Abstract
Although some Weissella species play beneficial roles in food fermentation and in probiotic products, others such as Weissella confusa are emerging Gram-positive pathogens in immunocompromised hosts. Weissella species are difficult to identify by conventional biochemical methods and commercial automated systems and are easily misidentified as Lactobacillus and Leuconostoc species. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is increasingly being used for bacterial identification. Little, however, is known about the effectiveness of MALDI-TOF MS in identifying clinical isolates of Weissella to the species level. In this study, we evaluated whether the MALDI-TOF MS Bruker Biotyper system could accurately identify a total of 20 W. confusa and 2 W. cibaria blood isolates that had been confirmed by 16s rRNA sequencing analysis. The MALDI-TOF Biotyper system yielded no reliable identification results based on the current reference spectra for the two species (all score values <1.7). New W. confusa spectra were created by randomly selecting 3 W. confusa isolates and external validation was performed by testing the remaining 17 W. confusa isolates using the new spectra. The new main spectra projection (MSP) yielded reliable score values of >2 for all isolates with the exception of one (score value, 1.963). Our results showed that the MSPs in the current database are not sufficient for correctly identifying W. confusa or W. cibaria. Further studies including more Weissella isolates are warranted to further validate the performance of MALDI-TOF in identifying Weissella species.
Keywords: Weissella confusa, Weissella cibaria, Bacteremia, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, 16S rRNA
Introduction
Members of Weissella species are found in various fermented food products and are considered to be potential probiotics (Nam et al., 2002; Fairfax et al., 2014; Kot et al., 2014; Fusco et al., 2015). However, some Weissella species, namely W. confusa, can cause opportunistic infections in immunocompromised hosts and have been found to be intrinsically resistant to vanomycin, making treatment difficult (Lee et al., 2011). W. confusa is most likely an underestimated pathogen because it shares similar staining and biochemical properties with other Gram-positive, catalase-negative bacteria such as Leuconostoc and Lactobacillus species (Facklam and Elliott, 1995; Shin et al., 2007; Schillinger et al., 2008; Lee et al., 2011; Fairfax et al., 2014).
Traditionally, correct identification of Weissella species relies on the implementation of 16S rRNA sequencing (Chelo et al., 2007; Lee et al., 2011). Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is increasingly being used to identify unusual bacterial pathogens including Weissella species isolated from clinical specimens (Lee et al., 2013; Chen et al., 2014; Fairfax et al., 2014; Hsueh et al., 2014). MALDI-TOF MS has the advantage of being faster and more cost-effective than the conventional 16S rRNA sequencing methods and is, therefore, expected to play a more important role in food and clinical laboratories (Steensels et al., 2011). Few studies, however, have addressed and tested the effectiveness of MALDI-TOF MS in identifying Weissella species.
In this study, we compared the accuracy of MALDI-TOF MS with that of 16S rRNA sequencing in identifying Weissella species.
Materials and Methods
Bacterial Isolates
A total of 147 isolates of Gram-positive rods, including Lactobacillus and Leuconostoc species, that had been recovered from patients with bloodstream infections at the National Taiwan University Hospital (a 2900-bed tertiary care center in northern Taiwan) during the period 2000–2014 were obtained from the hospital’s microbiology laboratory. These isolates had initially been identified as either Lactobacillus or Leuconostoc species by traditional or commercial automated identification systems as reported previously (Lee et al., 2011, 2015). Among these isolates, 22 isolates were identified as Weisella species by colony morphology, a positive Gram stain reaction, growth at 37°C, and negative reactions to pyrrolidonyl arylamidase and leucine aminopeptidase as well as by 16S rRNA sequencing analysis.
16S rRNA Gene Sequencing Analysis
Partial sequencing of up to 1475 base pairs of the 16S rRNA gene was performed for species identification of all 147 isolates of Lactobacillus and Leuconostoc species using the following primers: forward: 5′-AGAGTTTGATCCTGGCTCAG-3′; reverse: 5′-GGTTACCTTGTTACGACTT-3′ (Tsai et al., 2004). The results were compared with published sequences in the GenBank database using the BLASTN algorithm. The closest matches and GenBank accession numbers were obtained. L. lactis ATCC 19256 was used as the control strain in each test as described previously (Lee et al., 2011).
Performance of the MALDI Biotyper
For analysis of the 22 isolates of Weissella species by the MALDI-TOF Biotyper (Microflex LT; Bruker Daltonik GmbH, Bremen, Germany), the samples were prepared and analyzed as previously described (Verroken et al., 2010; Hsueh et al., 2014). Briefly, all isolates were incubated in Trypticase soy agar with 5% sheep blood (BAP) (Becton, Dickinson Microbiology Systems, Sparks, MD, USA) and incubated for 48 h at 37°C. Colonies were transferred to a 1.5-ml screw-cap Eppendorf tube containing 300 μl of distilled water and then mixed with 900 μl of ethanol by pipetting. The suspension was pelleted by centrifugation at 13,000 rpm for 2 min, evaporated to dryness, and then reconstituted in 50 μl of 70% formic acid. After incubation for 30 s, 50 μl of acetonitrile (Sigma–Aldrich) was added. The suspension was then centrifuged at 13,000 rpm for 2 min. Next, 1.0 μl of the supernatant was applied to a 96-spot polished steel target plate (Bruker Daltonik GmbH, Bremen, Germany) and dried. A saturated solution of 1.0 μl of MALDI matrix (α-cyano-4-hydroxycinnamic acid [HCCA]; Bruker Daltonik GmbH) was applied to each sample and dried. Measurements were performed with the Bruker microflex LT MALDI-TOF MS system (Bruker Daltonik GmbH) using FlexControl software with Compass Flex Series version 1.3 software and a 60-Hz nitrogen laser (337 nm wavelength). The spectra were collected in the linear positive mode in a mass range covering m/z 1,960–20,132. Spectra ranging from the mass-to-charge ratio (m/z) 2,000–20,000 were analyzed using Bruker Biotyper automation control and the Bruker Biotyper 3.1 software and library (database [DB] 5627 with 5,627 entries). Identification scores of ≥2.000 indicated species-level identification, scores of 1.700–1.999 indicated genus-level identification, and scores of <1.700 indicated no reliable identification. All isolates with discrepant identification results between the molecular and Bruker Biotyper methods were retested twice.
Cluster Analysis and Creation of New W. confusa and W. cibria Main Spectra Projection
Clustering analysis of 22 isolates of the two genetically identified Weissella species collected from NTUH was performed using ClinProTools 3.0 (Bruker Daltonics GmbH, Bremen, Germany) as previously described (Chen et al., 2014; Hsueh et al., 2014). Because the current MALDI-TOF database failed to make a correct identification of all W. confusa and W. cibaria isolates, spectra of 3 W. confusa isolates were randomly selected from the 20 isolates of W. confusa obtained from the NTUH for the creation of main spectra projections (MSPs; database entrance) using Bruker Biotyper MALDI-TOF MS software (Bruker Daltonics) as previously described (Chen et al., 2014; Hsueh et al., 2014). The spectra generated using the 3 isolates were blindly tested against those of the residual 17 W. confusa isolates.
Results
The MALDI-TOF Biotyper system with current reference spectra yielded no reliable identification results (score values ranged from 1.282 to 1.519) among the 20 W. confusa and 2 W. cibria isolates (Table 1). Using the spectra created by the 3 W. confusa isolates (Isolates 1, 2, and 3), we further tested the residual 17 W. confusa isolates. We found that all of the W. confusa isolates with the exception of one (Isolate 17, score value of 1.963) were identified correctly with a score >2 using the three newly created MSPs. The highest scores were achieved with MSP created by W. confusa Isolate 1 in the ten W. confusa isolates, MSP created by W. confusa Isolate 2 in the five W. confusa isolates and MSP created by W. confusa Isolate 3 in the W. confusa isolate.
Table 1.
Species identification of 22 isolates of Weissella species by 16S rRNA sequencing analysis and the MALDI-TOF Biotyper system using preexisting (DB 5627) and newly established databases.
| Isolate no. | Original identification results by conventional methods | 16S rRNA sequencing |
Identification by MALDI Biotyper based on database 5627 |
Identification by MALDI Biotyper based on newly developed database |
|||
|---|---|---|---|---|---|---|---|
| Results (maximal identity, %) | Accession number | Results | Score value | Results (best match) | Score value | ||
| 1∗ | Lactobacillus species | W. confusa (100) | KF245541.1 | NRI | 1.361 | – | – |
| 2∗ | Leuconostoc species | W. confusa (99.8) | EU807756.1 | NRI | 1.336 | – | – |
| 3∗ | Leuconostoc species | W. confusa (100) | GU138614.1 | NRI | 1.298 | – | – |
| 4 | Lactobacillus species | W. confusa (100) | KJ476186.1 | NRI | 1.391 | W. confusa | 2.054 |
| 5 | Lactobacillus species | W. confusa (100) | KF245541.1 | NRI | 1.448 | W. confusa | 2.073 |
| 6 | Lactobacillus species | W. confusa (100) | KF245541.1 | NRI | 1.29 | W. confusa | 2.250 |
| 7 | Lactobacillus species | W. confusa (100) | KF245541.1 | NRI | 1.39 | W. confusa | 2.243 |
| 8 | Lactobacillus species | W. confusa (100) | KF245541.1 | NRI | 1.43 | W. confusa | 2.501 |
| 9 | Lactobacillus species | W. confusa (100) | KF245541.1 | NRI | 1.338 | W. confusa | 2.376 |
| 10 | Lactobacillus species | W. confusa (100) | KJ476186.1 | NRI | 1.503 | W. confusa | 2.113 |
| 11 | Leuconostoc species | W. confusa (99.9) | KJ476186.1 | NRI | 1.303 | W. confusa | 2.395 |
| 12 | Leuconostoc species | W. confusa (100) | EU807756.1 | NRI | 1.35 | W. confusa | 2.101 |
| 13 | Leuconostoc species | W. confusa (100) | EU807756.1 | NRI | 1.362 | W. confusa | 2.04 |
| 14 | Leuconostoc species | W. confusa (99.6) | KJ476186.1 | NRI | 1.391 | W. confusa | 2.332 |
| 15 | Lactobacillus species | W. confusa (100) | GU138614.1 | NRI | 1.416 | W. confusa | 2.333 |
| 16 | Leuconostoc species | W. confusa (99.7) | GU138614.1 | NRI | 1.394 | W. confusa | 2.022 |
| 17 | Leuconostoc species | W. confusa (100) | GU138614.1 | NRI | 1.38 | W. confusa | 1.963 |
| 18 | Leuconostoc species | W. confusa (99.8) | KJ476186.1 | NRI | 1.282 | W. confusa | 2.368 |
| 19 | Leuconostoc species | W. confusa (100) | GU138614.1 | NRI | 1.413 | W. confusa | 2.302 |
| 20 | Lactobacillus species | W. confusa (100) | KJ476186.1 | NRI | 1.382 | W. confusa | 2.355 |
| 21 | Leuconostoc species | W. cibaria (100) | AB510747.1 | NRI | 1.333 | – | – |
| 22 | Leuconostoc species | W. cibaria (99.3) | AB510747.1 | NRI | 1.519 | – | – |
∗Isolates for creation of W. confusa spectra; NRI, no reliable identification.
The characteristic spectra of W. confusa (Isolate 8) and W. cibria (Isolate 22) are illustrated in Figure 1. The characteristic peaks for W. cibria were 3008.1, 3634.1, 4079.8, 4954.5, 5380.2, 6371.5, 7267.7, 7962.6, 9530.7, and 11028.2. The characteristic peaks for W. confusa were 3010.1, 3585.2, 4162.9, 4608.2, 4844.0, 5183.7, 6404.8, 7167.4, 7964.3, 9215.7, and 9686.8.
FIGURE 1.
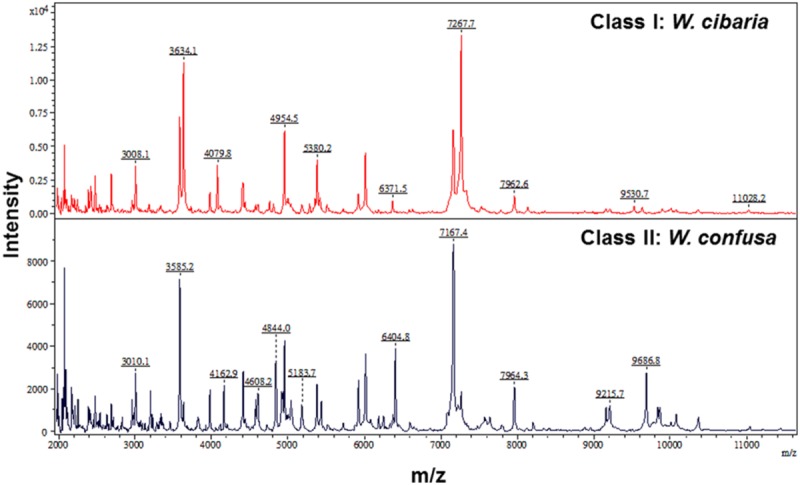
Two clusters of Weissella species spectra, i.e., cluster I (W. cibaria) and cluster II (W. confusa), analyzed by clustering analysis of MALDI-TOF MS results. The absolute intensities of the ions are shown on the y axis and the masses (m/z) of the ions are shown on the x axis. The m/z values represent the mass-to-charge ratio.
Discussion
We found that current reference spectra in the MALDI-TOF database were much less accurate than 16s rRNA sequencing in correctly identifying W. confusa and W. cibaria. Using the W. confusa MSPs created by our database, we were able to correctly identify all W. confusa isolates.
It is notoriously difficult to identify W. confusa. The 16S rRNA sequencing method is considered the standard identification method, although other methods have shown to be able to accurately identify W. confusa. A novel PCR-based identification method using amplified fragment length polymorphism (AFLP)-derived marker was shown to have 100% specificity in identifying W. confusa (Fusco et al., 2011). MALDI-TOF has also been shown to correctly identify the organism in blood samples (Lee et al., 2013; Fairfax et al., 2014). Despite satisfying results, it has to be kept in mind that the tested case numbers were small and only five Weissella species are listed in the MALDI-TOF database, namely W. confusa, W. halotolerans, W. kandleri, W. minor, and W. viridescens. Stevenson et al. (2010) reported that only one W. confusa isolate achieved a score value of 1.7, indicating an identification only at genus level. In our study, all of the W. confusa isolates that had been correctly identified by 16S rRNA sequencing failed to be identified with MALDI-TOF using the current database. The discrepancy between the two identification methods remains to be solved. Further studies comparing the results between 16S rRNA sequencing and MALDI-TOF are warranted.
The most well-studied food product in association with Weissella species is kimchi, a traditional Korean food. Lactic acid bacteria, belonging to the genera Leuconostoc, Lactobacillus, and Weissella, are considered to be the most likely organisms responsible for kimchi fermentation (Lee et al., 2005; Jung et al., 2013; Kwak et al., 2014), and studies have shown that W. confusa is involved in the early and middle stages of kimchi fermentation (Lee et al., 2005; Kim et al., 2012). W. confusa has also been detected in food products from other countries, such as fermented milk products in Ghana, sourdough products in Italy, fermented soy sauce in Taiwan, and in soy sauce brine in Malaysia (Corsetti et al., 2001; Akabanda et al., 2013; Wei et al., 2013; Sulaiman et al., 2014).
Although W. ceti is well known in veterinary medicine for causing weissellosis in rainbow trout (Snyder et al., 2015), to the best of our knowledge, only W. confusa has been linked to human invasive infections. Therefore, other Weissella species are probably safer alternatives to W. confusa as probiotics.
Bacteremia due to W. confusa is rare and tends to occur in severely immunocompromised hosts, such as patients who have received organ transplants (Harlan et al., 2011; Lee et al., 2011, 2013; Salimnia et al., 2011; Fairfax et al., 2014). Immunocompromised hosts undergoing abdominal surgery have also been shown to be at particular risk since abdominal surgery results in the breakdown of intestinal mucosa (Lee et al., 2011). Given the findings of our study, the incidence of bacteremia due to W. confusa is underestimated because of the difficulties in making a correct identification with commercial and biochemical methods. The findings also suggest that immunocompromised patients should refrain from consuming fermented foods.
In our previous study, the results of 16S rRNA sequencing analysis showed that 10 of 43 blood isolates of catalase-negative, vancomycin-resistant coccobacilli were actually W. confusa (Lee et al., 2011). In this study, all Weissella species were either identified as Lactobacillus or Leuconostoc species by commercial automatic systems or traditional biochemical methods. This finding is in concordance with a phylogenetical study by Chelo et al. (2007), who showed that Leuconostoc species and Lactobacillus species were close to W. confusa in neighbor-joining tree by 16S rRNA sequencing. The applications of 16S rRNA sequencing may lead to a better understanding of the incidence of bacteremia due to Weissella species.
During the study period, the actual number of bacteremia due to Weissella species might be under-estimated. In this study, we did not perform 16S rRNA sequencing on bacteremic isolates of Gram-positive cocci, including viridians group streptococci, Pediococcus, and Gemella species. Some Weissella species were misidentified as these organisms by conventional identification methods (Ruoff, 2002; Lee et al., 2011). In this study, 16S rRNA sequencing was only performed on bacteremic isolates of Lactobacillus and Leuconostoc species, which were the most closely relevant species and thus were considered the most possible bacteria W. confusa could be misidentified as (Chelo et al., 2007; Lee et al., 2011).
Conclusion
The MALDI Biotyper system is less accurate that 16S rRNA sequencing in correctly identifying Weissella species. W. confusa is an underestimated cause of bacteremia due to its tendency to be misidentified. More extensive epidemiologic studies with molecular methods are needed to reveal the true prevalence and incidence of invasive W. confusa infections. Also, studies involving more clinical Weissella isolates are needed to explore the effectiveness of the MALDI Biotyper system in identification of Weissella species.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Akabanda F., Owusu-Kwarteng J., Tano-Debrah K., Glover R. L., Nielsen D. S., Jespersen L. (2013). Taxonomic and molecular characterization of lactic acid bacteria and yeasts in nunu, a Ghanaian fermented milk product. Food Microbiol. 34 277–283. 10.1016/j.fm.2012.09.025 [DOI] [PubMed] [Google Scholar]
- Chelo I. M., Zé-Zé L., Tenreiro R. (2007). Congruence of evolutionary relationships inside the Leuconostoc-Oenococcus-Weissella clade assessed by phylogenetic analysis of the 16S rRNA gene, dnaA, gyrB, rpoC and dnaK. Int. J. Syst. Evol. Microbiol. 57(Pt 2) 276–286. 10.1099/ijs.0.64468-0 [DOI] [PubMed] [Google Scholar]
- Chen P. L., Lee T. F., Wu C. J., Teng S. H., Teng L. J., Ko W. C., et al. (2014). Matrix-assisted laser desorption ionization-time of flight mass spectrometry can accurately differentiate Aeromonas dhakensis from A. hydrophila, A. caviae, and A. veronii. J. Clin. Microbiol. 52 2625–2628. 10.1128/JCM.01025-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsetti A., Lavermicocca P., Morea M., Baruzzi F., Tosti N., Gobbetti M. (2001). Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 64 95–104. 10.1016/S0168-1605(00)00447-5 [DOI] [PubMed] [Google Scholar]
- Facklam R., Elliott J. A. (1995). Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin. Microbiol. Rev. 8 479–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfax M. R., Lephart P. R., Salimnia H. (2014). Weissella confusa: problems with identification of an opportunistic pathogen that has been found in fermented foods and proposed as a probiotic. Front. Microbiol. 5:254 10.3389/fmicb.2014.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco V., Quero G. M., Cho G. S., Kabisch J., Meske D., Neve H., et al. (2015). The genus Weissella: taxonomy, ecology and biotechnological potential. Front. Microbiol. 6:155 10.3389/fmicb.2015.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco V., Quero G. M., Stea G., Morea M., Visconti A. (2011). Novel PCR-based identification of Weissella confusa using an AFLP-derived marker. Int. J. Food Microbiol. 145 437–443. 10.1016/j.ijfoodmicro.2011.01.015 [DOI] [PubMed] [Google Scholar]
- Harlan N. P., Kempker R. R., Parekh S. M., Burd E. M., Kuhar D. T. (2011). Weissella confusa bacteremia in a liver transplant patient with hepatic artery thrombosis. Transpl. Infect. Dis. 13 290–293. 10.1111/j.1399-3062.2010.00579.x [DOI] [PubMed] [Google Scholar]
- Hsueh P. R., Lee T. F., Du S. H., Teng S. H., Liao C. H., Sheng W. H., et al. (2014). Bruker biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Nocardia, Rhodococcus, Kocuria, Gordonia, Tsukamurella, and Listeria species. J. Clin. Microbiol. 52 2371–2379. 10.1128/JCM.00456-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. Y., Lee S. H., Jin H. M., Hahn Y., Madsen E. L., Jeon C. O. (2013). Metatranscriptomic analysis of lactic acid bacterial gene expression during kimchi fermentation. Int. J. Food Microbiol. 163 171–179. 10.1016/j.ijfoodmicro.2013.02.022 [DOI] [PubMed] [Google Scholar]
- Kim B., Seo W. T., Kim M. G., Yun H. D., Cho K. M. (2012). Metagenomic lactic acid bacterial diversity during Mulkimchi fermentation based on 16S rRNA sequence. J. Korean Soc. Appl. Biol. Chem. 55 787–792. 10.1007/s13765-012-2185-3 [DOI] [Google Scholar]
- Kot W., Neve H., Heller K. J., Vogensen F. K. (2014). Bacteriophages of Leuconostoc, Oenococcus, and Weissella. Front. Microbiol. 5:186 10.3389/fmicb.2014.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S. H., Cho Y. M., Noh G. M., Om A. S. (2014). Cancer preventive potential of kimchi lactic acid bacteria (Weissella cibaria, Lactobacillus plantarum). J. Cancer Prev. 19 253–258. 10.15430/JCP.2014.19.4.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Heo G. Y., Lee J. W., Oh Y. J., Park J. A., Park Y. H., et al. (2005). Analysis of kimchi microflora using denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 102 143–150. 10.1016/j.ifoodmicro.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Lee M. R., Huang Y. T., Liao C. H., Lai C. C., Lee P. I., Hsueh P. R. (2011). Bacteraemia caused by Weissella confusa at a university hospital in Taiwan, 1997-2007. Clin. Microbiol. Infect. 17 1226–1231. 10.1111/j.1469-0691.2010.03388.x [DOI] [PubMed] [Google Scholar]
- Lee M. R., Tsai C. J., Liang S. K., Lin C. K., Huang Y. T., Hsueh P. R. (2015). Clinical characteristics of bacteraemia caused by Lactobacillus spp. and antimicrobial susceptibilities of the isolates at a medical centre in Taiwan, 2000–2014. Int. J. Antimicrob. Agents 46 439–445. 10.1016/j.ijantimicag.2015.06.017 [DOI] [PubMed] [Google Scholar]
- Lee W., Cho S. M., Kim M., Ko Y. G., Yong D., Lee K. (2013). Weissella confusa bacteremia in an immune-competent patient with underlying intramural hematomas of the aorta. Ann. Lab. Med. 33 459–462. 10.3343/alm.2013.33.6.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H., Ha M., Bae O., Lee Y. (2002). Effect of Weissella confusa strain PL9001 on the adherence and growth of Helicobacter pylori. Appl. Environ. Microbiol. 68 4642–4645. 10.1128/AEM.68.9.4642-4645.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoff K. L. (2002). Miscellaneous catalase-negative, gram-positive cocci: emerging opportunists. J. Clin. Microbiol. 40 1129–1133. 10.1128/JCM.40.4.1129-1133.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimnia H., Alangaden G. J., Bharadwaj R., Painter T. M., Chandrasekar P. H., Fairfax M. R. (2011). Weissella confusa: an unexpected cause of vancomycin-resistant gram-positive bacteremia in immunocompromised hosts. Transpl. Infect. Dis. 13 294–298. 10.1111/j.1399-3062.2010.00586.x [DOI] [PubMed] [Google Scholar]
- Schillinger U., Boehringer B., Wallbaum S., Caroline L., Gonfa A., Huch Née Kostinek M., et al. (2008). A genus-specific PCR method for differentiation between Leuconostoc and Weissella and its application in identification of heterofermentative lactic acid bacteria from coffee fermentation. FEMS Microbiol. Lett. 286 222–226. 10.1111/j.1574-6968.2008.01286.x [DOI] [PubMed] [Google Scholar]
- Shin J. H., Kim D. I., Kim H. R., Kim D. S., Kook J. K., Lee J. N. (2007). Severe infective endocarditis of native valves caused by Weissella confusa detected incidentally on echocardiography. J. Infect. 54 e149–e151. 10.1016/j.jinf.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Snyder A. K., Hinshaw J. M., Welch T. J. (2015). Diagnostic tools for rapid detection and quantification of Weissella ceti NC36 infections in rainbow trout. Lett. Appl. Microbiol. 60 103–110. 10.1111/lam.12365 [DOI] [PubMed] [Google Scholar]
- Steensels D., Verhaegen J., Lagrou K. (2011). Matrix assisted laser desorption ionization time of flight mass spectrometry for the identification of bacteria and yeast in clinical microbiological laboratory: a review. Acta Clin. Belg. 66 267–273. [DOI] [PubMed] [Google Scholar]
- Stevenson L. G., Drake S. K., Murray P. R. (2010). Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48 444–447. 10.1128/JCM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman J., Gan H. M., Yin W. F., Chan K. G. (2014). Microbial succession and the functional potential during the fermentation of chinese soy sauce brine. Front. Microbiol. 5:556 10.3389/fmicb.2014.00556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. C., Teng L. J., Hsueh P. R. (2004). Direct detection of bacterial pathogens in brain abscesses by polymerase chain reaction amplification and sequencing of partial 16S ribosomal deoxyribonucleic acid fragments. Neurosurgery 55 1154–1162. 10.1227/01.NEU.0000140842.37422.EE [DOI] [PubMed] [Google Scholar]
- Verroken A., Janssens M., Berhin C., Bogaerts P., Huang T. D., Wauters G., et al. (2010). Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Nocardia species. J. Clin. Microbiol. 48 4015–4021. 10.1128/JCM.01234-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C. L., Chao S. H., Tsai W. B., Lee P. S., Tsau N. H., Chen J. S., et al. (2013). Analysis of bacterial diversity during the fermentation of inyu, a high-temperature fermented soy sauce, using nested PCR-denaturing gradient gel electrophoresis and the plate count method. Food Microbiol. 33 252–261. 10.1016/j.fm.2012.10.001 [DOI] [PubMed] [Google Scholar]


