased with decreasing material stiffness.54 It was suggested that gels with higher stiffness pose an increased physical barrier and confinement, thus impairing cell proliferation in 3D environments. The higher the storage modulus G’, the denser the gel scaffold surrounding the cells, and the smaller the space for the cells to proliferate, spread and migrate.54
Quantification of cell viability via GFP fluorescence has been demonstrated in the literature previously. The half-life of GFP in NIH3T3 fibroblasts is 2.8 h,56 and decrease in fluorescence intensity has been shown to correlate well with traditional apoptosis and necrosis assays.57 GFP fluorescence in dead cells returned to background levels and was indistinguishable from the autofluorescence of the live cells.58 Thus, it has been demonstrated that the loss of GFP fluorescence parallels the loss of cell viability in NIH3T3 fibroblast cells.58 We examined cell viability via other ways too. First the GFP fluorescence of dead cells was evaluated and was shown to decrease rapidly (Figure S4, Supporting Information), in agreement with literature findings,58 demonstrating that the GFP fluorescence of the entrapped cells in Figure 4 and 5 comes from live cells. The Live/Dead cell viability assay was also used on NIH3T3 cells (without the GFP plasmid) encapsulated in 0.5 wt% and 1.0 wt% PR_g/E2 gels after 24 h and 5 days (Figure S5, Supporting Information). After 24 h, there was some cell death following the initial cell entrapment, as both green and red cells were observed. However, after 5 days only green cells were observed indicating that there was no additional death among cells that survived the initial entrapment.
Real-time RT-PCR for ECM mRNA
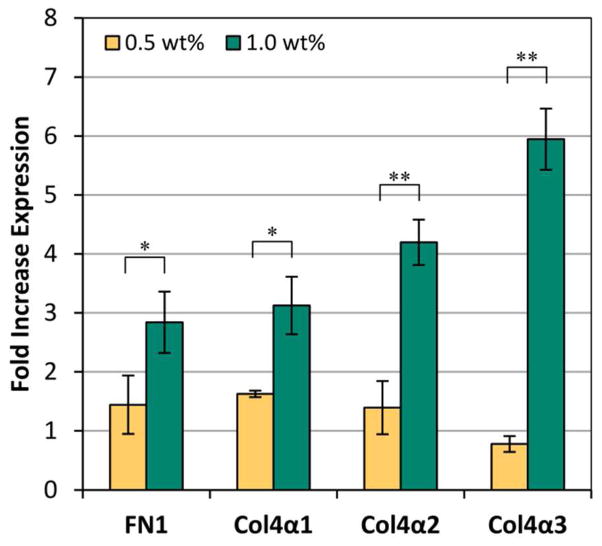
Real-time RTPCR was used to measure the expression of ECM mRNA produced by the NIH3T3/GFP cells entrapped in the 0.5 and 1.0 wt% PR_g/E2 hydrogels. After 5 days of culture, an increase in the expression of fibronectin and type VI collagen was observed for both gel concentrations (Figure 6). A previous study has also shown an increase in collagen ECM expression over time after osteogenic differentiation of dental pulp stem cells encapsulated in a matrix metal-loproteinase- sensitive, RGD-functionalized peptide-amphiphile scaffold.13 The mechanical properties of polymeric hydrogels have been shown before to influence ECM production in different ways, and increased cell proliferation has been frequently correlated with decreased ECM synthesis, as seen in our study.59–61 This is the first study though that shows that cells within the 1.0 wt% peptide-amphiphile hydrogel had significantly higher expression of fibronectin and type IV collagen (α1, α2, and α3 isoforms) compared to the 0.5 wt% peptide-amphiphile gels. It has previously been shown that the α3 isoform of type IV collagen plays a critical role in the assembly of stable collagen IV molecules and that increased expression of the α3 occurs simultaneously with collagen IV protein deposition.62,63 The significant increase in the α3 isoform of collagen IV in the 1.0 wt% gels over time and compared to the 0.5 wt% samples is likely indicative of more fibril collagen IV protein in the ECM of the cells entrapped in the 1.0 wt% gels.
Figure 6.
Real-time PCR results of ECM mRNA expression from the NIH3T3/GFP cells entrapped in 5 mol% PR_g - 95 mol% E2 gels. Fold increase in expression compares 3 h and day 5 expression of fibronectin (FN1) and collagen IV (Col4α1, Col4α2, Col4α3). Data are shown as mean ± standard error from four independent experiments (n=4) done in triplicates. Statistical analysis was performed using a one-sided t-test and significance is noted for bracketed data (* p < 0.05; ** p < 0.01).
Histology and immunohistochemistry
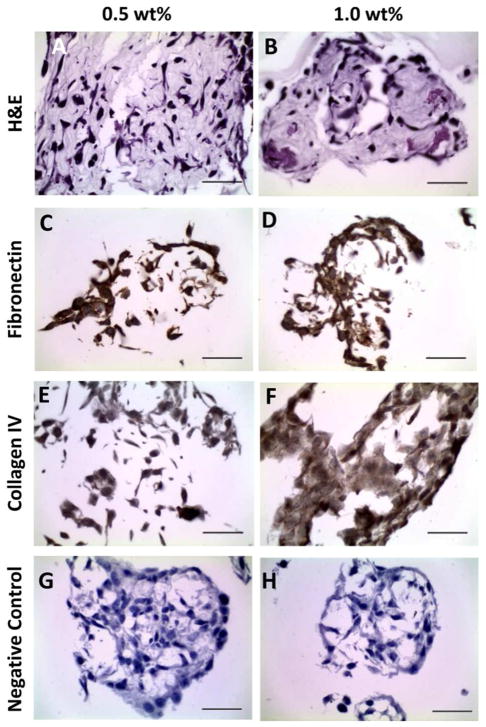
Hematoxylin and eosin (H&E) staining revealed that the NIH3T3/GFP fibroblasts in both the 0.5 wt% and 1.0 wt% gels were still associated with the PR_g/E2 peptide-amphiphile hydrogel after 5 days of culture (Figures 7A,B). Cell clusters were visible in both 0.5 wt% and 1.0 wt% gel, with larger clusters and higher density of cell clusters in the 0.5 wt% gels, consistent with the results of Figure 4. Immunohistochemistry revealed the presence of fibronectin (Figure 7C,D) and collagen IV (Figure 7E,F) ECM proteins. A negative control without primary antibodies (Figure 7G,H) showed that the fibronectin and collagen IV staining were specific. The 1.0 wt% gels appeared to have more collagen IV and fibronectin ECM proteins compared to the 0.5 wt% gels, consistent with the observed increase in mRNA expression shown in Figure 6.
Figure 7.
Histological and immunohistochemical evaluation of 1.35 x 106 NIH3T3/GFP cells/mL entrapped for 5 days in 0.5 wt% and 1.0 wt% hydrogels of 5 mol% PR_g - 95 mol% E2 peptide-amphiphiles. H&E stain (A,B), fibronectin stain (C,D), collagen IV stain (E,F), and a negative control without primary antibodies (G, H). The scale bar is 50 μm for all images.
CONCLUSIONS
A peptide-amphiphile hydrogel was designed using the fibronectin-mimetic PR_g peptide-amphiphile and the E2 diluent peptide-amphiphile to support 3D cell culture. The hydrogel microstructure and mechanical properties of 0.5 wt% and 1.0 wt% gels were studied and it was found that the elastic modulus increased with higher wt% gels. Even though the presence of entrapped cells decreased the elastic modulus of the gels, the values still fall within the range reported for soft tissue, supporting the potential relevance of these hydrogels as tissue engineering scaffolds. Both 0.5 and 1.0 wt% hydrogels were able to support 3D culture and proliferation of NIH3T3/GFP fibroblasts over a period of five days. While the 0.5 wt% gels supported significantly higher rates of proliferation after 5 days of culture, the 1.0 wt% gels supported significantly higher mRNA expression and production of ECM proteins. These results show that the PR_g/E2 peptideamphiphile hydrogels are an effective scaffold for 3D cell culture which has the potential to improve the design of ECM mimetic scaffolds for tissue engineering.
Supplementary Material
Acknowledgments
This work was supported by NSF/CBET-1253913. CMS was partially supported by a biotechnology training grant 5T32 GM08347.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
ASSOCIATED CONTENT
Supporting Information. Movies of rotating 3D representations of z-stack confocal images from entrapped NIH3T3/GFP cells in PR_g/E2 gels, ANOVA analysis p-values of fluorescent signal from entrapped NIH3T3/GFP cells in PR_g/E2 gels, rheology of PR_g/E2 gels as a function of cell load and time, higher magnification confocal images of NIH3T3/GFP cells entrapped in the PR_g/E2 gels, GFP fluorescence of NIH3T3/GFP dead cells, Live/Dead cell viability assay for NIH3T3 cells entrapped in the PR_g/E2 gels. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ma Z, Kotaki M, Inai R, Ramakrishna S. Potential of Nanofiber Matrix as Tissue-Engineering Scaffolds. Tissue Eng. 2005;11:101–109. doi: 10.1089/ten.2005.11.101. [DOI] [PubMed] [Google Scholar]
- 2.Chen M, Patra PK, Warner SB, Bhowmick S. Role of Fiber Diameter in Adhesion and Proliferation of NIH 3T3 Fibroblast on Electrospun Polycaprolactone Scaffolds. Tissue Eng. 2007;13:579–587. doi: 10.1089/ten.2006.0205. [DOI] [PubMed] [Google Scholar]
- 3.Badami AS, Kreke MR, Thompson MS, Riffle JS, Goldstein AS. Effect of Fiber Diameter on Spreading, Proliferation, and Differentiation of Osteoblastic Cells on Electrospun Poly(lactic Acid) Substrates. Biomaterials. 2006;27:596–606. doi: 10.1016/j.biomaterials.2005.05.084. [DOI] [PubMed] [Google Scholar]
- 4.Flemming RG, Murphy CJ, Abrams GA, Goodman SL, Nealey PF. Effects of Synthetic Micro- and Nano-Structured Surfaces on Cell Behavior. Biomaterials. 1999;20:573–588. doi: 10.1016/s0142-9612(98)00209-9. [DOI] [PubMed] [Google Scholar]
- 5.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking Cell-Matrix Adhesions to the Third Dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 6.Gluck JM, Delman C, Full S, Shemin RJ, Heydarkhan-Hagvall S. Stem Cell Extracellular Matrix Interactions in Three- Dimensional System via Integrins. J Regen Med. 2013;2:1000107. [Google Scholar]
- 7.Birgersdotter A, Sandberg R, Ernberg I. Gene Expression Perturbation in Vitro-A Growing Case for Three- Dimensional (3D) Culture Systems. Semin Cancer Biol. 2005;15:405–412. doi: 10.1016/j.semcancer.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Yokoi H, Kinoshita T, Zhang S. Dynamic Reassembly of Peptide RADA16 Nanofiber Scaffold. Proc Natl Acad Sci U S A. 2005;102:8414–8419. doi: 10.1073/pnas.0407843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinman HK, Martin GR. Matrigel: Basement Membrane Matrix with Biological Activity. Semin Cancer Biol. 2005;15:378–386. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Anderson SB, Lin CC, Kuntzler DV, Anseth KS. The Performance of Human Mesenchymal Stem Cells Encapsulated in Cell-Degradable Polymer-Peptide Hydrogels. Biomaterials. 2011;32:3564–3574. doi: 10.1016/j.biomaterials.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salinas CN, Anseth KS. The Enhancement of Chondrogenic Differentiation of Human Mesenchymal Stem Cells by Enzymatically Regulated RGD Functionalities. Biomaterials. 2008;29:2370–2377. doi: 10.1016/j.biomaterials.2008.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kokkoli E, Mardilovich A, Wedekind A, Rexeisen EL, Garg A, Craig JA. Self-Assembly and Applications of Biomimetic and Bioactive Peptide-Amphiphiles. Soft Matter. 2006;2:1015–1024. doi: 10.1039/b608929a. [DOI] [PubMed] [Google Scholar]
- 13.Galler KM, Cavender A, Yuwono V, Dong H, Shi S, Ph D, Schmalz G, Hartgerink JD, Souza RND. Self- Assembling Peptide Amphiphile Nanofibers as a Scaffold for Dental Stem Cells. Tissue Eng Part A. 2008;14:2051–2058. doi: 10.1089/ten.tea.2007.0413. [DOI] [PubMed] [Google Scholar]
- 14.Galler KM, Aulisa L, Regan KR, D’Souza RN, Hartgerink JD. Self-Assembling Multidomain Peptide Hydrogels: Designed Susceptibility to Enzymatic Cleavage Allows Enhanced Cell Migration and Spreading. J Am Chem Soc. 2010;132:3217–3223. doi: 10.1021/ja910481t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson JM, Kushwaha M, Tambralli A, Bellis SL, Camata RP, Jun HW. Osteogenic Differentiation of Human Mesenchymal Stem Cells Directed by Extracellular Matrix-Mimicking Ligands in a Biomimetic Self-Assembled Peptide Amphiphile Nanomatrix. Biomacromolecules. 2009;10:2935–2944. doi: 10.1021/bm9007452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosseinkhani H, Hosseinkhani M, Tian F, Kobayashi H, Tabata Y. Osteogenic Differentiation of Mesenchymal Stem Cells in Self-Assembled Peptide-Amphiphile Nanofibers. Biomaterials. 2006;27:4079–4086. doi: 10.1016/j.biomaterials.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 17.Hosseinkhani H, Hosseinkhani M, Kobayashi H. Proliferation and Differentiation of Mesenchymal Stem Cells Using Self-Assembled Peptide Amphiphile Nanofibers. Biomed Mater. 2006;1:8–15. doi: 10.1088/1748-6041/1/1/002. [DOI] [PubMed] [Google Scholar]
- 18.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Selective Differentiation of Neural Progenitor Cells by High-Epitope Density Nanofibers. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 19.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, Kessler JA. Self- Assembling Nanofibers Inhibit Glial Scar Formation and Promote Axon Elongation after Spinal Cord Injury. J Neurosci. 2008;28:3814–3823. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sur S, Pashuck ET, Guler MO, Ito M, Stupp SI, Launey T. A Hybrid Nanofiber Matrix to Control the Survival and Maturation of Brain Neurons. Biomaterials. 2012;33:545–555. doi: 10.1016/j.biomaterials.2011.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah RN, Shah NA, Del Rosario Lim MM, Hsieh C, Nuber G, Stupp SI. Supramolecular Design of Self- Assembling Nanofibers for Cartilage Regeneration. Proc Natl Acad Sci U S A. 2010;107:3293–3298. doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim DJ, Antipenko SV, Vines JB, Andukuri A, Hwang PTJ, Hadley NT, Rahman SM, Corbett JA, Jun H-W. Improved MIN6 B-Cell Function on Self-Assembled Peptide Amphiphile Nanomatrix Inscribed with Extracellular Matrix-Derived Cell Adhesive Ligands. Macromol Biosci. 2013;13:1404–1412. doi: 10.1002/mabi.201300155. [DOI] [PubMed] [Google Scholar]
- 23.Stendahl JC, Wang LJ, Chow LW, Kaufman DB, Stupp SI. Growth Factor Delivery from Self-Assembling Nanofibers to Facilitate Islet Transplantation. Transplantation. 2008;86:478–481. doi: 10.1097/TP.0b013e3181806d9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghanaati S, Webber MJ, Unger RE, Orth C, Hulvat JF, Kiehna SE, Barbeck M, Rasic A, Stupp SI, Kirkpatrick CJ. Dynamic in Vivo Biocompatibility of Angiogenic Peptide Amphiphile Nanofibers. Biomaterials. 2009;30:6202–6212. doi: 10.1016/j.biomaterials.2009.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beniash E, Hartgerink JD, Storrie H, Stendahl JC, Stupp SI. Self-Assembling Peptide Amphiphile Nanofiber Matrices for Cell Entrapment. Acta Biomater. 2005;1:387–397. doi: 10.1016/j.actbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Rexeisen EL, Fan W, Pangburn TO, Taribagil RR, Bates FS, Lodge TP, Tsapatsis M, Kokkoli E. Self- Assembly of Fibronectin Mimetic Peptide-Amphiphile Nanofibers. Langmuir. 2010;26:1953–1959. doi: 10.1021/la902571q. [DOI] [PubMed] [Google Scholar]
- 27.Mardilovich A, Craig JA, McCammon MQ, Garg A, Kokkoli E. Design of a Novel Fibronectin-Mimetic Peptide- Amphiphile for Functionalized Biomaterials. Langmuir. 2006;22:3259–3264. doi: 10.1021/la052756n. [DOI] [PubMed] [Google Scholar]
- 28.Craig JA, Rexeisen EL, Mardilovich A, Shroff K, Kokkoli E. Effect of Linker and Spacer on the Design of a Fibronectin-Mimetic Peptide Evaluated via Cell Studies and AFM Adhesion Forces. Langmuir. 2008;24:10282–10292. doi: 10.1021/la702434p. [DOI] [PubMed] [Google Scholar]
- 29.Shroff K, Pearce TR, Kokkoli E. Enhanced Integrin Mediated Signaling and Cell Cycle Progression on Fibronectin Mimetic Peptide Amphiphile Monolayers. Langmuir. 2012;28:1858–1865. doi: 10.1021/la203322t. [DOI] [PubMed] [Google Scholar]
- 30.Shroff K, Rexeisen EL, Arunagirinathan MA, Kokkoli E. Fibronectin-Mimetic Peptide-Amphiphile Nanofiber Gels Support Increased Cell Adhesion and Promote ECM Production. Soft Matter. 2010;6:5064–5072. [Google Scholar]
- 31.Behanna HA, Donners JJJM, Gordon AC, Stupp SI. Coassembly of Amphiphiles with Opposite Peptide Polarities into Nanofibers. J Am Chem Soc. 2005;127:1193–1200. doi: 10.1021/ja044863u. [DOI] [PubMed] [Google Scholar]
- 32.Webber MJ, Tongers J, Renault MA, Roncalli JG, Losordo DW, Stupp SI. Development of Bioactive Peptide Amphiphiles for Therapeutic Cell Delivery. Acta Biomater. 2010;6:3–11. doi: 10.1016/j.actbio.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Storrie H, Guler MO, Abu-Amara SN, Volberg T, Rao M, Geiger B, Stupp SI. Supramolecular Crafting of Cell Adhesion. Biomaterials. 2007;28:4608–4618. doi: 10.1016/j.biomaterials.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 34.Sur S, Matson JB, Webber MJ, Newcomb CJ, Stupp SI. Photodynamic Control of Bioactivity in a Nanofiber Matrix. ACS Nano. 2012;6:10776–10785. doi: 10.1021/nn304101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei Y, Gojgini S, Lam J, Segura T. The Spreading, Migration and Proliferation of Mouse Mesenchymal Stem Cells Cultured inside Hyaluronic Acid Hydrogels. Biomaterials. 2011;32:39–47. doi: 10.1016/j.biomaterials.2010.08.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li YJ, Chung EH, Rodriguez RT, Firpo MT, Healy KE. Hydrogels as Artificial Matrices for Human Embryonic Stem Cell Self-Renewal. J Biomed Mater Res A. 2006;79:1–5. doi: 10.1002/jbm.a.30732. [DOI] [PubMed] [Google Scholar]
- 37.Burdick JA, Anseth KS. Photoencapsulation of Osteoblasts in Injectable RGD-Modified PEG Hydrogels for Bone Tissue Engineering. Biomaterials. 2002;23:4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 38.Jung JP, Nagaraj AK, Fox EK, Rudra JS, Devgun JM, Collier JH. Co-Assembling Peptides as Defined Matrices for Endothelial Cells. Biomaterials. 2009;30:2400–2410. doi: 10.1016/j.biomaterials.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamgoue A, Ohayon J, Tracqui P. Estimation of Cell Young’s Modulus of Adherent Cells Probed by Optical and Magnetic Tweezers: Influence of Cell Thickness and Bead Immersion. J Biomech Eng. 2007;129:523–530. doi: 10.1115/1.2746374. [DOI] [PubMed] [Google Scholar]
- 40.Laurent VM, Henon S, Planus E, Fodil R, Balland M, Isabey D, Gallet F. Assessment of Mechanical Properties of Adherent Living Cells by Bead Micromanipulation: Comparison of Magnetic Twisting Cytometry vs Optical Tweezers. J Biomech Eng. 2002;124:408–421. doi: 10.1115/1.1485285. [DOI] [PubMed] [Google Scholar]
- 41.Dokukin ME, Guz NV, Sokolov I. Quantitative Study of the Elastic Modulus of Loosely Attached Cells in AFM Indentation Experiments. Biophys J. 2013;104:2123–2131. doi: 10.1016/j.bpj.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Guo X, Palmer AF, Das H, Guan J. High- Efficiency Matrix Modulus-Induced Cardiac Differentiation of Human Mesenchymal Stem Cells inside a Thermosensitive Hydrogel. Acta Biomater. 2012;8:3586–3595. doi: 10.1016/j.actbio.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 43.Almany L, Seliktar D. Biosynthetic Hydrogel Scaffolds Made from Fibrinogen and Polyethylene Glycol for 3D Cell Cultures. Biomaterials. 2005;26:2467–2477. doi: 10.1016/j.biomaterials.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 44.Liu SQ, Ee PLR, Ke CY, Hedrick JL, Yang YY. Biodegradable Poly(ethylene Glycol)-Peptide Hydrogels with Well-Defined Structure and Properties for Cell Delivery. Biomaterials. 2009;30:1453–1461. doi: 10.1016/j.biomaterials.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 45.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Substrate Modulus Directs Neural Stem Cell Behavior. Biophys J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 47.Janmey PA, Miller RT. Mechanisms of Mechanical Signaling in Development and Disease. J Cell Sci. 2011;124:9–18. doi: 10.1242/jcs.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wex C, Frochlich M, Brandstadter K, Bruns C, Stoll A. Experimental Analysis of the Mechanical Behavior of the Viscoelastic Porcine Pancreas and Preliminary Case Study on the Human Pancreas. J Mech Behav Biomed Mater. 2015;41:199– 207. doi: 10.1016/j.jmbbm.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 49.Kraning-Rush CM, Carey SP, Califano JP, Smith BN, Reinhart-King CA. The Role of the Cytoskeleton in Cellular Force Generation in 2D and 3D Environments. Phys Biol. 2011;8:015009. doi: 10.1088/1478-3975/8/1/015009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sengers BG, Taylor M, Please CP, Oreffo ROC. Computational Modelling of Cell Spreading and Tissue Regeneration in Porous Scaffolds. Biomaterials. 2007;28:1926– 1940. doi: 10.1016/j.biomaterials.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 51.Raeber GP, Lutolf MP, Hubbell JA. Molecularly Engineered PEG Hydrogels: A Novel Model System for Proteolytically Mediated Cell Migration. Biophys J. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Synthetic Matrix Metalloproteinase-Sensitive Hydrogels for the Conduction of Tissue Regeneration: Engineering Cell-Invasion Characteristics. Proc Natl Acad Sci U S A. 2003;100:5413– 5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mandal BB, Kundu SC. Cell Proliferation and Migration in Silk Fibroin 3D Scaffolds. Biomaterials. 2009;30:2956–2965. doi: 10.1016/j.biomaterials.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Bott K, Upton Z, Schrobback K, Ehrbar M, Hubbell JA, Lutolf MP, Rizzi SC. The Effect of Matrix Characteristics on Fibroblast Proliferation in 3D Gels. Biomaterials. 2010;31:8454–8464. doi: 10.1016/j.biomaterials.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 55.Friedl P, Brocker EB. The Biology of Cell Locomotion within Three-Dimensional Extracellular Matrix. Cell Mol Life Sci. 2000;57:41–64. doi: 10.1007/s000180050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halter M, Tona A, Bhadriraju K, Plant AL, Elliott JT. Automated Live Cell Imaging of Green Fluorescent Protein Degradation in Individual Fibroblasts. Cytometry A. 2007;71:827–834. doi: 10.1002/cyto.a.20461. [DOI] [PubMed] [Google Scholar]
- 57.Strebel A, Harr T, Bachmann F, Wernli M, Erb P. Green Fluorescent Protein as a Novel Tool to Measure Apoptosis and Necrosis. Cytometry. 2001;43:126–133. doi: 10.1002/1097-0320(20010201)43:2<126::aid-cyto1027>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 58.Steff A, Arguin C, Hugo P. Detection of a Decrease in Green Fluorescent Protein Fluorescence for the Monitoring of Cell Death : An Assay Amenable to High-Throughput Screening Technologies. Cytometry. 2001;243:237–243. doi: 10.1002/1097-0320(20011201)45:4<237::aid-cyto10024>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 59.Liao H, Munoz-Pinto D, Qu X, Hou Y, Grunlan MA, Hahn MS. Influence of Hydrogel Mechanical Properties and Mesh Size on Vocal Fold Fibroblast Extracellular Matrix Production and Phenotype. Acta Biomater. 2008;4:1161–1171. doi: 10.1016/j.actbio.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeong CG, Hollister SJ. Mechanical and Biochemical Assessments of Three-Dimensional Poly(1,8- Octanediol-co-Citrate) Scaffold Pore Shape and Permeability Effects on In Vitro Chondrogenesis Using Primary Chondrocytes. Tissue Eng Part A. 2010;16:3759–3768. doi: 10.1089/ten.tea.2010.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bryant SJ, Anseth KS. Hydrogel Properties Influence ECM Production by Chondrocytes Photoencapsulated in Poly (Ethylene Glycol) Hydrogels. J Biomed Mater Res. 2001;59:63–72. doi: 10.1002/jbm.1217. [DOI] [PubMed] [Google Scholar]
- 62.Lamande SR, Sigalas E, Pan TC, Chu ML, Dziadek M, Timpl R, Bateman JF. The Role of the α3(VI) Chain in Collagen VI Assembly. Expression of an α3(VI) Chain Lacking N-Terminal Modules N10-N7 Restores Collagen VI Assembly, Secretion, and Matrix Deposition in an α3(VI)- Deficient Cell Line. J Biol Chem. 1998;273:7423–7430. doi: 10.1074/jbc.273.13.7423. [DOI] [PubMed] [Google Scholar]
- 63.Hatamochis A, Mauch C, Chull M, Timplg R, Krieg T. Regulation of Collagen VI Expression in Fibroblasts. J Biol Chem. 1989;264:3494–3499. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.