Abstract
Background. Epidemiological studies suggest that, following infection with influenza virus, there is a short period during which a host experiences a lower susceptibility to infection with other influenza viruses. This viral interference appears to be independent of any antigenic similarities between the viruses. We used the ferret model of human influenza to systematically investigate viral interference.
Methods. Ferrets were first infected then challenged 1–14 days later with pairs of influenza A(H1N1)pdm09, influenza A(H3N2), and influenza B viruses circulating in 2009 and 2010.
Results. Viral interference was observed when the interval between initiation of primary infection and subsequent challenge was <1 week. This effect was virus specific and occurred between antigenically related and unrelated viruses. Coinfections occurred when 1 or 3 days separated infections. Ongoing shedding from the primary virus infection was associated with viral interference after the secondary challenge.
Conclusions. The interval between infections and the sequential combination of viruses were important determinants of viral interference. The influenza viruses in this study appear to have an ordered hierarchy according to their ability to block or delay infection, which may contribute to the dominance of different viruses often seen in an influenza season.
Keywords: temporary immunity, viral interference, A(H1N1)pdm09, seasonal influenza, hierarchy, ferret, influenza, pandemic, influenza B, A(H3N2)
(See the editorial commentary by Schultz-Cherry on pages 1690–1.)
Viral interference, whereby infection with one virus limits infection and replication of a second virus, has been described in humans [1–7]. Within populations, different respiratory viruses reach their epidemic peaks at different times [8–11]. In 2009, separate peaks of rhinovirus, influenza A(H1N1)pdm09 virus, and respiratory syncytial virus (RSV) infections were observed in Norway and France, although all 3 viruses were cocirculating at low levels [10–12]. Separate peaks of infection with different influenza virus (sub)types were also observed in 2009. Influenza A(H3N2) viruses were followed by A(H1N1)pdm09 viruses in South Africa [13], while A(H3N2), A(H1N1)pdm09, and influenza B viruses circulated sequentially in Beijing [14].
Viral interference, defined as a state of temporary immunity from infection, is proposed to be induced by viral infection [7]. By preventing infection with seasonal influenza viruses, influenza vaccination may prevent induction of temporary immunity that could protect against subsequent respiratory infections [7, 15, 16]. Epidemiological studies have shown that preventing seasonal influenza virus infection by vaccination increased the risk of infection with A(H1N1)pdm09 virus in Canada and Japan [17–19]. Similarly, in a randomized controlled trial, influenza vaccine recipients displayed higher rates of infection with noninfluenza respiratory viruses, compared with unvaccinated subjects [20, 21]. This effect is observed only within specific periods [7, 16], estimated to be weeks to months [20, 22]. The interval between initial infection and subsequent exposure may explain the different findings between epidemiological studies [7, 17].
Limited animal studies provide evidence for viral interference. Separation of a primary infection and a secondary challenge with different respiratory viruses by 3 days delayed shedding of the second virus in pigs and poultry [23, 24] or reduced mortality in chickens [25]. When infections of different respiratory viruses were separated by weeks in mice, morbidity and mortality were reduced [26]. Cross-protective adaptive immunity directed to common epitopes between influenza A virus subtypes has been associated with protection in the ferret and guinea pig models, with intervals of at least 21 days between infections [27–29].
Overall, epidemiological, modeling and animal studies provide observational and inferential support for the existence of viral interference. However, the duration and extent of this state of temporary immunity is unclear, as is the importance of the antigenic relationship between respiratory viruses. In this study, we used the ferret model of human influenza to investigate viral interference and its effect on the probability and kinetics of subsequent infection. We assessed the potential for viral interference between antigenically unrelated influenza virus types (A and B), for which few T and B lymphocyte epitopes are similar [30], and between influenza virus subtypes that share T and B lymphocyte epitopes (A[H3N2] and A[H1N1]pdm09 viruses). Of direct relevance to recent epidemiological reports, combinations of influenza viruses that circulated in 2009 and 2010 were assessed.
MATERIALS AND METHODS
Ferrets
Adult ferrets (weight, 500–1500 g) were housed at bioCSL under a Support Services Agreement with the Victorian Infectious Diseases Reference Laboratory. Ferrets were seronegative (hemagglutination inhibition [HI] titer, <10) to currently circulating influenza virus strains before use. Experiments were conducted with approval from the CSL Limited/Pfizer Animal Ethics Committee, in accordance with the National Health and Medical Research Council, Australia, code of practice for the care and use of animals for scientific purposes.
Viruses and Cells
A/Tasmania/2004/2009 (A[H1N1]pdm09), A/Perth/16/2009 A(H3N2) and B/Brisbane/1/2007 (B/Florida/4/2006-like; B/Yamagata lineage) viruses were passaged in the allantoic cavity of embryonated hen eggs and stored at −80°C. Infectious virus was measured by 50% tissue culture infectious doses (TCID50) assays [31], using hemagglutination as the read-out.
Virus Infection, Sampling, and Monitoring of Ferrets
Ferrets were anaesthetized (12.5 mg/kg ketamine and 2.5 mg/kg Ilium Xylazil-20 in a 1:1 [v/v] mixture; Troy Laboratories) and inoculated by dropwise delivery of 103.5 TCID50 of virus in 0.5 mL into the nostrils. After infection, ferrets were housed individually in a high-efficiency particulate arrestance (HEPA)-filtered isolation unit. Blood samples were obtained from ferrets before the primary virus infection and at the termination of the experiment. Animals were weighed and visually inspected, and their temperature was measured daily by using implanted temperature transponders fitted to identification chips (LifeChip Bio-Thermo, Digivet). Nasal wash specimens were collected and stored as previously described [32]. On the day of collection, RNA was extracted from 140 µL of nasal wash for real-time reverse transcription–polymerase chain reaction (RT-PCR) analysis.
Real-Time RT-PCR Quantification of Viral Load in Ferret Nasal Wash Samples
RNA was extracted from nasal wash as previously described [32]. A total of 4 µL RNA was assayed by real-time RT-PCR with influenza virus–specific primers and probes, using the SensiFAST Probe Lo-ROX One-Step Kit, according to the manufacturer's instructions (Bioline), on the 7500 Fast Real-Time system (Applied Biosystems). Primers and probes were from the Centers for Disease Control and Prevention (CDC) Influenza Virus Real-Time RT-PCR Influenza A (H1/H3/H1pdm09) Subtyping Panel and the CDC Influenza B Lineage Genotyping Panel, obtained from the Influenza Reagent Resource (available at: http://www.influenzareagentresource.org/).
Definitions of Infection Measurements and Statistical Analyses
Viral kinetics were assessed using real-time RT-PCR data. Infection was defined as a challenge virus load of >106 copies/100 µL of nasal wash for at least 1 measurement, blocking/prevention was defined as a challenge virus load of <106 copies/100 µL of nasal wash for all measurements, and coinfection was defined as primary virus and challenge virus loads of >106 copies/100 µL of nasal wash for at least 1 day.
For statistical analysis, ferrets infected with the challenge virus were split into 2 groups: those in which primary infection persisted following challenge and those in which primary infection subsided and was below the threshold for all days following challenge. Control ferrets were excluded. The times from challenge to (1) the start of shedding of challenge virus and (2) the peak of shedding were calculated for each ferret, and the group median value was determined. The difference in median values between groups was analyzed using the Wilcoxon rank sum test, with the significance level set at an α of 0.05.
Linear regression was performed on the number of days from challenge to the start/peak of shedding for the challenge virus versus the number of days for which the primary infection persisted following challenge, and a 95% confidence interval (CI) for the gradient was obtained; this indicates the interval that the start/peak of shedding for the challenge virus was delayed for each day that the primary infection persisted following challenge. The null hypothesis, that there is no delay, was tested, with the significance level set at an α of 0.05.
The duration of infection with the challenge virus was compared for ferrets first infected and challenged with heterosubtypic influenza A viruses (when challenge was successful) and corresponding control ferrets; the difference in the median value was analyzed using the Wilcoxon rank sum test, with the significance level set at an α of 0.05. Statistical analysis was conducted using R, version 2.15.1 [33].
RESULTS
Infection With Influenza Virus Can Prevent Subsequent Infection With a Different Influenza Type Within Short Intervals
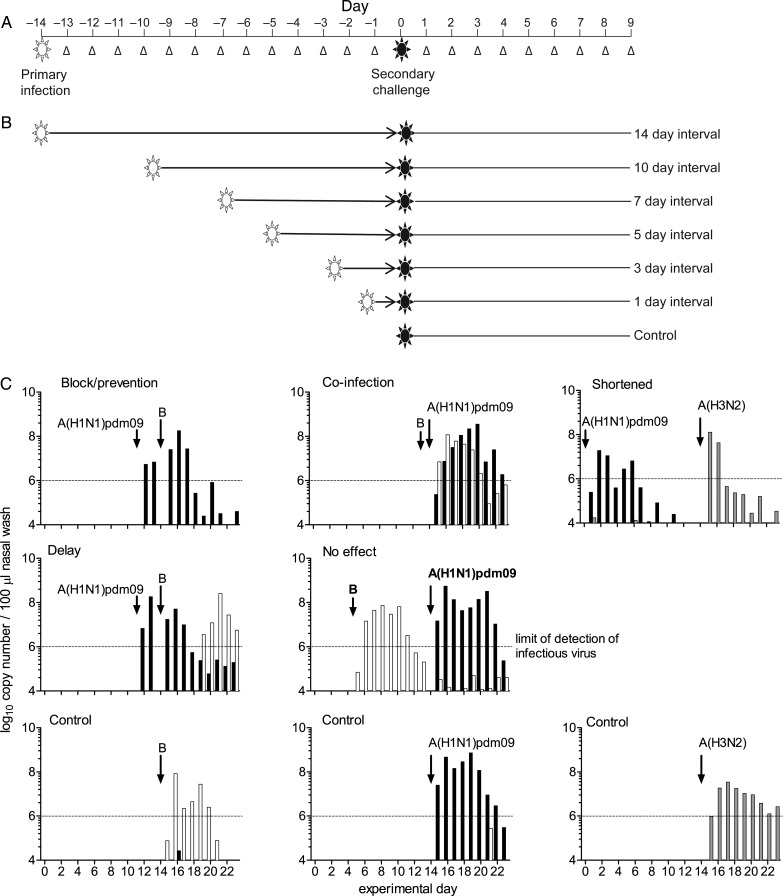
To investigate viral interference following infection with influenza virus, we systematically assessed the influence of a primary virus infection on the kinetics of a secondary virus challenge in the ferret model (Figure 1A). Two intervals (1 day and 3 days) represented the start and peak of virus shedding, respectively, in the upper respiratory tract (URT); one interval (5 days) corresponded to decreased virus shedding; and one interval (7 days) reflected the end of virus shedding. Seroconversion typically occurs 7–12 days following infection (data not shown); thus, the 7-day interval and 2 later intervals (10 days and 14 days) represented the adaptive immune response (Figure 1B).
Figure 1.
Experimental plan. A, Ferrets were infected intranasally with the primary virus and then challenged intranasally with a second virus. Virus shedding was assessed daily in nasal washings (Δ) as a measure of upper respiratory tract infection. B, Six intervals between primary infection and challenge were assessed. Primary infections were staggered to allow challenge of all animals on the same day, enabling direct comparison of virus shedding after challenge. C, Examples of virus shedding patterns after primary infection and challenge. Each graph represents data from an individual ferret. Shedding patterns were compared to those for controls that were infected with the challenge virus only on day 14 (bottom row).
We determined whether prior infection could prevent shedding of a different virus type. Test ferrets were first infected with influenza A virus and then challenged with influenza B virus, or vice versa (primary infection → challenge: A[H1N1]pdm09 → B, B → A[H1N1]pdm09, A[H3N2] → B, and B → A[H3N2]). Control ferrets had no primary infection.
Infectious virus shedding was defined by a real-time RT-PCR–determined value of 106 copies/100 µL of nasal wash for all assays (Flu A M, Flu B NS, H1 HA, and H3 HA), correlating to the minimum amount of detectable infectious virus in in vitro TCID50 assays (Supplementary Figure 1) and infectivity measured previously by transmission [32]. Various patterns of shedding were observed after challenge: (1) prevention of secondary infection, (2) coinfection, (3) shortened secondary infection, (4) delayed secondary infection, and (5) no effect as compared to the control group (Figure 1C). Individual virus shedding data for all ferrets are shown in Supplementary Figures 3–6. No clinical signs (weight loss, fever, or symptoms) were associated with any virus shedding pattern (data not shown). As virus shedding was measured for only 9 days after challenge, seroconversion could not reliably be used as a measure of infection (data not shown).
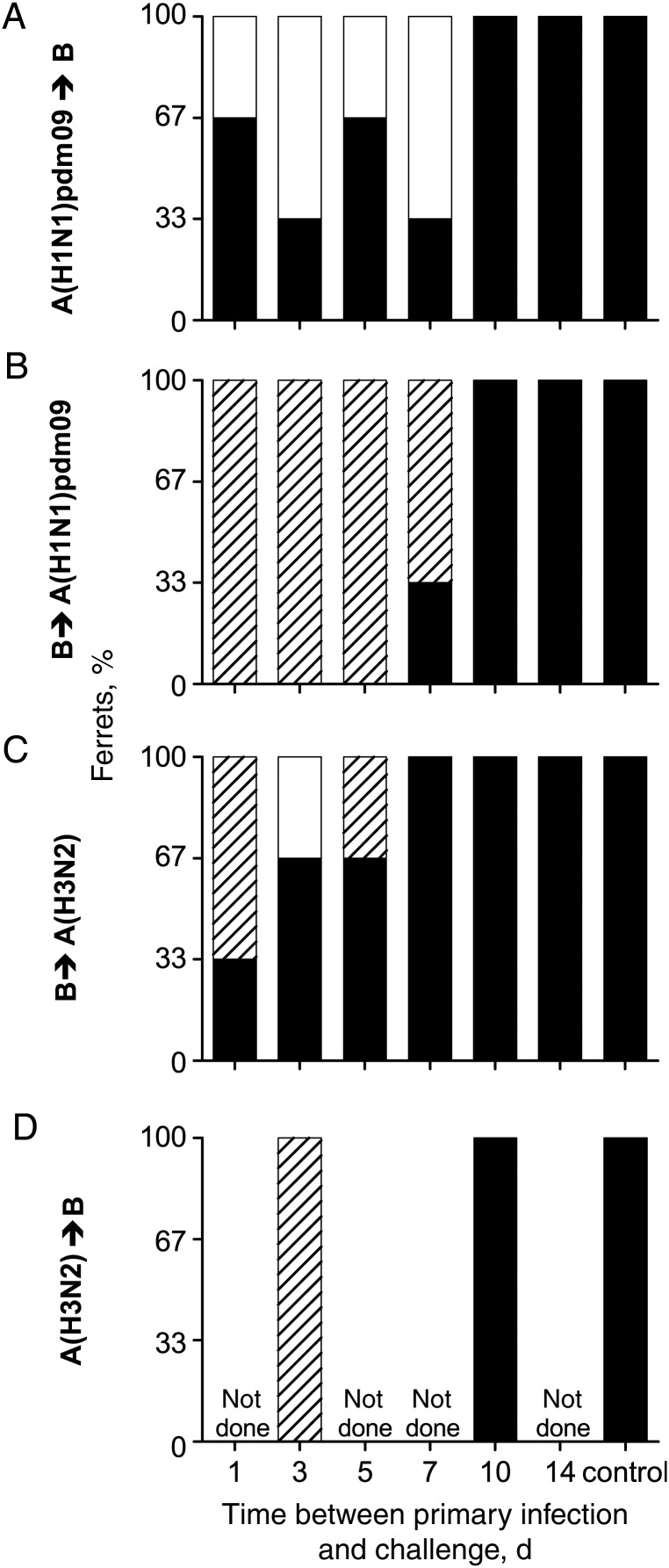
Different outcomes were observed with different combinations and sequences of virus infections (Figure 2, Supplementary Figure 2, and Table 1). Primary infection with A(H1N1)pdm09 virus 1 or 3 days prior prevented secondary infection with influenza B virus in half of the ferrets (Figure 2A and Table 1). In contrast, in all animals first infected with influenza B virus and then challenged with A(H1N1)pdm09 virus, coinfection with shedding of both viruses was observed (Figure 2B and Table 1). Prior infection with influenza B virus protected 1 ferret from subsequent infection with A(H3N2) virus, while 2 ferrets were coinfected (Figure 2C and Table 1). Ferrets first infected with A(H3N2) virus and then challenged with influenza B virus were not protected and shed both viruses after challenge (Figure 2D and Table 1).
Figure 2.
Shedding of the challenge virus is prevented, or coinfections may occur in short intervals. Ferrets were infected with virus and then challenged with a different virus type (primary virus → challenge virus). Data indicate the outcome of the challenge infection, in which ferrets were infected with challenge virus (black), protected from infection with challenge virus (white), or coinfected with challenge and primary virus (striped; n = 3–6).
Table 1.
Outcome of Challenge Infection for Virus Pairs Evaluated in the Study
| Virus Pair | Ferrets Protected, Proportion, by Interval Between Primary Infection and Challenge, d |
Control | |||||
|---|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 7 | 10 | 14 | ||
| A(H1N1)pdm09 → B | 1/3 | 2/3 | 1/3 | 2/3 | 0/3 | 0/3 | 0/3 |
| B → A(H1N1)pdm09 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
| B → A(H3N2) | 0/3 | 1/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/6 |
| A(H3N2) → B | ND | 0/3 | ND | ND | 0/3 | ND | 0/3 |
| A(H1N1)pdm09 → A(H3N2) | 0/3 | 3/3 | 3/3 | 3/3 | 2/3 | 0/3 | 0/6 |
| A(H3N2) → A(H1N1)pdm09 | 0/3 | 0/3 | 2/3 | 2/3 | 0/3 | 0/3 | 0/6 |
Data are no. of ferrets in which challenge infection was prevented from secondary challenge/no. evaluated.
Abbreviation: ND, not done.
Similar patterns were observed for 5- or 7-day intervals (Figure 2 and Table 1). Prior infection with A(H1N1)pdm09 virus prevented infection with influenza B virus in half of the ferrets (Figure 2A and Table 1). Interference was not observed if the order of viruses was reversed; coinfections occurred in the majority of animals (Figure 2B and Table 1). Prior infection with influenza B virus did not protect any ferrets from challenge with A(H3N2) virus 5 or 7 days later (Figure 2C and Table 1). For longer intervals of 10 or 14 days, no protection from challenge was observed in any groups (Figure 2 and Table 1).
Continued Shedding From the Primary Virus Infection Correlates With a Delay in Shedding Following Challenge With a Different Influenza Virus Type
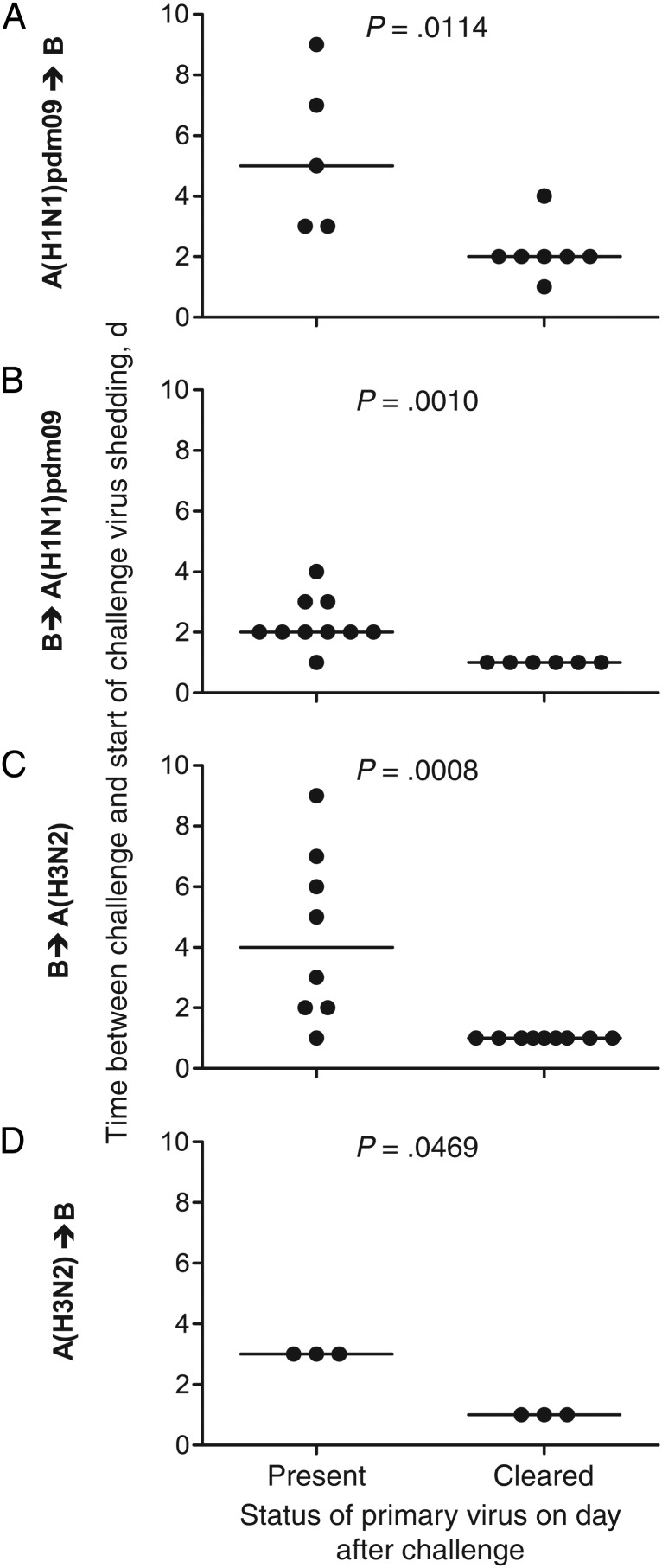
Because prevention of challenge virus infection occurred only for intervals of ≤7 days and primary virus was detected for 5–7 days (Supplementary Figures 3–6), a temporary state of immunity that affects challenge outcome may require the presence of primary virus. Animals shedding secondary virus after challenge (Figure 2) were grouped according to whether the primary infection virus could be detected in URT samples after challenge, as a proxy for the presence of virus on the day of challenge. The kinetics of challenge virus shedding were compared between the groups (Figures 3 and 4 and Supplementary Figure 2).
Figure 3.
The presence of primary virus shedding immediately after challenge can delay the start of shedding of the challenge virus between influenza virus types. Each ferret was categorized as shedding (present) or not shedding (cleared) primary virus on the day after challenge, and the interval to the start of challenge virus shedding was determined. Line indicates median values.
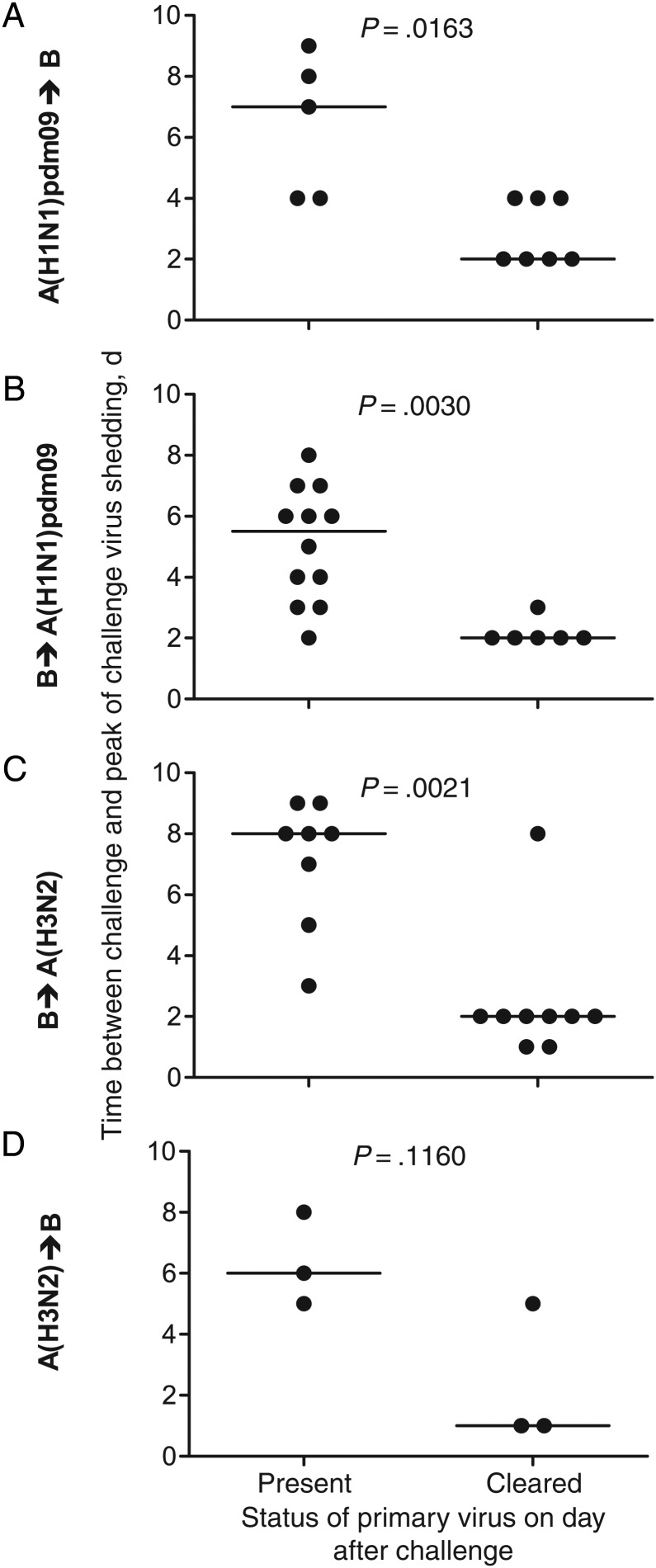
Figure 4.
The presence of primary virus shedding immediately after challenge can delay the peak of shedding of the challenge virus between influenza virus types. Each ferret was categorized as shedding (present) or not shedding (cleared) primary virus the day after challenge, and the interval to the peak of challenge virus shedding was determined. Line indicates median.
When A(H1N1)pdm09 or influenza B were the primary infection viruses, their continued shedding on the day of challenge correlated with a delay to the start (Figure 3A–C) and peak (Figures 4A–C) of shedding of the challenge viruses. Ongoing shedding of A(H1N1)pdm09 virus delayed the start of shedding of the influenza B challenge virus for a longer period (Figure 3A) than was observed when the order of the viruses was reversed (Figure 3B). Ferrets first infected with A(H3N2) virus displayed no significant delay to the start (Figure 3D) or peak (Figure 4D) of shedding of the challenge influenza B virus.
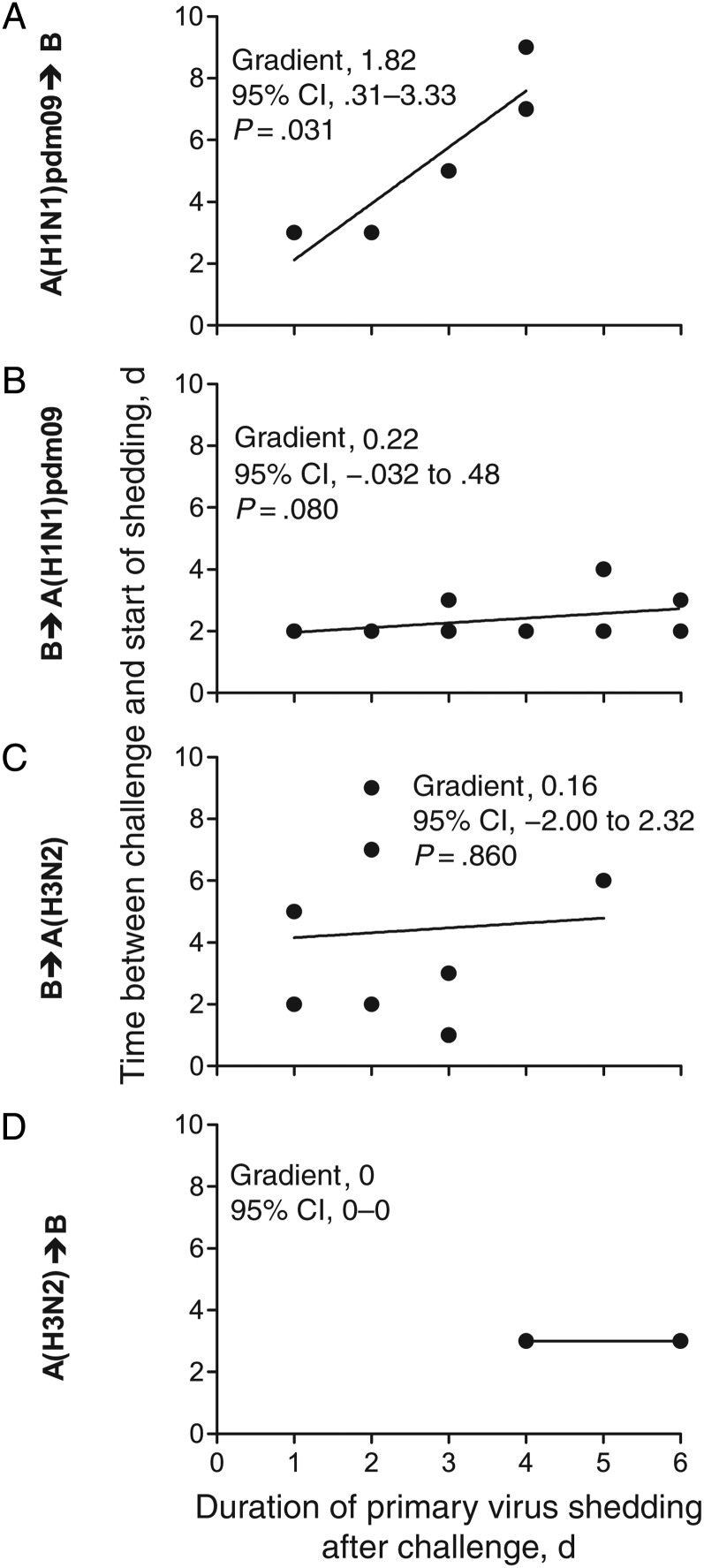
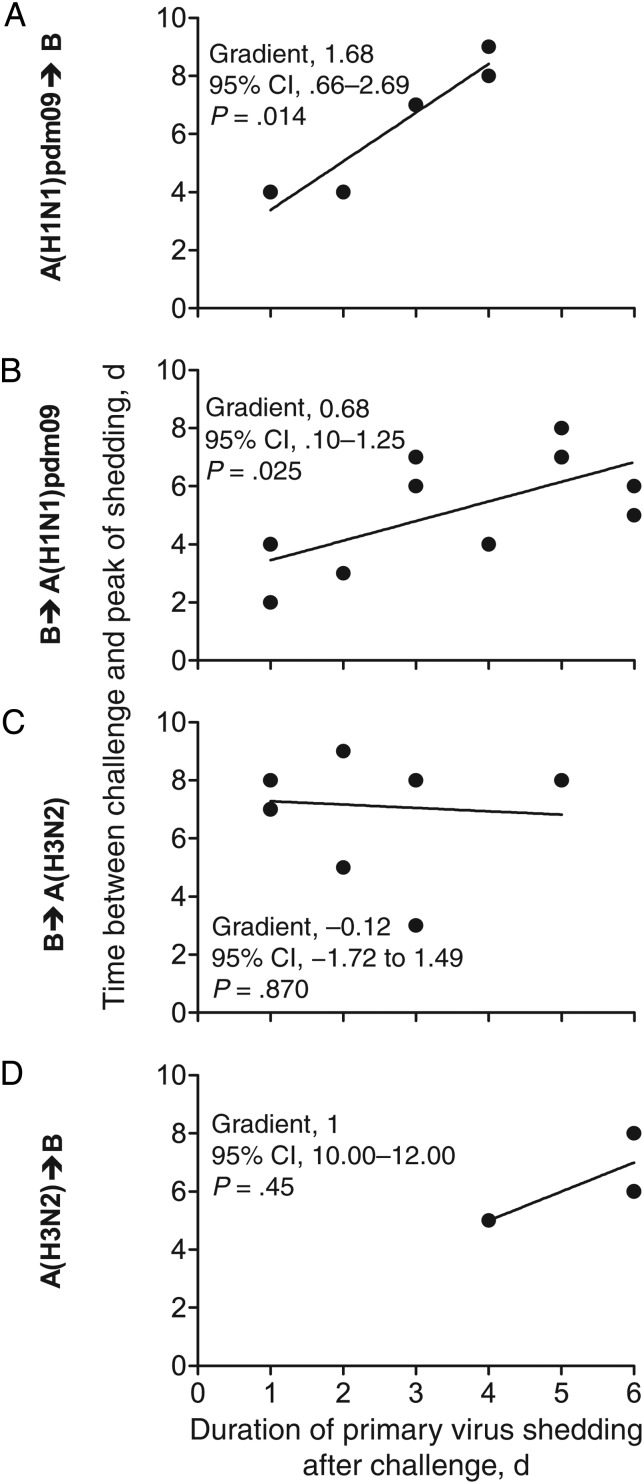
We next examined how the duration of primary virus shedding after challenge modified the challenge virus kinetics. When ferrets were first infected with A(H1N1)pdm09 virus, for every day that A(H1N1)pdm09 virus was detected following challenge, the start of B virus shedding was delayed by 1.82 days (95% CI, .31–3.33; P = .031; Figure 5A), and the peak of B virus shedding was delayed by 1.68 days (95% CI, .66–2.69; P = .014; Figure 6A). When ferrets were first infected with influenza B virus, this effect was reduced. For every day the primary influenza B virus infection continued after A(H1N1)pdm09 virus challenge, the peak of A(H1N1)pdm09 virus shedding was delayed by 0.68 days (95% CI, .10–1.25; P = .025; Figure 6B), with some evidence for a delay to start of A(H1N1)pdm09 virus shedding (Figure 5B). There was no evidence of delay when ferrets were first infected with influenza B virus and then challenged with A(H3N2) virus, or vice versa (Figure 5C and 5D and Figure 6C and 6D). These data show that temporary immunity depends on the continued presence of the primary virus and the respective virus (sub)types. There was no statistical support for an impact on the kinetics of the primary virus infection (shedding was not shortened), regardless of the timing of the secondary virus challenge (Supplementary Figures 3–6).
Figure 5.
The delay of the start of virus shedding after challenge is variable between influenza virus types. For each ferret that was shedding primary virus on the day after challenge, the number of days after challenge during which the primary virus shedding was still detected (x-axis) was plotted against the interval to the start of challenge infection (y-axis). A line of best fit yielded by linear regression analysis was plotted for every virus pair to determine the delay for an infection to begin. Abbreviation: CI, confidence interval.
Figure 6.
The delay of the peak of virus shedding after challenge is variable between influenza virus types. For each ferret that was shedding primary virus on the day after challenge, the number of days after challenge during which the primary virus shedding was still detected (x-axis) was plotted against the interval to the peak of challenge infection (y-axis). A line of best fit yielded by linear regression analysis was plotted for every virus pair to determine the delay for an infection to peak. Abbreviation: CI, confidence interval.
Infection With Influenza A Virus May Block, Delay, or Allow Coinfection With a Different Influenza A Virus Subtype
We assessed viral interference between influenza A viruses that have common antigenic epitopes. Ferrets were infected with A(H1N1)pdm09 virus and challenged with A(H3N2) virus, and vice versa (Supplementary Figures 2, 7, and 8). At the shortest interval of 1 day, there was no prevention of the challenge infection; coinfections occurred in all animals, irrespective of the order of infection (Supplementary Figure 9 and Table 1). However, infection with A(H1N1)pdm09 virus completely prevented infection with A(H3N2) virus 3–7 days later (and, for 2 of 3 ferrets, at a 10-day interval; Supplementary Figure 9A and Table 1). Infection with A(H3N2) virus 3 days prior to A(H1N1)pdm09 virus challenge resulted in coinfection (Supplementary Figure 9B and Table 1), while for intervals of 5 or 7 days, infection with A(H3N2) virus prevented infection with A(H1N1)pdm09 virus in 2 of 3 ferrets (Supplementary Figure 9B and Table 1). There was no significant delay to the start or peak of shedding for either combination of influenza A virus, but this may reflect the small number of ferrets (Supplementary Figure 10).
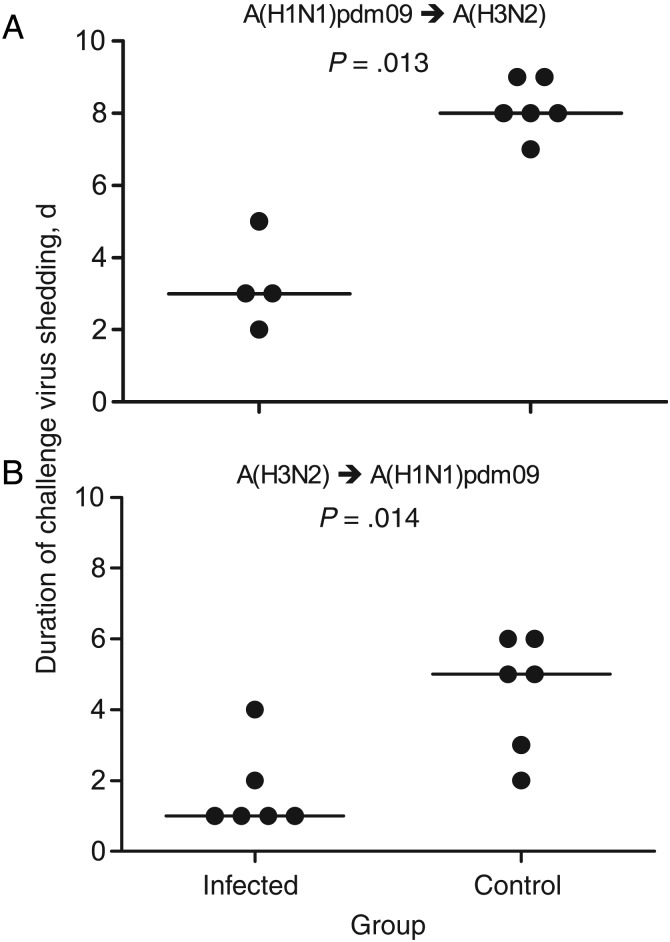
When ferrets were challenged 10 or 14 days after the primary infection, prior infection with A(H3N2) virus did not prevent infection with A(H1N1)pdm09 (Supplementary Figure 9B and Table 1). However, the challenge virus was cleared more rapidly (Figure 1C and Supplementary Figure 8); ferrets shed A(H1N1)pdm09 virus for a median duration of 1 day, compared with 5 days for control ferrets (Figure 7B). Similarly, ferrets that were first infected with A(H1N1)pdm09 virus and then infected with A(H3N2) virus 10–14 days later, shed A(H3N2) virus for a median of 3 days, compared with 8 days for control ferrets (Figure 7A).
Figure 7.
The duration of virus shedding is reduced when ferrets are infected with different influenza A subtypes within 10–14-day intervals. The number of days of shedding of the challenge virus was determined for each ferret within the 10–14-day intervals and for the control group.
For all animals first infected with A(H3N2) virus and then infected with A(H1N1)pdm09 virus, at all intervals, there was a statistically significant delay before reaching the peak of virus shedding of 0.3 days (Supplementary Figure 11D), but there was no statistical support for a delay to the start of shedding (Supplementary Figure 11B). No delays were seen when A(H1N1)pdm09 virus infection preceded A(H3N2) virus challenge (Supplementary Figure 11A and 11C).
DISCUSSION
Using a ferret model of human influenza, we demonstrated that infection with influenza virus can prevent or modify a subsequent infection with a different influenza virus (sub)type. Outcomes varied according to both virus combination and the interval between primary infection and challenge. Prior infection with both antigenically related and unrelated viruses provided protection from subsequent infection or modified the infection kinetics of the challenge virus. For relatively short exposure intervals, these effects required the continued presence of the primary virus following challenge. For heterosubtypic influenza A viruses, when primary infection and challenge were separated by at least 1 week, the challenge virus was rapidly cleared. The outcome depended on the virus combinations, indicating a hierarchy of influenza viruses inducing different levels of temporary immunity. In this study, the A(H1N1)pdm09 virus was most effective at inducing a temporary state of immunity, followed by influenza B virus, and, last, the A(H3N2) virus. These data may explain why viral interference is observed only in some epidemiological studies and during some influenza seasons [7, 8, 17], but further studies using other influenza strains are required to confirm these observations.
A key finding of this study is that coinfection was a consistent outcome in the ferret (5 of 6 virus pairs) when exposure to both influenza viruses occurred within 3 days. Human coinfections with influenza viruses are rarely detected [34–38], probably reflecting the low prospect of sequential exposure to different circulating strains within the short window of opportunity. Given that only a subset of laboratories routinely subtype influenza A viruses, there is a potential for missed identification of coinfections [36, 38]. Coinfections with influenza A viruses may give rise to novel reassortants, so understanding their true frequency and likelihood of occurrence is important for public health reasons.
Multiple mechanisms may contribute to establishing a temporary state of immunity. At intervals of <1 week, innate immune antiviral mechanisms may prevent or delay secondary viral infections. These mechanisms are antigen independent, so they may be common to both influenza A and B viruses. Upon infection with influenza virus, expression of type I interferons is upregulated in infected epithelial cells, and proinflammatory cytokines and chemokines are released from cells of the innate immune system [39]. Intrinsic antiviral factors, such as the IFITM and IFIT families and MxA, inhibit influenza virus fusion and replication by directly interacting with viral proteins or viral RNA [40, 41]. Recent studies have also demonstrated that nucleoprotein from influenza B virus can interact with nucleoprotein from influenza A virus and limit its polymerase activity [42, 43], suggesting that viral intermediates may also inhibit coinfections. Together, these mechanisms may limit the amount of virus shed by making epithelial cells refractory to infection, mopping up free virus, phagocytosing infected cells, or preventing/reducing viral replication. Many of these antigen-independent mechanisms reach their peak levels at the peak of virus shedding (L. Carolan and K. Laurie, unpublished observations); in this study, virus shedding peaked 2–3 days after infection and persisted for 5–6 days, consistent with the timing observed for temporary immunity. This timing also implies a lag of 1–2 days following primary infection before innate immunity and intrinsic antiviral factors reach maximum levels, providing a possible explanation for the observed window of coinfection.
Cross-reactive immune responses may limit infection by viruses that share epitopes [44]. A shortened period of virus shedding was seen only when ferrets were infected with consecutive influenza A viruses. Neutralizing antibodies were only detected 7–12 days after inoculation, consistent with our observation of shortened challenge infection 10 and 14 days after the first infection (data not shown). Previous studies, in which infections with heterosubtypic influenza A viruses were separated by 8 weeks, showed a similar shortening of the duration of virus shedding [28]. Cross-reactive epitopes between influenza A and B viruses have been reported in the fusion peptide of hemagglutinin (HA) and the enzymatic region of neuraminidase [30]. Generation of cross-reactive T cells has yet to be investigated, but we observed no protection, or change in kinetics, when ferrets were infected with influenza A virus and then challenged with influenza B virus 8 weeks later, and vice versa (data not shown). The rapid clearance observed in ferrets consecutively infected with influenza A viruses at 10- and 14-day intervals was not detected in ferrets consecutively infected with influenza A and B viruses, suggesting that cross-reactive lymphocytes do not play a major role in viral clearance in this experiment.
Understanding why some challenge viruses can still infect the respiratory epithelium of ferrets with an established influenza virus infection is of interest. Viral fitness, whereby one virus can outgrow another and even successfully be transmitted to a new host, may be important [45]. In another study, when ferrets were coinfected with A(H1N1)pdm09/A(H1N1) or A(H1N1)pdm09/A(H3N2) viruses, only the A(H1N1)pdm09 virus was transmitted to naive contact ferrets, showing a clear fitness advantage [46]. HA receptor specificity may also contribute. The A(H1N1)pdm09 virus in our study can infect cells of both the URT and lower respiratory tract, whereas the A(H3N2) and influenza B viruses used in this study infect only the URT following intranasal infection (data not shown). This suggests a broader receptor specificity for the HA of A(H1N1)pdm09, compared with the H3 or influenza B HA proteins. Experiments are planned to investigate these aspects on our model.
Our findings imply that temporary immunity between unrelated viruses lasts for 1 week or less in the ferret. While this period is short in comparison with observed interference in epidemiological or modeling studies [20, 22], individual level influences on susceptibility and transmissibility will predictably have consequences over longer time frames at the level of populations. We used a direct inoculation model to ensure that the challenge was controlled and that the dose of virus was consistent. A low dose of virus was used to mirror the kinetics of a natural infection, but the duration of temporary immunity may be different in an aerosol transmission model and more closely match those periods seen in humans. Live attenuated influenza vaccines (LAIVs) have been shown to induce the innate immune response [47], and thus the ability of LAIV to induce viral interference to homologous and hetero(sub)typic viruses in rapid time frames is also of interest.
Overall, our study provides insight into the phenomenon of viral interference and provides the first experimental evidence for a hierarchy among influenza viruses in inducing a temporary state of immunity, as previously suggested by surveillance studies [13, 14]. In humans, a lack of adaptive immunity in specific age groups was most likely the major mechanism behind the dominance of the A(H1N1)pdm09 virus upon its emergence in 2009 [48]. However, our study suggests that temporary immunity may also have contributed to the observed dominance of A(H1N1)pdm09 virus and higher levels of infection seen in many countries in 2009. Induction of a nonspecific localized temporary state of immunity may provide a strategy to control infection with emerging influenza viruses in the absence of an antigen-matched inactivated vaccine.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank L. Redman, N. Hearne, C. Agpasa, M. Spiteri, R. Murphy, A. Hageman, J. Moody, and P. Franchina from bioCSL, for excellent technical assistance with animal handling and husbandry; and A. C. Hurt, for a critical reading of the manuscript.
Financial support. This work was supported by the Australian Government, Department of Health (to the Melbourne WHO Collaborating Centre for Reference and Research on Influenza).
Potential conflicts of interest. K. L. L., T. A. G., L. A. C., M. A., A. K., and I. A. B. are affiliated with the WHO Collaborating Centre for Reference and Research on Influenza, which has or has had collaborative agreements with the International Federation of Pharmaceutical and Manufacturers Associations, Novartis Vaccines and Diagnostics, and CSL for work unrelated to this study. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Duffy CE. Interference between St. Louis encephalitis virus and equine encephalomyelitis virus (Western Type) in the chick embryo. Science 1944; 99:517–8. [DOI] [PubMed] [Google Scholar]
- 2.Henle W, Henle G. Interference of inactive virus with the propagation of virus of influenza. Science 1943; 98:87–9. [DOI] [PubMed] [Google Scholar]
- 3.Isaacs A, Lindenmann J. Virus interference I. The interferon. Proc R Soc Lond B Biol Sci 1957; 147:258–67. [PubMed] [Google Scholar]
- 4.Lindenmann J. From interference to interferon: a brief historical introduction. Philos Trans R Soc Lond B Biol Sci 1982; 299:3–6. [DOI] [PubMed] [Google Scholar]
- 5.Greer RM, McErlean P, Arden KE et al. Do rhinoviruses reduce the probability of viral co-detection during acute respiratory tract infections? J Clin Virol 2009; 45:10–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DaPalma T, Doonan BP, Trager NM, Kasman LM. A systematic approach to virus-virus interactions. Virus Res 2010; 149:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly H, Barry S, Laurie K, Mercer G. Seasonal influenza vaccination and the risk of infection with pandemic influenza: a possible illustration of nonspecific temporary immunity following infection. Eurosurveillance 2010; 15:pii:19722. [DOI] [PubMed] [Google Scholar]
- 8.Linde A, Rotzen-Ostlund M, Zweygberg-Wirgart B, Rubinova S, Brytting M. Does viral interference affect spread of influenza? Eurosurveillance 2009; 14:pii:19354. [PubMed] [Google Scholar]
- 9.Wang W, Cavailler P, Ren P et al. Molecular monitoring of causative viruses in child acute respiratory infection in endemo-epidemic situations in Shanghai. J Clin Virol 2010; 49:211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anestad G, Nordbo SA. Virus interference. Did rhinoviruses activity hamper the progress of the 2009 influenza A(H1N1) pandemic in Norway? Med Hypotheses 2011; 77:1132–4. [DOI] [PubMed] [Google Scholar]
- 11.Casalegno JS, Ottmann M, Bouscambert Duchamp M et al. Rhinoviruses delayed the circulation of the pandemic influenza A(H1N1) 2009 virus in France. Clin Microbiol Infect 2010; 16:326–9. [DOI] [PubMed] [Google Scholar]
- 12.Casalegno JS, Ottmann M, Bouscambert-Duchamp M, Valette M, Morfin F, Lina B. Impact of the 2009 influenza A(H1N1) pandemic wave on the pattern of hibernal respiratory virus epidemics, France, 2009. Eurosurveillance 2010; 15:pii:19485. [PubMed] [Google Scholar]
- 13.Van Kerkhove MD, Mounts AW. 2009 versus 2010 comparison of influenza activity in southern hemisphere temperate countries. Influenza Other Respir Viruses 2011; 5:375–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Wang Z, Ren L et al. Influenza A/H1N1 2009 Pandemic and Respiratory Virus Infections, Beijing, 2009–2010. PLoS One 2012; 7:e45807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathews JD, McCaw CT, McVernon J, McBryde ES, McCaw J. A biological model for influenza transmission: pandemic planning implications of asymptomatic infection and immunity. PLoS One 2007; 2:e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer GN, Barry SI, Kelly H. Modelling the effect of seasonal influenza vaccination on the risk of pandemic influenza infection. BMC Public Health 2011; 11:S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skowronski DM, De Serres G, Crowcroft NS et al. Association between the 2008–09 seasonal influenza vaccine and pandemic H1N1 illness during Spring-Summer 2009: four observational studies from Canada. PLoS Med 2010; 6:e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janjua NZ, Skowronski DM, Hottes TS et al. Seasonal influenza vaccine and increased risk of pandemic A/H1N1-related illness: first detection of the association in British Columbia, Canada. Clin Infect Dis 2010; 51:1017–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuchihashi Y, Sunagawa T, Yahata Y et al. Association between seasonal influenza vaccination in 2008–2009 and pandemic influenza A(H1N1) 2009 infection among school students from Kobe, Japan, April-June 2009. Clin Infect Dis 2012; 54:381–3. [DOI] [PubMed] [Google Scholar]
- 20.Cowling BJ, Fang VJ, Nishiura H et al. Increased risk of noninfluenza respiratory virus infections associated with receipt of inactivated influenza vaccine. Clin Infect Dis 2012; 54:1778–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowling BJ, Nishiura H. Virus interference and estimates of influenza vaccine effectiveness from test-negative studies. Epidemiology 2012; 23:930–1. [DOI] [PubMed] [Google Scholar]
- 22.Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature 2003; 422:428–33. [DOI] [PubMed] [Google Scholar]
- 23.Van Reeth K, Nauwynck H, Pensaert M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet Microbiol 1996; 48:325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa-Hurtado M, Afonso CL, Miller PJ et al. Virus interference between H7N2 low pathogenic avian influenza virus and lentogenic Newcastle disease virus in experimental co-infections in chickens and turkeys. Vet Res 2014; 45:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo SH, Webster RG. Cross-reactive, cell-mediated immunity and protection of chickens from lethal H5N1 influenza virus infection in Hong Kong poultry markets. J Virol 2001; 75:2516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walzl G, Tafuro S, Moss P, Openshaw PJM, Hussell T. Influenza virus lung infection protects from respiratory syncytial virus-induced immunopathology. J Exp Med 2000; 192:1317–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bodewes R, Kreijtz JH, Geelhoed-Mieras MM et al. Vaccination against seasonal influenza A/H3N2 virus reduces the induction of heterosubtypic immunity against influenza A/H5N1 virus infection in ferrets. J Virol 2011; 85:2695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurie K, Carolan L, Middleton D, Lowther S, Kelso A, Barr I. Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. J Infect Dis 2010; 202:1011–20. [DOI] [PubMed] [Google Scholar]
- 29.Steel J, Staeheli P, Mubareka S, Garcia-Sastre A, Palese P, Lowen AC. Transmission of pandemic H1N1 influenza virus and impact of prior exposure to seasonal strains or interferon treatment. J Virol 2010; 84:21–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terajima M, Babon JAB, Co MDT, Ennis FA. Cross-reactive human B cell and T cell epitopes between influenza A and B viruses. Virol J 2013; 10:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh DY, Barr IG, Mosse JA, Laurie KL. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J Clin Microbiol 2008; 46:2189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guarnaccia T, Carolan LA, Maurer-Stroh S et al. Antigenic drift of the pandemic 2009 A(H1N1) influenza virus in a ferret model. PLoS Pathog 2013; 9:e1003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R: A language and environment for statistical computing. Vienna, Austria: R Core Team, R Foundation for Statistical Computing, 2012. [Google Scholar]
- 34.Toda S, Okamoto R, Nishida T et al. Isolation of influenza A/H3 and B viruses from an influenza patient: confirmation of co-infection by two influenza viruses. Jpn J Infect Dis 2006; 59:142–3. [PubMed] [Google Scholar]
- 35.Takao S, Hara M, Kakuta O et al. Eleven cases of co-infection with influenza type A and type B suspected by use of a rapid diagnostic kit and confirmed by RT-PCR and virus isolation [in Japanese]. Kansenshogaku Zasshi 2005; 79:877–86. [DOI] [PubMed] [Google Scholar]
- 36.Falchi A, Arena C, Andreoletti L et al. Dual infections by influenza A/H3N2 and B viruses and by influenza A/H3N2 and A/H1N1 viruses during winter 2007, Cor- sica Island, France. J Clin Virol 2008; 41:148–51. [DOI] [PubMed] [Google Scholar]
- 37.Eshaghi A, Blair J, Burton L et al. Characterization of an influenza A and influenza B co-infection of a patient in a long-term care facility with co-circulating influenza A and influenza B. Int J Infect Dis 2009; 13:e127–8. [DOI] [PubMed] [Google Scholar]
- 38.Peacey M, Hall RJ, Sonnberg S et al. Pandemic (H1N1) 2009 and seasonal influenza A (H1N1) co-infection, New Zealand, 2009. Emerg Infect Dis 2010; 16:1618–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braciale TJ, Sun J, Kim TS. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol 2012; 12:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol 2012; 13:214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 2013; 13:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaru-ampornpan P, Narkpuk J, Wanitchang A, Jongkaewwattana A. Nucleoprotein of influenza B virus binds to its type A counterpart and disrupts influenza A viral polymerase complex formation. Biochem Biophys Res Commun 2014; 443:296–300. [DOI] [PubMed] [Google Scholar]
- 43.Wanitchang A, Narkpuk J, Jaru-ampornpan P, Jengarn J, Jongkaewwattana A. Inhibition of influenza A virus replication by influenza B virus nucleoprotein: an insight into interference between influenza A and B viruses. Virology 2012; 432:194–203. [DOI] [PubMed] [Google Scholar]
- 44.Greenbaum JA, Kotturi MF, Kim Y et al. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci U S A 2009; 106:20365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Butler J, Hooper KA, Petrie SM et al. Estimating the fitness advantage conferred by permissive neuraminidase mutations in recent oseltamivir-resistant A(H1N1)pdm09 influenza viruses. PLoS Pathog 2014; 10:e1004065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez DR, Sorrell E, Angel M et al. Fitness of pandemic H1N1 and seasonal influenza A viruses during co-infection. PLoS Curr 2009; 1:RRN1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer WA II, Chasond KD, Brighton M, Jaspers I. Live attenuated influenza vaccine strains elicit a greater innate immune response than antigenically-matched seasonal influenza viruses during infection of human nasal epithelial cell cultures. Vaccine 2014; 15:1761–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelly H, Peck HA, Laurie KL, Wu P, Nishiura H, Cowling BJ. The age-specific cumulative incidence of infection with pandemic influenza H1N1 2009 was similar in various countries prior to vaccination. PLoS One 2011; 6:e21828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.