Abstract
Color is an important agronomic trait of pears, and the anthocyanin content of fruit is immensely significant for pear coloring. In this study, an anthocyanin-activating R2R3-MYB transcription factor gene, PyMYB10.1, was isolated from fruits of red sand pear (Pyrus pyrifolia cv. Aoguan). Alignments of the nucleotide and amino acid sequences suggested that PyMYB10.1 was involved in anthocyanin regulation. Similar to PyMYB10, PyMYB10.1 was predominantly expressed in red tissues, including the skin, leaf and flower, but it was minimally expressed in non-red fruit flesh. The expression of this gene could be induced by light. Dual-luciferase assays indicated that both PyMYB10 and PyMYB10.1 activated the AtDFR promoter. The activation of AtDFR increased to a greater extent when combined with a bHLH co-factor, such as PybHLH, MrbHLH1, MrbHLH2, or AtbHLH2. However, the response of this activation depended on the protein complex formed. PyMYB10-AtbHLH2 activated the AtDFR promoter to a greater extent than other combinations of proteins. PyMYB10-AtbHLH2 also induced the highest anthocyanin accumulation in tobacco transient-expression assays. Moreover, PybHLH interacted with PyMYB10 and PyMYB10.1. These results suggest that both PyMYB10 and PyMYB10.1 are positive anthocyanin biosynthesis regulators in pears that act via the formation of a ternary complex with PybHLH. The functional characterization of PyMYB10 and PyMYB10.1 will aid further understanding of the anthocyanin regulation in pears.
Introduction
Pear is an economically important temperate fruit. Until to now, at least 22 primary species of Pyrus have been identified; however, only the four major species Pyrus bretschneideri, Pyrus pyrifolia, Pyrus ussuriensis, and Pyrus communis have been utilized for commercial fruit production [1].The sand pear (P. pyrifolia)is primarily cultivated in eastern Asia. In general, the sand pear can be divided into four types based on skin color: red, green, russet, and an intermediate color (russet and green). Most cultivated sand pear varieties are green and russet. Although red pears exist, their supply is inadequate [2].Thus, red color has recently become an important breeding objective for pear cultivars, especially in Asian countries such as China.
The red coloration of pears is mainly determined by the skin anthocyanin content [3]. Cyanidin-3-galactosideand peonidin-3-galactoside are the main anthocyanin components of pear skin[4, 5]. The anthocyanin biosynthetic pathway of pears is usually divided into two main sections: chalcone synthase (CHS), chalconeisomerase (CHI) and flavanone 3-hydroxylase (F3H) are in the early section, dihydroflavonol 4-reductase(DFR), anthocyanidin synthase (ANS) and UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT) are in the late section. Genes of these enzymes have been well characterized and they are primarily regulated by transcription factors at the transcriptional level [6, 7, 8].
MYB and basic helix loop helix (bHLH) transcription factor have been commonly identified as anthocyanin regulators in model plants and fruits, including grapes [9], apples [10], and Chinese bayberries [11]. Notably, MYB appears to play a key role in anthocyanin accumulation. MYB proteins, the largest transcription factor family in plants, are identified based on the number of the MYB conserved domain (R1-MYB, R2R3-MYB, or R1R2R3-MYB). Most MYB TFs involved in the regulation of anthocyanin pathway are R2R3-MYB TFs. With respect to their distinct functions, they are usually divided into two groups. One group consists of most known positive anthocyanin regulators, including PhAN2 and PhAN4 in petunias [12], PAP1 and PAP2 in Arabidopsis [13], MdMYB10 in apples [10], and VvMYBA1 in grapes [14].The members of the second group were served as anthocyanin repressors, including FaMYB1 in strawberry [15]; AtMYB6, AtMYB4,and AtMYB3 in Arabidopsis [16, 17, 18]; and AmMYB308 in Antirrhinum [19].Many researchers have shown that changes to these R2R3-MYB TFs can markedly affect phenotype. For example, red-fleshed apples are the result of a tandem repeats in the MdMYB10 promoter [20].Ectopic expression assays suggested that R2R3-MYB can work independently or together with bHLH in controlling anthocyanin biosynthesis. For example, the maize P factor has been demonstrated to activate a subset of anthocyanin biosynthetic genes independently of a bHLH coactivator [21].Conversely, the maize C1 factor has been shown to cooperate with R, to activate the promoter of DFR [22]. Based on the theoretical progress made in petunia, apple and Arabidopsis, the first R2R3-MYB TF in sand pear, PyMYB10, was isolated and reported to regulate anthocyanin synthesis in red-skinned pears [23]. However, the interactions between PyMYB10 and bHLH proteins are not well understood.
In the present study, a R2R3-MYB, PyMYB10.1, was isolated from the red sand pear (cv. Aoguan). PyMYB10.1 shares a high level of sequence homology with other known anthocyanin regulators. Phylogenetic results revealed that PyMYB10.1 and PyMYB10 are in the same clade. In addition, PyMYB10.1 and PyMYB10 are preferentially expressed in tissues where anthocyanin accumulates. A dual-luciferase assay indicatedthatPyMYB10.1 and PyMYB10could activate the AtDFR promoter in the presence of a bHLH co-factor. Furthermore, yeast two-hybrid and BiFC assays confirmed that PyMYB10.1 and PyMYB10 interact with PybHLH TF. Lastly, the in vitro transient expression of PyMYB10 and PyMYB10.1 induced differential accumulation of anthocyanin in the injection area of tobacco leaves when co-expressed with several bHLH TFs.
Materials and Methods
Plant materials
No specific permit was required for this experiment. The location is not protected in any way, and the study did not involve endangered or protected species.
All the tissues were collected from ‘Aoguan’ plants. Plants were grown at Aoguan FruitCorp. (lat: 116.2501, lng: 36.589; elevation: 31m). The skins were collected from ‘Aoguan’ on September 2012, 6 days after debagging. The samples were immediately frozen by liquid nitrogen, and then stored at -80°C for subsequent experiments.
Anthocyanin content analysis
Anthocyanins were extracted according to the method described by Pirie and Mullins [24] and Wang et al. [25]. Absorbance of the extracts was monitored at 553 and 600 nm. Anthocyanin content was calculated as described by Wang et al. [25]. Three replicates of each sample were analyzed.
Total RNA extraction and cDNA synthesis
Total RNA was extracted according to a modified cetyltrimethylammoniumbromide (CTAB) method [26], and then treated by DNase I (Fermentas, USA). cDNA synthesis was performed using the Revert Aid™ First Strand cDNA Synthesis kit (Fermentas, USA).
Isolation of PyMYB10.1
The PyMYB10.1 gene (accession numberKT748756) was cloned from ‘Aoguan’ cDNA using degenerate primers [27].Based on homology with the R2R3-MYBs related to anthocyanin biosynthesis in other species, PyMYB10.1 was selected for further study. A full-length cDNA of the PyMYB10.1 was subsequently obtained by 5’-RACE with the primer 5′-TCTTCCAGCAATTATTGACCACCTG-3′ and 3’-RACE with the primer 5′-GCAGGAAAAGCTGCAGACAGAGGTG-3′ using SMART™ RACE cDNA Amplification Kit (Invitrogen, USA).
Bioinformatic analysis
The phylogenetic tree was constructed utilized MEGA version 3.1 [28]. Multiple sequence alignments was performed with the Clustal W 2 (http://www.ebi.ac.uk/Tools/clustalw2/). The motif sequence and protein domains were identified using InterPro software.
Real-time quantitative PCR
qPCR DNA amplification was performed using the Light Cycler System (Bio-Rad, USA). All reactions were carried out in triplicate using a volume of 20μ Lreaction mixture containing 2μL of Master Mix (TaKaRa), 0.5 M of each primer, 2μl of diluted cDNA. The qPCR reaction programs were as follows: 95°C for 5 min; 40 cycles for 10 s at 95°C, 30 s at 56°C and 30 s at 72°C; and a final extension at 72°C for 3 min. The primers used for PyMYB10.1, PyMYB10 and PybHLH are listed in S1 Table. PyActin (accession number CN938023) was used as a constitutive control gene.
Tobacco transient-expression assay
The promoter of ArabidopsisDFR (TT3, AT5g42800) was subcloned into the vector pGreenII 0800-LUC [29]. The full-length CDS of the TFs PyMYB10,PyMYB10.1, MrbHLH1,MrbHLH2 and AtbHLH2 were subcloned into the vector pGreenII 0029 62-SK.The tobacco (N. tabacum) abaxial leaf surface was infiltrated with PyMYB10, PyMYB10.1, MrbHLH1, MrbHLH2and AtbHLH2either singly or in pairs. Tobacco (N. tabacum) was transformed with Agrobacterium. The TF-promoter interactions were measured based on the ratio of LUC activity to REN activity. Three days after the transformation, the LUC and REN activities were analyzed as described by Liu et al. [11]. Digital photos of infiltration area were taken 8 days after infiltration. The primers used for full-length TF amplification are listed in S2 Table. All the statistical analyses were performed using SPSS software.
Yeast two-hybrid assay (Y2H)
The PyMYB10 (GU253310) and PyMYB10.1ORFs were inserted into the pGADT7 vector, and the PybHLH (HM622265) ORF was inserted into the pGBKT7 vectors (BD Biosciences) [30]. These clones were then used to study the PyMYB10, PyMYB10.1and PybHLH interactions in the Y2H assay. The interactions of these proteins were detected using Matchmaker yeast two-hybrid system (BD Biosciences, USA). AH109 competent cells were co-transformed with the PyMYB10, PyMYB10.1 and PybHLH constructs. The co-transformants were initially selected on synthetic dropout medium lacking Leu and Trp (SD/−Leu/−Trp) and then streaked on quadruple-dropout medium deficient in Ade, His, Leu and Trp (SD/−Ade/−His/−Leu/−Trp). X-gal was further used to assess theβ-galactosidase activity to confirm positive interactions. The primers used to construct the plasmids are presented in S3 Table.
Bimolecular fluorescence complementation assay (BiFC)
The full-length CDS of PyMYB10 and PyMYB10.1or PybHLH were cloned into the binary YFP BiFC vectors35S-pSPYCE-YFPor35S-pSPYNE-YFP, respectively, resulting in the recombinant plasmids PyMYB10-YFPC, PyMYB10.1-YFPC and PybHLH-YFPN. The primers used to construct the plasmids are presented in S4 Table. Onion epidermis cells were transformed with Agrobacterium as described by Li et al. [31]. Two days after transformation, the YFP signals were examined in the transfected cells using a confocal microscope (Deerfield, IL, USA).
Results
Isolation of PyMYB10.1 from pears
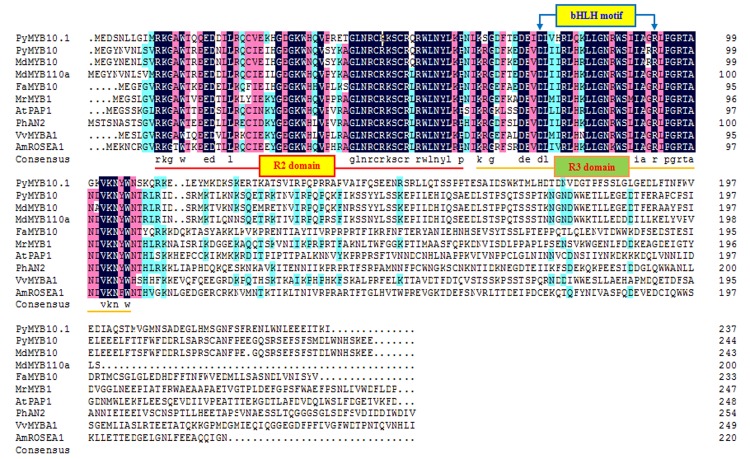
Using degenerate primers [27], a 249-bp R2R3-MYB fragment was isolated from ‘Aoguan’ cDNA samples. Then the 714-bp full-length ORF cDNA was obtained by RACE PCR. The predicted protein is 237 amino acids in length with a calculated molecular mass of 27.4kDa and an isoelectric point of 8.78.An alignment of the deduced amino acid sequence with known anthocyanin MYB regulators indicated high conserved at the R2R3-MYB domain particularly with PyMYB10, but the C-termini downstream of this region are more divergent (Fig 1).The R3-MYBdomain of the conserved N-terminal portion of the protein sequence contains the bHLH-binding region motif ([DE]Lx2[RK]x3Lx6Lx3R). At the protein level, the predicted amino acid sequence has 51%homology with PyMYB10 from pears, 49% homology with MdMYB10 from apples, and 44% homology with VvMYBA1 from grapes. It suggested that a new homologous gene of PyMYB10 was isolated from red sand pear, and we named this sequence PyMYB10.1.
Fig 1. Multiple alignments of PyMYB10.1 and anthocyanin R2R3-MYB regulators.
Identical amino acids are shaded in black and similar amino acids in pink or turquoise. The R2- and R3-MYB DNA-binding domains of selected MYB proteins are underlined. The bHLH binding motif is indicated with brackets. The GenBank accession numbers of R2R3-MYB proteins are as follows: Pyrus pyrifolia PyMYB10 (ADN52330); Malus x domestica MdMYB10(ACQ45201) andMdMYB110a (AB743999); Fragaria x ananassa FaMYB10(ABX79947); Petunia x hybrida PhAN2(AAF66727); Arabidopsis thaliana AtPAP1(AAG42001); Vitis vinifera VvMYBA1(BAD18977); and Antirrhinum majus AmROSEA1(ABB83826).
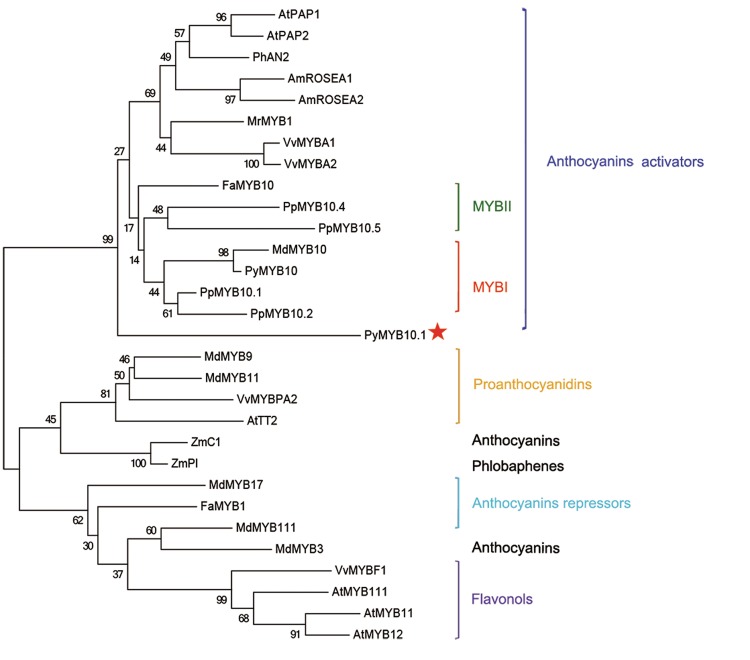
To further analyze the relationship between PyMYB10.1 and other R2R3-MYB transcription factors, a phylogenetic tree was constructed. A phylogenetic analysis revealed that PyMYB10.1 clusters with the pear anthocyanin MYB regulator, PyMYB10. PyMYB10.1 is phylogenetically close to the known anthocyanin MYBI type regulators, particularly those from Rosaceae, but is more distantly related to the Arabidopsis AtTT2 [32], which controls proanthocyanidin synthesis; Arabidopsis AtMYB11 and AtMYB12 [33], which control flavonol synthesis; and FaMYB1 from strawberries [15], which represses anthocyanin synthesis (Fig 2).These results suggest that PyMYB10.1 plays a role in regulating anthocyanin synthesis.
Fig 2. Phylogenetic relationship of PyMYB10.1 to other R2R3-MYBs.
A phylogenetic tree was constructed using the neighbor-joining method by the MEGA3 software. The reliability of the trees was tested using a bootstrapping method with 1000 replicates. PyMYB10.1 is indicated with an asterisk. Putative regulatory functions of the selected R2R3-MYB proteins are indicated. The GenBank accession numbers of some R2R3-MYB protein sequences are as follows: Antirrhinum majus AmROSEA2 (ABB83827); Prunus persica PpMYB10.1 (Ppa026640m), PpMYB10.2 (Ppa016711m), PpMYB10.4 (Ppa018744m), PpMYB10.5(Ppa024617m); Arabidopsis thaliana AtMYB11 (EFH52939), AtMYB12 (AEC10843), AtMYB111 (EFH41988), AtPAP2 (AAG42002), and AtTT2 (AED93980); Fragaria ananassa FaMYB1 (AAK84064); Malus domesticaMdMYB3 (AEX08668.1), MdMYB9 (ABB84757), MdMYB11 (AAZ20431), MdMYB17 (ADL36757), and MdMYB111 (ADL36754); Pyrus pyrifolia PyMYB10 (ADN52330); Morella rubra MrMYB1 (ADG21957); Zea mays ZmC1 (AAA33482),ZmPl (AAA19819);and Vitis vinifera VvMYBA2 (BAD18978), VvMYBF1 (ACV81697), and VvMYBPA2 (ACK56131).
PyMYB10, PyMYB10.1and PybHLH gene expression analysis
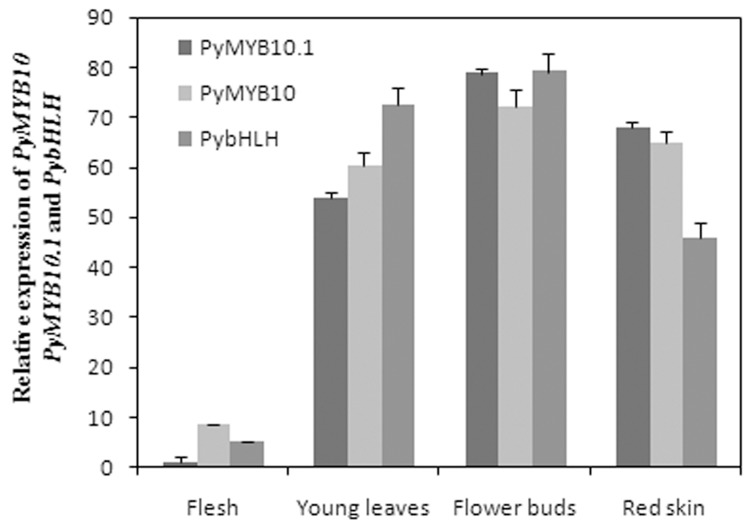
To determine the spatial expression patterns of PyMYB10 and PyMYB10.1 in ‘Aoguan’, the transcripts of both genes in young leaves, flower buds, skin and flesh was profiled. Both PyMYB10 and PyMYB10.1 mRNAs were detected in all four tissues, and higher levels were found in the anthocyanin-rich red tissues of young leaves, fruit skins and flower buds. Only extremely low levels of PyMYB10 and PyMYB10.1 were observed in non-red fruit flesh. Similarly to PyMYB10and PyMYB10.1, PybHLH was predominantly expressed in the anthocyanin-rich red tissues (Fig 3).
Fig 3. qPCR analysis of PyMYB10, PyMYB10.1 and PybHLH in different tissues of ‘Aoguan’ pears including young leaves, flower buds, red pericarps, and flesh.
PyActin was used as an internal control to normalize gene expression under identical conditions. Error bars represent means ± SE (n = 3).
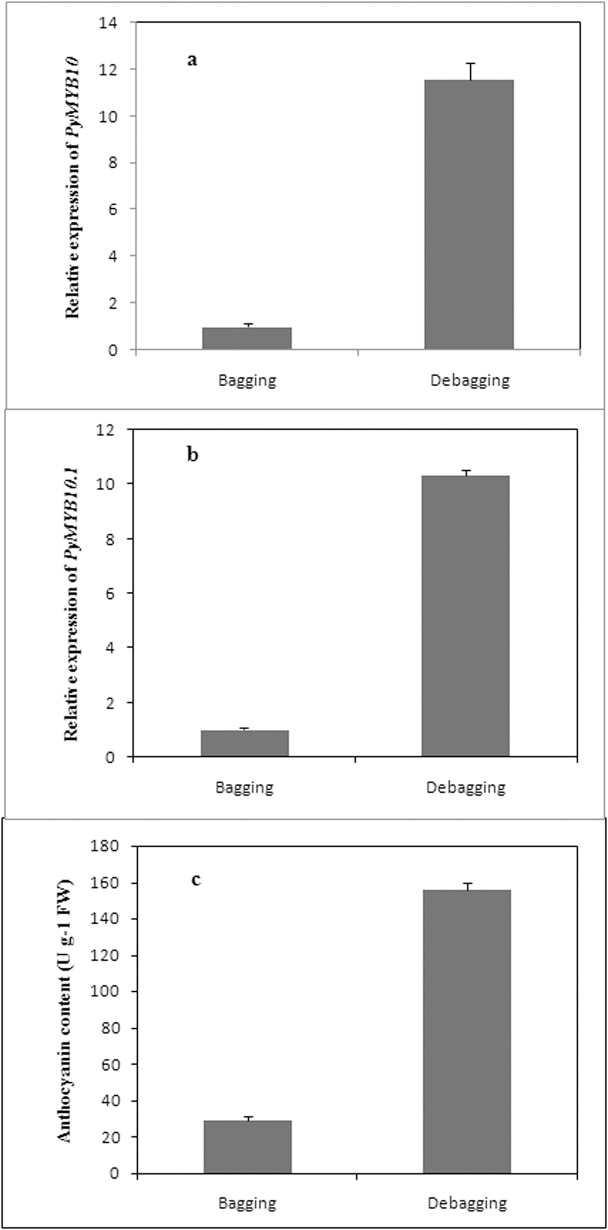
The expression levels of PyMYB10 and PyMYB10.1 in response to light treatment are shown in Fig 4. Compared to the bagged fruits, PyMYB10 and PyMYB10.1 were obviously upregulated in debagged fruit. The levels of the two genes were more than 11-foldhigher than in bagged fruit. Accordingly, the debagged fruits anthocyanin accumulation was obviously higher than bagged fruit (Fig 4).
Fig 4. Anthocyanin accumulation and expression analysis of PyMYB10 and PyMYB10.1 in ‘Aoguan’ pears in response to light.
(a) Expression analysis of PyMYB10 in ‘Aoguan’ pears during bagging and debagging treatments. (b) Expression analysis of PyMYB10.1 in ‘Aoguan’ pears during bagging and debagging treatments. (c) Anthocyanin contents in ‘Aoguan’ pears during bagging and debagging treatments. Fruits were collected 6 days after debagging. The fruits retained in the bags were sampled as controls. Error bars represent means ± SE (n = 3).
Protein–protein interactions of PyMYB10, PyMYB10.1 and PybHLHs
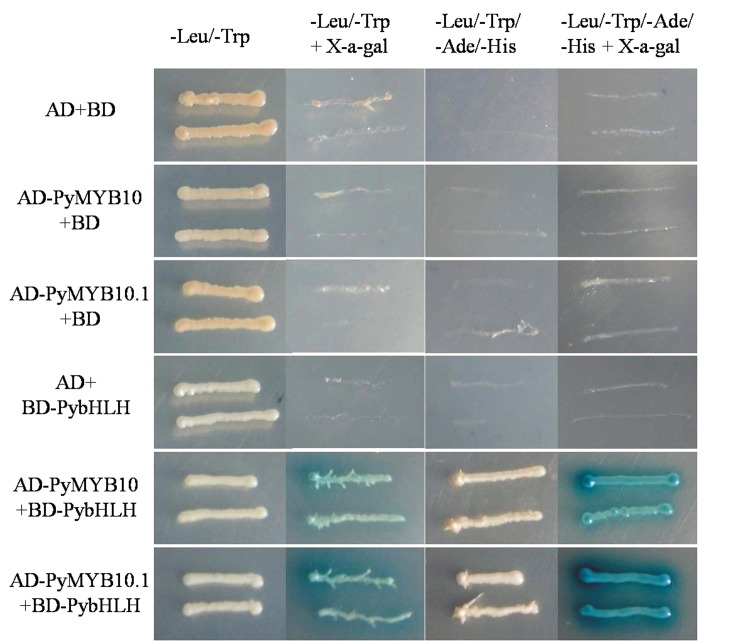
A motif ([DE]Lx2[RK]x3Lx6Lx3R) which is necessary for the interaction between MYB and bHLH proteins was found in the R3-MYB domains of PyMYB10 and PyMYB10.1 [34]. To investigate whetherPyMYB10 or PyMYB10.1interacts with the pear bHLH anthocyanin regulator PybHLH, we employed a GAL4-basedyeast two-hybrid assay. The autoactivation of PyMYB10 and PyMYB10.1 was investigated first. Yeast harboring pAD-GAL4 (AD) plus pBD-GAL4-PyMYB10 (BD-PyMYB10) or BD-PyMYB10.1 grew well on the quadruple-selection medium, while the negative control, which contained pBD-GAL4 (BD) and pAD-GAL4-PyMYB10 (AD-PyMYB10) or AD-PyMYB10.1, did not grow, indicating that PyMYB10and PyMYB10.1can auto-activate. Subsequently, AD-PyMYB10, AD-PyMYB10.1 and BD-PybHLH were introduced into yeast. As shown in Fig 5, yeast cells containing either the combination of PyMYB10and PybHLH or PyMYB10.1and PybHLH grew well on all synthetic dropout(SD) selective media. These Y2H results demonstrate that PyMYB10 and PyMYB10.1 can physically interact with PybHLH in vitro.
Fig 5. Interactions between PyMYB10 or PyMYB10.1 and PybHLH were detected through the yeast two-hybrid assay.
AH109 yeast cells containing plasmids AD+BD, AD-PyMYB10+BD, AD-PyMYB10.1+BD, AD+BD-PybHLH, AD-PyMYB10+BD-PybHLH, or AD-PyMYB10.1+BD-PybHLH were grown on double- and quadruple-selection media. The X-gal assay was performed to confirm positive interactions.
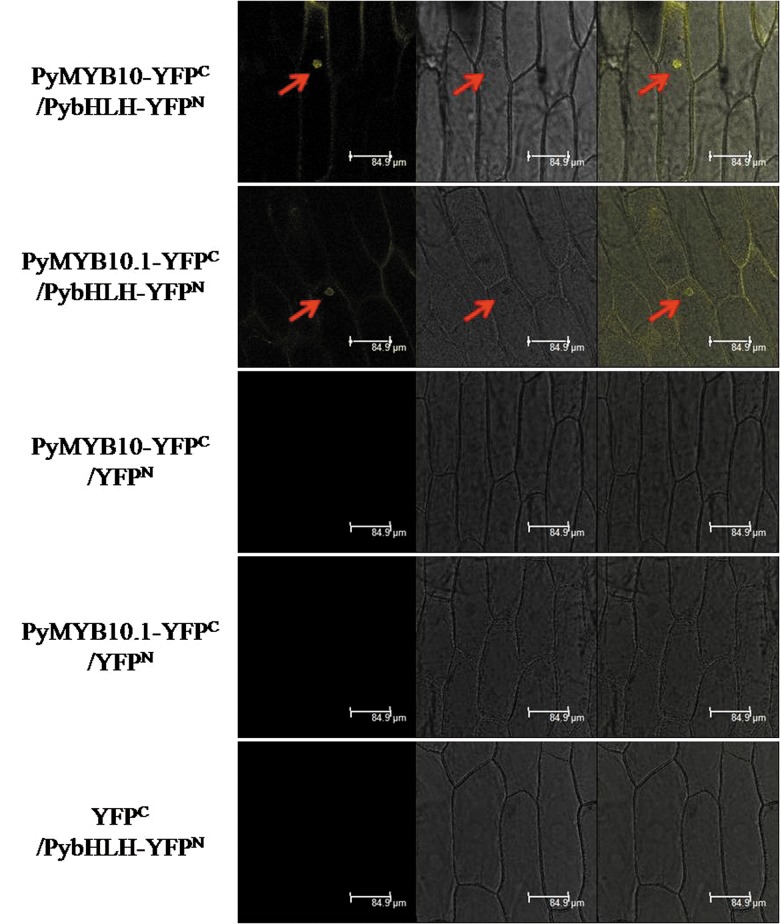
BiFC assays were further performed to investigate the interaction between PybHLH and PyMYB10or PyMYB10.1 in vivo. A plasmid harboring the N-terminal domain of YFP fused to the PyMYB10 (PyMYB10-NYFP) or PyMYB10.1 (PyMYB10.1-NYFP) cDNA and a plasmid containing the C-terminal domain of YFP fused to the PybHLH cDNA (PybHLH-CYFP) were transiently co-expressed in onion epidermis cells. Cells containing PyMYB10/PybHLH or PyMYB10.1/PybHLH were with a strong detectable fluorescence signal. Conversely, no fluorescence signal was detected in control cells (Fig 6).These BiFC results demonstrate that PyMYB10 and PyMYB10.1can interact with PybHLH in vivo.
Fig 6. Interactions between PyMYB10 or PyMYB10.1 and PybHLH were detected using the BiFC assay.
Onion epidermal cells were co-transfected with a mixture of Agrobacterium suspensions containing plasmids PyMYB10-YFPC + PybHLH-YFPN, PyMYB10.1-YFPC + PybHLH-YFPN, PyMYB10-YFPC + YFPN, PyMYB10.1-YFPC + YFPN, or YFPC + PybHLH-YFPN. YFP fluorescence signals were detected 48 h after transfection. Bar = 84.9 μm.
Transient luminescence assays of PyMYB10 and PyMYB10.1 activity
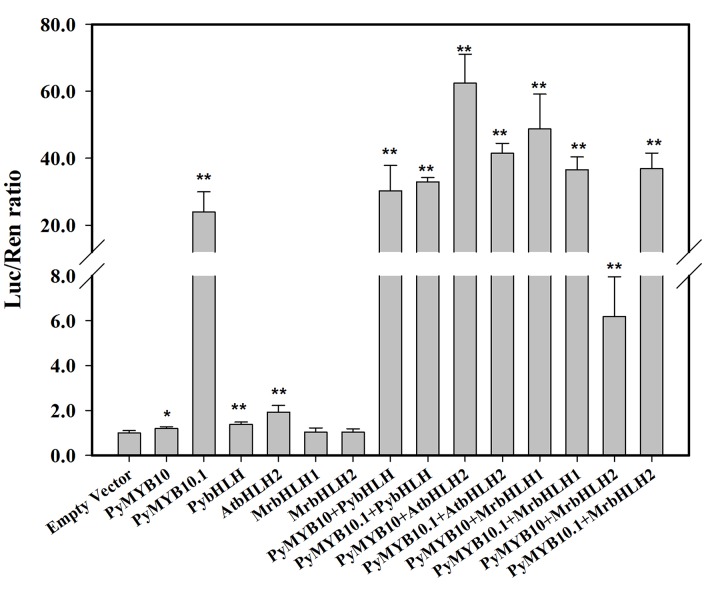
To assess transient PyMYB10 and PyMYB10.1activity,a transient luminescence assay was performed.PyMYB10and PyMYB10.1were infiltrated into N. benthamiana leaves with the AtDFR promoter. As shown in Fig 7, both PyMYB10 and PyMYB10.1 could induce the DFR promoter alone. Compared to control, the induction by PyMYB10 was 1.2-fold higher. But the induction by PyMYB10.1 was more obvious, approximately 20-fold higher than that by PyMYB10. The increased activities of PyMYB10 and PyMYB10.1 were observed when they were with abHLH co-factor, and the highest activity was observed when they were co-infected with AtbHLH2. In all combinations tested, the induction by PyMYB10+AtbHLH2 was the highest.
Fig 7. Activation of the AtDFR promoter by PyMYB10, PyMYB10.1 and anthocyanin-related bHLH transcription factors in a tobacco transient-expression assay.
LUC and REN activities were analyzed three days after transformation. Error bars represent means ± SE (n = 6).Asterisks, * and **, indicate significant differences (P<0.05 and P<0.01).
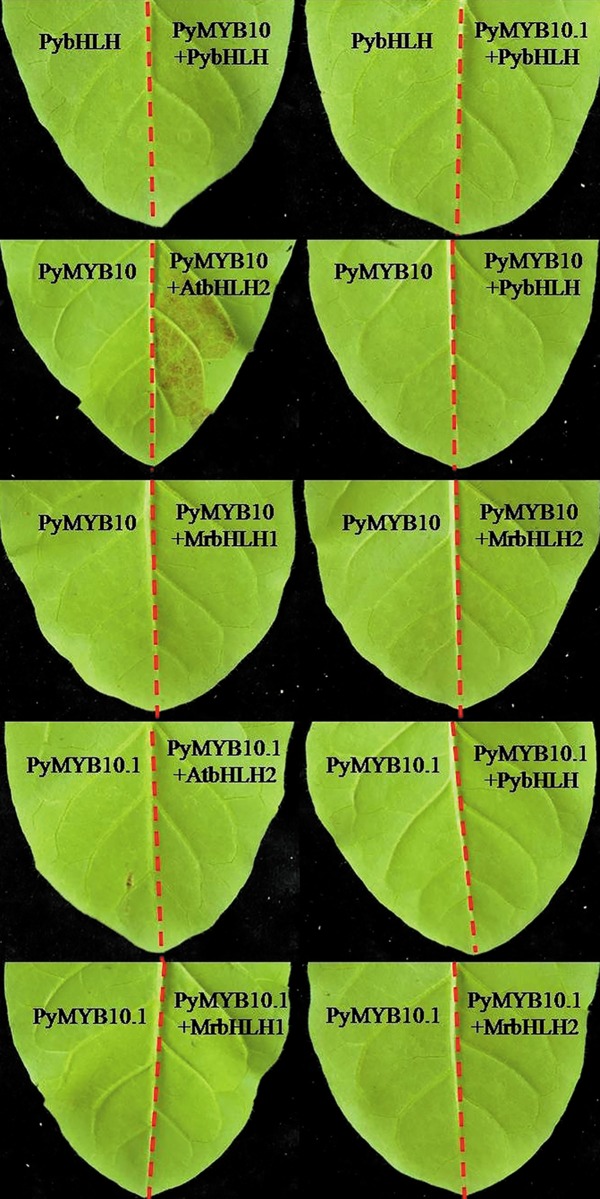
Patches of foliar anthocyanin production can reportedly be induced by the co-expression of R2R3-MYBs and bHLHs in Nicotiana tabacum leaves [11].In the present study, the induction of anthocyanin biosynthesis by PyMYB10 and PyMYB10.1was tested, and only PyMYB10 could induce a patch of anthocyanin when co-expressed with AtbHLH2 (Fig 8).
Fig 8. Patches of anthocyanin production in tobacco leaves infused with Agrobacterium tumefaciens.
Photos of infiltration areas were taken 8 days after transformation with PyMYB10 or PyMYB10.1 together with AtbHLH2, PybHLH, MrbHLH1, or MrbHLH2.
Discussion
Plant pigmentation is temporally and spatially regulated by the anthocyanin structural genes. The structural genes are primarily regulated at the transcriptional level. R2R3-MYBs play key roles in anthocyanin regulation through controlling the transcripts of anthocyanin structural genes. In recent years, numerous anthocyanin R2R3-MYB regulators in different fruit plants have been isolated and characterized. In the present study, a homologous of PyMYB10, PyMYB10.1, was cloned from ‘Aoguan’. A sequence analysis revealed that PyMYB10.1 shares a high sequence identity at R2R3-domain with anthocyanin-activating MYB transcription factors, particularly PyMYB10 from pears, but less homologous was detected at the full protein sequence. In a phylogenetic tree, PyMYB10.1 was closely related to anthocyanin-activating MYBI subgroup members, such as PyMYB10 [23], MdMYB10 [10] and FaMYB10 [35], suggesting that PyMYB10.1 may responsible for fruit anthocyanin regulation.
Previously, many R2R3-MYB TFs have been shown to display tissue-specific expression patterns that correlate strongly with anthocyanin accumulation. For example, the sweet potato IbMYB1 was predominantly expressed in red tuberous roots [36]. Similarly, high transcript levels of PyMYB10 and PyMYB10.1 were detected in anthocyanin-rich flower buds, young leaves and fruit skins. In contrast, low levels of PyMYB10 and PyMYB10.1transcripts were observed in the non-red fruit cortex. Therefore, the anthocyanin content must closely correlate with the PyMYB10 and PyMYB10.1transcript levels in a tissue-specific manner. Furthermore, the higher transcript levels of PybHLH in anthocyanin-rich tissues were also observed. It seems that both PyMYBs and PybHLHs act roles in the regulation of anthocyanin synthesis, like in other plants.
Environmental factors, such as temperature and light, have been suggested to regulate anthocyanin biosynthesis via the up- or down-regulation of these R2R3-MYB transcription factors. In the present study, we found that the transcripts of PyMYB10 and PyMYB10.1in pear skins were all up-regulated by sunlight, which is consistent with the expression ofMdMYB1 in apples [27] and MrMYB1 in Chinese bayberries [11]. Sunlight likely regulates pear anthocyanin synthesis by up-regulating the expression of PyMYB10 and PyMYB10.1.These results suggest that PyMYB10 and PyMYB10.1 likely act as anthocyanin activators in pears.
R2R3-MYBs regulate anthocyanin biosynthesis via activating the anthocyanin structural genes. Generally, the efficient induction of structural genes by R2R3-MYBs depends on the co-expression of a bHLH transcription factor, which jointly regulates target promoters. In apples, MdMYB10 could trans-activate the AtDFR promoter together with MdbHLH3 or MdbHLH33. The function of MdMYB10was weak in the absence of a bHLH co-factor[10].Similarly, the activity of the AtDFR promoter was higher when PyMYB10 and PyMYB10.1were co-expressed with a bHLH protein in tobacco than they were alone. Interestingly, the activity of the AtDFR promoter varied amongst the different combinations of R2R3-MYBs and bHLHs. For example, in apples, the combination of MdbHLH3 andMdMYB10resulted in a greater activation of AtDFR transcription compared with the combination of MdbHLH33and MdMYB10 [10]. As a result, these combinations led to different levels of anthocyanin. In the present study, the abilities of bHLH proteins with PyMYB10 or PyMYB10.1for the trans-activation of AtDFR depended on the species. In particular, the combination of PyMYB10 and AtbHLH2 showed trans-activation values more than ten-fold that of PyMYB10 and MrbHLH2. Clearly, the anthocyanin accumulation was detected in tobacco leaves co-infiltrated by the combination of PyMYB10 and AtbHLH2 but not by the combination of PyMYB10 and MrbHLH2. Therefore, the phenotypic differences may be partly attributed to the functional differences among these combinations. Surprisingly, the highly activation of AtDFR was detected in the combination of PyMYB10.1 and PybHLH, but none red pigmentation was induced. It has been reported that distinct anthocyanin structural genes may not be regulated by a single MYB protein or bHLH protein in several plants [37]. So, we speculated that the key structural genes that could not be regulated by PyMYB10.1 and PybHLH may exist. The anthocyanin structural genes should be further classified by experimental assays. Moreover, anthocyanin repressors may compete with anthocyanin activators [38]. Therefore, whether the negative regulators affect the anthocyanin accumulation process need to be clarified.
R2R3-MYBs are known to interact with bHLHs to control anthocyanin biosynthesis. In Arabidopsis, PAP1 could interact withEGL3. The motif ([DE]Lx2[RK]x3Lx6Lx3R) in the PAP1 R3-MYB domain is necessary for this interaction[17]. Similar results have shown that anthocyanin-related R2R3-MYBs containing this motif can interact with other heterologous EGL3-like bHLH proteins, such as MdMYB10 and MdbHLH3 from apples [10], VvMYC1 [39] and VvMYB5b from grapes [40], and BoMYB1 and BobHLH1 from purple cauliflower [41].In contrast, mutants of this motif abrogate the binding activity ofpurple cauliflowerBoMYB3 [41] and grape VvMYB5b [40] to BobHLH1 and VvMYC1, respectively. In the present study, a signature binding motif between MYB and bHLH proteins was also detected in both PyMYB10 and PyMYB10.1. Y2H and BiFC assays proved that both PyMYB10 and PyMYB10.1 interactwiththe recently identified pear bHLH TF PybHLH. Taking into account the expression patterns of PyMYB10, PyMYB10.1 and PybHLH, the MYB-bHLH regulatory complexes may play important roles in pear anthocyanin accumulation.
More than one R2R3-MYB TF is often present in a single plant species and determines tissue-specific anthocyanin accumulation. However, some R2R3-MYB proteins abundantly regulate anthocyanin biosynthesis in the same organs. For example, the R2R3-MYB TFsVvMYBA1 and VvMYBA2 from grape both appear to control fruit skin coloration. Mutations of these two genes removed their ability in activating anthocyanin biosynthesis, and deactivating both genes results in a white cultivar [42].Deficient AtPAP1 expression in Arabidopsis did not block the anthocyanin synthesis regulated by AtPAP2, AtMYB113 and AtMYB114 in vegetative tissues [13].In pears, we observed that both PyMYB10 and PyMYB10.1arepredominantly expressed in anthocyanin-rich tissues and up-regulated by light. In addition, all of these R2R3-MYBs form protein complexes with bHLHs to regulate anthocyanin structural genes. These results indicate that PyMYB10 and PyMYB10.1 may play redundant roles in pear anthocyanin regulation, as similarly reported in grapes and Arabidopsis. However, the anthocyanin regulatory ability of each protein significantly differed in the presence of a bHLH co-factor. Understanding the anthocyanin-activating functions of PyMYB10 and PyMYB10.1 will promote the biological breeding of red sand pears.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank Prof. Kunsong Chen, Xueren Yin and Dr. Xiaofen Liu from Zhejiang University for their kind help in transient luminescence assays.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant No.31201593).
References
- 1. Wu J, Wang ZW, Shi ZB, Zhang S, Ming R, Zhu SL, et al. (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23: 396–408. 10.1101/gr.144311.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tao B, Shu Q, Wang JJ, Zhang WB. (2004) Studies on frontiers and prospects of resources of red pears and its application. SouthwestChin J Agric Sci 47:409–412. [Google Scholar]
- 3. Steyn WJ, Wand SJE, Holcroft DM, Jacobs G.(2005) Red colour development and loss in pears. Acta Hortic 671:79–85. [Google Scholar]
- 4. Dayton DF. (1966) The pattern and inheritance of anthocyanin distribution in red pears. J Proc Am Soc Hortic Sci 89:110–116. [Google Scholar]
- 5. Dussi MC, Sugar D, Wrolstad RE. (1995) Characterizing and quantifying anthocyanins in red pears and effect of light quality on fruit color. J Am Soc Hortic Sci 120:785–789. [Google Scholar]
- 6. Fischer TC, Gosch C, Pfeiffer J, Halbwirth H, Halle C, Stich K et al. (2007) Flavonoid genes of pear (Pyrus communis). Trees 21:521–529. [Google Scholar]
- 7. Zhang X, Allan AC, Yi Q, Chen L, Li K, Shu Q et al. (2011) Differential gene expression analysis of yunnan red pear, PyrusPyrifolia, during fruit skin coloration. Plant Mol Biol Rep 29:305–314. [Google Scholar]
- 8. Yang YN, Zhao G, Yue WQ, Zhang SL, Gu C, Wu J. (2013) Molecular cloning and gene expression differences of the anthocyanin biosynthesis-related genes in the red/green skin color mutant of pear (Pyrus communis L.). Tree Genet Genomes 9:1351–1360. [Google Scholar]
- 9. Wu BH, Cao YG, Guan L, Xin HP, Li JH, Li SH. (2014) Genome-wide transcriptional profiles of the berry skin of two red grape cultivars (Vitis vinifera) in which anthocyanin synthesis is sunlight-dependent or -independent. PLOS One 9(8):e105959 10.1371/journal.pone.0105959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49(3):414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu XF, Yin XR, Allan AC, Lin-Wang K, Shi YN, Huang YJ et al. (2013) The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosynthetic genes in tobacco and Chinese bayberry (Myricarubra) during anthocyanin biosynthesis. Plant Cell Tiss Organ Cult 115:285–298. [Google Scholar]
- 12. Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. (2011) Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65:771–784. 10.1111/j.1365-313X.2010.04465.x [DOI] [PubMed] [Google Scholar]
- 13. Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827. [DOI] [PubMed] [Google Scholar]
- 14. Azuma A, Kobayashi S, Mitani N, Shiraishi M, Yamada M, Ueno T et al. (2008) Genomic and genetic analysis of MYB-related genes that regulate anthocyanin biosynthesis in grape berry skin. Theor Appl Genet 117(6):1009–1019. 10.1007/s00122-008-0840-1 [DOI] [PubMed] [Google Scholar]
- 15. Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A et al. (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28(3):319–332. [DOI] [PubMed] [Google Scholar]
- 16. Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J et al. (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis . EMBO J 19(22):6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. (2010) MYB transcription factors in Arabidopsis . Trends Plant Sci 15(10):573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- 18. Fornale S, Lopez E, Salazar-henao JE, Fernández-nohales P, Rigau J, Caparros-ruiz D. (2014) AtMYB7, a new player in the regulation of UV-Sunscreens in Arabidopsis thaliana . Plant Cell Physiol 55(3):507–516. 10.1093/pcp/pct187 [DOI] [PubMed] [Google Scholar]
- 19. Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K et al. (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10(2):135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R et al. (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183. 10.1105/tpc.108.059329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grotewold E, Chamberlin M, Snook M, Siame B, Butler L, Swenson J et al. (1998) Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10(5):721–740. [PMC free article] [PubMed] [Google Scholar]
- 22. Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E et al. (2006) A small family of MYB regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18:831–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng SQ, Wang YL, Y Song, Xu YT, Chen XS. (2010) Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232(1):245–255. 10.1007/s00425-010-1170-5 [DOI] [PubMed] [Google Scholar]
- 24. Pirie A, Mullins MG. (1976) Changes in anthocyanin and phenolics content of grapevine leaf and fruit tissues treated with sucrose, nitrate abscisic acid. Plant Physiol 58:468–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang HC, Huang XM, Hu GB, Huang HB.(2004) Studies on the relationship between anthocyanin biosynthesis and related enzymesin Litchi pericarp. Sci Agric Sin 37:2028–2032. [Google Scholar]
- 26. Cheng S, Puryear J, Cairney J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11:113–116. [Google Scholar]
- 27. Takos AM, Jaffe FW, Jacob SR, Bogs J, Robinson SP, Walker AR. (2006) Light induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142(3):1216–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumar S, Tamura K, Nei M. (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163. [DOI] [PubMed] [Google Scholar]
- 29. Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD et al. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang W, Sun W, Lv H, Luo M, Zeng S, Pattanaik S et al. (2013) A R2R3-MYB transcription factor from Epimedium sagittatum regulates the flavonoid biosynthetic pathway. PLOS One 8(8):e70778 10.1371/journal.pone.0070778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li M, Zhang Y, Zhang Z, Ji X, Zhang R, Liu D et al. (2013) Hypersensitive ethylene signaling and ZMdPG1expression lead to fruit softening and dehiscence. PLOS One 8:e58745 10.1371/journal.pone.0058745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. (2001) The Arabidopsis T T2gene encodes an R2R3MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. PlantCell 13(9):2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K et al. (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling.Plant J 50(4):660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40(1):22–34. [DOI] [PubMed] [Google Scholar]
- 35. Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK et al. (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae .BMC Plant Biol 10:50 10.1186/1471-2229-10-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mano H, Ogasawara F, Sato K, Higo H, Minobe Y. (2007) Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol 143:1252–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bruce W, Folkerts O, Garnaat C, Crasta O, Roth B, Bowen B. (2000) Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 12:65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matsui K, Umemura Y, Ohme-Takagi M. (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis . Plant J 55:954–967. 10.1111/j.1365-313X.2008.03565.x [DOI] [PubMed] [Google Scholar]
- 39. Hichri I, Heppel SC, Pillet J, Léon C, Czemmel S, Delrot S et al. (2010) The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. MolPlant 3:509–523. [DOI] [PubMed] [Google Scholar]
- 40. Hichri I, Deluc L, Barrieu F, Bogs J, Mahjoub A, Regad F et al. (2011)A single amino acid change within the R2 domain of the VvMYB5b transcription factor modulates affinity for protein partners and target promoters selectivity. BMC Plant Biol 11:117 10.1186/1471-2229-11-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chiu LW, Li L. (2012) Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta 236:1153–1164. [DOI] [PubMed] [Google Scholar]
- 42. Walker AR, Lee E, Bogs J, McDavid DAJ, Thomas MR, Robinson SP. (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49:772–785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.