Abstract
The floor plate (FP), a ventral midline structure of the developing neural tube, has differential neurogenic capabilities along the anterior-posterior axis. The midbrain FP, unlike the hindbrain and spinal cord floor plate, is highly neurogenic and produces midbrain dopaminergic (mDA) neurons. Canonical Wnt/beta–catenin signaling, at least in part, is thought to account for the difference in neurogenic capability. Removal of beta–catenin results in mDA progenitor specification defects as well as a profound reduction of neurogenesis.
To examine the effects of excessive Wnt/beta–catenin signaling on mDA specification and neurogenesis, we have analyzed a model wherein beta–catenin is conditionally stabilized in the Shh+ domain. Here, we show that the Foxa2+/Lmx1a+ domain is extended rostrally in mutant embryos, suggesting that canonical Wnt/beta–catenin signaling can drive FP expansion along the rostrocaudal axis. Although excess canonical Wnt/beta–catenin signaling generally promotes neurogenesis at midbrain levels, less tyrosine hydroxylase (Th)+, mDA neurons are generated, particularly impacting the Substantia nigra pars compacta. This is likely because of improper progenitor specification. Excess canonical Wnt/beta–catenin signaling causes downregulation of net Lmx1b, Shh and Foxa2 levels in mDA progenitors. Moreover, these progenitors assume a mixed identity to that of Lmx1a+/Lmx1b+/Nkx6-1+/Neurog1+ progenitors. We also show by lineage tracing analysis that normally, Neurog1+ progenitors predominantly give rise to Pou4f1+ neurons, but not Th+ neurons. Accordingly, in the mutant embryos, Neurog1+ progenitors at the midline generate ectopic Pou4f1+ neurons at the expense of Th+ mDA neurons. Our study suggests that an optimal dose of Wnt/beta–catenin signaling is critical for proper establishment of the mDA progenitor character. Our findings will impact embryonic stem cell protocols that utilize Wnt pathway reagents to derive mDA neuron models and therapeutics for Parkinson’s disease.
Keywords: midbrain dopaminergic neurons, floor plate, Wnt/beta–catenin, Lmx1b
Background
The midbrain floor plate (FP), but not the hindbrain or spinal cord FP, is highly neurogenic, and gives rise to midbrain dopaminergic (mDA) neurons (Andersson et al., 2006; Blaess et al., 2011; Bonilla et al., 2008; Hayes et al., 2011; Joksimovic et al., 2009a; Joksimovic et al., 2009b; Kittappa et al., 2007; Ono et al., 2007). The neurogenic character of the FP can be attributed to the presence of Wnts in the midbrain FP, but not in the hindbrain or spinal cord FP (Joksimovic and Awatramani, 2014). The role of Wnt signaling in the midbrain FP is thus significant and warrants further attention.
Several lines of evidence have suggested the importance of Wnts in midbrain FP neurogenesis and mDA neuron specification (Alves dos Santos and Smidt, 2011). Wnt1 mutants display a drastic deficit in mDA neuron numbers (Prakash et al., 2006; Yang et al., 2013), which is exacerbated in the Wnt1/Wnt5a double mutant (Andersson et al., 2006). The conditional removal of beta–catenin results in alterations in mDA neuronal patterning and neurogenesis, and significantly less mDA neurons (Chilov et al., 2010; Joksimovic et al., 2009b; Tang et al., 2009). In these mutants, the midbrain FP is less proliferative, partly loses expression of Otx2, Msx1, Neurog2 (Joksimovic et al., 2009b), and Lmx1a (Chilov et al., 2010; Joksimovic et al., 2009b), and maintains the expression of Shh, Spon1 and Lmx1b, thereby molecularly resembling the hindbrain FP (Joksimovic et al., 2009b). In contrast, FP-specific stabilization of beta–catenin results in increased proliferation, the induction of Lmx1a, Otx2, Msx1, and Neurog2, and suppression of Shh, Spon1 and Lmx1b in the hindbrain. In such mutants, the hindbrain and spinal cord were highly neurogenic, but ectopic mDA neurons were restricted to the rostral aspect of rhombomere 1 (Joksimovic et al., 2012; Joksimovic et al., 2009b).
Given the importance of Wnt/beta–catenin signaling for mDA specification and neurogenesis, and the tight spatiotemporal expression of Wnt1 at the midline, we sought to analyze the effect of excessive Wnt/beta–catenin signaling on the ventral midline. We found that in Shh::cre;Ctnnb1lox(ex3) embryos, the Foxa2 and Lmx1a domains are rostrally extended along the anterior-posterior axis. At midbrain levels, fewer Th+ neurons were observed, particularly in the developing Substantia Nigra pars compacta (SNpc) region. We found that overall proliferation and neurogenesis are increased, and therefore are unlikely to account for reduced Th+ mDA neurons in this mutant. We next reasoned that this reduction of Th+ mDA neurons was because of perturbations in mDA progenitor gene expression profiles. We determined that stabilization of beta–catenin leads to net downregulation of key mDA neuron determinants, particularly Lmx1b and Foxa2, and failure to repress lateral markers such as Nkx6-1 and Neurog1 from the midline. We also show by genetic inducible fate mapping that Neurog1+ progenitors normally give rise to Pou4f1+ neurons of the red nucleus. Thus, excessive Wnt/beta–catenin signaling results in a vacillating mDA progenitor pool (Lmx1a+/Lmx1b+/Nkx6-1+/Neurog1+), that generates at least two distinct neuron populations: Lmx1a+/Lmx1b+/Th+ and Lmx1a+/Lmx1b+/Pou4f1+ neurons. These studies further uncover the molecular logic underlying mDA specification and neurogenesis, and reveal that Wnt/beta–catenin signaling needs to be carefully titred to maintain optimal mDA progenitor characteristics and generate the correct numbers of mDA neurons.
Results
Stabilization of beta–catenin in the Shh domain results in excessive and ectopic Wnt/beta–catenin signaling
We first performed experiments to confirm that Shh::cre;Ctnnb1lox(ex3) embryos demonstrated excessive Wnt/beta–catenin signaling in the FP. We generated Shh::cre;Ctnnb1lox(ex3), Axin2lacZ embryos in which a NLS-lacZ reporter gene is expressed under the control of the Axin2 promoter region. Axin2 is a direct target of the Wnt/beta–catenin pathway and serves as an indicator of canonical Wnt/beta–catenin signaling (Jho et al., 2002; Lustig et al., 2002; van Amerongen et al., 2012). In the midbrain FP, X-gal labeling is markedly increased in Shh:cre;Ctnnb1lox(ex3), Axin2lacZ compared to Axin2lacZ controls (Figure 1A and C). In the hindbrain FP, where X-gal labeling is largely absent in controls (Figure 1B), prominent X-gal labeling is observed in mutants (Figure 1D). In the rostral hypothalamus, a marked increase in X-gal labeling is also observed in mutants (Supplemental Figure 1A and B). Thus, Shh::cre;Ctnnb1lox(ex3) embryos demonstrate excessive Wnt/beta–catenin signaling throughout the Shh domain.
Figure 1. Stabilization of beta–catenin in the Shh+ midbrain and hindbrain floor plate results in drastically increased Wnt/beta–catenin signaling.
(A–D) E10.5 coronal midbrain (A and C) and hindbrain (B and D) sections were labeled with X-gal to determine Axin2lacZ expression in control (Axin2lacZ; A and B) and mutant (Shh::cre;Ctnnb1lox(ex3),Axin2lacZ; C and D) embryos. (A and C) In the ventral midbrain, intense X-gal+ labeling is present in the mutant compared to the moderate X-gal labeling in the control. (B and D) In the ventral hindbrain, the mutant floor plate is characterized by a substantial number of Xgal+ cells in comparison to the corresponding region in the control, which has few to no Xgal+ cells. Black dotted lines delineate the midbrain and hindbrain parenchyma. mesenchymal cells surrounding the neural tube are Xgal+ in all sections shown. Scale bars: 50μm.
Ectopic Wnt/beta–catenin signaling results in a rostral expansion of the Foxa2 domain
In normal embryos, the Foxa2+/Lmx1a+ expression domain stops in a sharp boundary rostral to base of the zona limitans intrathalamica (ZLI). In contrast, Shh expression extends more rostrally, in a manner that is dependent on specific rostral enhancers (Geng et al., 2008; Jeong et al., 2006). In Shh::cre;Ctnnb1lox(ex3) embryos, Wnt/beta–catenin signaling is increased throughout the Shh domain (Figure 1C and D; Supplemental Figure 1B). In sagittal sections of Shh::cre;Ctnnb1lox(ex3) embryos, we observed a marked rostral expansion of Foxa2 beginning at E10.5 (Supplemental Fig. 2A, A′, B, and B′) and persisting through E12.5, where Foxa2 expression extends to the anterior hypothalamus (Figure 2B, B′ Bottom arrowhead, and C; Supplemental Figure 1D). Using the FP/ZLI border as a land mark (Figure 2A′ and B′ top arrowheads), we measured the distance from the ZLI to the rostral Foxa2 boundary (Figure 2A′ and B′ bottom arrowheads). Our measurements revealed a significant expansion of the rostral Foxa2 domain between the controls and mutants (Figure 2C). Likewise, at least by E12.5, the Lmx1a domain is extended rostrally, although at reduced levels in Shh::cre;Ctnnb1lox(ex3) embryos (Figure 2B″). These results demonstrate that Wnt/beta–catenin signaling could potentially drive the expansion of the Foxa2+/Lmx1a+ domain along the rostro-caudal axis.
Figure 2. Sustained Wnt/beta–catenin signaling results in a rostral expansion of the Foxa2 domain.
(A–C) E12.5 sagittal sections from control and Shh::cre;Ctnnb1lox(ex3) mutant embryos were co-labeled with Th (A and B), Foxa2 (A, A′, B, and B′), Lmx1a (A, A″, B, and B″). Mutant sections show an expanded diencephalic ventricle (A and B; see also Suppl. Fig 1) The number of Th+ mDA neurons (green) are severely reduced, particularly in rostral regions in the mutant (arrowheads) when compared to control. (A and B). Foxa2 expression extends rostrally, to the anterior hypothalamus (B and bottom arrowhead in B′) in contrast to the sharp Foxa2 limit in the posterior hypothalamus in controls (A and bottom arrowhead in A′). (A, A″, B and B″) In mutants, similar to the anterior shift of Foxa2, Lmx1a expression extends rostrally (B and bottom arrowhead in B″) albeit at reduced levels. (C) A significant rostral expansion (p< 0.001) of the Foxa2 domain is observed in mutants (544μm ± 4.62 s.e.m.) compared to controls (control 308μm ± 10.6 s.e.m.) when measured from the ZLI/FP boundary (Arrowheads in A′ and B′). ZLI, zona limitans intrathalamica; FP, floor plate. Anterior is to the left, posterior to the right, dorsal up, and ventral down. Scale bars: 100μm in A and B.
Sustained Wnt/beta–catenin signaling results in fewer mDA neurons
To determine whether excessive canonical Wnt/beta–catenin signaling affects mDA neuron production, we examined expression of multiple mDA neuron markers in Shh::cre;Ctnnb1lox(ex3) midbrain sagittal and coronal sections at E12.5, E14.5, and E18.5. Our initial analyses at E12.5 and E14.5 revealed a drastic reduction of Th in mutants compared to controls (Figures 2A and B; Fig. 3A and B; Supplemental Fig. 3E and F). This effect was especially pronounced in rostral midbrain sections (Figure 3E), where we observed up to a 75% reduction in Th+ neuron numbers. We also observed a reduction in the number of cells expressing Slc6a3 and Pitx3 (Supplemental Fig. 3A–F) in Shh::cre;Ctnnb1lox(ex3) embryos. Furthermore, Slc6a3 expression appears lower even in the remnant Th+ neurons (Supplemental Fig. 3E and F). At E18.5, fewer Th+ neurons are observed in the developing SNpc of mutants compared to controls, consistent with a previous study (Tang et al., 2010). In contrast, the Ventral Tegmental Area (VTA) does not appear drastically diminished (Figure 3D). Some ectopic Th+ neurons are also observed in the interpeduncular region, likely representing ectopic hindbrain FP derived mDA neurons (Joksimovic et al., 2012). Together, these results demonstrate that excessive Wnt/beta–catenin signaling leads to reduced mDA neuron numbers.
Figure 3. Constitutively active Wnt/beta–catenin signaling results in a severe reduction in numbers of Th+ neurons.
E12.5 (A and B) and E18.5 (C and D) coronal midbrain sections were labeled with Th antibody in control (A and C) and Shh::cre;Ctnnb1lox(ex3) mutant (B and D) embryos. (A and B) Note an apparent loss of Th+ cells in the mutant compared to the control at E12.5. (C and D) At E18.5, the developing Substantia Nigra pars compacta (SNpc; right arrows) is particularly affected, but the developing Ventral Tegmental Area is less affected (VTA; left arrows). (E) A significant decrease in the total number of Th+ cells per section at E12.5 is observed rostrally (p<0.001) and caudally (p<0.05) in the mutants (n=3; rostral 133 ± s.e.m 15; caudal 114 ± s.e.m 22) in comparison to the control (n=3; rostral 370 ± s.e.m 35; caudal 238 ± s.e.m 39). Scale bars: 50μm in A–D.
Excessive Wnt/beta–catenin signaling promotes neurogenesis
We next sought to determine if the reduction in Th+ neurons in Shh::cre;Ctnnb1lox(ex3) was the result of changes in mDA proliferation or neurogenesis. To assess the effect of Wnt/beta–catenin signaling on mDA progenitor proliferation and cell cycle exit, we performed short and long pulse BrdU analyses. First, we investigated whether stabilizing beta-catenin may selectively affect the fraction of progenitors in S-phase within either the Lmx1a+ or Nkx6-1+ domains. To accomplish this, we determined the Labeling Index (Omodei et al., 2008) at E10.5 and E12.5 by providing a short 30 minute BrdU pulse and measuring the percentage of BrdU+ cells over the total number of cells within either of the two domains. We found no differences in the fraction of progenitors in S-phase at E10.5 in either domain between control and mutant embryos (Figure 4A–C; percentage of BrdU+/DAPI+ in Lmx1a domain and BrdU+/DAPI+ in Nkx6-1 domain; n=3 each; DAPI not shown). However at E12.5, 30 minute BrdU labeling revealed an increase in the fraction of progenitors in S-phase within the Lmx1a domain in Shh::cre;Ctnnb1lox(ex3) when compared to controls (Figure 4D–F;). Concurrently at E12.5, a significant increase (p<0.001) in the total number of BrdU+ cells per section was observed in mutants (n=3; 284 ± 28 s.e.m) when compared to controls (n=3; 135 ± 10 s.e.m), consistent with previous observations that Wnt/beta–catenin signaling promotes progenitor proliferation (Joksimovic et al., 2009b; Tang et al., 2010).
Figure 4. Excessive Wnt/beta–catenin signaling results in increased proliferation and number of mDA progenitors exiting the cell-cycle.
(A–C) short pulse BrdU labeling at E10.5 reveals no change in total BrDU+ numbers (not shown) or the labeling index (BrdU+/DAPI+) of the Lmx1a and Nkx6-1 domains between controls and mutants. (D–F) Short pulse BrdU labeling at E12.5. (D–F) An increase (p<0.05) in the labeling index at E12.5 of Shh::cre;Ctnnb1lox(ex3) mutants (n=3; 53.4% ± 1.2 s.e.m) was observed when compared with controls (n=3; 37% ± 3.3 s.e.m) within the Lmx1a+ domain. A significant increase (p<0.001) in the total number of BrdU+ cells per section was observed in mutants (n=3; 284 ± 28 s.e.m) when compared to controls (n=3; 135 ± 10 s.e.m). (G–I) BrdU was administered at E12.5 and embryos harvested at E13.5 to assess the number of mDA progenitors that have exited the cell-cycle between these two time points. (G and H) Sections were labeled with Ki67 and BrdU antibodies. Dotted lines indicate the dopamine field determined by Lmx1a labeling on the same or adjacent sections (not shown). In this Lmx1a+ dopaminergic field, the cells were counted and quantified in I. (I) A significant increase (p<0.01) in the total number of BrdU+ cells per section was observed in mutants (n=3; 258.1 ± 8.3 s.e.m.) compared to controls (n=3; 184.4 ± 9.0 s.e.m.). In addition, an increase (p<0.05) in the number of post-mitotic, BrdU+/Ki67− cells was detected in mutants (n=3; 161.6 ± 5.8 s.e.m) compared to controls (n=3; 119.5 ± 9.2 s.e.m). The quit fraction (BrdU+;Ki67-/BrdU+ total) was not significantly altered at this stage. (L–N) E15.5 coronal midbrain sections were labeled with Lmx1a and HuC/D antibodies in the control (L) and mutant (M). (N) A significant increase (p<0.01) in the average number of Lmx1a+ cells per section is observed in the mutants (n=3; 789 ± 67 s.e.m.) in comparison to controls (n=3; 503 ± 51 s.e.m.). Scale bars: 50μm in A, B, D, and E; 100μm in G, H, J and K.
We also measured the net amount of neurogenesis by performing 24 hour BrdU pulse labeling experiments. In these experiments, the total number of cells that have exited the cell cycle between E12.5 and E13.5 (BrdU+;Ki67−) is significantly increased in the mutants (Figure 4G–I). The quit fraction is minimally altered at this stage when measured as BrdU+;Ki67-/BrdU+ total (control 0.646 ± 0.016 s.e.m.; mutant 0.626 ± 0.001 s.e.m.; p=.380) or when measured similar to Tang et al. as BrdU+;Ki67-/BrdU+;Ki67+ (control 2.37 ± 0.275 s.e.m.; mutant 1.88 ± 0.035 s.e.m.; p= 0.208). Thus, we observe more neurons exiting the cell cycle between E12.5 and E13.5 likely due to an increase in the number of progenitors at E12.5, but not due to an increase in the quit fraction. Finally, by E15.5, large numbers of Lmx1a+ cells can be seen in the mantle zone in mutants compared to controls (Figure 4J and K). Quantification of Lmx1a+ cells revealed a ~57% increase in mutants compared to controls, consistent with increased FP proliferation and neurogenesis (Figure 4L). This increased proliferation and neuronal production are consistent with a role for Wnt/beta–catenin signaling in promoting FP neurogenesis (Joksimovic et al., 2009b; Tang et al., 2010). Together, these data show that the reduction in numbers of Th+ neurons in Shh::cre;Ctnnb1lox(ex3) embryos is not due to a general decrease in FP neurogenesis.
Excessive Wnt/beta–catenin signaling results in vacillating mDA progenitors
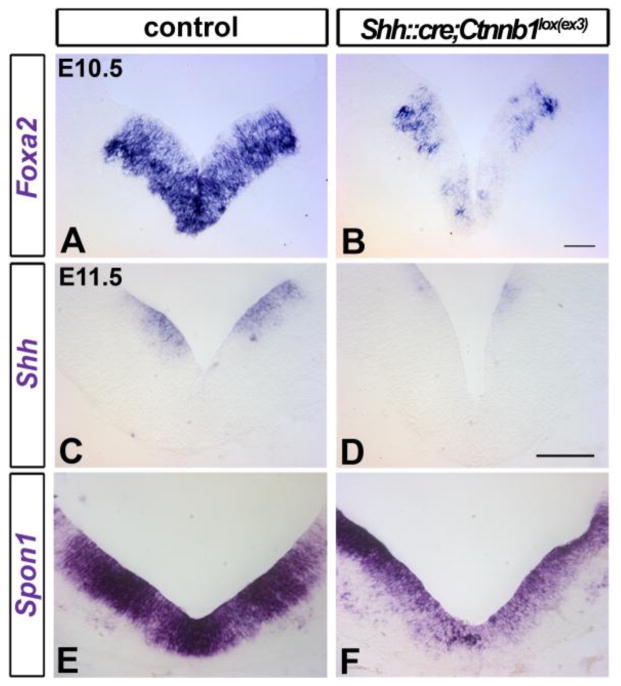
We next investigated whether the reduction in Th+ neurons in Shh::cre;Ctnnb1lox(ex3) mutants was due to disruptions in progenitor gene expression profiles. We surveyed the expression of key genes in mDA progenitors and neighboring ventral midbrain progenitor domains. First, we screened for early expression of Foxa2 and Shh which are known to be critical genetic networks for the regulation of mDA neuron development (Blaess et al., 2006; Chung et al., 2009; Ferri et al., 2007; Metzakopian et al., 2012; Perez-Balaguer et al., 2009) Although there is significant ectopic expansion of Foxa2 expression in the hypothalamus (Figure 2A′, B′ and C; Supplemental Figure 2A–B′), there is an apparent reduction of Foxa2 expression in the mDA ventricular zone (Figure 5A and B, Supplemental Figure 4A and B) consistent with a previous study (Tang et al., 2010). Similarly, we found that Shh is downregulated in the mutant ventral midbrain compared to the control which is consistent with our previous study (Figure 5C and D; Joksimovic et al., 2009b). Another key FP gene Spon1 is also downregulated in the mutant ventricular zone (Figure 5E and F).
Figure 5. Excessive Wnt/beta–catenin signaling results in downregulation of floor plate genes.
E10.5 (A and B) and E11.5 (C–F) coronal midbrain sections were labeled with Foxa2, Shh and Spon1 riboprobes in control (A, C, and E) and Shh::cre;Ctnnb1lox(ex3) mutant embryos (B, D, and F). Expression of Foxa2 (A and B), Shh (C and D), and Spon1 (E and F) is reduced in the mutants. Note that in most mutants at this age, and more prominent at later stages, a deeper indentation at the ventral midline is observed, and the angle of ventrolateral midbrain is altered with respect to the midline. Scale bars: 50μm in A and B; 100μm in C–F.
We then examined other factors known to be expressed within the mDA progenitor domain and important in mDA development. In the control midbrain, Lmx1b starts to be downregulated in mDA progenitors at ~E11.0 in a rostral-to-caudal manner (Anderegg et al., 2013; Andersson et al., 2006). However, Lmx1a does not appear to be significantly reduced at this time point. In E10.5 and E11.5 mutants, we observed that at the midbrain ventral midline, Lmx1b is prematurely downregulated relative to controls, whereas Lmx1a was not drastically altered (Figure 6A, B, K, L, M, and N). Lmx1b was also reduced in the hindbrain and spinal cord FP (Joksimovic et al., 2012). Wnt1, Wnt5a, and Aldh1a1 also appeared reduced in mutants compared to controls (Figure 6C–H). Neurog2+ cells were increased in mutants (Figure 6Q and R), consistent with the previously observed increase in FP neurogenesis (Figure 4).
Figure 6. Excessive Wnt/beta–catenin signaling results in vacillating mDA progenitors.
E10.5 (A–J), E11.5 (K–P) and E12.5 (Q and R) coronal midbrain sections were labeled with indicated antibodies (K–N, Q and R) or Lmx1b, Wnt1, Wnt5a, Aldh1a1, Nkx6-1 and Neurog1 riboprobes in control (A, C, E, G, I, and O) and Shh::cre;Ctnnb1lox(ex3) mutant embryos (B, D, F, H, J, and P). Lmx1b, Wnt1, Wnt5a, and Aldh1a1 are all downregulated (B, D, F, and H; in the case of Aldh1a1 n=5/7 mutants) while Lmx1a is minimally if at all reduced (N). Increased midline expression of Nkx6-1 (J and N) and Neurog1 (P) in mDA progenitors can be seen in the mutant embryos. In controls, we observed very low levels of Nkx6-1 expression (I) and occasional Neurog1+ cells (O) at the midline. Red (K, L, Q, and R) dotted lines indicate a separation between the ventricular (vz) and mantle zones (mz). White (I and J) and black (O and P) dotted lines delineate the dopaminergic field. Scale bars: 50 μm for A–J and 100 μm for K–R.
We next analyzed genes that are known to be expressed adjacent to the mDA progenitor domain. Neurog1 and Nkx6-1, which are adjacent to, but largely excluded from the Lmx1a/Lmx1b domain in controls (Figure 6I, M and O), appear within the Lmx1a/Lmx1b domain in mutants (Figure 6J, N and P); in the case of Neurog1, this is most evident in rostral midbrain sections. Ectopic midline Nkx6-1 expression is prominent at E10.5 and E11.5 (Figure 6J and N) but not at later time points (data not shown).
Our previous studies have shown that canonical Wnt/beta-catenin signaling is required for Lmx1a expression in mDA progenitors (Joksimovic et al., 2009b). Additionally, increasing Wnt/beta-catenin signaling in the hindbrain FP was sufficient to induce Lmx1a expression in this region (Joksimovic et al., 2012). Thus it was interesting that in the Shh::cre;Ctnnb1lox(ex3) mutants, Lmx1a was not induced in the Nkx6-1 domain despite the presence of Foxa2, and increased Wnt signaling in this region. We postulated that in addition to Foxa2 and canonical Wnt signaling, Lmx1b was also required for the induction Lmx1a expression. We therefore overexpressed Lmx1b (Figure 7B) in the ventral midbrain using a Shh::Cre;Lmx1bOE model (Li et al., 2010). In these embryos we observed a significant lateral expansion of Wnt1, although at the midline Wnt1 remained absent similar to controls (Figure 7C and D). Additionally, the Lmx1a domain was expanded well into the Nkx6-1 domain in mutants but not in controls (Figure 7E and F). Our interpretation of this data is that in addition to Fox genes (Ferri et al., 2007) and canonical Wnt signaling (Joksimovic et al., 2009b), Lmx1b is required for the induction of Lmx1a. This is consistent with the finding that Lmx1b binds to regions upstream of Lmx1a (Chung et al., 2009).
Figure 7. Maintenance of Lmx1b signaling alters ventral midbrain patterning.
E12.5 (A–F) coronal midbrain sections were labeled with Lmx1b (A and B), Wnt1 (C and D), and Lmx1a (E and F) riboprobes in control and Shh::Cre,Lmx1bOE mutant embryos. Lmx1b is expanded dorsally in mutants (arrowhead in B) when compared to control. Similarly, Wnt1 is increased in the ventricular zone (arrowhead in D) of the ShhCre::Lmx1bOE mutant. The Lmx1a expression boundary also expands dorsally as a consequence of increased Lmx1b expression in mutants (arrowhead in F) when compared with control. Scale bars: 50μm A–F.
Pou4f1+ neurons are derived from Neurog1+ progenitors
Having observed Neurog1 expanded beyond its lateral domain into the midline in Shh::cre;Ctnnb1lox(ex3) mutants (Figure 6P), we sought to determine the fate of ventral midbrain Neurog1+ progenitors in normal embryos. To do this, we performed a fate-mapping experiment by crossing a Neurog1::creERT2 mouse to Ai9, a Rosa26-tdTomato reporter line. Females harboring Neurog1::creERT2,Ai9 embryos were injected with tamoxifen at E9.75 and harvested at E12.75. In these embryos, several Pou4f1+/tdTomato+ cells were observed immediately lateral to the largely Pou4f1−/tdTomato–mDA neuron domain (Figure 8B–F). This data suggests that Neurog1+ progenitors give rise to Pou4f1+ red nucleus neurons consistent with our previous fate maps and models proposed by others (Blaess et al., 2011; Joksimovic et al., 2009a; Nakatani et al., 2007; Prakash et al., 2009).
Figure 8. In normal embryos, Neurog1+ progenitors give rise to Pou4f1+ red nucleus neurons.
(A) E10.5 coronal midbrain sections were labeled with Neurog1 riboprobe. Note that Neurog1 is expressed in the progenitor region (arrow) immediately adjacent to the predominantly Neurog1– midline (arrowhead) that corresponds to the position of mDA progenitors. (B–F) Lineage of Neurog1 progenitors in Neurog1::creERT2,Ai9 embryos. Tamoxifen was administered at E9.75 and embryos were harvested at E12.75. Consistent with the Neurog1 expression pattern (A), tdTomato+ cells can be seen in the region (Arrows; B and C) just lateral to the largely negative medial region (Arrowheads; B and C) where mDA progenitors and neurons are situated. Several of these Neurog1+ descendants are Pou4f1+ red nucleus neurons (arrows; C–F). C is a higher magnification of the right side of the ventral midline roughly corresponding to the area indicated by the arrow and arrowhead in B. D–F panels show high magnification confocal images of the region corresponding to the area indicated by the arrow in C. Separate channels and the merged image are shown in D–F. The section in B is counterstained with DAPI to visualize nuclei. Scale bars: 100μm in A; 200μm in B; 50μm in C; 10μm in D–F.
Pou4f1+ neurons are generated from the ventral midline in Shh::cre;Ctnnb1lox(ex3) mutant embryos
Considering the fact that a) Nkx6-1 is required for generation of Pou4f1+ neurons (Prakash et al., 2009) b) Neurog1+ progenitors normally give rise to Pou4f1+ neurons (Figure 8) and c) Nkx6-1 and Neurog1 are expressed in midline progenitors in mutants (Figure 6J and P), we next examined the expression of Pou4f1 in Shh::cre;Ctnnb1lox(ex3) embryos. Consistent with ectopic expression of Nkx6-1 and Neurog1 in the mutant mDA progenitor region, we detected large numbers of medially located Pou4f1+ cells in the mantle zone, particularly in rostral sections (Figure 9B, D, F, G, I, K, and M; Supplemental Fig.5C, D, C′, and D′). Most of these Pou4f1+ neurons express Lmx1b (Figure 9D) and Lmx1a (Figure 9K, U), suggesting that they are derived from the Lmx1a/Lmx1b mDA progenitor domain. Some of these ectopic Pou4f1+ neurons also expressed Lhx1, a marker normally not observed in midline regions (Supplemental Fig.5C′ and D′). In controls, a few Pou4f1+ cells were also medially located in the mantle zone predominantly at the rostral midbrain levels consistent with previous studies (Lahti et al., 2012). Few, if any, Pou4f1+ cells co-expressed Th in mutants (Figure 9F inset and Figure 9M and U; Supplemental Fig. 6B). Analysis of E15.5 embryos also revealed persistence of Pou4f1+ cells in ectopic medial locations in mutants compared to controls (data not shown). These results, together with recent studies (Andersson et al., 2006; Nakatani et al., 2010) suggest that depression of net Lmx1b/Foxa2 and possibly other factors by Wnt/beta–catenin signaling, prevents robust antagonism of Nkx6-1 and Neurog1 at the midline; this results in a mixed Lmx1a/Lmx1b+/Nkx6-1+/Neurog1+ progenitor pool and the genesis of both Th+ and Pou4f1+ neurons from the Lmx1a/Lmx1b progenitor domain.
Figure 9. Excessive Wnt/beta–catenin signaling increases the number of Pou4f1+ neurons within the midbrain dopaminergic field.
E12.5 (A–F) and E13.5 (H–U) coronal midbrain sections were labeled with indicated antibodies in control (A, C, E, H, J, L, and N–Q) and Shh::cre;Ctnnb1lox(ex3) mutant embryos (B, D, F, I, K, M, and R–U). Note that Pou4f1+/Lmx1b+ cells appear to be in excess in the mutant (arrowheads in B and D) comparing to the control (arrows in A and C) within the dopaminergic field while Th+ neurons are reduced in the mutant. White dotted lines (A–D) delineate the dopaminergic field. Images were taken from the same section triple-labeled for Pou4f1, Lmx1b, and Th and are shown in a two-color combination. Insets in E and F are high magnification confocal images of the ventral midline in control and mutants, respectively. (G) A significant increase (p<0.001) in a total number of Brn3a+ cells per section within the dopaminergic field is observed in the mutants (n=3; rostral 347 ± s.e.m 32; caudal 24 ± s.e.m 7) in comparison to the control (n=4; rostral 28 ± s.e.m 10; caudal 0.2 ± s.e.m 0.1). (H–M) Confocal images were taken from the same E13.5 section triple-labeled for Pou4f1, Lmx1a, and Th and shown in a two-color combination. At E13.5, increased numbers of Pou4f1+ cells persist at the ventral midline in the mutants (I). Note that these midline Pou4f1+ cells express Lmx1a (K) but not Th (M). (R–U) At the mutant midline, Pou4f1 (arrowheads in S) and Lmx1a (arrowheads in R) are expressed in the same cells (arrowheads in U), but do not express Th (blue). Scale bars: 50μm in A–F and H–M; 10μm in N–U.
Discussion
We show here that although Wnt/beta–catenin signaling increases the number of neurons produced from the Lmx1a domain, this excessive signaling results in gene expression changes in mDA progenitors that, in part, account for a decrease in numbers of Th+ mDA neurons. By examining gene expression in the mDA progenitor region, we find that excessive Wnt/beta–catenin signaling results in an ambivalent progenitor pool that generates both Th+ and Pou4f1+ neurons. The data presented here could have relevance for optimizing embryonic stem cell protocols that utilize Wnt pathway reagents to derive mDA neurons (Cajanek et al., 2009; Chung et al., 2009; Cooper et al., 2010; Fasano et al., 2010; Grealish et al., 2014; Kirkeby et al., 2012; Kriks et al., 2011; Xi et al., 2012).
Here, we report that excessive Wnt/beta–catenin signaling reduces the number of Th+ mDA neurons. In a previous study (Tang et al., 2010), this reduction in Shh::cre;Ctnnb1lox(ex3) mutants was partly attributed to a reduced rate of cell cycle exit between E11.5 and E12.5. The authors also reported increased proliferation at E12.5 and no detectable increase in cell death. Our data agree with these findings, in that the total number of Lmx1b+ postmitotic mDA neurons appears to be slightly reduced at E12.5 (Figure 9D), consistent with a decreased quit fraction between E11.5 and E12.5 (Tang et al., 2010). We also observed increased proliferation at E12.5. However, we extended this analysis by showing that between E12.5 and E13.5, there was an increase in the total numbers of postmitotic, mDA lineage neurons produced (Figure 4). In fact, by E15.5, the overall number of Lmx1a+ post mitotic neurons produced from the mDA domain was increased (Figure 4L). To reconcile these studies, we propose a model in which excessive Wnt/beta–catenin signaling initially increases proliferation of mDA progenitors and initially reduces the rate of cell cycle exit. As development proceeds, the surplus of accumulated mDA progenitors exits the cell cycle and produces excess Lmx1a+ neurons. Therefore, although excessive Wnt/beta–catenin signaling initially reduces cell cycle exit (Tang et al., 2010), it promotes overall neurogenesis (this study). Furthermore, since there is in fact an increase in Lmx1a+ mDA lineage neuron production, alterations in neurogenesis cannot explain the observed reduction of total Th+ neurons in these mutants.
Why are fewer Th+ neurons observed in the ventral midbrain of Shh::cre;Ctnnb1lox(ex3) mutants? During normal development, key mDA neuron determinants such as Lmx1b, Foxa2, and possibly other factors are required for repressing lateral, predominantly non–mDA progenitor fates (Andersson et al., 2006; Deng et al., 2011; Nakatani et al., 2010). Specifically, in the Nakatani et al. study, only transgenes containing both Foxa2 and Lmx genes were able to suppress Neurog1 expression. Thus, we postulate that downregulation of these key mDA genes at the ventral midline in Shh::cre;Ctnnb1lox(ex3) mutants likely results in an inability to repress the lateral progenitor program. This causes lateral markers such as Nkx6-1 and Neurog1 to be ectopically expressed at the ventral midline within the mDA progenitor pool. As a result, the mDA progenitor pool takes on a hybrid molecular profile comprising of both mDA and lateral progenitor markers (Lmx1a/Lmx1b+/Nkx6-1+/Neurog1+), rendering it bipotent. In normal embryos, Neurog1 progenitors give rise to Pou4f1+ cells. Accordingly, in the Shh::cre;Ctnnb1lox(ex3) mutants, Pou4f1+ cells are observed at the midline at the expense of Th+ cells. These Pou4f1+ cells co-express Lmx1a/Lmx1b, indicating their origin in the mDA progenitor domain. Thus, we posit that by down regulating Lmx1b and Foxa2, and potentially other unknown factors, excessive Wnt/beta–catenin signaling results in a vacillating progenitor phenotype (Lmx1blow, Foxa2low, Lmx1a+, Nkx6-1+, Neurog1+) that gives rise to many Pou4f1+ neurons at the expense of Th+ neurons. It remains to be determined whether these Pou4f1+ neurons persist.
Excess numbers of Pou4f1+ neurons have also been observed at the midline of FGF receptor loss-of-function mutants. In such mutants, early DA precursors are intermingled with Pou4f1+/Lmx1a+/FoxP1+ cells in the ventral midbrain (Lahti et al., 2012). In addition these mutants, show decreased expression of Wnt1, Wnt5a, and Aldh1a1 in the midbrain. The similarities between the Shh::cre;Ctnnb1lox(ex3) mutants, and the FGFR mutants open the possibility that Wnt/beta-catenin signaling and FGF signaling have an antagonistic interaction, which should be further investigated.
In Shh::cre;Ctnnb1lox(ex3) mutants, we find a more severe reduction in the SNpc compared to the VTA, consistent with the Tang et al study. Why might this be? One possibility is alterations in the timing of neurogenesis at E11.5 (Tang et al., 2010), a time when substantial SNpc neurons are born (Bye et al., 2012). Additionally, even at early embryonic ages, rostral regions are much more severely affected, compared to caudal regions. More Pou3f1 neurons are produced rostrally, instead of Th+ neurons. Since the SNpc is likely derived from rostral DA progenitors (Blaess et al., 2011; Hayes et al., 2011; Joksimovic et al., 2009a; Veenvliet et al., 2013), a more severe loss of SNpc neurons is therefore a predictable finding. Why rostral DA progenitors are more severely affected, however remains to be determined. It is interesting to note, than Wnt signaling displays a rostrallow caudalhigh gradient (Awatramani, unpublished results); therefore rostral SNpc progenitors may be more susceptible to elevation of Wnt signaling. Understanding the establishment and functional consequences of Wnt gradients remains to be further studied.
In summary, we have demonstrated that excessive Wnt/beta–catenin signaling results in rostral expansion of the Foxa2+/Lmx1a+ domain, suggesting that canonical Wnt signaling can drive FP expansion. Further, we have shown that excessive Wnt/beta–catenin signaling in the midbrain FP increases neuron production. Despite its pro-neurogenic capabilities, excessive Wnt/beta–catenin signaling downregulates key mDA determinants, including Foxa2 and Lmx1b, and thus alters the mDA progenitor identity to that of Lmx1blow, Foxa2low, Lmx1a+, Nkx6-1+, Neurog1+ progenitors. These vacillating progenitors give rise to Pou4f1+ neurons, and Th+ mDA neurons. These data suggest that Wnt/beta–catenin signaling levels need to be fine-tuned to achieve the proper molecular signature of mDA progenitors. In this regard, miR135a2, which is predicted to target multiple aspects of the Wnt cascade, may play an important role (Anderegg et al., 2013). Further studies will be required to decipher the modulation of the Wnt pathway, the targets of canonical Wnt/beta catenin signaling and potential interactions with FGF signaling, as well as the potential impact of Wnt/beta catenin signaling on the genesis of diverse mDA neuron subtypes (Di Salvio et al., 2010; Panman et al., 2014; Poulin et al., 2014; Veenvliet et al., 2013) - these will be important for optimally deriving mDA neurons from stem cells, a key goal for Parkinson’s disease modelling and therapeutics (Steinbeck and Studer, 2015).
Methods
Mouse strains and genotyping
Mice were maintained and sacrificed according to the protocols approved by the Northwestern University Animal Care and Use Committee. Male mice bearing a GFPcre fusion that was knocked in to the Shh locus (hereafter designated Shh::cre; (Harfe et al., 2004) were either crossed to female Ctnnb1lox(ex3) female mice (Harada et al., 1999) to obtain Shh::cre;Ctnnb1lox(ex3) embryos or to mice harboring the Rosa26Lmx1b/+ allele (Li et al., 2010). For fate mapping experiments, male mice bearing an Ngn1::creERT2 (Kim et al., 2011) allele were crossed to Ai9 female reporter mice (Madisen et al., 2010). 2 mg of tamoxifen per 40 g of body weight were injected intraperitoneally into pregnant females to induce cre activity. The morning of the day when a vaginal plug was detected was designated as E0.5.
RNA in situ hybridization, X-gal, and fluorescent immunohistochemistry
Embryos or whole brains were harvested and fixed in 0.2%–4% PFA in PBS for various amount of time depending upon embryonic ages, and sectioned at 20–40 μm. X-gal, immunohistochemistry, and mRNA in situ hybridization were performed as described (Joksimovic et al., 2009b). For fluorescent immunohistochemistry, tissue sections were post-fixed in 1%–4% PFA in PBS, rinsed in PBS, microwave treated with Antigen Unmasking Solution (Vector Laboratories), blocked in 5% donkey serum and 0.1% Triton X-100 in PBS, and incubated overnight at 4°C with primary antibodies diluted in blocking solution: rabbit Lmx1a (gift from Dr. Michael German; 1:1,000), guinea pig Lmx1a (1:20,000), rat BrdU (Serotec; 1:400), mouse Ki67 (BD Pharmingen; 1:200), guinea pig Lmx1b (gift from Dr. Thomas Muller and Dr. Carmen Birchmeier; 1:10,000), rabbit Lmx1b (custom made; 1:5,000), mouse Nkx6-1 (Developmental Studies Hybridoma Bank; 1:100), goat Foxa2 (Santa Cruz; 1:50), rat Slc6a3 (Santa Cruz; 1:50), goat anti Lhx1 (Santa Cruz; 1:50), Mouse Isl1 (DSHB; 1:100), guinea pig Neurog2 (1:10,000), rabbit Pou4f1 (gift from Dr. Eric Turner; 1:4,000), mouse Pou4f1 (Santa Cruz; 1:100), rabbit and sheep Th (Pel Freeze; 1:500 and 1:250, respectively), and mouse HuC/D (Molecular Probes; 1:100). Sections were rinsed in PBS and incubated with appropriate Alexa 488, 555, and 647 (Molecular Probes) or Cy3 and Cy5 (Jackson ImmunoResearch) secondary antibodies diluted 1:250 in blocking solution, rinsed in PBS, covered with DAPI (1 mg/mL; Sigma) in PBS, rinsed in PBS, and coverslipped followed by epifluorescent (Leica) or confocal microscopy (Zeiss LSM 510 META and Leica DM6000B). Images were processed in Adobe Photoshop CS2 and CS5.
BrdU labeling/detection
Pregnant dams were injected with BrdU (50 mg/Kg, Sigma) and harvested either 30 minutes or 24 hours later. BrdU immunostaining was performed as described above.
Statistical analysis
All results were expressed as mean ± standard error of the mean (s.e.m.) and analyses were performed using Microsoft Excel software. Two experimental groups were compared with Student’s two sample t test. Significant differences were taken at p 0.05. For the 30 minute BrdU pulse, a number of BrdU+ cells within the Lmx1a+ ventricular zone were counted on every third 20μm section throughout the midbrain. For Pou4f1, Th, and Lmx1a quantification, the cells were counted within the dopaminergic field on every third 20 μm section throughout the midbrain. To quantify the mDA progenitor cell cycle exit, BrdU was injected in pregnant mothers at E12.5 and embryos harvested at E13.5. The total number of BrdU+ cells and the number of BrdU+/Ki67 progenitors in the ventricular zone were counted within the Lmx1a domain on every third 20 μm section throughout the midbrain. Similar to Tang et al., mDA progenitor cell cycle exit was also measured by counting the number of BrdU+/Ki67− in the mantle zone over the number of BrdU+/Ki67+ cells in the ventricular zone of the Lmx1a domain on every third 20 μm section throughout the midbrain.
Supplementary Material
Highlights.
A mix of DA and red nucleus progenitors.
Increase in proliferation and neurogenesis.
Loss of SNpc population vs. VTA.
Acknowledgments
We thank Dr. Cliff Tabin for Shh::creGFP mice and Shh cDNA, and Dr. Michael German, Dr. Thomas Muller, Dr. Carmen Birchmeier, and Dr. Eric Turner for antibodies. R.A. was supported by NIHR21 (1R21NS072703-01), Northwestern Memorial Foundation (Paul Ruby Foundation for Parkinson’s Research), the Whitehall Foundation and the Brain Research Foundation. A.A. was supported by the NRSA NIH 1F31 NS065670-01A2. J.F.P. was supported by FRSQ (Fonds de Recherche du Quebec en Sante) and the MJFF (Michael J. Fox Foundation).
Footnotes
Authors’ contributions
N.N., M.J.P., and M.J. designed and conducted the experiments, prepared figures and participated in writing the manuscript. J.F.P. and A.A. helped to characterize the conditional mutants. M.T. provided the Ctnnb1lox(ex3) mouse strain. Y.C.M. generated and provided the guinea-pig Lmx1a and Neurog2 antibodies. R.A. supervised the study and wrote the manuscript. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Navid Nouri, Email: navidnouri2015@u.northwestern.edu.
Meera J. Patel, Email: meerapatel@uchicago.edu.
Milan Joksimovic, Email: milanjok@hotmail.com.
Jean-Francois Poulin, Email: j-poulin@northwestern.edu.
Angela Anderegg, Email: angela.anderegg@gmail.com.
M. Mark Taketo, Email: taketo@mfour.med.kyoto-u.ac.jp.
Yong-Chao Ma, Email: ma@northwestern.edu.
Rajeshwar Awatramani, Email: r-awatramani@northwestern.edu.
References
- Alves dos Santos MT, Smidt MP. En1 and Wnt signaling in midbrain dopaminergic neuronal development. Neural development. 2011;6:23. doi: 10.1186/1749-8104-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderegg A, Lin HP, Chen JA, Caronia-Brown G, Cherepanova N, Yun B, Joksimovic M, Rock J, Harfe BD, Johnson R, Awatramani R. An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS genetics. 2013;9:e1003973. doi: 10.1371/journal.pgen.1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Blaess S, Bodea GO, Kabanova A, Chanet S, Mugniery E, Derouiche A, Stephen D, Joyner AL. Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural Dev. 2011;6:29. doi: 10.1186/1749-8104-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- Bonilla S, Hall AC, Pinto L, Attardo A, Gotz M, Huttner WB, Arenas E. Identification of midbrain floor plate radial glia-like cells as dopaminergic progenitors. Glia. 2008 doi: 10.1002/glia.20654. [DOI] [PubMed] [Google Scholar]
- Bye CR, Thompson LH, Parish CL. Birth dating of midbrain dopamine neurons identifies A9 enriched tissue for transplantation into parkinsonian mice. Experimental neurology. 2012;236:58–68. doi: 10.1016/j.expneurol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Cajanek L, Ribeiro D, Liste I, Parish CL, Bryja V, Arenas E. Wnt/beta-catenin signaling blockade promotes neuronal induction and dopaminergic differentiation in embryonic stem cells. Stem Cells. 2009;27:2917–2927. doi: 10.1002/stem.210. [DOI] [PubMed] [Google Scholar]
- Chilov D, Sinjushina N, Saarimaki-Vire J, Taketo MM, Partanen J. beta-Catenin regulates intercellular signalling networks and cell-type specific transcription in the developing mouse midbrain-rhombomere 1 region. PLoS ONE. 2010;5:e10881. doi: 10.1371/journal.pone.0010881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Leung A, Han BS, Chang MY, Moon JI, Kim CH, Hong S, Pruszak J, Isacson O, Kim KS. Wnt1-lmx1a forms a novel autoregulatory loop and controls midbrain dopaminergic differentiation synergistically with the SHH-FoxA2 pathway. Cell stem cell. 2009;5:646–658. doi: 10.1016/j.stem.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper O, Hargus G, Deleidi M, Blak A, Osborn T, Marlow E, Lee K, Levy A, Perez-Torres E, Yow A, Isacson O. Differentiation of human ES and Parkinson’s disease iPS cells into ventral midbrain dopaminergic neurons requires a high activity form of SHH, FGF8a and specific regionalization by retinoic acid. Mol Cell Neurosci. 2010;45:258–266. doi: 10.1016/j.mcn.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Andersson E, Hedlund E, Alekseenko Z, Coppola E, Panman L, Millonig JH, Brunet JF, Ericson J, Perlmann T. Specific and integrated roles of Lmx1a, Lmx1b and Phox2a in ventral midbrain development. Development. 2011;138:3399–3408. doi: 10.1242/dev.065482. [DOI] [PubMed] [Google Scholar]
- Di Salvio M, Di Giovannantonio LG, Acampora D, Prosperi R, Omodei D, Prakash N, Wurst W, Simeone A. Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP. Nature neuroscience. 2010;13:1481–1488. doi: 10.1038/nn.2661. [DOI] [PubMed] [Google Scholar]
- Fasano CA, Chambers SM, Lee G, Tomishima MJ, Studer L. Efficient derivation of functional floor plate tissue from human embryonic stem cells. Cell stem cell. 2010;6:336–347. doi: 10.1016/j.stem.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ferri AL, Lin W, Mavromatakis YE, Wang JC, Sasaki H, Whitsett JA, Ang SL. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134:2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- Geng X, Speirs C, Lagutin O, Inbal A, Liu W, Solnica-Krezel L, Jeong Y, Epstein DJ, Oliver G. Haploinsufficiency of Six3 fails to activate Sonic hedgehog expression in the ventral forebrain and causes holoprosencephaly. Dev Cell. 2008;15:236–247. doi: 10.1016/j.devcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grealish S, Diguet E, Kirkeby A, Mattsson B, Heuer A, Bramoulle Y, Van Camp N, Perrier Anselme L, Hantraye P, Björklund A, Parmar M. Human ESC-Derived Dopamine Neurons Show Similar Preclinical Efficacy and Potency to Fetal Neurons when Grafted in a Rat Model of Parkinson’s Disease. Cell stem cell. 2014;15:653–665. doi: 10.1016/j.stem.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. Embo J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Hayes L, Zhang Z, Albert P, Zervas M, Ahn S. Timing of Sonic hedgehog and Gli1 expression segregates midbrain dopamine neurons. The Journal of comparative neurology. 2011;519:3001–3018. doi: 10.1002/cne.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joksimovic M, Anderegg A, Roy A, Campochiaro L, Yun B, Kittappa R, McKay R, Awatramani R. Spatiotemporally separable Shh domains in the midbrain define distinct dopaminergic progenitor pools. Proceedings of the National Academy of Sciences of the United States of America. 2009a;106:19185–19190. doi: 10.1073/pnas.0904285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joksimovic M, Awatramani R. Wnt/beta-catenin signaling in midbrain dopaminergic neuron specification and neurogenesis. Journal of molecular cell biology. 2014;6:27–33. doi: 10.1093/jmcb/mjt043. [DOI] [PubMed] [Google Scholar]
- Joksimovic M, Patel M, Taketo MM, Johnson R, Awatramani R. Ectopic Wnt/beta–catenin signaling induces neurogenesis in the spinal cord and hindbrain floor plate. PLoS One. 2012;7:e30266. doi: 10.1371/journal.pone.0030266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joksimovic M, Yun BA, Kittappa R, Anderegg AM, Chang WW, Taketo MM, McKay RD, Awatramani RB. Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nature neuroscience. 2009b;12:125–131. doi: 10.1038/nn.2243. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Hori K, Wyckoff A, Dickel LK, Koundakjian EJ, Goodrich LV, Johnson JE. Spatiotemporal fate map of neurogenin1 (Neurog1) lineages in the mouse central nervous system. J Comp Neurol. 2011;519:1355–1370. doi: 10.1002/cne.22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkeby A, Grealish S, Wolf DA, Nelander J, Wood J, Lundblad M, Lindvall O, Parmar M. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell reports. 2012;1:703–714. doi: 10.1016/j.celrep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Kittappa R, Chang WW, Awatramani RB, McKay RD. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS biology. 2007;5:e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, Yang L, Beal MF, Surmeier DJ, Kordower JH, Tabar V, Studer L. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti L, Peltopuro P, Piepponen TP, Partanen J. Cell-autonomous FGF signaling regulates anteroposterior patterning and neuronal differentiation in the mesodiencephalic dopaminergic progenitor domain. Development. 2012;139:894–905. doi: 10.1242/dev.071936. [DOI] [PubMed] [Google Scholar]
- Li Y, Qiu Q, Watson SS, Schweitzer R, Johnson RL. Uncoupling skeletal and connective tissue patterning: conditional deletion in cartilage progenitors reveals cell-autonomous requirements for Lmx1b in dorsal-ventral limb patterning. Development. 2010;137:1181–1188. doi: 10.1242/dev.045237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, Behrens J. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzakopian E, Lin W, Salmon-Divon M, Dvinge H, Andersson E, Ericson J, Perlmann T, Whitsett JA, Bertone P, Ang SL. Genome-wide characterization of Foxa2 targets reveals upregulation of floor plate genes and repression of ventrolateral genes in midbrain dopaminergic progenitors. Development. 2012;139:2625–2634. doi: 10.1242/dev.081034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani T, Kumai M, Mizuhara E, Minaki Y, Ono Y. Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev Biol. 2010;339:101–113. doi: 10.1016/j.ydbio.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Nakatani T, Minaki Y, Kumai M, Ono Y. Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development. 2007;134:2783–2793. doi: 10.1242/dev.02870. [DOI] [PubMed] [Google Scholar]
- Omodei D, Acampora D, Mancuso P, Prakash N, Di Giovannantonio LG, Wurst W, Simeone A. Anterior-posterior graded response to Otx2 controls proliferation and differentiation of dopaminergic progenitors in the ventral mesencephalon. Development. 2008;135:3459–3470. doi: 10.1242/dev.027003. [DOI] [PubMed] [Google Scholar]
- Ono Y, Nakatani T, Sakamoto Y, Mizuhara E, Minaki Y, Kumai M, Hamaguchi A, Nishimura M, Inoue Y, Hayashi H, Takahashi J, Imai T. Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- Panman L, Papathanou M, Laguna A, Oosterveen T, Volakakis N, Acampora D, Kurtsdotter I, Yoshitake T, Kehr J, Joodmardi E, Muhr J, Simeone A, Ericson J, Perlmann T. Sox6 and Otx2 control the specification of substantia nigra and ventral tegmental area dopamine neurons. Cell reports. 2014;8:1018–1025. doi: 10.1016/j.celrep.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Perez-Balaguer A, Puelles E, Wurst W, Martinez S. Shh dependent and independent maintenance of basal midbrain. Mechanisms of development. 2009;126:301–313. doi: 10.1016/j.mod.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Poulin JF, Zou J, Drouin-Ouellet J, Kim KY, Cicchetti F, Awatramani RB. Defining midbrain dopaminergic neuron diversity by single-cell gene expression profiling. Cell reports. 2014;9:930–943. doi: 10.1016/j.celrep.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash N, Brodski C, Naserke T, Puelles E, Gogoi R, Hall A, Panhuysen M, Echevarria D, Sussel L, Weisenhorn DM, Martinez S, Arenas E, Simeone A, Wurst W. A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development. 2006;133:89–98. doi: 10.1242/dev.02181. [DOI] [PubMed] [Google Scholar]
- Prakash N, Puelles E, Freude K, Trumbach D, Omodei D, Di Salvio M, Sussel L, Ericson J, Sander M, Simeone A, Wurst W. Nkx6–1 controls the identity and fate of red nucleus and oculomotor neurons in the mouse midbrain. Development. 2009;136:2545–2555. doi: 10.1242/dev.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbeck JA, Studer L. Moving stem cells to the clinic: potential and limitations for brain repair. Neuron. 2015;86:187–206. doi: 10.1016/j.neuron.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Miyamoto Y, Huang EJ. Multiple roles of {beta}-catenin in controlling the neurogenic niche for midbrain dopamine neurons. Development. 2009;136:2027–2038. doi: 10.1242/dev.034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Villaescusa JC, Luo SX, Guitarte C, Lei S, Miyamoto Y, Taketo MM, Arenas E, Huang EJ. Interactions of Wnt/beta-catenin signaling and sonic hedgehog regulate the neurogenesis of ventral midbrain dopamine neurons. J Neurosci. 2010;30:9280–9291. doi: 10.1523/JNEUROSCI.0860-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Veenvliet JV, Dos Santos MT, Kouwenhoven WM, von Oerthel L, Lim JL, van der Linden AJ, Koerkamp MJ, Holstege FC, Smidt MP. Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development. 2013;140:3373–3384. doi: 10.1242/dev.094565. [DOI] [PubMed] [Google Scholar]
- Xi J, Liu Y, Liu H, Chen H, Emborg ME, Zhang SC. Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem cells. 2012;30:1655–1663. doi: 10.1002/stem.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Brown A, Ellisor D, Paul E, Hagan N, Zervas M. Dynamic temporal requirement of Wnt1 in midbrain dopamine neuron development. Development. 2013;140:1342–1352. doi: 10.1242/dev.080630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.