Abstract
Animal personality and coping styles are basic concepts for evaluating animal welfare. Struggling response of piglets in so-called backtests early in life reflects their coping strategy. Behavioral reactions of piglets in backtests have a moderate heritability, but their genetic basis largely remains unknown. Here, latency, duration and frequency of struggling attempts during one-minute backtests were repeatedly recorded of piglets at days 5, 12, 19, and 26. A genome-wide association study for backtest traits revealed 465 significant SNPs (FDR ≤ 0.05) mostly located in QTL (quantitative trait locus) regions on chromosome 3, 5, 12 and 16. In order to capture genes in these regions, 37 transcripts with significant SNPs were selected for expressionQTL analysis in the hypothalamus. Eight genes (ASGR1, CPAMD8, CTC1, FBXO39, IL19, LOC100511790, RAD51B, UBOX5) had cis- and five (RANGRF, PER1, PDZRN3, SH2D4B, LONP2) had trans-expressionQTL. In particular, for PER1, with known physiological implications for maintenance of circadian rhythms, a role in coping behavior was evidenced by confirmed association in an independent population. For CTC1 a cis-expression QTL and the consistent relationship of gene polymorphism, mRNA expression level and backtest traits promoted its link to coping style. GWAS and eQTL analyses uncovered positional and functional gene candidates for coping behavior.
Consistent behavior of animals along lifetime and/or across situations is an expression of an individual reaction pattern, temperament or animal personality1. In addition to behavioral and evolutionary sciences these concepts of animal personality gains interest with regard to animal welfare and ethical animal husbandry. In fact, animal welfare concerns play a significant role in the design of modern pig production systems. Another idea in this context is ‘coping’, which comprises all behavioral and physiological reactions in response to challenging, aversive situation. Coping styles evolved to form general adaptive response patterns with the “proactive or active pattern” characterized by a fight-or-flight response, high levels of aggression, and territorial control, where as the “reactive or passive pattern” shows a conservation-withdrawal response, immobility, and low aggression2,3,4. Both patterns differ in their behavioral, physiological and immunological characteristics. Between coping style and cortisol levels, the major indicator of stress, inverse relationships were shown in human and animals studies5,6,7,8. The behavior patterns of pigs living in confinement can vary according to several factors, but heredity is known to be a key factor in predicting temperament9. Defining phenotypes according to molecular features would promote the knowledge of functional traits like behaviour in both human and animal research. Knowledge of the genetic variations and molecular mechanisms which affect behavior facilitate the use of genetic or genomic selection is an alternative approach to increase the adaptability and reduce aggressive behavior of animals10,11. Indicators of behavior and aggressiveness in pigs include the backtest12, open-field test13, human approach test14, and counting skin lesions (lesion scoring)15,16. The backtest is a well-established, standardized test that has primarily been studied in relation to piglet behavior and other physiologic characteristics12,17,18,19. The backtest generates a clear behavioral response and provides insight into the behavioral flexibility of piglets20. The genetic parameters of the backtest like heritability were reported to vary from 0.10 to 0.5 depending on the sample size or test conditions21,22,23.
Several genomic regions and gene variants associated with stress response, aggression, and depression have been reported in both humans and animal models24,25,26. Pigs share numerous physiologic and genomic similarities with humans and therefore provide a tractable model in which to study the genetic determination of behavioral, physiological, and metabolic traits27. In fact, behavioral quantitative trait loci (QTL) have been mapped to stress10, feeding behavior28, maternal behavior29, and behavioral indices under healthy and disease conditions30 in porcine models.
Variation of complex traits is largely due to polymorphisms affecting regulatory sequences rather than coding sequences. Expression-QTL (eQTL) analysis integrates gene expression levels and genome-wide genotyping information to find genetic variation association with change in gene expression. Estimation of eQTL involves the handling of transcript abundances as phenotypes in linkage or association studies. Knowledge of the position of analyzed genes and markers enable differentiating cis and trans eQTL, with the first indicating a regulatory variation in the gene whose transcript level is recorded itself, whereas trans eQTL suggest a polymorphism elsewhere in the genome affecting the expression of the target gene31. Moreover, signals revealed by genome wide association studies (GWAS) are often located in regions with high linkage disequilibrium (LD) harboring several genes. ExpressionQTL analysis in relevant tissue point to regulatory variation and provide additional evidence for the role of genes within LD regions for a particular trait of interest. We have previously shown that the integration of GWAS revealing (clusters of) trait associated markers (regions) with eQTL analyses of trait-dependent expressed genes facilitates highlighting of genes related to complex traits like muscle and meat properties32. The genetic variants associated with coping behaviors in pigs remain largely unstudied. Here, we describe the first GWAS with subsequent eQTL analysis conducted to identify genetic variants responsible for coping behavior in two independent herds of pig.
Results
A total of 294 German Landrace piglets were genotyped with the Illumina PorcineSNP60 BeadChip consisting of 62,163 loci. After filtering, 48,909 SNPs were used for further analysis. Backtest parameters (latency, duration, frequency of struggling at four time points) and coping type (high-reactive (HR), indistinct (IR), low-reactive (LR)) were used for association study (Table 1). 465 SNPs were significantly associated with at least one of the backtest traits. We measured cortisol levels, and the correlation coefficients of coping behavior and cortisol levels ranged between |0.11–0.20| and indicated significant correlation (p < 0.05, N = 290). However, no significant association between coping groups and cortisol levels was found. HR and LR showed a tendency of higher cortisol levels than IR groups.
Table 1. Median, minimum (Min), and maximum (Max) of the backtest raw data for latency until the first response, duration of response and frequency of response in piglets (N = 294) at 5, 12, 19 and 26 days (d) of age.
| Trait | Median | Min | Max | |
|---|---|---|---|---|
| Latency (L) until the first response (in seconds) | ||||
| L at 5 days | (Ld5) | 10.5 | 1 | 60 |
| L at 12 days | (Ld12) | 14 | 1 | 60 |
| L at 19 days | (Ld19) | 13 | 1 | 60 |
| L at 26 days | (Ld26) | 16 | 1 | 60 |
| Duration (D) of all response (in seconds) | ||||
| D at 5 days | (Dd5) | 19 | 0 | 53 |
| D at 12 days | (Dd12) | 17 | 0 | 47 |
| D at 19 days | (Dd19) | 18.5 | 0 | 49 |
| D at 26 days | (Dd26) | 15 | 0 | 48 |
| Frequency (F) of response (number) | ||||
| F at 5 days | (Fd5) | 3 | 0 | 8 |
| F at 12 days | (Fd12) | 2 | 0 | 5 |
| F at 19 days | (Fd19) | 2 | 0 | 7 |
| F at 26 days | (Fd26) | 2 | 0 | 6 |
The backtest traits were highly correlated. Latency showed negative correlation with duration and positive with frequency of struggling at each time point (rs = |0.70–0.82|; p < 0.0001). The correlation between each time point of latency, duration and frequency of struggling ranged between rs = |0.37–0.57|, rs = |0.51–0.66| and rs = |0.47–0.66|, respectively, with p < 0.0001. The summation of total latency (tL), total duration (tD), and total frequency (tF) and coping type were correlated with each other with highly significant coefficients of correlation ranging between |0.80–0.88|.
Genome-wide association analysis of first response latency
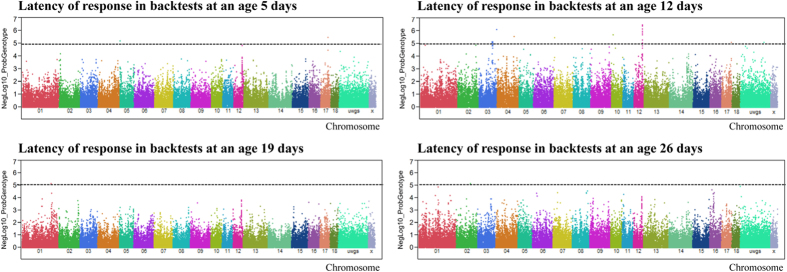
Manhattan plots of genome-wide association analyses of latency (L) at age 5 (Ld5), 12 (Ld12), 19 (Ld19), and 26 (Ld26) days are shown in Fig. 1. No SNPs were significantly associated with latency at days 5, 19, or 26 at a false discovery rate (FDR) of ≤5%, but 82 SNPs were significantly associated with Ld12, explaining about 7% of latency variance (Supplementary Table 1). Most of the significantly associated markers (N = 33, 40%) were located at 53–56 Mb of porcine chromosome (sus scrofa chromosome, SSC) 12. The most prominent gene located within this region is period circadian clock 1 (PER1). The top five markers significantly associated with Ld12 are shown in Table 2. Sixteen out of the 82 SNPs associated with Ld12 were located on SSC3 between 110 and 112 Mb.
Figure 1. Genome-wide association with latency of response in backtests at ages of 5, 12, 19 and 26 days.
Black lines at negative log10 (NegLog10) of 5 corresponds to FDR ≤ 0.05.
Table 2. Markers significantly associated with backtest traits including latency (L) until the first response, duration (D) of responses and frequency (F) of responses in piglets at 5, 12, 19 and 26 days (d) of age.
| SNP ID | Backtest | −log10 | % variance | FDR | SSC (Mb) | Gene |
|---|---|---|---|---|---|---|
| ALGA0090009 | Dd5 | 6.15 | 9.5 | 0.018 | 16 (31.0) | HCN1 |
| ALGA0112274 | Dd5 | 6.02 | 9.3 | 0.018 | ||
| ALGA0090013 | Dd5 | 5.86 | 9.0 | 0.018 | 16 (31.0) | HCN1 |
| DIAS0001557 | Dd5 | 5.69 | 8.8 | 0.018 | 12 (55.9) | RANGRF |
| M1GA0008168 | Dd5 | 5.50 | 8.5 | 0.018 | 5 (106.4) | |
| ALGA0067468 | Dd12 | 5.63 | 8.7 | 0.035 | 13 (3.4) | |
| ALGA0067470 | Dd12 | 5.63 | 8.7 | 0.035 | 13 (3.3) | |
| ASGA0102838 | Dd12 | 5.50 | 8.5 | 0.035 | 12 (55.6) | |
| ASGA0104602 | Dd12 | 5.49 | 8.5 | 0.035 | 2 (15.5) | |
| ASGA0055092 | Dd12 | 5.35 | 8.3 | 0.035 | 12 (55.8) | PER1 |
| ASGA0055092 | Dd19 | 7.45 | 11.3 | 0.001 | 12 (55.8) | PER1 |
| ASGA0102838 | Dd19 | 7.38 | 11.2 | 0.001 | 12 (55.6) | |
| ALGA0066975 | Dd19 | 6.59 | 10.1 | 0.003 | 12 (55.7) | |
| ALGA0121951 | Dd19 | 6.57 | 10.1 | 0.003 | 12 (55.9) | |
| ALGA0095323 | Dd19 | 6.34 | 9.7 | 0.004 | 17 (50.7) | |
| DIAS0001557 | Dd26 | 6.23 | 9.6 | 0.019 | 12 (55.9) | RANGRF |
| ASGA0055076 | Dd26 | 6.07 | 9.3 | 0.019 | 12 (55.2) | ATP1B2 |
| DIAS0000983 | Dd26 | 5.86 | 9.0 | 0.019 | 5 (7.3) | |
| ALGA0107834 | Dd26 | 5.78 | 8.9 | 0.019 | 3 (139.8) | ASGR1 |
| ASGA0085214 | Dd26 | 5.55 | 8.6 | 0.019 | 12 (54.6) | |
| ALGA0115736 | Fd5 | 6.33 | 9.9 | 0.021 | 2 (1.5) | liprin-alpha-1-like |
| ASGA0102974 | Fd5 | 5.65 | 8.8 | 0.041 | ||
| ASGA0076061 | Fd5 | 5.57 | 8.7 | 0.041 | 17 (32.2) | LOC100511790 |
| CASI0009812 | Fd5 | 5.36 | 8.4 | 0.045 | 4 (92.9) | |
| ASGA0040214 | Fd5 | 5.23 | 8.2 | 0.045 | 8 (141.7) | |
| ALGA0056876 | Fd12 | 6.60 | 8.9 | 0.007 | 10 (10.6) | |
| ASGA0102838 | Fd12 | 6.54 | 10.1 | 0.007 | 12 (55.6) | |
| ASGA0055092 | Fd12 | 6.21 | 9.6 | 0.010 | 12 (55.8) | PER1 |
| ALGA0066975 | Fd12 | 5.85 | 9.1 | 0.011 | 12 (55.9) | |
| ALGA0066986 | Fd12 | 5.82 | 9.0 | 0.011 | 12 (56.1) | |
| ALGA0115736 | Fd19 | 5.53 | 8.6 | 0.033 | 2 (1.5) | liprin-alpha-1-like |
| ASGA0102838 | Fd19 | 5.47 | 8.6 | 0.033 | 12 (55.6) | |
| ASGA0055092 | Fd19 | 5.46 | 8.5 | 0.033 | 12 (55.8) | PER1 |
| H3GA0056700 | Fd19 | 5.31 | 8.3 | 0.033 | ||
| ALGA0066975 | Fd19 | 5.23 | 8.2 | 0.033 | 12 (55.9) | |
| ALGA0096195 | Fd26 | 5.19 | 8.1 | 0.043 | 17 (64.1) | |
| ALGA0104320 | Fd26 | 5.11 | 8.0 | 0.043 | 1 (22.5) | |
| ALGA0107609 | Fd26 | 5.11 | 8.0 | 0.043 | 1 (22.3) | |
| ASGA0094670 | Fd26 | 5.11 | 8.0 | 0.043 | 1 (22.5) | |
| ASGA0055092 | Fd26 | 5.10 | 8.0 | 0.043 | 12 (55.8) | PER1 |
| ALGA0066975 | Ld12 | 6.47 | 9.9 | 0.006 | 12 (55.9) | |
| ASGA0055092 | Ld12 | 6.39 | 9.8 | 0.006 | 12 (55.8) | PER1 |
| ALGA0121951 | Ld12 | 6.34 | 9.7 | 0.006 | 12 (55.9) | |
| ALGA0066986 | Ld12 | 6.12 | 9.4 | 0.006 | 12 (56.1) | |
| ASGA0099599 | Ld12 | 6.12 | 9.4 | 0.006 | 12 (55.1) |
Genome-wide association analysis of response duration
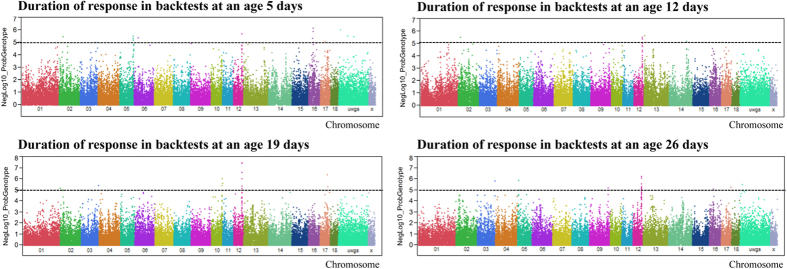
The top five markers significantly associated with Dd5, Dd12, Dd19 and Dd26 are shown in Table 2. Struggle duration at day 5 (Dd5) was significantly associated with 68 SNPs at FDR ≤ 5% (Fig. 2, Supplementary Table 2). These markers explained, on average, 7% of the phenotypic variance. The most prominent SNPs associated with Dd5 are located on SSC16 (31–32 Mb), SSC12 (55–56 Mb), and SSC5 (106–108 Mb). Two SNPs (ALGA0090009 and ALGA0090013) are located in a gene responsible for activating cyclic nucleotide-gated potassium channel 1 (HCN1) and are located on SSC16.
Figure 2. Genome-wide association with duration of response in backtests at ages of 5, 12, 19 and 26 days.
Black lines at negative log10 (NegLog10) of 5 corresponds to FDR ≤ 0.05.
We found that Dd12 was significantly associated with 100 SNPs (FDR ≤ 5%) (Fig. 2, Supplementary Table 2). At SSC12 from 55–56 Mb there were 13 markers of interest and on SSC13 and 14 there were 12 and 10, respectively. Two SNPs (ALGA0090009 and ALGA0090013) on SSC16 were also found to be positively associated with struggle duration on Dd12.
Fifty-six SNPs were significantly (FDR ≤ 5%) associated with struggle duration at Dd19 (Fig. 2, Supplementary Table 2). Twenty-three of the 56 were located between 53 and 56 Mb of SSC12. Seven SNPs were located at 72–75 Mb on SSC10 and 4 SNPs were on SSC17.
Thirty-seven of the 101 SNPs associated with struggle duration on day 26 were found on SSC12 (Fig. 2, Supplementary Table 2). SNPs associated with Dd26 were also found on SSC 3, 5, 9, 13, 16, and 17.
Genome-wide association analysis of struggle bout frequency
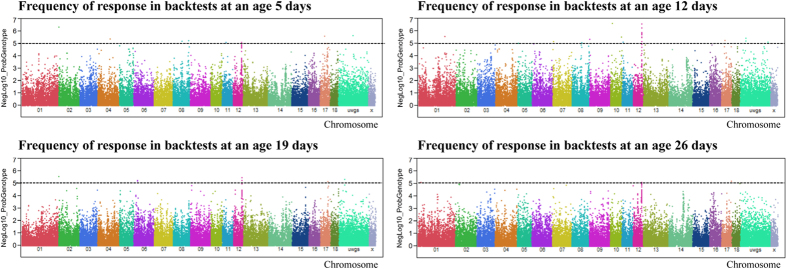
Struggling bout frequency (F) at ages 5 (Fd5), 12 (Fd12), 19 (Fd19), and 26 (Fd26) days was significantly associated with 96, 124, 49, and 80 SNPs, respectively, at FDR ≤ 5% (Fig. 3, Supplementary Table 3). These markers explained, on average, 7% of the phenotypic variance. The top five markers significantly associated with Fd5, Fd12, Fd19 and Fd26 are shown in Table 2. Most of the SNPs associated with the frequency of struggling bouts at day 5 were found on SSC17, SSC12, SSC5, and SSC8. One SNP is located directly in the transcript of liprin-alpha-1-like (LOC100738060) on SSC2 with p < 1 × 10−6. This SNP was also significantly associated with the frequency of struggling bouts at days 12 and 19. At day 12, the frequency of struggling bouts was prominently associated with SNPs between 52 and 56 Mb on SSC12. ALGA0056876, on SSC10, was most strongly associated (p < 1 × 10−6) with Fd12. The markers associated with Fd19 were located on SSC12 (53–56 Mb), SSC6 (25 Mb), and SSC17 (50–56 Mb). The SNPs (ASGA0055092, ALGA0066975, ALGA0121951) located on SSC12 were within PER1. We also found that Fd26 was significantly associated with SNPs on SSC17 (37 and 64 Mb), SSC1 (22 Mb), and SSC12 (53–56 Mb).
Figure 3. Genome-wide association with frequency of response in backtests at ages of 5, 12, 19 and 26 days.
Black lines at negative log10 (NegLog10) of 5 corresponds to FDR ≤ 0.05.
Genome-wide association analysis of total latency, duration, frequency, and coping type
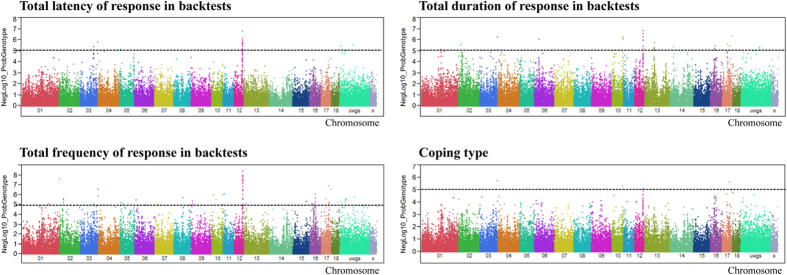
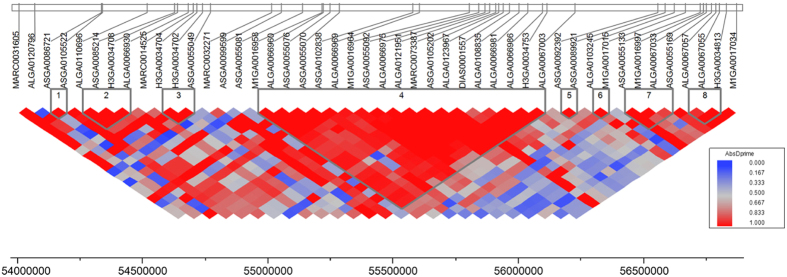
The top ten markers significantly associated with total latency (tL), total response duration (tD), and total response frequency (tF) across testing at each age are shown in Table 3. Total latency, total duration, and total frequency were associated with 26, 40, and 69 SNPs, respectively, with p < 1 × 10−5 and FDR < 5% (Fig. 4, Supplementary Table 4). Eighteen of the 26 SNPs on SSC12 localized to WRAP53, PER1, CTC1, RANGRF and one on SSC3 localized to ASGR1. Twelve of the 40 SNPs associated with total duration localized to SSC12 and 9 of the 40 SNPs to SSC13. Finally, 22 of the 69 SNPs associated with total frequency localized to SSC12, with additional SNPs on SSC17 and SSC16. Together, the most prominent regions associated with total latency, total duration, and total frequency were located between 53 and 56 Mb on SSC12. The LD structure of SNPs located on SSC12 at 54–56 Mb is shown in Fig. 5. LD block 4 was the largest block identified, and was located between 55 and 56 Mb.
Table 3. Markers significantly associated with backtest traits of total latency until the first response (tL), total duration of all responses (tD), total frequency of all responses (tF) and backtest type in piglets.
| SNP_ID | Trait | −log 10 | % variance | FDR | SSC (Mb) | Gene |
|---|---|---|---|---|---|---|
| DIAS0001557 | tL | 6.79 | 10.4 | 0.0073 | 12 (56.0) | RANGRF |
| ASGA0099599 | tL | 6.10 | 9.4 | 0.0101 | 12 (55.1) | |
| H3GA0034753 | tL | 5.96 | 9.2 | 0.0101 | 12 (56.1) | LOC100739126 |
| ALGA0066960 | tL | 5.83 | 9.0 | 0.0101 | 12 (55.2) | WRAP53 |
| M1GA0016958 | tL | 5.83 | 9.0 | 0.0101 | 12 (55.2) | WRAP53 |
| ALGA0066969 | tL | 5.83 | 9.0 | 0.0101 | 12 (55.6) | |
| ALGA0107834 | tL | 5.81 | 8.9 | 0.0101 | 3 (139.8) | ASGR1 |
| ASGA0055092 | tL | 5.70 | 8.8 | 0.0111 | 12 (55.8) | PER1 |
| ASGA0102838 | tL | 5.57 | 8.6 | 0.0111 | 12 (55.6) | |
| ALGA0027884 | tL | 5.56 | 8.6 | 0.0111 | ||
| ASGA0055092 | tF | 8.36 | 13.3 | 0.0002 | 12 (55.8) | PER1 |
| ALGA0066975 | tF | 7.99 | 12.8 | 0.0002 | 12 (55.9) | |
| ALGA0121951 | tF | 7.84 | 12.6 | 0.0002 | 12 (55.9) | |
| H3GA0034753 | tF | 7.79 | 12.5 | 0.0002 | 12 (56.1) | LOC100739126 |
| ASGA0102838 | tF | 7.57 | 12.2 | 0.0002 | 12 (55.6) | |
| ALGA0115736 | tF | 7.57 | 12.1 | 0.0002 | 2 (1.5) | liprin-alpha-1-like |
| ASGA0105202 | tF | 7.48 | 12.0 | 0.0002 | 12 (55.9) | CTC1 |
| MARC0073387 | tF | 7.48 | 12.0 | 0.0002 | 12 (55.9) | |
| M1GA0016964 | tF | 7.33 | 11.8 | 0.0002 | 12 (55.8) | |
| ALGA0066960 | tF | 7.16 | 11.5 | 0.0003 | 12 (55.2) | WRAP53 |
| ASGA0055092 | tD | 6.84 | 10.5 | 0.0039 | 12 (55.8) | PER1 |
| ASGA0102838 | tD | 6.58 | 10.1 | 0.0039 | 12 (55.6) | |
| ALGA0096195 | tD | 6.38 | 9.8 | 0.0039 | 17 (64.1) | |
| ALGA0066975 | tD | 6.27 | 9.6 | 0.0039 | 12 (55.9) | |
| MARC0035809 | tD | 6.25 | 9.6 | 0.0039 | 10 (72.9) | |
| ALGA0066986 | tD | 6.25 | 9.6 | 0.0039 | 12 (56.1) | |
| ALGA0107834 | tD | 6.24 | 9.6 | 0.0039 | 3 (139.8) | ASGR1 |
| ASGA0049082 | tD | 6.10 | 9.4 | 0.0044 | 10 (73.9) | |
| ALGA0060087 | tD | 6.07 | 9.3 | 0.0044 | 10 (73.6) | |
| ALGA0112457 | tD | 6.03 | 9.3 | 0.0044 | 6 (31.8) | LONP2 |
| ALGA0107834 | coping type | 5.72 | 8.8 | 0.0546 | 3 (139.8) | ASGR1 |
| ALGA0095323 | coping type | 5.63 | 8.6 | 0.0546 | 17 (50.7) | |
| MARC0035809 | coping type | 5.34 | 8.2 | 0.0712 | 10 (72.9) | |
| DIAS0001557 | coping type | 5.11 | 7.9 | 0.0807 | 12 (56.0) | RANGRF |
| ALGA0112457 | coping type | 5.03 | 7.7 | 0.0807 | 6 (31.8) | LONP2 |
| ALGA0096195 | coping type | 4.95 | 7.6 | 0.0807 | 17 (64.1) | |
| ASGA0055092 | coping type | 4.90 | 7.6 | 0.0807 | 12 (55.8) | PER1 |
| MARC0062446 | coping type | 4.76 | 7.4 | 0.0807 | 18 (2.9) | |
| ASGA0076061 | coping type | 4.65 | 7.2 | 0.0807 | 17 (32.2) | C17H20orf26 |
| ASGA0102838 | coping type | 4.62 | 7.2 | 0.0807 | 12 (55.6) |
Figure 4. Genome-wide association with total latency, duration, frequency of response in backtests and backtest type.
Black lines at negative log10 (NegLog10) of 5 corresponds to FDR ≤ 0.05.
Figure 5. Linkage disequilibrium (LD) of single nucleotide polymorphisms (SNPs) located on SSC12 between 54 and 56 Mb that were significantly associated with backtest results.
Color indicates the level of linkage disequilibrium between loci as depicted in the key. LD block 4 was the largest block located between 55 and 56 Mb.
Ten SNPs were associated with coping type at FDR < 8%. Manhattan plots of genome-wide association analyses and the markers associated with coping type are shown in Fig. 4 and Table 3. These SNPs are located on SSC3, SSC6, SSC12, and some non-annotate in genome.
Replication in second herd of crossbred piglets
Genotyping and association analysis of the most significant SNPs in PER1 (ASGA0055092) and CTC1 (ASGA0105202) in the first herd with 794 crossbred piglets from independent herds revealed significant association of ASGA0055092 in PER1 with all backtest traits except Fd19; marker ASGA0105202 in CTC1 was significantly associated with Fd12 only (Table 4).
Table 4. Markers which significantly associated with backtest traits were used for association study in a second herd for validation; latency (L), duration (D) and frequency (F) of responses in piglets were recorded at 12 and 19 days (d) of age as well as the totalizing data of both ages (tD, tL and tF).
| Trait | Sample Size | p-vaule | FDR | SNP | Gene |
|---|---|---|---|---|---|
| Dd12 | 794 | 0.185 | 0.185 | ASGA0105202 | CTC1 |
| Dd19 | 791 | 0.583 | 0.583 | ||
| tD | 791 | 0.190 | 0.190 | ||
| Fd12 | 794 | 0.017 | 0.017 | ||
| Fd19 | 794 | 0.718 | 0.718 | ||
| tF | 794 | 0.493 | 0.493 | ||
| Ld12 | 794 | 0.346 | 0.346 | ||
| Ld19 | 791 | 0.849 | 0.849 | ||
| tL | 791 | 0.657 | 0.657 | ||
| Dd12 | 788 | 0.015 | 0.030 | ASGA0055092 | PER1 |
| Dd19 | 785 | 0.003 | 0.006 | ||
| tD | 785 | 0.001 | 0.003 | ||
| Fd12 | 788 | 0.006 | 0.011 | ||
| Fd19 | 788 | 0.191 | 0.381 | ||
| tF | 788 | 0.009 | 0.017 | ||
| Ld12 | 788 | 0.007 | 0.015 | ||
| Ld19 | 785 | 0.035 | 0.069 | ||
| tL | 785 | 0.013 | 0.026 |
eQTL identification
We detected eQTLs of 37 genes all containing SNPs significantly associated with backtest traits in the hypothalamus, an organ whose secretions are often responsible for behavioral traits (Supplementary Table 5). 326 eQTL (association of 304 SNPs with abundance of 13 transcripts) were identified at FDR ≤ 5% (Supplementary Table 6). Eight transcripts (ASGR1, CPAMD8, CTC1, FBXO39, IL19, LOC100511790, RAD51B, and UBOX5) contained cis eQTL (average p = 4 × 10−7). Another five transcripts (RANGRF, PER1, PDZRN3, SH2D4B, and LONP2) harbored trans eQTL (average p = 1.2 × 10−6). Representation of the cis and trans eQTL is shown in Fig. 6. In total, 38 out of the 465 SNPs that were significantly associated with the 13 backtest traits were also significantly associated with transcript levels of CTC1, UBOX5, RAD51B, IL19, CPAMD, ASGR1 and FBXO39. Interestingly, all of them were cis eQTL.
Figure 6. Genome-wide association for the abundance of candidate transcripts in porcine hypothalamus, which were significantly associated with backtest traits.
Manhattan plots of genome-wide association analyses of transcript abundance of CTC1, FBXO39, PER1, RANGRF, IL19, RAD51B, UBOX5 and LOC10051790 are shown. Black lines at negative log10 (NegLog10) of 5 corresponds to FDR ≤ 0.05.
Discussion
The struggling response of piglets in a backtest early in life reflects the coping strategy and is a heritable and repeatable indicator of behavioral characteristics and of coping style also at later age. Some studies reported phenotypic and genetic correlation of backtests with other performance traits17,23. With a much larger data set of 3555 animals, of which the animals used here represent a subset, it was previously shown that the correlations among the backtest traits frequency, duration and latency of struggles were moderate to high at a single time point (rs = |0.63–0.78|) and moderate between each trait at different time points (rs = |0.19–0.44|); genetic correlations were high for all traits and time points (rg > 0.8); this strongly supports the backtest traits as reliable parameters of personality and coping style33. Cortisol levels in plasma have been shown to be both directly and indirectly related to behavioral, physiological, and metabolic disease15,34. Our previous study detected a major quantitative trait locus at the position of the glucocorticoid receptor gene (NR3C1). SNP c.1829C>T, leading to a p.Ala610Val substitution in the ligand binding domain, showed a large effect on cortisol level and adrenal weight35. The genome-wide association study did not reveal any association of this QTL with coping style. Accordingly, we did not find significant association between cortisol levels and coping style in pigs as did Reimert et al.19. Backtest data were recorded early in life (<26 days) and cortisol level much later (180 days) in our study. The lack of association might be due to experimental reasons: the time gap may obscure a possible association between corticoid plasma concentrations and backtest traits. However, we have previously shown that the repeated backtests reflect personality and coping strategy with a moderate intra-individual consistency and heritability33, making the time gap less relevant and suggesting that basal cortisol levels and coping strategies are at best loosely functionally linked properties.
To our knowledge this is the first report of a genome-wide association study (GWAS) for backtest traits related to coping style in pigs. Most of the associated markers displayed peaks of closely linked loci that point to particular genomic regions. We identified 893 associations (465 SNPs) for 13 backtest traits at FDR ≤ 5%. The genomic region most frequently associated with backtest traits is located on SSC12 between 55 and 56 Mb, a region with high LD. Because of the large LD block, we decided to analyse eQTL effects of transcripts mapping to this regions in hypothalamus, which has implications for behavioral traits. The hypothalamus releases neurochemical signals to the paraventricular nucleus of the thalamus with in turn affects behavior36. Direct links between hypothalamic gene expression and aggressive behavior were shown in chicken37. Expression-QTL studies have been reported to detect cis-eQTL signals for more than 20% of catalogued GWAS hits for neurological disorders38, corroborating its relevance to our study. We identified candidate genes positioned in genomic regions with numerous trait-associated SNPs and with cis eQTL (ASGR1, CPAMD8, CTC1, FBXO39, IL19, LOC100511790, RAD51B, UBOX5) or trans eQTL (RANGRF, PER1, PDZRN3, SH2D4B, LONP2). Some SNPs associated with backtest traits are located in transcripts with plausible functions like LOC100739126 (NDEL1), WRAP53, C17H20orf26, HCN1, FCHSD2, SHQ1, and LOC100738060 (liprin-alpha-1-like) but were not associated with altered transcript levels.
The 6 markers associated with all 13 backtest traits were ASGA0055092, ASGA0105202, H3GA0034753, ALGA0066975, ALGA0121951, and MARC0073387, located on SSC12 (55–56 Mb) within CTC1, PER1, and LOC100739126 (nuclear distribution protein nudE-like 1-like), i.e. genes with cis eQTL, trans-eQTL or plausible functions, respectively.
In fact, these SNPs were also associated with the transcript levels of CTC1 in hypothalamus as cis eQTL. CTC1 (CTS telomere maintenance complex component 1) encodes a component of the CST complex, which protects telomeres from degradation. Polymorphisms of CTC1 are causal for cerebroretinal microangiopathy39. The possible role of CTC1 for behavior remains unclear, though there are indications of impact on the central and peripheral neurons. We found that the allele A of ASGA0105202 in CTC1 is significantly more frequent in LR than HR; the allele is associated with higher expression of CTC1, and CTC1 is higher expressed in in LR than HR. This consistent tri-angular relationship of polymorphism of the gene, variation of the phenotype and expression of the gene evidences CTC1 as a gene contributing to genetic variation of coping behavior.
Indeed, eQTL complements the GWAS for backtest traits not only by pointing to the identical genomic regions but also to particular genes within these regions of high LD. ExpressionQTL, in particular cis eQTL, provides additional evidence for the association of the genes, as these genes harbor SNPs that are associated with the backtest traits and also with their own expression level, which is in turn associated with the backtest traits. Trait-associated SNPs are more likely to be eQTLs, in line with the role of regulatory SNPs in phenotypic variation. Consequently, the application of eQTL information can enhance discovery of trait-associated SNPs for complex phenotypes38,40.
The GWAS revealed an association between backtest traits and SNPs located in PER1 with appealing implications for behavioral traits. Period circadian clock 1 (PER1) is a member of the period family genes and encodes components of circadian rhythm responsible for locomotion, metabolism, and behavior. The circadian rhythm pathway plays a key role in the maintenance of various endocrine, physiological, and behavioral functions. PER1-deficient mice were shown to display alterations in glucocorticoid rhythmicity41, innate routine behavior42 and feeding behavior43. Interestingly, a polymorphism in the human PER1 was associated with corticoid induced expression and mutant mice showed increased alcohol drinking in response to social defeat linking stress response and circadian rhythmicity - both being adaptations to exogenous factors44. In our study transcripts levels of PER1 were regulated by a trans eQTL indicating that a yet to be identified non-regulatory polymorphism of PER1 might be causative; while polymorphisms at the sites of the trans eQTL affect the expression of PER1. The SNP rs10720116 analyzed here is a synonymous coding transition obviously in linkage disequilibrium with the anticipated underlying SNP. The lack of association of cortisol levels and backtest traits in particular further suggests that a putative causal polymorphism does not affect the glucocorticoid induction of PER1. The association of PER1 with backtest traits was shown here in two independent groups of animals, strongly promoting PER1 as a gene playing a significant role in coping behavior and linking coping behavior and a circadian rhythm-related gene shown to be involved in response to exogenous stimuli. Notable, no other circadian rhythm-related genes or SNP close to were detected by this GWAS.
The genes associated with backtest traits are related to neurogenesis and neuronal activity. For example, the nuclear distribution factor E-like gene regulates developmental neurogenesis and cortical neuronal positioning45,46. Genetic analyses revealed an association of nuclear distribution factor E (NDE1, or NudE) and its ortholog NDE-like 1 (NDEL1, or Nudel) with mental disorders47,48.
WRAP53 is located on SSC12 (55–56 Mb), a region associated with backtest traits, and play important roles in telomere functions affecting the cell cycle49,50. WRAP53 (WD repeat containing, antisense to TP53) has implications for the survival of motor neurons51. It encodes an essential component of the telomerase holoenzyme complex, is involved in the construction and maintenance of Cajal bodies, sub-organelles mediating RNA modifications.
Hyperpolarization-activated and cyclic nucleotide-gated (HCN) channels are encoded by four genes (HCN1-4) and are widely expressed throughout the heart and central nervous system. HCN1 is the predominant isoform known to be expressed in the hippocampus, neocortex, and cerebellar cortex52. HCN channels are involved in neuron excitation and synaptic activity in individual neurons and neuronal networks53. It has been reported that HCN1 channels constrain learning and memory and are involved in many diseases such as epilepsy, Alzheimer’s disease, and peripheral neuralgia54,55,56. Similarly, Liprin-α proteins are major protein constituents of synapses and contribute to synaptic transmission in different regions of the brain57,58. Knockdown of Liprin-α was reported to lead to a defect in presynaptic development and function59. The SNPs located in both HCN and Liprin-α were associated with coping style in this study but not with their transcript levels.
Other genes harboring cis eQTL, such as UBOX5 and FBXO39, encode U box and F box protein and act as protein-ubiquitin ligases60,61. RAD51 family members function in both mitotic and meiotic homologous recombination and DNA repair. RAD51B has been shown to be associated with breast cancer and Werner syndrome, which features progressive neurodegeneration62,63. The SNPs located in these 3 genes associated with coping behavior and also with their transcripts levels as cis eQTL.
An additional region of interest was SSC3 (139.8–139.9 Mb), with 3 SNPs (ALGA0107834/ASGR1, ALGA0066945, and ALGA0066946) associated with 8–13 backtest traits. The transcript levels of ASGR1 were also associated with ALGA0066945 and ALGA0066946. ASGR1 encodes a subunit of the asialoglycoprotein receptor and mediates the capture and endocytosis of galactose- (Gal) and N-acetylgalactosamine- (GalNAc) terminating glycoproteins. ASGR1 is highly expressed in hepatocytes64 and much weaker in other tissues including thyroid65 and peripheral blood monocytes66. Asialoglycoprotein receptors are involved in thyroglobulin transport, providing a link to thyroid gland hormones with implications for behavior67. We found that ASGR1 is expressed in the hypothalamus and demonstrated that its transcripts levels are associated with SNP markers, which are also associated with coping style in pigs.
It was recently suggested that an imbalance of the immune system plays an important role in psychological stress and behavior37,68. SNPs located within the CPAMD8 and IL9 genes, both related to immune function, were associated with coping behavior and transcript levels (cis eQTL) in this study. CPAMD8 is a member of the complement 3/alpha(2)-macroglobulin (C3/alpha(2)M) family and has functions in innate immunity69. It has been demonstrated that psychological stress enhances the production of immunosuppressive cytokines including IL19 via activation of beta-adrenoreceptors, which may impact stress-related disease processes68. Recently, it was demonstrated that the putative cis-acting haplotype of the IL19 gene is associated with schizophrenia70. Together, the SNPs located in the CPAMD8 and IL9 genes were found to be associated with coping style, suggesting a relationship between the immune system and behavior.
Our results represent the first genome-wide SNP-based association analysis for behavioral traits related to coping type. Hypothalamic genes were identified within QTL regions showing considerable linkage disequilibrium using eQTL. The most highly associated region was SSC12 (53–56 Mb) harboring cis eQTL signals (CTC1 and FBXO39) and trans eQTL signals (PER1 and RANGRF). Other cis eQTL signals included ASGR1 located on SSC3, IL19 and CPAMD8 located on SSC2, and UBOX5 and LOC10051179 located on SSC17. SNPs located on HCN1 (SSC16) and Liprin-α like (SSC2), which have been linked to neuropathological disorders, showed no cis or trans eQTL signals. The genes identified in this study are known to be involved in telomere function signaling, immune function, ubiquitin system as well as the circadian rhythm pathway and neurotransmitter receptors, i.e. central nervous affairs related to behavior, offering perspectives to derive markers for breeding prediction. In particular, for CTC1 and PER1 with known physiological implications in the central nervous system, their role in behavior was evidenced by cis-eQTL and confirmed association in an independent population.
Methods
Ethics statement
All experiments were performed in accordance with relevant regulations and guidelines. Animals were provided by the Leibniz Institute for Farm Animal Biology (FBN). Animal care and tissue collection procedures followed the guidelines of the German Law of Animal Protection, and the experimental protocol was approved by the Animal Care Committee of the Leibniz Institute for Farm Animal Biology and the State Mecklenburg-Western Pomerania (Landesamt für Landwirtschaft, Lebensmittelsicherheit und Fischerei; LALLF M-V/TSD/7221.3-2.1-020/09). The experimental protocol was carried out in accordance with the approved guidelines for safeguarding good scientific practice at the institutions in the Leibniz Association.
Animals, sample collection and trait measurement
German Landrace (DL) piglets (N = 294) were subjected to a backtest as previously described33. Briefly, we repeatedly performed a 1-min-backtest in domestic piglets at the ages of 5, 12, 19, and 26 days. Animals were put on their backs in a special V-shaped device. The test lasted for exactly 60 s and started as soon as the piglet was lying immobile in a supine position. The latency (L, time to first struggle), duration (D, cumulative time of struggling within the 1-min testing period), and frequency (F) of struggling bouts were recorded. The results from each parameter at each age tested were added together to reflect a summation of total latency (tL), total duration (tD), and total frequency (tF) for each individual piglet. Piglets were classified as high (HR), indistinct (IR), or low (LR) in reactivity, according to the following criteria: the latency of HR was ≤5 s and LR ≥ 35 s, the duration of HR was ≥25 s and LR ≤ 5 s, and the frequency of HR was ≥4 and LR ≤ 171.
Cortisol measurement
Total cortisol secretion was measured in the morning via the collection of 50 mL trunk blood from each pig during exsanguinations at about 180 days of age. Blood was collected in a plastic tube containing 1 mL of 0.5 M EDTA, stored on ice until plasma isolation, then stored long-term at −80 °C. Total plasma cortisol levels were determined using an enzyme-linked immunosorbent assay (DRG, Marburg, Germany) in duplicate according to the manufacturer’s protocol. The intra- and inter-assay coefficients of variation were ≤7.0% and 9.8%, respectively. Spearman’s rank correlation coefficient (Spearman’s rho, rs) and association between cortisol levels and coping behavior were also calculated.
SNP genotyping
Genotyping was performed using the PorcineSNP60 BeadChip (Illumina Inc., San Diego, CA, USA) per manufacturer’s SNP Infinium HD assay protocol. In brief, 200 ng of DNA were amplified, fragmented, and hybridized to the PorcineSNP60 BeadChip containing 62,163 locus-specific 50-mers. Intensity data were measured and quality scores and genotypes were derived using the GenomeStudio software (Illumina Inc.). Call rates (<99%), minor-allele frequency (<5%), and deviation from Hardy-Weinberg equilibrium (p < 0.0001) were considered to fix the final set of markers for GWAS. The average call rate for all samples was 99.8 ± 0.2. The markers of the 60 K chip were mapped to the porcine reference genome, Sscrofa 10.2 (http://www.ensembl.org/).
Genome wide association of coping behavior
The backtest parameters, latency and duration data were not normally distributed. These traits were converted into classes enabling Poisson distribution analysis. According to latency and duration, animals were assigned to four L-classes: class 1: 1–10 s; class 2: 11–20 s; class 3: 21–30 s; class 4: 31–60 s and five D-classes: class 1: 0 s; class 2: 1–10 s; class 3: 11–20 s; class 4:21–30 s; class 5: 31–60 s.
All backtest traits (latency, duration, and frequency) at ages 5, 12, 19, and 26 days, as well as the classification of piglets (coping type) based on backtest data, were associated with SNPs by generalized linear mixed models (Proc Glimmix) using JMP Genomics (SAS Institute, Cary, NC, USA). The genetic similarity matrix between individuals was first computed as identity by descent of each pair for the k matrix and used as a random effect. Using top principal components as covariates enabled correction for stratification in the population72,73. Additionally, genotype and gender were used as fixed effects, day of birth was used as a random effect, and piglet weight was used as a covariate. A genotype test for association of SNP alleles was also performed. To correct for multiple testing, a false discovery rate (FDR) of 5% was used.
Replication in a second herd of cross-bred piglets
In total 794 cross-bred animals of the breeds German Landrace (DL) and Large White (DE) were used. The data were collected from research farm of the Institute of Animal Breeding and Husbandry of the University Kiel. The 1-min-backtests were repeatedly performed at an age of 12 and 19 days. The most significant SNPs in PER1 (ASGA0055092) and CTC1 (ASGA0105202) were used for genotyping by pyrosequencing. For association analysis generalized linear mixed models (Proc Glimmix; JMP Genomics (SAS Institute, Cary, NC, USA)) were used with the kindship matrixes of individuals considered as random effect, genotype and gender as fixed effects. To correct for multiple testing, a false discovery rate (FDR) was used.
Quantitative PCR
We measured the levels of 37 hypothalamus transcripts (Supplementary Table S5) from 184 animals with tissue samples available using quantitative PCR (qPCR) with the Fluidigm BioMark HD System. The genes selected for analysis harbor SNPs represented on the Illumina PorcineSNP60 BeadChip that showed significant association with backtest traits. Samples for transcript abundance measurements were taken immediately after exsanguinations at about 180 day of age and snap frozen in liquid nitrogen until use. For qPCR, pre-amplification sample mixtures were prepared containing 2.7 μL of the diluted cDNA and primers (Supplementary Table S5), 3 μL SSO Evagreen supermix (BioRad), and 0.3 μL 20× sample-loading reagent. Separately, an assay mixture was prepared for each primer pair and this included 2.25 μL assay loading reagent, 0.25 μL 100 μM forward and reverse primer, and 2.5 μl 2× assay-loading reagent. The dynamic array was first primed with control line fluid and then loaded with the sample and assay mixtures via the appropriate inlets using an IFC controller. The array chip was placed in the BioMark Instrument for PCR at 95 °C for 10 min, followed by 30 cycles at 95 °C for 15 s and 60 °C for 1 min. Each sample was investigated in duplicate. The data were analyzed with real-time PCR analysis software in the BioMark HD instrument (Fluidigm Corporation, San Francisco, CA). Data were normalized using HPRT1 and RPS11 as internal controls.
eQTL detection
The expression levels of 37 transcripts from the hypothalamus of 184 individuals representing a subset of those used for backtests were subjected to a mixed-model analysis of variance using JMP Genomics (Proc Mixed; SAS Institute, Rockville, MD). As in the GWAS of backtest traits, an IBD matrix between individuals was used as a random effect and top principal components as covariates corrected for stratification in the population. Additionally, genotype, gender, and the batch of RT-PCR were used as fixed effects, and weight at slaughter and average mean of housekeeping gene CT values were used as covariates. To correct for multiple testing, a false discovery rate (FDR) at 5% was used. We defined an eQTL as ‘cis’ if an associated SNP was located within an area less than 1 Mb from the gene. All other eQTL were considered as ‘trans’.
Additional Information
How to cite this article: Ponsuksili, S. et al. Integrated Genome-wide association and hypothalamus eQTL studies indicate a link between the circadian rhythm-related gene PER1 and coping behavior. Sci. Rep. 5, 16264; doi: 10.1038/srep16264 (2015).
Supplementary Material
Acknowledgments
The authors thank Annette Jugert, Hannelore Tychsen and Joana Bittner for excellent technical help. This work was partly funded by the Federal Ministry of Education and Research (BMBF; www.bmbf.de) as part of the PHENOMICS project (support code: 0315536G; www.phaenomics.auf.uni-rostock.de). The Leibniz Institute for Farm Animal Biology (FBN) provided own matched funding.
Footnotes
Author Contributions S.P. and K.W. conceived the study. S.P. analysed the data and drafted the manuscript; M.Z., B.P. and J.K. performed backtests and data collection. M.Z., B.P., E.M., N.T., J.K., S.M. and K.W. discussed and contributed to data interpretation and helped in drafting the manuscript.
References
- Dingemanse N. J. & Réale D. Natural selection and animal personality. Behaviour 142, 1159–1184 (2005). [Google Scholar]
- Koolhaas J. M. Coping style and immunity in animals: making sense of individual variation. Brain Behav Immun 22, 662–667 (2008). [DOI] [PubMed] [Google Scholar]
- Koolhaas J. M., de Boer S. F., Coppens C. M. & Buwalda B. Neuroendocrinology of coping styles: towards understanding the biology of individual variation. Front Neuroendocrinol 31, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- Koolhaas J. M. et al. Coping styles in animals: current status in behavior and stress-physiology. Neurosci Biobehav Rev 23, 925–935 (1999). [DOI] [PubMed] [Google Scholar]
- Bohnen N., Nicolson N., Sulon J. & Jolles J. Coping style, trait anxiety and cortisol reactivity during mental stress. J Psychosom Res 35, 141–147 (1991). [DOI] [PubMed] [Google Scholar]
- O’Donnell K., Badrick E., Kumari M. & Steptoe A. Psychological coping styles and cortisol over the day in healthy older adults Psychoneuroendocrinology 33, 601–611 (2008). [DOI] [PubMed] [Google Scholar]
- Geverink N. A., Schouten W. G. P., Gort G. & Wiegant V. M. Individual differences in aggression and physiology in peri-pubertal breeding gilts. Appl Anim Behav Sci 77, 43–52 (2002). [Google Scholar]
- Hessing M. J., Hagelsø A. M., Schouten W. G., Wiepkema P. R. & van Beek J. A. Individual behavioral and physiological strategies in pigs. Physiol Behav 55, 39–46 (1994). [DOI] [PubMed] [Google Scholar]
- Frädrich H. A comparison of behaviour in the Suidae. The Behaviour of Ungulates and its Relation to Management 1, 133–143 (1974). [Google Scholar]
- Désautés C. et al. Genetic linkage mapping of quantitative trait loci for behavioral and neuroendocrine stress response traits in pigs. J Anim Sci 80, 2276–2285 (2002). [DOI] [PubMed] [Google Scholar]
- Kramer M. et al. Accuracy of direct genomic values for functional traits in Brown Swiss cattle. J Dairy Sci 97, 1774–1781 (2014). [DOI] [PubMed] [Google Scholar]
- Hessing M. J. C. et al. Individual behavioural characteristics in pigs Applied Animal Behaviour Science 37, 285–295 (1993). [DOI] [PubMed] [Google Scholar]
- Spoolder H. A. M., Burbidge J. A., Lawrenceb A. B., Simminsa P. H. & Edwardsb S. A. Individual behavioural differences in pigs: intra-and inter-test consistency. Applied Animal Behaviour Science 49, 185–198 (1996). [Google Scholar]
- Thodberg K., Jensen K. H. & M. S. H. A general reaction pattern across situations in prepubertal gilts. Applied Animal Behaviour Science 63, 103–119 (1999). [Google Scholar]
- Muráni E. et al. Association of HPA axis-related genetic variation with stress reactivity and aggressive behaviour in pigs. BMC Genet 11, 74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. P. et al. Genetic validation of postmixing skin injuries in pigs as an indicator of aggressiveness and the relationship with injuries under more stable social conditions. J Anim Sci 87, 3076–3082 (2009). [DOI] [PubMed] [Google Scholar]
- Camerlink I., Ursinus W. W. & Bolhuis J. E. Struggling to survive: early life challenges in relation to the backtest in pigs. J Anim Sci 92, 3088–3095 (2014). [DOI] [PubMed] [Google Scholar]
- Geverink N. A., Heetkamp M. J., Schouten W. G., Wiegant V. M. & Schrama J. W. Backtest type and housing condition of pigs influence energy metabolism. J Anim Sci 82, 1227–33 (2004). [DOI] [PubMed] [Google Scholar]
- Reimert I., Bolhuis J. E., Kemp B. & Rodenburg T. B. Social support in pigs with different coping styles. Physiol Behav 129, 221–229 (2014). [DOI] [PubMed] [Google Scholar]
- Bolhuis J. E., Schouten W. G. P., Schrama J. W. & Wiegant V. M. Individual coping characteristics, aggressiveness and fighting strategies in pig. Animal Behaviour 69, 1085–1091 (2005). [Google Scholar]
- Rohrer G. A., Brown-Brandl T., Rempel L. A., Schneider J. F. & Holl J. Genetic analysis of behavior traits in swine production. Livest. Sci 157, 28–37 (2013). [Google Scholar]
- Scheffler K., Stamer E., Traulsen I. & Krieter J. Genetic analysis of the individual pig behaviour in backtests and human approach tests. Appl Anim Behav Sci 160, 38–45 (2014). [Google Scholar]
- Velie B. D., Maltecca C. & Cassady J. P. Genetic relationships among pig behavior, growth, backfat, and loin muscle area. J Anim Sci 87, 2767–2773 (2009). [DOI] [PubMed] [Google Scholar]
- Flint J. Analysis of quantitative trait loci that influence animal behavior. J Neurobiol 54, 46–77 (2003). [DOI] [PubMed] [Google Scholar]
- Juhasz G. et al. Brain galanin system genes interact with life stresses in depression-related phenotypes. Proc Natl Acad Sci USA 111, 1666–1673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisan M. P. Genotype-phenotype associations in understanding the role of corticosteroid-binding globulin in health and disease animal models. Mol Cell Endocrinol 316, 35–41 (2010). [DOI] [PubMed] [Google Scholar]
- Bode G. et al. The utility of the minipig as an animal model in regulatory toxicology. J Pharmacol Toxicol Methods 62, 196–220 (2010). [DOI] [PubMed] [Google Scholar]
- Houston R. D., Haley C. S., Archibald A. L. & Rance K. A. A QTL affecting daily feed intake maps to Chromosome 2 in pigs. Mamm Genome 16, 464–470 (2005). [DOI] [PubMed] [Google Scholar]
- Quilter C. R. et al. Porcine maternal infanticide as a model for puerperal psychosis. Am J Med Genet B Neuropsychiatr Genet 144, 862–868 (2007). [DOI] [PubMed] [Google Scholar]
- Reiner G. et al. Mapping of quantitative trait loci affecting behaviour in swine. Anim Genet 40, 366–376 (2009). [DOI] [PubMed] [Google Scholar]
- Wimmers K., Murani E. & Ponsuksili S. Functional genomics and genetical genomics approaches towards elucidating networks of genes affecting meat performance in pigs. Brief Funct Genomics 9, 251–258 (2010). [DOI] [PubMed] [Google Scholar]
- Ponsuksili S., Murani E., Trakooljul N., Schwerin M. & Wimmers K. Discovery of candidate genes for muscle traits based on GWAS supported by eQTL-analysis. Int J Biol Sci 10, 327–337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebunke M., Repsilber D., Nürnberg G., Wittenburg D. & Puppe B. The backtest in pigs revisited – An analysis of intra-situationalbehaviour. Appl. Anim. Behav. Sci. 169, 17–25 (2015). [Google Scholar]
- Muráni E. et al. Differential mRNA expression of genes in the porcine adrenal gland associated with psychosocial stress. J Mol Endocrinol 46, 165–174 (2011). [DOI] [PubMed] [Google Scholar]
- Murani E., Reyer H., Ponsuksili S., Fritschka S. & Wimmers K. A substitution in the ligand binding domain of the porcine glucocorticoid receptor affects activity of the adrenal gland. PLoS One 7, e45518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac G. J. Placing the paraventricular nucleus of the thalamus within the brain circuits that control behavior. Neuroscience & Biobehavioral Reviews 56, 315–329 (2015). [DOI] [PubMed] [Google Scholar]
- Brunberg E., Jensen P., Isaksson A. & Keeling L. Feather pecking behavior in laying hens: hypothalamic gene expression in birds performing and receiving pecks. Poult Sci 90, 1145–1152 (2011). [DOI] [PubMed] [Google Scholar]
- Ramasamy A. et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 17, 1418–1428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polvi A. et al. Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am J Hum Genet 90, 540–549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae D. L. et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6, e1000888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann R., Touma C., Palme R., Albrecht U. & Steinlechner S. Impaired daily glucocorticoid rhythm in Per1 (Brd) mice. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 192, 769–775 (2006). [DOI] [PubMed] [Google Scholar]
- Bechstein P. et al. The clock gene Period1 regulates innate routine behaviour in mice. Proc Biol Sci 281, 20140034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. et al. Characterizing dynamic changes in the human blood transcriptional network. PLoS Comput Biol 6, e1000671 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L. et al. Effects of the circadian rhythm gene period 1 (per1) on psychosocial stress-induced alcohol drinking. Am J Psychiatry 168, 1090–1098 (2011). [DOI] [PubMed] [Google Scholar]
- Pei Z. et al. The expression and roles of Nde1 and Ndel1 in the adult mammalian central nervous system. Neuroscience 271, 119–136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T. et al. Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263–277 (2004). [DOI] [PubMed] [Google Scholar]
- Bakircioglu M. et al. The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. Am J Hum Genet 88, 523–535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingason A. et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry 16, 17–25 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M. & Blackburn E. H. The telomere syndromes. Nat Rev Genet 13, 693–704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Belcher M. & van der Harst P. Healthy aging and disease: role for telomere biology? Clin Sci 120, 427–440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi S. et al. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol 8, e1000521. (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster A. L. et al. Quantitative analysis and subcellular distribution of mRNA and protein expression of the hyperpolarization-activated cyclic nucleotide-gated channels throughout development in rat hippocampus. Cereb Cortex 17, 702–712 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch E. E. HCN channels: function and clinical implications. Neurology 80, 304–310 (2013). [DOI] [PubMed] [Google Scholar]
- Herrmann S., Stieber J. & Ludwig A. Pathophysiology of HCN channels. Pflugers Arch 454, 517–522 (2007). [DOI] [PubMed] [Google Scholar]
- Reid C. A., Phillips A. M. & Petrou S. HCN channelopathies: pathophysiology in genetic epilepsy and therapeutic implications. Br J Pharmacol 165, 49–56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Alonso J. L., Halliwell J. V. & Colino A. ZD 7288 inhibits T-type calcium current in rat hippocampal pyramidal cells. Neurosci Lett 439, 275–280 (2008). [DOI] [PubMed] [Google Scholar]
- Spangler S. A. & Hoogenraad C. C. Liprin-alpha proteins: scaffold molecules for synapse maturation. Biochem Soc Trans 35, 1278–1282 (2007). [DOI] [PubMed] [Google Scholar]
- Spangler S. A. et al. Differential expression of liprin-α family proteins in the brain suggests functional diversification. J Comp Neurol 519, 3040–3060 (2011). [DOI] [PubMed] [Google Scholar]
- Spangler S. A. et al. Liprin-α2 promotes the presynaptic recruitment and turnover of RIM1/CASK to facilitate synaptic transmission. J Cell Biol 201, 915–928 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev 18, 2573–2580 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson C. A new gun in town: the U box is a ubiquitin ligase domain. Sci STKE 116, pe4 (2002). [DOI] [PubMed] [Google Scholar]
- Jeppesen D. K., Bohr V. A. & Stevnsner T. DNA Repair Deficiency in Neurodegeneration. Prog Neurobiol 94, 166–200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr N. et al. Genome-wide association study identifies a common variant in RAD51B associated with male breast cancer risk. Nat Genet 44, 1182–1184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. et al. A new splice variant of the major subunit of human asialoglycoprotein receptor encodes a secreted form in hepatocytes. PLoS One 5, e12934 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifico F., Liguoro D., Acquaviva R., Formisano S. & Consiglio E. Thyroglobulin binding and TSH regulation of the RHL-1 subunit of the asialoglycoprotein receptor in rat thyroid. Biochimie 81, 493–496 (1999). [DOI] [PubMed] [Google Scholar]
- Harris R. L., van den Berg C. W. & Bowen D. J. ASGR1 and ASGR2, the genes that encode the asialoglycoprotein receptor (Ashwell Receptor), are expressed in peripheral blood monocytes and show interindividual differences in transcript profile. Mol Biol Int 2012, 283974 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinò M. & McCluskey R. T. Role of thyroglobulin endocytic pathways in the control of thyroid hormone release. Am J Physiol Cell Physiol 279, 1295–1306 (2000). [DOI] [PubMed] [Google Scholar]
- Curtin N. M., Mills K. H. & Connor T. J. Psychological stress increases expression of IL-10 and its homolog IL-19 via beta-adrenoceptor activation: reversal by the anxiolytic chlordiazepoxide. Brain Behav Immun 23, 371–379 (2009). [DOI] [PubMed] [Google Scholar]
- Li Z. F., Wu X. H. & Engvall E. Identification and characterization of CPAMD8, a novel member of the complement 3/alpha2-macroglobulin family with a C-terminal Kazal domain. Genomics 83, 1083–1093 (2004). [DOI] [PubMed] [Google Scholar]
- Okazaki S. et al. Association analysis of putative cis-acting polymorphisms of interleukin-19 gene with schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry, 151–156 (2014). [DOI] [PubMed] [Google Scholar]
- Zebunke M., Nuernberg G., Repsilber D. & Puppe B. The backtest in pig-individual variation in behaviour or behavioural strategy? KTBL-Schrift 503, 188–197 (2013). [Google Scholar]
- Price A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- Zhu X., Zhang S., Zhao H. & Cooper R. S. Association mapping, using a mixture model for complex traits. Genet Epidemiol 23, 181–196 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.