Introduction
The Collaborative Cross (CC) and the complementary Diversity Outbred (DO) population were conceived as a platform for the next generation of studies of the genetic basis for complex traits in mouse (Churchill et al. 2004). The CC promised to combine the strengths of existing panels of recombinant inbred (RI) lines [e.g., BXD (Taylor et al. 1973), LXS (Williams et al. 2004)] and inbred strains (Ghazalpour et al. 2012)—phenotypic variation, replication, and integration of multiple phenotypes—with the genetic randomization and absence of population structure provided by populations such as the heterogeneous stock (Valdar et al. 2006b). This was to be accomplished by performing many iterations of the “funnel” breeding scheme illustrated in Fig. 1: three generations of outcrossing followed by sibling mating to create RI lines with contributions from all eight founder strains. Because of the large number of novel pairwise and higher order allele combinations generated by the factorial breeding scheme and the genetic diversity of the founder strains, phenotypic variability in the resulting set of RI lines was expected to span and exceed that in the founder strains.
Fig. 1.
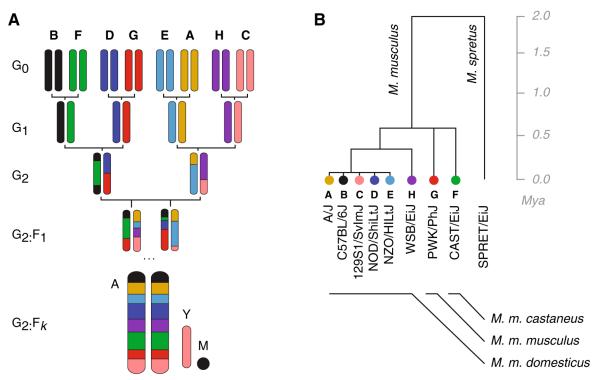
Breeding scheme of the Collaborative Cross (CC) and phylogenetic relationships between founder strains. a A representative CC breeding funnel. In each mating, the dam is shown on the left and the sire on the right. Because the positions in the funnel are non-exchangeable, each ordering of founder strains at the G0 generation defines a unique realization of the breeding scheme for the autosomes (marked “A”). The origin of the uniparentally inherited, non-recombining Y chromosome and mitochondrial genome (marked “M”) can always be predicted from the funnel order. Founder strains in this and other figures in the article are denoted by single-letter codes and by a color code. b Schematic phylogeny of the eight CC founder strains, with color key. The three M. musculus subspecies began to diverge approximately 0.5 million years ago (Mya); their branching order is not well resolved. The five classical inbred strains are primarily of M. m. domesticus origin, as is the wild-derived WSB/EiJ. M. m. musculus and M. m. castaneus are represented by PWK/PhJ and CAST/EiJ, respectively. Mus spretus, represented here by the inbred strain SPRET/EiJ, diverged from M. musculus approximately 2 Mya and is shown only as an outgroup; it is not a founder strain of the CC
Simulations (Valdar et al. 2006a) suggested that 500 lines would be required to achieve good resolution in haplotype association-mapping studies. But widespread genomic incompatibility—the biological basis for which remains mostly unexplored—has limited the number of extant lines to ~150 at time of writing. Nonetheless, the CC has begun to fulfill its promise as a source of extreme phenotypic variability and associated candidate loci (Iraqi et al. 2014). Within just the past year, CC lines have been shown to be highly variable for traits related to both normal physiology and disease, including gene expression in healthy liver (Aylor et al. 2011; Weiser et al. 2014), allergic airway inflammation (Kelada et al. 2014), lymphocyte counts (Phillippi et al. 2014), susceptibility to melanoma (Ferguson et al. 2014), and susceptibility to viral pathogens including influenza (Ferris et al. 2013) and Ebola virus (Rasmussen et al. 2014). Some phenotypic outliers constitute new disease models on their own: CC011/Unc, for example, is the first mouse line to spontaneously develop inflammatory bowel disease in the absence of chemical treatment or infection (Rogala et al. 2014). Up-to-date information on the status of the CC population is available at http://www.csbio.unc.edu/CCstatus/.
The CC and DO (discussed further in the article by Bogue et al. in this issue) clearly provide an exciting avenue for dissecting the genetic and molecular networks underlying of complex traits such as behavior (Chesler et al. 2014). The ultimate goal of genetic mapping is to identify the sequence variants (or combinations thereof) which are causative for variation in the trait of interest. In this article, we review databases, analysis tools, and other informatics resources relevant to this goal. The genomes of CC and DO mice can be expressed, to a very good approximation, as mosaics of segments inherited from these founders. Founder strains were chosen to span most of the pool of standing genetic variation available within laboratory strains and represent all three subspecies of the house mouse, M. m. domesticus, M. m. musculus, and M. m. castaneus (Fig. 1b). Five (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, NZO/HILtJ) are so-called “classical laboratory strains.” classical laboratory strains are descended from a small population of “fancy mice” of European and Japanese origin within the last 100 years (Wade et al. 2002; Petkov et al. 2004; Yang et al. 2007, 2011; Didion et al. 2013). They are all relatively closely related and share long tracts of pairwise identity by descent (IBD; see Appendix), but these patterns of relatedness are not consistent across the genome as a result of the strains’ somewhat convoluted ancestry. More than 90 % of the genomes of classical laboratory strains are of M. m. domesticus descent; the remainder is mostly of M. m. musculus origin with a smaller contribution from M. m. castaneus. The remaining three founder strains—CAST/EiJ (M. m. castaneus), PWK/PhJ (M. m. musculus), and WSB/EiJ (M. m. domesticus)—are “wild-derived.” These strains are descended from wild-caught mice by repeated back-crossing or sibling mating. They are genetically distinct from classical laboratory mice, although it is now well-known that, due to both introgression (see Appendix) in the wild and contamination in the laboratory, not all wild-derived strains are “pure” representatives of their respective subspecies (Yang et al. 2011). Most segregating variation in the CC and DO, both within and across subspecies, is contributed by the wild-derived strains.
All pairs of CC lines or DO individuals are expected to be equally related at the genome-wide level. Local relatedness may deviate from the genome-wide expectation due to sampling effects, breeding errors, and the complex patterns of kinship and admixture among the founder strains (see Appendix). Key to understanding fine-scale genetic variation in these populations, then, is a deep characterization of the founder genomes. The resources presented in this review first place genetic variation in the founder strains of the CC in the context of the phylogeny of the mouse at three scales: variation between subspecies, variation within populations, and variation between sister strains or within inbred lines. We then discuss computational tools developed for the analysis of CC and DO genomes and for genetic mapping in these populations. Together, these databases and tools provide an integrated and comprehensive view of the polymorphisms segregating in the CC and DO.
Primary data sources
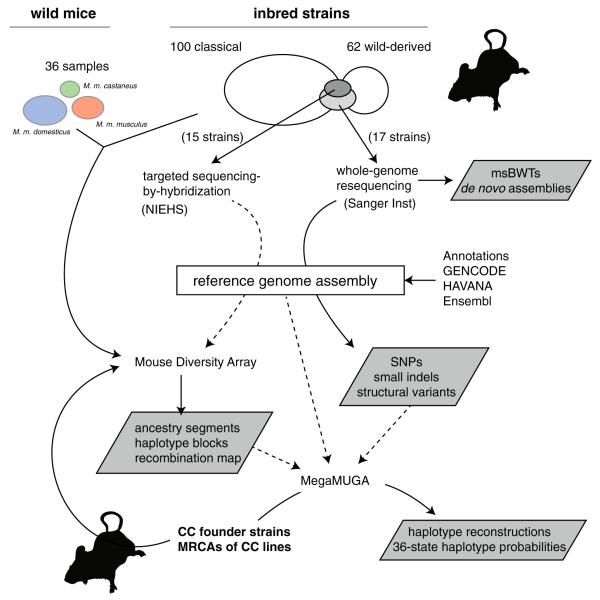
The resources discussed in this review are derived from three large primary datasets: whole-genome resequencing data for 17 mouse strains from the Sanger Mouse Genomes Project; genotypes from the 600,000-marker Mouse Diversity Array (MDA)(Yang et al. 2009) for 198 strains and 273 incipient CC lines; and genotypes from the 77808-marker Mega Mouse Universal Genotyping Array (MegaMUGA, discussed later), for 2–8 obligate ancestors of each of 69 available CC lines. The MDA platform was itself designed on the basis of a prior study which identified variants in 11 strains using sequencing-by-hybridization (Frazer et al. 2007). Likewise, the MegaMUGA platform was designed mostly using information from the MDA dataset and the Sanger Mouse Genomes Project. The relationships between these primary datasets and the resources derived from them are illustrated in Fig. 2. We describe the Sanger and MDA datasets in more detail below.
Fig. 2.
Primary datasets and informatics resources derived from them. Solid lines indicate direct analyses; dashed lines indicate information propagated from one experiment to the design of another experiment or assay. Mouse silhouettes (from http://phylopic.org/) indicate input of mouse samples of known or unknown ancestry. Derived informatics resources are boxed in gray. Note the dependency of most of these resources on the reference genome assembly
Sanger Mouse Genomes Project
The most comprehensive available catalog of sequence variation in laboratory mice is the Wellcome Trust Sanger Institute’s Mouse Genomes Project (Keane et al. 2011). The Sanger Institute performed deep whole-genome sequencing on the Illumina platform of 17 commonly used mouse strains, including all eight CC founder strains and SPRET/EiJ (of Mus spretus origin), and identified 57 million segregating SNPs, 9 million indels, and 0.3 million structural variants relative to the C57BL/6J reference genome (build GRCm38). Since the initial release in 2011, Sanger scientists have continued to update the database as variant-calling methods improve and more sequence data become available. As of June 2015, alignments and variant calls from 11 more classical laboratory strains and two more wild-derived strains (ZALENDE/EiJ and LEWES/EiJ) have been made available.
Mouse Diversity Array dataset
The first high-density genotyping array developed for mouse was the MDA (Yang et al. 2009). This array, developed on the Affymetrix platform, queried 623,124 SNPs selected to capture the full spectrum of genetic diversity present in current stocks of laboratory mice, including both classical laboratory and wild-derived inbred strains. MDA also contains 916,269 invariant genomic probes selected to tag functional elements of the genome and detect copy-number differences. A total of 100 classical laboratory strains, 62 wild-derived strains, and 36 wild mice were genotyped on MDA at the Jackson Laboratory and the University of North Carolina at Chapel Hill to characterize patterns of haplotype diversity in Mus musculus (Yang et al. 2011; Didion et al. 2012). Those genotypes are now available for public browsing and download at http://cgd.jax.org/datasets/popgen/diversityarray/yang2011.shtml. The MDA was also applied to incipient CC lines. First, 474 mice from the third generation of the CC breeding scheme (denoted G2:F1, Fig. 1), the first generation at which alleles from all eight founders are represented in single genomes, were genotyped in order to construct a new standard recombination map (Liu et al. 2014) which guided the design of future marker panels. Second, more than 300 incipient CC lines (the “pre-CC” population) were genotyped for a group of proof-of-principle studies which demonstrated the utility of the CC for high-resolution genetic mapping (Aylor et al. 2011; Kelada et al. 2012; Ferris et al. 2013).
Microarray genotypes necessarily have lower resolution than whole-genome sequence and are subject to ascertainment bias (Clark et al. 2005)—in the case of MDA, undersampling of minor alleles from M. m. musculus and M. m. castaneus (Yang et al. 2007, 2009). However, the broad scope of the MDA dataset makes it an extremely valuable resource for understanding both broad and fine-scale patterns of diversity in mouse.
We emphasize that all of these resources ultimately depend on the mouse reference genome assembly (Waterston et al. 2002) and both automatic [GENCODE, (Harrow et al. 2006)] and manual [HAVANA, (Wilming et al. 2008)] sequence annotations. The genome assembly and annotations are made available to the community via many online genome browsers, the most popular of which are hosted by the University of California at Santa Cruz [UCSC, (Karolchik et al. 2014)] and Ensembl (Flicek et al. 2013). Use of a single haploid reference sequence as an anchor for all studies of genetic variation in mouse offers many practical advantages. But the dependency on a reference genome requires several assumptions about the nature of genetic variation which may be violated in practice—the strongest of which is that of genomic collinearity (i.e., conserved marker order) between strains. We consider the implications of these assumptions in the Discussion section.
Databases of genetic variation in founder strains
Sanger Mouse SNP/Indel Viewer
All SNP, indel, and structural variants (including copy-number variants) from the Sanger Mouse Genomes Project are publicly available at http://www.sanger.ac.uk/resources/mouse/genomes/. SNPs and small indels were annotated for predicted functional consequences using the Ensembl Variant Effect Predictor (McLaren et al. 2010). Users can search for variants by genomic coordinate, gene, strain of origin, variant type, and predicted functional consequence. As of June 2015, search results are linked directly to a viewer for the underlying read alignments. The complete dataset—including read alignments (BAM format) and variant calls (VCF format)—is available for download for computational users.
Mouse Phenome Database
The Jackson Laboratory’s Mouse Phenome Database provides a web interface to several catalogs of sequence variation in inbred strains, including the Sanger dataset and the Jax-UNC MDA dataset (http://phenome.jax.org/db/q?rtn=snp/home). Several additional CNV datasets besides the Sanger structural variant calls are available. Users can query by region, by gene, or by strain, and can filter results according to polymorphism in strains of interest.
Mouse Phylogeny Viewer
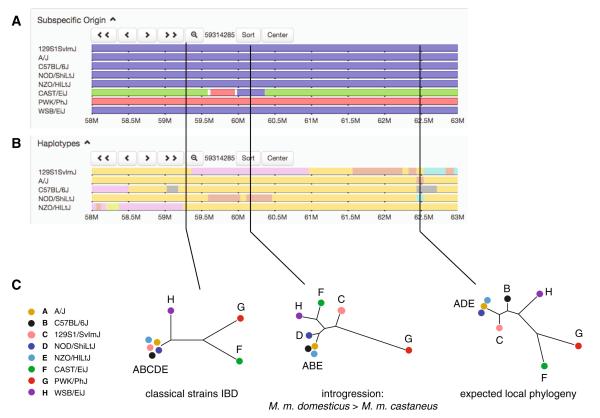
The Jax-UNC MDA dataset provides a rich resource for understanding high-level patterns of relatedness among laboratory mice. The time to the most recent common ancestor (MRCA) of a pair of haplotypes originating in different M. musculus subspecies is approximately 500,000 years ago (Boursot et al. 1993; Geraldes et al. 2008), on the same order as the divergence time between human and chimp. Much polymorphism in mouse thus segregates between, not within, subspecies. Using wild-caught mice from the home ranges of each of the three M. musculus subspecies, Yang et al. trained a model to classify genomic segments according to subspecies of origin in 162 laboratory strains (Yang et al. 2011). Subspecific origin assignment confirmed the presumed ancestry of most wild-derived strains (including CC founders) but also revealed the existence of widespread inter-subspecific introgression (see Appendix) in both classical laboratory and wild-derived strains. This has important implications for the CC and DO: even for loci at which all eight founder alleles (i.e., all three subspecies) are nominally present, genetic diversity may be lower than expected in the presence of introgression. An example is the middle of chr2 (Fig. 3): due to the introgression of a M. m. domesticus segment into CAST/EiJ, no M. m. castaneus haplotype is present at this locus in the CC. Subspecific origin tracks are browsable at http://msub.csbio.unc.edu/.
Fig. 3.
Exploring local haplotype diversity in the Collaborative Cross founder strains. a Subspecific origin tracks from the Mouse Phylogeny Viewer for a five Mbp interval on chromosome 2. Segments are colored according to the subspecies from which they were most likely inherited: blue for M. m. domesticus, red for M. m. musculus, and green for M. m. castaneus. As expected, the five classical laboratory strains are of mostly M. m. domesticus ancestry, but an introgression tract (see Appendix), from M. m. musculus into the classical strain NZO/HILtJ, is visible in the distal portion of the interval. b Fine-scale haplotype block maps for the five classical laboratory strains. (The three wild-derived founder strains are excluded from this analysis because each has a private haplotype, shared with none of the other founder strains, in almost every genomic interval. See Appendix.) Strains with the same haplotype at a given position are assigned the same color, but colors are recycled along the length of the window. c Local phylogenetic trees reflect the varying ancestry of the CC founder strains along the genome. From left: an interval in which all five classical strains share a haplotype identical-by-descent (IBD, see Appendix); an interval in which CAST/EiJ clusters within a class of classical inbred strains, reflecting introgression (probably due to breeding errors in the laboratory); and an interval whose phylogeny is consistent with the genome-wide expected relationship between strains (see Fig. 1).In the absence of epistasis, allele effects at a QTL should be concordant with the local phylogenetic tree: for instance, in the middle interval, the effect of the PWK/PhJ allele should differ from that of any of the other seven alleles, and the effects of the other seven should be similar to each other
Mouse Imputation Resource
In contrast to wild-derived strains, classical inbred strains are descended from a small founder population in the recent past. The genomes of individuals in such a population are related chiefly by recombination, with little contribution from mutation: each individual’s genome is a mosaic of segments sampled from a pool of founder haplotypes. At this scale, the natural unit of genetic analysis is the haplotype block, the minimal segment which is inherited unbroken by recombination (see Appendix). Yang et al. applied the four-gamete test (Hudson et al. 1985; see Appendix) to MDA genotypes from 100 classical laboratory strains to demarcate haplotype blocks in M. m. domesticus (Wang et al. 2010; Yang et al. 2011). The median number of haplotypes at a given locus is only 5, and 97 % of loci can be mapped onto ten or fewer haplotypes. Wang et al. combined these haplotype blocks with whole-genome resequencing data from 12 strains to generate high-confidence imputed genotypes at 12 million loci (Wang et al. 2012a, b). Haplotype blocks and imputation results are available for browsing and download at http://msub.csbio.unc.edu/ and http://csbio.unc.edu/imputation/, respectively.
An important observation from the MDA studies was that the genetic diversity available within classical inbred strains of mice is not uniformly distributed across the genome. Local patterns of haplotype sharing between strains may depart from global estimates of their relatedness (i.e., from their genealogy). The effective number of independent haplotypes among the five classical laboratory strains in the CC varies from 1 (i.e., regions of IBD) to five along the genome (CCC et al. 2012) (Fig. 3). This information is critical for interpretation of QTL-mapping studies. First, haplotype blocks inform expectations of allele effects at QTL. Second, accurate identification of candidate causative variants depends on knowledge of local haplotype structure: if two founder strains share a haplotype at a QTL peak but their respective alleles have opposing effects at that QTL, the number of candidate causative variants is immediately reduced. Finally, patterns of haplotype sharing between CC and non-CC strains aid in rational comparison of results from CC and non-CC crosses.
Genotyping and haplotype inference
A critical step in any genetic mapping study is to express the genotypes of individuals in the mapping population as mosaics of parental haplotypes. This requires obtaining genotypes at informative markers spaced along the genome at adequate density to capture most of the recombination events between founder chromosomes which have accumulated during breeding. Traditional experimental designs such as the F2 intercross comprise only two parental genotypes and a single generation of informative meioses; for practical sample sizes, the number of recombinations is small enough that panels of hundreds to a few thousand markers are sufficient to reconstruct progeny haplotypes with little uncertainty. Multiparental populations such as the CC and DO pose two challenges for genotyping: first, the highly recombinant structure of chromosomes in later generations requires a much denser marker panel; and second, the presence of more than two parental haplotypes means that multiple biallelic markers are required to discriminate between parents at a given locus. A custom genotyping platform, the MegaMUGA, was designed to address these challenges in the CC. Probabilistic methods based on hidden Markov models can be applied to these genotypes to recover the mosaic structure of the genome in a CC or DO individual. The resulting haplotype probabilities (or “dosages”) are used as input to association-mapping software.
MegaMUGA
The MegaMUGA is a custom Illumina Infinium genotyping microarray designed specifically to support the Collaborative Cross (CCC et al. 2012). Its content, which we describe in detail below, is optimized for the identification of founder contribution and detection of residual heterozygosity among CC strains at any stage of inbreeding.
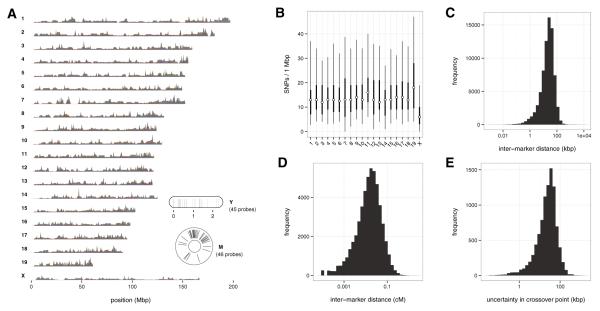
The vast majority of the 77,808 oligonucleotide probes on MegaMUGA were designed to assay traditional biallelic SNPs. Target SNPs were selected to be distributed across the entire genome, including the mitochondria and the Y chromosome, with an average physical spacing of 33 kbp. For the autosomes, these probes were distributed as evenly as possible across a new sex-averaged linkage map (Liu et al. 2014) for the mouse with a slight excess of probes in the telomeric regions to facilitate detection of recombination events in the distal chromosomes. The majority of target SNPs, about 65,000, were chosen because they were maximally informative—that is, had high minor-allele frequencies and covered many strain distribution patterns—in the CC and DO. An additional 14,000 were chosen to assay variants segregating in wild mice of all three subspecies (M. m. domesticus, M. m. musculus, M. m. castaneus); 750 were chosen that segregate within Mus spretus-derived strains; 150 were chosen to differentiate between C57BL/6J and C57BL/6N; 102 were selected for detecting transgenes and other engineered constructs; and a final subset of about 100 were designed to target specific loci of experimental interest, such as the X-chromosome controlling element (Xce) locus (Calaway et al. 2013). The genomic distribution of MegaMUGA markers is shown in Fig. 4.
Fig. 4.
The MegaMUGA genotyping array. a Genomic distribution of 77,808 probes, represented as density. Gray, all probes; blue, red, and green, probes diagnostic for M. m. domesticus, M. m. musculus, or M. m. castaneus ancestry, respectively. Probes for the mitochondria and for the male-specific region of the Y chromosome are displayed in the inset. b Median, inner interquartile range (25th–75th percentile), and outer interquartile range (5th–95th percentile) of probe density in 1 Mbp windows, per chromosome. c Distribution of physical distance between probes on the same chromosome (mean 33 kbp, median 23 kbp). d Distribution of genetic distance between probes on the same chromosome (mean 0.0019 cM, median 0.0063 cM). e A key measure of performance of a genotyping array in the CC is the resolution at which it can identify crossover events between founder strain haplotypes; that is, the number of markers required to confidently detect a transition from one haplotype to another. The distribution of uncertainty in crossover point for 9424 accumulated recombination events in 69 CC lines is shown (mean 35.8 kbp, median 26.7 kbp)
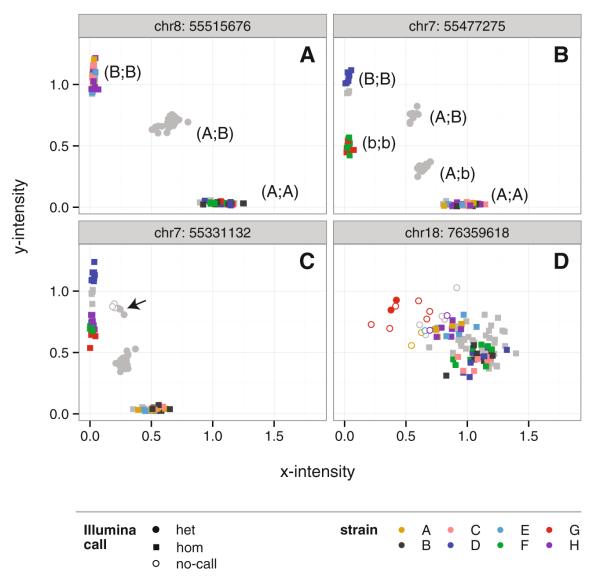
Standard genotype-calling methods for microarrays such as MegaMUGA are designed for biallelic markers and attempt to classify each sample as belonging to one of four states (reference allele, alternate allele, heterozygous, or missing/“no-call”) based on probe hybridization intensity signals (Fig. 5). For truly biallelic markers with no off-target variation within the probe sequence (Fig. 5a), this classification recovers all available information. Illumina’s proprietary calling algorithm cannot accommodate multiallelic markers (Fig. 5b) or marker-sample combinations for which the genotype state is uncertain (Fig. 5c, d). At such markers the continuous hybridization intensity values capture more information than the discrete genotype calls. Several tools for exploring MegaMUGA genotypes are hosted by the CC Status website (http://csbio.unc.edu/CCstatus/). Discrete genotype calls, either from the standard Illumina algorithm or a more flexible algorithm which accommodates multiallelic markers, can be downloaded by genomic region and by sample via Dump Genotypes. The Cluster Browser displays 2D hybridization intensity signals at specific markers similar to the plots shown in Fig. 5. The PCA Tool performs principal components analysis over hybridization intensities from multiple markers to reveal local haplotype clusters even in the absence of confident genotype calls at any single marker.
Fig. 5.
Interpretation of genotyping array data in the CC. Four markers, all designed to probe biallelic SNPs, are shown. Samples are either inbred CC founder strains (colored points) or F1s between founder strains (gray points). Open shapes indicate samples flagged as “no-call” (missing) by the Illumina software. a A marker which performs as designed: homozygous samples fall in two clusters representing the two possible homozygous states (A;A) and (B;B), while samples heterozygous (A;B) for the target SNP fall in an intermediate cluster. Homozygotes (filled squares) and heterozygotes (filled circles) are both called correctly by the Illumina software. b A multiallelic marker: homozygous samples fall in three clusters representing three homozygous states (A;A), (B;B), and (b;b) due to off-target sequence variation in or near the probe sequence. Both possible heterozygous states (A;B) and (A;b) are correctly called heterozygous by the Illumina software, but information is lost by collapsing five states to three. c Another multiallelic marker, but with lower calling accuracy by Illumina: samples in one of the two heterozygous clusters (arrowhead) are arbitrarily called as heterozygous (filled circle) or no-call (open circle). d A poorly performing marker: samples collapse into the middle of the plot, and Illumina calls are almost completely arbitrary. However, samples of the same genotype are loosely clustered in 2D space, albeit with poor discrimination. Haplotype reconstruction on the basis of intensity rather than genotype calls preserves this information
The MegaMUGA platform will be succeeded in July 2015 by a new array, GigaMUGA, also available through Neogen Inc. GigaMUGA will offer approximately double the marker density of MegaMUGA (~143,000 markers, including 66,000 markers carried over from MegaMUGA) for equal or lesser cost per sample. Like its predecessor, GigaMUGA is designed to be maximally informative in crosses derived from the CC founder strains, but will also include markers designed to discriminate between closely related laboratory strains (de Villena, personal communication).
Haplotype reconstructions for CC lines
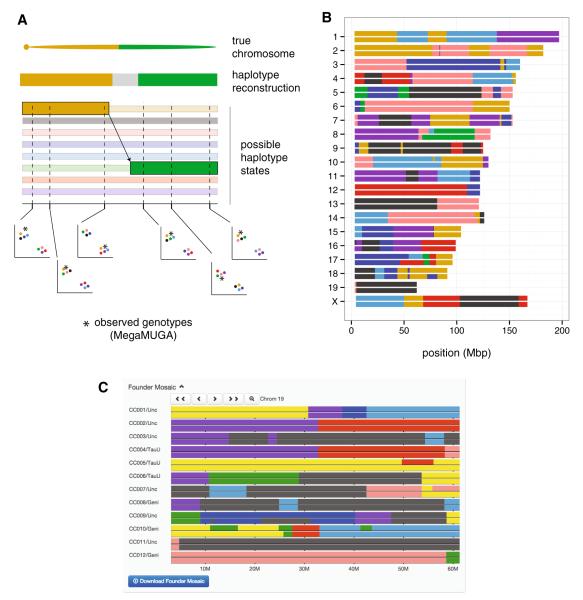
Formally, the genome of an individual from an admixed population is a mosaic of segments inherited from its ancestors. Ancestry inference on such an admixed individual refers to the problem of partitioning the individual’s genome into haplotype blocks labeled with the contributing ancestor (see Appendix). We call the most likely representation of this ancestry mosaic an individual’s haplotype reconstruction or haplotype mosaic. For the CC, the pool of ancestral haplotypes is restricted to the eight founder strains—in contrast to natural populations, in which the pool of founder haplotypes is not known a priori. Figure 6 shows an example haplotype reconstruction for line CC011/Unc. Segments are colored according to their founder strain of origin. Since haplotype blocks are, by definition, the minimal segments of the genome inherited without recombination, they represent the fundamental unit for genetic mapping: at most one independent test of genotype–phenotype association can be performed per haplotype block. Obtaining haplotype reconstructions is thus the first analysis step in QTL-mapping studies in the CC and DO.
Fig. 6.
Ancestry inference by a hidden Markov model (HMM) in CC lines. a Schematic of the HMM procedure. (Only the eight homozygous states are shown for simplicity, but the full model has an additional 28 states representing the possible heterozygous combinations.) The true underlying chromosome is recombinant for the A/J and CAST/EiJ haplotypes. Probability of each of eight possible haplotypes is estimated as a function of observed 2D genotyping array intensities (asterisk, unknown sample; colored circles, CC founder strains) along the genome. Information is shared across markers, and the MegaMUGA array is designed to discriminate between all eight founder strains in any 3-marker window. The transition from the A/J to the CAST/EiJ haplotype—representing a crossover event—occurs between the third and fourth markers, but its exact position remains uncertain (gray region) in the final haplotype reconstruction. b Example haplotype reconstruction of a CC line, CC011/Unc, from MegaMUGA genotypes of three obligate ancestors. The line is still segregating for regions on chromosomes 8, 10, 14, 17, and 18. c Screen capture from the interactive Collaborative Cross Viewer showing haplotype mosaics for 12 CC lines in an interval on chromosome 19
There are numerous methods for inferring ancestor mosaics given the genotypes of an individual and a set of ancestral haplotypes. In this review, we restrict our attention to methods designed to take genotyping microarray data as input. All use a hidden Markov model (HMM; see Appendix) approach to estimate probability of descent from each ancestor at each locus along the genome given observed genotype data (Fig. 6a). The first such algorithm for inferring ancestry in outbred model organism populations with known ancestors was HAPPY (Mott et al. 2000), a package for QTL mapping designed for mouse outbred stocks. Improved methods for ancestry inference in recombinant inbred strains have been designed for the Collaborative Cross; in particular GAIN (Liu et al. 2010), which combines the HMM framework with knowledge of the pedigree to efficiently infer ancestry probabilities.
An important assumption of HAPPY, GAIN, and other existing methods is that genotype calls have little error: low-performing markers and markers with off-target variation must be excluded from the input data. This assumption has two important disadvantages. First, it requires filtering out a substantial fraction of markers; and second, it ignores the extra information content of multiallelic markers. Therefore, Fu et al. (2012) developed an HMM-based method for inferring ancestry without first converting the probe intensity data into genotype calls (Fig. 5). This method works by minimizing the distance, in the 2D intensity space, between a target individual and one or more of its ancestors. Markers with poor discrimination between alleles (as in Fig. 5d) need not be excluded; uncertainty in genotype is accommodated naturally by the probabilistic framework of the HMM. The extra information provided by multiallelic markers is rescued, reducing ascertainment bias (Didion et al. 2012).
The ancestry-inference procedure for CC and DO samples models the underlying diploid genotype of a sample as one of 36 possible states: eight homozygous states and 28 (unphased) heterozygous states. The distinction regarding phase is important: transition penalties in the HMMs of Fu et al. (2012) and HAPPY (Mott et al. 2000) suppress gratuitous haplotype switching, but do not explicitly account for phase. Switch errors are possible in intervals over which both of a sample’s chromosomes are recombinant. In general, some pedigree information is required to avoid switch errors, as in GAIN (Liu et al. 2010). At each locus along the genome, the model estimates the posterior probability of each state given marker information. This allows for explicit representation of both heterozygosity and uncertainty in ancestry: for instance, having the (129S1/SvImJ)/(C57BL6/J) heterozygous genotype with probability 0.98 is not the same as having the 129/129 or B6/B6 state each with probability 0.49.
Haplotype reconstructions for CC lines are available for browsing and download via the CC viewer (Fig. 6c) (http://www.csbio.unc.edu/CCstatus/index.py?run=CCV). Thirty-six-state haplotype probabilities suitable for genetic mapping are available for download at http://www.csbio.unc.edu/CCstatus/index.py?run=AvailableLines: use check-boxes to select lines of interest, and click “More info” to see a table with links to haplotype probabilities as well as breeding performance and pedigree information. A complete data package containing 36-state haplotype probabilities (intensity-based) and consensus genotype calls (from the Illumina calling software) from the MegaMUGA array for all CC lines in distribution is available for download at http://csbio.unc.edu/CCstatus/gstemp/AllImageHapAndGenotypeFiles.zip.
Haplotype reconstruction is unambiguous for a single sample but more complicated for a (incompletely inbred) CC line. The probabilistic reconstructions reported for CC lines represent an average across 2–8 obligate ancestors of that line, and genotype calls at the link above represent the consensus call across the obligate ancestors.
Genetic mapping in the CC and DO
Statistical methods for genetic mapping in traditional designs such as the backcross or F2 intercross, in which all members of the mapping population are equally related, are well established. The simplest model, known as Haley–Knott regression (Haley et al. 1992), amounts to regression of the phenotype value on genotype probabilities (“dosages”) at each locus. In this and related methods, genotype is modeled as a fixed effect and residual variation is assumed to be independent across individuals and across loci—that is, phenotype values are assumed to be uncorrelated across individuals conditional on genotype at a QTL. Because many more loci are typically tested than there are individuals in the mapping population (an “n ⪢ p problem”), only a limited number of QTL can be identified with any certainty. An appropriate statistical significance threshold is established either via a multiple-testing correction (e.g., Benjamini et al. 1995) or by permutation (Churchill et al. 1994).
Mapping in multiparental populations such as the CC is complicated by two factors. First, the presence of eight possible alleles at any locus (instead of two) increases the difficulty of assigning paternal haplotype and leads to a very large parameter space (eight possible homozygous plus 28 possible heterozygous states, ignoring phase) which cannot be exhaustively explored in samples of practical size. For genome-wide association scans, it is only statistically practical to model additive effects of the eight founder alleles.
Second, relatedness between individuals in a multiparental population typically varies. Although the population structure in the CC (CCC et al. 2012) and DO (Svenson et al. 2012) is in principle much weaker than, for example, in the Hybrid Mouse Diversity Panel (Bennett et al. 2010), residual correlation between unlinked loci (often termed “long-range” linkage disequilibrium) is an inevitable result of breeding in relatively small closed populations (see Appendix). These correlations may give rise to false-positive associations between genotype and phenotype when a simple statistical model which ignores relatedness is used (Valdar et al. 2009).
The most popular methods for mapping in such populations extend the simple linear model described previously by adding a random effect whose covariance across individuals is parameterized by the observed (from genotype data) or expected (from pedigree) kinship structure in the population (Kang et al. 2008; Lippert et al. 2011; see also Appendix); these have been reviewed by Gonzales et al. (2014). An alternative method, proposed by Valdar et al. (2009), uses bootstrapping and resample model averaging in the context of a fixed-effects model to control false-positive rate in QTL mapping in the presence of population structure.
Below we discuss software available for QTL mapping in the CC, DO, and related experimental designs. Importantly, all of these require, as input, haplotype probabilities derived from a platform such as MegaMUGA.
QTL mapping in the CC
Two software packages have been applied to map QTL in the pre-CC and CC: BAGPIPE and BAGPHENOTYPE. They implement a fixed-effects model based on HAPPY (Mott et al. 2000) in which the phenotype value is regressed on a vector of haplotype probabilities for each of the eight founders, and can model both additive and dominance effects. Experimental (e.g., batch) and biological (e.g., sex) covariates can be modeled as either fixed or random effects. Significance levels are estimated by unrestricted permutation (Churchill et al. 1994). BAGPIPE (http://valdarlab.unc.edu/software/bagpipe) is suitable for single-locus mapping for normally distributed traits in the absence of gross population structure. BAGPHENOTYPE (http://valdarlab.unc.edu/bagphenotype.html) implements resample model averaging and model selection for multiple-locus models described in Valdar et al. (2009). It also allows the mapping of traits with a non-normal distribution (for instance, binary traits) via the generalized linear model. Although BAGPHENOTYPE is no longer under active development, its features are being merged into BAGPIPE. Both packages are written in R and Perl and run from a command line.
Penalized and Bayesian alternatives
The statistical models described so far model genotype–phenotype association at each locus (or small group of loci) independently and apply post hoc criteria to control false-positive rate. An alternative family of models instead fits a single model for all loci simultaneously, using a penalized regression method—e.g., LASSO or ridge regression—to limit the number of spurious associations identified. Such methods, although widely used in agricultural genetics under the guise of “genomic prediction” (recently reviewed in Daetwyler et al. (2013)), have not yet been applied to the CC or DO. Penalized regression can be framed as a partially Bayesian approach (Gelman et al. 2007). A fully Bayesian method applicable to multiparental populations, dubbed Diplo effect, was recently proposed by Zhang et al. (2014). The Bayesian hierarchical framework flexibly and intuitively models dependencies between (possibly many) model parameters as well as uncertainty in their values. Diplo effect explicitly models uncertainty in haplotype reconstruction and uses shrinkage to achieve well-behaved estimates of non-additive allele effects. The model was shown to outperform methods similar to BAGPIPE in the presence of uncertainty in founder haplotype assignment. However, this advantage comes at a cost of greatly increased computation time.
QTL mapping in the DO
The DOQTL package for R (Gatti et al. 2014), incorporates both HMM-based haplotype reconstruction (from Mega-MUGA genotypes) and QTL mapping. DOQTL implements a mixed-effects model with a kinship matrix estimated from reconstructed haplotype probabilities. Assuming that sequencing data are available for founders of the population, such as the Sanger Mouse Genomes Project mentioned above, DOQTL is also able to impute the genomes of the DO or other outbred sample, and use this imputed genome to conduct single-marker association mapping in the style of human GWAS. This R package and its reference manual are available publically at http://www.bioconductor.org/packages/release/bioc/html/DOQTL.html.
Other experimental designs
As CC lines with extreme phenotypes are identified, the most efficient experimental designs for follow-up studies will be intercrosses or backcrosses between lines at phenotypic extremes, or between lines and founder strains. An early example is a study by Rogala et al. (2014), in which a CC line which develops an autoimmune colitis (CC011/Unc) was backcrossed to a colitis-resistant strain (C57BL/6J) to map loci associated with colitis susceptibility. Using MegaMUGA genotypes from CC founders (publicly available at http://csbio.unc.edu/CCstatus/index.py?run=GeneseekMM) in combination with the haplotype reconstruction for CC011/Unc, a subset of non-redundant MegaMUGA markers was identified which was expected to be informative in the cross. The experiment was then analyzed as a standard backcross using R/qtl (Broman et al. 2003) to successfully identify three QTL with both additive and epistatic effects. We expect that this approach will be broadly applicable to CC-derived backcrosses or intercrosses.
An important design consideration for such studies is which CC strains and/or founder strains to choose. The choice depends both on the phenotype distribution among potential parental strains and on the genetic architecture of the trait. One means for identifying useful strain combinations is to survey several F1 crosses, potentially including reciprocal crosses. This design can be expressed as an incomplete (“sparse”) diallel. A Bayesian method for analysis of diallel experiments has recently been published by Lenarcic et al. (2012). The model estimates the broad-sense heritability of the trait of interest, and decomposes that heritability into strain-specific (i.e., additive, dominance), cross-specific (i.e., non-additive), and parent-of-origin components. These estimates can be used to inform the design of downstream experiments.
Resources for next-generation sequencing
The first analysis step in most next-generation sequencing experiments—whether for quantification (e.g., mRNA-seq, CHIP-seq) or variant discovery (DNA-seq)—is alignment of reads to a (haploid) reference sequence. Fidelity and efficiency of read alignment decrease with increasing genetic distance between the sequenced organism and the reference genome. This biases analysis of heterozygous samples: reads from the more divergent haplotype are more likely to be lost than reads from the less divergent haplotype. The ideal alignment reference is thus one which incorporates as much prior knowledge about the sequenced template as possible, including its ploidy. Conveniently, alignment to a diploid reference is implicitly allele specific. Divergence between the two parental haplotypes which introduces bias in the case of naive haploid alignment instead increases power for allele-specific diploid alignment.
Software for allele-specific read alignment
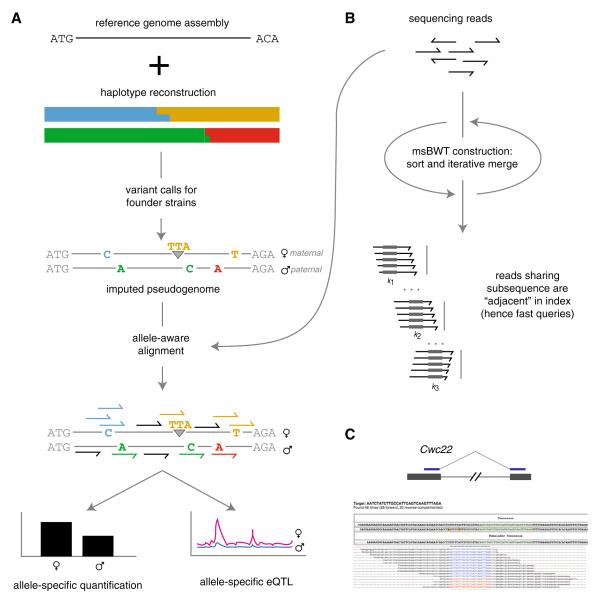
Two software pipelines have been developed specifically to mitigate alignment bias in the CC and DO. Both take as input a reference genome and an individual-specific list of known variant sites relative to that reference to produce an improved, imputed, diploid reference sequence which we term a pseudogenome. Reads are then aligned to the pseudogenome rather than the off-the-shelf reference (Fig. 7a). Post-processing steps can take advantage of both the improved overall alignment quality and allele specificity. The principal challenge to pseudogenome alignment is maintaining a common coordinate system across pseudogenomes, since inclusion of indels in pseudogenomes breaks one-to-one correspondence between base pairs (Fig. 7a).
Fig. 7.
Resources for next-generation sequencing in the CC. a Allele-specific diploid alignment pipeline. An individual’s haplotype mosaic is combined with a catalog of variants in the founder strains to create an imputed diploid pseudogenome for allele-specific read alignment. Reads overlapping a variant site can be assigned to a parental chromosome (colored reads); reads not overlapping a variant remain unassigned (black reads). b The msBWT, a compressed and searchable data structure for alignment-free analyses of next-generation sequencing reads. c Direct evidence for a splice junction in a transcript of the Cwc22 gene, in a msBWT of 100 bp mRNA-seq reads from whole brain of a CAST/EiJ mouse. A query with a 40-bp fragment spanning two exons returned 48 reads containing exactly that sequence on the forward cDNA strand (blue highlights) or reverse cDNA strand (red highlights). In a dataset of 90 million reads, the query took <1 s
The first software suite for this purpose (in mouse), developed in the McMillan group at the University of North Carolina, is modtools + lapels + suspenders (Huang et al. 2014) (http://www.csbio.unc.edu/CCstatus/index.py?run=Pseudo). Known variant sites are preprocessed from variant call format (VCF) to a list of atomic “sequence edit” instructions (represented in a MOD file), from which a pseudogenome is constructed by modtools. Alignments to one (in the haploid or inbred case) or more (in the diploid case) pseudogenomes are first processed by lapels, which annotates each read with the “sequence edit” instructions it overlaps and projects it back into the reference coordinate system. Then suspenders uses these tags to assign each read to zero or more pseudogenomes. Allele-specific quantification by counting of reads which may overlap multiple variant sites provides greatly improved accuracy and precision versus counting aligned bases over single variant sites (Baker et al. 2015; Crowley et al. 2015). The Seqnature suite (Munger et al. 2014) (https://github.com/jaxcs/Seqnature) developed in the Churchill group at the Jackson Laboratory is similar, and is tailored to RNA-seq in the DO.
Construction of an individualized pseudogenome for a sample requires prior knowledge of variant sites in that sample’s genome. In, for instance, an F1 cross between strains for which whole-genome sequencing data are available, imputing the pseudogenome is trivial. Genomes of recombinant individuals (e.g., CC or DO) can be expressed as mosaics of founder haplotypes on the basis of genotyping (discussed previously), and a pseudogenome stitched together accordingly. However, the sequencing data itself are likely to contain information sufficient to recover the founder mosaic without preliminary genotyping. If reads are aligned not to an individualized diploid pseudogenome but instead to haploid pseudogenomes of all eight possible founders, a probabilistic algorithm could in principle be used to simultaneously estimate the probability of descent from each founder at each locus, and provide allele-specific read quantification.
Imputed genomes for founder strains and CC lines
MOD files and pseudogenomes for the 17 strains resequenced by the Sanger Mouse Genomes Project, including the eight founders of the CC and DO, are available for download at http://www.csbio.unc.edu/CCstatus/index.py?run=Pseudo. Pseudogenomes for all 69 available CC lines have also been constructed on the basis of haplotype mosaics derived from microarray genotyping, as discussed previously. Imputation has been performed for both the NCBI build 37 and GRCm38 reference assemblies.
Allele-specific gene expression in CC founders
In order to explore variation in regulation of gene expression among CC founders, the Center for Integrated Systems Genetics at the University of North Carolina profiled gene expression in four tissues in a full diallel cross between CAST/EiJ, PWK/PhJ and WSB/EiJ. Expression was measured by very deep RNA-seq in whole brain, and by microarray in brain, lung, liver, and kidney (Crowley et al. 2015). The lapels + suspenders pipeline was used for allele-specific read alignment. The diallel design allows simultaneous estimation of additive, dominant, parent-of-origin, and sex effects on both total and allele-specific gene expression (Fig. 7a). Gene-wise results for 31,259 genes are browsable and searchable by gene name or Ensembl ID via the GECCO (Gene Expression in the Collaborative Cross) viewer at http://csbio.unc.edu/gecco/.
Tools for alignment-free analyses of sequencing data
The vast majority of next-generation sequencing experiments in mouse have read alignment to a reference genome as their first step. However, the primary data from any sequencing experiment are the reads themselves. Recognition that the raw reads are information-rich has led to the development of alignment-free algorithms for error correction (among many others, Chaisson and Pevzner 2008), abundance estimation (Patro et al. 2014), and de novo assembly (for example, Grabherr et al. 2011). Alignment-free approaches invert the usual approach to a sequencing experiment: rather than interpreting the reads through the lens of the reference genome (after alignment), the reference genome is interpreted through the lens of the reads. These approaches attempt to exploit the information present in short reads without making any claim about the specific position in the template genome from which the reads originated—an important distinction for reads which cannot be mapped uniquely in the reference assembly.
Holt and McMillan have recently extended a data structure for string compression, the multi-string Burrows–Wheeler transform (msBWT) (Bauer et al. 2013), to next-generation sequencing data (Holt et al. 2014). A msBWT is a compressed, indexed representation of raw, unaligned sequence reads which allows fast queries for specific sequences over very large datasets (Fig. 7b). Whole-genome resequencing reads from the Sanger Mouse Genomes Project plus RNA-seq reads from the diallel experiment have been converted to msBWT for public access at http://www.csbio.unc.edu/CEGSseq/index.py?run=MsbwtTools. Users can query the datasets for the presence of specific sequences, and retrieve the raw reads containing those sequences (Fig. 7c). For instance, to demonstrate the expression of a gene of interest, a user could count how many reads contain subsequences unique to that transcript, such as a subsequence spanning a splice junction.
The msBWT and its associated FM-index also have straightforward extensions to targeted de novo assembly via de Bruijn graphs, and this application is an area of active research.
Discussion and outlook
The approximately 150 Collaborative Cross lines extant in colonies at Tel Aviv University (Tel Aviv, Israel), Geniad Llc (Perth, Australia), and the University of North Carolina (Chapel Hill, NC, USA) are the fruits of a 12-year collaboration between dozens of scientists, students, staff and institutions worldwide. The scale and complexity of the project motivated the development of a suite of informatics resources and experimental tools which are now widely applicable to the CC, its sister population the Diversity Outbred, and other mouse populations. The tools and databases discussed in this review characterize the genetic diversity in the CC, DO, and their founder strains at several evolutionary scales by integrating data from many sources. The Mouse Phylogeny Viewer provides a detailed view of fine-scale patterns of both relatively distant (subspecies of origin within M. musculus) and relatively recent (haplotype blocks passing the four-gamete test within M. m. domesticus) ancestry and population structure in inbred strains and wild mice. Although not per se a CC resource, the Sanger Mouse Genomes Project provides a deep catalog of nucleotide-level variation between the CC founder strains. The CC Viewer allows exploration of local similarity within the CC population by expressing the genomes of CC lines as mosaics of founder haplotypes.
These haplotype mosaics form the basis of genetic analysis and data integration in the CC and DO. In contrast to natural or commercial outbred populations, the founder haplotypes of these multiparental populations (and similar populations in other model organisms) are known and well characterized by sequencing. This presents a tremendous advantage in the search for causal variants of complex traits: provided a genomic segment in an experimental animal can be assigned to a founder haplotype using a few tagging markers, the remaining known variants can be imputed with essentially complete certainty. Annotations such as inferred subspecies ancestry can likewise be projected onto CC and DO genomes once the haplotype mosaic is known. Two software packages, Seqnature and modtools + lapels + suspenders, combine haplotype mosaics with the Sanger variant catalog to perform allele-specific read alignment in next-generation sequencing experiments. A growing list of tools for genetic mapping, including BAGPIPE and DOQTL, takes haplotype mosaics as input in order to map quantitative traits in a fixed-effects or mixed-effects framework.
It is therefore important to understand the relationship between the “average genome” of a CC line, as reflected in its haplotype reconstruction, and the genomes of individual members of that CC line. Although all CC lines assigned “distributable” status have reached >90 % homozygosity, a line remains a dynamic entity. The haplotype reconstructions available in the CC Viewer are averages over a group of individuals who were obligate ancestors (MRCAs) of a line, and represent a snapshot of the line at some point in time between 1 and 5 years (median 3 years) in the past. Present-day CC mice will be more homozygous than the line’s haplotype reconstruction reflects simply due to additional generations of inbreeding and drift accumulated since the MRCAs. Some portion of the regions which were segregating in the MRCAs are almost certain to have fixed during subsequent generations. In this sense, the haplotype reconstruction for a CC line represents a worst-case estimate of residual heterozygosity: it indicates which regions may still be segregating in the line, not which regions are segregating in a group of individuals sampled from that line in the present day. Continued inbreeding will mitigate the impact of residual heterozygosity. However, severe bottleneck events, such as re-derivation of a line in a new facility or initiation of a new breeding colony from a small number of breeding pairs, may create distinct sub-lines which have fixed different alleles at loci which were segregating in the MRCAs. This is no different than the process of sub-line divergence within widely used strains such as the 129 (Cook et al. 2002) or NOD (Simecek et al. 2015) strain groups. Bottlenecks within (nearly) inbred strains can have important phenotypic consequences if they affect causal loci (Rogala et al. 2014; Simecek et al. 2015). Users of the CC should be aware of these considerations when designing experiments and interpreting results.
Although the final number of CC lines is far short of the total envisioned in early discussions (Churchill et al. 2004; Valdar et al. 2006a), the massive extinction during inbreeding provides a unique opportunity to study the mechanisms of intra-genomic incompatibility resulting from admixture between three subspecies along a gradient of genetic isolation. Existing studies of inter-subspecific incompatibility in mouse have so far been limited to pairwise comparisons between M. m. musculus and M. m. domesticus (Forejt et al. 1974; Good et al. 2008) or M. m. castaneus and M. m. domesticus (Orth et al. 1998), either in wild individuals or simple F2 or backcross designs. The CC is the first population of mice in which alleles from all three subspecies may each be present, in homozygosity, over a large fraction within the same genome. As a result of the CC’s balanced factorial breeding scheme such heterosubspecific combinations are expected to be distributed almost uniformly across the genome. The CC thus provides a unique platform for exploring the space of Bateson–Dobzhansky–Muller incompatibilities (Dobzhansky et al. 1936) in mouse. Detailed knowledge of the subspecies contributions to CC genomes, obtained by integrating CC lines’ haplotype mosaics with data from the Mouse Phylogeny Viewer, will be critical to this effort.
Most of the resources discussed in this review ultimately depend on the mouse reference genome. A high-quality, well-annotated reference assembly for any model organism is extremely valuable for the research community. In addition to the genomic sequence itself, a reference genome provides a backbone for annotation and a common coordinate system to anchor genetic maps. Population surveys by microarray genotyping and next-generation sequencing project all genetic variation back onto the reference genome. Predictions about the molecular and organismal phenotypic consequences of genetic variants are likewise based on an annotation derived from the reference sequence. The assumption that most genetic variation can be discovered and defined against a fixed, haploid reference sequence is convenient—and practically useful—but comes at a price. First, large-scale differences in genome content, such as large copy-number variants, are difficult to reconcile to the reference genome. Despite being the most variable fraction of mammalian genomes (Bailey et al. 2002, 2004; She et al. 2008), such variants are highly underrepresented relative to SNPs and small indels in the databases listed in this review. A dramatic example is the male-specific region of the Y chromosome, which differs in size by hundreds of kilobases between inbred strains (Soh et al. 2014). Second, variation in repetitive sequence, including microsatellites, transposable elements, and centromeric sequences, is difficult or impossible to characterize by microarray or short-read sequencing. The Sanger Mouse Genomes Project reported that 13–23 % of the genome is “inaccessible” for SNP and small indel discovery (Keane et al. 2011) by next-generation sequencing with standard methods in any given strain. Finally, differences in sequence organization such as inversions and translocations break collinearity between the genome of an individual and the reference assembly. However, the algorithms underlying many of the databases in this review, including HMMs used for haplotype reconstruction in CC lines, assume collinearity with the reference.
The shortcomings of a single, linear reference genome per species are well appreciated, and richer reference data structures are an active area of research (Church et al. 2015). An alternative is de novo assembly of the genomes of commonly used strains. The Sanger Mouse Genomes Project is using a combination of long-insert “jumping” libraries and optical mapping to build de novo assemblies for 18 laboratory strains including the CC founders. Assembled full-length chromosomes are available on pre-publication release as of June 2015 (ftp://ftp-mouse.sanger.ac.uk/REL-1504-Assembly/). Comparison of strain-specific assemblies to each other and to the reference assembly will provide a much fuller picture of large-scale structural variation between strains. Ab initio gene prediction, integrating both genomic and transcriptome sequence to build strain-specific gene models, is on the horizon. The use of true strain-specific genomes for read alignment, rather than the reference genome or imputed pseudogenomes, will pose new analytical challenges. It will also offer the opportunity to capture biological signals which are not apparent in the present framework.
One remaining gap in the CC infrastructure is the lack of a centralized, public platform for sharing and integrating phenotype data on CC lines. The Mouse Phenome Database (http://phenome.jax.org/) (Grubb et al. 2014) serves this purpose for the strains in the Hybrid Mouse Diversity Panel, and GeneNetwork (http://www.genenetwork.org/webqtl/) provides access to an extensive catalog of phenotypes for more than a dozen advanced intercross and recombinant inbred panels (Williams et al. 2001). These sites have become mainstays in the mouse genetics community and now provide both access to raw data and browser-based tools for data exploration. Accumulation of phenotype data across experiments was a major goal of the original CC design (Churchill et al. 2004); we encourage the CC user community to establish a central “data hub” for this purpose. The Mouse Phenome Database would be a natural choice: it already provides a controlled vocabulary for representing phenotype measurements and enforces correct strain nomenclature to facilitate accurate comparisons across studies. Effective integration of phenotypic and genetic data, facilitated by the databases and analytical tools presented in this review, is critical to realizing the promise of the CC as it exists today.
Acknowledgments
The development of the CC population and related tools at the UNC Systems Genetics Core Facility was supported by Grants from the National Institutes of Health (U01CA134240, P50MH090338, P50HG006582, and U54AI081680); Ellison Medical Foundation (Grant AG-IA-0202-05), and National Science Foundation (Grants IIS0448392 and IIS0812464). Essential support was provided by the Dean of the UNC School of Medicine, the UNC Mutant Mouse Regional Resource Center (U42OD010924), the Lineberger Comprehensive Cancer Center at UNC (U01CA016086 from the National Cancer Institute), and the University Cancer Research Fund from the state of North Carolina. Development of the Mouse Diversity Array was supported by NIH Grant P50GM076468. Support for development of the MegaMUGA array was provided by Neogen Corporation, Lincoln, NE. APM was supported by Grants T32GM067553 and F30MH103925. The authors thank Darla Miller, the UNC Systems Genetics Group, and the Center for Genome Dynamics at the Jackson Laboratory for helpful discussions; and Fernando Pardo-Manuel de Villena and Leonard McMillan for their mentorship and for their comments on this manuscript.
Appendix. terms and definitions
Relatedness
Relatedness in the genetic sense refers to the proportion of alleles shared between two individuals. The degree to which two individuals are genetically related depends on the number of common ancestors they share and the number of generations which have elapsed since they shared them. A pedigree describes the expected relatedness between individuals: first-degree relatives (parents or siblings) share, on average, half of their alleles; second-degree relatives (grandparents) one-fourth; and so on. With dense genotype data, we can instead compute realized relatedness as the proportion of shared, unlinked alleles.
Using dense genotypes, we can define relatedness both at the genome-wide and at the local scale. In the presence of admixture or introgression (see below), local relatedness in different regions of the genome may deviate from the genome-wide average.
Population structure
A population is “structured” when it has experienced deviations from random mating, or equivalently, when it is divided into subpopulations with restricted genetic exchange between them. In a structured population, some groups of individuals are more closely related to (share more alleles with) each other than with other groups. Geography and mating behavior generate at least some degree of structure in most natural populations. Population structure in laboratory mouse strains is widespread: for instance, the 129 and C57BL strain groups form a genetic cluster distinct from so-called “Swiss mice” including FVB/NJ, the NOD substrains, and ICR outbred stock (Beck et al. 2000). Failure to account for population structure can lead to false-positive QTL in genetic mapping of complex traits.
Linkage disequilibrium (LD)
Two loci are said to be in LD if the frequencies of pairwise genotypes depart from those expected if alleles were sampled randomly at each locus. LD is decreased by recombination, and therefore generally decreases with time and with physical distance between loci. Unlinked markers are expected to be in linkage equilibrium, but non-random mating can produce “long-range” LD between unlinked loci in structured populations.
Haplotype block
A haplotype block is a chromosomal segment in which there is no evidence for recombination during the history of a sample of individuals. Within a block, individuals in a population can be collapsed into one of a small (relative to the population size) number of ancestral haplotypes (Wall et al. 2003). LD is relatively high between loci within a block, but relatively low between loci in adjacent blocks.
Although many schemes have been proposed for defining haplotype blocks, the one discussed in this review is the four-gamete test (Hudson et al. 1985). Consider two loci A and B with alleles A,a and B,b, respectively. There are four possible haploid genotypes (gametes)—AB, aB, Ab, and ab—and if all four are observed in a sample, recombination between A and B must have occurred at least once in the past.
Haplotype blocks are a useful means of investigating patterns of genetic diversity at intermediate timescales since a common ancestor, such as among classical inbred strains of mice (Yang et al. 2011). But because recombination events accumulate and LD decreases with time, haplotype blocks shared between two individuals with a common ancestor far in the past—for example, a wild-derived inbred strain and a classical laboratory strain—will be very short. For this reason, haplotype blocks were not inferred for the wild mice and wild-derived strains in Yang et al. (2011).
Identity by descent (IBD)
A chromosomal segment is shared identical-by-descent between two individuals if it was inherited from their common ancestor without recombination. The notion of IBD is closely related to the haplotype block.
Admixture
Admixture refers to inter-breeding between individuals from populations which were previously genetically isolated from one another. Admixture facilitates gene flow between populations, and in the process creates heterogeneity of relatedness across the genome.
Introgression
Introgression refers to the introduction of a chromosomal segment from one population into a separate, genetically distinct population. It is often used to describe gene flow between species or subspecies which can still form fertile hybrids. Unlike admixture, which describes ongoing inter-breeding, introgression describes events which are episodic in nature. In this review, we refer to genetic exchange between mouse subspecies, which do not interbreed in the wild except at narrow hybrid zones (Ursin 1952), as introgression.
Ancestry inference
Broadly speaking, an ancestry-inference procedure steps along the genome of an individual and attempts to assign each segment to one of a few ancestral clusters. These clusters may represent ancestral population groups, for samples from natural populations, or founder haplotypes in laboratory populations. Examples of ancestry inference discussed in this review include assignment of subspecific origin in wild mice (Yang et al. 2011), which labels genomic regions with one of three subspecies; and haplotype reconstruction on the CC and DO (Fu et al. 2012), which assigns genomic regions to one of those populations’ 8 founder strains.
Hidden Markov model (HMM)
A hidden Markov model is a probabilistic model which describes how an observed sequence can be generated from an underlying, unknown sequence of “hidden states” (Baum and Petrie 1966; Rabiner 1989). Efficient algorithms can be used to “decode” the sequence of hidden states given an observed sequence. In this review, we discuss HMMs in which the observed sequences are genotypes along a chromosome, and the hidden states are founder haplotypes.
Footnotes
URLs
BAGPIPE. http://valdarlab.unc.edu/software/bagpipe
BAGPHENOTYPE. http://valdarlab.unc.edu/bagphenotype.html
Collaborative Cross Status website. http://www.csbio.unc.edu/CCstatus/
Collaborative Cross Viewer. http://www.csbio.unc.edu/CCstatus/index.py?run=CCV
DOQTL. http://www.bioconductor.org/packages/release/bioc/html/DOQTL.html
GECCO gene expression browser. http://csbio.unc.edu/gecco/
MDA genotypes for 100 inbred strains. http://cgd.jax.org/datasets/popgen/diversityarray/yang2011.shtml
MegaMUGA genotypes for CC founder strains. http://csbio.unc.edu/CCstatus/index.py?run=GeneseekMM
modtools + lapels + suspenders pipeline. http://www.csbio.unc.edu/CCstatus/index.py?run=Pseudo
Mouse Imputation Resource. http://csbio.unc.edu/imputation/
Mouse Phylogeny Viewer. http://msub.csbio.unc.edu/
Sanger Mouse Genomes Project. http://www.sanger.ac.uk/resources/mouse/genomes/
Searchable index of sequencing reads from CC founder strains. http://www.csbio.unc.edu/CEGSseq/index.py?run=MsbwtTools
Seqnature. https://github.com/jaxcs/Seqnature
References
- Aylor DL, Valdar W, Foulds-Mathes W, et al. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 2011;21:1213–1222. doi: 10.1101/gr.111310.110. doi:10.1101/gr.111310.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. doi:10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Baertsch R, Kent WJ, et al. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004;5:R23. doi: 10.1186/gb-2004-5-4-r23. doi:10.1186/gb-2004-5-4-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Kajita S, Walker M, et al. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 2015;11:e1004916. doi: 10.1371/journal.pgen.1004916. doi:10.1371/journal.pgen.1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer MJ, Cox AJ, Rosone G, et al. Lightweight algorithms for constructing and inverting the BWT of string collections. Theor Comput Sci. 2013;483:134–148. doi:10.1016/j.tcs.2012.02.002. [Google Scholar]
- Baum LE, Petrie T. Statistical inference for probabilistic functions of finite state Markov chains. Ann Math Stat. 1966;37:1554–1563. [Google Scholar]
- Beck JA, Lloyd S, Hafezparast M, et al. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. doi:10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, et al. Controlling the false-discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. doi:10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursot P, Auffray JC, Britton-Davidian J, Bonhomme F, et al. The evolution of house mice. Annu Rev Ecol Syst. 1993;24:119–152. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA, et al. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Calaway JD, Lenarcic AB, Didion JP, et al. Genetic architecture of skewed X inactivation in the laboratory mouse. PLoS Genet. 2013;9:e1003853. doi: 10.1371/journal.pgen.1003853. doi:10.1371/journal.pgen.1003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCC et al. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. doi:10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. doi:10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, et al. Out of the bottleneck: the Diversity Outcross and Collaborative Cross mouse populations in behavioral genetics research. Mamm Genome. 2014;25:3–11. doi: 10.1007/s00335-013-9492-9. doi:10.1007/s00335-013-9492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DM, Schneider VA, Steinberg KM, et al. Extending reference assembly models. Genome Biol. 2015;16:13. doi: 10.1186/s13059-015-0587-3. doi:10.1186/s13059-015-0587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW, et al. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. doi:10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Clark AG, Hubisz MJ, Bustamante CD, et al. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. doi:10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MN, Bolivar V, McFadyen MP, Flaherty L, et al. Behavioral differences among 129 substrains: implications for knockout and transgenic mice. BehavNeurosci. 2002;116:600–611. doi:10.1037/0735-7044.116.4.600. [PubMed] [Google Scholar]
- Crowley JJ, Zhabotynsky V, Sun W, et al. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat Genet. 2015 doi: 10.1038/ng.3222. doi:10.1038/ng.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler HD, Calus MPL, Pong-Wong R, et al. Genomic prediction in animals and plants: simulation of data, validation, reporting, and benchmarking. Genetics. 2013;193:347–365. doi: 10.1534/genetics.112.147983. doi:10.1534/genetics.112.147983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion JP, Yang H, Sheppard K, et al. Discovery of novel variants in genotyping arrays improves genotype retention and reduces ascertainment bias. BMC Genom. 2012;13:34. doi: 10.1186/1471-2164-13-34. doi:10.1186/1471-2164-13-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion JP, de Villena FP-M, et al. Deconstructing Mus gemischus: advances in understanding ancestry, structure, and variation in the genome of the laboratory mouse. Mamm Genome. 2013;24:1–20. doi: 10.1007/s00335-012-9441-z. doi:10.1007/s00335-012-9441-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, et al. Studies on hybrid sterility. II Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B, Ram R, Handoko HY, et al. Melanoma susceptibility as a complex trait: genetic variation controls all stages of tumor progression. Oncogene. 2014 doi: 10.1038/onc.2014.227. doi:10.1038/onc.2014.227. [DOI] [PubMed] [Google Scholar]
- Ferris MT, Aylor DL, Bottomly D, et al. Modeling host genetic regulation of influenza pathogenesis in the Collaborative Cross. PLoS Pathog. 2013;9:e1003196. doi: 10.1371/journal.ppat.1003196. doi:10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. doi:10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forejt J, Ivanyi P, et al. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.) Genet Res. 1974;24:189–206. doi: 10.1017/s0016672300015214. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. doi:10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Fu C-P, Welsh CE, de Villena FP-M, McMillan L, et al. Inferring ancestry in admixed populations using microarray probe intensities. Proceedings of the ACM conference on bioinformatics, computational biology and biomedicine—bCB’12; New York. ACM Press; 2012. pp. 105–112. [Google Scholar]
- Gatti DM, Svenson KL, Shabalin A, et al. Quantitative trait locus mapping methods for Diversity Outbred mice. 2014;G3(4):1623–1633. doi: 10.1534/g3.114.013748. doi:10.1534/g3.114.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Hill J, et al. Data analysis using regression and multilevel/hierarchical models. Cambridge University Press; Cambridge: 2007. [Google Scholar]
- Geraldes A, Basset P, Gibson B, et al. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol Ecol. 2008;17:5349–5363. doi: 10.1111/j.1365-294X.2008.04005.x. doi:10.1111/j.1365-294X.2008.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Rau CD, Farber CR, et al. Hybrid Mouse Diversity Panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm Genome. 2012;23:680–692. doi: 10.1007/s00335-012-9411-5. doi:10.1007/s00335-012-9411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales NM, Palmer AA, et al. Fine-mapping QTLs in advanced intercross lines and other outbred populations. Mamm Genome. 2014;25:271–292. doi: 10.1007/s00335-014-9523-1. doi:10.1007/s00335-014-9523-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Dean MD, Nachman MW, et al. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics. 2008;179:2213–2228. doi: 10.1534/genetics.107.085340. doi:10.1534/genetics.107.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. doi:10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb SC, Bult CJ, Bogue MA, et al. Mouse phenome database. Nucleic Acids Res. 2014;42:D825–D834. doi: 10.1093/nar/gkt1159. doi:10.1093/nar/gkt1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA, et al. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. doi:10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S41–S49. doi: 10.1186/gb-2006-7-s1-s4. doi:10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt J, McMillan L, et al. Merging of multi-string BWTs with applications. Bioinformatics. 2014;30:3524–3531. doi: 10.1093/bioinformatics/btu584. doi:10.1093/bioinformatics/btu584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Holt J, Kao C-Y, et al. A novel multi-alignment pipeline for high-throughput sequencing data. Database. 2014;2014:bau057. doi: 10.1093/database/bau057. doi:10.1093/database/bau057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL, et al. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqi FA, Athamni H, Dorman A, et al. Heritability and coefficient of genetic variation analyses of phenotypic traits provide strong basis for high-resolution QTL mapping in the Collaborative Cross mouse genetic reference population. Mamm Genome. 2014;25:109–119. doi: 10.1007/s00335-014-9503-5. doi:10.1007/s00335-014-9503-5. [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. doi:10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, et al. The UCSC genome browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. doi:10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. doi:10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelada SNP, Aylor DL, Peck BCE, et al. Genetic analysis of hematological parameters in incipient lines of the Collaborative Cross. 2012;G3 2:157–165. doi: 10.1534/g3.111.001776. doi:10.1534/g3.111.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelada SNP, Carpenter DE, Aylor DL, et al. Integrative genetic analysis of allergic inflammation in the murine lung. Am J Respir Cell Mol Biol. 2014;51:436–445. doi: 10.1165/rcmb.2013-0501OC. doi:10.1165/rcmb.2013-0501OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic AB, Svenson KL, Churchill GA, Valdar W, et al. A general Bayesian approach to analyzing diallel crosses of inbred strains. Genetics. 2012;190:413–435. doi: 10.1534/genetics.111.132563. doi:10.1534/genetics.111.132563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Liu Y, et al. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–835. doi: 10.1038/nmeth.1681. doi:10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- Liu EY, Zhang Q, McMillan L, et al. Efficient genome ancestry inference in complex pedigrees with inbreeding. Bioinformatics. 2010;26:i199–i207. doi: 10.1093/bioinformatics/btq187. doi:10.1093/bioinformatics/btq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EY, Morgan AP, Chesler EJ, et al. High-resolution sexspecific linkage maps of the mouse reveal polarized distribution of crossovers in male germline. Genetics. 2014;197:91–106. doi: 10.1534/genetics.114.161653. doi:10.1534/genetics.114.161653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. doi:10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, et al. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA. 2000;97:12649–12654. doi: 10.1073/pnas.230304397. doi:10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger SC, Raghupathy N, Choi K, et al. RNA-seq alignment to individualized genomes improves transcript abundance estimates in multiparent populations. Genetics. 2014;198:59–73. doi: 10.1534/genetics.114.165886. doi:10.1534/genetics.114.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth A, Adama T, Din W, Bonhomme F, et al. Natural hybridization between two subspecies of the house mouse, Mus musculus domesticus and Mus musculus castaneus, near Lake Casitas, California. Genome. 1998;41:104–110. doi: 10.1139/g97-109. [DOI] [PubMed] [Google Scholar]
- Patro R, Mount SM, Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol. 2014;32:462–646. doi: 10.1038/nbt.2862. doi:10.1038/nbt. 2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. doi:10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillippi J, Xie Y, Miller DR, et al. Using the emerging Collaborative Cross to probe the immune system. Genes Immun. 2014;15:38–46. doi: 10.1038/gene.2013.59. doi:10.1038/gene.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner LR. A tutorial on hidden Markov models and selected applications in speech recognition. Proc IEEE. 1989;77:257–286. [Google Scholar]
- Rasmussen AL, Okumura A, Ferris MT, et al. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014 doi: 10.1126/science.1259595. doi:10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogala AR, Morgan AP, Christensen AM, et al. The Collaborative Cross as a resource for modeling human disease: CC011/Unc, a new mouse model for spontaneous colitis. Mamm Genome. 2014;25:95–108. doi: 10.1007/s00335-013-9499-2. doi:10.1007/s00335-013-9499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Cheng Z, Zöllner S, et al. Mouse segmental duplication and copy number variation. Nat Genet. 2008;40:909–914. doi: 10.1038/ng.172. doi:10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simecek P, Churchill GA, Yang H, et al. Genetic analysis of substrain divergence in NOD mice. 2015;G3(5):771–775. doi: 10.1534/g3.115.017046. doi:10.1534/g3.115.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh YQS, Alföldi J, Pyntikova T, et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159:800–813. doi: 10.1016/j.cell.2014.09.052. doi:10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, et al. High-resolution genetic mapping using the mouse Diversity Outbred population. Genetics. 2012;190:437–447. doi: 10.1534/genetics.111.132597. doi:10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BA, Heiniger HJ, Meier H, et al. Genetic analysis of resistance to cadmium-induced testicular damage in mice. Proc Soc Exp Biol Med. 1973;143:629–633. doi: 10.3181/00379727-143-37380. [DOI] [PubMed] [Google Scholar]
- Ursin E. Occurrence of voles, mice, and rats (Muridae) in Denmark, with a special note on a zone of intergradation between two subspecies of the house mouse (Mus musculus L.) Vid Medd Dansk Naturhist Foren. 1952;114:217–244. [Google Scholar]
- Valdar W, Flint J, Mott R, et al. Simulating the Collaborative Cross: power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics. 2006a;172:1783–1797. doi: 10.1534/genetics.104.039313. doi:10.1534/genetics.104.039313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006b;38:879–887. doi: 10.1038/ng1840. doi:10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- Valdar W, Holmes CC, Mott R, Flint J, et al. Mapping in structured populations by resample model averaging. Genetics. 2009;182:1263–1277. doi: 10.1534/genetics.109.100727. doi:10.1534/genetics.109.100727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CM, Kulbokas EJ, Kirby AW, et al. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. doi:10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- Wall JD, Pritchard JK, et al. Haplotype blocks and linkage disequilibrium in the human genome. Nat Rev Genet. 2003;4:587–597. doi: 10.1038/nrg1123. doi:10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- Wang J, Moore KJ, Zhang Q, et al. Genome-wide compatible SNP intervals and their properties. Proceedings of the first aCM international conference on bioinformatics and computational biology—bCB’10; New York. ACM Press; 2010. p. 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JR, de Villena FP-M, Lawson HA, et al. Imputation of single-nucleotide polymorphisms in inbred mice using local phylogeny. Genetics. 2012a;190:449–458. doi: 10.1534/genetics.111.132381. doi:10.1534/genetics.111.132381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JR, de Villena FP-M, McMillan L, et al. Comparative analysis and visualization of multiple collinear genomes. BMC Bioinform. 2012b;13(Suppl 3):S13. doi: 10.1186/1471-2105-13-S3-S13. doi:10.1186/1471-2105-13-S3-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. doi:10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weiser M, Mukherjee S, Furey TS, et al. Novel distal eQTL analysis demonstrates effect of population genetic architecture on detecting and interpreting associations. Genetics. 2014;198:879–893. doi: 10.1534/genetics.114.167791. doi:10.1534/genetics.114.167791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Gu J, Qi S, Lu L, et al. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2001;2:46. doi: 10.1186/gb-2001-2-11-research0046. doi:10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Bennett B, Lu L, et al. Genetic structure of the LXS panel of recombinant inbred mouse strains: a powerful resource for complex trait analysis. Mamm Genome. 2004;15:637–647. doi: 10.1007/s00335-004-2380-6. doi:10.1007/s00335-004-2380-6. [DOI] [PubMed] [Google Scholar]
- Wilming LG, Gilbert JGR, Howe K, et al. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. doi:10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, de Villena FPM, et al. On the subspecific origin of the laboratory mouse. Nat Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. doi:10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- Yang H, Ding Y, Hutchins LN, et al. A customized and versatile high-density genotyping array for the mouse. Nat Methods. 2009;6:663–666. doi: 10.1038/nmeth.1359. doi:10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 2011;43:648–655. doi: 10.1038/ng.847. doi:10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang W, Valdar W, et al. Bayesian modeling of haplotype effects in multiparent populations. Genetics. 2014;198:139–156. doi: 10.1534/genetics.114.166249. doi:10.1534/genetics.114.166249. [DOI] [PMC free article] [PubMed] [Google Scholar]