Abstract
Dysregulation of pyramidal cell network function by the soma- and axon-targeting inhibitory neurons that contain the calcium-binding protein parvalbumin (PV) represents a core pathophysiological feature of schizophrenia. In order to gain insight into the molecular basis of their functional impairment, we used laser capture microdissection (LCM) to isolate PV-immunolabeled neurons from layer 3 of Brodmann’s area 42 of the superior temporal gyrus (STG) from postmortem schizophrenia and normal control brains. We then extracted ribonucleic acid (RNA) from these neurons and determined their messenger RNA (mRNA) expression profile using the Affymetrix platform of microarray technology. Seven hundred thirty-nine mRNA transcripts were found to be differentially expressed in PV neurons in subjects with schizophrenia, including genes associated with WNT (wingless-type), NOTCH, and PGE2 (prostaglandin E2) signaling, in addition to genes that regulate cell cycle and apoptosis. Of these 739 genes, only 89 (12%) were also differentially expressed in pyramidal neurons, as described in the accompanying paper, suggesting that the molecular pathophysiology of schizophrenia appears to be predominantly neuronal type specific. In addition, we identified 15 microRNAs (miRNAs) that were differentially expressed in schizophrenia; enrichment analysis of the predicted targets of these miRNAs included the signaling pathways found by microarray to be dysregulated in schizophrenia. Taken together, findings of this study provide a neurobiological framework within which hypotheses of the molecular mechanisms that underlie the dysfunction of PV neurons in schizophrenia can be generated and experimentally explored and, as such, may ultimately inform the conceptualization of rational targeted molecular intervention for this debilitating disorder.
Keywords: cerebral cortex, gene expression profiling, laser capture microdissection, microRNA
INTRODUCTION
Cellular heterogeneity characterizes the organization of the cerebral cortex, where functionally and structurally distinct classes of neurons mediate specific aspects of information processing (Markram et al., 2004; Soltesz, 2005). The fact that collectively these distinct populations of neurons are able to seamlessly regulate the flow of information and thereby the accurate generation of perceptual and cognitive constructs critically depends on neuronal type–specific physiological properties and connectional architecture. For instance, the inhibitory neurons that contain parvalbumin (PV) exhibit distinct fast-spiking nonaccommodating firing properties, and, synaptically, they comprise basket and chandelier neurons, which target the cell body and the axon initial segment of pyramidal neurons, respectively. Together these physiological and connectional characteristics endow PV neurons with the unique ability to time and sync the oscillatory firing and synchronization of cortical pyramidal cell networks in the gamma frequency band (i.e., 30–100 Hz), which is thought to be the electrophysiological substrate that mediates a wide range of higher-order perceptual and cognitive functions (Singer et al., 1990; Uhlhaas et al., 2009; Wang, 2010).
Converging lines of evidence derived from postmortem human brain studies suggest that PV neurons in the cerebral cortex are functionally aberrant in schizophrenia (Lewis et al., 2011). First, immunohistochemical visualization and quantification of γ-aminobutryic acid (GABA) transporter GAT-1-immunoreactive axon cartridges of chandelier neurons revealed a decrease in the density of these terminals by ~ 40% in subjects with schizophrenia (Pierri et al., 1999; Woo et al., 1998). Second, the amount of the 67-kDa isoform of the GABA-synthesizing enzyme glutamic acid decarboxylase (GAD)67 is decreased by ~50% in the terminals of basket neurons (Curley et al., 2011; Curley & Lewis, 2012). Consistent with this finding, in pyramidal neurons, the expression of the GABAA receptor alpha 1 subunit, which is preferentially localized to receptors postsynaptic to axon terminals furnished by basket neurons, is also decreased (Glausier & Lewis, 2011). Third, the expression of the mRNA for PV has been shown to be decreased by ~ 30% (Hashimoto et al., 2003). Finally, in ~45% of PV neurons, the mRNA for GAD67 is undetectable (Hashimoto, et al., 2003). In light of these findings and given the well-known role of PV neurons in the generation of gamma band oscillation, it is particularly relevant that, clinically, gamma oscillation generation has been consistently shown to be impaired in patients with schizophrenia (Gonzalez-Burgos et al., 2010; Spencer et al., 2004; Uhlhaas & Singer, 2010). In other words, gamma band oscillation disturbances in schizophrenia patients may represent a read-out of the dysregulation of pyramidal cell networks that results, at least in part, from the dysfunction of PV neurons.
In the accompanying article, we report microarray findings implicating the molecular mechanisms of pyramidal cell dysfunction in layer 3 of the superior temporal gyrus (STG) in schizophrenia. It has been speculated that functional disturbances of pyramidal neurons may in part be a consequence of the dysfunction of the PV neurons that innervate them (Homayoun & Moghaddam, 2007; Lisman et al., 2008; Woo et al., 2010). In an attempt to gain insight into the molecular underpinnings of the disturbances of the PV neurons that are presynaptic to these pyramidal neurons, we combined immunolaser capture microdissection (LCM) with Affymetrix (xx, xx) microarray to profile the mRNA expression of neurons immunoreactive for PV in layer 3 of the STG in schizophrenia. We identified 739 differentially expressed genes, including those that regulate WNT and NOTCH signaling, many apoptotic genes, and genes that regulate cell cycle. Furthermore, when comparing with the result of the accompanying study, only 89 (12%) of these 739 genes were also differentially expressed in pyramidal neurons, suggesting that the patholophysiology of schizophrenia appears to be predominantly neuronal type specific. Finally, we identified 15 microRNAs (miRNAs) that were differentially expressed in schizophrenia and the predicted targets of these miRNAs included the dysregulated pathways identified by microarray analysis. Together, findings of this study provide a neurobiological framework within which experiments can be designed to better understand the molecular basis of the dysfunction of PV neurons, and, as such, this methodological approach may ultimately inform the conceptualization and development of targeted molecular intervention for this devastating disorder.
METHODS AND STATISTICS
Postmortem Human Brain Tissue
Liquid nitrogen vapor fresh-frozen blocks, approximately 3 mm thick and containing Brodmann’s area 42 of the STG, were obtained from the Harvard Brain Tissue Resource Center (HBTRC) at McLean Hospital in Belmont, Massachusetts, and matched for age, sex, and postmortem interval (PMI) (Table 1), following procedures detailed in the accompanying article.
Table 1.
Subjects included in this study.
| Group | Sex | Age (years) | PMI | pH | Smoker | Cause of death | Antipsychotic |
|---|---|---|---|---|---|---|---|
| Control | F | 79 | 15.0 | 6.59 | Yes | Cardiac arrest | N/A |
| Control | M | 22 | 21.5 | 6.75 | Yes | Myocardial infarction | N/A |
| Control | M | 75 | 20.3 | 6.35 | Yes | Unknown | N/A |
| Control | M | 80 | 15.5 | 6.26 | Yes | Myocardial infarction | N/A |
| Control | F | 58 | 21.1 | 6.79 | No | Myocardial infarction | N/A |
| Control | M | 61 | 17.0 | 6.64 | Yes | Unknown | N/A |
| Control | F | 71 | 20.5 | 6.89 | Unknown | Unknown | N/A |
| Control | F | 90 | 12.7 | 6.10 | Unknown | Lung cancer | N/A |
| Mean±SD | 4M/4F | 67.0±20.9 | 17.9±3.3 | 6.54±0.28 | |||
| Schizophrenia | F | 93 | 6.92 | 6.13 | Unknown | Renal failure | Perphenazine |
| Schizophrenia | M | 55 | 21.40 | 6.51 | Yes | Myocardial infarction | Perphenazine |
| Schizophrenia | F | 55 | 22.00 | 5.90 | Yes | Cancer | Clozapine, olanzapine |
| Schizophrenia | M | 36 | 17.97 | 6.45 | Unknown | Cardiac arrest | Clozapine |
| Schizophrenia | M | 62 | 10.75 | 6.50 | Yes | Chronic obstructive pulmonary disease Lung cancer |
Clozapine |
| Schizophrenia | F | 92 | 17.80 | 6.34 | Unknown | Cardiomyopathy | Unknown |
| Schizophrenia | M | 56 | 21.83 | 6.75 | No | Car accident | Olanzapine |
| Schizophrenia | F | 88 | 13.33 | 6.65 | No | Unknown | Thiothixene |
| Mean±SD | 4M/4F | 67.1±21.2 | 16.5±5.6 | 6.4±0.28 |
Note. F = female; M = male; PMI = postmortem interval in hours; N/A = not applicable.
Immuno-LCM
A detailed methodology for tissue preparation, LCM, and RNA processing has been described elsewhere (Pietersen et al., 2009, 2011). Briefly, sections of 8 μm were cut on a cryostat and mounted onto slides. Mounted sections were incubated in an anti-PV antibody (1:10 dilution; mouse, Sigma-Aldrich, St. Louis, MO, USA) for 7 minutes and in peroxidase AffiniPure donkey anti-mouse immunoglobulin G (IgG) secondary antibody (1:10 dilution; Jackson ImmunoResearch Laboratories, West Grove, PA, USA) for 7 minutes, together with an RNase Inhibitor (40U/μL; Roche, Basel, Switzerland). Neurons were ultimately visualized with the NovaRED substrate-chromogen (12 minutes; Vector; Figure 1A). For each of the subjects, approximately 300 PV-immunolabeled neurons were captured onto a CapSure HS LCM cap (Life Technologies, xx, NY, USA). To corroborate the identity of the captured neurons, we used quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) to verify the presence of the mRNA for PVALB, the gene that encodes PV, and the absence of the mRNAs for three markers that are not expressed by PV neurons: calbindin (CALB1; a marker for a nonoverlapping subset of inhibitory neurons), calcium/calmodulin-dependent protein kinase II (CAMKII; a marker for pyramidal neurons), and glial fibrillary acidic protein (GFAP; a marker for astrocytes). Sequences of primers used for qRT-PCR can be found in (Supplementary Table S1 to be found online at http://informahealthcare.com/doi/abs/10.3109/01677063.2013.878339).
Figure 1.
Identification of PV neurons, representative virtual gels of amplified RNA products, and heatmap of 739 differentially expressed genes. (A) Photomicrograph of PV-immunoreactive neurons. Scale bar = 10 μm. (B) Representative virtual gels showing the sizes of products after two rounds of linear amplification of RNA extracted from PV-immunoreactive neurons from a normal control (C) and a schizophrenia (S) subjects. (C) Heatmap of the 739 differentially expressed genes in schizophrenia compared with normal control subjects, showing the segregation of genes based on diagnosis under the stringency criteria of fold-change >1.2, FDR-adjusted p<0.05.
Affymetrix-Based Microarray Gene Expression Profiling
RNA isolation, amplification, labeling, and hybridization were performed as described in the accompanying article. Validation of microarray data by qRT-PCR was performed on 16 genes (10 randomly selected and 6 selected from the most differentially expressed pathways) according to the approach described by Miron et al. (Miron et al., 2006). Microarray data have been deposited into the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo/).
miRNA Profiling Using Megaplex miRNA TaqMan Arrays
Total RNA was extracted and miRNA profiling was conducted as described in the accompanying article.
Data Analysis
Microarray Data Analysis
Each array was scanned twice and the Affymetrix Microarray Suite 5.1 software averaged the two images to compute an intensity value for each probe cell within each probe set. For the quality control step, we employed the dChip (http://www.biostat.harvard.edu/complab/dchip) and Partek software’s built-in function (Partek, MO, USA). We then normalized all data with Partek’s standard normalization method (i.e., data have a mean of zero and a variance of one, and each column for each sample was divided by the average of all control samples). Principal component analysis revealed the contribution of batch effect, but not age, sex, PMI, or antipsychotic medication treatment, to the observed gene expression variance. As a result, an analysis of covariance (ANCOVA) was performed with diagnosis as the main effect and batch effect (scan date) as covariate (Simunovic et al., 2009), with false discovery rate (FDR) set at 10%. Differentially expressed genes were visualized by performing unsupervised hierarchical clustering as stringency of the filtering criteria (fold-change and p-value) was systematically varied in order to determine a representative gene list for pathway analysis (Supplementary Table S2 to be found online at http://informahealthcare.com/doi/abs/10.3109/01677063.2013.878339).
To explore the biological significance of our expression data, we performed pathway analysis with two Web-based algorithms, Ingenuity Pathway Analysis (www.ingenuity.com; Ingenuity Systems, xx, CA, USA) and MetaCore (www.genego.com; GeneGo, xx, CA, USA), to map the differentially expressed genes onto biological functions and signaling pathways. With Ingenuity, the significance was determined via a right-tailed Fischer’s exact test, whereas MetaCore makes use of their algorithm for hypergeometric distribution to identify pathways overrepresented with significant genes.
miRNA Data Analysis
Primary analysis of the acquired signal data was performed in SDS and RQ Manager (Applied Biosystems, xx, CA, USA). Quality control analysis and differential expression analysis was performed using the qRT-PCR package for miRNA arrays (Applied) in Bioconductor. Significant enrichment of specific Gene Ontology categories or pathways was estimated by hypergeometric tests or chi-square tests. Enriched KEGG (Kyoto Encyclopedia of Genes) pathways overrepresented by potential miRNA target genes were determined with the p-value obtained by hypergeometric tests and adjusted by multiple test correction (Kanehisa & Goto, 2000; Kanehisa et al., 2006, 2010).
RESULTS
Identity of Laser-Captured Neurons
The presence of the mRNA for PV (mean Ct ± SD= 23.2 ± 1.7) and the absence (Ct > 40) of the mRNAs for CALB1, CAMKII, and GFAP in our laser-captured samples were confirmed by qRT-PCR, supporting the cellular purity of our samples.
Affymetrix Platform–Based Microarray Gene Expression Profiling
Approximately 1–7 ng of total RNA was extracted from captured PV-immunolabeled neurons. For many of the samples, the amount of total RNA obtained was too small for the determination of RNA quality indicator (RQI). Even in cases that furnished sufficient RNA for possible determination for these values, it would have required the consumption of most of the available sample. Therefore, in order to preserve as much RNA as possible for linear amplification and downstream microarray and qRT-PCR applications, RQI determination was circumvented. Nevertheless, total RNA extracted from homogenized cortical tissue from all of the subjects used in this study was available and appears to be of superb quality (Table 2). Importantly, as shown in Figure 1B, the sizes of the linearly amplified products typically extended into the 6 kb range, suggesting that mRNA integrity was well preserved. Finally, the efficiency of microarray hybridization appeared to be adequate in terms of probe intensity and percentage of present calls, and these parameters were highly comparable between the schizophrenia and normal control groups, with average (± SD) probe intensity being 76.3 ± 2.9 and 75.4 ± 3.7, respectively, and average (± SD) percent present calls 27.59 ± 2.18 and 28.67 ± 5.37, respectively (Table 2). Overall, these percentages of present calls, which are virtually identical to those reported in the accompanying paper, are as expected lower than what have been reported in previously published schizophrenia microarray studies performed on RNA extracted from homogenized cortex (typically in the range of 40–45%), which contains a much greater number of RNA species in significantly larger quantities. As discussed in the accompanying article, our data, however, are comparable in magnitude to those reported in a recent microarray study of laser-dissected hippocampal subfields in schizophrenia (mean ± SD = 31.8 ± 4.9) (Benes et al., 2008) and to those described in previous microarray studies based on laser-captured cells from clinical samples or other single cells specimens (Luzzi et al., 2001; Mahadevappa & Warrington, 1999).
Table 2.
Quantity and quality of RNA.
| Group | RIN of total RNA extracted from homogenized cortexa | Amount of RNA before/after amplification (μg) | A260/A280 | Percent present |
|---|---|---|---|---|
| Control | 8.2 | 0.0013/27.94 | 2.85 | 19.9 |
| Control | 7.9 | 0.003/22.90 | 2.80 | 30.7 |
| Control | 8.9 | 0.002/38.75 | 2.75 | 26.6 |
| Control | 8.8 | 0.005/31.92 | 2.83 | 34.7 |
| Control | 9 | 0.003/21.17 | 2.79 | 28.7 |
| Control | 8.4 | 0.004/26.12 | 2.65 | 33.8 |
| Control | 8 | 0.0012/23.14 | 2.87 | 32.5 |
| Control | 8.8 | 0.003/22.59 | 2.45 | 22.4 |
| Mean±SD | 8.50±0.43 | 0.0033±0.0019/29.98±11.00 | 2.79±0.07 | 28.67±5.37 |
| Schizophrenia | 8.9 | 0.0011/25.89 | 2.76 | 26.5 |
| Schizophrenia | 8 | 0.007/29.36 | 2.85 | 30.7 |
| Schizophrenia | 7.9 | 0.003/19.01 | 2.53 | 28.1 |
| Schizophrenia | 9.2 | 0.004/24.78 | 2.62 | 28.2 |
| Schizophrenia | 8.5 | 0.007/28.06 | 2.70 | 31.8 |
| Schizophrenia | 8.2 | 0.002/25.20 | 2.84 | 27.3 |
| Schizophrenia | 8.8 | 0.002/25.42 | 2.75 | 28.0 |
| Schizophrenia | 8.4 | 0.002/36.16 | 2.68 | 25.0 |
| Mean±SD | 8.48±0.46 | 0.0035±0.0038/26.07±4.97 | 2.72±0.03 | 27.59±2.18 |
RNA integrity number (RIN) of total RNA extracted from homogenized cortex determined by an Agilent bioanalyzer.
We systematically varied the stringency criteria of fold-change and false discovery rate (FDR)-adjusted p-value and then used hierarchical clustering to visualize how well the differentially expressed genes segregated according to diagnostic groups (Figure 1C and Table S2). Consequently, based on the criteria of a fold-change of 1.2 and P < 0.05, ANCOVA revealed 739 genes that were differentially expressed in schizophrenia (Supplementary Table S3 to be found online at http://informahealthcare.com/doi/abs/10.3109/01677063.2013.878339). Among these genes, 55% and 45% were down-regulated and up-regulated, respectively, and of the 47 most significantly affected genes, as defined by fold-change >2 and P<0.01, all were down-regulated (Table 3). This finding is in contrast to the observation made in pyramidal neurons, as described in the accompanying article, in which the majority (61%) of genes that were differentially expressed in schizophrenia were found to be up-regulated.
Table 3.
The most significantly affected genes in PV neurons in schizophrenia.
| Gene title | Gene symbol | FDR-adjusted p-value | Fold-change (S vs. C)a |
|---|---|---|---|
| Dual specificity phosphatase 4 | DUSP4 | 0.00738662 | −4.52455 |
| Family with sequence similarity 87, member A/Family with sequence similarity | FAM87A/FAM87B | 0.0399046 | −4.22443 |
| Transmembrane protein 41B | TMEM41B | 0.00773543 | −3.10579 |
| Early growth response 1 | EGR1 | 0.0435856 | −2.90765 |
| Discs, large homolog 5 (Drosophila) | DLG5 | 0.00551372 | −2.56638 |
| SCY1-like 3 (S. cerevisiae) | SCYL3 | 0.0300174 | −2.55763 |
| Spermatid perinuclear RNA binding protein | STRBP | 0.0283199 | −2.52468 |
| LIM homeobox 6 | LHX6 | 0.0210674 | −2.52408 |
| Quinoid dihydropteridine reductase | QDPR | 0.00667599 | −2.47499 |
| Septin 2 | SEPT2 | 0.00153254 | −2.45578 |
| Glycosyltransferase 25 domain containing 2 | GLT25D2 | 0.0240631 | −2.44144 |
| H2A histone family, member Y | H2AFY | 0.0459498 | −2.426 |
| Methionine sulfoxide reductase B2 | MSRB2 | 0.0487961 | −2.42584 |
| Hypothetical protein LOC285771 | LOC285771 | 1.23E-05 | −2.4246 |
| G1 to S phase transition 2 | GSPT2 | 0.0313829 | −2.39153 |
| KIAA0391 | KIAA0391 | 0.0203425 | −2.38239 |
| NMDA receptor regulated 1 | NARG1 | 0.0232586 | −2.34326 |
| Aminopeptidase puromycin sensitive | NPEPPS | 0.0137793 | −2.32083 |
| Chromosome 11 open reading frame 24 | C11orf24 | 0.0240455 | −2.29815 |
| Dual specificity phosphatase 6 | DUSP6 | 0.01487 | −2.28725 |
| Potassium large conductance calcium-activated channel, subfamily M, beta member | KCNMB4 | 0.034706 | −2.28311 |
| ATP-binding cassette, subfamily G (WHITE), member 2 | ABCG2 | 0.0326947 | −2.28077 |
| FERM and PDZ domain containing 2/FERM and PDZ domain containing 2 like 1/FERM and PDZ domain containing 2 like 2 | FRMPD2/FRMPD2L1/FRMPD2L2 | 0.0424632 | −2.27124 |
| tripartite motif-containing 33 | TRIM33 | 0.0052884 | −2.26592 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 8 | ADAMTS8 | 0.00228409 | −2.25771 |
| Zyg-11 homolog B (C. elegans) | ZYG11B | 0.0269688 | −2.2506 |
| Collagen, type XXVII, alpha 1 | COL27A1 | 0.0173052 | −2.24339 |
| Neurexin 3 | NRXN3 | 0.0453457 | −2.22702 |
| Fatty acid desaturase 1 | FADS1 | 0.0197786 | −2.20793 |
| RAD51-like 3 (S. cerevisiae) | RAD51L3 | 0.0461984 | −2.18879 |
| chromosome Y open reading frame 15B | CYorf15B | 0.0319824 | −2.18731 |
| FEZ family zinc finger 2 | FEZF2 | 0.0294616 | −2.18306 |
| ADP-ribosylation factor-like 5A | ARL5A | 0.0204707 | −2.14259 |
| WD repeat domain 8 | WDR8 | 0.0226652 | −2.12477 |
| Chromosome 19 open reading frame 12 | C19orf12 | 0.0343692 | −2.12217 |
| hCG2003663 | hCG_2003663 | 0.0228243 | −2.10932 |
| Tetraspanin 5 | TSPAN5 | 0.00677307 | −2.10383 |
| Testis specific, 14 | TSGA14 | 0.0348805 | −2.0858 |
| Zinc finger protein 281 | ZNF281 | 0.0173262 | −2.07372 |
| WD repeat and SOCS box-containing 1 | WSB1 | 0.0252365 | −2.05327 |
| Ring finger protein 175 | RNF175 | 0.0461219 | −2.05126 |
| PDZ and LIM domain 7 (enigma) | PDLIM7 | 0.0435179 | −2.03901 |
| Kazrin | RP1–21O18.1 | 0.0154391 | −2.03602 |
| CSRP2 binding protein | CSRP2BP | 0.0374447 | −2.03196 |
| Phosphatase and tensin homolog | PTEN | 0.00340582 | −2.01107 |
| N-acylsphingosine amidohydrolase (nonlysosomal ceramidase) 2B | ASAH2B | 0.0259562 | −2.00806 |
| Ectodermal-neural cortex (with BTB-like domain) | ENC1 | 0.045941 | −2.00162 |
S = schizophrenia; C = control.
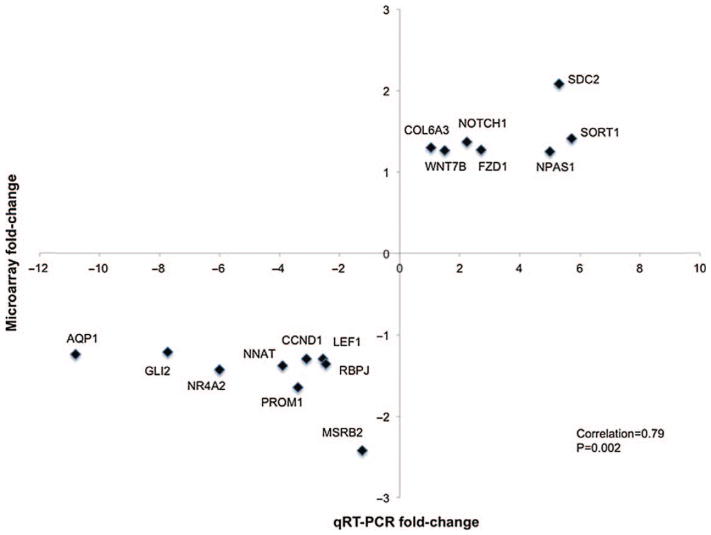
Finally, 16 genes were chosen for global validation of microarray data by qRT-PCR, using the approach laid out by Miron et al. (Miron et al., 2006); we found a statistically highly significant correlation of 0.79 (p = 0.002) between microarray and qRT-PCR data (Figure 2). Correlation analyses between the expression of these genes and PMI, age, and chlorpromazine equivalent dosage (CED) revealed no evidence of association between gene expression changes and these potential confounds.
Figure 2.
Correlation analysis comparing fold-changes of selected genes determined by microarray and qRT-PCR. Comparison of fold-changes of randomly selected genes (N = 10) and genes selected from the most significantly affected pathways (N = 6) determined by microarray and qRT-PCR. AQP1, aquaporin 1; CCND1, cyclin D1; COL6A3, collagen, type VI, alpha 3; FZD1, frizzled family receptor 1; GLI2, glioma-associated oncogene family zinc finger 2; LEF1, lymphoid enhancer-binding factor 1; MSRB2, methionine sulfoxide reductase B2; NNAT, neuronatin; NOTCH1, notch1; NPAS1, neuronal PAS domain protein 1; NR4A2, nuclear receptor subfamily 4, group A, member 2; PROM1, prominin 1; RBPJ, recombinant signal binding protein for immunoglobulin kappa J region; SDC2, syndecan 2; SORT1, sortilin; WNT7B, wingless-type MMTV integration site family, member 7B.
Altered Signaling Pathways Suggest Cell Cycle and Apoptosis Dysregulation in Schizophrenia
The GeneGo networks and pathways that were overrepresented in this data set are summarized in Table 4. For the most part, the functional gene networks and canonical pathways that were altered in schizophrenia differ from those found to be differentially regulated in pyramidal neurons. For instance, the most significantly affected pathways in PV neurons included the WNT (Table 5a), NOTCH (Table 5b), and prostaglandin E2 (PGE2) (Table 5c) signaling cascades. Interestingly, although all three of these pathways are known to be involved in a multitude of cellular regulatory functions, one of their common targets involve cell survival, cell cycle regulation, and apoptosis (Figure 3). Genes that regulate these events were also found to be differentially altered in pyramidal neurons in schizophrenia, suggesting potentially shared pathophysiological features between the two cell types, although the associated signaling pathways appear to be neuronal type specific.
Table 4.
GeneGo pathways and process networks and Go processes significantly overrepresented in schizophrenia and corresponding p-values.
| GeneGo pathways | |
|
| |
| PGE 2 pathways in cancer | 7.90267E-07 |
| Apoptosis and survival FAS signaling cascades | 2.58101E-05 |
| Signal transduction_Erk Interactions: Inhibition of Erk | 9.72639E-05 |
| Development_NOTCH-induced EMT | 0.000118194 |
| Development_Hedgehog and PTH signaling pathways in bone and cartilage development | 0.000147372 |
| Development_Neurotrophin family signaling | 0.000215176 |
| Development_Notch Signaling Pathway | 0.000304476 |
| Reproduction_GnRH signaling | 0.00047633 |
| Apoptosis and survival_p53-dependent apoptosis | 0.000650394 |
| Development_IGF-1 receptor signaling | 0.000680001 |
|
| |
| GeneGo process networks | |
|
| |
| Signal transduction_WNT signaling | 9.75398E-05 |
| DNA damage_Checkpoint | 0.000204291 |
| Cell cycle_Meiosis | 0.00106629 |
| Development_Hemopoiesis, Erythropoietin pathway | 0.001626138 |
| Signal Transduction_TGF-beta, GDF and Activin signaling | 0.003883838 |
| Cell cycle_G1-S Interleukin regulation | 0.003922786 |
| Development_Neurogenesis in general | 0.007096267 |
| Development_Hedgehog signaling | 0.008957277 |
| Reproduction_Spermatogenesis, motility and copulation | 0.009948942 |
| Development_EMT_Regulation of epithelial-to-mesenchymal transition | 0.010964709 |
|
| |
| Go processes | |
|
| |
| Forebrain development | 7.28258E-07 |
| Nervous system development | 9.15548E-07 |
| Ossification | 9.36578E-07 |
| Neurogenesis | 1.14942E-06 |
| Epithelial to mesenchymal transition | 3.41059E-06 |
| Response to organic substance | 4.07396E-06 |
| Cell differentiation | 4.48892E-06 |
| Cellular developmental process | 5.54542E-06 |
| Generation of neurons | 1.02798E-05 |
| Cell morphogenesis involved in differentiation | 1.17357E-05 |
Table 5a.
Significantly affected genes in WNT signaling pathway.
| Gene title | Gene symbol | p-Value | Fold-change (S vs. C)a |
|---|---|---|---|
| Wingless-type MMTV integration site family, member 7B | WNT7B | 0.04 | 1.26 |
| Frizzled family receptor 1 | FZD1 | 0.04 | 1.27 |
| Phosphatase and tensin homolog | PTEN | 0.003 | −2.0 |
| Dishevelled associated activator of morphogenesis 1 | DAAM1 | 0.003 | −1.78 |
| Phosphoinositide-3-kinase, regulatory subunit 2 (beta) | PIK3R2 | 0.02 | 1.26 |
| Paired box 6 | PAX6 | 0.01 | 1.24 |
| Lymphoid enhancer-binding factor 1 | LEF1 | 0.007 | −1.30 |
| Sp1 transcription factor | SP1 | 0.01 | 1.22 |
| Cyclin A2 | CCNA2 | 0.02 | −1.49 |
| Cyclin D1 | CCND1 | 0.03 | −1.30 |
| Plasminogen activator, urokinase receptor | PLAUR | 0.04 | 1.36 |
| Epidermal growth factor receptor | EGFR | 0.02 | 1.36 |
S = schizophrenia; C = control.
Table 5b.
Significantly affected genes in NOTCH signaling pathway.
| Gene title | Gene symbol | p-Value | Fold-change (S vs. C)a |
|---|---|---|---|
| Tumor protein 63 | TP63 | 0.04 | −1.33 |
| Notch homolog 1, translocation-associated (Drosophila) | NOTCH1 | 0.03 | 1.37 |
| Recombination signal binding protein for immunoglobulin kappa J region | RBPJ | 0.003 | −1.36 |
| Retinoblastoma binding protein 8 | RBBP8 | 0.04 | −1.45 |
| Histone cluster 1, H4h | HIST1H4H | ||
| Mitogen-activated protein kinase kinase kinase 1 | MAP3K1 (MEKK1) | 0.03 | −1.26 |
| Cyclin D1 | CCND1 | 0.03 | −1.30 |
| Ring finger protein 1 | RING1 | 0.03 | −1.40 |
S = schizophrenia; C = control.
Table 5c.
Significantly affected genes in PGE2 signaling pathway.
| Gene title | Gene symbol | p-Value | Fold-change (S vs. C)a |
|---|---|---|---|
| Epidermal growth factor receptor | EGFR | 0.02 | 1.36 |
| E twenty-six (ETS)-like transcription factor 1 | ELK1 | 0.04 | 1.63 |
| Cyclin D1 | CCND1 | 0.03 | −1.30 |
| Early growth response 1 | EGR1 | 0.04 | −2.90 |
| Lymphoid enhancer-binding factor 1 | LEF1 | 0.007 | −1.30 |
| Nuclear receptor subfamily 4, group A, member 2 | NR4A2 | 0.02 | −1.43 |
| Adenylate cyclase 1 (brain) | ADCY1 | 0.04 | −1.80 |
S = schizophrenia; C = control.
Figure 3.
Schematic diagram showing the convergence of WNT and NOTCH signaling onto cell cycle regulation. Modified GeneGo pathway diagram showing that aberrant WNT and NOTCH signaling in schizophrenia may contribute to cell cycle dysregulation (see text for details). FZD1, frizzled family receptor 1; EGFR, epidermal growth factor receptor; LEF1, lymphoid enhancer-binding factor 1; TCF, transcription factor 1; RBPJ, recombination signal binding protein for immunoglobulin kappa J region.
Canonical WNT signaling exerts neuroprotective effects via β-catenin- and glycogen synthase kinase (GSK)-dependent activation of transcription factors, such as lymphocyte enhancer factor (LEF1) (Chen et al., 2001; Ishitani et al., 2003). Our findings that Frizzled family receptor 1 (FZD1) and WNT7B were up-regulated by 1.27-fold (p =0.04) and 1.26-fold (p = 0.04), respectively, suggest that in schizophrenia WNT signaling is activated. As a result, one would predict that LEF1 would also be up-regulated, but instead it was down-regulated by 1.3-fold in the schizophrenia subjects (p = 0.007). This may be because in addition to being a target of WNT signaling activation, LEF1 is also regulated by other signaling mechanisms, among them NOTCH and PGE2 signaling pathways (Das et al., 2008; Rodriguez et al., 2009). Hence, the down-regulation of LEF1 may represent the net consequence of the extensive crosstalk between WNT, NOTCH, PGE2, and possible other signaling cascades (Carlson et al., 2008; Hayward et al., 2005; Letamendia et al., 2001).
Among the WNT-responsive target genes are those that regulate cell cycle, such as cyclin D1 (CCND1) (Niehrs & Acebron, 2012; Shtutman et al., 1999). We found that CCND1 were down-regulated by 1.30-fold in PV neurons in schizophrenia (p = 0.03), which may represent a downstream consequence of decreased LEF1 expression. CCND1 is a key regulator of G1/S checkpoint in the cell cycle and breaching of this checkpoint has been associated with apoptosis (Copani et al., 2001; Liu & Greene, 2001; Park et al., 1997; Wang et al., 2010). These findings are consistent with results of a recent microarray study showing that LEF1 and the gene that encodes another cyclin D, cyclin D2 (CCND2), were down-regulated in hippocampal inhibitory neurons in subjects with schizophrenia (Benes, 2011; Benes et al., 2007).
In addition to WNT signaling, we found that NOTCH signaling was also dysregulated in subjects with schizophrenia. Specifically, NOTCH1 was found to be up-regulated by 1.37-fold (p = 0.03), which may represent a compensatory change in response to cellular injury. As a result of NOTCH activation, transcription complexes, of which RBPJ (recombination signal binding protein for immunoglobulin kappa J region) is a major component, are converted from transcription suppressors to activators, a key process that mediates the biological effects of NOTCH1 signaling. In our samples, we found that RBPJ expression was down-regulated by 1.36-fold in PV neurons in schizophrenia (p = 0.003). As a result, downstream gene transcription that normally results from NOTCH activation may be attenuated, which, together with abnormal WNT signaling discussed above, may further contribute to the aberrant expression of CCND1 (Guo et al., 2009; Ling et al., 2010; Miele & Osborne, 1999; Yang et al., 2004).
Finally, dysregulated PGE2 signaling can stimulate epidermal growth factor receptor (EGFR) via transactivation and other signaling cascades, such as insulin-like growth factor (IGF) signaling, thereby modulating the downstream transcription of genes associated with cell cycle and apoptosis (Anderson et al., 2002; Carro et al., 2003; Chun & Langenbach, 2011; Han & Wu, 2005; Kaur & Sanyal, 2010). In fact, our data indicate that EGFR was up-regulated by 1.36-fold (p = 0.02) and IGF signaling was also found to be differentially regulated in schizophrenia (p = 0.0007; Table 4).
miRNA Profiling by Megaplex miRNA TaqMan Arrays
miRNA expression levels were normalized to the endogenous sno-RNA MammU6, whose levels were unchanged between control and schizophrenia samples for both of the Human miRNA A and B Cards (Applied Biosystems, Grand Island, NY, USA). Out of the 754 miRNAs interrogated, 15 were found to be differentially expressed in the schizophrenia subjects (Table 6).
Table 6.
Differentially expressed miRNAs in schizophrenia.
| miRNA assay name | p-Value | Log2 fold-change |
|---|---|---|
| hsa-miR-151-3p-002254 | 0.03 | −2.38 |
| hsa-miR-338-5P-002658 | 0.007 | −3.56 |
| hsa-miR-106a-4395280 | 0.05 | 2.00 |
| hsa-miR-197–4373102 | 0.04 | −3.36 |
| hsa-miR-342-3p-4395371 | 0.008 | −3.93 |
| hsa-miR-518f-4395499 | 0.02 | −1.88 |
| hsa-miR-1274b-002884 | 0.0005 | −7.45 |
| hsa-miR-151-3p-002254 | 0.007 | −2.01 |
| hsa-miR-195–4373105 | 0.04 | −2.35 |
| hsa-miR-197–4373102 | 0.001 | −3.31 |
| hsa-miR-218–4373081 | 0.0002 | 4.22 |
| hsa-miR-342-3p-4395371 | 0.03 | 2.77 |
| hsa-miR-34a-4395168 | 0.05 | −1.21 |
| hsa-miR-361-5p-4373035 | 0.007 | −5.03 |
| hsa-miR-520c-3p-002400 | 0.02 | −2.23 |
DISCUSSION
PV-containing fast-spiking inhibitory neurons time and sync the activity of pyramidal cell networks in gamma frequency. Functional disturbances of these neurons, which have long been implicated as a core pathophysiological event in schizophrenia, can disrupt information processing and integration mediated by gamma oscillation of pyramidal cell networks, contributing to the symptoms and cognitive deficits of schizophrenia (Benes & Berretta, 2001; Gonzalez-Burgos et al., 2010; Lewis et al., 2005, 2012; Spencer, 2009; Uhlhaas & Singer, 2010; Woo et al., 2010). Therefore, understanding the molecular mechanisms that underlie the disturbances of these neurons will have important clinical significance. In this context, we have identified the mRNA and miRNA expression profiles of the population of PV neurons that, collectively, were presynaptic to the pyramidal neurons examined in the accompanying study. Our data suggest that genes associated with WNT, NOTCH, and PGE2 signaling, cell cycle regulation, and apoptosis were among the most differentially expressed in this illness.
Methodological Considerations
To assess the cellular purity of our samples, we confirmed the presence of the mRNA for PV and the absence of the mRNAs for CALB1, CAMKII, and GFAP, which are cell type–specific markers for calbindin-containing inhibitory neurons, pyramidal neurons, and astrocytes, respectively. However, we still cannot exclude the possibility that elements of these cells or that of other cell types that were present but not readily visualized in the immediate vicinity of the PV-immunolabeled neurons, such as the axon terminals of pyramidal neurons, processes of astrocytes or perineuronal oligodendrocytes, etc., and thus any RNA these elements might contain could have been inadvertently included during laser capture. This may explain findings such as the 1.57-fold down-regulation of BDNF (Table S2), as BDNF mRNA is not believed to be present in PV neurons but its existence in our samples was confirmed by qRT-PCR. However, definitively addressing this potential confound is not technically straightforward; it would require the comparison of the “background” gene expression pattern, determined by performing transcriptomic analysis on RNA extracted from neuropil that is immediately adjacent to PV-immunolabeled neurons, with findings of this study.
Because the PV immunolabeling procedures were significantly lengthier than the Nissl staining protocol employed in the accompanying study for pyramidal cell identification, we captured a smaller number of PV neurons (~ 300) in order to minimize RNA degradation by reducing the duration of time at room temperature between laser capture and RNA extraction. Accordingly, the amount of RNA extracted per sample was relatively small; this limited the number of genes we were able to assay for qRT-PCR validation, including the number of housekeeping genes to which samples were normalized. Aside from these considerations, additional limitations that are inherent in this combined LCM and gene expression profiling approach are discussed in the accompanying article. Because of these constraints, potential confounds and caveats, we consider this study proof-of-principle and emphasize the fact that our findings need to be confirmed in the future in a different and ideally larger cohort of subjects.
Cell Cycle and Apoptosis Dysregulation in PV Neurons in Schizophrenia
Converging lines of evidence from postmortem studies in the last two decades strongly suggest that functional disturbances of PV neurons play a key role in the pathogenesis of schizophrenia (Benes & Berretta, 2001; Gonzalez-Burgos et al., 2010; Lewis et al., 2005, 2012; Spencer, 2009; Uhlhaas & Singer, 2010; Woo et al., 2010). Specifically, inhibitory inputs of PV neurons to pyramidal cells may be decreased (Hashimoto et al., 2003), which may in turn contribute to gamma oscillation impairment. Although the precise pathophysiological events that lead to the dysfunction of PV neurons remain unknown, there has been increasing evidence suggesting that oxidative injury may be a major culprit (Behrens & Sejnowski, 2009; Bitanihirwe & Woo, 2011; Do et al., 2009; Kulak et al., 2013; Nakazawa et al., 2011; Powell et al., 2011).
Injury of PV neurons does not necessarily lead to cell death, at least not in large scale, as the number of neurons in the neocortex appears to be largely unaltered in subjects with schizophrenia (Hashimoto et al., 2003; Selemon & Goldman-Rakic, 1999; Woo et al., 1997), although cell loss may still occur in other brain regions, e.g., limbic cortices such as the anterior cingulate cortex or the hippocampus (Benes et al., 1991, 1998). In this context, our findings may reflect a rather complex molecular snapshot of signaling changes orchestrated to counter the deleterious effects of cellular injury and thereby attenuate apoptosis. In fact, many of the most significantly affected signaling pathways, such as the WNT, NOTCH, and PGE2 signaling cascades, are known to regulate cell cycle and apoptosis. Consistent with this, genes that regulate cell cycle have previously been shown to be dysregulated in schizophrenia in other microarray studies, although the directions of changes in the expression of specific genes are not always consistent between our findings (derived from RNA extracted from homogeneous PV neurons) and the findings of these previous studies (derived from RNA extracted from homogenized postmortem cortical tissue, biopsied olfactory neuroepithelial samples, or patient-derived fibroblasts) (Fan et al., 2012; Katsel et al., 2008).
The roles of WNT and NOTCH signaling in early brain development by promoting neuronal differentiation and patterning and attenuating apoptosis are well established (Mason et al., 2005; Oishi et al., 2004). Increasing evidence, however, suggests that these pathways also regulate various cellular processes in postmitotic neurons in the adult brain and that dysregulation of these pathways contributes to the pathophysiology of neurological disorders (Ables et al., 2011; Ahmad-Annuar et al., 2006; Budnik & Salinas, 2011; Chen et al., 2006; Freese et al., 2010; Gogolla et al., 2009; Jensen et al., 2012a; Malaterre et al., 2007; Miele & Osborne, 1999). For instance, down-regulation of NOTCH signaling has been associated with neurodegenerative disorders (e.g., Alzheimer’s disease), in part by promoting apoptosis (Ables et al., 2011). Likewise, dysregulated WNT signaling can lead to neuronal apoptosis by disturbing cell cycle homeostasis (Caricasole et al., 2003). Our data suggest that in schizophrenia disturbances of WNT and NOTCH signaling may contribute to the injury of PV neurons by compromising the intricate balance of cell cycle regulation. Finally, consistent with our observation of dysregulated PGE2 signaling, there has been evidence in the literature suggesting that the level of PGE2, an inflammatory marker and a product of COX2 (cyclooxygenase-2), is increased in patients with schizophrenia (Kaiya et al., 1989; Martinez-Gras et al., 2011; Muller, 2010). In the context of all of these findings, it would be of interest to investigate if oxidative stress may in fact lead to aberrant WNT, NOTCH, and PGE2 signaling in PV neurons. This can be done in vivo, using established animal models (Cabungcal et al., 2013a, 2013b), or in vitro, such as in neurons differentiated from induced pluripotent stem cells derived from either sporadic schizophrenia patients or those with a specific genetic background (e.g., 22q11.2 deletion).
Connectional Plasticity of PV Neurons in Schizophrenia
It has long been known that WNT and NOTCH signaling orchestrates nervous system development and patterning by regulating neurogenesis, axonal growth, synaptogenesis, and apoptosis (Danesin & Houart, 2012; Tiberi et al., 2012). Increasing evidence suggests that both of these signaling cascades may also play an important role in activity-dependent synaptic plasticity in the adult brain (Ables et al., 2011; Inestrosa & Arenas, 2010; Park & Shen, 2012; Sahores & Salinas, 2011). For instance, it has been shown that WNT signaling may regulate synaptic strength in the neuromuscular junction in Caenorhabditis elegans by posttranslationally regulating the accumulation of acetylcholinergic receptors in an experience-dependent manner (Jensen et al., 2012a, 2012b). In mammals, WNT7A signaling has been shown to regulate presynaptic glutamate release, the size of postsynaptic dendritic spines, and long-term potentiation (Chen et al., 2006; Ciani et al., 2011), and the effects of WNT7A on synaptic plasticity appear to be experience dependent (Gogolla et al., 2009; Hall et al., 2000). Of further interest, specific WNT proteins may differentially regulate the formation and stability of excitatory versus inhibitory synapses and hence the balance of excitation and inhibition within neural circuits (Ciani et al., 2011; Cuitino et al., 2010). Similarly, NOTCH signaling has also been shown to regulate long-term potentiation and depression in the hippocampus (Conboy et al., 2007; Wang et al., 2004). Finally, the functional activity of cortical circuitry in terms of excitatory and inhibitory balance is also regulated by additional factors, such as LHX6 (LIM homeobox 6). LHX 6 is a homeoprotein that selectively regulates the specification and differentiation of somatostatin- and PV-containing neurons in development (Liodis et al., 2007), both of which are believed to play a role in the pathophysiology of schizophrenia (Lewis et al., 2008, 2011). Furthermore, reduced LHX6 expression is known to disturb cortical network activity balance and can lead to seizures (Neves et al., 2012). In this study, we found that LHX6 was down-regulated by 2.45-fold (p = 0.02) in PV neurons in schizophrenia, raising the possibility that decreased expression of this gene may further contribute to dysregulated pyramidal cell network firing in schizophrenia.
miRNA Expression Dysregulation in PV Neurons in Schizophrenia
The number of miRNAs identified in this study is relatively small, but in the same order of magnitude as found in the accompanying study and in previous studies investigating miRNA expression in the cerebral cortex in schizophrenia (Beveridge et al., 2008; Miller et al., 2012; Moreau et al., 2011; Santarelli et al., 2011). Pathway analysis of the predicted target genes of the differentially expressed miRNAs revealed signaling pathways that overlap with those identified by microarray mRNA expression profiling (Supplementary Table 4). These data raise an interesting possibility that the pathophysiology of PV cell dysfunction may in part be mediated by the concerted dysregulation of gene network functions as a result of the altered expression of a relatively small number of miRNAs.
CONCLUSION
Findings of this study point to the involvement of WNT and NOTCH signaling in the pathophysiology of PV neurons in schizophrenia. Of interest, in a recent study, these signaling pathways were also found to be among the most significantly affected in neurons differentiated from induced pluripotent stem cells from patients with schizophrenia (Brennand et al., 2011). The extensive crosstalk between these pathways and a large number of other signaling cascades means that aberrant activity of these pathways can have a wide array of biological consequences, among them dysregulation of cell cycle and apoptosis. Our findings, however, do not tell us what specific upstream events may be responsible for the altered functioning of these signaling pathways; toward this end, available evidence in the literature suggests that insult to PV neurons in pathological conditions, such as oxidative stress (Behrens & Sejnowski, 2009; Bitanihirwe & Woo, 2011; Do et al., 2009; Kulak et al., 2013; Nakazawa et al., 2011; Powell et al., 2011), may play a role. This hypothesis can be tested in the laboratory, for example, by studying the expression profiles of PV neurons in either in vitro (Kinney et al., 2006; Steullet et al., 2006) or in vivo (Cabungcal et al., 2006; Powell et al., 2011) systems under the condition of oxidative stress to see if signaling pathways identified in the present study may be affected. This “reverse translational approach” of generating experimentally testable neurobiological hypotheses from postmortem gene expression profiling findings will lead to increased understanding of neuronal type–specific pathophysiology of schizophrenia and may ultimately inform the conceptualization of targeted molecular intervention that can fundamentally restore the functional integrity of the underlying cortical circuits.
Supplementary Material
Table 5d.
Significantly affected genes associated with apoptosis and survival.
| Gene title | Gene symbol | p-Value | Fold-change (S vs. C)a |
|---|---|---|---|
| Apoptosis and survival_BAD phosphorylation | |||
| Epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | EGFR | 0.02 | 1.59 |
| Phosphoinositide-3-kinase, regulatory subunit 2 (beta) | PI3KR2 | 0.04 | 1.23 |
| Protein kinase, cAMP-dependent, catalytic, gamma | PRKACG | 0.02 | 1.25 |
| Protein tyrosine phosphatase, non-receptor type 11 | PTPN11 | 0.03 | −1.11 |
| SHC (Src homology 2 domain containing) transforming protein 1 | SHC1 | 0.03 | 1.16 |
| DNA damage_Role of SUMO in p53 regulation | |||
| CREB binding protein | CBP | 0.01 | 1.31 |
| SMT3 suppressor of mif two 3 homolog 1 (S. cervisiae) | SUMO-1 | 0.03 | 1.17 |
| Ubiquitin-like modifier activating enzyme 1 | UBA1 | 0.04 | 1.14 |
| Other genes involved in DNA damage and oxidative stress | |||
| HLA-B (major histocompatibility complex, class I, B) associated transcript 3 | BAT3/BAG6 | 0.01 | −1.21 |
| Calpain 9 | CAPN9 | 0.05 | 1.15 |
| Calpain 10 | CAPN10 | 0.02 | 1.14 |
| Clusterin | CLU | 0.02 | 2.02 |
S = schizophrenia; C = control.
Acknowledgments
This study was supported by grants P50MH080272 (Boston CIDAR: Vulnerability to Progression in Schizophrenia) and R01MH076060 from the National Institutes of Health.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ables JL, Breunig JJ, Eisch AJ, Rakic P. Not(ch) just development: Notch signalling in the adult brain. Nat Rev Neurosci. 2011;12:269–283. doi: 10.1038/nrn3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad-Annuar A, Ciani L, Simeonidis I, Herreros J, Fredj NB, Rosso SB, et al. Signaling across the synapse: A role for Wnt and Dishevelled in presynaptic assembly and neurotransmitter release. J Cell Biol. 2006;174:127–139. doi: 10.1083/jcb.200511054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MF, Aberg MA, Nilsson M, Eriksson PS. Insulin-like growth factor-I and neurogenesis in the adult mammalian brain. Brain Res Dev Brain Res. 2002;134:115–122. doi: 10.1016/s0165-3806(02)00277-8. [DOI] [PubMed] [Google Scholar]
- Behrens MM, Sejnowski TJ. Does schizophrenia arise from oxidative dysregulation of parvalbumin-interneurons in the developing cortex? Neuropharmacology. 2009;57:193–200. doi: 10.1016/j.neuropharm.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM. Regulation of cell cycle and DNA repair in post-mitotic GABA neurons in psychotic disorders. Neuropharmacology. 2011;60:1232–1242. doi: 10.1016/j.neuropharm.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Benes FM, Berretta S. GABAergic interneurons: Implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology. 2001;25:1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- Benes FM, Kwok EW, Vincent SL, Todtenkopf MS. A reduction of nonpyramidal cells in sector CA2 of schizophrenics and manic depressives. Biol Psychiatry. 1998;44:88–97. doi: 10.1016/s0006-3223(98)00138-3. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Subburaju S, Walsh JP. Circuitry-based gene expression profiles in GABA cells of the trisynaptic pathway in schizophrenics versus bipolars. Proc Natl Acad Sci USA. 2008;105:20935–20940. doi: 10.1073/pnas.0810153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci USA. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, McSparren J, Bird ED, SanGiovanni JP, Vincent SL. Deficits in small interneurons in prefrontal and cingulate cortices of schizophrenic and schizoaffective patients. Arch Gen Psychiatry. 1991;48:996–1001. doi: 10.1001/archpsyc.1991.01810350036005. [DOI] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Bitanihirwe BK, Woo TUW. Oxidative stress in schizophrenia: An integrated approach. Neurosci Biobehav Rev. 2011;35:878–893. doi: 10.1016/j.neubiorev.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik V, Salinas PC. Wnt signaling during synaptic development and plasticity. Curr Opin Neurobiol. 2011;21:151–159. doi: 10.1016/j.conb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabungcal JH, Nicolas D, Kraftsik R, Cuenod M, Do KQ, Hornung JP. Glutathione deficit during development induces anomalies in the rat anterior cingulate GABAergic neurons: Relevance to schizophrenia. Neurobiol Dis. 2006;22:624–637. doi: 10.1016/j.nbd.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Cabungcal JH, Steullet P, Kraftsik R, Cuenod M, Do KQ. Early-life insults impair parvalbumin interneurons via oxidative stress: Reversal by N-acetylcysteine. Biol Psychiatry. 2013a;73:574–582. doi: 10.1016/j.biopsych.2012.09.020. [DOI] [PubMed] [Google Scholar]
- Cabungcal JH, Steullet P, Morishita H, Kraftsik R, Cuenod M, Hensch TK, et al. Perineuronal nets protect fast-spiking interneurons against oxidative stress. Proc Natl Acad Sci USA. 2013b;110:9130–9135. doi: 10.1073/pnas.1300454110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caricasole A, Copani A, Caruso A, Caraci F, Iacovelli L, Sortino MA, et al. The Wnt pathway, cell-cycle activation and beta-amyloid: Novel therapeutic strategies in Alzheimer’s disease? Trends Pharmacol Sci. 2003;24:233–238. doi: 10.1016/s0165-6147(03)00100-7. [DOI] [PubMed] [Google Scholar]
- Carlson ME, Silva HS, Conboy IM. Aging of signal transduction pathways, and pathology. Exp Cell Res. 2008;314:1951–1961. doi: 10.1016/j.yexcr.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro E, Trejo JL, Nunez A, Torres-Aleman I. Brain repair and neuroprotection by serum insulin-”like growth factor I. Mol Neurobiol. 2003;27:153–162. doi: 10.1385/MN:27:2:153. [DOI] [PubMed] [Google Scholar]
- Chen J, Park CS, Tang SJ. Activity-dependent synaptic Wnt release regulates hippocampal long term potentiation. J Biol Chem. 2006;281:11910–11916. doi: 10.1074/jbc.M511920200. [DOI] [PubMed] [Google Scholar]
- Chen S, Guttridge DC, You Z, Zhang Z, Fribley A, Mayo MW, et al. Wnt-1 signaling inhibits apoptosis by activating beta-catenin/T cell factor-mediated transcription. J Cell Biol. 2001;152:87–96. doi: 10.1083/jcb.152.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun KS, Langenbach R. The prostaglandin E2 receptor, EP2, regulates survivin expression via an EGFR/STAT3 pathway in UVB-exposed mouse skin. Mol Carcinog. 2011;50:439–448. doi: 10.1002/mc.20728. [DOI] [PubMed] [Google Scholar]
- Ciani L, Boyle KA, Dickins E, Sahores M, Anane D, Lopes DM, et al. Wnt7a signaling promotes dendritic spine growth and synaptic strength through Ca(2) (+)/Calmodulin-dependent protein kinase II. Proc Natl Acad Sci U S A. 2011;108:10732–10737. doi: 10.1073/pnas.1018132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy L, Seymour CM, Monopoli MP, O’Sullivan NC, Murphy KJ, Regan CM. Notch signalling becomes transiently attenuated during long-term memory consolidation in adult Wistar rats. Neurobiol Learn Mem. 2007;88:342–351. doi: 10.1016/j.nlm.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Copani A, Uberti D, Sortino MA, Bruno V, Nicoletti F, Memo M. Activation of cell-cycle-associated proteins in neuronal death: A mandatory or dispensable path? Trends Neurosci. 2001;24:25–31. doi: 10.1016/s0166-2236(00)01663-5. [DOI] [PubMed] [Google Scholar]
- Cuitino L, Godoy JA, Farias GG, Couve A, Bonansco C, Fuenzalida M, et al. Wnt-5a modulates recycling of functional GABAA receptors on hippocampal neurons. J Neurosci. 2010;30:8411–8420. doi: 10.1523/JNEUROSCI.5736-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curley AA, Arion D, Volk DW, Asafu-Adjei JK, Sampson AR, Fish KN, et al. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: Clinical, protein, and cell type-specific features. Am J Psychiatry. 2011;168:921–929. doi: 10.1176/appi.ajp.2011.11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curley AA, Lewis DA. Cortical basket cell dysfunction in schizophrenia. J Physiol. 2012;590(Pt 4):715–724. doi: 10.1113/jphysiol.2011.224659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesin C, Houart C. A Fox stops the Wnt: Implications for forebrain development and diseases. Curr Opin Genet Dev. 2012;22:323–330. doi: 10.1016/j.gde.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Das AV, Bhattacharya S, Zhao X, Hegde G, Mallya K, Eudy JD, et al. The canonical Wnt pathway regulates retinal stem cells/progenitors in concert with Notch signaling. Dev Neurosci. 2008;30:389–409. doi: 10.1159/000178017. [DOI] [PubMed] [Google Scholar]
- Do KQ, Cabungcal JH, Frank A, Steullet P, Cuenod M. Redox dysregulation, neurodevelopment, and schizophrenia. Curr Opin Neurobiol. 2009;19:220–230. doi: 10.1016/j.conb.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Fan Y, Abrahamsen G, McGrath JJ, Mackay-Sim A. Altered cell cycle dynamics in schizophrenia. Biol Psychiatry. 2012;71:129–135. doi: 10.1016/j.biopsych.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Freese JL, Pino D, Pleasure SJ. Wnt signaling in development and disease. Neurobiol Dis. 2010;38:148–153. doi: 10.1016/j.nbd.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glausier JR, Lewis DA. Selective pyramidal cell reduction of GABA(A) receptor alpha1 subunit messenger RNA expression in schizophrenia. Neuropsychopharmacology. 2011;36:2103–2110. doi: 10.1038/npp.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogolla N, Galimberti I, Deguchi Y, Caroni P. Wnt signaling mediates experience-related regulation of synapse numbers and mossy fiber connectivities in the adult hippocampus. Neuron. 2009;62:510–525. doi: 10.1016/j.neuron.2009.04.022. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Burgos G, Hashimoto T, Lewis DA. Alterations of cortical GABA neurons and network oscillations in schizophrenia. Curr Psychiatry Rep. 2010;12:335–344. doi: 10.1007/s11920-010-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Ye J, Dai J, Li L, Chen F, Ma D, et al. Notch-1 regulates Akt signaling pathway and the expression of cell cycle regulatory proteins cyclin D1, CDK2 and p21 in T-ALL cell lines. Leuk Res. 2009;33:678–685. doi: 10.1016/j.leukres.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Hall AC, Lucas FR, Salinas PC. Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT-7a signaling. Cell. 2000;100:525–535. doi: 10.1016/s0092-8674(00)80689-3. [DOI] [PubMed] [Google Scholar]
- Han C, Wu T. Cyclooxygenase-2-derived prostaglandin E2 promotes human cholangiocarcinoma cell growth and invasion through EP1 receptor-mediated activation of the epidermal growth factor receptor and Akt. J Biol Chem. 2005;280:24053–24063. doi: 10.1074/jbc.M500562200. [DOI] [PubMed] [Google Scholar]
- Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward P, Brennan K, Sanders P, Balayo T, DasGupta R, Perrimon N, et al. Notch modulates Wnt signalling by associating with Armadillo/beta-catenin and regulating its transcriptional activity. Development. 2005;132:1819–1830. doi: 10.1242/dev.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homayoun H, Moghaddam B. NMDA receptor hypofunction produces opposite effects on prefrontal cortex interneurons and pyramidal neurons. J Neurosci. 2007;27:11496–11500. doi: 10.1523/JNEUROSCI.2213-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inestrosa NC, Arenas E. Emerging roles of Wnts in the adult nervous system. Nat Rev Neurosci. 2010;11:77–86. doi: 10.1038/nrn2755. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Ninomiya-Tsuji J, Matsumoto K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:1379–1389. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M, Brockie PJ, Maricq AV. Wnt signaling regulates experience-dependent synaptic plasticity in the adult nervous system. Cell Cycle. 2012a;11:2585–2586. doi: 10.4161/cc.21138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M, Hoerndli FJ, Brockie PJ, Wang R, Johnson E, Maxfield D, et al. Wnt signaling regulates acetylcholine receptor translocation and synaptic plasticity in the adult nervous system. Cell. 2012b;149:173–187. doi: 10.1016/j.cell.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiya H, Uematsu M, Ofuji M, Nishida A, Takeuchi K, Nozaki M, et al. Elevated plasma prostaglandin E2 levels in schizophrenia. J Neural Transm. 1989;77:39–46. doi: 10.1007/BF01255817. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(Database issue):D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, et al. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34(Database issue):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsel P, Davis KL, Li C, Tan W, Greenstein E, Kleiner Hoffman LB, et al. Abnormal indices of cell cycle activity in schizophrenia and their potential association with oligodendrocytes. Neuropsychopharmacology. 2008;33:2993–3009. doi: 10.1038/npp.2008.19. [DOI] [PubMed] [Google Scholar]
- Kaur J, Sanyal SN. PI3-kinase/Wnt association mediates COX-2/PGE(2) pathway to inhibit apoptosis in early stages of colon carcinogenesis: Chemoprevention by diclofenac. Tumour Biol. 2010;31:623–631. doi: 10.1007/s13277-010-0078-9. [DOI] [PubMed] [Google Scholar]
- Kinney JW, Davis CN, Tabarean I, Conti B, Bartfai T, Behrens MM. A specific role for NR2A-containing NMDA receptors in the maintenance of parvalbumin and GAD67 immunoreactivity in cultured interneurons. J Neurosci. 2006;26:1604–1615. doi: 10.1523/JNEUROSCI.4722-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak A, Steullet P, Cabungcal JH, Werge T, Ingason A, Cuenod M, et al. Redox dysregulation in the pathophysiology of schizophrenia and bipolar disorder: Insights from animal models. Antioxid Redox Signal. 2013;18:1428–1443. doi: 10.1089/ars.2012.4858. [DOI] [PubMed] [Google Scholar]
- Letamendia A, Labbe E, Attisano L. Transcriptional regulation by Smads: Crosstalk between the TGF-beta and Wnt pathways. J Bone Joint Surg Am. 2001;83-A(Suppl 1)(Pt 1):S31–S39. [PubMed] [Google Scholar]
- Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2011;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Morris HM. Cell and receptor type-specific alterations in markers of GABA neurotransmission in the prefrontal cortex of subjects with schizophrenia. Neurotox Res. 2008;14:237–248. doi: 10.1007/BF03033813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- Ling H, Sylvestre JR, Jolicoeur P. Notch1-induced mammary tumor development is cyclin D1-dependent and correlates with expansion of pre-malignant multipotent duct-limited progenitors. Oncogene. 2010;29:4543–4554. doi: 10.1038/onc.2010.186. [DOI] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman JE, Coyle JT, Green RW, Javitt DC, Benes FM, Heckers S, et al. Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci. 2008;31:234–242. doi: 10.1016/j.tins.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DX, Greene LA. Neuronal apoptosis at the G1/S cell cycle checkpoint. Cell Tissue Res. 2001;305:217–228. doi: 10.1007/s004410100396. [DOI] [PubMed] [Google Scholar]
- Luzzi V, Holtschlag V, Watson MA. Expression profiling of ductal carcinoma in situ by laser capture microdissection and high-density oligonucleotide arrays. Am J Pathol. 2001;158:2005–2010. doi: 10.1016/S0002-9440(10)64672-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevappa M, Warrington JA. A high-density probe array sample preparation method using 10- to 100-fold fewer cells. Nat Biotechnol. 1999;17:1134–1136. doi: 10.1038/15124. [DOI] [PubMed] [Google Scholar]
- Malaterre J, Ramsay RG, Mantamadiotis T. Wnt-Frizzled signalling and the many paths to neural development and adult brain homeostasis. Front Biosci. 2007;12:492–506. doi: 10.2741/2077. [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Martinez-Gras I, Perez-Nievas BG, Garcia-Bueno B, Madrigal JL, Andres-Esteban E, Rodriguez-Jimenez R, et al. The anti-inflammatory prostaglandin 15d-PGJ2 and its nuclear receptor PPARgamma are decreased in schizophrenia. Schizophr Res. 2011;128:15–22. doi: 10.1016/j.schres.2011.01.018. [DOI] [PubMed] [Google Scholar]
- Mason HA, Rakowiecki SM, Raftopoulou M, Nery S, Huang Y, Gridley T, et al. Notch signaling coordinates the patterning of striatal compartments. Development. 2005;132:4247–4258. doi: 10.1242/dev.02008. [DOI] [PubMed] [Google Scholar]
- Miele L, Osborne B. Arbiter of differentiation and death: Notch signaling meets apoptosis. J Cell Physiol. 1999;181:393–409. doi: 10.1002/(SICI)1097-4652(199912)181:3<393::AID-JCP3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron M, Woody OZ, Marcil A, Murie C, Sladek R, Nadon R. A methodology for global validation of microarray experiments. BMC Bioinformatics. 2006;7:333. doi: 10.1186/1471-2105-7-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MP, Bruse SE, David-Rus R, Buyske S, Brzustowicz LM. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol Psychiatry. 2011;69:188–193. doi: 10.1016/j.biopsych.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller N. COX-2 inhibitors as antidepressants and antipsychotics: Clinical evidence. Curr Opin Investig Drugs. 2010;11:31–42. [PubMed] [Google Scholar]
- Nakazawa K, Zsiros V, Jiang Z, Nakao K, Kolata S, Zhang S, et al. GABAergic interneuron origin of schizophrenia pathophysiology. Neuropharmacology. 2011;62:1574–1583. doi: 10.1016/j.neuropharm.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Shah MM, Liodis P, Achimastou A, Denaxa M, Roalfe G, et al. The LIM homeodomain protein Lhx6 regulates maturation of interneurons and network excitability in the mammalian cortex. Cereb Cortex. 2012;23:1811–1823. doi: 10.1093/cercor/bhs159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C, Acebron SP. Mitotic and mitogenic Wnt signalling. EMBO J. 2012;31:2705–2713. doi: 10.1038/emboj.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Kamakura S, Isazawa Y, Yoshimatsu T, Kuida K, Nakafuku M, et al. Notch promotes survival of neural precursor cells via mechanisms distinct from those regulating neurogenesis. Dev Biol. 2004;276:172–184. doi: 10.1016/j.ydbio.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Park DS, Morris EJ, Greene LA, Geller HM. G1/S cell cycle blockers and inhibitors of cyclin-dependent kinases suppress camptothecin-induced neuronal apoptosis. J Neurosci. 1997;17:1256–1270. doi: 10.1523/JNEUROSCI.17-04-01256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Shen K. WNTs in synapse formation and neuronal circuitry. EMBO J. 2012;31:2697–2704. doi: 10.1038/emboj.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierri JN, Chaudry AS, Woo TU, Lewis DA. Alterations in chandelier neuron axon terminals in the prefrontal cortex of schizophrenic subjects. Am J Psychiatry. 1999;156:1709–1719. doi: 10.1176/ajp.156.11.1709. [DOI] [PubMed] [Google Scholar]
- Pietersen CY, Lim MP, Macey L, Woo TUW, Sonntag KC. Neuronal type-specific gene expression profiling and laser-capture microdissection. Methods Mol Biol. 2011;755:327–343. doi: 10.1007/978-1-61779-163-5_28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen CY, Lim MP, Woo T-UW. Obtaining high quality RNA from single cell populations in human postmortem brain tissue. J Vis Exp. 2009:30. doi: 10.3791/1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SB, Sejnowski TJ, Behrens MM. Behavioral and neurochemical consequences of cortical oxidative stress on parvalbumin-interneuron maturation in rodent models of schizophrenia. Neuropharmacology. 2011;62:1322–1331. doi: 10.1016/j.neuropharm.2011.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez DA, Tapia JC, Fernandez JG, Torres VA, Munoz N, Galleguillos D, et al. Caveolin-1-mediated suppression of cyclooxygenase-2 via a beta-catenin-Tcf/Lef-dependent transcriptional mechanism reduced prostaglandin E2 production and survivin expression. Mol Biol Cell. 2009;20:2297–2310. doi: 10.1091/mbc.E08-09-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahores M, Salinas PC. Activity-mediated synapse formation a role for Wnt-Fz signaling. Curr Top Dev Biol. 2011;97:119–136. doi: 10.1016/B978-0-12-385975-4.00011-5. [DOI] [PubMed] [Google Scholar]
- Santarelli DM, Beveridge NJ, Tooney PA, Cairns MJ. Upregulation of dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol Psychiatry. 2011;69:180–187. doi: 10.1016/j.biopsych.2010.09.030. [DOI] [PubMed] [Google Scholar]
- Selemon LD, Goldman-Rakic PS. The reduced neuropil hypothesis: A circuit based model of schizophrenia. Biol Psychiatry. 1999;45:17–25. doi: 10.1016/s0006-3223(98)00281-9. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D’Amico M, Pestell R, et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic F, Yi M, Wang Y, Macey L, Brown LT, Krichevsky AM, et al. Gene expression profiling of substantia nigra dopamine neurons: Further insights into Parkinson’s disease pathology. Brain. 2009;132(Pt 7):1795–1809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer W, Gray C, Engel A, Konig P, Artola A, Brocher S. Formation of cortical cell assemblies. Cold Spring Harb Symp Quant Biol. 1990;55:939–952. doi: 10.1101/sqb.1990.055.01.088. [DOI] [PubMed] [Google Scholar]
- Soltesz I. Diversity in the neuronal machine. New York: Oxford University Press; 2005. [Google Scholar]
- Spencer KM. The functional consequences of cortical circuit abnormalities on gamma oscillations in schizophrenia: Insights from computational modeling. Front Hum Neurosci. 2009;3:33. doi: 10.3389/neuro.09.033.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer KM, Nestor PG, Perlmutter R, Niznikiewicz MA, Klump MC, Frumin M, et al. Neural synchrony indexes disordered perception and cognition in schizophrenia. Proc Natl Acad Sci U S A. 2004;101:17288–17293. doi: 10.1073/pnas.0406074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steullet P, Neijt HC, Cuenod M, Do KQ. Synaptic plasticity impairment and hypofunction of NMDA receptors induced by glutathione deficit: Relevance to schizophrenia. Neuroscience. 2006;137:807–819. doi: 10.1016/j.neuroscience.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Tiberi L, Vanderhaeghen P, van den Ameele J. Cortical neurogenesis and morphogens: Diversity of cues, sources and functions. Curr Opin Cell Biol. 2012;24:269–276. doi: 10.1016/j.ceb.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Uhlhaas PJ, Pipa G, Lima B, Melloni L, Neuenschwander S, Nikolic D, et al. Neural synchrony in cortical networks: History, concept and current status. Front Integr Neurosci. 2009;3:17. doi: 10.3389/neuro.07.017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas PJ, Singer W. Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci. 2010;11:100–113. doi: 10.1038/nrn2774. [DOI] [PubMed] [Google Scholar]
- Wang L, Lockstone HE, Guest PC, Levin Y, Palotas A, Pietsch S, et al. Expression profiling of fibroblasts identifies cell cycle abnormalities in schizophrenia. J Proteome Res. 2010;9:521–527. doi: 10.1021/pr900867x. [DOI] [PubMed] [Google Scholar]
- Wang XJ. Neurophysiological and computational principles of cortical rhythms in cognition. Physiol Rev. 2010;90:1195–1268. doi: 10.1152/physrev.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chan SL, Miele L, Yao PJ, Mackes J, Ingram DK, et al. Involvement of Notch signaling in hippocampal synaptic plasticity. Proc Natl Acad Sci U S A. 2004;101:9458–9462. doi: 10.1073/pnas.0308126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo TUW, Miller JL, Lewis DA. Schizophrenia and the parvalbumin-containing class of cortical local circuit neurons. Am J Psychiatry. 1997;154:1013–1015. doi: 10.1176/ajp.154.7.1013. [DOI] [PubMed] [Google Scholar]
- Woo TUW, Spencer K, McCarley RW. Gamma oscillation deficits and the onset and early progression of schizophrenia. Harv Rev Psychiatry. 2010;18:173–189. doi: 10.3109/10673221003747609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo TUW, Whitehead RE, Melchitzky DS, Lewis DA. A subclass of prefrontal gamma-aminobutyric acid axon terminals are selectively altered in schizophrenia. Proc Natl Acad Sci U S A. 1998;95:5341–5346. doi: 10.1073/pnas.95.9.5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Klein R, Tian X, Cheng HT, Kopan R, Shen J. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev Biol. 2004;269:81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.