Abstract
Environmental DNA (eDNA) sampling is a powerful tool for detecting invasive and native aquatic species. Often, species of conservation interest co-occur with other, closely related taxa. Here, we developed qPCR (quantitative PCR) markers which distinguish westslope cutthroat trout (Oncorhynchus clarkii lewsi), Yellowstone cutthroat trout (O. clarkii bouvieri), and rainbow trout (O. mykiss), which are of conservation interest both as native species and as invasive species across each other’s native ranges. We found that local polymorphisms within westslope cutthroat trout and rainbow trout posed a challenge to designing assays that are generally applicable across the range of these widely-distributed species. Further, poorly-resolved taxonomies of Yellowstone cutthroat trout and Bonneville cutthroat trout (O. c. utah) prevented design of an assay that distinguishes these recognized taxa. The issues of intraspecific polymorphism and unresolved taxonomy for eDNA assay design addressed in this study are likely to be general problems for closely-related taxa. Prior to field application, we recommend that future studies sample populations and test assays more broadly than has been typical of published eDNA assays to date.
Introduction
Environmental DNA (eDNA) sampling is the search for genetic material in the environment (e.g., water or soil) to infer species presence [1]. This approach is particularly well suited to detecting aquatic species when they are rare, such as small populations of endangered native species or new invasions of introduced species (e.g., recent reviews include [2–4]). Many recent applications of eDNA sampling have focused on one to several taxa, using cost-effective species-diagnostic mitochondrial markers (i.e., species-specific PCR; e.g., [5,6]). The ideal taxon-specific marker will have both high specificity (amplifying only the DNA of the target taxon) and broad generality (amplifying the DNA of all populations of the target taxon across its range). Attaining both goals, however, can be challenging [7]. Achieving specificity, for example, has been problematic when attempting to distinguish among closely related taxa. For example, Wilcox et al. [7] found that insufficient specificity in a quantitative PCR (qPCR) assay could result in reduced detectability for the target species in mixed samples of closely related chars (salmonid fishes in the genus Salvelinus). Conversely, Fukumoto et al. [8] were unable to reliably distinguish the eDNA from two closely related and hybridizing salamanders (Andrias spp.) using qPCR, and resorted to additional sequencing to confirm species identity. Alternatively, achieving generality in an eDNA assay is difficult for taxa with substantial phylogenetic structure because some populations or clades may exhibit polymorphisms that reduce assay sensitivity. For example, Goldberg et al. [9] designed species-specific hydrolysis markers (TaqMan) to detect invasive New Zealand mudsnail (Potamopyrgus antipodarum). After developing their assay, a new haplotype with a polymorphism within the locus of one of the assay primers was discovered. They confirmed that this polymorphism did not affect assay sensitivity, but point out that if this polymorphism had been on the 3’ end of an amplification primer there could have been false negative results from environmental samples with low quantities of DNA. Detecting these polymorphisms requires examination of samples of a target taxon from across its range (e.g., [10]).
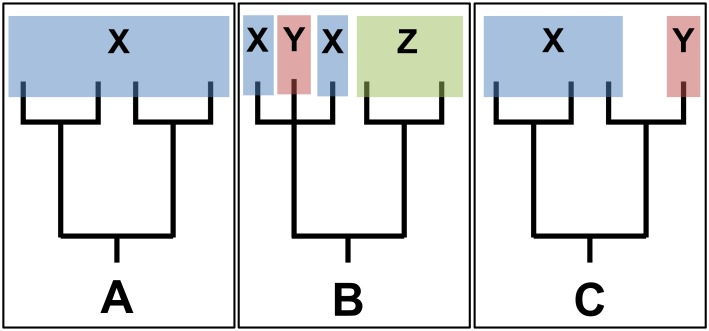
Additional challenges in building eDNA assays arise when there are conflicts between taxonomy and phylogenetic relationships, or when phylogenetic relationships are unresolved. Developing an assay with generality within a taxon may be difficult or impossible when divergent lineages are assigned to a single taxon (Fig 1A). Conversely, there may be few or no diagnostic loci that distinguish among taxa lacking phylogenetic divergence (e.g., paraphyly; Fig 1B). These scenarios are not mutually exclusive—some taxa may comprise markedly divergent lineages (polyphyly), yet also lack divergence from other named taxa (Fig 1C).
Fig 1. Environmental DNA marker development may be difficult or impossible when the target taxon’s taxonomy is unresolved or has poor concordance with true phylogeny.
Colored boxes show recognized taxa and black lines show the phylogenetic relationships among populations. (A) Taxon X includes multiple, divergent lineages. It is important that all of these lineages are sampled during marker validation to insure intraspecific generality. Even extensive sampling of a single lineage (red circle) is not sufficient and will lead to ascertainment bias and low marker generality. (B) Recognized taxa X and Y are not monophyletic (i.e., they are both paraphyletic groups), which may make distinguishing between them using sequence data impossible. (C) Recognized taxon X includes divergent lineages (polyphyly), but some of those lineages may also not be distinguishable from recognized taxon Y.
We confronted these issues in developing qPCR assays for eDNA detection of three salmonid taxa native to western North America: rainbow trout (RBT; Oncorhynchus mykiss) and two subspecies of cutthroat trout (O. clarkii), westslope cutthroat trout (WCT; O. c. lewisi) and Yellowstone cutthroat trout (YCT; O. c. bouvieri). Rainbow trout are the sister species to cutthroat trout (genetic distance = 7.3–8.7% in NADH2), whereas WCT and YCT (genetic distance = 2.7%; [11]) are subspecies both found in interior western North America [12]. In portions of the Columbia River basin, RBT and WCT naturally co-occur, but introductions of all three taxa outside their historical ranges have led to far greater recent sympatry [12]. Despite global increases in their distributions, the distribution of native populations of these three taxa have declined [13–15]. Consequently, identifying the current distributions of both native and introduced populations is a priority for management agencies [13,16,17]. Environmental DNA sampling could be a useful tool to detect the presence of each taxon, whether at the leading edge of an invasion or a remnant native population. Because RBT, WCT, and YCT are closely related and commonly co-occur, highly specific eDNA assays are critical for robust detection, particularly where the target taxon is rare and one or both of the others are common [7]. To address these challenges, we examined DNA sequences and samples from across the historical ranges of the two cutthroat trout subspecies and from an array of native populations and hatchery stocks of rainbow trout, and used an automated pipeline to generate and test candidate assays for each taxa.
Methods
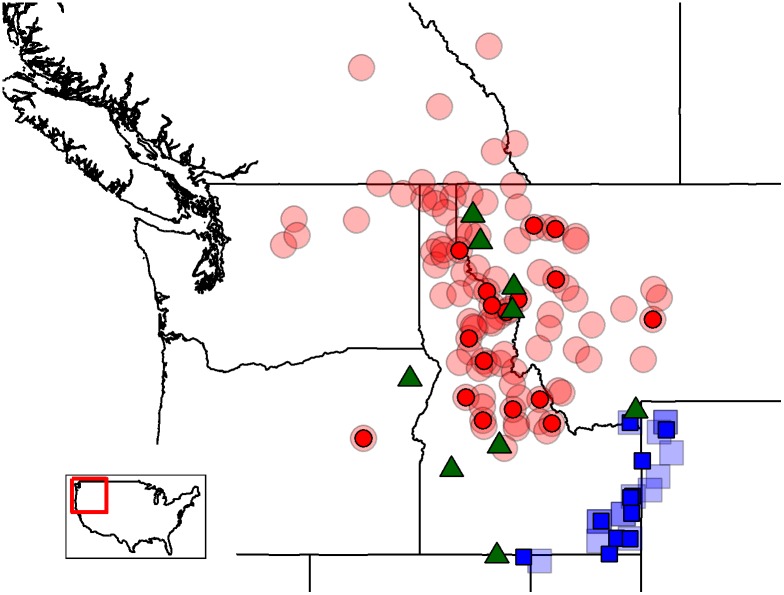
Sequence data for assay development
We assembled sequence data for our target taxa and three other congeners—Chinook salmon (O. tshawytscha), coho salmon (O. kisutch), and sockeye salmon (O. nerka)–to design qPCR assays in the NADH region of the mitogenome. We used NADH because it tends to show relatively high sequence divergence and is one of the most commonly archived mitochondrial sequences for salmonids. For YCT, we obtained sequences from a 3,483-bp region of the NADH subunits 1 and 2 for 17 individuals from across its range ([18]; GenBank accession numbers: EU186781.1 –EU186797.1; Table 1 and Fig 2). We obtained these same gene regions from whole mitogenomes for five RBT from Pacific North America (GenBank accession numbers: DQ288268.1 –DQ288271.1, L29771.1) and each of the Pacific salmon congeners listed above (GenBank accession numbers: AF392054.1 and NC_002980.1 for Chinook, EF126369.1 for coho, and EF055889.1 for sockeye). We used previously unpublished sequences from 96 WCT from across the species’ range (S1 Text). All tissue samples used in this study were stored in ethanol, lysis buffer, or dried on chromatography paper. All samples used in this study were provided by collaborators from previous studies conducted under appropriate scientific sampling permits or from collections under Montana Fish, Wildlife and Parks Scientific Collectors Permits 12–2001, 14–2001, and 19a-2009 issued to M. K. Young (Table 1, Fig 2). Sampling sites were accessed via public land and did not require special permission. Sampling of protected species was allowed under U.S. Fish and Wildlife Service Federal Fish and Wildlife Permit TE220826-0 issued to M. K. Young. Animals were captured via backpack electrofishing, a small fin tissue sample was collected, and then the animal was released at the place of capture in accordance with a protocol approved under these scientific sampling permits. DNA was extracted from these samples using QIAGEN DNeasy Blood and Tissue Kit following the manufacturer’s protocol.
Table 1. Source populations for fish used in assay design and testing.
| Species | Souce | Purpose |
|---|---|---|
| Yellowstone cutthroat trout | Clear Cr (Idaho, USA) | Sequence data; assay testing |
| Barnes Cr (Idaho, USA) | Sequence data; assay testing | |
| Goose Cr (Nevada, USA) | Sequence data; assay testing | |
| Cottonwood Cr, (Idaho, USA) | Sequence data; assay testing | |
| Upper Blackfoot R (Montana, USA) | Sequence data; assay testing | |
| Badger Cr (Idaho, USA) | Sequence data; assay testing | |
| Yellowstone L (Wyoming, USA) | Sequence data; assay testing | |
| Bonneville cutthroat trout | Bear L (Utah, USA) | Sequence data; assay testing |
| Bear R (Idaho, USA) | Sequence data; assay testing | |
| Glenwood FH (Utah, USA)* | Sequence data; assay testing | |
| Harkness Cr (Idaho, USA) | Sequence data; assay testing | |
| Rainbow trout | Skookumchuck R FH (Washington, USA)* | Sequence data |
| Gulkana R (Alaska, USA)* | Sequence data | |
| Dworshak FH (Idaho, USA)* | Sequence data | |
| Ennis FH—Arlee (Montana, USA)* | Assay testing | |
| Ennis FH—Eagle L (Montana, USA)* | Assay testing | |
| Ennis FH—Fish L (Montana, USA)* | Assay testing | |
| Ennis FH—Shasta (Montana, USA)* | Assay testing | |
| Ennis FH—McConaughy (Montana, USA)* | Assay testing | |
| Dry Cr (Idaho, USA) | Assay testing | |
| Sawtooth FH (Idaho, USA)* | Assay testing | |
| Shack Cr (Idaho, USA) | Assay testing | |
| Wallowa FH (Idaho, USA)* | Assay testing | |
| Bobtail Cr (Montana, USA) | Assay testing | |
| Silver Butte Fisher R (Montana, USA) | Assay testing | |
| E.F. Lolo Cr (Montana, USA) | Assay testing | |
| McCormick Cr (Montana, USA) | Assay testing | |
| Red Canyon (Montana, USA) | Assay testing | |
| Westslope cutthroat trout | John Day R (Oregon, USA) | Sequence data; assay testing |
| NF Elkhorn Cr (Idaho, USA) | Sequence data; assay testing | |
| Split Cr (Idaho, USA) | Sequence data; assay testing | |
| Withington Cr (Idaho, USA) | Sequence data; assay testing | |
| Duck Cr (Idaho, USA) | Sequence data; assay testing | |
| Heller Cr (Idaho, USA) | Sequence data; assay testing | |
| Rampike Cr (Idaho, USA) | Sequence data; assay testing | |
| Ditch Cr (Idaho, USA) | Sequence data; assay testing | |
| Crooked Fork Cr (Idaho, USA) | Sequence data; assay testing | |
| Pete King Cr (Idaho, USA) | Sequence data; assay testing | |
| Osier Cr (Idaho, USA) | Sequence data; assay testing | |
| Albert Cr (Montana, USA) | Sequence data; assay testing | |
| Fourmile Cr (Montana, USA) | Sequence data; assay testing | |
| Youngs Cr (Montana, USA) | Sequence data; assay testing | |
| Wounded Buck Cr (Montana, USA) | Sequence data; assay testing | |
| Twentyfivemile Cr (Montana, USA) | Sequence data; assay testing | |
| Flat Cr (Idaho, USA) | Sequence data | |
| Meadow Cr (Idaho, USA) | Sequence data | |
| French Cr (Idaho, USA) | Sequence data | |
| Baldy Cr (Idaho, USA) | Sequence data | |
| Twisp R (Washington, USA) | Sequence data | |
| Buck Cr (Washington, USA) | Sequence data | |
| American R (Idaho, USA) | Sequence data | |
| Falls Cr (Washington, USA) | Sequence data | |
| Hungery Cr (Idaho, USA) | Sequence data | |
| Cayuse Cr (Idaho, USA) | Sequence data | |
| Indian Grave Cr (Idaho, USA) | Sequence data | |
| Gravey Cr (Idaho, USA) | Sequence data | |
| Beaver Cr (Montana, USA) | Sequence data | |
| Moose Cr (Montana, USA) | Sequence data | |
| Deer Cr (Montana, USA) | Sequence data | |
| EF Bull R (Montana, USA) | Sequence data | |
| Ketchikan Cr (Montana, USA) | Sequence data | |
| McGuire Cr (Montana, USA) | Sequence data | |
| Martin Cr (Montana, USA) | Sequence data | |
| McCabe Cr (Montana, USA) | Sequence data | |
| Miller Cr (Montana, USA) | Sequence data | |
| Norton Cr (Montana, USA) | Sequence data | |
| Ontario Cr (Montana, USA) | Sequence data | |
| North Cr (Montana, USA) | Sequence data | |
| Tyler Cr (Montana, USA) | Sequence data | |
| Wilkes Cr (Montana, USA) | Sequence data | |
| Canuck Cr (Idaho, USA) | Sequence data | |
| Ball Cr (Idaho, USA) | Sequence data | |
| West Gold Cr (Idaho, USA) | Sequence data | |
| Beaver Cr (Idaho, USA) | Sequence data | |
| Mokins Cr (Idaho, USA) | Sequence data | |
| NF St. Joe R (Idaho, USA) | Sequence data | |
| Skin Cr (Idaho, USA) | Sequence data | |
| Slowey Cr (Montana, USA) | Sequence data | |
| Dry Wolf Cr (Montana, USA) | Sequence data | |
| Tributary of Armstrong Cr (Idaho, USA) | Sequence data | |
| Bad Luck Cr (Idaho, USA) | Sequence data | |
| Cedar Cr (Washington, USA) | Sequence data | |
| Ninemile Cr (Washington, USA) | Sequence data | |
| Scotchman Gulch (Montana, USA) | Sequence data | |
| Kraft Cr (Montana, USA) | Sequence data | |
| Leiberg Cr (Idaho, USA) | Sequence data | |
| N. Grouse Cr (Idaho, USA) | Sequence data | |
| SF Red R (Idaho, USA) | Sequence data | |
| Yoosa Cr (Idaho, USA) | Sequence data | |
| Ross Cr (Montana, USA) | Sequence data | |
| EF Emerald Cr (Idaho, USA) | Sequence data | |
| Bitter Cr (B.C., Canada) | Sequence data | |
| Blairmore Cr (B.C., Canada) | Sequence data | |
| Crazy Cr (B.C., Canada) | Sequence data | |
| Hartley Cr (B.C., Canada) | Sequence data | |
| Monk Cr (B.C., Canada) | Sequence data | |
| Pack R (Idaho, USA) | Sequence data | |
| Sheep Cr (Montana, USA) | Sequence data | |
| Toby Cr (B.C., Canada) | Sequence data | |
| Truman Cr (Montana, USA) | Sequence data | |
| Twin Cr (Idaho, USA) | Sequence data | |
| Waiparous Cr (B.C., Canada) | Sequence data | |
| Werner Cr (Montana, USA) | Sequence data | |
| Bostwick Cr (Montana, USA) | Sequence data | |
| Avalanche Cr (Montana, USA) | Sequence data | |
| Fish Cr (Montana, USA) | Sequence data | |
| NF Dupuyer Cr (Montana, USA) | Sequence data | |
| Sawmill Cr (Montana, USA) | Sequence data | |
| Fourmile Cr (Montana, USA) | Sequence data | |
| NF Teton R (Montana, USA) | Sequence data | |
| Thayer Cr (Montana, USA) | Sequence data | |
| Buffalo Cr (Montana, USA) | Sequence data | |
| Bluff Cr (Idaho, USA) | Sequence data | |
| Trail Cr (Idaho, USA) | Sequence data | |
| Floodwood Cr (Idaho, USA) | Sequence data | |
| Jacobs Ladder Cr (Idaho, USA) | Sequence data | |
| Blackbird Cr (Idaho, USA) | Sequence data | |
| Mill Cr (Idaho, USA) | Sequence data | |
| Morse Cr (Idaho, USA) | Sequence data | |
| MF Little Timber Cr (Idaho, USA) | Sequence data | |
| Big Cr (Idaho, USA) | Sequence data | |
| Boundary Cr (Idaho, USA) | Sequence data | |
| Colson Cr (Idaho, USA) | Sequence data | |
| Warm Springs Cr (Idaho, USA) | Sequence data | |
| Chinook salmon | SF Salmon R (Idaho, USA)* | Assay testing |
| Pahsimeroi R (Idaho, USA)* | Assay testing | |
| Clearwater R (Idaho, USA)* | Assay testing | |
| Coho salmon | Nez Perce Tribal FH (Idaho, USA)* | Assay testing |
| Sockeye salmon | Redfish L (Idaho, USA)* | Assay testing |
Locations marked with an asterisk are not shown in Fig 2. Sequencing; sequences from fish were used for initial assay design, Assay testing; extracted tissue samples from fish were used for assay testing.
Fig 2. Source locations for rainbow trout (green triangles), westslope cutthroat trout (red circles), and Yellowstone/Bonneville cutthroat trout (both subspecies indicated with blue squares) sampled from the wild for assay development (large, light-colored shapes) and for assay testing (small, dark-colored shapes).
The source locations for one rainbow trout sample obtained from Alaska (USA), all hatchery-derived fish, and the Pacific salmon congeners used in assay testing are not shown here, but are listed in Table 1.
Primer development
We used the DECIPHER package [19] in R v. 3.0.1 [20] to generate candidate primer sets for the NADH gene region for each species using the sequence data from above. We visually compared alignments of these candidate primers with consensus sequences of all Oncorhynchus species in MEGA5 [21], and adjusted primer length to optimize annealing temperature in Primer Express v. 3 (Life Technologies).
Candidate primer sets (n = 3, 4, and 5 for WCT, YCT, and RBT respectively) were tested against extracted DNA from tissues of three individuals of each Oncorhynchus taxon in 20-μl reactions composed of 0.1 ng of DNA (extracted from tissue using QIAGEN DNEasy Blood and Tissue Kit following the manufacturer’s protocol and quantified on a NanoPhotometer; IMPLEN), 1X concentration of SYBR Green PCR Mastermix (Life Technologies) and each primer at 150 nM. We used cycling conditions of 95°C/10 min [95°C/15 s, 60°C/60 s] × 45 cycles on a StepOne Plus Real-time PCR Instrument (Life Technologies), followed by a melt curve from 65°C to 95°C in 0.3°C increments (to test for primer dimer formation). Candidate primer sets for all three target species were tested both with and without an induced mismatch (single base pair mismatch with both target and non-target sequences) in the reverse primer six nucleotides from the 3' end to increase target specificity (i.e., reduce hybridization with non-target sequences [22]). To determine if this induced mismatch influenced sensitivity, we compared amplification curves of primer sets both with and without the mismatch. In all cases, there was no detectable difference in cycle threshold (Ct) when using DNA from the target taxon, but an increased delay in amplification when using DNA from non-target taxa, so we used primer sets with the induced mismatch for probe design.
Probe development and assay optimization
We used PrimerExpress 3.0 (Life Technologies) to design eight hydrolysis probes (TaqMan-MGB probes) for the seven most target-specific primer sets (n = 2, 2, and 4 probes and 2, 2, and 3 primer sets for WCT, YCT, and RBT respectively). We optimized primer concentrations for primer-limiting reactions to increase assay specificity and ease of future multiplexing. This was done by independently varying final primer concentrations to 100, 300, 600, and 900 nM (n = 16 combinations; probe concentration held at 250 nM). We used the lowest primer concentrations that also resulted in the lowest Ct value while maintaining high end-point fluorescence relative to the highest primer concentration for each assay for further testing. Screening of the assays with probes were done in 15-μl reaction volumes composed of ~ 0.1 ng of DNA, a final concentration of 1X Environmental Mastermix 2.0 (Life Technologies), and optimized concentrations of the primers and probe (Table 2) on a StepOne Plus Real-time PCR Instrument (Life Technologies) using the same cycling conditions as above (except without a final melt curve step).
Table 2. Sequences for each of the validated taxon-specific qPCR assays.
| Species | Oligo | Sequence | Rxn [] (nM) | Amplicon ln |
|---|---|---|---|---|
| westslope cutthroat | F | 5’-CCTAAAACTATTTATTAAAGAACCAGTTCG-3’ | 100 | 88 |
| R | 5’-AAGTGTAAGGGCGAGTCTRGGG-3’ | 900 | ||
| P | 6FAM-5’-CCACCTCCTCTCCCT-3’ -MGBNFQ | 250 | ||
| Yellowstone cutthroat | F | 5’-CGACCTTCCACCTCCTCC-3’ | 600 | 152 |
| R | 5’-AGCTAGACTGGATAGCTCAAGC-3’ | 900 | ||
| P | 6FAM-5’-CTCGCCACACCTATACT-3’ -MGBNFQ | 250 | ||
| rainbow trout | F | 5’-AGTCTCTCCCTGTATATCGTC-3’ | 300 | 102 |
| R | 5’-GATTTAGTTCATGAAGTTGCGTGAGTA-3’ | 600 | ||
| P | 6FAM-5’-CCAACAACTCTTTAACCATC-3’ -MGBNFQ | 250 |
Target species, oligonucleotide (forward primer; F, reverse primer R, probe; P), oligonucleotide sequence, and optimized reaction concentration of the oligonucleotide (nM), and amplicon length (bp). The induced mismatch (mismatch between both target and non-target taxa to increase specificity) in each reverse primer is underlined.
We screened these assays against the same individuals above for specificity, as well as samples of three commonly sympatric salmonids: brook trout (Salvelinus fontinalis), bull trout (Salvelinus confluentus), and brown trout (Salmo trutta; Morrell Creek, Montana, USA; N 47.1779 W -113.4699) to select assays for final screening. All assays that did not amplify DNA from non-target species were tested for generality by screening against tissue samples from across the northwestern U.S. range of each of the three target taxa (n = 30 each), including collections from seven RBT hatchery strains and samples covering most of the range of each subspecies of cutthroat trout (Table 1 and Fig 2). The panel for YCT also included 11 individuals of the closely related Bonneville cutthroat trout (BCT; O. c. utah; [11]). We also tested for non-target template competition [7] by amplifying mixed samples in which the DNA of a target taxon made up 1% and DNA of the other five Oncorhynchus made up 99% of the sample (i.e., approximately 0.001 ng of target DNA and 0.099 ng of non-target DNA).
Assay sensitivity
To determine the amplification efficiency and limit of detection for a final set of three taxon-specific assays, we performed standard curve experiments for each. For each assay, a synthetic template was prepared by ordering a synthetic gene from Integrated DNA Technologies that included the target amplicon sequence. The lyophilized gene was resuspended in sterile TE, linearized with a Pvu1 restriction digest, purified, and quantified on a Quibit 2.0 fluorometer (ThermoFisher Scientific; see [23] for details). From this stock, a five-level dilution series (6 250, 1 250, 250, 50, and 10 copies/4 μl) was prepared in sterile TE. We ran six replicates of each dilution series for each assay to determine the standard curve slope (amplification efficiency) and limit of detection (lowest concentration with >95% amplification success [24]).
Results
We optimized a final set of three assays—one for each target taxon (Table 2). These assays had no amplification of non-target DNA, except that the YCT assay also amplified DNA from closely-related Bonneville cutthroat trout (Table 3). Assays were not influenced by non-target template competition (i.e., all mixed samples had amplification of the target DNA). However, the RBT and WCT assays did have reduced amplification efficiency or failed amplification for some individuals within each taxon (Table 3).
Table 3. Summary of results of the validated taxon-specific qPCR assays.
| Assay | Problematic polymorphism |
|---|---|
| Yellowstone cutthroat | Bonneville cutthroat trout also amplify |
| westslope cutthroat | Rare polymorphism caused some Youngs Cr individuals not to amplify |
| rainbow trout | Delayed amplification for individuals from Eagle Lake (O. mykiss aquilarium) |
Except for the noted problematic polymorphisms, each assay was taxon-specific and amplified all individuals of the target taxon with similar efficiency.
Yellowstone cutthroat trout
The YCT panel of 30 fish for assay testing included both YCT and BCT individuals, which are recognized as separate subspecies, but which have little genetic divergence across much of their range [11]. DNA from all of these individuals amplified, but amplification efficiency was substantially decreased (~ 10 Ct amplification curve delay) for BCT individuals from Harkness Creek and Glenwood Fish Hatchery (n = 3 and 2 individuals, respectively; six BCT individuals from Bear Lake and Bear River all amplified efficiently). The mtDNA haplotypes of individuals sampled from these Harkness Creek and Glenwood Fish Hatchery populations have three base-pair polymorphisms within the reverse primer binding region of our assay (7, 16, and 19 bp from the 3’ end of the primer [18]). These base-pair differences reduce primer hybridization, resulting in the substantially increased Ct for these individuals. Based on the standard curve experiments, this assay had an amplification efficiency of 95.5% (standard curve y-intercept = 43.4, r 2 = 0.965) and limit of detection of 10 mtDNA copies/rxn.
Westslope cutthroat trout
The marker for WCT was designed with a degenerate base in the reverse primer to account for a known intraspecific polymorphism. All of the 30 individuals in the screening panel for this assay amplified with the same efficiency. However, after assay design, we discovered another polymorphism within the probe-binding region of this assay in a sequence from a single fish captured in Youngs Creek (tributary to the South Fork Flathead River, Montana, USA) which we had not previously noted. We tested the assay against ten individuals from this population. Six individuals amplified as expected, but we observed complete amplification failure of the remaining four fish. There was 100% amplification success in an additional 20 fish screened from within the same river basin (ten each from two other streams < 100 km downstream in the Flathead River basin, Montana, USA). Based on the standard curve experiments, this assay had an amplification efficiency of 96.3% (standard curve y-intercept = 40.4, r 2 = 0.971) and limit of detection of 50 mtDNA copies/rxn.
Rainbow trout
The marker for RBT amplified all 30 individuals in the screening panel; however, the two individuals from the Eagle Lake hatchery strain (Table 1) had reduced amplification efficiency (~ 2 Ct delay in amplification). This commonly stocked strain is sourced from an isolated lake population and may represent its own subspecies (O. mykiss aquilarum; [25]). Testing against additional fish from this same hatchery strain (n = 5) found this Ct delay to be consistent across fish from the Eagle Lake strain, indicating the presence of an intraspecific polymorphism in one of the primers (confirmed by additional sequencing to be due to a polymorphism on the 3’ end of the reverse primer; n = 5 Eagle Lake individuals, S1 Text). Based on the standard curve experiments, this assay had an amplification efficiency of 95.9% (standard curve y-intercept = 40.7, r 2 = 0.998) and limit of detection of 10 mtDNA copies/rxn.
Discussion
We developed taxon-specific eDNA markers for two subspecies of cutthroat trout and for rainbow trout. These markers did not result in cross-amplification among these three taxa or other congeneric (Oncorhynchus) or confamilial (Salmonidae) species, even when target DNA constituted a minute fraction of the DNA in laboratory mixtures. Using software that models PCR chemistry in conjunction with induced base-pair mismatches and primer-limiting reactions allowed us to distinguish sequences that differ at only a few loci between RBT, WCT, and YCT.
Our emphasis on obtaining samples from throughout most of their respective geographic ranges enabled us to design assays capable of detecting many populations of the target taxa. Nevertheless, we discovered intraspecific polymorphisms among populations of two taxa that reduced assay sensitivity and could lead to less consistent or failed detections in field samples (i.e. reduced intraspecific generality), and a lack of specificity in one assay that could lead to ambiguity with regard to which subspecies of cutthroat trout was present.
Our RBT marker efficiently amplified fish from multiple coastal rainbow trout (O. mykiss irideus) and Columbia redband trout/steelhead (O. mykiss gairdneri) strains. Reduced sensitivity (~2 Ct amplification curve delay) was only found in a single hatchery strain (Eagle Lake) that may represent its own subspecies [25]. This issue is expected when a single taxon (O. mykiss) includes multiple, divergent lineages (Fig 1A). In the case of WCT, the polymorphism in the Youngs Creek sample which prevented amplification of some individuals could not be predicted based on previous phylogeographic work [11,26]. It was only with a very large sample size (> 100 individuals sequenced or tested with the assay) that this polymorphism was found. The rarity of this polymorphism means that this assay is still broadly applicable across the range of WCT, but highlights what we suspect is a general problem: for broadly distributed taxa composed of several evolutionary lineages and represented by hundreds of populations, it may be difficult or impossible to develop a single eDNA assay that identifies all members of that taxon. It also suggests that some current assays may be of regional rather than global utility, or useful when confronted with limited mixtures of taxa, but not in other circumstances.
Broad-scale sampling prior to marker development, however, does not solve the issue of unresolved taxonomies that may influence both the ability of an assay to distinguish between closely related taxa and to have generality within a taxon. For example, YCT and BCT are recognized as separate subspecies [27], but their phylogenetic distinctiveness is ambiguous. Campbell et al. [18] and Loxterman & Keeley [11] were able to genetically separate these taxa only in portions of their ranges, and identified a third group (the southern BCT clade) as having diverged from Yellowstone and northern BCT ~ 1.0–1.6 million years ago. Perhaps unsurprisingly, our assay reliably detected eDNA from the YCT and northern BCT lineages (n = 19 and 6 individuals respectively). Individuals from Harkness Creek and the Glenwood Fish Hatchery represent individuals from the southern BCT clade, which displayed reduced amplification efficiency when screened with the YCT assay. This may indicate that there is insufficient phylogenetic divergence to distinguish two recognized taxa (Yellowstone and some lineages of BCT [20]; Fig 1B), and is also likely a case of grouping divergent lineages under a single recognized taxon (i.e., polyphyly; northern and southern clades within BCT; Fig 1C).
Although several recent eDNA studies have sampled broadly for marker development (e.g., [9]), we suspect the tradeoff between interspecific specificity and intraspecific generality has been under-appreciated when building assays to separate closely related sympatric species. The first step to addressing this issue lies in determining the range and frequency of intraspecific polymorphisms across populations of the target taxa. This is simply a restatement of a maxim of phylogeography: good phylogenetic assessments require comprehensive geographic sampling [28]. This type of broad sampling is more likely to produce truly general assays, and may reveal previously undetected, cryptic diversity. This truth has also been recognized for other molecular species identification tools (i.e. barcoding [10,29]). Moreover, assays need to be tested locally on collected tissue samples and, if possible, eDNA samples taken from areas with confirmed presence of the target species prior to application. Additionally, it may be useful to apply multiple markers at different loci to reduce the risk of false negatives (failure to detect a species when present) due to rare polymorphisms and to separate closely related species. Finally, eDNA studies have increasingly incorporated occupancy models that account for imperfect detection [30–32]. These are a useful tool for accounting for imperfect detection due to eDNA degradation and variation in eDNA capture, but may also reduce false negative inference due to rare polymorphisms that results in reduced detection probabilities.
Supporting Information
(DOCX)
(XLS)
Acknowledgments
We thank Matthew Laramie and Matthew Campbell for providing tissue samples for assay testing. Funding for this work was provided by the U.S. Forest Service. TW is supported by a NSF Graduate Research Fellowship (Grant No. DGE-1313190).
Data Availability
Data generated for this study are within the paper and the supporting S1 Text. Sequence data from previous studies can be accessed using GenBank accession numbers listed in the paper.
Funding Statement
Region 1 of the U.S. Forest Service provided funding for this work. TMW is supported by a fellowship from the National Science Foundation (Grant No. DGE-1313190). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jerde CL, Mahon AR, Chadderton WL, Lodge DM. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv Lett. 2011;4: 150–157. 10.1111/j.1755-263X.2010.00158.x [DOI] [Google Scholar]
- 2. Pedersen MW, Overballe-Petersen S, Ermini L, Sarkissian C Der, Haile J, Hellstrom M, et al. Ancient and modern environmental DNA. Philos Trans R Soc Biol. 2015;370 Available: 10.1098/rstb.2013.0383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bohmann K, Evans A, Gilbert MTP, Carvalho GR, Creer S, Knapp M, et al. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol. Elsevier Ltd; 2014; 1–10. 10.1016/j.tree.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 4. Thomsen PF, Willerslev E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv. Elsevier Ltd; 2014; 10.1016/j.biocon.2014.11.019 [DOI] [Google Scholar]
- 5. Biggs J, Ewald N, Valentini A, Gaboriaud C, Dejean T, Griffiths RA, et al. Using eDNA to develop a national citizen science-based monitoring programme for the great crested newt (Triturus cristatus). Biol Conserv. Elsevier Ltd; 2015;183: 19–28. 10.1016/j.biocon.2014.11.029 [DOI] [Google Scholar]
- 6. Sigsgaard EE, Carl H, Møller PR, Thomsen PF. Monitoring the near-extinct European weather loach in Denmark based on environmental DNA from water samples. Biol Conserv. Elsevier Ltd; 2015;183: 46–52. 10.1016/j.biocon.2014.11.023 [DOI] [Google Scholar]
- 7. Wilcox TM, McKelvey KS, Young MK, Jane SF, Lowe WH, Whiteley AR, et al. Robust detection of rare species using environmental DNA: the importance of primer specificity. PLoS One. 2013;8: e59520 10.1371/journal.pone.0059520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fukumoto S, Ushimaru A, Minamoto T. A basin-scale application of environmental DNA assessment for rare endemic species and closely related exotic species in rivers: a case study of giant salamanders in Japan. Crispo E, editor. J Appl Ecol. 2015; n/a–n/a. 10.1111/1365-2664.12392 [DOI] [Google Scholar]
- 9. Goldberg CS, Sepulveda A, Ray A, Baumgardt J, Waits LP. Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshw Sci. 2013;32: 792–800. 10.1899/13-046.1 [DOI] [Google Scholar]
- 10. Young MK, McKelvey KS, Pilgrim KL, Schwartz MK. DNA barcoding at riverscape scales : assessing biodiversity among fishes of the genus Cottus (Teleostei) in northern Rocky Mountain streams. Mol Ecol Resour. 2013;13: 583–95. 10.1111/1755-0998.12091 [DOI] [PubMed] [Google Scholar]
- 11. Loxterman JL, Keeley ER. Watershed boundaries and geographic isolation: patterns of diversification in cutthroat trout from western North America. BMC Evol Biol. BioMed Central Ltd; 2012;12: 38–53. 10.1186/1471-2148-12-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Behnke R. Native trout of western North America Am Fish Soc Monogr. 1992;6. [Google Scholar]
- 13. Shepard BB, May BE, Urie W. Status and Conservation of Westslope Cutthroat Trout within the Western United States. North Am J Fish Manag. 2005;25: 1426–1440. 10.1577/M05-004.1 [DOI] [Google Scholar]
- 14. Nehlsen W, Williams JE, Lichatowich JA. Pacific Salmon at the Crossroads: Stocks at Risk from California, Oregon, Idaho, and Washington. Fisheries. 1991;16: 4–21. [Google Scholar]
- 15. Kruse CG, Hubert WA, Rahel FJ. North American Journal of Fisheries Management Status of Yellowstone Cutthroat Trout in Wyoming Waters. North Am J Fish Manag. 2000;20: 693–705. [DOI] [Google Scholar]
- 16. Muhlfeld CC, Albeke SE, Gunckel SL, Writer BJ, Shepard BB, May BE. Status and Conservation of Interior Redband Trout in the Western United States. North Am J Fish Manag. 2015;35: 31–53. 10.1080/02755947.2014.951807 [DOI] [Google Scholar]
- 17. Gresswell RE. Biology, Status, and Management of the Yellowstone Cutthroat Trout. North Am J Fish Manag. 2011;31: 782–812. 10.1080/02755947.2011.608980 [DOI] [Google Scholar]
- 18. Campbell MR, Kozfkay CC, Meyer KA, Powell MS, Williams RN. Historical Influences of Volcanism and Glaciation in Shaping Mitochondrial DNA Variation and Distribution in Yellowstone Cutthroat Trout across Its Native Range Historical Influences of Volcanism and Glaciati. Trans Am Fish Soc. 2011;140: 91–107. [Google Scholar]
- 19. Wright ES, Yilmaz LS, Ram S, Gasser JM, Harrington GW, Noguera DR. Exploiting extension bias in polymerase chain reaction to improve primer specificity in ensembles of nearly identical DNA templates. Environ Microbiol. 2013; n/a–n/a. 10.1111/1462-2920.12259 [DOI] [PubMed] [Google Scholar]
- 20.(R Core Development Team). R: A language and environment for statistical computing [Internet]. Vienna, Austria: Foundation for Statistical Computing; 2013. Available: http://www.r-project.org/ [Google Scholar]
- 21. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28: 2731–9. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wright ES, Yilmaz LS, Ram S, Gasser JM, Harrington GW, Noguera DR. Exploiting extension bias in polymerase chain reaction to improve primer specificity in ensembles of nearly identical DNA templates. Environ Microbiol. 2013;6: 1354–1365. 10.1111/1462-2920.12259 [DOI] [PubMed] [Google Scholar]
- 23. Wilcox TM, McKelvey KS, Young MK, Jane SF, Lowe WH, Whiteley AR, et al. Robust Detection of Rare Species Using Environmental DNA: The Importance of Primer Specificity. PLoS One. 2013;8: e59520 10.1371/journal.pone.0059520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bustin S a, Benes V, Garson J a, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55: 611–22. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 25. Moyle PB. Inland Fishes of California. University of California Press; 2002. [Google Scholar]
- 26. Drinan DP, Kalinowski ST, Vu N V., Shepard BB, Muhlfeld CC, Campbell MR. Genetic variation in westslope cutthroat trout Oncorhynchus clarkii lewisi: implications for conservation. Conserv Genet. 2011;12: 1513–1523. 10.1007/s10592-011-0249-2 [DOI] [Google Scholar]
- 27. Page LM, Héctor Espinosa-Pérez LTF, Gilbert CR, Lea RN, Mandrak NE, Mayden RL, et al. Common and Scientific Names of Fishes from the United States, Canada, and Mexico American Fisheries Society; 2013. [Google Scholar]
- 28. Avise JC. Phylogeography. Harvard University Press; 2000. [Google Scholar]
- 29. Bergsten J, Bilton DT, Fujisawa T, Elliott M, Monaghan MT, Balke M, et al. The effect of geographical scale of sampling on DNA barcoding. Syst Biol. 2012;61: 851–69. 10.1093/sysbio/sys037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mackenzie DI, Nichols JD, Lachman GB, Droege S, Andrew J, Langtimm CA. Estimating Site Occupancy Rates When Detection Probabilities Are Less Than One. Ecology. 2002;83: 2248–2255. [Google Scholar]
- 31. Rees HC, Bishop K, Middleditch DJ, Patmore JRM, Maddison BC, Gough KC. The application of eDNA for monitoring of the Great Crested Newt in the UK. Ecol Evol. 2014; 1–10. 10.1002/ece3.1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmidt BR, Kéry M, Ursenbacher S, Hyman OJ, Collins JP. Site occupancy models in the analysis of environmental DNA presence/absence surveys: A case study of an emerging amphibian pathogen. Methods Ecol Evol. 2013;4: 646–653. 10.1111/2041-210X.12052 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(XLS)
Data Availability Statement
Data generated for this study are within the paper and the supporting S1 Text. Sequence data from previous studies can be accessed using GenBank accession numbers listed in the paper.