Abstract
Background. We previously reported that infants in Kenya were infected with Epstein-Barr virus (EBV) at <6 months of age, suggesting that mothers were the likely source of transmissible virus to the infant. In this study, we investigated whether breast milk contained infectious EBV and the role of malaria in EBV shedding in breast milk.
Methods. Breast milk samples were obtained from Kenyan mothers at postpartum weeks 6, 10, 14, and 18 and analyzed for presence of infectious EBV.
Results. We found that the prevalence of EBV DNA and the mean EBV load were significantly higher at 6 weeks and decreased through postpartum week 18 (P < .0001). High EBV load in breast milk correlated with mothers who had Plasmodium falciparum malaria at delivery. To determine whether viral DNA was encapsidated, breast milk samples were treated with DNAse before DNA extraction. Sixty percent of samples were DNAse resistant, suggesting that the viral DNA in breast milk was encapsidated. Next, we exposed peripheral blood mononuclear cells to breast milk supernatant, which resulted in the generation of EBV-positive lymphoblastoid cell lines, indicating that the virus in breast milk was infectious.
Conclusions. Our data suggest that breast milk contains infectious EBV and is a potential source of viral transmission to infants living in malaria-endemic regions.
Keywords: breast milk, EBV transmission, malaria, Kenya
Both Epstein-Barr virus (EBV) and holoendemic malaria are etiologic cofactors in the development of endemic Burkitt lymphoma (eBL), the most common childhood cancer in sub-Saharan Africa [1–4]. The seminal prospective study conducted by de-Thé in Uganda in the1970s indicated that children with higher EBV antibody titers were more likely to develop eBL [2]. This observation led de-Thé [2] to posit that perinatal infection with EBV (ie, infection before 6 months of age) was a risk factor for eBL. More recently, we reported that infants from a region of Kenya where transmission of malaria is high were more likely to be infected with EBV earlier in life than infants from a region where malaria transmission is low [5]. Strikingly, 35% of infants from the region of high transmission were infected before 6 months of age, suggesting that malaria is enhancing EBV transmission in early infancy.
EBV, a strict human pathogen, is thought to be transmitted primarily through contact with saliva [6, 7]. For example, early studies indicated that there was cell-free virus in the saliva and that exposure to saliva resulted in generation of EBV-positive lymphoblastoid cell lines [7, 8]. An alternative source of EBV transmission for infants could be breast milk. EBV DNA was detected in breast milk in a subset of both healthy mothers [9] and human immunodeficiency virus (HIV)–infected mothers [10], but neither study went further to examine whether there was infectious virus in the breast milk. A study of Japanese infants indicated that there was no difference in the seroprevalence of EBV at 12 months of age in infants born to mothers who were breast-fed or who were exclusively bottle-fed, suggesting that breast milk was not a source of EBV transmission in children [11]. However, comparison of EBV transmission among geographically different populations is challenging because the prevalence of EBV infection in infants varies significantly on the basis of geography [5].
Our study showing that infants from a malaria high transmission region were infected with EBV before 6 months of age supported the long-standing hypothesis of Guy de-Thé [2], but left open the question of why these infants were infected in the perinatal period. In this prospective study, we investigated whether breast milk could be a source of EBV transmission and the role of malaria in EBV shedding in breast milk. Pregnant women were recruited from a malaria-endemic and eBL high-risk region of Kenya and longitudinally followed through delivery and the postpartum period, and EBV was assayed in breast milk. High levels of infectious EBV were detected in the breast milk, and this correlated with mothers who had malaria at delivery. Our results indicate that breast milk could be a potential source of transmission of EBV from mothers to infants in malaria-endemic and eBL high-risk regions.
METHODS
Study Site and Population
Participants for the study were recruited from Chulaimbo Subdistrict Hospital, which serves a rural population in Kisumu County, Kenya. Details of recruitment and follow-up have been previously described [12]. HIV-negative pregnant women (n = 175) were longitudinally evaluated in an active monthly antenatal follow-up visits (up to 4 per mother) through the time of delivery. Following delivery, 94 mothers who delivered at a health facility, had no blood transfusion within 24 hours of delivery, and had a normal vaginal delivery with term and live-born singletons were further followed during the postpartum period, and, where possible, breast milk samples were collected at approximately postpartum weeks 6 (n = 79), 10 (n = 79), 14 (n = 76), and 18 (n = 59). No breast milk was collected from mothers with bilateral mastitis at the time of visit or recurrent mastitis. If mothers had unilateral mastitis, breast milk was collected from the unaffected side.
Approval of this study was obtained from the KEMRI and UMU ethical review boards. Written informed consent was obtained from the study participants before any sample collection.
Breast Milk Specimen Collection and Processing
Breast milk specimens (3–5 mL) were collected using aseptic technique. Milk was manually expressed into sterile tubes, after discarding the first several drops. Breast milk samples were refrigerated and processed within 3 hours of collection. Whole milk was stored at −80°C until analysis.
Measurement of EBV in Breast Milk
Whole milk was centrifuged at 3900 RCF for 5 minutes at 4°C to separate the lipid, supernatant, and cell debris. The lipid layer was discarded, and the clear supernatant was aspirated into a separate sterile tube and stored at −80°C. DNA was extracted by use of a QiaAmp DNA Mini Kit (Qiagen, Valencia, California) in accordance with the manufacturer's protocol. DNA purity and quantity was assessed with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, Delaware). EBV DNA was detected and quantified by quantitative polymerase chain reaction (qPCR) as described previously [5, 13]. EBV load was expressed as copies/mL breast milk.
DNAse I Treatment of Breast Milk
A subset of breast milk supernatants (n = 40) was DNAse I treated according to the manufacturer's instructions (Invitrogen, Carlsbad, California). DNA extraction was done using the QiaAmp DNA Mini kit protocol (Qiagen). DNA purity and quantity were determined with a NanoDrop 2000 spectrophotometer (Thermo Scientific). EBV DNA was then assessed by qPCR.
Proliferation Assay
Peripheral blood mononuclear cells (PBMCs) were stained with e450 Cell Proliferation Dye (eBioscience, San Diego, California) as per the manufacturer's protocol. Following staining, PBMCs were suspended in complete Roswell Park Memorial Institute (RPMI) medium, mixed 1:1 with Kenyan breast milk samples or with phosphate-buffered saline as a negative control, and placed at 37°C for 2 hours. PBMCs were then washed by centrifugation and plated at 5 × 105 cells/mL in complete RPMI medium containing 1 µg/mL cyclosporine A. Cultures were harvested 7 days after breast milk exposure, treated with Fc block (BD Biosciences, San Jose, California) for 20 minutes, and stained for 20 minutes with PerCp-Cy5.5–conjugated anti-CD19 monoclonal antibody (Biolegend, San Diego, California). For the proliferation analysis, cells were run on a BD LSR-Fortessa flow cytometer equipped with FACS Diva software (BD Biosciences, San Jose, California). The percentage of CD19+ B cells and the mean fluorescence intensity (MFI) of e450 were analyzed with FlowJo v10 (TreeStar, Ashland, Oregon). A total of 20 individual Kenyan breast milk samples were used for proliferation analysis, with PBMC from 2 US donors being exposed to each breast milk sample.
Generation of Lymphoblastoid Cell Lines (LCLs)
A total of 5 × 105 PBMCs were suspended in 250 µL of complete RPMI medium and added to 250 µL of breast milk supernatant or to medium alone, incubated at 37°C for 2 hours, subsequently washed by centrifugation, and resuspended in 500 µL of complete RPMI medium containing 1 µg/mL cyclosporine A. PBMCs were maintained for the long term by weekly removal of the top 50% of culture medium and replacing it with fresh complete RPMI medium containing 1 µg/mL cyclosporine A. At the time that numerous prototypical LCL clumps were observed and wells reached approximately 80% confluence, cells were split. A standard PCR for EBV genome that distinguishes EBV types, as described below, was used to confirm that each cell line was EBV positive.
Cell Imaging
Cell aggregation was observed microscopically with a Nikon Eclipse Ti inverted microscope and imaging system, using NIS-Elements software (Nikon, Tokyo, Japan). All cell images were taken at 10 times the original magnification, and the automatic exposure function was used to select the exposure time for each image.
EBV Typing
The EBNA-3C region was amplified in DNA extracted from breast milk supernatant, using the following primers: 5′-AGAAGGGGAGCGTGTGTTG-3′ and 5′-GGCTCGTTTTTGACGTCGG-3′ (Integrated DNA Technologies) [14]. Probes were designed to distinguish between type 1 and type 2 EBV in the EBNA-3C region. The probe sequence for type 1 was 5′-FAM-TGTGACACTGACGA-T(BHQ-1)-GAATCTTCGGTGGTTTCAACAT-SpacerC3-3′ and for type 2 was: 5′-CAL Fluor Orange 560-CCGTGTGACTGGAAGACATGGCACTCC-BHQ-1-3′ (Biosearch Technologies). The real-time PCR conditions were as follows: 95°C for 3 minutes followed by 45 cycles of 95°C for 10 seconds and 55°C for 30 seconds. All reactions were performed with the iTaq Universal Probes Supermix (BioRad). To create a standard curve, plasmids were generated from the type 1 cell line B95-8 and the type 2 cell line Jijoye, using the TOPO TA cloning kit with PCR2.1 TOPO (Life Technologies) in accordance with the manufacturer's instructions. The number of EBV copies was normalized on the basis of the number of copies per milliliter of breast milk. The EBV genome in the LCL generated following infection of PBMCs with breast milk was typed by PCR for a region of EBNA3c as previously described [14].
Data Analysis
All statistical analyses were performed using Stata, IC software (13.1, StataCorp, College Station, Texas), setting a 2-tailed α to reject the null hypothesis at 0.05. All statistical assumptions were tested prior to interpretation of effects. Data tables present categorical data as percentages, while continuous data are presented using means with standard errors. EBV load, our primary outcome variable, was log transformed and then zero-filled for statistical evaluation, using Heckman's 2-step regression modeling with robust standard error estimates [15] to accommodate the longitudinal experimental design. Heckman's analysis is a 2-step process in which a bivariate model evaluates the effects of predictors on a dichotomous outcome (zero vs nonzero), with ordinary least squares regression methods evaluating the effects of predictors on the normally distributed continuously scaled outcomes (in this case, the nonzero log-transformed EBV load). We included β coefficients comparing baseline (week 6) to each subsequent period (10, 14, and 18 weeks), as well as indicators for malaria status at birth and ever during pregnancy in both models. Our selection model also incorporated the mother's anemia status during pregnancy, as an indicator of stress, which might potentially modulate EBV shedding in breast milk and contribute to nonzero EBV loads.
RESULTS
Characteristics of Study Participants
The study participants from whom these samples were collected have been described previously [12]. Briefly, the cohort consisted of 175 HIV-negative pregnant women who were longitudinally evaluated during active monthly antenatal follow-up visits (up to 4 visits per mother) conducted through the time of delivery. Following delivery, 94 mothers with term, live-born singletons who delivered at health facilities and had at least 1 breast milk sample collected at approximately postpartum weeks 6, 10, 14, and 18 were included in this analysis. The sociodemographic and obstetric characteristics have been described previously [12]. Briefly, the mean age (±SD) of the women was 22.3 ± 5.6 years, 39% were primigravidae, and the majority (58%) had at least an upper primary level of education. All pregnant women (100%) were EBV seropositive, 49.7% had moderate anemia at enrollment, and 33% were qPCR positive for P. falciparum malaria at enrollment. Fifty-four percent of the pregnant women had malaria, defined as at least 1 positive result of a qPCR for P. falciparum anytime during pregnancy, whereas 19% had detectable P. falciparum malaria at delivery.
EBV DNA Prevalence and Viral Load in Breast Milk at Postpartum Weeks 6, 10, 14, and 18
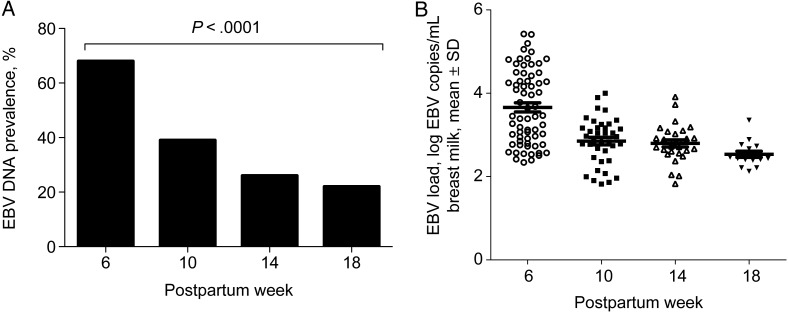
To determine whether EBV DNA could be detected in breast milk in various postpartum visits, EBV DNA was measured by qPCR. At 6 weeks, 68% of samples (54 of 79) had detectable EBV DNA. When we examined the prevalence of EBV DNA over time, we observed that the prevalence of EBV DNA in breast milk was significantly higher at 6 weeks and decreased through 18 weeks (6 weeks, 68% [54 of 79]; 10 weeks, 39% [31 of 79]; 14 weeks, 26% [20 of 76]; 18 weeks, 22% [13 of 59]; χ2 = 33.4; P < .0001; Figure 1A). We also compared the detected (nonzero) EBV load of the EBV-positive breast milk samples. The mean EBV load was 3.62 log copies/mL (range, 2.33–5.42 log copies/mL) at 6 weeks, 2.81 log copies/mL (range, 1.82–3.64 log copies/mL) at 10 weeks, 2.70 log copies/mL (range, 1.82–3.72 log copies/mL) at 14 weeks, and 2.56 log copies/mL (range, 2.12–3.35 log copies/mL) at 18 weeks (Figure 1B). EBV load was log transformed and then zero-filled for statistical evaluation using Heckman's [15] 2-step regression modeling with robust standard error estimates to accommodate the longitudinal experimental design of our study. We found significant differences in EBV load in breast milk for all 3 periods (10, 14, and 18 weeks) relative to 6 weeks in both the selection and continuous equations (P < .0001).
Figure 1.
A, Prevalence of Epstein-Barr virus (EBV) DNA in breast milk at postpartum weeks 6 (n = 79), 10 (n = 79), 14 (n = 76), and 18 (n = 59). The prevalence of EBV DNA was significantly higher at 6 weeks and decreased through 18 weeks (P < .0001). B, EBV loads in breast milk at approximately postpartum weeks 6, 10, 14, and 18. The mean ± SE EBV load of detectable (nonzero) samples was 3.62 ± 0.127 log copies/mL at 6 weeks, 2.81 ± 0.091 log copies/mL at 10 weeks, 2.70 ± 0.099 log copies/mL at 14 weeks, and 2.56 ± 0.089 log copies/mL at 18 weeks.
Higher EBV Load in Breast Milk Correlated With Mothers Who had P. falciparum Malaria Infection at Delivery
We examined whether P. falciparum infection at any time during pregnancy had an influence in EBV shedding in breast milk. We observed statistically higher viral loads in breast milk specimens from mothers who had malaria at delivery in both the selection and continuous Heckman's [15] equations (P < .02). To address whether there were other potential variables that could account for increased shedding of EBV, we asked whether mothers who had anemia in the model as an indicator of stress that might potentially modulate EBV shedding in breast milk. However, anemia status did not contribute to our analysis of the dichotomous portion (zero vs nonzero EBV load; P = .92).
EBV in Breast Milk Is Encapsidated
An important question to ask was whether the viral DNA detected in the breast milk samples represented DNA from lysed EBV-positive cells or was due to DNA isolated from infectious virions. To assess this possibility, a subset of EBV-positive breast milk samples (n = 40), collected at approximately postpartum week 6 was treated with DNAse I prior to DNA extraction. The strategy was to discriminate encapsidated virions from naked viral DNA, based on the differential degradation by DNAse I, given that the capsid would protect intact virions from DNAse digestion. qPCR was then used to detect EBV after DNAse I treatment. DNAse I–resistant EBV was observed in 24 of 40 (60%) of the samples that were EBV PCR positive prior to DNAse I treatment, suggesting that the virus DNA in breast milk supernatant was encapsidated.
Breast Milk Contains Transforming Virus
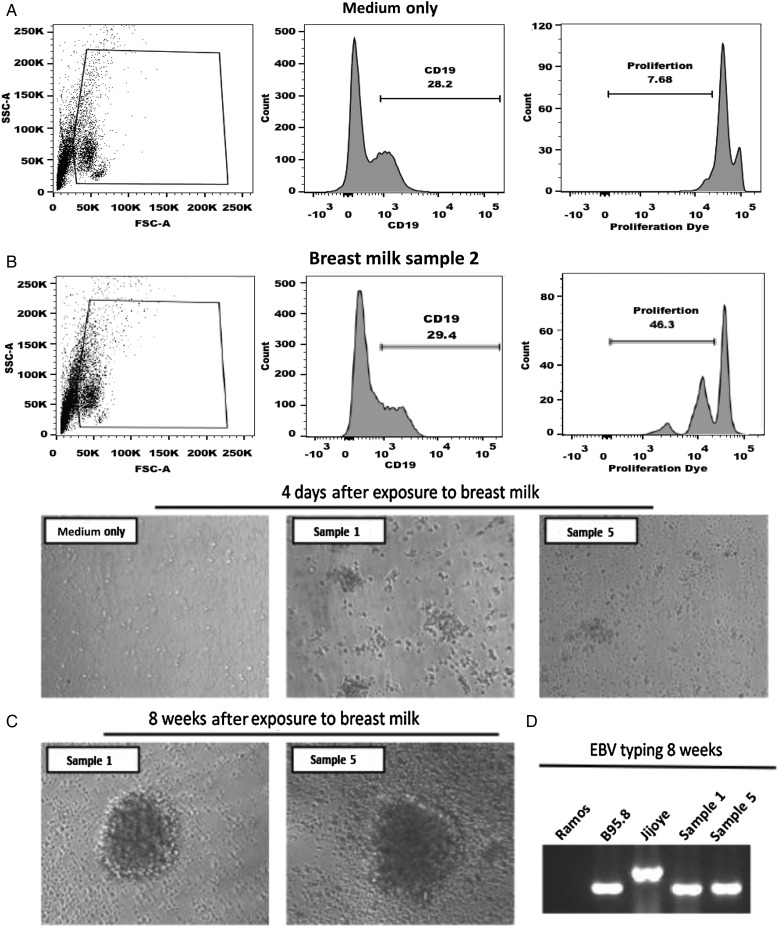
To assess whether EBV in breast milk is infectious, 20 Kenyan breast milk samples collected at approximately postpartum week 6 that were EBV PCR positive were assessed for their capacity to induce B-cell proliferation in the short term and in the long term by generation of EBV-transformed LCL. PBMCs were labeled with the e450 proliferation dye and then exposed to breast milk supernatant or medium alone. Cells were cultured for 7 days and then analyzed by flow cytometry to determine the percentage of CD19+ B cells that proliferated. Two donors of PBMCs were tested per each breast milk sample. Shown in Figure 2A is a representative example from donor 2. By 7 days after infection, 29.3% of cells were CD19+, with 46.3% undergoing at least 1 round of proliferation, as indicated by a decrease in MFI. In contrast, among PBMCs treated with medium alone, only 7.68% had undergone only 1 round of proliferation. Table 1 shows the results from 20 breast milk samples, with the mean proliferation (±standard error of the mean [SEM]) induced by exposure to breast milk being 22.6% ± 3.1% for donor 1 and 23.3% ± 2.8% for donor 2. As another indicator of transformation, cells were also monitored for blast formation at 4 days (Figure 2B) and then at 8 weeks after infection (Figure 2C). LCLs were derived from 10 of 20 breast milk samples tested, but no LCLs were generated when PBMCs were cultured with medium alone. To confirm EBV infection in the LCLs, 10 samples were typed for EBV strain by PCR (Figure 2D), and all LCLs were EBV type 1.
Figure 2.
Peripheral blood mononuclear cells (PBMCs) were labeled with e450 proliferation dye and exposed to breast milk collected at approximately postpartum week 6 or medium for 2 hours. Subsequently, cells were washed, cultured for 7 days, and analyzed for proliferation via flow cytometry. B cells were initially gated and analyzed for proliferation, as measured by diluting fluorescence intensity of e450 proliferation dye. A, Representative histograms of B-cell proliferation for 20 individual breast milk samples collected at approximately postpartum week 6; 2 PBMC donors per breast milk sample were tested. B, Prototypical Epstein-Barr virus (EBV)-induced cell aggregation was observed microscopically 4 days after exposure to breast milk. Representative images are shown. C and D, Following exposure to breast milk, PBMCs were cultured for 8 weeks and examined for generation of EBV lymphoblastoid cell lines. C, Microscopic images of 2 lymphoblastoid cell lines generated form PBMCs following exposure to breast milk collected at approximately postpartum week 6 are shown. D, Breast milk lymphoblastoid cell lines were analyzed by polymerase chain reaction (PCR) analysis of the EBNA3C encoding region to determine EBV type. The EBV-negative cell line Ramos served as a negative control, and B95.8 and Jijoye cell lines were used as positive controls for type 1 and type 2 EBV, respectively. PCR product sizes were as expected for EBV type 1 and type 2.
Table 1.
Copies of Epstein-Barr Virus (EBV) Type 1 and Type 2 and Percentage Proliferation Following Exposure to Breast Milk Samples Collected at Postpartum Week 6
| Sample | EBV Type(s) | EBV Copies per PBMC, No.a |
Proliferating B Cells, % |
||
|---|---|---|---|---|---|
| Type 1 | Type 2 | Donor 1 | Donor 2 | ||
| Medium only | ND | ND | ND | 6.5b | 6.0b |
| Breast milk | |||||
| 1 | 1, 2 | 209.90 | 201.93 | 10.6 | 8.29 |
| 2 | 1, 2 | 297.86 | <1c | 60.5 | 46.3 |
| 3 | 1, 2 | 477.02 | <1c | 59.4 | 51.9 |
| 4 | 1, 2 | 121.38 | 81.15 | 17.6 | 14.1 |
| 5 | 1, 2 | 117.14 | <1c | 24.1 | 9.4 |
| 6 | 1, 2 | 443.12 | 106.90 | 14.8 | 18.7 |
| 7 | 1, 2 | 77.55 | 114.60 | 10.9 | 30.5 |
| 8 | 1 | 384.07 | ND | 16.7 | 20.1 |
| 9 | 1 | 240.48 | ND | 31.0 | 27.7 |
| 10 | 1 | 397.20 | ND | 17.1 | 25.3 |
| 11 | 1, 2 | 871.77 | 119.77 | 20.6 | 22.0 |
| 12 | 1, 2 | 333.04 | 1966.68 | 16.5 | ND |
| 13 | 1 | 88.87 | ND | 12.3 | 16.6 |
| 14 | 1, 2 | 408.16 | 444.15 | 20.7 | 15.6 |
| 15 | 1, 2 | 166.06 | <1c | 19.1 | 22.3 |
| 16 | 1, 2 | 155.33 | <1c | 16.8 | 14.3 |
| 17 | 1, 2 | 5566.59 | <1c | 26.1 | 38.9 |
| 18 | 1 | 778.66 | ND | 14.6 | 15.9 |
| 19 | 1 | 550.37 | ND | 32.2 | 26.1 |
| 20 | 1, 2 | 472.78 | 267.95 | 25.9 | 35.2 |
Abbreviations: ND, not detected; PBMC, peripheral blood mononuclear cell.
a Copies of type 1 and type 2 in 750 µL of breast milk cultured with PBMCs for 2 hours.
b Data are averaged from 2 independent experiments.
c Samples in which EBV genome was detected but fell below the standard curve are indicated as <1 copy/µL DNA.
Breast Milk Contains Both Strains of EBV
We next determined whether EBV type 1 or type 2 was present in the breast milk samples collected at approximately postpartum week 6. We found that 14 of 20 samples contained both EBV type 1 and type 2, while 6 of 20 contained type 1 alone. No samples were detected that contained EBV type 2 alone. We also quantified the viral load difference between EBV type 1 and type 2 and found that the mean EBV type 1 load (±SEM) was 810 ± 354 copies/mL, while that for type 2 was 551 ± 191 copies/mL (P < .0001, by the unpaired t test).
DISCUSSION
We have previously reported that primary EBV infection in infants from Kisumu can occur in infants <6 months of age [5]. A cohort study in this same region of Kenya was then conducted and actively followed up pregnant women during pregnancy through the postpartum period. We hypothesized that malaria during pregnancy could cause EBV reactivation, leading to a high EBV load in circulation and shedding of the virus, which could subsequently enhance the risk of EBV infection at an early age. Our initial findings from this cohort study revealed a significantly higher frequency of elevated EBV loads—an indicator of EBV reactivation—in pregnant women with P. falciparum malaria, compared with women without evidence of P. falciparum infection [12]. In the present study, we examined samples from this same cohort and observed EBV DNA in the breast milk; that the virus in the breast milk was DNAse I resistant, indicating it was present as virions; and that EBV DNA–positive breast milk induced B-cell proliferation and long-term LCLs, indicating that the virus was infectious. This suggests that breast milk could be a potential source of virus transmission to infants in a malaria-endemic region. Mothers were more likely to shed EBV in breast milk if they had malaria at birth, pointing to a potential role for malaria in enhancing EBV transmission and resulting in EBV infection before 6 months of age.
The present study is the first to examine longitudinal breast milk samples obtained from EBV-seropositive mothers from approximately postpartum weeks 6–18 in an area where malaria is endemic and the eBL risk is high. The frequency of EBV detection and the EBV load were significantly higher at postpartum week 6, with declining prevalence over time. These findings suggest that EBV shedding in breast milk is greatest during the first few weeks of the postpartum period and that breast-feeding infants could be exposed to a higher EBV load soon after delivery. The observed declining trend of both frequency and viral load over time could be as a result of a rebound of immunity especially cell-mediated immunity during the postpartum period [16, 17].
Previous cross-sectional studies reported an EBV DNA prevalence in breast milk of 45% in women tested in Zimbabwe [10] and 46% in British Columbia women [9], suggesting that EBV shedding in breast milk can occur. Still unexplored by these studies was whether the EBV DNA in breast milk was encapsidated or whether it represented naked DNA released by cellular lysis of EBV-infected cells. To assess this possibility, an assay that relies on DNAse I treatment, which selectively degrades naked viral DNA, was used in this study. EBV in breast milk supernatant was found to resist degradation by DNAse I, suggesting that the virus was encapsidated and present as virions. The origin of cell-free virus in breast milk is largely unknown. We observed evidence of viral transformation (eg, blast formation, clumping, and proliferation) of CD19+ B cells after short-term exposure of PBMCs to breast milk. Conclusive evidence that the virus in the breast milk was infectious was obtained through the generation of EBV-infected LCL. Of note, the presence of encapsidated DNA in breast milk is not sufficient for infectivity of EBV; instead, transformation of lymphocytes by EBV also requires the presence of the viral envelope. These data are the first to demonstrate that EBV in breast milk is infectious and capable of transforming B cells and supports the hypothesis that breast milk could be a source for EBV transmission to infants.
EBV can be grouped into 2 major types, type 1 and type 2, based on variances in the sequence of the genes that code for EBV nuclear antigen 2 (EBNA-2), EBNA-3a, EBNA-3b, and EBNA-3c [18, 19]. EBV type 1, the dominant type worldwide, immortalizes B cells much more efficiently than EBV-2, which is common in sub-Saharan Africa [20, 21]. Detection of EBV type 2 has been observed in both blood samples [22] and in saliva [23]. However, whether both EBV strains can be detected in breast milk was previously unknown. In this study, we detected both EBV type 1 and type 2 in breast milk but only EBV type 1 in the LCLs generated from breast milk samples. This could be indicative of either the poor transforming ability of EBV type 2 or the lower levels of EBV type 2 found in breast milk samples, compared with EBV type 1.
A study conducted in Japan found comparable rates of EBV transmission in breast-fed versus formula-fed children at the age of 12–23 months, suggesting that breast-feeding does not add any potential risk for mother-to-child transmission of EBV [11]. In a more recent study based in the United States, differences in EBV seroprevalence in US children suggested other factors, possibly including genetics or other aspects of the family environment [24]. However, geographical differences in age at primary EBV infection exist, with infants in sub-Saharan Africa getting infected earlier in life than their counterparts in the developed world, where infections occur in adolescent and adulthood [5, 25]. We recently documented EBV infections that occurred before 6 months of age among children living in a malaria-holoendemic area of western Kenya [5]. Thus, the findings from Japan [11] would not account for the differences in age of primary infection between Japan and Kenya and cannot be extrapolated to our cohort. Our observation that the presence of malaria at delivery increased the likelihood of EBV detection in breast milk suggests an important role for P. falciparum malaria in enhancing shedding of EBV in breast milk.
In summary, we observed EBV DNA in the breast milk; that the virus in the breast milk was DNAse I resistant, indicating it was present as virions; and that EBV DNA–positive breast milk induced B-cell proliferation and long-term EBV-transformed LCLs, indicating that the virus was infectious. Moreover, P. falciparum infection plays a role in enhancing EBV shedding in breast milk and suggests that breast milk could be a potential source of virus transmission to infants in a malaria-endemic and eBL high-risk region.
Notes
Acknowledgments. We thank the Chulaimbo Subdistrict Hospital, for allowing us to use their facilities to perform this study; Philip Bondo, our clinical officers, medical officers, and data entry and field staff involved in the project; and the mothers, for their participation in this study. The manuscript was submitted for publication with the permission of the Director of the Kenya Medical Research Institute.
Financial support. This work was supported by the National Cancer Institute (CA102667 to R. R. and D43 training grant NCI 153707 to I. I. D.) and the National Institute of Allergy and Infectious Diseases (AI 098511 to A. E. D.), National Institutes of Health; and Burroughs Wellcome (CAMS 1006818 to A. E. D.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Dalldorf G. Lymphomas of African children. JAMA 1963; 183:619–20. [DOI] [PubMed] [Google Scholar]
- 2.de-The G. Is Burkitt's lymphoma related to perinatal infection by Epstein-Barr virus? Lancet 1977; 1:335–8. [DOI] [PubMed] [Google Scholar]
- 3.Morrow RH. Epidemiological evidence for the role of falciparum malaria in the pathogenesis of Burkitt's lymphoma. IARC Sci Pub 1985; 60:177–86. [PubMed] [Google Scholar]
- 4.Rainey JJ, Mwanda WO, Wairiumu P, Moormann AM, Wilson ML, Rochford R. Spatial distribution of Burkitt's lymphoma in Kenya and association with malaria risk. Trop Med Int Health 2007; 12:936–43. [DOI] [PubMed] [Google Scholar]
- 5.Piriou E, Asito AS, Sumba PO et al. . Early age at time of primary Epstein-Barr virus infection results in poorly controlled viral infection in infants from Western Kenya: clues to the etiology of endemic Burkitt lymphoma. J Infecti Dis 2012; 205:906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen JI. Epstein-Barr virus infection. N Engl J Med 2000; 343:481–92. [DOI] [PubMed] [Google Scholar]
- 7.Niederman JC, Miller G, Pearson HA, Pagano JS, Dowaliby JM. Infectious mononucleosis. Epstein-Barr-virus shedding in saliva and the oropharynx. N Engl J Med 1976; 294:1355–9. [DOI] [PubMed] [Google Scholar]
- 8.Yao QY, Rickinson AB, Epstein MA. A re-examination of the Epstein-Barr virus carrier state in healthy seropositive individuals. Int J Cancer 1985; 35:35–42. [DOI] [PubMed] [Google Scholar]
- 9.Junker AK, Thomas EE, Radcliffe A, Forsyth RB, Davidson AG, Rymo L. Epstein-Barr virus shedding in breast milk. Am J Med Sci 1991; 302:220–3. [DOI] [PubMed] [Google Scholar]
- 10.Gantt S, Carlsson J, Shetty AK et al. . Cytomegalovirus and Epstein-Barr virus in breast milk are associated with HIV-1 shedding but not with mastitis. AIDS 2008; 22:1453–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusuhara K, Takabayashi A, Ueda K et al. . Breast milk is not a significant source for early Epstein-Barr virus or human herpesvirus 6 infection in infants: a seroepidemiologic study in 2 endemic areas of human T-cell lymphotropic virus type I in Japan. Microbiol Immunol 1997; 41:309–12. [DOI] [PubMed] [Google Scholar]
- 12.Daud I, Ogolla S, Amolo A et al. . Plasmodium falciparum infection is associated with epstein–barr virus reactivation in pregnant women living in malaria holoendemic area of Western Kenya. Matern Child Health J 2015; 19:606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moormann AM, Chelimo K, Sumba OP et al. . Exposure to holoendemic malaria results in elevated Epstein-Barr virus loads in children. J Infect Dis 2005; 191:1233–8. [DOI] [PubMed] [Google Scholar]
- 14.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol 1993; 67:2014–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heckman JJ. Sample selection bias as a specification error. J Econ Soc 1979; 47:153–61. [Google Scholar]
- 16.Shimaoka Y, Hidaka Y, Tada H et al. . Changes in cytokine production during and after normal pregnancy. Am J Reprod Immunol 2000; 44:143–7. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe M, Iwatani Y, Kaneda T et al. . Changes in T, B, and NK lymphocyte subsets during and after normal pregnancy. Am J Reprod Immunol 1997; 37:368–77. [DOI] [PubMed] [Google Scholar]
- 18.Rowe D, Heston L, Metlay J, Miller G. Identification and expression of a nuclear antigen from the genomic region of the Jijoye strain of Epstein-Barr virus that is missing in its nonimmortalizing deletion mutant, P3HR-1. Proc Natl Acad Sci USA 1985; 82:7429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sample J, Young L, Martin B et al. . Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol 1990; 64:4084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rickinson AB, Young LS, Rowe M. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J Virol 1987; 61:1310–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young LS, Yao QY, Rooney CM et al. . New type B isolates of Epstein-Barr virus from Burkitt's lymphoma and from normal individuals in endemic areas. J Gen Virol 1987; 68(Pt 11):2853–62. [DOI] [PubMed] [Google Scholar]
- 22.Sixbey J, Chesney PJ, Shirley P, Buntin D, Resnick L. Detection of a second widespread strain of Epstein-Barr virus. Lancet 1989; 334:761–5. [DOI] [PubMed] [Google Scholar]
- 23.Walling DM, Brown AL, Etienne W, Keitel WA, Ling PD. Multiple Epstein-Barr virus infections in healthy individuals. J Virol 2003; 77:6546–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Condon LM, Cederberg LE, Rabinovitch MD et al. . Age-specific prevalence of Epstein-Barr virus infection among Minnesota children: effects of race/ethnicity and family environment. Clin Infect Dis 2014; 59:501–8. [DOI] [PubMed] [Google Scholar]
- 25.Biggar RJ, Henle G, Bocker J, Lennette ET, Fleisher G, Henle W. Primary Epstein-Barr virus infections in African infants. II. Clinical and serological observations during seroconversion. Int J Cancer 1978; 22:244–50. [DOI] [PubMed] [Google Scholar]