Abstract
Abnormal composition of intestinal bacteria —“dysbiosis”— is characteristic of Crohn’s disease. Disease treatments include dietary changes and immunosuppressive anti-TNFα antibodies as well as ancillary antibiotic therapy but their effects on microbiota composition are undetermined. Using shotgun metagenomic sequencing, we analyzed fecal samples from a prospective cohort of pediatric Crohn’s disease patients starting therapy with enteral nutrition or anti-TNFα antibodies and reveal the full complement and dynamics of bacteria, fungi, archaea and viruses during treatment. Bacterial community membership was associated independently with intestinal inflammation, antibiotic use, and therapy. Antibiotic exposure was associated with increased dysbiosis, whereas dysbiosis decreased with reduced intestinal inflammation. Fungal proportions increased with disease and antibiotic use. Dietary therapy had independent and rapid effects on microbiota composition distinct from other stressor-induced changes and effectively reduced inflammation. These findings reveal that dysbiosis results from independent effects of inflammation, diet, and antibiotics and shed light on Crohn disease treatments.
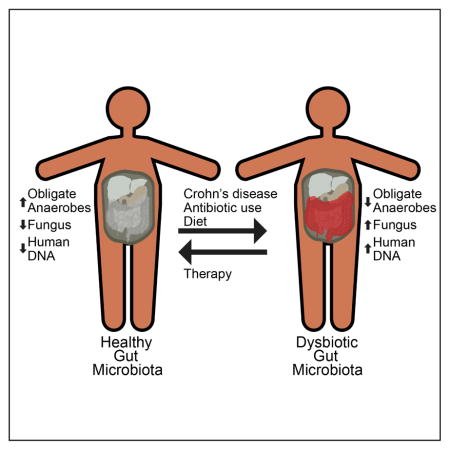
Graphical abstract
INTRODUCTION
The human gut microbiota is densely populated by microbes from all three domains of life together with their viruses (Hoffmann et al., 2013; Human Microbiome Project, 2012; Samuel et al., 2007). Crohn’s disease results from a pathologic interaction between the mucosal immune system and the environment, particularly the microbes residing in the gut lumen (Sartor, 2006, 2008) and is characterized by altered—or “dysbiotic”--gut bacterial composition (Huttenhower et al., 2014; Khor et al., 2011; Sartor, 2006, 2008). Numerous human genetic loci encoding proteins involved in host immune responses have been linked to Crohn’s disease (Jostins et al., 2012), but the impact of these genes on the dysbiosis associated with Crohn’s disease is limited (Knights et al., 2014). Rather, dysbiosis is hypothesized to be a response of the microbes, particularly bacteria, to environmental stressors such as the host inflammatory response (Huttenhower et al., 2014) and/or the production of electron acceptors that facilitate anaerobic respiration (Winter and Baumler, 2014). Dysbiosis is commonly characterized by an expansion of Proteobacteria and a decrease in Firmicutes, along with a decrease in community richness (Nagalingam and Lynch, 2012). Much less is known about the responses of other domains of microbial life to environmental stressors. Similarly, it is unknown whether dysbiosis resulting from inflammation is rapidly reversible. Patients with Crohn’s disease are exposed to antibiotics and dietary changes which are likely to affect the microbiota, but the influence of these factors and their interactions with each other are incompletely understood.
Antibiotics are often used as an ancillary therapy for Crohn’s disease (Khan et al., 2011). However, the main therapies for Crohn’s disease includes episodic or chronic immunosuppression with corticosteroids, antimetabolite agents, or antibodies directed against host immune proteins such as tumor necrosis factor α (anti-TNF) (Borrelli et al., 2006; Grover et al., 2014; Rutgeerts et al., 2012). The anti-TNF medications are administered parenterally and are not expected to alter the gut microbiota composition directly. An alternative therapy, used predominantly in children, is the defined formula diet, also known as enteral nutrition therapy. Both elemental and polymeric formulae, containing, respectively, amino acids and intact protein, have proved efficacious in treating symptoms and intestinal inflammation in Crohn’s disease in addition to supporting nutritional needs for growth and weight maintenance (Borrelli et al., 2006; Grover et al., 2014). The efficacy of these diets is greatest when used as the exclusive source of nutrition(Grover et al., 2014; Lee et al., 2015). Dietary therapy has the advantage of avoiding immunosuppression but is difficult to maintain long term. If the mechanism of action of dietary-based therapies were understood, it might be possible to develop less restrictive diets that deliver the same therapeutic benefit. One hypothesis is that diet therapy alters the composition of the gut microbiota in a manner that contributes to the therapeutic benefit, though data supporting this are limited (Gerasimidis et al., 2014; Kaakoush et al., 2015).
We reasoned that longitudinal characterization of the gut microbiome of pediatric Crohn’s disease patients initiating therapy would allow us to characterize the concurrent effects of intestinal inflammation, antibiotics, and diet on gut microbial community structure. We conducted a longitudinal study of 90 children with Crohn’s disease who were initiating treatment with either a defined formula diet or anti-TNF therapy, and compared them to 26 healthy control children. We tracked symptoms, mucosal inflammation, and changes in the gut microbiome over an eight-week study period. The gut microbiome was quantified using shotgun metagenomic DNA sequence analysis of longitudinal samples. Dysbiosis was quantified as the distance of each sample from the centroid of samples from healthy controls. Communities partitioned into two distinct clusters based on the bacterial composition, as seen previously (Frank et al., 2007; Gevers et al., 2014), one of which overlapped the healthy controls. The dysbiotic community was associated with increases in specific fungi, prior antibiotic therapy and higher concentration of human DNA in feces. By tracking the microbiota composition over the course of therapy, we found that dysbiosis was reduced in response to decreased bowel inflammation, and that inflammation, antibiotic exposure, and diet independently influenced different taxa. Fungi were elevated with disease and antibiotic use, but diminished with diet therapy. Thus while dysbiosis in the gut is common in Crohn’s disease, the response of the gut microbiome depends on the environmental stressor.
RESULTS
Clinical outcomes in patients treated with a defined formula diet versus anti-TNF
Ninety children initiated one of the study therapies (52 anti-TNF; 22 exclusive enteral nutrition (EEN); 16 partial enteral nutrition with ad lib diet (PEN)) (Supplemental methods and (Lee et al., 2015). EEN and PEN treated children consumed approximately 90% and 53% of daily calories from dietary formulas, respectively. Inflammation was quantified by measuring fecal calprotectin (FCP). Response to therapy was defined as a reduction of FCP to below 250mcg/g because this measure is associated with diminished mucosal inflammation (Lin et al., 2014). Reduction in FCP below 250 mcg/g was more common among those receiving anti-TNF (62%) and EEN (45%) than PEN (10%)(Lee et al., 2015).
Microbial community patterns in Crohn’s disease and healthy controls
Adequate stool samples were available from 86 individuals to conduct shotgun metagenomic analysis. Samples were collected at four time points: baseline, 1 week, 4 weeks, and 8 weeks into therapy (Table S1A). We also compared stool samples from 26 healthy children (Table 1) collected in a prior study (Wu et al., 2011), but analyzed by shotgun metagenomic sequencing here. None of the healthy children had received antibiotics in the prior 6 months. DNA was prepared from whole stool, and sequenced using the Illumina HiSeq paired-end method. After filtering out low quality, human and contaminating reads, we were left with 6.5×1011 bases of microbial DNA sequence for analysis.
Table 1.
Characteristics of healthy controls and patients with Crohn’s disease^ in the two clusters based on bacterial taxonomy*
| Healthy Controls (n=26) | Near Cluster (n=57) | Far Cluster (n=28) | P value | Adjusted Odds Ratio (95% CI)† | |
|---|---|---|---|---|---|
| Age (years) | 14.3 (7.9–19.9) | 12.6 (10.7–15.1) | 13.9 (10.1–15.5) | 0.58 | |
| Female sex | 12 (46%) | 21 (37%) | 13 (46%) | 0.48 | |
| White race | 17 (65%) | 52 (91%) | 23 (82%) | 0.29 | |
| Canadian | 0 (0%) | 23 (40%) | 9 (32%) | 0.49 | |
| Prior bowel surgery | 0 (0%)§ | 3 (5%) | 3 (11%) | 0.38 | |
| Duration of Crohn’s disease (years) | N/A | 0.1 (0.0–1.1) | 0.2 (0.1–1.0) | 0.87 | |
| Disease location | |||||
| Ileum | N/A | 52 (91%) | 23 (82%) | 0.29 | |
| Cecum | N/A | 39 (68%) | 24 (86%) | 0.12 | |
| Colorectal | N/A | 54 (95%) | 27 (96%) | >0.99 | |
| Upper gastrointestinal | N/A | 33 (58%) | 16 (57%) | >0.99 | |
| Phenotype | |||||
| Penetrating | N/A | 2 (4%) | 2 (7%) | 0.60 | |
| Stricturing | N/A | 5 (9%) | 1 (4%) | 0.66 | |
| Perianal disease | N/A | 14 (25%) | 8 (29%) | 0.79 | |
| Symptoms | |||||
| PCDAI | N/A | 32.5 (20–45) | 30 (21.25–40) | 0.73 | |
| Bleeding | N/A | 16 (28%) | 8 (29%) | >0.99 | |
| Consistency | 0.14 | ||||
| Formed | N/A# | 20 (35%) | 4 (14%) | ||
| Partially formed | N/A# | 21 (37%) | 14 (50%) | ||
| Unformed | 0 (0%) | 16 (28%) | 10 (38%) | ||
| Stools per day | 0.59 | ||||
| 0–2 | 25 (96%) | 26 (46%) | 11 (39%) | ||
| 3–5 | 1 (4%) | 27 (47%) | 13 (46%) | ||
| 6–8 | 0 (0%) | 2 (4%) | 3 (11%) | ||
| >8 | 0 (0%) | 2 (4%) | 1 (4%) | ||
| Medications | |||||
| Mesalamine | N/A | 28 (49%) | 14 (50%) | >0.99 | |
| Oral corticosteroids | N/A | 14 (25%) | 18 (64%) | 0.001 | 8.2 (1.7–39.6) |
| Rectal corticosteroids | N/A | 0 (0%) | 1 (4%) | 0.33 | |
| Thiopurine | N/A | 7 (12%) | 1 (4%) | 0.26 | |
| Antibiotics within prior 6 months | 0 (0%) | 21 (37%) | 23 (82%) | <0.001 | 8.4 (1.9–38.1) |
| Current antibiotic use | 0 (0%) | 9 (16%) | 15 (54%) | 0.001 | |
| Abundance of fungi (quartiles) | <0.001 | 7.1 (2.8–18.5)‡ | |||
| 1st | 9 (35%) | 18 (32%) | 1 (4%) | Ref | |
| 2nd | 10 (38%) | 16 (29%) | 1 (4%) | 0.8 (0.04–18.0) | |
| 3rd | 7 (2%) | 16 (29%) | 5 (18%) | 9.2 (0.7–124.2) | |
| 4th | 0 (0%) | 6 (11%) | 21 (75%) | 111.5 (7.1–1759.1) | |
| Abundance of human DNA (quartiles) | <0.001 | ||||
| 1st | 14 (54%) | 14 (25%) | 0 (0%) | ||
| 2nd | 8 (31%) | 16 (29%) | 3 (11%) | ||
| 3rd | 4 (15%) | 18 (32%) | 6 (21%) | ||
| 4th | 0 (0%) | 8 (14%) | 19 (68%) | ||
| Fecal calprotectin (mcg/g) | N/A | 822.5 (348–1458 | 748.5 (342.5–1881) | 0.89 | |
One subject was missing a baseline stool sample, therefore 85 patients were included in the analysis
Continuous variables reported as medians and interquartile range. Unadjusted p values from Fisher exact test or Wilcoxon rank sum test comparing children with Crohn’s disease in the near and far clusters, respectively
Adjusted odds ratios derived from logistic regression using backwards elimination starting with all variables with unadjusted p value <0.2 and retaining all variables with a p value <0.1
Diarrhea was an exclusion criteria for healthy control children
One child had prior appendectomy
Quartiles as a continuous variable
N/A – Not applicable
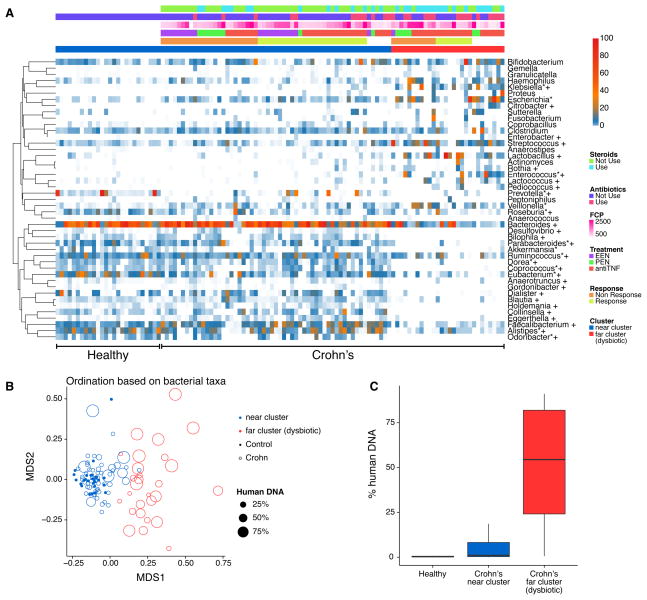
We quantified the bacterial taxonomic composition using MetaPhlAn (Segata et al., 2012). Figure 1 shows a comparison of bacterial lineages in the healthy control subjects and pediatric Crohn’s disease cohort prior to initiation of therapy (“baseline” in the below). Figure 1A shows taxonomic proportions for each sample at baseline with metadata summarized above the heat map (all time points are shown in Figure S1A).
Figure 1.
Bacterial composition in samples from children with Crohn’s disease and healthy controls. (A) A heatmap demonstrating relative abundance of bacterial taxa prior to therapy according to presence or absence of Crohn’s disease, cluster assignment, use of corticosteroids and antibiotics, FCP concentration, and response to therapy. Metadata is indicated by the color code at the top of the figure. White cells indicate missing data. Taxa that were statistically different in abundance between Crohn’s disease and healthy controls are identified by *; taxa that were statistically different in abundance between the two Crohn’s disease clusters are identified by † (q<0.05). FCP in this and subsequent figures indicates Fecal Calprotectin. Samples were ordered by the metadata (healthy versus Crohn’s samples, and cluster 1 versus cluster 2, then other forms of metadata). (B) Multidimensional scaling (MDS) analysis of samples from children with Crohn’s disease and healthy controls. Bacterial taxa present were quantified by MetaPhlAn, distances were calculated using binary Jaccard Index, and samples were plotted based on MDS. Samples from healthy controls are shown by the filled circles, Crohn’s disease as open circles. Clusters were defined by partitioning around medoids with estimation of number of clusters (PAMK), and are colored blue (healthy associated) and red (dysbiotic). The size of the dot is scaled by the proportion of human DNA in the sample. (C) Percentage of human DNA reads in each metagenomic sequence sample. Near cluster (blue, associated with healthy controls) and far cluster (red, dysbiotic) refer to the groups shown in 1B.
After filtering out very low abundance genera, comparison of median relative abundances showed 14 of 45 genera differed between children with Crohn’s disease and healthy controls (FDR controlled p (q)<0.05 by Wilcoxon rank sum test; Table S1B). The Crohn’s patients had reduced relative abundance of Prevotella, Eubacterium, Odoribacter, Akkermansia, Roseburia, Parabacteroides, Alistipes, Coprococcus, Dorea and Ruminococcus and increased abundance of Escherichia, Klebsiella, Enterococcus and Veillonella. Random forest was used to identify bacterial lineages that best distinguished healthy children from Crohn’s patients with active disease, and predicted Crohn’s disease and normal with 86% prediction accuracy. Mostly similar lineages (Prevotella, Odoribacter, Eubacterium, Escherichia and Faecalibacterium) were found to distinguish the groups.
Community patterns were analyzed using PAMK (partitioning around medoids with estimation of number of clusters) to find the optimal number of clusters, and visualized after multidimensional scaling (MDS) (Figure 1B). Samples from patients with active Crohn’s disease partitioned into two clusters (Figure S1B), one that was near to the healthy controls (near cluster) and one that clustered separately (far dysbiotic cluster), paralleling previous studies (Frank et al., 2007; Gevers et al., 2014). Thirty of 45 genera differed in abundance (q<0.05) between the two clusters (Table S1C). The far dysbiotic cluster was characterized by an increased relative abundance of Streptococcus, Klebsiella, Lactobacillus and reduced relative abundance of Faecalibacterium, Parabacteroides, Dorea, Blautia, Holdemania, Collinsella, Coprococcus, Odoribater, Prevotella, Bacteroides, Dialister, Eubacterium, Alistipes and Ruminococcus. Random forest with bacterial abundance had 92% prediction accuracy for predicting these two clusters. The top five most predictive genera were Blautia, Faecalibacterium, Dialister, Lactobacillus and Bacteroides.
Reduced diversity was observed in both clusters relative to healthy subjects, and in the dysbiotic far cluster compared to the near cluster (see Supplemental Results). An analysis using phylogenetic attributions at the species level yielded similar conclusions (Figure S1C and S1D).
In summary, baseline samples clustered into two groups, with the far dysbiotic cluster being more distant from the healthy controls and characterized by altered bacterial composition and lower diversity.
High levels of human DNA in stool correlated with dysbiosis
In processing the metagenomic DNA from stool, we observed that the proportion of human DNA varied widely (Figure 1C). The percentage of human reads was low in the healthy controls (mean=0.87%, max=10.1%, minimum=0.05%) relative to patients with active Crohn’s disease (mean=16.6%, maximum=94.2%, minimum=0.01%; p<5×10−11), with over 50% of the sequence reads mapped to human in 49 Crohn’s disease samples. Selected DNA samples were tested for human DNA content using QPCR to detect human beta-tubulin-coding sequences, revealing that the amounts detected were positively correlated with the calls from metagenomic sequencing (r=0.81 Pearson’s correlation, p= 9.3×10−11). We hypothesized that the source of human DNA was either epithelial cells or blood shed into feces associated with disease activity.
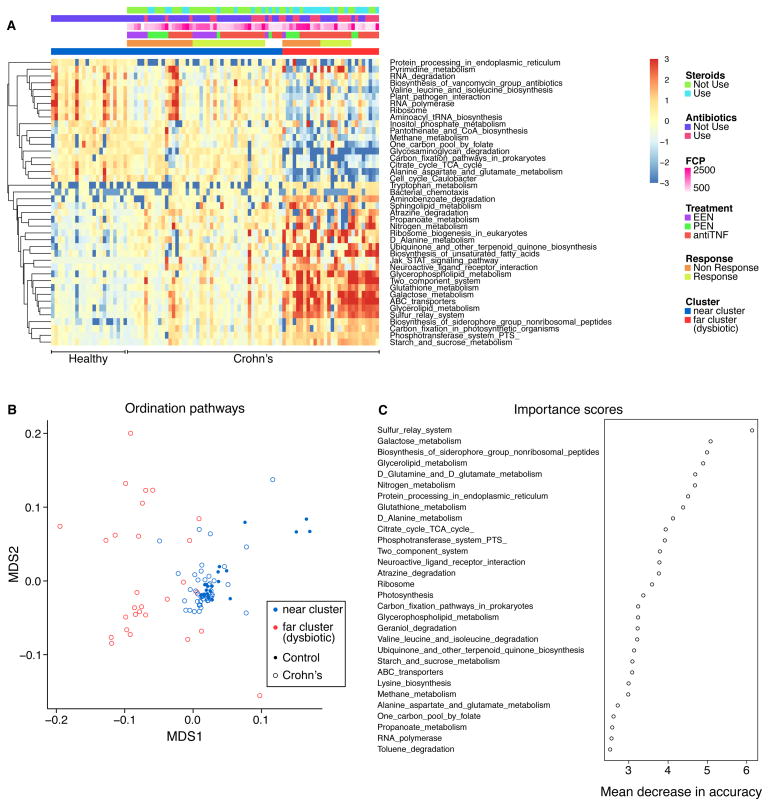
Analysis of fecal microbial gene pathways
The gene content of the gut microbiota was compared to assess associations between gene function and dysbiosis (Figure 2A) using HUMAnN (Abubucker et al., 2012) Differences were found between IBD and healthy controls in 42 of 163 pathways examined (Table S1D). Crohn’s samples clustered into two groups based on gene pathway data, one of which more closely resembled the healthy controls and one more dysbiotic (Figure 2B and Table S1E). The clusters separated based on pathway data tracked closely with the near and far clusters defined by bacterial taxonomic representation (Figure 2B, red and blue colors).
Figure 2.
Comparison of gene pathways present in samples from Crohn’s disease subjects and healthy controls at baseline assessed by shotgun metagenomics. (A) Heatmap of pathways that differed significantly (q value <0.05) between healthy controls and Crohn’s disease subjects at baseline. Each row was normalized by z-score. B) Cluster analysis based on multidimensional scaling using the pathway data. Colors indicate samples that are members of the near cluster (overlapping healthy controls) or far cluster (dysbiotic) as defined by the analysis of bacterial taxa. Controls are defined by filled circles, Crohn’s disease samples by open circles. (C) Analysis using the machine learning algorithm Random Forest. Gene pathways that most strongly distinguish patients with Crohn’s disease from healthy controls were identified. Importance scores were derived from the loss in accuracy measured when each indicated pathway was removed from the analysis. The units on the x-axis indicate mean decrease in accuracy.
To guide interpretation, we analyzed the Spearman correlation between bacterial abundance and gene pathway enrichment in these data (Figure S2A). Pathway data partitioned the bacteria into two notable groups, one including gammaproteobacteria together with Streptococcus and Enterococcus, and a second group dominated by Bacteroides and multiple anaerobic Bacteroidetes and Firmicutes typical of healthy gut. Below we propose relationships between specific gene pathways and persistence in the presence of different environmental stressors. However, enrichment may also be due to passenger effects, where outgrowth of specific organisms resulted in enrichment in pathways that were not themselves major contributors to differential fitness.
Random forest was used to identify those pathways that best discriminated between healthy children and the active Crohn’s disease cohort at baseline (Figure 2C and Figure S2B and 2C). Samples membership in the two categories could be predicted with 87% accuracy using gene pathway data, comparable to the partitioning achieved using bacterial taxonomic data. The top six most notable pathways, higher in the Crohn’s samples, were sulfur relay systems, galactose metabolism, biosynthesis of siderophores, glycerolipid metabolism, glutamine/glutamate metabolism, and nitrogen metabolism (Table S1D). Similarly, 94 of 163 gene pathways were differentially abundant between the two clusters, 51 with greater representation in the more dysbiotic far cluster (Table S1E). Random forest using gene pathway abundance had 87% prediction accuracy for classification of the two bacterial clusters (near and far dysbiotic) defined by taxonomic proportions. The five most predictive pathways were glycerophospholipid metabolism, aminobenzoate degradation, sulfur relay system and glutathione metabolism which were increased in the far dysbiotic cluster and selenocompound metabolism which was decreased in the far cluster. Thirty-three of 42 pathways that were significantly different between Crohn’s disease and healthy controls were also significantly different between the two clusters of Crohn’s disease, suggesting that they are related primarily to the composition of the dysbiotic microbiota rather than being intrinsic alterations associated with IBD per se.
Fungal community structure
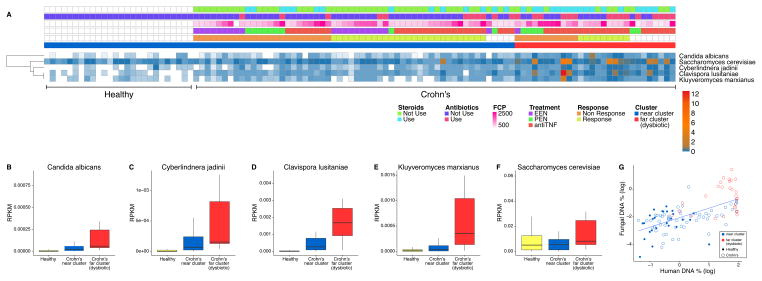
We characterized the fungal reads in the metagenomic data by alignment to sequenced fungal genomes, followed by extensive filtering to remove artifacts (described in Supplemental procedures and Tables S1F and G). Five fungal taxa were detected in the samples--Saccharomyces cerevisiae, Clavispora lusitaniae (also known as Candida lusitaniae), Cyberlindnera jadinii (also known as Pichia jadinii and Candida utilis), Candida albicans, and Kluyveromyces marxianus.
Figure 3A shows the representation of fungal taxa in healthy controls and Crohn’s disease samples at baseline, and the relationship to metadata. All of the fungal taxa were more represented in the samples from Crohn’s disease subjects, particularly those in the more dysbiotic far cluster as defined by the bacterial taxonomic analysis in Figure 1 (Figure 3 B–F). Abundance of the different fungi were highly correlated with each other (Spearman correlation 0.66–0.84, Table S1H). Higher fungal proportions in samples also correlated with higher levels of human DNA (Figure 3G). Random Forest analysis based on fungal data had 92% prediction accuracy for classification of normal and baseline Crohn’s disease samples. The most predictive fungus was Clavispora lusitaniae. Thus the five yeasts detected are positively associated with Crohn’s disease, particularly in the setting of greater bacterial dysbiosis.
Figure 3.
Fungal representation in samples from Crohn’s disease subjects and healthy controls at baseline. (A) Heatmap summarizing the abundance of fungal taxa present in healthy controls (left side of figure) and children with Crohn’s disease (right side of figure). Metadata is indicated by the color code at the top of the figure. White cells indicate missing data. Bar graphs (B)–(F) show the relative abundance of the five main taxa detected. The y-axis shows reads per kilobase of target DNA per million reads in the sample (RPKM). (G) Graph showing correlation between human DNA % and fungal DNA %. Points are colored by membership in the near or far clusters based on the bacterial taxonomic data. Samples from healthy subjects are shown filled, Crohn’s disease by open circles.
Some correlations were observed between bacteria and fungi in the cohort, but these varied between the healthy controls and the two clusters of patients with Crohn’s disease (Supplemental Results and Figure S3A–3C).
Analysis of archaea
Archaea comprise a third domain of life, and are primarily represented in the gut by Methanobrevibacter smithii (Hoffmann et al., 2013; Human Microbiome Project, 2012; Samuel et al., 2007). We aligned the full set of non-human sequence reads to the M. smithii genome, and found that most of the pediatric subjects studied had little or no detectable colonization. No difference was observed in mean values for the Crohn’s disease (baseline time point) and healthy cohorts (p=0.81 for comparison of mean proportions), paralleling a previous study (Chehoud et al., 2015).
Analysis of viral representation
We investigated the representation of DNA viruses in the patient samples after alignment of sequence reads to databases of viral genomes. No virus was identified that was significantly enriched in Crohn’s disease samples versus healthy controls. A recent report proposed that tailed phages (Caudovirales) were enriched in viral concentrates from stool of IBD patients (Norman et al., 2015). We thus investigated the representation of bacteriophage sequences in our data. The analysis was limited by use of whole stool instead of purified viral particles, and by the high natural phage diversity that results in difficulty in identification in metagenomic data. No significant difference was found in the abundance of Caudovirales between healthy controls and Crohn’s disease samples at the baseline time point (p=0.98; Wilcoxon rank sum test), or for comparison of the near and far cluster as defined by the bacterial taxonomic composition (p=0.35; Wilcoxon rank sum test).
Two subjects on anti-TNF therapy showed substantial representation of animal cell viruses in stool samples (anellovirus and bocavirus). To investigate this further, viral particles were purified from the two samples and sequenced, which validated detection of each. These findings raise the question of whether the anti-TNF therapy itself may have promoted these viral blooms, similar to blooms in solid organ transplant recipients receiving immunosuppression (De Vlaminck et al., 2013; Young et al., 2015).
Exploring the baseline clusters defined by bacterial abundance in a multivariate model
We next explored factors associated with membership in the dysbiotic far cluster at baseline (Figure 1B). In the multivariable model, use of antibiotics within the preceding 6 months, current use of corticosteroids, and abundance of fungi were independently associated with membership in the far cluster (Table 1). These data indicate that at least part of the previously described dysbiosis in IBD may be due to antibiotic and corticosteroid exposure and that the dysbiosis extends beyond the bacteria to include fungi.
Given the association with antibiotics, we further defined the bacterial taxa that distinguished Crohn’s disease from healthy controls by analyzing only the subset of patients not treated with antibiotics (Table S1I). Separately we analyzed the association of antibiotic use and taxonomic representation within the Crohn’s disease cohort (Table S1J). Bacterial genera associated with Crohn’s disease in the absence of antibiotic use were almost completely different from genera associated with antibiotic use in the Crohn’s disease cohort (discussed further below).
Clustering did not predict response to therapy
Rates of response to treatment were not significantly different among patients in the near or far dysbiotic cluster. Twenty-seven patients (53%) in the near cluster and 9 (45%) patients in the far dysbiotic cluster achieved a reduction in FCP below 250mcg/g (p=0.60). Among anti-TNF treated patients the response rates were 66.7% and 60%, respectively (p=0.74). Too few EEN treated patients were available for meaningful comparison.
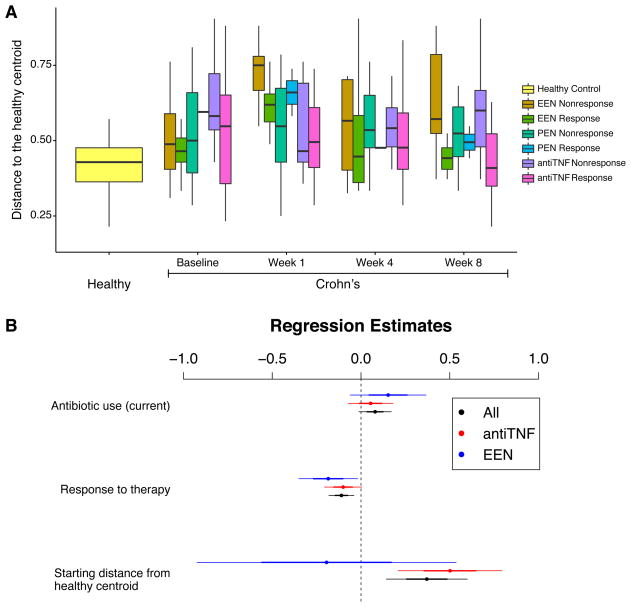
Dynamics of dysbiosis with treatment
We categorized sequences from each fecal sample by its Jaccard distance from the centroid of the healthy control samples, providing a quantitative measure of dysbiosis, and then analyzed longitudinal dynamics. One of the proposed mechanisms of action of diet-based therapy is to alter the gut microbiota composition. We thus assessed the bacterial composition at baseline and 1 week into therapy (Figure 4A). The microbiota composition among the EEN-treated group changed within 1 week of therapy, moving significantly farther from centroid of the healthy controls (relative to baseline p=0.005 overall, p=0.02 among responders, p=0.14 among non-responders). One week after initiation of EEN, abundance of 6 of the 40 genera examined were nominally different at a threshold of p=0.05 (Haemophilus, Alistipes, Streptococcus, Dialister, Dorea, and Gordonibacter; not significant after correction for multiple comparisons; Table S1K). A similar pattern was not seen among the PEN treated patients (p=0.83) or anti-TNF treated patients (p=0.02, note anti-TNF treated group moved closer to healthy centroid within 1 week), suggesting that either there is a dose-dependent effect of enteral nutrition or that fully removing regular table food from the diet influences microbiota composition within one week.
Figure 4.
Change in the microbiota composition among children with Crohn’s disease treated with EEN, PEN, and anti-TNF therapy. (A) Characterization of the bacterial taxonomic composition based on distance from the centroid of healthy controls. Boxes show median and first and third quartile. The x-axis shows the group and time point, the y-axis shows the distance from the centroid of the healthy controls. The groups compared are shown to the right. (B) Plot of regression coefficients and their confidence intervals. The dependent variable used is distance to the healthy centroid. Covariates included antibiotic use, response to therapy defined as reduction in FCP to less than 250mcg/g, and the starting distance from the healthy centroid. Regression coefficients are shown as dots, one standard deviation is shown as the thin lines, and two standard deviations as the thick lines.
We next used linear regression to examine the independent effects of reduction in fecal calprotectin, antibiotic exposure and the degree of dysbiosis at baseline on the final state of the microbial community (Figure 4B). Given the different microbial response to the three therapies during the first week, analyses were conducted combined and stratified by treatment. At the end of the study, responders (i.e. those with reduction in FCP) were closer to the centroid of the healthy controls than nonresponders after adjusting for antibiotic use and initial degree of distance from the centroid (p=0.003). Similar patterns were seen for anti-TNF therapy (p=0.06) and EEN (p=0.06). There were too few responders to PEN for meaningful analysis. Analysis at the species level yielded generally similar results, but with a more pronounced difference in the composition of responders and non-responders to EEN after 1 week of therapy (p=0.025), a difference that persisted at week 8 (Figure S4 and S4B).
The resolution of dysbiosis was not complete among responders. Among the anti-TNF-treated patients who were responders, 11 taxa differed in abundance from healthy controls at baseline q<0.05). At the end of follow-up, 9 of these taxa (82%) remained significantly different at a nominal p<0.05 and 6 (55%) were significant at a q<0.05 (Klebsiella, Prevotella, Escherichia, Odoribacter, Enterococcus, and Fusobacterium). Thus, clinical response was associated with evolution of communities toward healthy structure (Figure 4B), consistent with the observation that human DNA levels diminished with successful therapy (q=0.0002; quantile regression), but normal community structure was not usually fully restored (Figure 4A). This may be due to continuing environmental stress from the host inflammatory response as indicated by mildly elevated FCP despite response to treatment. An analysis at the species level yielded generally similar conclusions (Figure S4A and S4B). Fungal colonization was not reduced with successful therapy (p=0.35; quantile regression).
To explore the independent effect of inflammation on the composition of the gut microbiota, we created separate robust quantile (75%) regression models adjusted for antibiotic use and treatment modality with fold-change in abundance of individual taxa as the dependent variable. Response to therapy defined as final FCP<250mcg/g was associated with decrease in Actinomyces (p=0.0002, q=0.01) and increase in Lactococcus (p=0.002, q=0.046) and Roseburia (p=0.006, q=0.084). (Table S1L). Multiple gene pathways were also associated with antibiotic use, treatment, and response (Table S1M).
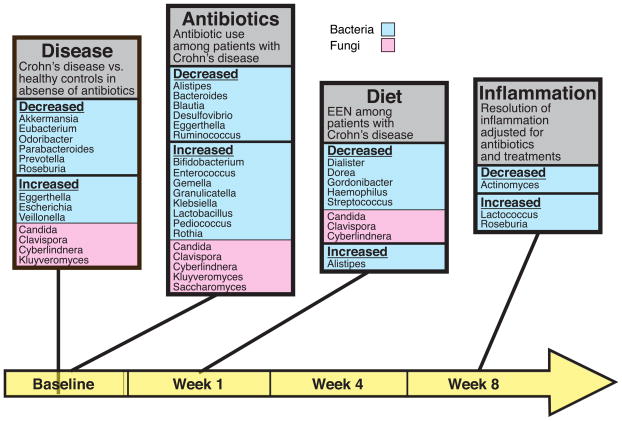
Distinctive responses of genus-level taxa are summarized in Figure 5, which also indicates the time points queried. Specific bacteria lineages showed increases or decreases in abundance associated with 1) health or Crohn’s disease at baseline, 2) antibiotic use in the Crohn’s disease cohort at baseline, 3) diet therapy in the Crohn’s cohort at week 1, and resolution of inflammation (corrected for antibiotic use) in the Crohn’s cohort at week 8.
Figure 5.
Bacterial and fungal genera associated with environmental stressors. Microbial genera are shown that differed in four comparisons: Crohn’s disease versus healthy controls at baseline (“Disease”); antibiotic use at baseline in the Crohn’s disease cohort (“Antibiotics”); diet (EEN) or anti-TNF therapy at week 1 (Diet”); and reduction of inflammation or not at the end of the study at week 8 (“Inflammation”). The time line is shown along the bottom (yellow). Bacterial lineages are shown in light blue, fungal lineages in pink. Data for the diagrams are in Tables S1I–L, and S1 N–Q. Taxa shown were significantly associated after adjustment for multiple comparisons for Crohn’s disease versus healthy controls, for antibiotic use comparisons, and for all fungal comparisons. The bacterial taxa shown for the effect of EEN and resolution of inflammation were significant at a nominal p value<0.05 (i. e. without correction for multiple comparisons).
Distinctive responses were also seen associated with fungal colonization (Figure 5 and Tables S1N–Q). At baseline, Candida, Clavispora, Cyberlindnera, and Kluyveromyces were all enriched in Crohn’s samples versus healthy controls (Table S1N). Comparisons between antibiotic treated versus not treated patients showed enrichment of all these fungi plus Saccharomyces as well (Table S1O). In contrast, after one week of diet therapy, Candida, Clavispora, and Cyberlindnera were all reduced in abundance (Table S1P). Similarly, abundance of Clavispora Cyberlindnera, and Kluyveromyces were significantly reduced from baseline to week 8 only in the EEN treated group (q<0.05) and not the anti-TNF treated group. At week 8, overall response to therapy after adjusting for treatment and antibiotic use was not associated with a change in fungal abundance (Table S1Q).
DISCUSSION
Here we assess the concurrent effects of inflammation, diet, and antibiotic use on the the gut microbiome in pediatric Crohn’s disease. The environmental stresses experienced by Crohn’s disease patients were associated with changes in microbial taxonomy that mostly differed among the stressors studied (Figure 5). We observed that patients with Crohn’s disease can be categorized into two groups defined by the composition of the bacterial populations, paralleling previous studies based on 16S rRNA gene tag sequencing (Frank et al., 2007; Gevers et al., 2014; Tong et al., 2013). We further showed that dysbiosis involved differences in microbial gene representation, increases in fungal representation, and higher levels of human DNA in stool. Antibiotic exposure, which is common in this population, has also been identified as a risk factor for new onset Crohn’s disease (Card et al., 2004; Margolis et al., 2010) and was strongly associated with the dysbiosis observed here. EEN further altered the composition of the gut microbiota, possibly due to elimination of regular table food, despite having a favorable effect on gut inflammation. By our definition, the effect of diet qualifies as dysbiosis, though we note that the changes were distinct from the other stressors studied (Figure 5). Finally, effective therapy with either anti-TNF or EEN reduced, but did not completely eliminate, the dysbiosis present at baseline. Thus our data demonstrate that diet, antibiotics and inflammation each independently influence different components of the microbial community.
We described two clusters of patients based on microbiota composition, though the data can be characterized as either a continuum or a dichotomous grouping. The two clusters were associated with clinical features such as antibiotic use and human DNA, but did not appear to predict response to therapy. Thus, the biologic and clinical significance of the clusters remains to be determined.
There were very few taxa whose abundance was associated with more than one stressor. However, the genus Alistipes was increased with EEN, but diminished by antibiotic treatment, raising the possibility that antibiotics antagonize the beneficial effects of EEN.
The independent effects of antibiotics, diet and inflammation likely reflect different mechanisms. Antibiotics are direct toxins to bacteria and may facilitate outgrowth of fungi. Changes in diet provide novel substrates supporting bacterial growth. Inflammation may select for bacterial taxa able to live in the setting of oxidative stress. The normal oxygen gradient in the colon influences the composition of the gut microbiota with a higher abundance of Proteobacteria in the microaerobic environment of the mucosal surface (Albenberg et al., 2014). Disruption of the epithelium and bleeding due to active Crohn’s disease is expected to lead to greater intraluminal oxygen, which in turn would favor outgrowth of taxa belonging to the Proteobacteria phylum. In addition, some Enterobacteriacaea are known to exploit compounds produced during inflammation as terminal electron acceptors, promoting their outgrowth (Winter and Baumler, 2014).
We observed a rapid change in the composition of the gut microbiota within one week of initiating EEN, similar to that observed in other small studies of defined formula diets (Gerasimidis et al., 2014) and with alterations in whole food diets (David et al., 2014; Wu et al., 2011). However, this study was unique in the ability to compare PEN to EEN. A similar change in the microbiota was not observed in children receiving PEN despite administration of nearly the same amount of enteral formula, suggesting that the exclusion of table foods was the primary determinant in changing the gut microbiota and perhaps mediating the increased effectiveness of EEN (Lee et al., 2015). After 1 week of EEN, the composition of the microbiota differed between ultimate responders and non-responders, suggesting, measures of the microbiota may be used to predict response to therapy.
Genes from the category “biosynthesis of siderophore group nonribosomal peptides” (Figure 2a) were more represented in the more dysbiotic the far cluster. This may be due to the response of the gut microbiota to blood in the gut lumen, as indicated by the association with human DNA in stool. Siderophores scavenge iron from the environment. Iron has been linked to an invasive phenotype in bacteria, thereby promoting inflammation (Nairz et al., 2010). Thus, blood associated with active IBD may deliver iron to the gut microbiota, promoting dysbiosis.
The effect of environmental stress on the gut microbiota extends into the Eukaryotic domain. Others have documented increased richness of fungi in patients with Crohn’s disease using older methods, but have not linked this directly to specific fungi, environmental stressors or linked changes in the bacterial microbiota (Ott et al., 2008; Richard et al., 2015). In a separate study using sequencing of fungal gene tags, we found that Cyberlindnera jadinii (also known as Pichia jadinii or Candida utilis) was proportionally increased in Crohn’s patients (Chehoud et al., 2015), confirming the detection of this lineage here. In this study four of the five fungi detected (Candida, Clavispora, Cyberlindnera, and Kluyveromyces) showed increased representation in Crohn’s disease samples at baseline in the absence of antiboitic use. Independently, antibiotic use was also associated with increased colonization by all five fungi. Diet therapy showed the opposite behavior--after one week, fungal colonization was diminished (Figure 5). Thus increased fungal colonization is associated independently with disease and antibiotic treatment, and diminished with EEN. The reduction with EEN may be a consequence of lower consumption of fungi in food (Saccharomyces and Cyberlindnera are both found in foods), or due to a change in the microbial environment in the gut with EEN, paralleling the change in bacteria. The importance for pathogenesis is unknown, though we note that anti-Saccharomyces antibiodies are used as a biomarker in IBD (Peeters et al., 2001; Prideaux et al., 2012; Quinton et al., 1998), and fungi have shown to exacerbate colitis in mouse models (Iliev et al., 2012).
Cyberlindnera and Candida have been positively associated with bacteria including Streptococcus and Lactobacillus. Positive correlation of Candida and Streptococcus has been reported for oral samples from lung transplant recipients (Bittinger et al., 2014), an interaction proposed in medically important mixed biofilms (Metwalli et al., 2013). Possibly antibiotic exposure promotes formation of a mixed community in the inflamed gut, paralleling studies in mice (Dollive et al., 2013).
The presence of fungi may also have contributed to the metabolic genomic signature associated with IBD (Figures 2a and 2c). Since bacteria do not have phospholipid membranes as in eukaryotic organisms, it may be that the predominance of genes associated with “glycerolipid metabolism” and “glycerol phospholipid metabolism” (Figure 2c) in Crohn’s disease is due to the abundance of fungi in the dysbiotic far cluster. Indeed, with the exception of actinomycetes group, triacylglycerols in bacteria have rarely been described (Alvarez and Steinbuchel, 2002). These same pathways might also be involved in the catabolism of fatty acid lipids through either peroxisomal or mitochondrial beta-oxidation, again, a feature of eukaryotic organisms like yeast (Strijbis and Distel, 2010) and not bacteria.
The bacterial microbiota among responders became more similar to the healthy controls than the microbiota of non-responders with both anti-TNF and EEN. Anti-TNF therapy is administered parenterally and is not expected to alter the gut microbiome directly, indicating that dysbiosis is likely a consequence of inflammation. However, the bacterial dysbiosis was only partially resolved among responders and fungal dysbiosis did not resolve. Perhaps complete resolution would occur with longer follow-up along with a reduction in the stressors to the gut microbiota described here. In turn, complete resolution of bacterial and or fungal dysbiosis might have a favorable impact on Crohn’s disease (Rajca et al., 2014; Samuel et al., 2010).
In conclusion, we document that dysbiosis of Crohn’s disease extends beyond bacteria to include fungi. The dysbiosis results from a combination of inflammation, antibiotic exposure, and dietary changes, each exerting different impacts on the gut microbiota composition. EEN caused further departure of the composition of the gut bacteria from the pattern of healthy controls within one week, likely due to the elimination of table foods, but was effective in reducing inflammation. The extent of dysbiosis diminished with reduction of inflammation by anti-TNF or EEN, consistent with inflammation contributing directly to the dysbiosis. Gene pathway analysis suggested that the gut microbiota responds to gut environmental stressors through the modification of metabolism. Thus, while dysbiosis in general is common to Crohn’s disease, the nature of the dysbiosis is unique to each environmental stressor. Future studies should explore whether these effects interact in ways that influence outcome.
METHODS
Metagenomic analysis
Details on human subjects are reported in (Lee et al., 2015) and in the Supplemental methods. We enrolled children under age 22 with active disease defined as a Pediatric Crohn’s Disease Activity Index (PCDAI) score greater than 10 who were initiating therapy with either EEN, PEN or anti-TNF agent. Therapy was chosen by the attending physician. We excluded children with an ostomy, treatment with probiotics in the prior 2 weeks, receiving more than 50% of daily calories from parenteral nutrition, who used anti-TNF therapy within 8 weeks prior to starting EEN or PEN or who used a EEN or PEN within 1 week of starting anti-TNF therapy. Patients had stool samples collected for determining FCP concentration and microbiome characterization at baseline, 1 week, 4 weeks, and 8 weeks following initiation of the treatment. Reduction in inflammation was assessed with FCP concentration which is correlated with endoscopic findings of mucosal inflammation and decreases following initiation of medications in active Crohn’s disease(Lewis, 2011; Summerton et al., 2002). For this study, we defined response to therapy as a reduction in FCP to ≤250 μg/g among those with baseline FCP >250 μg/g.
Stool samples (n=366) were stored for up to 48 hr on ice, then frozen at −80°C. DNA was purified from each sample using the MoBio PowerSoil kit. Libraries for DNA sequencing were prepared using the TruSeq method, and sequence was acquired using the Illumina HiSeq method (Illumina, San Diego, CA).
Reads were preprocessed based on quality scores using the FASTX suite with default parameters. This yielded 8.7×1011 total bases of sequence. The amount of sequence collected for each sample is summarized in Table S1A. Using BMtagger, we next identified and removed human reads from the quality filtered reads. The bacterial abundance was quantified by MetaPhlAn. We filtered out the samples with non-human reads < 30000 and bacterial genera with max abundance < 1% across all samples or present in <10% of all samples. The distance between samples was measured by the Jaccard index. The pamk function in R package fpc was used to find the optimal clustering of bacterial data. Gene pathway abundance was measured by HUMAnN. We filtered out low abundance gene pathways with max abundance < 0.1% across all samples or present in <10 samples. We used the Bray-Curtis distance to measure the distance between samples for the pathway data. The R package quantreg was used for quantile regression with quantile set to 75%. We used the log fold change of abundance at week 8 over baseline as the response variable and treatments, antibiotic use, response status as the covariates in the quantile regression. To avoid dividing by the zero value or taking log of the zero value, we added the smallest non-zero abundance value to the abundance before calculating the log fold change. All p values in the above analysis were adjusted by the FDR (Benjamini-Hochberg) method for multiple comparisons except where noted.
Human DNA was quantified using ABI Taqman Gene Expression Assay Hs00742533_s1. Instant JChem was used for structure database management, search and prediction, Instant JChem 5.9.4, (2012-02-10), ChemAxon (http://www.chemaxon.com). Viral sequences were detected by BLAST alignment to the NCBI viral database. Archaeal sequences were detected by alignment to the M. smithii genome. Fungal sequences were detected and quality filtered as described in Supplemental procedures: Filtering procedure for analysis of fungal alignments.
Supplementary Material
Highlights.
Inflammation, antibiotics, and diet independently affect microbiota in Crohn’s disease.
Antibiotics are associated with bacterial dysbiosis and increased fungi.
Dysbiosis decreases with reduction in intestinal inflammation.
Diet has an independent and rapid effect on gut microbiota composition.
Acknowledgments
This work was supported by Project UH3DK083981; the Crohn’s and Colitis Foundation of America, Penn Digestive Disease Center (P30 DK050306); The Joint Penn-CHOP Center for Digestive, Liver, and Pancreatic Medicine; The Penn-CHOP Microbiome Program, S10RR024525; UL1RR024134, and K24-DK078228; and the University of Pennsylvania Center for AIDS Research (CFAR) P30 AI 045008.
Footnotes
ACCESSION NUMBERS
All sequence data used in this study is available at the NCBI SRA under SRP057027.
AUTHOR CONTRIBUTIONS
JL, GW and FB designed the study, JL, RB, AO, AG, DL, LA, EG, LN, AB, GW and FB collected samples, JL, CH, KB, RH, AB, GW and FB acquired sequence data, all authors analyzed the data.
Supplemental information includes eight figures, one table (containing 17 subtables), Supplemental Methods, and Supplemental Results, and can be found with this article online at xxx.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources, National Institutes of Health, or Pennsylvania Department of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS computational biology. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063. e1058. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez HM, Steinbuchel A. Triacylglycerols in prokaryotic microorganisms. Appl Microbiol Biotechnol. 2002;60:367–376. doi: 10.1007/s00253-002-1135-0. [DOI] [PubMed] [Google Scholar]
- Bittinger K, Charlson ES, Loy E, Shirley DJ, Haas AR, Laughlin A, Yi Y, Wu GD, Lewis JD, Frank I, et al. Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome biology. 2014;15:487. doi: 10.1186/s13059-014-0487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli O, Cordischi L, Cirulli M, Paganelli M, Labalestra V, Uccini S, Russo PM, Cucchiara S. Polymeric diet alone versus corticosteroids in the treatment of active pediatric Crohn’s disease: a randomized controlled open-label trial. Clin Gastroenterol Hepatol. 2006;4:744–753. doi: 10.1016/j.cgh.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Card T, Logan RF, Rodrigues LC, Wheeler JG. Antibiotic use and the development of Crohn’s disease. Gut. 2004;53:246–250. doi: 10.1136/gut.2003.025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflammatory bowel diseases. 2015 doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, Okamoto J, Snyder TM, Cornfield DN, Nicolls MR, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollive S, Chen YY, Grunberg S, Bittinger K, Hoffmann C, Vandivier L, Cuff C, Lewis JD, Wu GD, Bushman FD. Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PloS one. 2013;8:e71806. doi: 10.1371/journal.pone.0071806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimidis K, Bertz M, Hanske L, Junick J, Biskou O, Aguilera M, Garrick V, Russell RK, Blaut M, McGrogan P, et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn’s disease during enteral nutrition. Inflammatory bowel diseases. 2014;20:861–871. doi: 10.1097/MIB.0000000000000023. [DOI] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell host & microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover Z, Muir R, Lewindon P. Exclusive enteral nutrition induces early clinical, mucosal and transmural remission in paediatric Crohn’s disease. Journal of gastroenterology. 2014;49:638–645. doi: 10.1007/s00535-013-0815-0. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PloS one. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project C. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush NO, Day AS, Leach ST, Lemberg DA, Nielsen S, Mitchell HM. Effect of exclusive enteral nutrition on the microbiota of children with newly diagnosed Crohn’s disease. Clinical and translational gastroenterology. 2015;6:e71. doi: 10.1038/ctg.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan KJ, Ullman TA, Ford AC, Abreu MT, Abadir A, Marshall JK, Talley NJ, Moayyedi P. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. The American journal of gastroenterology. 2011;106:661–673. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]
- Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, Tyler AD, van Sommeren S, Imhann F, Stempak JM, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome medicine. 2014;6:107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Baldassano RN, Otley AR, Albenberg L, Griffiths AM, Compher C, Chen EZ, Li H, Gilroy E, Nessel L, et al. Comparative Effectiveness of Nutritional and Biological Therapy in North American Children with Active Crohn’s Disease. Inflammatory bowel diseases. 2015 doi: 10.1097/MIB.0000000000000426. [DOI] [PubMed] [Google Scholar]
- Lewis JD. The utility of biomarkers in the diagnosis and therapy of inflammatory bowel disease. Gastroenterology. 2011;140:1817–1826. e1812. doi: 10.1053/j.gastro.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JF, Chen JM, Zuo JH, Yu A, Xiao ZJ, Deng FH, Nie B, Jiang B. Meta-analysis: fecal calprotectin for assessment of inflammatory bowel disease activity. Inflammatory bowel diseases. 2014;20:1407–1415. doi: 10.1097/MIB.0000000000000057. [DOI] [PubMed] [Google Scholar]
- Margolis DJ, Fanelli M, Hoffstad O, Lewis JD. Potential association between the oral tetracycline class of antimicrobials used to treat acne and inflammatory bowel disease. The American journal of gastroenterology. 2010;105:2610–2616. doi: 10.1038/ajg.2010.303. [DOI] [PubMed] [Google Scholar]
- Metwalli KH, Khan SA, Krom BP, Jabra-Rizk MA. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS pathogens. 2013;9:e1003616. doi: 10.1371/journal.ppat.1003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalingam NA, Lynch SV. Role of the microbiota in inflammatory bowel diseases. Inflammatory bowel diseases. 2012;18:968–984. doi: 10.1002/ibd.21866. [DOI] [PubMed] [Google Scholar]
- Nairz M, Schroll A, Sonnweber T, Weiss G. The struggle for iron - a metal at the host-pathogen interface. Cell Microbiol. 2010;12:1691–1702. doi: 10.1111/j.1462-5822.2010.01529.x. [DOI] [PubMed] [Google Scholar]
- Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott SJ, Kuhbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scandinavian journal of gastroenterology. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- Peeters M, Joossens S, Vermeire S, Vlietinck R, Bossuyt X, Rutgeerts P. Diagnostic value of anti-Saccharomyces cerevisiae and antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease. The American journal of gastroenterology. 2001;96:730–734. doi: 10.1111/j.1572-0241.2001.03613.x. [DOI] [PubMed] [Google Scholar]
- Prideaux L, De Cruz P, Ng SC, Kamm MA. Serological antibodies in inflammatory bowel disease: a systematic review. Inflammatory bowel diseases. 2012;18:1340–1355. doi: 10.1002/ibd.21903. [DOI] [PubMed] [Google Scholar]
- Quinton JF, Sendid B, Reumaux D, Duthilleul P, Cortot A, Grandbastien B, Charrier G, Targan SR, Colombel JF, Poulain D. Anti-Saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut. 1998;42:788–791. doi: 10.1136/gut.42.6.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajca S, Grondin V, Louis E, Vernier-Massouille G, Grimaud JC, Bouhnik Y, Laharie D, Dupas JL, Pillant H, Picon L, et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflammatory bowel diseases. 2014;20:978–986. doi: 10.1097/MIB.0000000000000036. [DOI] [PubMed] [Google Scholar]
- Richard ML, Lamas B, Liguori G, Hoffmann TW, Sokol H. Gut fungal microbiota: the yin and yang of inflammatory bowel disease. Inflammatory bowel diseases. 2015;21:656–665. doi: 10.1097/MIB.0000000000000261. [DOI] [PubMed] [Google Scholar]
- Rutgeerts P, Van Assche G, Sandborn WJ, Wolf DC, Geboes K, Colombel JF, Reinisch W, Kumar A, Lazar A, Camez A, et al. Adalimumab induces and maintains mucosal healing in patients with Crohn’s disease: data from the EXTEND trial. Gastroenterology. 2012;142:1102–1111. e1102. doi: 10.1053/j.gastro.2012.01.035. [DOI] [PubMed] [Google Scholar]
- Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, Latreille P, Kim K, Wilson RK, Gordon JI. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel S, Loftus EV, Jr, Sandborn WJ. The effects of itraconazole on inflammatory bowel disease activity in patients treated for histoplasmosis. Alimentary pharmacology & therapeutics. 2010;32:1207–1209. doi: 10.1111/j.1365-2036.2010.04444.x. [DOI] [PubMed] [Google Scholar]
- Sartor RB. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- Sartor RB. Therapeutic correction of bacterial dysbiosis discovered by molecular techniques. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16413–16414. doi: 10.1073/pnas.0809363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nature methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijbis K, Distel B. Intracellular acetyl unit transport in fungal carbon metabolism. Eukaryot Cell. 2010;9:1809–1815. doi: 10.1128/EC.00172-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton CB, Longlands MG, Wiener K, Shreeve DR. Faecal calprotectin: a marker of inflammation throughout the intestinal tract. Eur J Gastroenterol Hepatol. 2002;14:841–845. doi: 10.1097/00042737-200208000-00005. [DOI] [PubMed] [Google Scholar]
- Tong M, Li X, Wegener Parfrey L, Roth B, Ippoliti A, Wei B, Borneman J, McGovern DP, Frank DN, Li E, et al. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PloS one. 2013;8:e80702. doi: 10.1371/journal.pone.0080702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Baumler AJ. Dysbiosis in the inflamed intestine: chance favors the prepared microbe. Gut microbes. 2014;5:71–73. doi: 10.4161/gmic.27129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, Haas AR, Abbas A, Frye L, Christie JD, et al. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2015;15:200–209. doi: 10.1111/ajt.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.