Abstract
Auxinic herbicides (e.g. dicamba) are extensively used in agriculture to selectively control broadleaf weeds. Although cultivated species of Brassicaceae (e.g. Canola) are susceptible to auxinic herbicides, some biotypes of Sinapis arvensis (wild mustard) were found dicamba resistant in Canada. In this research, dicamba tolerance from wild mustard was introgressed into canola through embryo rescue followed by conventional breeding. Intergeneric hybrids between S. arvensis (2n = 18) and B. napus (2n = 38) were produced through embryo rescue. Embryo formation and hybrid plant regeneration was achieved. Transfer of dicamba tolerance from S. arvensis into the hybrid plants was determined by molecular analysis and at the whole plant level. Dicamba tolerance was introgressed into B. napus by backcrossing for seven generations. Homozygous dicamba-tolerant B. napus lines were identified. The ploidy of the hybrid progeny was assessed by flow cytometry. Finally, introgression of the piece of DNA possibly containing the dicamba tolerance gene into B. napus was confirmed using florescence in situ hybridization (FISH). This research demonstrates for the first time stable introgression of dicamba tolerance from S. arvensis into B. napus via in vitro embryo rescue followed by repeated backcross breeding. Creation of dicamba-tolerant B. napus varieties by this approach may have potential to provide options to growers to choose a desirable herbicide-tolerant technology. Furthermore, adoption of such technology facilitates effective weed control, less tillage, and possibly minimize evolution of herbicide resistant weeds.
Introduction
Auxinic herbicides were the first synthetic herbicides discovered and have been in use for several decades in agriculture. These herbicides have been a favorite choice among farmers worldwide as they selectively control broadleaf weeds. As such, auxinic herbicides cannot be used in broadleaf crops. Despite seven decades of extensive use, only 32 auxinic herbicide-resistant biotypes have been documented worldwide [1]; a relatively low number compared to herbicides with other modes of action such as acetolactate synthase (ALS)-inhibitors or triazines. Due to intense selection pressure, Sinapis arvensis (wild mustard) biotypes evolved resistance to auxinic herbicides in western Canadian cereal fields [2]. These auxinic herbicide-resistant S. arvensis biotypes were found to be highly resistant to both picloram and dicamba (104 fold) [2]. Following extensive morphological, physiological, biochemical, and molecular characterization, these S. arvensis auxinic herbicide-resistant biotypes have been used as a model species to study the mechanism of resistance and the mode of action of auxinic herbicides [3–5]. Genetic analysis of auxinic herbicide-resistant S. arvensis suggests that the resistance trait behaves as a single dominant Mendelian gene [6,7]. Furthermore, we recently reported the identification of morphological and molecular markers-linked to the dicamba resistance trait in S. arvensis [8].
Brassica crops are among the oldest crop species in agronomic history, with records of their cultivation dating back to 1500 BC [9]. Among the approximately 350 genera within Brassicaceae are numerous economically important oil and vegetable crops. Canola, a Brassica napus variety developed for its low eurcic acid, is one of the world’s most important oilseed crops [10]. Seed yield in B. napus crops is highly sensitive to weed competition, and poor weed management results in significant reduction in yield [11]. The successful introduction and adoption of glyphosate- and glyphosate-resistant canola varieties reflects the canola industry’s demand and need for simple and effective weed management. Despite its initial success, the long term viability of cropping glyphosate-resistant canola has come under scrutiny as reports of glyphosate-resistant weeds have become increasingly prevalent in the last decade [1]. One of the primary reasons for the development of glyphosate resistance in weeds is due to the sole and continual use of glyphosate in glyphosate–resistant cropping systems [12]. The continuous use of herbicides with the same mode of action creates selection pressure and results in the evolution of herbicide-resistant biotypes in weeds naturally found in these crop fields. The availability of cultivars resistant to herbicides with different mechanisms of action will allow farmers to rotate herbicides between crops thereby breaking selection pressure and effectively minimizing the possibility of evolving herbicide resistance in nearby weed species.
Dicamba is an important auxinic herbicide in monocotyledonous cropping systems such as corn and other cereals. Dicamba is used both as a pre-emergence and post emergence herbicide for selectively controlling broadleaf weeds. During the past several decades, the demand for dicamba in agriculture has remained consistent despite the introduction of herbicides (glyphosate, triazines, and ALS-inhibitors) with different modes of action primarily because of its selectivity, efficacy, and low application costs [13]. Selectivity for broadleaf weeds is also a major limitation in the use of dicamba as it cannot be used in broadleaf crops such as canola. The availability of dicamba-resistant canola cultivars will not only offer canola farmers an alternative low-cost herbicide option for crop protection, but may reduce the evolution of weeds resistant to current herbicides.
A number of intergeneric hybrids were developed previously between S. arvensis and B. napus [14–16]. Hu et al. [17] reported that intergeneric hybrids between B. napus and S. arvensis could be produced by using protoplast fusion. The numbers of hybrid progeny reported in these studies were low and most of the progeny were sterile. The overall goal of our research was to transfer dicamba tolerance from S. arvensis to B. napus by means of conventional breeding. The specific objectives were to: a) produce intergeneric hybrids between dicamba-resistant S. arvensis and B. napus via embryo rescue b) introgress dicamba tolerance into B. napus through backcrossing and c) demonstrate the stable introgression of dicamba tolerance into B. napus.
Methods
Production of hybrids from auxinic-herbicide resistant S. arvensis and B. napus crosses
Dicamba-resistant S. arvensis and -susceptible B. napus were raised from seed in six inch plastic pots containing Promix (Plant Products, Brampton, Canada). All plants were grown in a growth chamber under a 16-h photoperiod with a light intensity of 350 μmoles.s-1m-2 and 22/15°C day/night temperatures. Plants were irrigated as required and fertilized weekly with 20-8-20 fertilizer. To confirm dicamba resistance, plants of S. arvensis were treated by spraying with dicamba (Banvel, BASF, USA) at 200 g acid equivalent (ae)/ha at the three- to four-leaf stage (procedure for dicamba application below).
Intergeneric F1 hybrids between dicamba-resistant S. arvensis and B. napus were produced through reciprocal crosses following the procedures described by Jugulam et al. [7]. Embryo rescue was required as hybrid seeds failed to develop. To optimize hybrid plant regeneration, two embryo rescue protocols, i.e., embryo culture and silique culture, were used according to Mithila and Hall [18]. Established plantlets were transferred to soil and were grown in the growth chamber.
Assessment of transfer of dicamba tolerance into hybrids
Molecular assay
We developed a genetic map describing AFLP (amplified fragment length polymorphism) molecular markers closely-linked to dicamba tolerance in S. arvensis [8]. The two closest markers that were identified and sequenced were 1.58 and—6.35 map units flanking the resistance locus, and were designated M5 and M2, respectively [8]. To confirm the presence of marker closely-linked to dicamba tolerance in the hybrid progeny, PCR was conducted using primers generated from the M5 molecular marker (forward primer: GGCCGCGAGACATTGGTGA and reverse primer: TCTCTCGTGACCCTTTACAATTAG ). The expected amplicon size was 225 bp. Genomic DNA was extracted from the leaf tissue of hybrid progeny and their parental plants using DNeasy® Plant Mini (QIAGEN, Mississauga, ON, Canada). Thirty cycles of PCR were conducted using the following conditions: initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 30 seconds, 56°C for 30 sec and 72°C for 45 sec with a final extension at 72°C for 7 min. Dicamba tolerance was determined by the presence of the expected 225 bp amplification product.
Whole-plant assay
Dicamba-resistant S. arvensis, susceptible B. napus and hybrid plants were grown and treated with dicamba 200 g ae ha-1 at the three- to four- leaf stage using a motorized hood sprayer. The sprayer was equipped with a flat-fan nozzle (8002 E) and calibrated to a spray rate of 200 l/ha. One and two weeks post-spray, seedlings were visually rated for injury, indicated by foliar epinasty (downward curlingof plant parts and a typical symptom of auxinic herbicides) and senescence. Hybrid plants were classified dicamba-tolerant or -susceptible by comparing the injury response with those of dicamba-resistant S. arvensis and–susceptible B. napus plants.
Assessment of fertility of hybrids
Pollen fertility/viability and stigma receptivity of dicamba-tolerant hybrids were assessed. Acetocarmine stain was used to test pollen viability in the hybrid progeny. Upon anthesis, the flowers were collected and the pollen was transferred onto a microscope slide, stained with 1% acetocarmine, and observed under a compound microscope (40X). Stigma receptivity of the hybrid plants was tested by performing crosses with wild-type B. napus as the pollen donor.
Repeated backcrosses to introgress dicamba tolerance from hybrids into B. napus
Backcrosses were performed using the dicamba-tolerant intergeneric hybrid as the donor parent and B. napus as the recurrent parent. BC3F1 plants were grown and treated with three doses of dicamba; i.e. 200, 400 or 600 g ae ha-1 at the three- to four- leaf stage (as described above). However, progeny from all seven backcross generation were tested with 200 g ae ha-1 dicamba. Chi square analysis was performed by SAS (version 9.3; Cary, NC) to determine the goodness of fit to a 1:1 segregation, i.e. tolerance: susceptible. After seven backcrosses, homozygous dicamba-tolerant B. napus plants were identified through three subsequent generations of self-pollination. Dicamba tolerance was confirmed via whole plant screening with dicamba spray and PCR analysis of the tolerant plants.
DNA ploidy of hybrids and backcross progeny
The DNA ploidy of the initial intergeneric hybrids and their backcrossed progeny were assessed by flow cytometry following the protocol of Kron et al. [19]. Dicamba-resistant S. arvensis (diploid) and B. napus (tetraploid) served as controls.
Florescent in situ hybridization
Fluorescent in situ hybridization (FISH) was performed to confirm the integration of the dicamba tolerance gene into the nuclear genome of the dicamba-tolerant B. napus plants. Homozygous dicamba-tolerant B. napus line (11-12-8) seeds were germinated on wet filter paper at room temperature in the dark for 2–3 days. Root-tips (5–7 mm) were harvested and fixed in Carnoy’s I fixative (3 parts 95% ethanol: 1 part acetic acid) for a minimum of one hour and no longer than sixteen hours. For chromosome preparation, the meristematic area of the root tip was excised and macerated in 45% acetic acid on a glass slide, then subsequently mounted with a glass coverslip. The eraser end of a wooden pencil was used to tap and dispersed the root tissue evenly. Slides were observed under a phase contrast microscope and those containing cells with full chromosome complements were stored at -80°C.
The fluorescent probe, produced from the 225 bp PCR product previously amplified, was labeled with Cy3 fluorochrome dCTP. Coverslips were removed from the frozen slide with a razor blade, and slides were placed in a 70% and 95% ethanol series to dehydrate the tissue, then air dried. RNA contamination was removed by incubating slides at 37°C for 60 min with RNase solution diluted to 100 μg/ml in 2X saline-sodium citrate (SSC) buffer. Tissues on slides were washed in 2X SSC, dehydrated in the ethanol series (70% and 95%), and air dried. The hybridization mixture was prepared with 50% de-ionized formamide, 10% dextran sulfate, 2X SSC and probe DNA. The hybridization solution was denatured at 75°C for 10 min and cooled on ice prior to being applied to chromosome preparations at a concentration of 100 ng of probe DNA per slide. The coverslips on each slide was sealed with liquid cement and incubated for 10 min at 80°C to denature chromosomal DNA. Hybridization occurred over 16 hours at 37°C in a sealed humidity chamber. Subsequently, slide covers were removed and slides were washed for 10 min in 2X SSC at room temperature, followed by two 15- min washes in 2X SSC at 37°C, concluding with a final wash in 4X SSC plus 0.2% Tween 20 surfactant for 10 min before air drying. The slides were mounted with Vectashield mounting media (Vector Labs) containing 4, 6-diamino-2-phenylindole (DAPI). DAPI (excitation at 360 nm and emission at 460 nm) fluoresces blue when bound to DNA. Cy3 fluorochrome (excitation at 550 nm and emission at 570 nm) fluoresces orange/red. The fluorescence signals were visualized using a Leica DM 5500 fluorescent microscope.
Results
Production of hybrids between S. arvensis and B. napus
Among two embryo rescue protocols used as described by Mithila and Hall [18], strategy II (i.e. culturing of immature siliques on medium prior to embryo rescue) was best for producing B. napus x S. arvensis hybrids (Table 1). Hybrids were produced only when B. napus was used as the maternal parent. The progeny exhibited several morphological traits of both the parents e.g. leaf shape, stem, plant height and flower color (Fig 1).
Table 1. Hybrid production between Brassica napus and Sinapis arvensis: frequency of embryo regeneration and hybrid plant establishment upon direct and reciprocal crosses followed by in vitro culture and embryo rescue.
| Cross Combination | # of buds pollinated | # of siliques cultured | # of embryos excised | # of embryos germinated | # of hybrids obtained |
|---|---|---|---|---|---|
| Strategy I | |||||
| 1. B. napus X S. arvensis | 200 | - | 60 | 2 | 2 |
| 2. S. arvensis X B. napus | 200 | - | 30 | 0 | 0 |
| Strategy II | |||||
| 1. B. napus X S. arvensis | 150 | 110 | 55 | 9 | 9 |
| 2. S. arvensis X B. napus | 150 | 120 | 10 | 0 | 0 |
Strategy I: Siliques harvested 10–12 days after pollination, embryos excised and cultured on media. Strategy II: Immature siliques harvested 3–5 days after pollination, cultured on media for 2 weeks and after which embryos were harvested and cultured on media.
Fig 1. Production of hybrids between B. napus and S. arvensis.

A and B represent B. napus and S. arvensis plants, respectively. C is a triploid hybrid plant produced by crossing B. napus and S. arvensis.
Assessment of dicamba tolerance in hybrids
PCR results demonstrated successful amplification of the expected 225 bp fragment, closely-linked to dicamba tolerance in S. arvensis and hybrids (S1 Fig). Sixty-seven percent of the hybrid plants were found to contain the marker closely-linked to dicamba tolerance and also survived dicamba (200 g ae ha-1) application.
Assessment of fertility and DNA ploidy of hybrids and backcross progeny
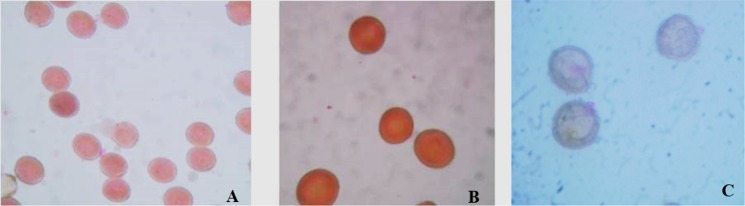
Pollen/stigma fertility tests demonstrated that among the hybrids that were found tolerant to dicamba, only 20% and 50% were male and female fertile, respectively. When stained with acetocarmine, viable pollen appeared stained and spherical in shape; conversely, non-viable pollen was colorless (Fig 2). The male fertile hybrids were maintained in vitro by clonal propagation. Flow cytometry data indicated the the F1 hybrids were all triploids with an estimated ploidy range of 3.1 to 3.2 (triploid); while the parents S. arvensis and B. napus had an estimated ploidy of 1.88 to 1.9, (diploid) and 4 to 4.01 (tetraploid) (S1 Table), respectively. However, the progeny from the initial few backcrosses consisted of either tetraploids, pentaploids, or anueploids and a large number of plants from backcross four onwards were found to be tetraploids (S1 Table).
Fig 2. Pollen viability test (stained with 1% acetocarmine).
A and B are viable pollen of B. napus and hybrid, respectively; whereas, C represents non-viable hybrid pollen. Pictures were taken at 40X magnification.
Assessment of dicamba tolerance and segregation in backcrosses progeny
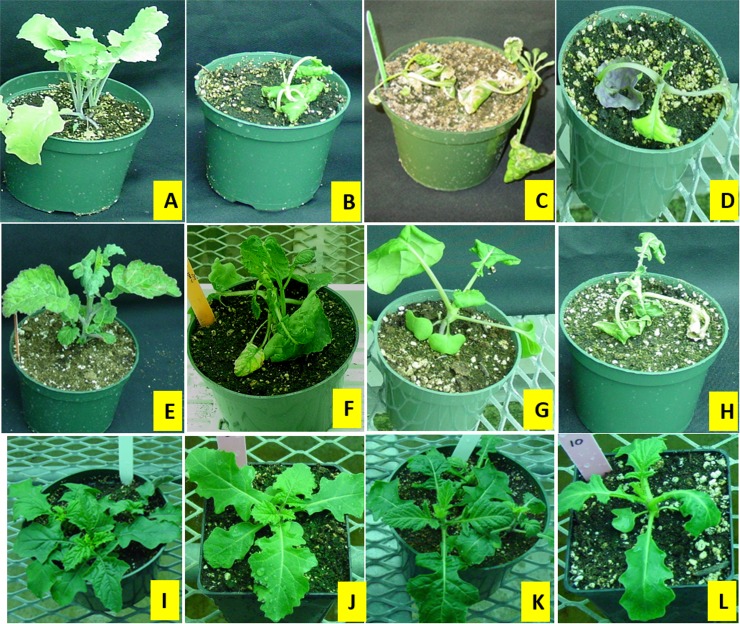
The dose-response of BC3F1 hybrids suggests that some of the hybrid plants were tolerant to dicamba up to 400 g ae ha-1 with their response comparable to dicamba-resistant S. arvensis treated with the same dose (Fig 3) and others were severely injured and eventually, died. However, the tolerant hybrids were found slightly injured at 600 g ae ha-1 dicamba (Fig 3). The progeny from other backcrosses showed varying responses following 200 g ae ha-1 dicamba application. Some seedlings exhibited severe epinasty one week post spray and eventually died while others showed only minor dicamba injury initially and recovered. When progeny from each backcross were treated with dicamba, based on the probabilities, plants in backcrosses 5, 6 and 7 appear to segregate 1:1 for R: S (Table 2 and Fig 4) as expected for a single dominant Mendelian trait.
Fig 3. Plant response to dicamba (200, 400 or 600 g ae ha-1) 2 weeks after treatment.
Top and bottom row represent B. napus and S. arvensis, respectively. The middle row shows the response of dicamba-tolerant BC3F1 hybrids. A, E and I are untreated plants; B, F and J illustrate plant response to 200 g ae ha-1; C, G and K represent plant response to 400 g ae ha-1; whereas, D, H, L indicate plant response to 600 g ae ha-1.
Table 2. Response of backcross progeny upon dicamba treatment and evaluation of fertility of the progeny.
Segregation of resistant (R) and susceptible (S) plants in backcross progeny following treatment with 200 g ae/ha of dicamba.
| Backcross | Segregation of plants | χ 2 | P-value | Fertility of R plants | |
|---|---|---|---|---|---|
| R | S | ||||
| BC1F1 | 5 | 0 | - | - | Plants were pollen fertile and female sterile |
| BC2F1 | 4 | 5 | 0.11 | 0.7389 | Only 1 R plant was pollen fertile |
| BC3F1 | 30 | 9 | 11.3 | 0.0008 | 17 R plants were pollen fertile |
| BC4F1 | 9 | 26 | 8.25 | 0.0041 | All R plants were pollen fertile |
| BC5F1 | 19 | 15 | 0.47 | 0.4927 | All R plants were pollen fertile |
| BC6F1 | 17 | 13 | 0.53 | 0.4652 | All R plants were pollen fertile |
| BC7F1 | 17 | 16 | 0.03 | 0.8618 | All R plants were pollen fertile |
χ 2:Chi-square values are the results of tests for goodness of fit to a 1:1 (R:S) segregation model. P-value significance is determined at < 0.05.
Fig 4. Plant response to dicamba (200 g ae ha-1) 2 weeks after treatment.
A, B, represent B. napus (epinasty) and S. arvensis (no epinasty), respectively. C and D segregation of dicamba-susceptible (epinasty) and–tolerant (no epinasty) plants in BC6F1 generation, respectively.
Identifying homozygous dicamba-tolerant B. napus lines
A homozygous dicamba-tolerant BC7F3 line (11-12-8) was identified when all the progeny tested from this line were tolerant to dicamba spray (200 g ae ha-1). Furthermore, PCR analysis of all the sprayed plants in line 11-12-8 showed the presence and amplification of the M5 molecular marker (225 bp; S2 Fig). The estimated DNA ploidy (based on flow cytometry data) of this line (S1 Table) was similar to that of B. napus suggesting that the amount of genetic material transferred from S. arvensis to B. napus was minimal.
Fluorescent in situ hybridization
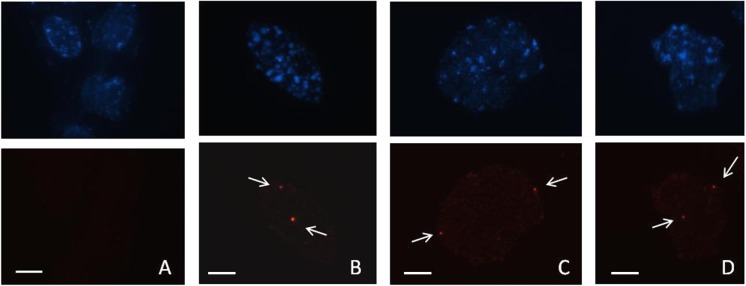
A short probe (225 bp) linked to the M5 molecular marker directly labeled with a Cy3 fluorochrome was detected in the hybridized B. napus line (11-12-8) chromosome suggesting the dicamba-tolerant gene was integrated into the nuclear genome of B. napus. Fluoresence of the Cy3 probe, indicated by reddish- orange dots (Fig 5), was clearly visible in individual root cells of the dicamba-tolerant B. napus line. No fluorescence was observed in the nuclear material of the dicamba-susceptible B. napus control (Fig 5A). Within each intact cell, two spatially separated red fluorescing points were observed indicating the presence of the transferred gene in a homozygous state on two homologous chromosomes (Fig 5B–5D).
Fig 5. Fluorescence in situ hybridization of a 225 bp introgressed from S. arvensis labeled with a Cy3 fluorochrome.
Panel A is the prophase stage cells from B. napus. Panels B-D are prophase stage cells from the root-tips of B. napus BC7F3 line (11-12-8). The top row in each panel is visualized with a filter for DAPI where DNA fluoresces blue. The bottom panel is the same cell visualized with a filter that detects Cy3 fluorescence; each cell in BC7F3 line (11-12-8) has two red/orange fluorescing dots (arrows) suggesting a homologous insertion. Bar = 1 μm.
Discussion
Successful gene transfer from intergeneric and interspecific crosses using conventional plant breeding techniques requires overcoming several barriers, including pollen stigma incompatibility, embryo/endosperm imbalance, sterility, and a lack of homologous chromosome pairing between the species. Cytogenetic analyses of B. napus and S. arvensis hybrids by Mizusima [20, 21] suggested that partial homologies can be established on the basis of allosyndetic pairing. In this research we produced fertile intergeneric hybrids between dicamba-susceptible B. napus and dicamba-resistant S. arvensis and conducted a series of backcrosses to produce a dicamba-tolerant B. napus line.
The BC1F1 plants showed a dose-dependent response to dicamba application (Fig 3), possibly because there may be some other genes from B. napus influencing the phenotype response upon treatment with dicamba. The observed Mendelian segregation ratios for dicamba tolerance or susceptibility in the later generations of backcrossed progeny (BC5F1, BC6F1, BC7F1) followed expected segregation of a single dominant trait and confirms earlier work of Jasieniuk et al. [6] where dicamba resistance in S. arvensis was found to be controlled by a single dominant nuclear allele. However, the segregation of dicamba tolerance or susceptibility in initial backcross progeny (i.e. BC1F1 to BC4F1), did not fit single gene inheritance configuration (Table 2). This could be because in BC3F1 there were more aneuploids than tetraploids (S1 Table). Dicamba tolerance was also confirmed by the presence of the M5 molecular marker (which was found closely linked to dicamba resistance in S. arvensis; Mithila et al. [8]) in all dicamba-tolerant progeny from homozygous BC7F3 line (11-12-8), suggesting a true-breeding dicamba-tolerant line. These homozygous BC7F3 plants that survived dicamba application (200 g ae/ha) also completed their life cycle with normal seed production (data not shown). However, future studies are required to assess fitness of these plants in terms of biomass and seed production in comparison with dicamba-susceptible B. napus.
FISH analysis confirmed the transfer of the wild-mustard DNA segment associated with dicamba tolerance into our B. napus line. Prophase stage cells from B. napus root-tips were well suited for retaining the intact chromosome complement and were used routinely in our FISH analysis. Cells at metaphase stage did not give us a well spread full complement of chromosomes most likely as the nuclear envelope is fragmented and the chromosomes tended to be dispersed. We were able to successfully use a very short 225 nucleotide probe directly labeled with Cy3 fluorochrome to detect the transfer of our herbicide resistant gene into the nuclear DNA into our introgressed B. napus line (Fig 5B–5D). Due to limited seed source and lack of good root-tip preparations, we were unable to perform FISH analysis with the donor resistant wild mustard. However, absence of fluorescent signal in the negative control, dicamba-susceptible canola (Fig 5A), confirms the specificity of the probe to the dicamba-tolerant trait that was introgressed. Unlike indirect labeled probes, the use of a direct labeled probe allowed for faster hybridization as a secondary detection molecule was not required to visualize the fluorescent signal. Furthermore, consistent signal presence and improved signal to background ratios have been reported with the direct method of labeling [22]. In contrast to the majority of relevant literature where FISH has been used to detect large (3–60 kb) or high copy inserts such as ribosomal RNA, our approach of FISH to detect a relatively small insert (0.3 kb) is unique. Large DNA fragments produce stronger signals but often contain repetitive sequences that make it difficult to detect specific target loci [23]. The decision to detect a small insert based off an AFLP marker ensured greater specificity. Furthermore, it has been suggested that a short probe (220–600 bp) allows for better penetration to the DNA packaged within the chromosome and may explain our success [24]. With respect to the use of Cy3 fluorochrome tagging, Cy3 is superior to classical dyes such as Fluorescein-isothiocyanate (FITC) and Tetramethylrhodamine (TRITC) as it has significantly brighter fluorescence and is stable to photobleaching [25]. The combination of a directly labeled short nucleotide probe along with the use of Cy3 fluorochrome enabled the successful visualization of a segment of DNA potentially consisting of dicamba-tolerance that was transferred from S. arvensis into B. napus. Furthermore, future studies using high-density single nucleotide polymorphism (SNP) genotyping array [26] can be applied to detect the introgressed DNA segment from S. arvensis into B. napus.
As the donor biotype of S. arvensis was resistant to the field use rate of dicamba (500 g ae/ha), the homozygous dicamba-tolerant line of B. napus is promising for field evaluation for the performance of the trait introgressed. Further studies evaluating agronomic performance of B. napus lines possessing dicamba-tolerance will determine the potential use of this technology as a weed management strategy in agriculture. Recently, cost-effective dicamba and 2,4-D-tolerant transgenic crop varieties were developed by multinational agro-based industries. The dicamba-tolerant soybean was developed by introducing a gene coding for an enzyme dicamba O-demethylase from a soil bacterium Pseudomonas maltophilia that can rapidly metabolize dicamba [27]. Development of dicamba-tolerant maize with insertion of dicamba O-demethylase gene provided high level of tolerance to dicamba [28]. Furthermore, 2,4-D-tolerant corn was developed by introducing an enzyme aryloxyalkanoate dioxygenase from yet another soil bacterium Ralstonia eutropha that can cleave 2,4-D into non-herbicidal form [29].
Although, glyphosate-tolerant B. napus cultivars have been in use across N. America for the greater part of the last few decades, continuous use of herbicides with a similar mechanism of action creates selection pressure thereby facilitating the evolution of herbicide-resistant weeds. Therefore, the availability of high performing crop cultivars resistant to herbicides of different mechanisms of actions will allow for the rotation of herbicides, a crucial and effective strategy to minimize the development of herbicide-resistant weeds. Furthermore, dicamba-resistant biotype of S. arvensis used for the generation of our dicamba-tolerant B. napus is highly resistant to dicamba and picloram but not to mecoprop, another auxinic herbicide [3]. Therefore, if and when there is any occurrence of volunteer dicamba-tolerant B. napus, mecoprop can still be used to control such volunteers [30].
Conclusions
In conclusion, this research demonstrated for the first time the transfer of dicamba-tolerant trait from S. arvensis into a B. napus line. We have presented a proof of concept breeding scheme to transfer dicamba-tolerant trait from a weed relative into crop species. More importantly, our FISH assay using the combination of a directly labeled short nucleotide probe along with Cy3 fluorochrome facilitated the successful visualization of a segment of DNA potentially consisting of dicamba-tolerance that was transferred from S. arvensis into B. napus. Herbicide-resistant agronomic weeds offer valuable source of germplasm for breeding programs. The backcross breeding scheme outlined in this research for introgressing dicamba tolerance is not limited to B. napus and can be applied to all agricultural and horticultural crops, and their related species. Finally, development and adoption of herbicide-tolerant technology facilitates effective weed control, less tillage, and possibly minimizes evolution of herbicide resistant weeds.
Supporting Information
(DOCX)
Lane 1: 100 bp ladder; 2: reaction without a template; 3: S. arvensis dicamba-resistant; 4: B. napus; 6, 7, 9, 10, 11 are hybrids generated via embryo rescue: 6, 10, 11 represent dicamba-tolerant hybrids, whereas, 7, 9 are dicamba-susceptible; 5 and 8: empty wells.
(TIF)
Lane 1: 100bp ladder; Lane 2: S. arvensis- dicamba susceptible; 3: wild-type B. napus; 4–13: Ten BC7F3 plants; 14, 15: S. arvensis-dicamba resistant; 16: reaction without a template.
(TIF)
Acknowledgments
This work was supported by a grant to JCH from the Natural Science and Engineering Research Council of Canada and BASF (NC, USA) through NSERC-CRD grant is gratefully acknowledged.
Abbreviations
- AFLP
amplified fragment length polymorphism
- BC
backcross
- DAPI
4, 6-diamino-2-phenylindole
- FISH
fluorescent in situ hybridization
- FITC
fluorescein-isothiocyanate
- SSC
saline-sodium citrate
- TRITC
tetramethylrhodamine
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a grant to JCH from the Natural Science and Engineering Research Council of Canada and BASF Company (NC, USA) through NSERC-CRD grant is gratefully acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Heap IM, International survey of herbicide resistant weeds,http://www.weedscience.org/summary/MOASummary.asp/ [accessed 23 December 2014].
- 2. Heap IM, Morrison IN: Resistance to auxin-type herbicides in wild mustard (Sinapis arvensis L) populations in western Canada Annual Meeting of Weed Science Society of America. 1992, Abstract # 32, pp. 164. [Google Scholar]
- 3. Penuik MG, Romano ML, Hall JC: Physiological investigations into the resistance of wild mustard (Sinapis arvensis L) biotype to auxinic herbicide. Weed Res 1993, 33: 431–440. [Google Scholar]
- 4. Hall JC, Webb SR, Deshpande S: An overview of auxinic herbicide resistance: wild mustard as a case study In Molecular Genetics and Evolution of Pesticide Resistance. Edited by Brown TM. Washington, DC: American Chemical Society; 1996: 28–43. [Google Scholar]
- 5. Zheng HG, Hall JC: Understanding auxinic herbicide resistance in Sinapis arvensis L.: Physiological, biochemical and molecular genetic approaches. Weed Sci 2001, 49: 276–281. [Google Scholar]
- 6. Jasieniuk M, Morrison IN, Brule-Babel AL: Inheritance of dicamba resistance in wild mustard (Brassica kaber) . Weed Sci 1995, 43: 192–195. [Google Scholar]
- 7. Jugulam M, McLean MD, Hall JC: Inheritance of picloram and 2,4-D resistance in wild mustard (Brassica kaber). Weed Sci 2005, 53: 417–423. [Google Scholar]
- 8. Mithila J, McLean MD, Chen S, Hall JC: Development of near-isogenic lines and identification of markers linked to auxinic herbicide resistance in wild mustard (Sinapis arvensis). Pest Manag Sci 2012, 68: 548–556. 10.1002/ps.2289 [DOI] [PubMed] [Google Scholar]
- 9. Prakash S, Hinata K: Taxonomy, cytogenetics and origin of crop Brassicas, a review. Opera Botanica 1980, 55: 3–57. [Google Scholar]
- 10. Raymer PL: Canola: An emerging oilseed crop In Trends in new crops and new uses. Edited by Janick J, Whipkey A. Alexandria, VA, USA: ASHS Press; 2002: 122–126. [Google Scholar]
- 11. Chandler JM, Hamill AS, Thomas AG: Crop losses due to weeds in Canada and the United States. Weed Science Society of America Annual Meetings, Champaign, IL: 1984. [Google Scholar]
- 12. Powles SB, Preston C: Evolved glyphosate resistance in plants: Biochemical and genetic basis of resistance. Weed Technol 2006, 20: 282–289. [Google Scholar]
- 13. Mithila J, Hall JC, Johnson WG, Kelley KB, Reichers DE: Evolution of resistance to auxinic herbicides: Historical perspectives, mechanism of resistance and implications of broadleaf weed management in agronomic crops. Weed Sci 2011, 59: 445–457. [Google Scholar]
- 14. Bing DJ, Downey RK, Rakow FW: An evaluation of the potential intergeneric gene transfer between Brassica napus and Sinapis arvensis . Plant Breeding 1995, 114: 481–484. [Google Scholar]
- 15. Inomata N: Intergeneric hybridization between Brassica napus and Sinapis arvensis and their crossability, Eucarpia Cruciferae Newsletter 1998, 13: 22–23. [Google Scholar]
- 16. Kerlan MC, Chevre AM, Eber F: Interspecific hybrids between a transgenic rapeseed (B. napus) and related species: Cytogenetical characterization and detection of the transgene. Genome 1993, 36: 1099–1106. [DOI] [PubMed] [Google Scholar]
- 17. Hu Q, Anderson SB, Dixelius C, Hansen LN: Production of fertile intergeneric somatic hybrids between Brassica napus and Sinapis arvensis for the enrichment of the rapeseed gene pool. Plant Cell Rep 2002, 21: 147–152. [Google Scholar]
- 18. Mithila J, Hall JC: Transfer of auxinic herbicide resistance from Brassica kaber to Brasssica juncea and Brassica rapa through embryo rescue. In Vitro Cell Dev Biol—Plant 2013, 49: 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kron P, Suda J, Husband BC: Application of flow cytometry to evolutionary and population biology. Annu Rev Ecol Evol Syst 2007, 38: 847–76. [Google Scholar]
- 20. Mizushima U: Karyogenetic studies of species and genus hybrids in the tribe Brassiceae of Cruciferae. Tohoku J Agricul Res 1950, 1: 1–14. [Google Scholar]
- 21. Mizushima U: Genome analysis in brassica and allied genera In Brassica crops and wild allies biology and breeding. Edited by Tsunada T, Hinata K, Gomez-Campo G. Tokyo, Japan: Scientific Press; 1980, 89–106. [Google Scholar]
- 22. Kato A, Vega JM, Han F, Lamb JC, Birchler JA: Advances in plant chromosome identification and cytogenetic techniques. Curr Opin Plant Biol 2005, 8: 148–154. [DOI] [PubMed] [Google Scholar]
- 23. Figueroa MD, Bass HW: A historical and modern perspective on plant cytogenetics. Brief. Funct. Genomics 2010, 9: 95–102. 10.1093/bfgp/elp058 [DOI] [PubMed] [Google Scholar]
- 24. Schwarzacher T: Fluorescent in situ hybridization to detect transgene integration into plant genomes In Methods in Molecular Biology, Transgenic Wheat, Barley and Oats. Edited by Jones HD, Shewry PR. Totowa, NJ, USA: Humana Press; 2008, 227–246. [DOI] [PubMed] [Google Scholar]
- 25. Moter A, Gobel UB: Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J Microbiol Methods 2000, 41: 85–112. [DOI] [PubMed] [Google Scholar]
- 26. Mason AS, Batley J, Bayer PE, Hayward A, Cowling WA, Nelson MN: High-resolution molecular karyotyping uncovers pairing between ancestrally related Brassica chromosomes. New Phytologis 2014, 202:964–974. [DOI] [PubMed] [Google Scholar]
- 27. Behrens M, Mutlu N, Chakraborty S, Dumitru R, Jiang WZ, LaVallee BJ, et al. : Dicamba resistance: enlarging and preserving biotechnology-based weed management strategies. Science 2007, 316:1185–1188. [DOI] [PubMed] [Google Scholar]
- 28. Cao M, Sato SJ, Behrens M, Jiang WZ, Clemente TE, Weeks DP: Genetic Engineering of Maize (Zea mays) for High-Level Tolerance to Treatment with the Herbicide Dicamba. J Agric. Food Chem. 2011, 59:5830–5834. 10.1021/jf104233h [DOI] [PubMed] [Google Scholar]
- 29. Wright TR, Shan G, Walsh TA, Lira JM, Cui C, Song P, et al. Robust crop resistance to broadleaf and grass herbicides provided by aryloxyalkanoate dioxygenase transgenes. Proc Natl Acad Sci, USA. 2010, 107: 20240–20245. 10.1073/pnas.1013154107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beckie HJ, Seguin-Swartz G, Nair H, Warwick SI, Johnson E: Multiple herbicide-resistant canola can be controlled by alternative herbicides. Weed Sci 52: 152–157. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Lane 1: 100 bp ladder; 2: reaction without a template; 3: S. arvensis dicamba-resistant; 4: B. napus; 6, 7, 9, 10, 11 are hybrids generated via embryo rescue: 6, 10, 11 represent dicamba-tolerant hybrids, whereas, 7, 9 are dicamba-susceptible; 5 and 8: empty wells.
(TIF)
Lane 1: 100bp ladder; Lane 2: S. arvensis- dicamba susceptible; 3: wild-type B. napus; 4–13: Ten BC7F3 plants; 14, 15: S. arvensis-dicamba resistant; 16: reaction without a template.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.