Abstract
The field of electron tomography has benefited greatly from manual and semi-automated approaches to marker-based tilt-series alignment that have allowed for the structural determination of multitudes of in situ cellular structures as well as macromolecular structures of individual protein complexes. The emergence of complementary metal-oxide semiconductor detectors capable of detecting individual electrons has enabled the collection of low dose, high contrast images, opening the door for reliable correlation-based tilt-series alignment. Here we present a set of automated, correlation-based tilt-series alignment, contrast transfer function (CTF) correction, and reconstruction workflows for use in conjunction with the Appion/Leginon package that are primarily targeted at automating structure determination with cryogenic electron microscopy.
Keywords: Cryo-electron tomography, Marker-free tilt-series alignment, Single particle tomography, Automated processing, Transmission electron microscopy, Protomo
1. Introduction
With the recent advent of direct electron detectors, cryo-electron microscopy (cryo-EM) has risen in popularity as a technique for high resolution, near-native structural biology determination. The in situ 3D structural determination method of cryo-electron tomography (cryo-ET) is the next frontier for high-throughput, high-resolution structure determination by EM. Electron tomography, particularly cryo-ET, has been the method of choice for determining cellular structure at resolutions up to 2 nm for over a decade (Zhang, 2013). The increase in computational resources over the same period of time has prompted the emergence of single particle tomography (SPT) as a popular method of quaternary, tertiary, and even secondary structure determination, with subvolume averaging and classification allowing for classification of specimens with substantial heterogeneity in topology and molecular structure (Briggs, 2013; Fernández, 2012). In recent years software suites have been developed to accommodate large SPT datasets using high performance CPU and GPU algorithms (Castaño-Díez et al., 2012; Galaz-Montoya et al., 2015, p. 2). Currently two research labs have used SPT to resolve highly symmetric and pseudo-symmetric protein complexes at resolutions better than one nanometer (Schur et al., 2015, p. 8, 2013; Tran et al., 2012).
At SPT resolutions near 1 nm, the primary resolution limits are the accuracy of tilt-series alignment, the accuracy of defocus estimation for CTF correction (Fernández et al., 2006; Xiong et al., 2009), the beam-induced motion of objects on the grid (Li et al., 2013; Scheres, 2014), and the signal to noise ratio (SNR) of the specimen (Lenz, 1954; Spence, 2013), which is dictated by the cumulative dose of a tilt-series (Grant and Grigorieff, 2015; Spear et al., 2015 this issue)). The movie mode features and improved detector quantum efficiency of direct electron cameras greatly increases the SNR of low dose images relative to CCD cameras (McMullan et al., 2014; Ruskin et al., 2013), allowing for lower-dose tilt-series to be collected. Compared to CCDs, lower-dose tilt-series collected with direct electron cameras preserve higher resolution specimen features and allow for measurable Thon rings in low tilt images. With this increase in SNR, per-image defocus estimation becomes feasible and correlation-based tilt-series alignment becomes more accurate. In comparison to tilt-series alignment with bead tracking, alignment by specimen correlation is robust against movement of beads during tilt-series collection.
1.1. Advantages of correlation-based tilt-series alignment
The most commonly used method for tilt-series alignment, bead tracking (Amat et al., 2008; Bernard Heymann et al., 2008; Kremer et al., 1996; Nickell et al., 2005), relies on the beads remaining stationary throughout the entire tilt-series exposure. If during imaging any of the tracked beads move relative to the specimen or to each other, then the calculated geometric relationships between tilt images may be inaccurate up to the beam-induced motion of the beads. We have observed beam-induced motion of gold beads in high tilt angle images at low dose and up to 10 nm at high dose (Fig. 1A,B). While this beam-induced bead motion can be almost completely corrected for within each image by frame aligning using direct electron cameras, the motion between tilt images is left uncorrected in conventional bead-tracking alignment workflows. For tilt-series that exhibit significant beam-induced bead motion and where the CTF can be determined, the resolution limit for bead tracking-based alignments is on the order of the relative bead-to-specimen movement between tilt images plus the tilt image alignment error. It is possible to align tilt-series that have been acquired with beads that show beam-induced motion by using correlation-based alignment methods. A potential solution with Protomo is to adjust the alignment search area so as to exclude aligning on fiducials.
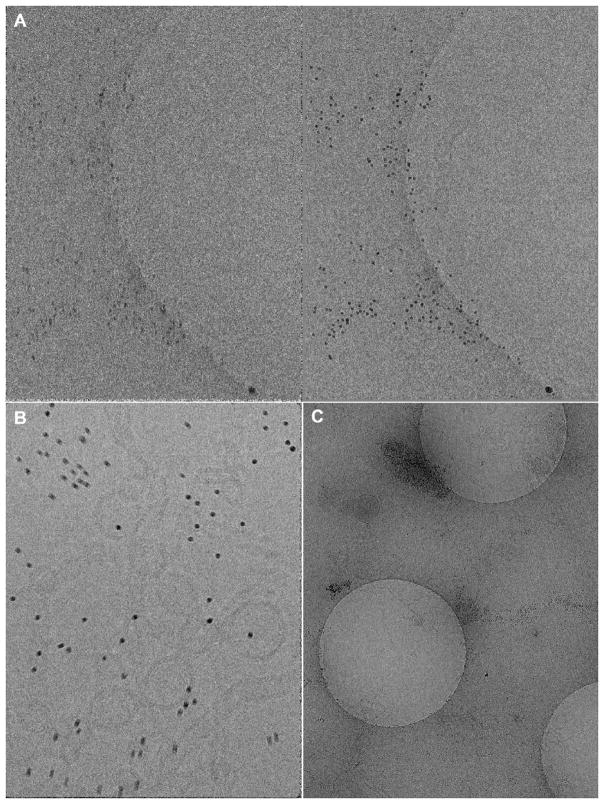
Fig. 1.
Beam-induced bead motion and bead aggregation. (A) A tilt-series exposure taken at a tilt angle of 51 degrees with a dose of 2.34 e−/Å2 after a cumulative dose of 60 e−/Å2, where (left) is before frame alignment while (right) is after. Some beam-induced beam motion can be observed. (B) A zero degree, frame uncorrected tilt exposure with a dose of 57.5 e−/Å2. Clear anisotropic beam-induced bead motion on the order of 1 to 10 nanometers can be observed. (C) Hole exposure showing nearly complete gold bead aggregation outside of holes. The central hole is (A). Gold colloidal beads are 10nm in diameter in all images.
In addition to the theoretical resolution limits of marker-based tilt-series alignment described above, there are scenarios where specimen preparation with colloidal gold beads is non-ideal or impossible. In some cases gold beads aggregate in grid holes, requiring significant manual identification during bead tracking. In other instances gold beads aggregate outside of grid holes, effectively eliminating their intended function (Fig 1C). Calibrating the concentration of gold beads can require significant efforts during preparation and does not always guarantee a uniform distribution. Additionally, reconstruction with fiducial markers can introduce artifacts in neighboring densities (Frank et al., 1987).
To circumvent the limitations imposed by bead-tracking for non-ideal samples, whether for SPT or non-SPT studies such as structural determination in cells, one must align tilt-series using a non-marker based alignment procedure. To these ends, we introduce an implementation of the correlation-based tilt-series software Protomo (Winkler and Taylor, 2006) from within the Appion package (Lander et al., 2009) with the goal of providing a standardized, high-throughput alternative to existing marker-based alignment methods.
The software describes herein can be used in conjunction with the Leginon tilt-series acquisition application (Suloway et al., 2009). The workflows provided in this paper allow for accurate, robust tilt-series alignment while broadening the types of samples that can be imaged by cryo-ET.
2. Workflow
The Appion-Protomo implementation is designed using the already existent Appion API including the Python backbone, MySQL interface, and PHP based web tools. Together, these provide an interface between all microscope images in the database and arrays of processing suites. The workflows presented here are accessible from the main Appion image processing webpage for any tilt-series in the database.
The general philosophy for aligning a set of tilt-series from a data collection session is to first align a single representative tilt-series from the session, then use the same parameters to align each tilt-series in the session in parallel. Protomo requires the user to provide an extensive parameter file to define the alignment parameters for a given tilt-series alignment iteration. We have automated this process. All file generation, database queries, and file management operations are controlled by the provided scripts as the user works through the four main Appion-Protomo workflows: File Preparation, Coarse Alignment, Refinement, and Reconstruction. Once the user identifies working alignment and reconstruction parameters, those parameter files can then be used to align and reconstruct each tilt-series in the session though the modular batch workflows.
2.1. Alignment quality metric
The Protomo refinement workflow is iterative, with each successive iteration’s geometry model and image transformations being used as starting values. The goal of a correlation-based tilt-series alignment workflow is to iteratively converge on a set of geometric transformations, including a model for the tilt azimuth, tilt elevation, and/or stage orientation together with shifts rotation, and/or scaling of each image. In addition to supplying new geometric transformations for a completed iteration, Protomo records residuals for the applied image transformations (Winkler and Taylor, 2006). In particular, residuals for transformations in the x direction and in the y direction, a residual for rotation, and a residual for scaling for each image in a tilt-series are provided. The residuals describe the degree to which an individual image must be distorted in order to match the model. As such, it is influenced by the accumulated dose between images, the accuracy of alignment between images, and the motion of the correlated objects, modulated by the filters applied before alignment. An optimally aligned tilt-series will have the majority of the x and y residuals below 1%, scaling residuals below 1%, and rotation residuals below 1°; i.e. the amount of additional correction to the transformation between the tilt image and the reference projection image required to produce perfectly correlated images should be below 1% for translations and scaling and below 1° for rotations.
To assist the user in assessing the convergence of a tilt-series alignment, a metric has been defined based on the residuals and their standard deviations:
where CCMS stands for Combined Correction Mean plus Standard deviation and X are the image correction factors for shifts, rotations, and scaling. To put the rotation CCMS value on equal footing with the shift and scaling CCMS values, one degree of CCMS(rotations) is scaled to 2% in CCMS(shifts/scalings) so that a single scaled sum CCMS value can be outputted for each iteration by Appion-Protomo. The goal of an Appion-Protomo alignment run then becomes minimizing each CCMS value to below 2.0%, ideally below 1.5%, while reaching convergence in the geometry model refinements at image samplings of 1 or 2. Fig. 2 shows an example plot of the CCMS(shifts) vs. iteration number together with plots of the correction factors in x and y for two iterations.
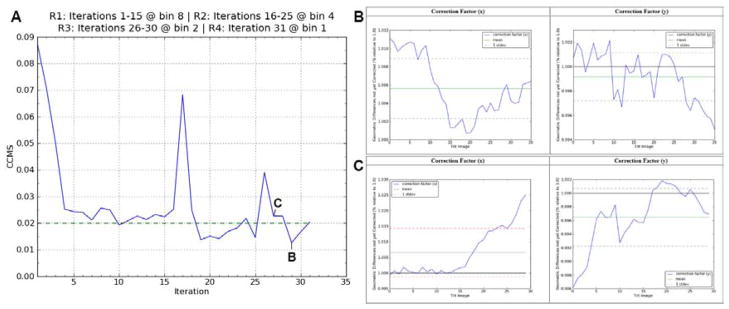
Fig. 2.
Quality assessment plots. (A) A plot of the CCMS(shift) values vs. iteration number for a real tilt-series. CCMS plots include a convergence baseline (dotted). The spikes in the CCMS plots happen after switching from higher to lower image sampling. This often causes the subsequent one or two iterations to over-estimate the shift, rotation and/or scaling values while the angle refinements are converging to their new and more accurate estimates. (B) Plots of the x and y correction factors for iteration 29 in (A) together with their mean and standard deviations. This tilt-series with a CCMS value of 1.26% is well aligned by the criteria displayed here. (C) Plots of the x and y correction factors for iteration 27 for comparison. Notice that the high tilt angle images on the right of the Correction Factor (x) plot are well above 1%, causing the standard deviation to produce an unacceptable CCMS(shift) value of 2.27%. Correction factor plots include color-coded mean and standard deviation lines (solid and dotted lines, respectively) where the color represents whether the value is above or below pre-defined convergence values (red or green, respectively). Tilt-series and reconstruction videos for iterations (B) and (C) are shown in Movies S1 and S2, respectively.
The CCMS is an objective metric for evaluating whether a tilt-series alignment has converged and so is used throughout this workflow as a simple way to assess the quality of alignments. The user should note that the CCMS values are dependent on image preprocessing parameters performed before the alignment. An overly sampled or lowpass filtered image set may produce artificially low CCMS values.
2.2. Semi-automated individual tilt-series processing
To run a Protomo alignment manually, a parameter file and a tilt angle file need to be supplied by the user. These files specify the image sampling, preprocessing parameters, mask, bandpass filters, correlation peak search options, geometry options, reconstruction options, directory paths, raw image paths, image tilt angles, and stage orientation. We automated the generation of the required files by providing an alignment webpage in the Appion user interface that includes default, data-based presets for all required parameters along with detailed help pop-up windows for each parameter (Movie S3). Thus, file generation and file management are performed automatically, which standardizes the alignment process and avoids error-prone manual intervention.
The first step of any Appion-Protomo run is File Preparation where the Leginon/Appion database is queried to retrieve raw tilt images, and metadata is queried to construct a Protomo tilt file. In the individual tilt-series alignment workflow, file preparation is performed seamlessly in conjunction with the second step, Coarse Alignment. The Coarse Alignment step takes advantage of the new Protomo grid search parameters, available in Protomo 2.4.X or greater, to estimate initial rotation and translation transforms for each image, eliminating the need for the user to manually align the images before refinement. Once the user has run Coarse Alignment and has confirmed on the subsequent summary webpage that the results are not divergent, then a full Refinement is ready to be run.
The next step in a typical tilt-series processing session is to proceed to Refinement, in which area matching is used to refine image alignments and the model for the tilt geometry. In our tests we have found that in order to minimize the chance of there being a divergent tilt-series alignment, refinement is best performed iteratively while gradually increasing the sampling rate. For this reason, the Refinement step introduces rounds to the alignment workflow. In a typical refinement, a user would refine for multiple iterations and look for CCMS values below 2%. If CCMS values do not reach this threshold, users might adjust the lowpass filter, the estimated specimen thickness, or other critical Protomo parameters. The Appion-Protomo web pages provide a convenient interface in which to vary these parameters and monitor the results. For each round the user can change the sampling factor and the number of iterations, along with all variable Protomo parameters. Once the user has identified a well-aligned iteration, a reconstruction can be made (Movie S3).
The Reconstruction step allows the user to create a weighted back-projection reconstruction. The CCMS values provide a convenient way for determining which set of parameters yield the best alignment. The Appion-Protomo web pages allow the user to reconstruct from any iteration and with or without filtering. A lowpass filter is recommended for visualization, segmentation, or non-SPT purposes (Movie S3).
Two preemptive and real-time exception-catching algorithms for circumventing the most common Protomo errors have been implemented. During File Preparation, overly shifted raw images at high angles due to poor estimation of image shift between tilts are removed from consideration for processing. During alignments, if the resampled search area is too large causing the search to be out of bounds, the search area will be automatically reduced until alignment can proceed.
At each step in the workflow the user is presented with the vast majority of Protomo parameters along with file management and depiction options for the summary webpages. All required parameters are filled in with default values based on information in the database. After each Refinement iteration a CCMS plot is made, native Protomo correction factor plots are made, and a correlation peak video is made to show how well each image aligned to the respective projection image. For each iteration the user can optionally generate an aligned tilt-series video and a z slice-through video of a preliminary reconstruction (Movies S1, S2, and S3). Depictions are presented on Appion summary webpages available after each alignment step. All videos are available for download in mp4 format for presentation purposes.
2.3. Batch tilt-series processing
Once a single representative tilt-series has been processed to or near convergence, then the user can proceed to the Batch Tilt-Series Alignment webpage. We have found that in most cases, once appropriate Protomo parameters have been found, they will work well for other tilt-series in the same session. The Batch Tilt-Series Alignment workflows allow the user to process in parallel any number of tilt-series in a given session sequentially through a set of File Preparation, Coarse Alignment, Refinement, Reconstruction, and/or CTF Correction steps. The batch workflows depend on Protomo parameter files being formatted identically to those created during the individual tilt-series processing. In the case where Appion-Protomo was used for an individual tilt-series alignment, the parameter files will have already been populated with appropriate parameters and be in the correct format.
After a batch tilt-series alignment, a batch summary webpage will be available for the user to quickly analyze the alignment of each tilt-series processed. The user is first presented with a grid of CCMS plots for each tilt-series. Upon clicking on one of the CCMS plots, the user is taken to an individual refinement iteration webpage formatted similarly to the Refinement summary page in the individual tilt-series processing workflow. The use of CCMS plots allows for an immediate assessment of tilt-series alignment quality while the nested layout of the webpages allows the user to easily navigate through potentially thousands of tilt-series refinement iterations (Movie S4).
2.4. CTF correction
To facilitate CTF correction, the ctfphaseflip program in IMOD (Xiong et al., 2009) has been wrapped into the Appion-Protomo workflows to optionally correct for CTF at any point in tilt-series processing. Provided that the defocus has been accurately estimated within Appion for the majority of the images in a tilt-series, it is recommended that CTF correction be performed before alignment unless doing so appreciably reduces the contrast of the images or unless the tilt azimuth is significantly refined during alignment, because the CTF correction strips run parallel to the calculated tilt axis. The function implemented here requires that the user has previously run image defocus estimation on each image in a given tilt-series through the Appion CTF estimation interface such that the defocus and defocus confidence values have been inserted into the database. The CTF correction algorithm first removes the low confidence defocus values, then removes defocus values that exceed one standard deviation from the tilt-series defocus mean, and finally replaces those values with linearly interpolated values between the remaining high confidence defocus values in the tilt-series (Fig. 3). Confident defocus values are those where the modeled CTF correlates better than 50% with the observed noise and envelope-subtracted power spectrum. This has been standardized in Appion across supported defocus estimation packages (Sheth et al., 2015).
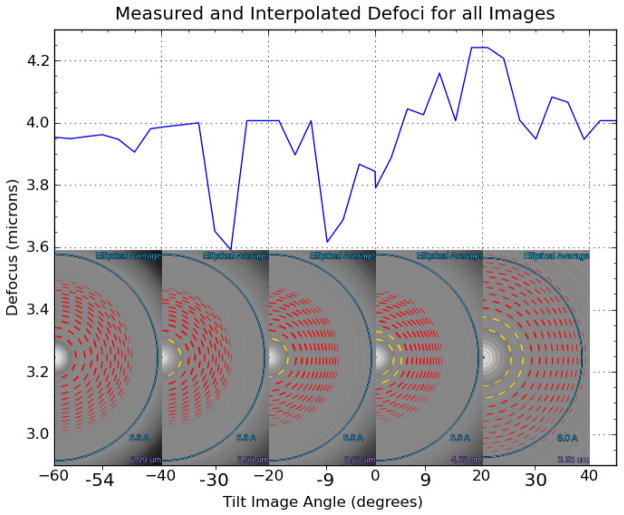
Fig. 3.
Defocus estimation and interpolation for the tilt-series in Fig. 2. The inset images show the estimated CTF curves and defocus values for a select number of tilt angle images, where yellow inner rings indicate a confidently estimated defocus value. Images for which the defocus could not be confidently estimated (e.g. inset, left) have their defocus values interpolated based on the confidently estimated defocus values over the entire tilt-series (blue line). Defocus values were estimated using ACE (Mallick et al., 2005).
2.5. Fully automated batch processing
The workflows described above allow for a fine-grained approach to getting the best alignments possible for all tilt series. However, in some cases such an exhaustive approach may not be necessary, and instead a standard refinement scheme can be used. A Fully Automated batch tilt-series alignment option is available which utilizes a sampling limited, highly iterative approach driven by the quality assessment statistics from each set of iterations. The Fully Automated option provides the user with a restricted set of Coarse Alignment, Refinement, Reconstruction, and CTF Correction parameters alongside additional automation and convergence parameters. By default, this option will sequentially search image geometry phase space for five rounds at a binning of 8, 6, 4, 2, and 1 for 20 iterations each, or until convergence is reached. If convergence is not reached, tilt axis elevation and scaling factors will be included in another set of 20 iteration rounds at a binning of 4, 2, and 1, using the best geometry values from the previous 40 iterations. Once convergence is reached at a binning of 2 or 1, five additional iterations will be performed at a binning of 1 and the best iteration will be reconstructed. The default convergence criteria is CCMS(X) <= 0.015. In this Fully Automated mode, depiction videos are only produced for reconstructed iterations.
The Fully Automated option is designed to make it simple for any user to perform exhaustive tilt-series geometry searches with no user intervention from microscope to tomogram. Aside from the high-throughput capabilities, the benefit of a fully automated approach founded atop an objective alignment metric is the accessibility of a reliable tilt-series alignment algorithm for novice users.
2.6. Screening mode
To facilitate microscope parameter determination prior to a data collection session using Leginon, an option has been included to screen tilt-series in near real-time. Screening Mode constantly waits for tilt-series N+1 to appear in the Leginon/Appion database before processing tilt-series N. Tilt-series are processed automatically through the File Preparation and Coarse Alignment steps, then depiction videos are produced in parallel. Only Coarse Alignment is performed so that processing can keep up with data collection. In our tests we found that the processing time required for screening a single tilt-series is roughly equivalent to the acquisition time for a tilt-series. Thus in Screening Mode a user can view a coarse alignment of tilt-series N as tilt-series N+1 finishes collecting. We envision that this mode will allow for more accurate empirical determination of optimal defocus values, dose per image, angular range, and angular increment, thereby increasing the likelihood of collecting alignable tilt-series.
3.1. Case study #1: AAV-DJ tilt-series session
To assess the throughput of the Appion-Protomo alignment algorithms described above, a tilt-series session of the 25 nm-diameter, icosahedrally symmetric viral capsid, AAV-DJ, was collected and processed.
3.1.1. Specimen preparation and data collection
Virus-like particles of AAV-DJ were expressed in insect cells from a baculovirus construct as previously described (Lerch et al., 2012). Empty capsids were purified as before using three rounds of CsCl density gradient ultracentrifugation, followed by heparin affinity chromatography, eluting with a NaCl gradient. Capsids were then diluted in 50 mM Hepes, 25 mM MgCl2, 25 mM NaCl, pH=7.4.
10 nm colloidal gold beads were diluted in buffer at a 1:10 ratio, then resuspended with the sample. 3 μl of sample was pipetted onto Quantifoil R2/2 200 mesh grids that had been glow discharged for 5 s with a Gatan Solarus plasma cleaner. An FEI Vitrobot Mark IV was used to blot grids for 3 s at 4 °C and 100% humidity before plunge freezing into liquid ethane. Data acquisition was performed automatically using Leginon on an FEI Titan Krios at 300 keV at a nominal magnification of 18,000x. Images were collected on a Direct Electron DE-20 in movie mode with a pixel size of 2.03 Å. Tilt-series were collected from 0° to 45°, then 0° to −60° at 3° increments. Electron dose scaled with the cosine of the tilt angle. Each exposure image consisted of 5 to 14 frames collected every 100 ms. Of the 42 tilt-series reported on here, two were collected with a nominal underfocus of 3.5 μm and a total dose of 79 e−/Å2, three at 5 μm and 79 e−/Å2, 22 at 5 μm and 58 e−/Å2, and 15 at 4 μm and 70–100 e−/Å2.
3.1.2 Results
To demonstrate the ease of use and performance of the Appion-Protomo individual tilt-series workflow as described in section 2.2., a single tilt-series was processed using only the default settings on the Appion alignment webpages as shown in Movie S3. The alignment reached convergence at iteration 29 after gradually increasing the sampling rate as shown in Fig. 2. The alignment took a total of nine hours on a single multicore workstation, with the depiction video generation consuming roughly half of that time. The resulting tilt-series alignment and reconstruction can be seen in Movies S1 and S2, respectively.
From this tilt-series a total of 229 virus particles were manually extracted from along the tilt axis. The particles were aligned and averaged using the automated SPT processing module in EMAN2 (Galaz-Montoya et al., 2015, p. 2). Figure 4 shows the resulting average alongside a 30 Å lowpass filtered map of AAV-DJ from Lerch, 2012. The resolution of this reconstruction is visually no better than 30 Å, due in large part to the low number of particles used. Though only 229 particles were used in this reconstruction, we estimate that >20,000 particles are present in the full dataset. Efforts to process the full SPT dataset together through subvolume averaging and classification are ongoing.
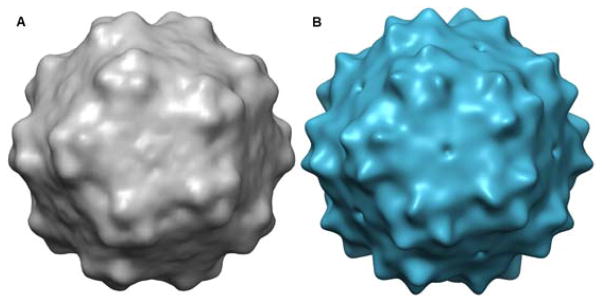
Fig. 4.
AAV-DJ reconstructions. (A) SPT reconstruction of AAV-DJ from 229 particles from a central tilt axis slice of a single tilt-series. (B) Single particle cryo-EM reconstruction of AAV-DJ lowpass filtered to 30 Å. UCSF Chimera was used for visualization (Pettersen et al., 2004).
The 42 tilt-series were processed in parallel using the Fully Automated workflow with default settings and parameter files from the individual tilt-series processing workflow described above. Tilt-series were aligned for between 60 to 160 iterations over the course of two days while gradually increasing the sampling rate. Three-quarters of the images in the dataset had confidently estimated defocus values. CTF correction was not performed before tilt-series alignment due to a decrease in contrast, and therefore alignability, of the CTF corrected images. The total processing time per tilt-series was between 24 and 48 hours. Of the 42 tilt-series processed, 21 aligned to convergence while about one-third aligned to very near convergence as described in section 2.1.
3.2. Case study #2: ETomo tutorial dataset
To provide a comparison between the correlation-based alignment method used in Protomo as implemented in Appion-Protomo with the widely used marker-based alignment method used in eTomo, we have chosen to apply the Appion-Protomo implementation to the freely available eTomo tutorial dataset (O’Toole, 2003). The double tilt-series is of a cross-section of the Chlamydomonas basal body with fiducials added roughly uniformly to the top and bottom and recorded with a pixelsize of 20.2 Å. The dataset contains raw tilt images for each axis together with fiducially aligned models for each axis. For this largely qualitative comparison we chose the first axis to perform an Appion-Protomo alignment.
3.2.1. Results
The tilt-series was left unbinned during alignment due to its relatively small size (61 images at 512×512) and was aligned for 75 iterations through 5 rounds of refinement parameters following Coarse Alignment. In each round the lowpass filters were lowered to include increasingly more information in the alignment (Fig. 5, left). Tilt axis elevation refinement was turned on at iteration 15, magnification scaling was turned on at iteration 25, tilt azimuth refinement and stage orientation was turned off at iteration 55, and the correlation mode was switched from mutual correlation to phase-only correlation at iteration 56 (Fig. 5). The alignment was automated and took less than five hours on a single multicore workstation with approximately half of that time used to create preliminary reconstructions and depiction videos.
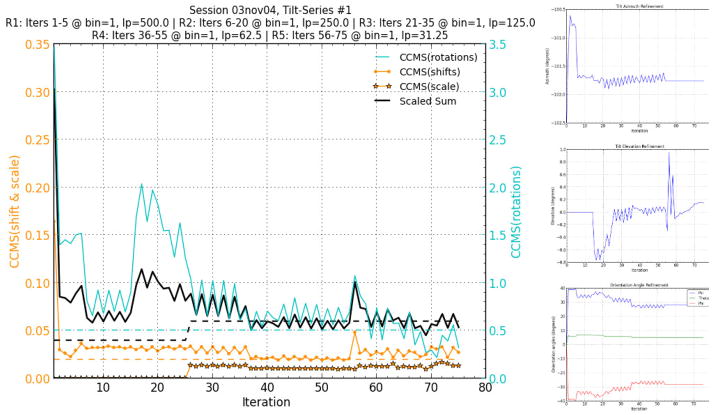
Fig. 5.
Full CCMS plot and geometry model refinement plots for the Protomo alignment of the first axis of the eTomo tutorial tilt-series. The CCMS values and their scaled sum for each iteration are shown together with recommended values as dotted lines (left). Lowpass (lp) units are angstroms. Angle refinement plots for the tilt azimuth, tilt elevation, and stage orientation are shown with convergence in later iterations (right).
As seen in Figure 5, the scaled sum of the CCMS values reached a minimum at iteration 69 while the angle refinements reached convergence. The search area chosen for Protomo refinement by area matching intentionally excluded alignment to the fiducials present on the top and bottom of the section. In this way, we were able to compare fiducial-less alignment with fiducial based alignment on the same tilt-series data. It is noted that excluding the fiducials from each back-projection alignment image actually hinders the alignment in correlation-based methods due to there still being fiducials in the respective tilt image to which the corresponding back-projection image is aligned. One potential benefit of this alignment technique is that the specimen, in this case the basal body cross-section, is being used for alignment instead of the potentially mobile fiducials. The resulting Protomo reconstruction of iteration 69 exhibits the same resolvable information as the eTomo fiducial-based alignment and reconstruction (Fig. 6, Movie S5). It is important to note that the orientation of the Protomo reconstruction volume presented in Figure 6 and Movie S5 is representative of the orientation of the imaged specimen on the grid. Protomo does not by default orient the normal of the grid parallel to the normal of the reconstruction volume as this would introduce an additional interpolation.
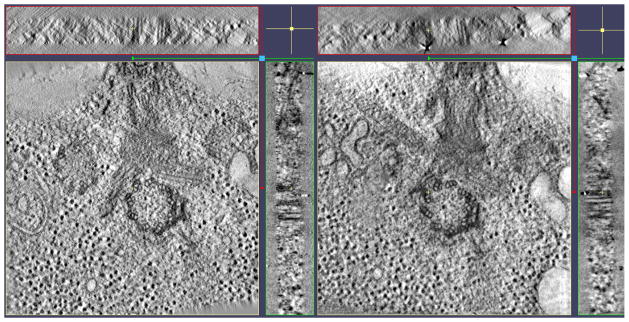
Fig. 6.
Similarly located x, y, and z slice-throughs of the eTomo fiducial alignment (left) and the Protomo non-fiducial correlation-based alignment (right). The resolution and level of detail in each reconstruction is comparable. Note that the grid of this particular sample was tilted and/or bent causing the slab in the reconstruction to be positioned at slight angles as seen in the x and y slice-throughs on the right. 3dmod was used for visualization (Kremer et al., 1996).
4. Discussion
The workflows presented here reduce the required technical knowledge of the user significantly and reduce the time from the microscope to a well-aligned tilt-series using correlation-based alignment by an order of magnitude. Prior to the development of Appion-Protomo, a user without knowledge of Protomo syntax, file management, and reliable alignment parameter determination would spend an inordinate amount of time 1) creating and parsing Protomo tilt and parameter files, 2) editing parameter files in search of working values, 3) diagnosing Protomo error messages, 4) manually starting new refinement iterations, 5) manually restarting failed refinement iterations with different parameters, 6) converting parameters from real-space values to Protomo values, 7) viewing a tilt-series alignment for a given iteration, 8) creating and viewing tomogram reconstructions on a per-iteration basis, 9) viewing and interpreting raw alignment statistics, 10) determining well aligned iterations, and 11) determining whether an alignment has converged. These problems have all been solved in the Appion-Protomo implementation, significantly reducing the amount of time required for an average user to produce well aligned tilt-series from a data collection session (Table 1).
Table 1.
Estimated time required for a novice Protomo user to produce a well aligned tilt-series using Protomo manually (middle) and using the Appion-Protomo implementation (right). The time required to process additional tilt-series is limited by the user’s increase in expertise while using Protomo manually, whereas the time required for the Appion-Protomo implementation scales only with the number of processors available to perform alignments.
| Task | User Time Required Manually using Protomo | User Time Required using Appion-Protomo |
|---|---|---|
| Creating Protomo files | Minutes to hours | None |
| Optimizing Protomo files/parameters | Days to months | Hours to days |
| Diagnosing Protomo errors | Minutes to months | None |
| Manually refining each iteration | Hours to days | None |
| Manually restarting failed iterations | Hours to days | None |
| Parameter unit conversion | Minutes to hours | None |
| Viewing refined tilt-series per iteration | Minutes | None |
| Creating and viewing tomograms per iteration | Minutes to hours | None |
| Viewing and interpreting alignment statistics | Minutes to hours | None |
| Determining well aligned iterations | Minutes to hours | None |
| Determining alignment convergence | Hours to days | None |
| Processing time | Hours to days | Hours to days |
| Total time required | Days to months | Hours to days |
We have shown that with the current implementation, the amount of time and effort required to confidently align tilt-series by cross correlation methods using Protomo is comparable to current semi-automated marker-based alignment methods. There is, however, still potential for future improvement. The Appion-Protomo alignment and reconstruction scripts currently work on individual multiprocessor workstations, yet are fully capable of being run on high performance computing clusters. Additionally, while Protomo is fully capable of processing dual-axis tilt-series (Winkler and Taylor, 2013), the potential of implementing fully automated dual-axis tilt-series alignment in Appion has yet to be realized, yet is possible. Other potential improvements include uploading tilt-series alignment runs to the database, the full support of raw image formats in addition to the mrc file format (Cheng et al., 2015), and the implementation of automated 3D particle picking from tomograms into the Appion infrastructure.
With the significant increases in performance, robustness, and ease of use of correlation-based tilt-series alignment described here, we hope that the workflows provided will increase the scope of resolvable specimens and the accessibility of cryo-ET.
5. Software
The Appion-Protomo scripts, which include the web interface files and the Python scripts, are available for free in the Appion package (release version 3.2+) which can be downloaded from http://nramm.nysbc.org/downloads/.
Protomo version 2.4.1 was used in the development of this software, which relies on the newly introduced command line grid search parameters for Coarse Alignment. This Appion-Protomo implementation is intended to be used with tilt-series images and metadata that has been inserted into the Leginon/Appion database. It is recommended that tilt-series be automatically collected using the Leginon acquisition software before using Appion-Protomo. In future Appion releases it will be possible to upload tilt-series from the Appion web interface. File Preparation and CTF Correction steps require access to the database.
5.1. Dependencies
Python 2.6+ is required to run the generated commands. Protomo 2.4+ is required for coarse alignments, refinements, and back projections. I3 is required for processing reconstructions. IMOD (Kremer et al., 1996) is required for CTF correction. EMAN2 (Tang et al., 2007, p. 2) is required for post-processing reconstructions. Matplotlib and PyLab are required for generation of static plots. ImageMagick is required for depiction videos. FFmpeg is required for HTML5 video output. NumPy, SciPy, and the Python Imaging Library are used throughout. Many of these packages are also required by Appion and are installed when Appion and Leginon are installed.
Supplementary Material
Example tilt-series videos of the example AAV-DJ tilt-series from the case study from the iterations shown in Fig. 2. Left is iteration 27, right is iteration 29. It is important to note the quality of the alignment even though there is a considerable amount of ice contamination in the images. Scale bar, tilt angle, and tilt image number are included on the bottom. Each video was automatically generated after each refinement and was downloaded using the ‘Download Video’ link seen in Movie S3. The images are binned by 8.
Example slice-through videos of preliminary reconstructions of the tilt-series seen in Movie S1. Left is iteration 27, right is iteration 29. Scale bar and z-slice are included on the bottom. Each video was automatically generated after each refinement and was downloaded using the ‘Download Video’ link seen in Movie S3. The differences in brightness is caused by the per-image normalization during video processing due to the high contrast gold beads. The reconstruction is unfiltered and binned by 8.
An overview of the Appion-Protomo individual tilt-series workflow and webpage layouts. Information for every option is documented from within the webpages. The user is guided between webpages at each step in the workflow.
An overview of the Appion-Protomo batch tilt-series webpage layouts. Information for every option is documented from within the webpages. The user is guided between webpages at each step in the workflows.
A z slice-through of video of an unfiltered, unbinned Protomo back-projection reconstruction of iteration 69 of the eTomo tutorial dataset. The outlined box that appears partway through the video shows the search area that Protomo used to create preliminary back-projections for alignment purposes. The search area excluded gold beads from the alignment. Note that the reconstruction slab is tilted as described in Figure 5.
Acknowledgments
We thank Michael Chapman for providing AAV-DJ for tilt-series collection. We thank John M. Spear for assisting with AAV-DJ grid preparation and screening. We thank Hanaa Hariri for the Sar1-GUV sample and grid preparation used in the tilt-series in Figure 1. We thank Sargis Dallakyan for including Protomo in the Appion auto-installation script. We thank Amber Herold, Anchi Cheng, Hanspeter Winkler, Kenneth Taylor, and David Mastronarde for helpful discussions. This work was supported in part by a National Institutes of Health grant (GM086892) and the Department of Physics at Florida State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amat F, Moussavi F, Comolli LR, Elidan G, Downing KH, Horowitz M. Markov random field based automatic image alignment for electron tomography. J Struct Biol; The 4th International Conference on Electron Tomography The 4th International Conference on Electron Tomography; 2008. pp. 260–275. [DOI] [PubMed] [Google Scholar]
- Bernard Heymann J, Cardone G, Winkler DC, Steven AC. Computational resources for cryo-electron tomography in Bsoft. J Struct Biol; The 4th International Conference on Electron Tomography The 4th International Conference on Electron Tomography; 2008. pp. 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA. Structural biology in situ — the potential of subtomogram averaging. Curr Opin Struct Biol Theory and simulation / Macromolecular assemblies. 2013;23:261–267. doi: 10.1016/j.sbi.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Castaño-Díez D, Kudryashev M, Arheit M, Stahlberg H. Dynamo: A flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J Struct Biol. 2012;178:139–151. doi: 10.1016/j.jsb.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Cheng A, Henderson R, Mastronarde D, Ludtke SJ, Schoenmakers RHM, Short J, Marabini R, Dallakyan S, Agard D, Winn M. MRC2014: Extensions to the MRC format header for electron cryo-microscopy and tomography. J Struct Biol. 2015 doi: 10.1016/j.jsb.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández JJ. Computational methods for electron tomography. Micron. 2012;43:1010–1030. doi: 10.1016/j.micron.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Fernández JJ, Li S, Crowther RA. CTF determination and correction in electron cryotomography. Ultramicroscopy. 2006;106:587–596. doi: 10.1016/j.ultramic.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Frank J, McEwen BF, Radermacher M, Turner JN, Rieder CL. Three-dimensional tomographic reconstruction in high voltage electron microscopy. J Electron Microsc Tech. 1987;6:193–205. doi: 10.1002/jemt.1060060210. [DOI] [Google Scholar]
- Galaz-Montoya JG, Flanagan J, Schmid MF, Ludtke SJ. Single particle tomography in EMAN2. J Struct Biol. 2015;190:279–290. doi: 10.1016/j.jsb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer Visualization of Three-Dimensional Image Data Using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P-W, Lyumkis D, Potter CS, Carragher B. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz F. Zur Streuung mittelschneller Elektronen in kleinste Winkel. Z Für Naturforschung A. 1954;9:185–204. [Google Scholar]
- Lerch TF, O’Donnell JK, Meyer NL, Xie Q, Taylor KA, Stagg SM, Chapman MS. Structure of AAV-DJ, a Retargeted Gene Therapy Vector: Cryo-Electron Microscopy at 4 Å Resolution. Structure. 2012;20:1310–1320. doi: 10.1016/j.str.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan G, Faruqi AR, Clare D, Henderson R. Comparison of optimal performance at 300 keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy. 2014;147:156–163. doi: 10.1016/j.ultramic.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell S, Förster F, Linaroudis A, Net WD, Beck F, Hegerl R, Baumeister W, Plitzko JM. TOM software toolbox: acquisition and analysis for electron tomography. J Struct Biol. 2005;149:227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- O’Toole E. ETomo Tutorial for IMOD Version 4.7. 2003 http://bio3d.colorado.edu/imod/doc/etomoTutorial.html.
- Ruskin RS, Yu Z, Grigorieff N. Quantitative characterization of electron detectors for transmission electron microscopy. J Struct Biol. 2013;184:385–393. doi: 10.1016/j.jsb.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schur FKM, Hagen WJH, de Marco A, Briggs JAG. Determination of protein structure at 8.5 Å resolution using cryo-electron tomography and sub-tomogram averaging. J Struct Biol. 2013;184:394–400. doi: 10.1016/j.jsb.2013.10.015. [DOI] [PubMed] [Google Scholar]
- Schur FKM, Hagen WJH, Rumlová M, Ruml T, Müller B, Kräusslich HG, Briggs JAG. Structure of the immature HIV-1 capsid in intact virus particles at 8. A resolution. Nature. 2015;517:505–508. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- Sheth LK, Piotrowski AL, Voss NR. Visualization and quality assessment of the contrast transfer function estimation. J Struct Biol. 2015 doi: 10.1016/j.jsb.2015.06.012. [DOI] [PubMed] [Google Scholar]
- Spear JM, Noble AJ, Xie Q, Sousa D, Chapman MS, Stagg SM. Frame Based Optimization of Direct Electron Detector Collected Images in Single-Particle CryoEM. J Struct Biol. 2015 doi: 10.1016/j.jsb.2015.09.006. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JCH. High-Resolution Electron Microscopy. 4 2013. [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol Software tools for macromolecular microscopy. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S. Structural Mechanism of Trimeric HIV-1 Envelope Glycoprotein Activation. PLoS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler H, Taylor KA. Marker-free dual-axis tilt series alignment. J Struct Biol. 2013;182:117–124. doi: 10.1016/j.jsb.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler H, Taylor KA. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy. 2006;106:240–254. doi: 10.1016/j.ultramic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Xiong Q, Morphew MK, Schwartz CL, Hoenger AH, Mastronarde DN. CTF determination and correction for low dose tomographic tilt series. J Struct Biol. 2009;168:378–387. doi: 10.1016/j.jsb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. Correlative cryo-electron tomography and optical microscopy of cells. Curr Opin Struct Biol Protein-carbohydrate interactions / Biophysical methods. 2013;23:763–770. doi: 10.1016/j.sbi.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Example tilt-series videos of the example AAV-DJ tilt-series from the case study from the iterations shown in Fig. 2. Left is iteration 27, right is iteration 29. It is important to note the quality of the alignment even though there is a considerable amount of ice contamination in the images. Scale bar, tilt angle, and tilt image number are included on the bottom. Each video was automatically generated after each refinement and was downloaded using the ‘Download Video’ link seen in Movie S3. The images are binned by 8.
Example slice-through videos of preliminary reconstructions of the tilt-series seen in Movie S1. Left is iteration 27, right is iteration 29. Scale bar and z-slice are included on the bottom. Each video was automatically generated after each refinement and was downloaded using the ‘Download Video’ link seen in Movie S3. The differences in brightness is caused by the per-image normalization during video processing due to the high contrast gold beads. The reconstruction is unfiltered and binned by 8.
An overview of the Appion-Protomo individual tilt-series workflow and webpage layouts. Information for every option is documented from within the webpages. The user is guided between webpages at each step in the workflow.
An overview of the Appion-Protomo batch tilt-series webpage layouts. Information for every option is documented from within the webpages. The user is guided between webpages at each step in the workflows.
A z slice-through of video of an unfiltered, unbinned Protomo back-projection reconstruction of iteration 69 of the eTomo tutorial dataset. The outlined box that appears partway through the video shows the search area that Protomo used to create preliminary back-projections for alignment purposes. The search area excluded gold beads from the alignment. Note that the reconstruction slab is tilted as described in Figure 5.