Abstract
The extent to which synthetic biology can be used to expand genetic information systems compatible with natural enzymes and cells will depend on the extent to which multiple and contiguous non-natural nucleobase pairs fit within the standard double helical conformations of DNA. Toward this goal, two non-standard nucleobases (Z, 6-amino-5-nitro-2(1H)-pyridone and P, 2-amino-imidazo[1,2-a]-1,3,5-triazin-4(8H)one) were designed to form a Z:P pair with a standard “edge on” Watson-Crick geometry, but with rearranged hydrogen bond donor and acceptor groups. Here, we present the crystal structures of two self-complementary 16-mer oligonucleotides containing Z:P pairs. The first contained two consecutive Z:P nucleobase pairs and was found to crystallize within a host-guest complex in B-form. The second contained six consecutive Z:P pairs; it was found to crystallize as an A-form DNA duplex, although it can adopt B-form in solution as inferred from circular dichroism spectra. Although Z:P pairs have some structural properties that are similar to those of G:C pairs, unique features include stacking of the nitro group on Z with the adjacent heterocyclic nucleobase ring in A-DNA. In both B-and A-DNA, major groove widths associated with the Z:P pairs are approximately 1 Å wider than those of comparable G:C pairs potentially due to the presence of the nitro group in Z. Thus, our structural studies suggest that multiple and consecutive Z:P pairs are readily accommodated in DNA duplex structures recognized by natural polymerases, and therefore the GACTZP synthetic genetic system has the requisite properties to expand sequence space.
Keywords: Non-natural base pairs, DNA structure, host-guest complex, synthetic biology, A-DNA, B-DNA
INTRODUCTION
Originally defined as simply the field seeking to create artificial life,1 “synthetic biology” became more narrowly construed in the 1970s to mean the use of emerging recombinant DNA technologies to rearrange and modify natural genetic pieces.2 Today, synthetic biologists are no longer constrained to exploiting or manipulating the natural nucleotides and amino acids found in modern terran life, which are merely products of prebiotic chemistry, adaptation, accident, and evolutionary “drift”. Instead, several kinds of nucleotide analogues have been reported that form non-standard nucleobase pairs “orthogonal” to the standard T:A and C:G pairs when they are incorporated into DNA.3-14 Further, in many cases, these extra deoxyribonucleotides can direct the synthesis of non-standard RNA containing extra ribonucleotides thereby increasing the number of codons that, in turn, can encode proteins containing additional amino acids.15,16 These developments have led to a new view of synthetic biology, adumbrated by Eric Kool,9 which seeks to create the properties that we value in life (including reproduction, adaptation, and evolution) using molecular platforms that combine natural and non-natural synthetic components.
In some cases, however, molecular behaviors peculiar to the added synthetic genetic components can limit that combination. For example, Romesberg has shown that two nonstandard nucleobases, d5SCIS and dNaM,8,17,18 designed to pair by hydrophobic and geometric complementarity, can be maintained in duplex DNA both in vitro by PCR polymerases and in living Escherichia coli cells for as long as nine hours.19 However, a potentially undesirable property of d5SCIS:dNaM within duplex DNA is that it intrinsically does not pair in the designed “edge-on” geometry, but rather by inter-strand “stacking” of the two hydrophobic species, which introduces significant distortion within the duplex DNA backbone.20 Only when bound within the active site of an engineered DNA polymerase is the d5SCIS:dNaM pair forced to resemble a the geometrical properties of a Watson-Crick pair.21 One might therefore predict that inclusion of d5SCIS:dNaM pairs at multiple or consecutive sites would have negatively impact the overall DNA conformation, especially given the known toxicity of intercalative DNA binding agents, which distort the DNA structure and prevent protein binding with deleterious consequences for gene regulation and subsequent transcriptional events.
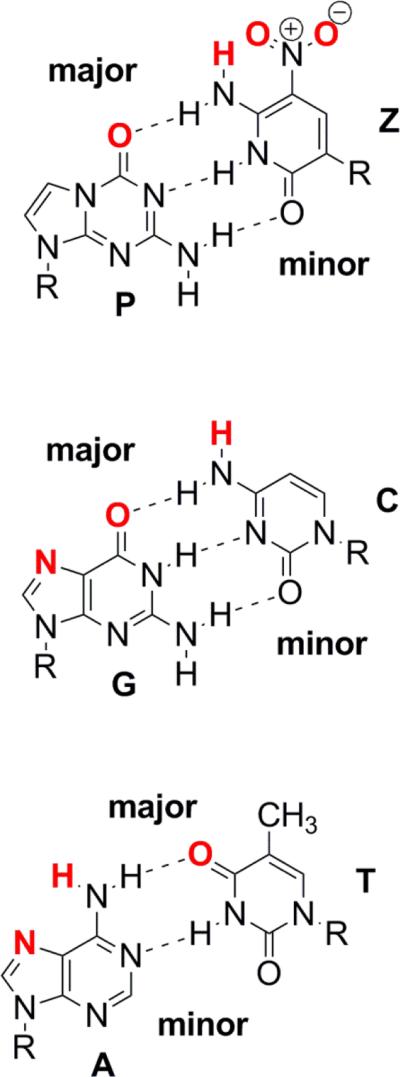
A class of artificially expanded genetic information systems (AEGIS) does not differ so greatly from those found in natural biology.13 Rather than pairing via hydrophobic interactions, these retain the hydrogen bonding that joins the standard A:T and G:C pairs. The pairing is “orthogonal” to the pairing of standard bases because the hydrogen bonding units are shuffled. For example, the small and 6-amino-5-nitro-2(1H)-pyridone heterocycle (trivially named Z) presents a donor-donor-acceptor pattern of hydrogen bonding units to a complementary large 2-amino-imidazo[1,2-a]-1,3,5-triazin-4(8H)one heterocycle (trivially named P), which presents an acceptor-acceptor-donor pattern of hydrogen bonding units (Figure 1).22 Further, Z brings to nucleic acids a functional moiety (the nitro group) that is not found in any natural encoded biopolymer, a functional group that may contribute to the ability of GACTZP DNA to display especially effective binding power following in vitro selection.23 Superficially, Z:P resembles the C-G nucleobase pair,24 but nevertheless it has been shown that DNA containing up to four consecutive Z:P units can be amplified successfully with high fidelity.24,25
Figure 1.
Chemical structures of Z:P, G:C, and A:T paired nucleobases with atoms that can hydrogen bond in the major or minor groove highlighted in red text.
Although X-ray crystallography has in the past been used to examine DNA containing single non-standard pairs,18,20,21,26 no data inform us about how the double-helical structure of DNA might be perturbed by the presence multiple non-natural nucleobase pairs, particularly DNA containing contiguous non-natural pairs. We therefore sought to obtain a series of X-ray structures for 16-bp oligonucleotide duplexes containing contiguous Z:P paired nucleotides. Our initial strategy was to employ a “host-guest” system in which the Z:P-containing oligonucleotides were co-crystallized with the N-terminal fragment of the Moloney murine leukemia virus reverse transcriptase (MMLV-RT).27-29 This system has most recently been used to obtain structures of double-helical DNA containing a thymine dimer30 and DNA bound to a Co(III)-substituted form of bleomycin.31
Here, we present the 1.8 Å crystal structure of a 16-bp, self-complementary oligonucleotide containing only two consecutive Z:P pairs in B-form DNA. For comparative analysis, we also determined the crystal structures at 1.78 Å and 1.68 Å of host-guest complexes including oligonucleotides with consecutive G:C and A:T pairs in place of the Z:P pairs, respectively. A 16-bp, self-complementary oligonucleotide containing six consecutive Z:P nucleobase pairs was found to crystallize in the absence of the host; its 1.98 Å crystal structure is reported here as determined by Br SAD phasing. These structural studies provide a basis for analyzing the structural properties of Z:P nucleobase pairs in the context of both A- and B-DNA and their resultant helical properties.
EXPERIMENTAL PROCEDURES
Synthesis and purification of Z-P containing oligonucleotides
Standard phosphoramidites (Bz-dA, Ac-dC, dmf-dG, dT, and 5-Br-dU-CE) and controlled pore glass (CPG) having standard nucleosides were purchased from Glen Research (Sterling, VA). AEGIS phosphoramidites (dZ and dP) were obtained from Firebird Biomolecular Sciences LLC (Alachua, FL). All oligonucleotides containing Z and P (2P, 3/6ZP, 3/6ZP Br1 and 3/6ZP Br2, see Figure 1 and Table 1) were synthesized on an ABI 394 DNA Synthesizer following standard phosphoramidite chemistry, as previously reported.32 The CPGs carrying the synthetic oligonucleotides were treated with 1 M DBU in anhydrous acetonitrile (2.0 mL) at room temperature for 24 hours to remove the NPE group from the Z nucleobase. Then, the CPGs were filtered and dried. The CPGs carrying 2P and 3/6ZP (without 5-Br-dU) were treated with concentrated ammonium hydroxide at 55 °C for 16 hours, while the CPGs carrying 3/6ZP Br1 and 3/6ZP Br2 (with 5-Br-dU) were treated with concentrated ammonium hydroxide at room temperature for 24 hours. After removal of ammonium hydroxide, the oligonucleotides containing Z and P were purified on ion-exchange HPLC, and then desalted using Sep-Pac® Plus C18 cartridges (Waters). Fully standard oligonucleotides were purchased from Midland Certified Reagent Co. (Midland, Texas) in desalted form and used without further purification.
Table 1.
Oligonucleotides used in this study
| Name | # stacked ZPs/ total ZPs | Sequence |
|---|---|---|
| 2P | 2/4 | 5′-CTTATPPTAZZATAAG |
| 3/6ZP | 6/6 | 5′-CTTATPPPZZZATAAG |
| 3/6 ZP Br1 | 6/6 | 5′-CTTATPPPZZZATAAG |
| 3/6 ZP Br2 | 6/6 | 5′-CTTATPPPZZZATAAG |
| AT | 5′-CTTATAAATTTATAAG | |
| GC | 5′-CTTATGGGCCCATAAG |
Crystallization of ZP containing and other oligonucleotides
Sixteen base pair self-complementary oligonucleotides containing two or six consecutive Z:P nucleobase pairs were designed to be compatible with our host-guest crystallization. The host in this system is the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase, which was purified as previously described,33 and the guests are the various oligonucleotides. Oligonucleotide sequences screened for crystallization are compiled in Table 1. The oligonucleotides were resuspended in buffer containing 10 mM HEPES pH 7.0 and 10 mM MgCl2 to give a final concentration of 2.5 mM duplex DNA and then annealed by heating to 70 °C followed by slow cooling to room temperature prior to crystallization. A 2.9 mM stock solution of the protein was diluted to 1.4 mM using 50 mM MES pH 6.0 and 0.3 M NaCl. This 1.4 mM sub-stock was then further diluted to 0.65 mM in 100 mM HEPES pH 7.5 and 0.3 M NaCl.
Each oligonucleotide was pre-complexed with the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase with final concentrations of 0.71 and 0.46 mM, respectively, as previously described, and then subjected to self-nucleation or microseeding experiments. For microseeding, vapor diffusion hanging drops included 1 μL each of protein-DNA microseeds diluted in reservoir solution containing 7 % PEG 4000, 5 mM magnesium acetate and 50 mM N-(2-acetamido)iminodiacetic acid (ADA) at pH 6.5 and 1 μL of the protein-DNA complex solution, suspended over the reservoir solution. Using a similar strategy to that described above, self-complementary 16-mer duplex DNA oligonucleotides including either G:C or A:T pairs replacing the Z:P pairs found in the 3/6ZP oligonucleotide (Table 1) were crystallized as host-guest complexes.
The oligonucleotide including 6 consecutive Z:P pairs (3/6 Z:P) did not crystallize under our normal host-guest complex conditions and was then subjected to further screening of crystallization conditions, specifically the Natrix screening kit (Hampton Research, Inc), by using the ARI Gryphon crystallization robot. For this experiment, the protein and DNA concentrations were reduced to 0.23 mM and 0.35 mM, respectively. Promising crystals were obtained for this oligonucleotide; however, we noted a large amount of precipitant present in conditions that included 10 mM magnesium acetate, 50 mM MES pH 5.6, and 2.5 M ammonium sulfate. Subsequent crystallizations including just the oligonucleotide, diluted to 0.35 mM in the same buffer that was used to dilute the protein described above, produced crystals under similar conditions. Two different oligonucleotides including 5-bromouracil replacing Ts in the sequence were synthesized (Table 1) for phasing purposes. Both of these oligonucleotides crystallized under similar conditions that included 10 mM magnesium acetate, 50 mM MES pH 5.6, and 1.7-2.0 M ammonium sulfate. The crystal used for the structure determination was that of the Br1 sequence, which crystallized in 10 mM magnesium acetate, 50 mM MES pH 5.6, and 1.7 M ammonium sulfate and was cryo-cooled in a solution including the reservoir with 20% glycerol added.
Data collection, structure determination, and crystallographic refinement
A Br single wavelength anomalous dispersion experiment was performed to obtain phases for the 3/6 ZP-Br structure. Approximately 10-fold redundant data were collected to 1.98 Å at the APS SBC 19-ID beamline (Table 2) for a crystal of the Br1 sequence including two Br atoms from 5-bromouracil replacing thymines in the sequence as shown in Table 1. Initial Patterson searching and phasing calculations were done by using HKL3000.34 Specifically, a single Br site was identified in SHELXD35 and refined in SHELXE36. Initial phases were then calculated to 2.45 Å using MLPHARE, figure of merit 0.56, and improved and extended to 2.25 Å by solvent flattening using DM.37-39
Table 2.
Crystallographic Data
| Dataset | 2P-HG | AT-HG | GC-HG | 3/6 ZP-Br |
|---|---|---|---|---|
| Data statistics | ||||
| a (Å) | 54.638 | 54.636 | 54.623 | 42.019 |
| b (Å) | 145.359 | 145.27 | 145.38 | 42.019 |
| c (Å) | 46.878 | 46.802 | 46.801 | 140.472 |
| Space group | P21212 | P21212 | P21212 | P3221 |
| Wavelength (Å) | 0.97911 | 0.97933 | 0.97933 | 0.91963 |
| Resolution (Å) | 30.26-1.8 | 29.05-1.68 | 28.71-1.78 | 50-1.98 |
| Total observations | 199682 | 183063 | 184656 | 105588 |
| Unique reflections | 41412 | 43118 | 36612 | 10571 |
| Completeness (%) | 99.8 (96.7) | 99.3 (99.3) | 99.9 (100) | 99.5 (98.8) |
| Rmerge (%) | 5.4 (42.0) | 3.0 (43.1) | 4.0 (38.8) | 6.9 (76.8) |
| Rpim | 2.6 (23.5) | 1.7 (23.0) | 2.0 (19.2) | 2.2 (33.9) |
| I/σ | 24.3 (3.4) | 24.8 (3.4) | 22.7 (4.3) | 29.5 (2.3) |
| Refinement statistics | ||||
| R value (%) | 21.0 | 21.6 | 21.6 | 21.2 |
| R free (%) | 23.7 | 23.9 | 23.7 | 23.8 |
| RMSD bonds (Å) | 0.005 | 0.006 | 0.006 | 0.003 |
| RMSD angles (°) | 1.031 | 1.115 | 1.064 | 0.829 |
| Atoms | ||||
| Protein/DNA | 2019/331 | 1982/325 | 1968/325 | 668 |
| water | 201 | 195 | 195 | 76 |
| Average B-factors | ||||
| Protein/DNA | 29.51/54.70 | 28.05/45.38 | 28.68/56.39 | 23.22/22.20* |
| water | 31.96 | 30.09 | 31.08 | 29.0 |
2P, AT, GC, and 3/6 ZP refer to the DNA sequences (Table 1), HG designates a host-guest complex. Values in parentheses are for the highest resolution shell of the data: 2P-HG (1.83-1.80 Å), AT-HG (1.71-1.68 Å), GC-HG (1.82-1.78 Å), and 3/6 ZP (2.01-1.98 Å).
B-factors shown are for the A/B chains of 3/6 ZP rather than protein/DNA.
This initial experimentally phased electron density map was of high quality and allowed automated building of the DNA structure by NAUTILUS,40 albeit with A:T pairs modeled in place of Z:P pairs. Following initial refinement of the structure generated by NAUTILUS in REFMAC5,41 positive peaks were observed in the Fo-Fc electron density maps consistent with missing chemical moieties present within the Z and P nucleotides. While maintaining the coordinate system, additional functional groups were added to the core NAUTILUS generated structure using the Molefacture Plug-in for Visual Molecular Dynamics (VMD) Graphics Viewer.42 Upon generation of the complete coordinates, each Z or P nucleobase was optimized for ten steps using GAMESS ab initio molecular quantum chemistry,43 with a 6-31G44 basis set. Each idealized nucleobase analog was then placed back into the model to form the completed DNA structure, including the novel functionality of the Z and P bases. Parameter files and linking statements were created for refinement in PHENIX.45 Addition of solvent molecules and manual model adjustment was done iteratively in COOT46 followed by refinement in PHENIX with maximum likelihood targets and isotropic B-factors. Addition of solvent molecules and manual model adjustment was done iteratively in COOT followed by refinement in PHENIX with maximum likelihood targets and isotropic B-factors.
Data were collected for the 2P host-guest complex crystal at the APS SBC 19-BM beamline to Bragg spacings of 1.8 Å and for the GC and AT host-guest complex crystals at the APS GM/CA 23-ID-D beamline to Bragg spacings of 1.78 Å and 1.7 Å, respectively (Table 2). The host-guest crystal structures were determined by molecular replacement as implemented in PHASER47 within the CCP4 suite of programs48 using the model of the N-terminal fragment of MMLV RT as the search model. This approach provides unbiased electron density for the DNA complexed to the protein. The protein model and associated water molecules were first adjusted in COOT and then refined initially in REFMAC and later in PHENIX to improve the electron density, and the DNA was subsequently modeled into Fo-Fc electron density maps. In building the DNA model, the first three nucleobase pairs were modeled and refined, then the next two pairs, and finally the remaining three pairs.
For the 2P structure, the Z:P nucleobase pairs were initially modeled as G:C pairs and then subsequently replaced by superimposing the base coordinates for either Z or P generated for the 3/6 ZP structure on the common atoms manually in COOT. The parameter and linking files were created for refinement in PHENIX. As the asymmetric unit includes one protein molecule and half of the DNA molecule, the DNA can be modeled as 8 pairs of duplex DNA or a single 16-mer strand of DNA. To ensure that the phosphodiester bond between bases 8 and 9 is appropriately connected, the DNA was modeled as a single 16-mer strand in the final round of the refinement, and the 8-mer duplex was then regenerated by symmetry.
Coordinates have been deposited for the 2P, GC, AT, and 3/6 ZP structures (Table 1) with PDB identifiers, 4XO0, 4XPE, 4XPC, and 4XNO, respectively.
Circular dichroism experiments
The DNA sequences including 3/6 ZP (5’- CTTATPPPZZZATAAG -3’), AT sequence (5’ CTTATAAATTTATAAG 3’) and GC sequence (5’ CTTATGGGCCCATAAG 3’) were analyzed by circular dichroism to determine the helical form of the oligonucleotides buffered at neutral pH in low salt. For CD analysis, the 2.5 mM stocks of these DNA sequences were diluted to 5 μM with buffer containing 10 mM HEPES pH 7.0 and 10 mM MgCl2. The CD spectra for DNA sequences were collected on a Jasco J-810 CD instrument at 25 °C, at a rate of 50 nm/min and a wavelength increment of 0.1 nm. Ellipticity, Ø (mdegrees) was recorded for the DNA sequences from a wavelength of 320 to 220 nm. The spectra were the average of five scans corrected for ellipticity readings obtained for buffer (10 mM HEPES pH 7.0, 10 mM MgCl2) by itself.
RESULTS AND DISCUSSION
Crystallization and structure determinations of ZP-containing oligonucleotides
Expansion of genetic alphabets that combine standard and non-natural nucleobase pairs in duplex DNA will be an essential element in the evolution of efforts in synthetic biology. The need to maintain (or at least prevent radical divergence from) existing canonical structures, however, represents an important constraint upon the incorporation of synthetic nucleotides into DNA if existing nucleic acid binding proteins or enzymes are to act on them. Of course, even canonical structures have considerable diversity, and the special properties of stacked A:T versus G:C nucleobase pairs in duplex DNA are well documented.49 Thus, characterizing the structural characteristics of DNA containing multiple and/or contiguous non-natural nucleobase pairs as compared to natural nucleobase pairs has considerable importance.
The focus of this study is the structural properties of stacked Z:P nucleobase pairs (Figure 1), which provide novel hydrogen-bonding arrangements between novel purine and pyrimidine-like bases. Accordingly, 16-mer self-complementary oligonucleotides containing either 4 or 6 Z:P pairs in total, with 2 or 6 consecutive Z:P pairs, respectively, were subjected to crystallization trials (Table 1). Initial crystallization screening involved the use of the host-guest system that the Georgiadis laboratory developed for the crystallization and analysis of novel DNA structures and DNA-ligand complexes. In this system, the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase (MMLV-RT) serves as the host and a self-complementary 16-mer duplex DNA oligonucleotide as the guest. The host interacts with the three terminal nucleobase pairs on either end through minor groove hydrogen-bonding while the central 10 nucleobase pairs are free of interactions with either protein or other DNA molecules. The crystal lattice results from packing of complexes that include two protein molecules and one 16-mer oligonucleotide, while the asymmetric unit, the unique repeating unit in the crystal lattice, includes only one protein molecule and 8 nucleobase pairs of DNA.
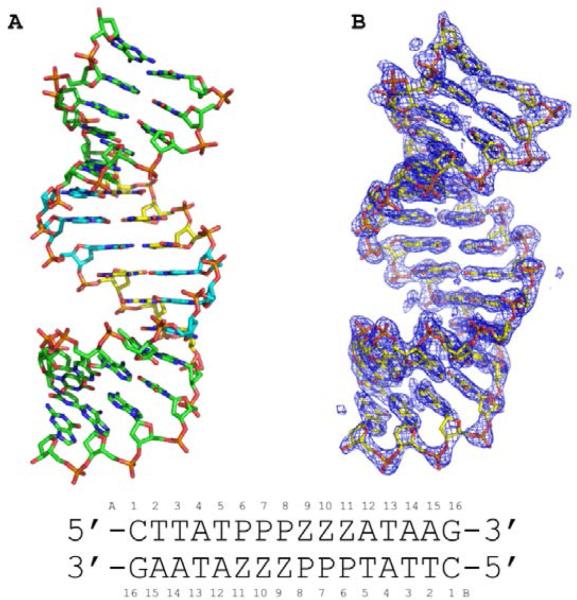
The oligonucleotide including two consecutive Z:P nucleobase pairs (2P) but not the oligonucleotide including six contiguous Z:P pairs (3/6 ZP) crystallized in the host-guest system. As compared to other oligonucleotides crystallized as host-guest complexes, the 2P host-guest complex produced much smaller crystals that took longer to grow. The 2P host-guest crystal was phased by molecular replacement using the protein structure as the search model and refined to 1.8 Å resolution. The 3/6 ZP oligonucleotide was subjected to a high-throughput crystallization screen as a complex with the N-terminal fragment of MMLV-RT. Crystals obtained for the oligonucleotide were found to grow without the host protein under conditions including relatively high salt concentrations (~ 2 M ammonium sulfate). As no structural model was available for molecular replacement phasing of the 3/6 ZP structure, oligonucleotides including 5-bromouracil in place of thymine were synthesized and crystallized (Table 1) for experimental phasing purposes. The crystals obtained for the brominated oligonucleotides were actually larger and diffracted to higher resolution than the non-brominated crystals. A Br SAD phasing experiment was performed for the 3/6 ZP Br1 crystals producing a 2.25 Å experimental electron density map of excellent quality as shown in Figure 2, and the structure was refined to 1.98 Å.
Figure 2.
Crystal structure of 16-mer oligonucleotide containing six consecutive Z:P pairs (3/6 ZP) is shown in (A) as a stick rendering with C, green for non-Z:P pairs, yellow for Z, and cyan for P, O in red, nitrogen in blue, phosphorous in orange. (B) The experimental electron density map derived from Br SAD phasing is shown as a blue mesh contoured at 1.5 σ with the final refined structural model as a stick model. The sequence of the oligonucleotide is shown along with numbering scheme employed in the coordinate file.
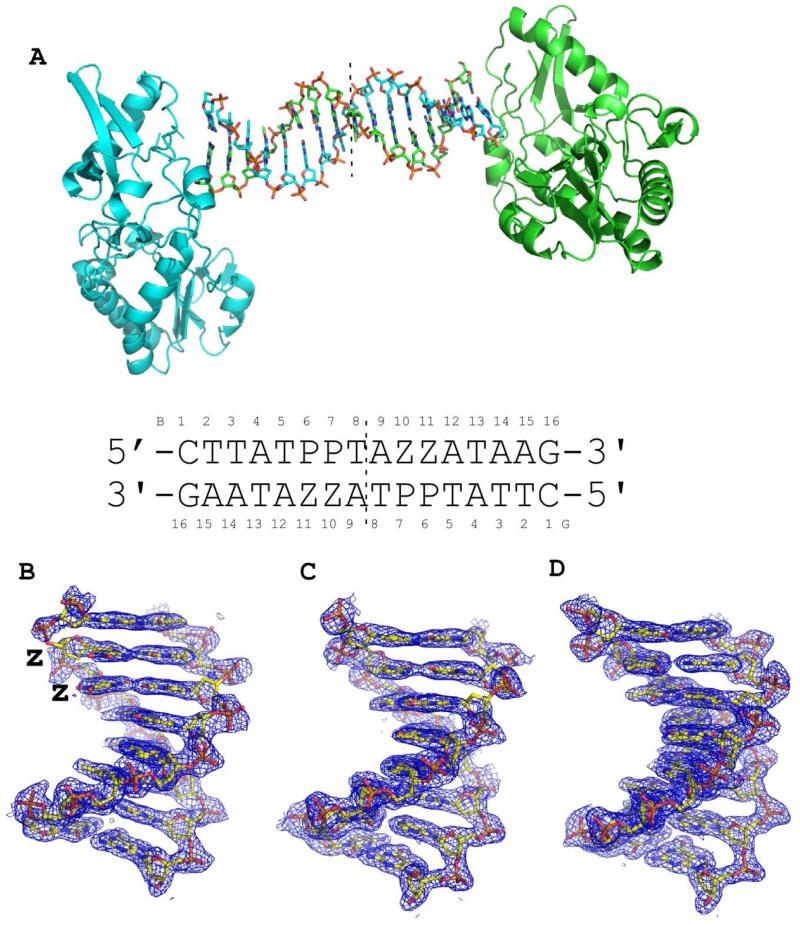
The host-guest system readily accommodates B-form duplex DNA oligonucleotides that are 16 bps in length. It was therefore of interest to determine whether oligonucleotides analogous to 3/6 ZP containing either AT or GC pairs in place of the ZP pairs would crystallize as host-guest complexes (Table 1). Both the AT and GC oligonucleotides crystallized as host-guest complexes; the GC crystals were much smaller and took longer to grow than the AT oligonucleotides, which grew rapidly and to large size (0.2-0.3 mm in each dimension). Both the AT and GC structures were determined to 1.68 Å and 1.78 Å, respectively, by molecular replacement. Final 2Fo-Fc electron density maps are shown in Figure 3 for the DNA in each structure.
Figure 3.
Crystal structures of host-guest complexes including self-complementary 16 base pair oligonucleotides. (A) The host guest complex includes two protein molecules, shown as cartoon renderings in cyan and green, and a 16 base pair oligonucleotide duplex, each strand shown as a stick rendering C, cyan or green, O, red, N, blue, and phosphorous in orange. The complex depicted is that of the host-guest complex for the oligonucleotide shown with two consecutive Z:P nucleobase pairs (2P). Within our crystals, the asymmetric unit includes only half of the complex depicted and thus the equivalent of 8 nucleobase pairs and one protein molecule, indicated by the dashed line. Final 2Fo-Fc electron density maps are shown as blue mesh renderings contoured at 1.0 σ for 8 nucleobase pairs from self-complementary oligonucleotides 2P (B), GC (C), and AT (D). Sequences of GC and AT oligonucleotides are provided in Table 1.
Characterization of the helical properties of ZP-containing oligonucleotides
The 3/6 ZP oligonucleotide crystallized as A-form DNA in a lattice involving molecular packing interactions typically observed in other A-DNA structures.50 In this case, one end of the duplex forms hydrogen bonds in the minor groove with Z:P pairs located in the middle of another DNA duplex. Specifically, O2 of C1 hydrogen bonds to N2 of P7 and N2 of G16 hydrogen bonds to O2 of Z10 (see Figure 2 for numbering scheme). As analyzed by the software package 3DNA,51 the helical form of this structure is classified as A-form excluding the first three dinucleotide steps and the 13th dinucleotide step, which includes the 5-bromouracil. All six contiguous Z:P-containing dinucleotide steps are therefore classified as A-DNA.
Other oligonucleotides known to crystallize under high salt conditions in A-form are GC-rich and involve similar hydrogen-bonding interactions between the ends and G:C pairs in the middle of the oligonucleotides. The fact that 3/6 ZP contains 8 A:T pairs and yet still crystallized as A-form suggests that Z:P pairs may have an even higher propensity to adopt A-form DNA than GC-rich sequences. In a survey of A-DNA structures, the oligonucleotide sequences were G:C-rich with most containing fewer than 2 A:T nucleobase pairs. In fact, the structure most similar to 3/6 ZP in terms of length is that of a GC-rich 14 base pair oligonucleotides (PDB ID: 4OKL), which includes only 2 central A:T nucleobase pairs.
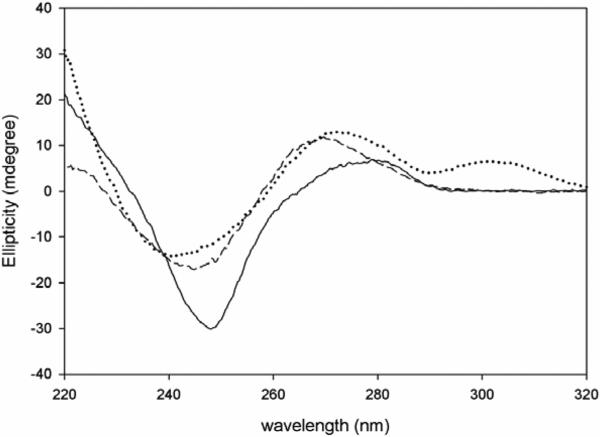
Given that 3/6 ZP crystallized as A-DNA in a high salt condition, it was of interest to determine its helical form in solution under low salt conditions. The CD spectra of all three sequences (AT, GC and 3/6 ZP) exhibit right-handed B-like spectra as shown in Figure 4, with subtle differences in the peak positions and heights pertaining to differences in the primary sequence of DNA. The spectra for the A:T-rich sequence exhibits a negative peak at 248 nm and a positive long wavelength peak at about 279 nm, typical of right handed B-DNA. The GC-rich sequence on the other hand has a broad negative peak centered at approximately 245 nm. The positive peak for the GC-rich sequence shifts to 270 nm instead of 279 nm observed for the AT rich sequence. The 3/6 ZP sequence behaves more similarly to the G:C-rich sequence with its broad negative peak at 241 nm and a positive peak at 273 nm. However, the spectral features of 3/6 ZP differ slightly from the control GC sequence. The positive peak for the 3/6 ZP sequence never reaches zero ellipticity, and at wavelengths greater than 290 nm, it attains another shorter positive peak extending from 290-320 nm. Overall, 3/6 ZP does appear to exist as B-DNA in solution, with CD spectral features most similar to GC.
Figure 4.
Characterization of oligonucleotides by circular dichroism (CD). The ellipticity is plotted versus the wavelength for 3/6 ZP (dotted line), GC (dashed line), and AT (solid line). All of the oligonucleotides have CD spectra indicative of right-handed B-form DNA.
In contrast to the 3/6 ZP, 2P, AT, and GC oligonucleotides (Table 1) crystallized in the host-guest system and exhibit B-form throughout, excluding the terminal dinucleotide steps, as analyzed by 3DNA.51 The AT and GC oligonucleotides are analogous to 3/6 ZP in that the 5'-P3Z3 sequence is replaced with 5'-G3C3 or 5'-A3T3, maintaining the positioning of purine and pyrimidine-like bases within the oligonucleotides. In theory, given that the 3/6 ZP oligonucleotide like the AT and GC oligonucleotides appears to adopt B-form under low salt conditions in solution as assessed by CD, it should have been possible to obtain host-guest crystals. The fact that it does not crystallize as a host-guest complex may be consistent with altered dynamic or other properties associate with the Z:P pairs.
Structural properties of ZP nucleobase pairs in A- and B-DNA
Of particular interest are the structural properties associated with the unique NO2 group in Z in the context of A- or B-DNA. This nitro group was introduced initially to manage the chemical properties of the system presenting a hydrogen bond donor-donor-acceptor pattern (from the major to the minor groove).22 Subsequently, the nitro group was found to confer binding potential on GACTZP libraries that appears to be absent in standard GACT libraries.23 As anticipated, N4, N3, and O2 of Z hydrogen bond to O6, N1, and N2 of P, respectively, with distances of 2.7-3.0 Å in 2P and 3/6 ZP structures.
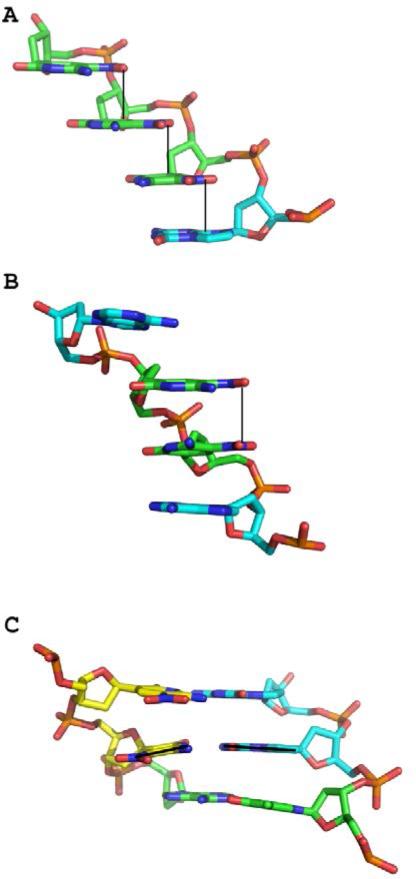
In the A-DNA structure, the Z nucleobase, including its NO2 group, is planar; this appears to facilitate stacking interactions between the NO2 of one Z nucleobase and the pyrimidine (or purine) ring of the adjacent nucleobase (Figure 5A). One oxygen atom of the nitro group in these stacked planar Z nucleobases is within hydrogen-bonding distance (~2.7 Å) of N4 of the pyrimidine-like ring. In contrast, in B-DNA, the NO2 group does not stack with adjacent bases but is nearly planar in both Z nucleobases with similar hydrogen-bonding between a nitro oxygen and N4 to that observed in 3/6ZP (Figure 5B). Accordingly, the electron density for the Z-NO2 groups in B-DNA is not as well ordered as in A-DNA (Figure 2), consistent with the more constrained conformation observed in the stacking interactions in A-form DNA. Potentially, the favorable stacking interactions of the Z-NO2 groups contribute to the ability of the 16-mer oligonucleotide including only six Z:P pairs to crystallize as A-form DNA. The fact that Z is a C-glycoside with a carbon-carbon linkage to its deoxyribose sugar rather than nitrogen-carbon linkage found in natural bases may also be a contributing factor.
Figure 5.
Structural characteristics of Z:P nucleobase pairs. (A) In A-DNA, the Z-NO2 stack over the pyrimidine or purine ring of the adjacent nucleobase emphasized by solid lines as shown in this stick renderings C, green for Z, cyan for P, O in red, N in blue, and phosphorous in orange for P8Z9Z10Z11, A chain. (B) In B-DNA, the Z-NO2 groups are positioned roughly over one another but do not stack with nucleobase rings. Rather, the pyrimidine rings stack with adjacent nucleobase rings as shown in this stick rendering for bases for A9Z10Z11A12 for the G chain. (C) The central Z:P pair, with a black line indicating the trajectory of each nucleobase, exhibits a large buckle angle as shown in this stick rendering for T5P6P7 (B chain) and Z10Z11A12 (G chain). C atoms are indicated in yellow for Z, cyan for P, and green for A or T, with other atoms as indicated above.
To characterize the DNA parameters of Z:P pairs in A-DNA, we analyzed the local base pair step parameters, local base pair helical parameters, and the groove widths for the central Z:P pairs in our structure and compared them to those of the GC-rich 4OKL structure (5’-CCCCGGTACCGGGG) using the program 3DNA. Average values were calculated for five Z:P or P:Z dinucleotide steps for our structure and 10 GC or CG dinucleotide steps from 4OKL. As previously noted, A:T pairs are found infrequently in A-DNA structures and thus are not available for comparison. As shown in Table 3, the average rise, roll, and twist values for Z:P dinucleotide steps are similar to the values observed for GC dinucleotide steps and the average parameters for A-DNA reported by Lu et al.51 However, the average slide value for Z:P pairs of −2.08 Å in A-DNA is slightly larger than observed in G:C pairs in 4OKL (−1.95 Å) and on average in A-DNA structures −1.53 +/− 0.34 Å. This finding is consistent with the preferential stacking of the NO2 group with the adjacent pyrimidine or purine ring.
Otherwise, local base pair helical parameters including H-rise, inclination, H-twist, and X-displacement are all within the average range observed in A-DNA structures and are similar to those observed for G:C pairs in 4OKL. Within the central Z:P pairs of our structure, the average major groove width of 18.92 Å is approximately 1 Å wider the groove width associated with the G:C pairs (18.0 Å) in the 4OKL structure. Widening of the major groove may result from the presence of the Z-NO2 in the major groove. The minor groove widths for both Z:P and G:C pairs in A-DNA are more similar than the major groove widths with average values of 16.52 and 16.92 Å, respectively.
Similarly, the properties of the Z:P nucleobase pairs in B-DNA were assessed and compared to A:T and G:C pairs at the same positions in the DNA sequence in our host-guest structures. There are four Z:P pairs within six nucleotides (5’-PPTAZZ) in the 16-mer sequence crystallized; however, only half of this sequence and its complementary sequence are unique as the other half is related by crystallographic symmetry. These sequences are referred to as B-ZP, B-GC, and B-AT in contrast to those in the A-DNA structures, which are A-ZP and A-GC. In this case, the three dinucleotide steps containing ZP pairs and the analogous dinucleotide steps from the AT and GC oligonucleotide structures were analyzed. The average local base pair step parameters and local base pair helical parameters for B-ZP all fall within the range observed for B-DNA structures. Average values for each of these parameters for B-ZP are more similar to those calculated for B-GC than for B-AT. As was true for A-ZP, the major groove width of B-ZP of 18.7 Å is significantly (~ 0.7 Å) wider than that of B-GC (17.97 Å). However, it is slightly narrower than B-AT, which has an average major groove width of 19.07 Å. The minor groove width for B-ZP of 12.67 Å is similar to that of B-GC (12.43 Å) and significantly wider than that of B-AT (9.67 Å). AT-rich sequences are known to exhibit deep, narrow minor grooves, which bind to a number of small molecules.
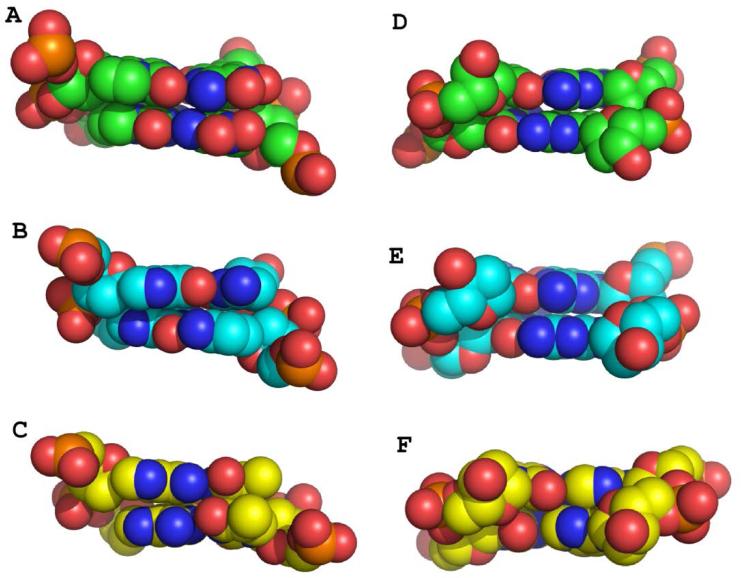
Local base pair parameters including shear, stretch, stagger, buckle, propeller, and opening were also analyzed using 3DNA for A-ZP pairs and found to be fairly uniform in value. Propeller values of −10.76 ° and −13.68 ° are the largest observed values and occur at the junctions between the PZ or ZP and natural nucleobase pairs. For B-ZP, the pair Z6P11 exhibits an unusually large buckle angle of −14.25 ° (Figure 5C), which is much larger than buckle angles of −1.4 and 5.38 ° exhibited for equivalent A:T or G:C base pairs, respectively. This large buckle angle for Z6P11 may contribute to the offset hydrogen-bonding interactions observed for this base pair. In the major groove, the Z:P pair provides novel major groove hydrogen bonding opportunities with four electronegative atoms, two provided by the NO2 group, as compared to three present in either A:T or G:C pairs (Figure 6A-C). In the minor groove, Z:P pairs present the same hydrogen bonding pattern of electronegative atoms as the GC base pair (Figure 6D-F), with all three nucleobase pairs including O2 and N3 atoms.
Figure 6.
Major and minor groove views of Z:P, G:C, and A:T pairs in B-DNA. Dinucleotides for Z10Z11/P6P7 (A) and the equivalent pairs for GC (B) and AT (C) host-guest complexes are shown as Van der Waals sphere renderings for the major groove with C in green for Z:P, cyan for GC, and yellow for AT. Minor groove views are shown for Z:P (D), GC (E), and AT (F).
Thus, our structural analysis indicates that Z:P nucleobase pairs are accommodated in both A- and B-DNA and overall exhibit DNA parameters that are quite similar to those observed for G:C pairs in the same helical form. The most significant impact of Z:P pairs on the structure of the DNA duplexes is widening of the major groove in both A and B-DNA as compared to values calculated for similar GC regions. This finding suggests that the NO2 group which faces the major groove has a widening effect. We speculate that stacked Z:P nucleobase pairs have a higher propensity to form A-DNA than equivalent sequences with G:C pairs based on the inability to crystallize the 3/6 ZP sequence in our host-guest system, which has a strong selection for B-form oligonucleotides, and the fact that the AT-rich 3/6 ZP sequence still crystallized in A-form while other examples of A-DNA are more G:C-rich. However, the CD spectrum of the 3/6 ZP oligonucleotide in a low salt buffering solution has features consistent with B-DNA and is similar overall to that obtained for the equivalent GC sequence. In conclusion, our analysis suggests that Z and P nucleotides will be useful in expanding the genetic alphabet, as contiguous Z:P pairs are well tolerated in standard helical forms of DNA.
ACKNOWLEDGEMENTS
We thank Dr. Michael McLeish (IUPUI) for providing assistance and instrumentation for the CD experiments. Results shown in this report are in part derived from work performed at Argonne National Laboratory, Structural Biology Center, beamlines 19-ID and 19-BM, at the Advanced Photon Source. We thank Z. Otwinowski and W. Minor for helpful discussions while at the SBC beamline and Marianne Cuff for assistance during data collection. Results were also derived from data collected at the GM/CA beamline 23-ID-D. We thank Rushlan Sanishvili for assistance during data collection at GM/CA.
Footnotes
This work was partially supported by grants R01DK061666 (NGJR) and R01GM111386 (SAB) from the National Institutes of Health, HDTRA1-13-1-0004 from the Defense Threat Reduction Agency, NNX14AK37G from the National Aeronautics and Space Administration (SAB), and the Templeton World Charitable Foundation (SAB). Data for this work were collected at GM/CA at the Advanced Photon Source, which has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357.
REFERENCES
- 1.Leduc S. La biologie synthétique. A. Poinat; Paris: 1912. [Google Scholar]
- 2.Szybalski W. In: Control of Gene Expression. Kohn A, Shatkay A, editors. Plenum Press; New York: 1974. [Google Scholar]
- 3.Sismour AM, Benner SA. Synthetic biology. Expert Opin Biol Ther. 2005;5:1409. doi: 10.1517/14712598.5.11.1409. [DOI] [PubMed] [Google Scholar]
- 4.Rappaport HP. The 6-thioguanine/5-methyl-2-pyrimidinone base pair. Nucleic Acids Res. 1988;16:7253. doi: 10.1093/nar/16.15.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Switzer C, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA. J Am Chem Soc. 1989;111:8322. [Google Scholar]
- 6.Piccirilli JA, Krauch T, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990;343:33. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 7.Ishikawa M, Hirao I, Yokoyama S. Synthesis of 3-(2-deoxy-β-d ribofuranosyl)pyridin-2-one and 2-amino-6-(N,N-dimethylamino)-9-(2-deoxy-β-d-ribofuranosyl)purine derivatives for an unnatural base pair. Tetrahedron Letters. 2000;41:3931. [Google Scholar]
- 8.Tae EL, Wu Y, Xia G, Schultz PG, Romesberg FE. Efforts toward expansion of the genetic alphabet: replication of DNA with three base pairs. J Am Chem Soc. 2001;123:7439. doi: 10.1021/ja010731e. [DOI] [PubMed] [Google Scholar]
- 9.Kool ET. Replacing the nucleobases in DNA with designer molecules. Acc Chem Res. 2002;35:936. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]
- 10.Geyer CR, Battersby TR, Benner SA. Nucleobase pairing in expanded Watson-Crick-like genetic information systems. Structure. 2003;11:1485. doi: 10.1016/j.str.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Henry AA, Romesberg FE. Beyond A, C, G and T: augmenting nature's alphabet. Curr Opin Chem Biol. 2003;7:727. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Minakawa N, Kojima N, Hikishima S, Sasaki T, Kiyosue A, Atsumi N, Ueno Y, Matsuda A. New base pairing motifs. The synthesis and thermal stability of oligodeoxynucleotides containing imidazopyridopyrimidine nucleosides with the ability to form four hydrogen bonds. J Am Chem Soc. 2003;125:9970. doi: 10.1021/ja0347686. [DOI] [PubMed] [Google Scholar]
- 13.Benner SA. Understanding nucleic acids using synthetic chemistry. Acc Chem Res. 2004;37:784. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 14.Hirao I, Harada Y, Kimoto M, Mitsui T, Fujiwara T, Yokoyama S. A two-unnatural-base-pair system toward the expansion of the genetic code. J Am Chem Soc. 2004;126:13298. doi: 10.1021/ja047201d. [DOI] [PubMed] [Google Scholar]
- 15.Leal NA, Kim HJ, Hoshika S, Kim MJ, Carrigan MA, Benner SA. Transcription, Reverse Transcription, and Analysis of RNA Containing Artificial Genetic Components. ACS Synth Biol. 2014 Aug 19; doi: 10.1021/sb500268n. Epub ahead of date. [DOI] [PubMed] [Google Scholar]
- 16.Bain JD, Switzer C, Chamberlin AR, Benner SA. Ribosome-mediated incorporation of a non-standard amino acid into a peptide through expansion of the genetic code. Nature. 1992;356:537. doi: 10.1038/356537a0. [DOI] [PubMed] [Google Scholar]
- 17.Seo YJ, Hwang GT, Ordoukhanian P, Romesberg FE. Optimization of an unnatural base pair toward natural-like replication. J Am Chem Soc. 2009;131:3246. doi: 10.1021/ja807853m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malyshev DA, Pfaff DA, Ippoliti SI, Hwang GT, Dwyer TJ, Romesberg FE. Solution structure, mechanism of replication, and optimization of an unnatural base pair. Chemistry. 2010;16:12650. doi: 10.1002/chem.201000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Correa IR, Jr., Romesberg FE. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509:385. doi: 10.1038/nature13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Romesberg FE, Marx A. Structural insights into DNA replication without hydrogen bonds. J Am Chem Soc. 2013;135:18637. doi: 10.1021/ja409609j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Dwyer TJ, Ordoukhanian P, Romesberg FE, Marx A. KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry. Nat Chem Biol. 2012;8:612. doi: 10.1038/nchembio.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutter D, Benner SA. Expanding the genetic alphabet: non-epimerizing nucleoside with the pyDDA hydrogen-bonding pattern. J Org Chem. 2003;68:9839. doi: 10.1021/jo034900k. [DOI] [PubMed] [Google Scholar]
- 23.Sefah K, Yang Z, Bradley KM, Hoshika S, Jimenez E, Zhang L, Zhu G, Shanker S, Yu F, Turek D, Tan W, Benner SA. In vitro selection with artificial expanded genetic information systems. Proc Natl Acad Sci U S A. 2014;111:1449. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Chen F, Chamberlin SG, Benner SA. Expanded genetic alphabets in the polymerase chain reaction. Angew Chem Int Ed Engl. 2010;49:177. doi: 10.1002/anie.200905173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Chen F, Alvarado JB, Benner SA. Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J Am Chem Soc. 2011;133:15105. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson H, Gao YG, Bauer C, Roberts C, Switzer C, Wang AH. 2′-Deoxyisoguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis. Biochemistry. 1998;37:10897. doi: 10.1021/bi980818l. [DOI] [PubMed] [Google Scholar]
- 27.Cote ML, Georgiadis MM. Structure of a pseudo-16-mer DNA with stacked guanines and two G-A mispairs complexed with the N-terminal fragment of Moloney murine leukemia virus reverse transcriptase. Acta Crystallogr D Biol Crystallogr. 2001;57:1238. doi: 10.1107/s090744490100943x. [DOI] [PubMed] [Google Scholar]
- 28.Cote ML, Yohannan SJ, Georgiadis MM. Use of an N-terminal fragment from moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing G-A mispairs. Acta Crystallogr D Biol Crystallogr. 2000;56:1120. doi: 10.1107/s0907444900008246. [DOI] [PubMed] [Google Scholar]
- 29.Montano SP, Cote ML, Roth MJ, Georgiadis MM. Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus. Nucleic Acids Res. 2006;34:5353. doi: 10.1093/nar/gkl693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh I, Jian Y, Li L, Georgiadis MM. The structure of an authentic spore photoproduct lesion in DNA suggests a basis for recognition. Acta Crystallogr D Biol Crystallogr. 2014;70:752. doi: 10.1107/S1399004713032987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodwin KD, Lewis MA, Long EC, Georgiadis MM. Crystal structure of DNA-bound Co(III) bleomycin B2: Insights on intercalation and minor groove binding. Proc Natl Acad Sci U S A. 2008;105:5052. doi: 10.1073/pnas.0708143105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z, Hutter D, Sheng P, Sismour AM, Benner SA. Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern. Nucleic Acids Res. 2006;34:6095. doi: 10.1093/nar/gkl633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun D, Jessen S, Liu C, Liu X, Najmudin S, Georgiadis MM. Cloning, expression, and purification of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase: crystallization of nucleic acid complexes. Protein Sci. 1998;7:1575. doi: 10.1002/pro.5560070711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 36.Sheldrick GM. Zeitschrift für Kristallographie/International journal for structural, physical, and chemical aspects of crystalline materials. 2002;217:644. [Google Scholar]
- 37.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:470. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowtan K, Main P. Miscellaneous algorithms for density modification. Acta Crystallogr D Biol Crystallogr. 1998;54:487. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- 39.Cowtan KD, Zhang KY. Density modification for macromolecular phase improvement. Prog Biophys Mol Biol. 1999;72:245. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 40.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, Dupuis M, Montgomery JA. General atomic and molecular electronic structure system. J Comput Chem. 1993;14:1347. [Google Scholar]
- 44.Ditchfield R, Hehre WJ, Pople JA. Self Consistent Molecular Orbital Methods. IX. An Extended Gaussian-Type Basis for Molecular-Orbital Studies of Organic Molecules. The Journal of Chemical Physics. 1971;54:724. [Google Scholar]
- 45.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.Neidle S. Nucleic Acid Structure. Oxford University Press; New York: 1999. [Google Scholar]
- 50.Wahl MC, Sundaralingam M. Crystal structures of A-DNA duplexes. Biopolymers. 1997;44:45. doi: 10.1002/(SICI)1097-0282(1997)44:1<45::AID-BIP4>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 51.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]