Abstract
Thermal processes are widely used in small molecule chemical analysis and metabolomics for derivatization, vaporization, chromatography, and ionization, especially in gas chromatography mass spectrometry (GC/MS). In this study the effect of heating was examined on a set of 64 small molecule standards and, separately, on human plasma metabolite extracts. The samples, either derivatized or underivatized, were heated at three different temperatures (60, 100, and 250 °C) at different exposure times (30 s, 60 s, and 300 s). All the samples were analyzed by liquid chromatography coupled to electrospray ionization mass spectrometry (LC/MS) and the data processed by XCMS Online (xcmsonline.scripps.edu). The results showed that heating at an elevated temperature of 100 °C had an appreciable effect on both the underivatized and derivatized molecules, and heating at 250 °C created substantial changes in the profile. For example, over 40% of the molecular peaks were altered in the plasma metabolite analysis after heating (250 °C, 300s) with a significant formation of degradation and transformation products. The analysis of 64 small molecule standards validated the temperature-induced changes observed on the plasma metabolites, where most of the small molecules degraded at elevated temperatures even after minimal exposure times (30 s). For example, tri- and diorganophosphates (e.g., adenosine triphosphate and adenosine diphosphate) were readily degraded into a mono-organophosphate (e.g., adenosine monophosphate) during heating. Nucleosides and nucleotides (e.g., inosine and inosine monophosphate) were also found to be transformed into purine derivatives (e.g., hypoxanthine). A newly formed transformation product, oleoyl ethyl amide, was identified in both the underivatized and derivatized forms of the plasma extracts and small molecule standard mixture, and was likely generated from oleic acid. Overall these analyses show that small molecules and metabolites undergo significant time-sensitive alterations when exposed to elevated temperatures, especially those conditions that mimic sample preparation and analysis in GC/MS experiments.
Small molecule analysis has become increasingly important across a broad range of research areas, including drug discovery,1 microbiology,2 plant physiology,3 nutrition,4 and the environmental sciences.5 It has been applied to examine pathophysiological processes,6 biomarker discovery,7,8 and unknown biological pathways.9 Over the past decade, metabolomic sample preparation, analytical techniques, and data analysis platforms have been maturing rapidly, with the most widely used analytical techniques being nuclear magnetic resonance spectroscopy (NMR) and hyphenated methods such as gas and liquid chromatography mass spectrometry (GC/MS and LC/MS). Due to their high sensitivity and resolution, MS-based technologies have become increasingly popular. GC/MS, in particular, combines excellent separation with sensitive mass detection. Also, the extensive and reproducible fragmentation patterns in the electron ionization (EI) source used in GC/MS can readily identify or classify small molecule metabolites. In contrast, LC/MS provides softer ionization; electrospray ionization (ESI) or atmospheric pressure chemical ionization (APCI) facilitate intact molecular ion formation and metabolite characterization with tandem mass spectrometry (MS/MS). Together, GC/MS and LC/MS/MS, when combined with the existing libraries (e.g., NIST,10 HMDB,11 LipidMaps,12 and METLIN13) have been increasingly applied to metabolomics and small molecule analysis in general.
One of the most significant challenges in generating accurate molecular profiles is maintaining the stability of molecules. Many small molecules, metabolites, or their intermediates are labile and can potentially undergo degradation during sample handling, including storage, extraction, and analysis. Temperature control is one of the key aspects of metabolomic experiments. For example, an NMR study showed that urine sample storage at different temperatures, i.e., room temperature (22 °C), refrigeration (4 °C), and low temperature freezing (−80 °C), can affect metabolite stability over time.14 In another study, plasma samples left at room temperature for 2.5 h had a significant impact on choline compound stability,15 albeit both studies were based on the chemical analysis of a limited set of metabolites. While lower temperatures are typically used to store compounds (as described above), the use of heat can have an even greater impact on the thousands of metabolites present in a biological sample. For example, derivatization at elevated temperatures is widely used in GC/MS analysis, where the reactions are usually conducted at 60 °C for several hours.16,17 In newborn screening, reactions between the derivatization reagent and amino acids from dried blood spots are heated to 60–80 °C for over 30 min.18,19 High temperatures are also used during the desolvation (vaporization temperature: 200–350 °C) and ionization processes (e.g., APCI) for LC/MS analysis, though the residence time is short. More importantly, heating is widely used in GC/MS analysis to facilitate gas phase generation and separation of metabolites from a sample. In a standard GC/MS method recommended in the user guideline of the GC/MS Metabolomics Library,16 the GC oven is ramped by 10 °C/min from 60 °C (1 min initial time) to 325 °C (10 min final time), resulting in a 37.5 min run time at elevated temperatures. Also, high temperatures are used in the injector (250 °C) and MS transfer line (290 °C). Interestingly, at least half of the metabolite peaks typically elute after 17 min when the calculated oven temperature is over 200 °C.20
To date, very little information is available on the effect of heating on small molecules and metabolites. In a previous GC/MS study analyzing human plasma metabolites, about 40% of the observed peaks remained unidentified even after annotation using a metabolite library.21 This relatively low level of identification is a common problem in global untargeted GC/MS metabolomic studies. While the lack of a comprehensive GC/MS library for human metabolites could explain this, it is possible that some of the metabolite peaks are formed due to derivatization byproducts or unknown metabolites formed at the high temperatures.
Since many small molecules contain polar functional groups and are thermally labile at the temperatures required for their vaporization and separation, derivatization prior to GC analysis is needed to extend the application of GC-based methods. The majority of GC methods derivatize with an oximation reagent followed by silylation,17,22 or alternatively, chloroformates are used.23 The derivatization step is designed to protect the small molecules from degrading at high GC/MS temperatures. However, to date, no direct experimental evidence is available to confirm this hypothesis.
Furthermore, the derivatization efficiency can vary greatly between small molecules, as shown by Koek et al.24 When using the most common derivatization agents, ethoxyamine hydrochloride and N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA), the derivatization efficiency of the amino acids alanine and tryptophan was 100% and 30%, respectively, and for fumaric acid and fructose 6-phosphate it was 60% and 45%, respectively.24 Therefore, a substantial portion of those small molecule standard mixtures contained the nonderivatized form when injected onto the GC/MS. Given the broad use of GC/MS, it is important to investigate the fate and transformation of those small molecules during the heating process in the sample preparation and instrumental analysis.
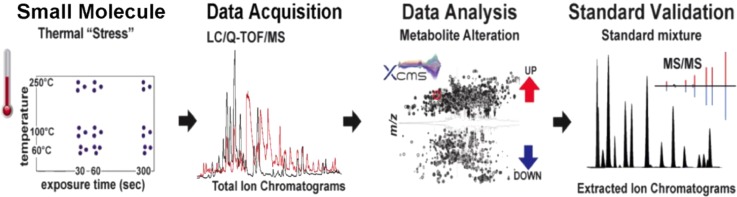
In this study, we designed a set of experiments (Figure 1) to examine the global changes that occur at three different temperatures commonly used for small molecule sample preparation and analysis (60, 100, and 250 °C). Specifically, we examined the effect of heating on the stability of small molecules by observing the global metabolomic profile (using HPLC-ESI-QTOF/MS) of human plasma metabolite extracts and a standard mixture of 64 small molecules including amino acids, purines, sugars, sugar phosphates, and free fatty acids.
Figure 1.
Global molecular profiling approach used to investigate heating on small molecule stability.
Experimental Section
Sample Preparation and Derivatization
The suppliers of all the chemicals and solvents were fully described in Supporting Information (SI) Text S1. A stock of standard human serum was obtained from Sigma-Aldrich (Catalog No.: H4522, human male AB plasma, USA origin, sterile-filtered). Plasma metabolites were extracted using a solvent mixture of acetonitrile and methanol. Aliquots of 200 μL serum was extracted with 800 μL ice-cold acetonitrile:methanol (1:1, v/v). The mixture was sonicated for 5 min and incubated for 1 h at −20 °C to precipitate the protein, which was followed by 15 min centrifugation at 13,000 rpm at 4 °C. The resulting supernatant was removed and combined together as a stock sample for the following experiments. Subsequently, ∼ 240 μL of the sample, which was equivalent to ∼60 μL plasma, was aliquoted into amber silylated GC vials and evaporated to dryness in a vacuum concentrator. The derivatization step was modified from several previous studies17,22,25 and was accepted as the standard method in the derivatization of GC/MS based metabolomics methods. In brief, ∼ 100 μL of toluene (dried over anhydrous sodium sulfate) was added to the dry sample, mixed for 1 min and dried again in the vacuum concentrator for 20 min. To the dried metabolite sample, 50 μL of fresh methoxylamine (MOX) reagent (20 mg/mL) was added and the samples incubated in the oven at 60 °C for 2 h. Subsequently, 100 μL of BSTFA with 1% (v/v) trimethylchlorosilane (TCMS) was rapidly added to each sample and the mixture was incubated for 1 h at 60 °C to form trimethylsilylated (TMS) derivatives.
Prior to heating, all the plasma small molecule metabolites were extracted and evaporated to dryness using a vacuum concentrator. All the vials were capped with a Teflon coated GC inlet septum which showed high temperature resistance and little background during heating. To minimize possible oxidation during heating, the vials were flushed with high purity nitrogen. For the underivatized plasma metabolites, heating at three different temperatures at 60, 100, and 250 °C was conducted. To observe the temporal change, samples were collected after heating for 30, 60, and 300 s. For the derivatized plasma metabolites, 250 °C at 30, 60, and 300 s was used. For the standard mixture, only 250 °C at 60 and 300s was used to compare against room temperature (25 °C) for both the underivatized and derivatized mixture because fewer features were observed at 60 and 100 and 250 °C 30 s. At each temperature and time point, triplicate biological replicates together with a blank control were prepared. After heating, 60 μL acetonitrile/H2O (1:1, v/v) was spiked into each vial and vortex-mixed after cooling down. Then 15 min centrifugation at 13,000 rpm at 4 °C was used to precipitate the insoluble particles and the supernatants were transferred to microvolume inserts prior to analysis.
Sample Analysis
The analysis of samples using HPLC-ESI-QTOF/MS in both reversed phase chromatography ESI positive mode (RP-ESI (+)) and hydrophilic interaction chromatography ESI negative mode (HILIC- ESI (−)) was detailed in Text S-2. The data analysis and quality control were described in Text S-3 and S-4.
Results and Discussion
Global Profiling of Heated Plasma Metabolites
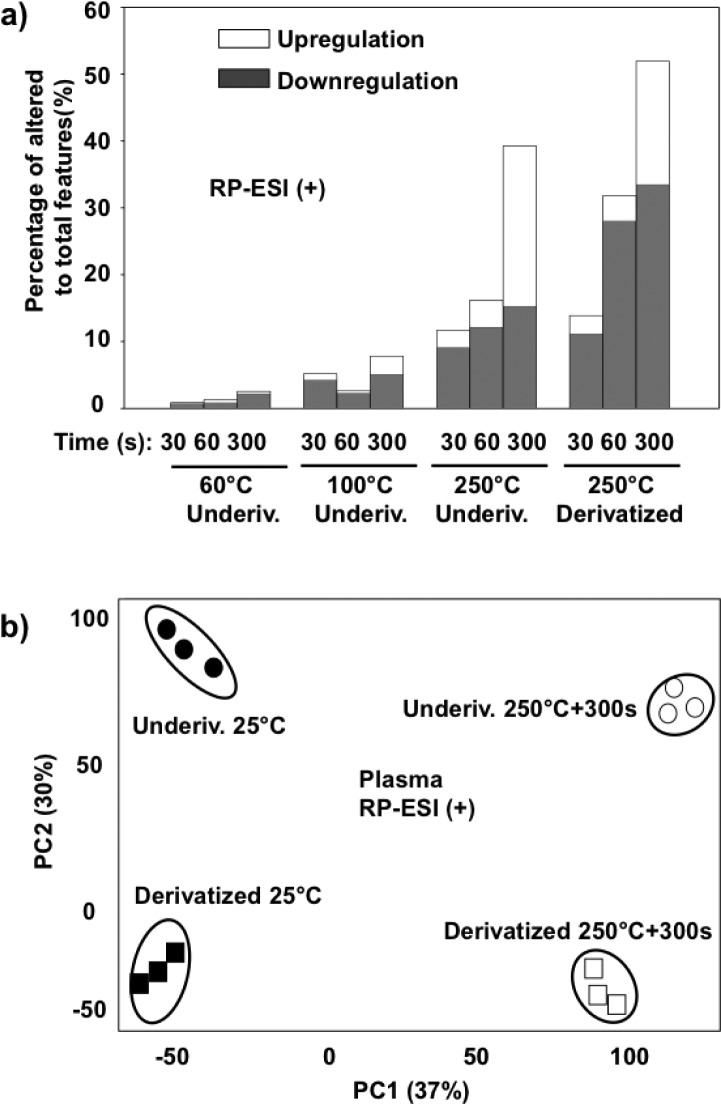
As shown in Figures 1 and 2, separate sets of plasma extracts were each heated at 60 °C, 100 °C, and 250 °C for three different time periods (30 s, 60 s, and 300 s). Generally it was observed that heating at high temperatures has a significant effect on the small molecules, as reflected in the percentage of total altered features in the plasma metabolites (Figure 2-a and S-1 (a)). Specifically, relatively few altered features were observed for the samples heated at 60 °C during all heating periods compared to the control at room temperature, 25 °C. At 100 °C heating for 300 s, 18% of the total aligned features were altered. As shown in Figure S-2 (b), small molecule metabolites heated at 100 °C for 300 s were also clearly separated from other conditions in the PCA plot. However, no significant alterations were observed for the molecules exposed to the shorter heating periods (30 s and 60 s). At a heating temperature of 250 °C, significantly higher number of features were altered (Figure 2-a and S-1 (a)). The percentages of altered features were 14%, 21%, and 51%, for 30 s, 60 s, and 300 s exposure time, respectively, most of which were observed by RP-ESI (+). As shown in Figure 1, the total ion chromatography (TIC) of the plasma metabolites heated at 250 °C was visually distinct from the 25 °C control. The greatest increase in altered features was observed in the sample heated for 300 s and the features were widely distributed along the entire gradient on the C18-RP column (Figure 3-a).
Figure 2.
Percentage of altered (both upregulated and downregulated) features compared to the total features in the underivatized and derivatized plasma small molecule metabolites heated at 60 °C, 100 °C, and 250 °C for 30 s, 60 s, and 300 s. The subsequent analyses were performed using (a) RP-ESI (+) and overall results displayed with (b) principal component analysis scores plots of 25 and 250 °C for 300 s heated metabolites. All the upregulated and downregulated features were from pairwise comparison of each tested condition with room temperature (25 °C) and filtered using p < 0.01.
Figure 3.
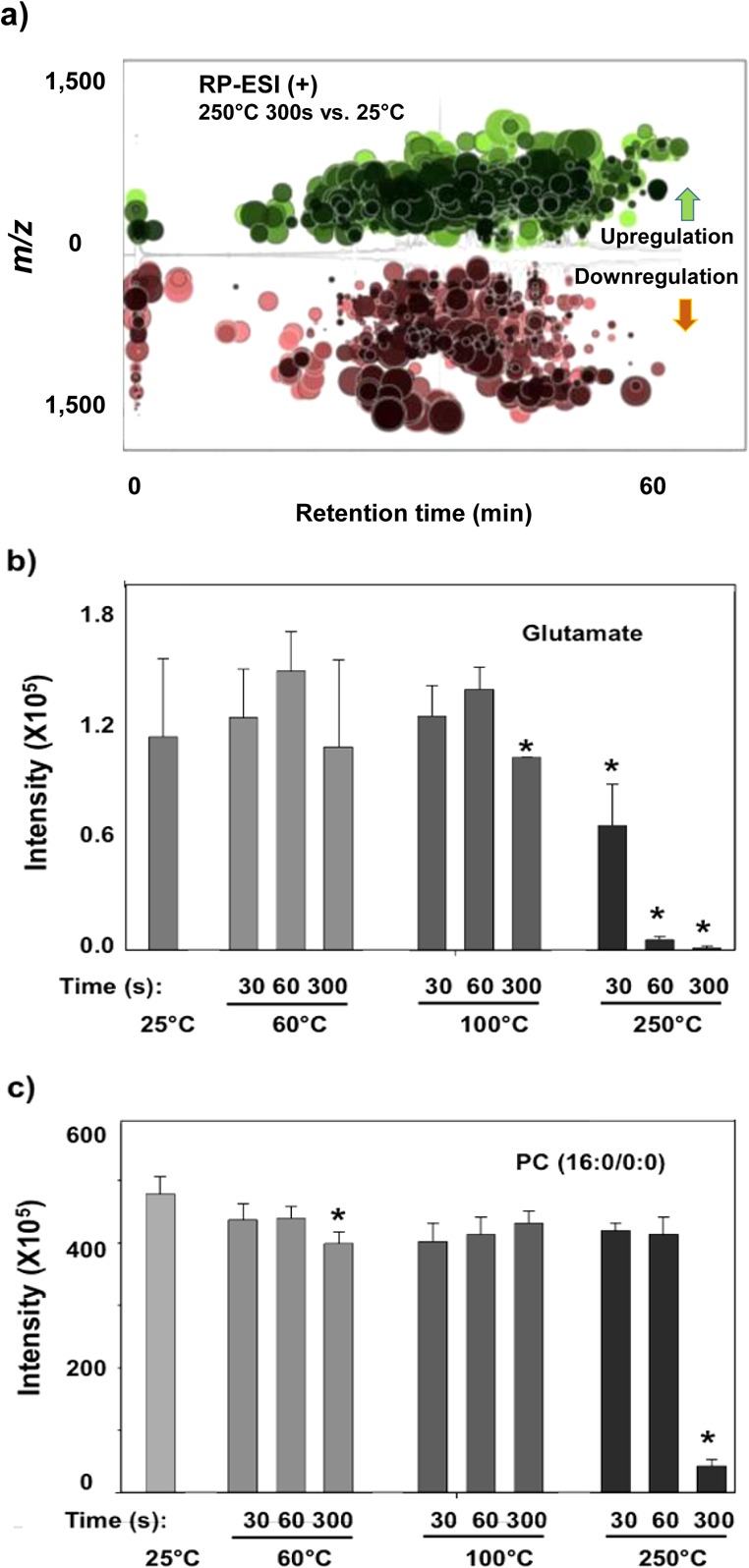
(a) Cloud plot showing thousands of altered features (represented by “bubbles”) from the underivatized plasma metabolites heated at 250 °C for 300 s (filtered by fold change >1.5 and p < 0.01). (b and c) Response change of glutamate and PC(16:0/0:0) heated at different temperatures and durations. The results and the standard deviation were obtained from triplicate analysis. “*” represent the samples which are significantly different from 25 °C with p < 0.01.
Interestingly, the numbers of upregulated features increased dramatically with the longer heating period and over 50% of the total altered features were upregulated. The majority of altered features were not previously observed, suggesting the formation of new molecules at high temperature. These newly formed metabolites may represent either the breakdown byproducts or de novo chemically produced small molecules under the high temperature. This finding could also explain the presence of many unknown peaks in the GC/MS analysis of the plasma extracts.
Several metabolites were further analyzed to validate the observed temperature-induced alterations. Glutamate, linoleic acid, and PC (16:0/0:0) were selected due to their high abundance and important roles in biochemistry. As shown in Figures 3b and c and S-3, these molecules degraded to different extents. While no significant change was observed at 60 and 100 °C during three heating periods, most of glutamate and PC (16:0/0:0) were absent after heating at 250 °C for 300 s. Glutamate seems to be among the most labile compounds. Approximately half degraded within 30 s, with complete degradation after 60 s. This finding is consistent with previous results that demonstrated thermal degradation of amino and organic acids.26 Linoleic acid gradually degraded under 250 °C heating, and ∼50% disappeared after heating for 60 s (Figure S3). PC (16:0/0:0) did not show a significant decrease after heating at 250 °C for 60 s; though it decreased rapidly after 300 s.
Global Profiling of Heated Plasma Metabolites after Derivatization
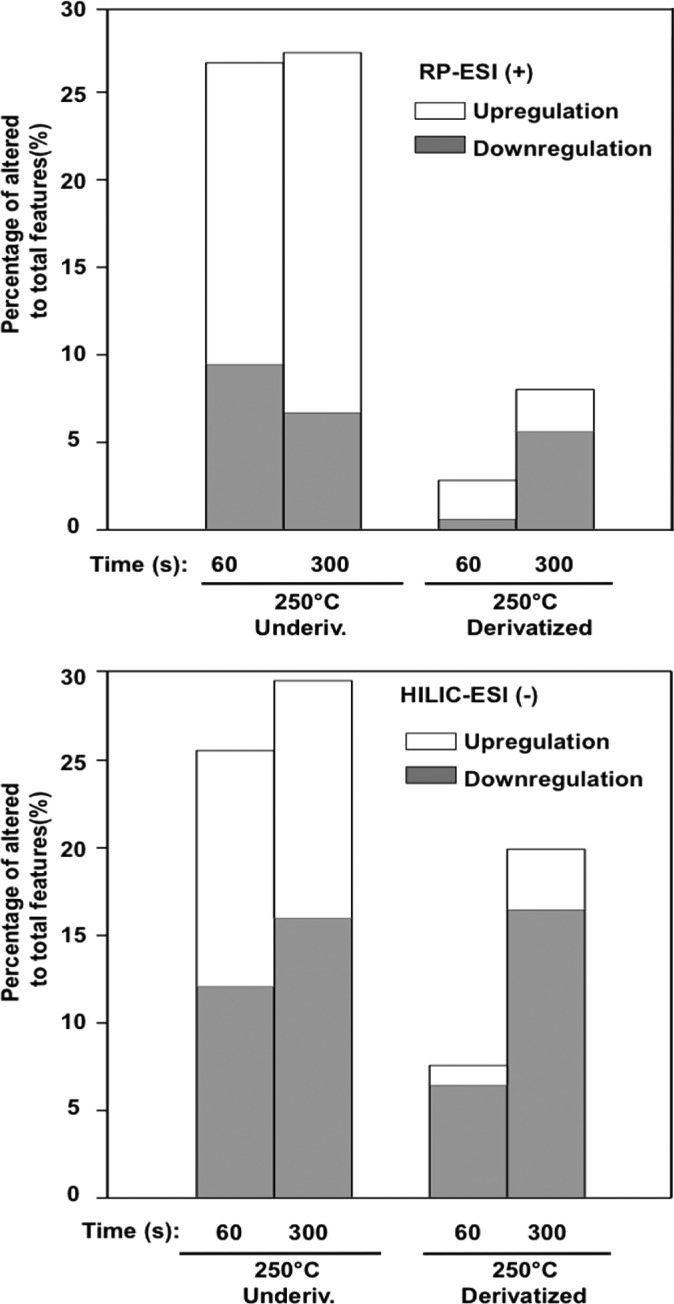
To explore the effect of derivatization on the plasma metabolome, the dry extracts were derivatized by the most widely used method, silylation, and then heated at 250 °C at different times. Overall, the profile of the derivatized small molecules was altered greatly after heating at 250 °C and the derivatization did not effectively protect the metabolites. As shown in Table S2, the percentages of the altered features to the total features were 27%, 41%, and 65% for 30 s, 60 s, and 300 s of heating at 250 °C, respectively. Furthermore, the derivatized small molecules showed a similar time course effect to the underivatized plasma at 250 °C. The PCA scores plot in Figure S-2 (d) shows that the feature variance could be well explained by two principal components and the plasma profiles were well grouped depending on sample preparation (heating and derivatization factor). Derivatization did not prevent temperature-induced changes. However, it was interesting to find that the percentage of the upregulated features (∼35% in Table S2) decreased in total in the derivatized plasma metabolites, compared with the underivatized plasma metabolites exposed to the same heating effect. This suggests that derivatization might decrease the de novo formation of new compounds from the breakdown or chemical reaction between metabolites.
Degradation of Underivatized and Derivatized Small Molecule Standards
Since many of the central carbon metabolites showed low signal intensities from the plasma sample, we prepared a mixture of 64 small molecules to examine the heating effect using similar experimental conditions. As shown in Figure 4 and Table S-3, heating greatly alters the global profile for both the underivatized and derivatized small molecule mixtures. PCA revealed that the underivatized standards were distinct from each other when treated with different temperatures at different times (25 °C control, 250 °C 60 s, and 250 °C 300 s) (See Figure S-4 (b)). The RP-ESI (+) and HILIC-ESI (−) data sets increased by 52% and 57% for the 60 s and 300 s heating at 250 °C, respectively. Approximately half of the altered features increased across the retention time window between 10 and 40 min on the C18 column (Figure S-4 (a)). It should also be noted that analysis of these samples using LC-ESI-MS provided an important alternative for examining the heating effect in GC/MS, since GC/MS requires heating to facilitate thermal vaporization.
Figure 4.
Percentage of altered (both upregulated and downregulated) features to the total features in the underivatized and derivatized 64 small molecule mixture heated with 250 °C for 30, 60, and 300 s using both the RP-ESI (+) and HILIC-ESI (−) methods.
Overall, heating at high temperature (250 °C) greatly altered the profile of both the standards and plasma metabolites after derivatization. However, more changes were observed from the plasma extracts, likely due to its higher complexity with thousands of metabolites. For example, it is known that many plasma lipids have not yet been well characterized.27 The derivatization efficacy for those metabolites is not known, and their breakdown byproducts could affect the global profiling of the plasma extracts.
It is worth noting that derivatization efficiency was also investigated to examine the effect on the global profile. The silylation derivatization method used has been widely applied in GC/MS based global metabolomics17,20 and in the quantification of many types of metabolites.24 The derivatization efficiency was estimated by comparing the samples before and after derivatization at room temperature (25 °C). As shown in Table S-1, the derivatization efficiency differed greatly among small molecules. Some of the small molecules could be efficiently derivatized, such as ATP, nucleotide sugar, free fatty acids, and metabolites associated with glycolysis. However, the derivatization efficiency was less than 50% for some molecules in the pathways of purine metabolism (e.g., adenosine and uric acid) and amino acid synthesis (e.g., tyrosine and cysteine sulfate). Some molecules, including UMP, xanthine, and arginine, could not be derivatized at all. This result was consistent with a previous study in which the derivatization efficiency significantly differed between compounds.24
Degradation of Small Molecule Standards
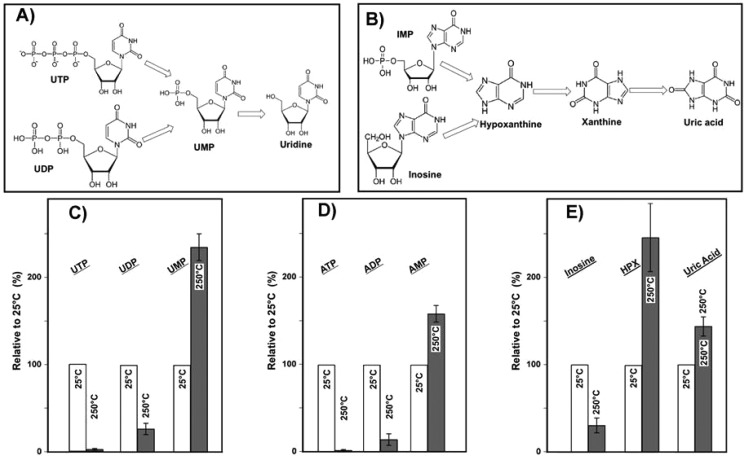
Most of the selected small molecule standards degraded rapidly at 250 °C, as summarized in Table S-1. The small organic acids (e.g., citric acid and malate), amino acid intermediates (e,g., aminoadipic acid and saccharopine), amides (e.g., glutamate), and several energy currency metabolites (e.g., NAD+, NADH, phospheoenol pyruvate, and acetyl-CoA) disappeared after 60 s of heating at 250 °C, suggesting that those molecules were particularly unstable. Consistent with the observation in the plasma extracts, fatty acids tended to be more stable. For the triphosphate nucleotides (See Figure 5C–E and Table S-1), such as ATP, UTP, and GTP, no detectable concentration was left after heating for 60 s, which was followed by similar degradation of their diphosphate metabolites.
Figure 5.
Proposed thermal degradation pathways for: (A) UTP, UDP, UMP, and uridine and (B) IMP, inosine, hypoxanthine, xanthine, and uric acid. Additionally, the relative increases of: (C) UTP, UDP, and UMP; D) ATP, ADP, and AMP; and (E) inosine, hypoxanthine (HPX), and uric acid heated at 250 °C for 60 s compared with 25 °C are shown. Error bars show standard deviation (based on triplicates).
We also observed that some molecules initially increased and then decreased over time. For example, the relative abundance of several monophosphates (i.e., AMP, CMP, GMP, and UMP) experienced an increase during the first 60 s and then decreased dramatically up to 300 s (Figures 5-C and D and Table S-1), suggesting that triphosphates and diphosphates could be degraded into monophosphate analogues as suggested in Figure 5-A. The level of uridine also increased after heating the mixture for 60 s at 250 °C (∼1.4 times higher than at 25 °C, Table S-1) and did not experience any significant drop after 300 s of heating, suggesting that the parent compounds such as UMP, UDP, or UMP break down to uridine. Though it is known that triphosphates are relatively unstable, this study for the first time showed triphosphates could be converted sequentially to monophosphates or even nucleosides during the heating process. The triphosphate metabolites such as ATP play an important role in the biological pathways, and dysregulation has been related with many diseases (e.g., Parkinson’s disease).28 For example, the ratio between intercellular ATP and AMP has been used to estimate the malfunction of the mitochondria dysfunction during aging and pathology.29 This study showed that conversion from ATP to AMP during sample treatment should be considered in future studies and very careful steps should be conducted to avoid the breakdown of ATP.
Another interesting observation was the increase of several tested molecules, including hypoxanthine and uric acid during heating. For example, the level of hypoxanthine after heating for 300 s at 250 °C was 2.8 times higher than the 25 °C control. Several molecules could contribute as the sources of this increase. Small molecules such as inosine (decreased over 70% after 60 s at 250 °C, Figure 5-e) and inosine monophosphate (IMP), both containing hypoxanthine, could potentially breakdown to generate hypoxanthine during the heating process. The deamination of adenine could also be a possible source for hypoxanthine.30 The steady concentration of xanthine and slight increase of uric acid after heating might be from other conversion processes such as oxidation of hypoxanthine. A possible pathway is shown in Figure 5-B. To further validate these changes, we conducted thermal sensitivity experiments on individual compounds of ATP, inosine, and IMP at 150 and 250 °C with higher temporal resolution (30 s, 60 s, 120 s, and 300 s). As shown in Figure S5, ATP was readily degraded to ADP and AMP at both 150 and 250 °C, which could be further degraded into adenosine and adenine at 250 °C. Again, a gradual increase of hypoxanthine was observed in the heating of inosine and IMP at 250 °C (no change at 150 °C). In general, the results in this study using both metabolite mixtures and individual compounds consistently showed the interconversion of many small molecules during heating, ultimately adding to the complexity of the global metabolomic experimental results.
Identification of de Novo Byproducts Formed during Heating
Many upregulated features were observed for both the heated plasma metabolite and the standard mixture, some of which were not detected in the control samples at 25 °C, suggesting the formation of new products during heating. To help characterize and identify the de novo generated byproducts during heating, we have performed targeted MS/MS for several of the most significantly altered features. Sixty targeted precursor ions were examined in both ESI (+) and ESI (−), and the resulting MS/MS fragments were searched. Most of the targeted MS/MS fragments gave no hits in the METLIN database or provided a very low match score, suggesting the heating byproducts were not common human metabolites. Interestingly, one possible byproduct was identified as oleoyl ethyl amide from the 250 °C heated standard mixture and the plasma metabolites and was further confirmed with a pure standard. As shown in Figure S-6, it was rapidly formed in both underivatized and derivatized samples and was observed in high abundance after 60 s of heating at 250 °C with a clear time-dependent trend. Also, this byproduct was identified in the heated underivatized and derivatized samples at 250 °C for 300 s and there was no detection in all heated blank samples used as controls. It is unknown how this byproduct is formed during heating; it is possible that the oleic acid could react with ethylamine, a breakdown product of other metabolites. For other unidentified features in heated plasma metabolites, a very interesting pattern was observed in many targeted MS/MS fragments. As shown in Figure S-7, a typical neutral loss of 45.057 mass units and m/z of 72.08 and 90.09 was observed. The m/z of 45.057 and 72.08 could be possibly C2H7N and C4H10N, respectively. One possible source of those ions was from the breakdown of peptides. Under heated conditions, peptides can potentially degrade and interfere with the global metabolomics profiling, which could contribute to the complexity of the data interpretation.
Conclusion
In this study the combined effect of heating and chemical derivatization on the human plasma extracts and a set of small molecule standards was investigated using global molecular profiling. The results show that heating had a significant effect on both sets of samples. Alterations began to be observed on plasma metabolites at 100 °C for 300 s. Most importantly, over 40% of the total features were altered after heating at 250 °C for 300 s and approximately half of these altered features were upregulated, suggesting that new compounds are being formed during the heating process. Furthermore, the protective effect of derivatization was not found to be significant. To validate these observations, we examined a standard mixture of 64 small molecules. The mixture profile was considerably altered, with the majority of molecules degraded within 60 s of being heated at 250 °C. For example, tri- and diorganophosphates (i.e., ATP and ADP) readily degraded to mono-organophosphate (AMP). Nucleotides (i.e., inosine and inosine monophosphate) were found to transform into purine derivatives (i.e., hypoxanthine). Oleoyl ethyl amide was one of the commonly identified byproducts observed in both the underivatized and derivatized mixtures after heating, which could be formed via reaction(s) involving oleic acid.
These results indicate that heating at elevated temperature (e.g., 250 °C) significantly alters the molecular profile and even moderate temperatures can produce significant degradation/transformation. These alterations are especially consequential in experiments involving minutes of exposure to elevated temperatures, such as those used in GC/MS systems, indicating that a significant amount of the spectral data generated in GC/MS experiments may correspond to thermal degradation products. These results may also provide an explanation as to why mass spectral databases (such as NIST) have been computationally challenging to interpret, as a significant amount of data may be coming from thermal degradation of the standards.
Acknowledgments
This study was funded by NIH Grants R01 GM114368 and P01 A1043376-02S1.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.5b03003.
Description of chemical materials, LC/MS analysis, data analysis, and QA/QC; calculated derivatization efficiency and response ratio of the 64 tested compounds; number of features of heated raw and derivatized plasma metabolites and of heated underivatized and derivatized small molecule mixtures; percentage of altered features compared to the total features in the underivatized and derivatized plasma metabolites; principal component analysis scores plot for underivatized and derivatized plasma metabolites; response change of linoleic acid heated at different temperatures and heating times in the underivatized plasma metabolites; cloud plot showing altered metabolite features, and PCA scores plot of underivatized metabolite mixture; formation of adenine, adenosine, AMP, and ADP from single 10 μM ATP heating, and formation of hypoxanthine (HPX) from single 10 μM inosine and IMP heating; identification of oleoyl ethyl amide as a possible byproduct during heating; examples of MS/MS fragments in the targeted MS/MS (m/z 400.3148, 272.254, and 300.2855) using RP-ESI (+) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Kell D. B. Drug Discovery Today 2006, 11, 1085–1092 10.1016/j.drudis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Allen J.; Davey H. M.; Broadhurst D.; Heald J. K.; Rowland J. J.; Oliver S. G.; Kell D. B. Nat. Biotechnol. 2003, 21, 692–696 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- Fiehn O.; Kopka J.; Dormann P.; Altmann T.; Trethewey R. N.; Willmitzer L. Nat. Biotechnol. 2000, 18, 1157–1161 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Lodge J. K. Proc. Nutr. Soc. 2010, 69, 95–102 10.1017/S0029665109991704. [DOI] [PubMed] [Google Scholar]
- Bundy J. G.; Davey M. P.; Viant M. R. Metabolomics 2009, 5, 3–21 10.1007/s11306-008-0152-0. [DOI] [Google Scholar]
- Patti G. J.; Yanes O.; Shriver L. P.; Courade J. P.; Tautenhahn R.; Manchester M.; Siuzdak G. Nat. Chem. Biol. 2012, 8, 232–234 10.1038/nchembio.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. H.; Dejea C. M.; Edler D.; Hoang L. T.; Santidrian A. F.; Felding B. H.; Ivanisevic J.; Cho K.; Wick E. C.; Hechenbleikner E. M.; Uritboonthai W.; Goetz L.; Casero R. A. Jr.; Pardoll D. M.; White J. R.; Patti G. J.; Sears C. L.; Siuzdak G. Cell Metab. 2015, 21, 891–897 10.1016/j.cmet.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatine M. S.; Liu E.; Morrow D. A.; Heller E.; McCarroll R.; Wiegand R.; Berriz G. F.; Roth F. P.; Gerszten R. E. Circulation 2005, 112, 3868–3875 10.1161/CIRCULATIONAHA.105.569137. [DOI] [PubMed] [Google Scholar]
- Peyraud R.; Kiefer P.; Christen P.; Massou S.; Portais J. C.; Vorholt J. A. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 4846–4851 10.1073/pnas.0810932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok V. I.; Linstrom P. J.; Reed J. J.; Zenkevich I. G.; Brown R. L.; Mallard W. G.; Stein S. E. Journal of chromatography. A 2007, 1157, 414–421 10.1016/j.chroma.2007.05.044. [DOI] [PubMed] [Google Scholar]
- Wishart D. S.; Knox C.; Guo A. C.; Eisner R.; Young N.; Gautam B.; Hau D. D.; Psychogios N. Nucleic acids research 2009, 37, D603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud M.; Fahy E.; Cotter D.; Brown A.; Dennis E. A.; Glass C. K.; Merrill A. H. Jr.; Murphy R. C.; Raetz C. R.; Russell D. W.; Subramaniam S. Nucleic Acids Res. 2007, 35, D527–532 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A.; O’Maille G.; Want E. J.; Qin C.; Trauger S. A.; Brandon T. R.; Custodio D. E.; Abagyan R.; Siuzdak G. Ther. Drug Monit. 2005, 27, 747–751 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Saude E. J.; Sykes B. D. Metabolomics 2007, 3, 19–27 10.1007/s11306-006-0042-2. [DOI] [Google Scholar]
- Pinto J.; Domingues M. R.; Galhano E.; Pita C.; Almeida Mdo C.; Carreira I. M.; Gil A. M. Analyst 2014, 139, 1168–1177 10.1039/c3an02188b. [DOI] [PubMed] [Google Scholar]
- Agilent. Agilent G1676AA Fiehn GC/MS Metabolomics RTL Library. http://www.chem.agilent.com/Library/usermanuals/Public/G1676-90001_Fiehn.pdf, 2013.
- Dunn W. B.; Broadhurst D.; Begley P.; Zelena E.; Francis-McIntyre S.; Anderson N.; Brown M.; Knowles J. D.; Halsall A.; Haselden J. N.; Nicholls A. W.; Wilson I. D.; Kell D. B.; Goodacre R. Human Serum Metabolome, C. Nat. Protoc. 2011, 6, 1060–1083 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- Febriani A. D.; Sakamoto A.; Ono H.; Sakura N.; Ueda K.; Yoshii C.; Kubota M.; Yanagawa J. Pediatr. Int. 2004, 46, 5–9 10.1111/j.1442-200X.2004.01825.x. [DOI] [PubMed] [Google Scholar]
- Wada M.; Kuroki M.; Minami Y.; Ikeda R.; Sekitani Y.; Takamura N.; Kawakami S.; Kuroda N.; Nakashima K. Biomed. Chromatogr. 2014, 28, 810–814 10.1002/bmc.3142. [DOI] [PubMed] [Google Scholar]
- Palazoglu M.; Fiehn O.. Metabolite Identification in Blood Plasma Using GC/MS and the Agilent Fiehn GC/MS Metabolomics RTL Library; http://www.chem.agilent.com/Library/applications/5990-3638en_lo%20CMS.pdf, 2009.
- Psychogios N.; Hau D. D.; Peng J.; Guo A. C.; Mandal R.; Bouatra S.; Sinelnikov I.; Krishnamurthy R.; Eisner R.; Gautam B.; Young N.; Xia J.; Knox C.; Dong E.; Huang P.; Hollander Z.; Pedersen T. L.; Smith S. R.; Bamforth F.; Greiner R.; McManus B.; Newman J. W.; Goodfriend T.; Wishart D. S. PLoS One 2011, 6, e16957. 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E. C.; Pasikanti K. K.; Nicholson J. K. Nat. Protoc. 2011, 6, 1483–1499 10.1038/nprot.2011.375. [DOI] [PubMed] [Google Scholar]
- Qiu Y.; Su M.; Liu Y.; Chen M.; Gu J.; Zhang J.; Jia W. Anal. Chim. Acta 2007, 583, 277–283 10.1016/j.aca.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Koek M. M.; Muilwijk B.; van der Werf M. J.; Hankemeier T. Anal. Chem. 2006, 78, 1272–1281 10.1021/ac051683+. [DOI] [PubMed] [Google Scholar]
- Fiehn O. TrAC, Trends Anal. Chem. 2008, 27, 261–269 10.1016/j.trac.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N.; Quitain A. T.; Kang K.; Daimon H.; Fujie K. Ind. Eng. Chem. Res. 2004, 43, 3217–3222 10.1021/ie020733n. [DOI] [Google Scholar]
- Bicalho B.; David F.; Rumplel K.; Kindt E.; Sandra P. Journal of chromatography. A 2008, 1211, 120–128 10.1016/j.chroma.2008.09.066. [DOI] [PubMed] [Google Scholar]
- Perier C.; Vila M. Cold Spring Harbor Perspect. Med. 2012, 2, a009332. 10.1101/cshperspect.a009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.; Li J. Comprehensive Physiology 2015, 5, 667–686 10.1002/cphy.c140041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunstmann B.; Osiewacz H. D. Aging Cell 2008, 7, 651–662 10.1111/j.1474-9726.2008.00412.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.