Abstract
A variety of lines of evidence support the idea that neutral evolutionary processes (genetic drift, mutation) have been important in generating cranial differences between Neandertals and modern humans. But how do Neandertals and modern humans compare with other species? And how do these comparisons illuminate the evolutionary processes underlying cranial diversification? To address these questions, we used 27 standard cranial measurements collected on 2524 recent modern humans, 20 Neandertals and 237 common chimpanzees to estimate split times between Neandertals and modern humans, and between Pan troglodytes verus and two other subspecies of common chimpanzee. Consistent with a neutral divergence, the Neandertal versus modern human split-time estimates based on cranial measurements are similar to those based on DNA sequences. By contrast, the common chimpanzee cranial estimates are much lower than DNA-sequence estimates. Apparently, cranial evolution has been unconstrained in Neandertals and modern humans compared with common chimpanzees. Based on these and additional analyses, it appears that cranial differentiation in common chimpanzees has been restricted by stabilizing natural selection. Alternatively, this restriction could be due to genetic and/or developmental constraints on the amount of within-group variance (relative to effective population size) available for genetic drift to act on.
Keywords: developmental constraint, genetic drift, Homo neanderthalensis, Homo sapiens, Pan troglodytes, stabilizing natural selection
1. Introduction
Neandertals are especially important for understanding human evolution, because, along with ‘Denisovans', they are the closest relatives of our own, modern human,1 evolutionary lineage [1,2]. ‘Classic’ Neandertals appear in the fossil record at approximately 130 ka and persist until approximately 40 ka [3–5]. While on present data the Neandertal geographical range appears to have been centred in Europe, it extended as far south as Israel and east as southern Siberia [4,6]. Modern humans were initially present in Africa [7–11], or just outside, but they began to populate the rest of the world during 60–45 ka [12,13]. The Neandertal and modern human evolutionary lineages diverged at approximately 500 ka [2,4,14–19], with subsequent limited gene flow between them when modern humans encountered Neandertals during their expansion from Africa [2,14,20].
A variety of lines of evidence support the idea that neutral evolutionary processes2 have been important in generating cranial differences between Neandertals and modern humans [21,22]. On the one hand, statistical tests based on theoretical predictions from quantitative and population genetics fail to detect deviations from neutrality for cranial differences between Neandertals and modern humans [23]; and cranial divergence between Neandertals and modern humans appears to have followed a sort of ‘morphological clock’ analogous to the ‘molecular clock’ [17]. These findings build on pioneering work that showed patterns of cranial differentiation among recent modern human groups were consistent with neutral divergence [24,25]. On the other hand, detailed functional analyses fail to support adaptive hypotheses such as climatic adaptation [26] and anterior dental loading (‘teeth as tools’) [27,28] for Neandertal cranial morphology. Finally, the piecemeal appearance of Neandertal features documented by the fossil record [4,29,30] seems more consistent with neutrality than with existing adaptive hypotheses.
But how do Neandertals and modern humans compare with other species? And how do these comparisons illuminate the evolutionary processes underlying cranial diversification? We would like to know if neutral evolutionary processes played a more important role in the cranial diversification of Neandertals and modern humans than they did in the cranial diversification of other taxa.
Common chimpanzees (Pan troglodytes) provide a useful starting point for placing the Neandertal and modern human results in a broader comparative context, because, along with bonobos (P. paniscus), they are the extant species most closely related to humans. Of particular interest are comparisons of the western subspecies of common chimpanzee (P. t. verus) with the central (P. t. troglodytes) and eastern (P. t. schweinfurthii) subspecies, because the lineage leading to the western subspecies split from the lineages leading to the other two subspecies about the same time that the lineages leading to Neandertals and modern humans split from each other [14,15,31–33]. Holding split time constant leads to more straightforward comparisons, because multiple studies have documented an inverse relationship between the rate of morphological divergence and split time [34–39].
Here, we use morphological split-time estimates and analyses of the amount of mutational variance required to produce between- and within-group patterns of morphological variation to assess the relative importance of neutral evolutionary process and natural selection in shaping patterns of cranial variation in Neandertals, modern humans and common chimpanzees.
2. Material and methods
(a). Data
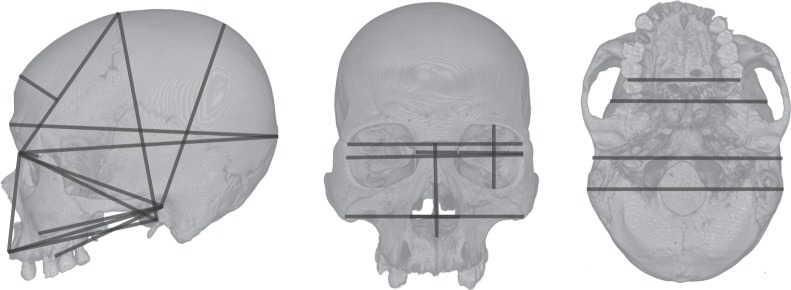
We base our analyses on 27 standard cranial measurements (figure 1; electronic supplementary material, table S1) collected on 2524 recent modern humans, 20 Neandertals and 237 common chimpanzees. The measurements were collected on the actual specimens with callipers. The recent modern humans come from 30 globally distributed groups, and the common chimpanzees come from three subspecies (electronic supplementary material, table S2). Owing to the fragmentary nature of fossil specimens, the Neandertal sample contains missing data, but we only use this sample to estimate the Neandertal mean for each measurement. Additional details about the Homo sample [23,40–42], Pan sample [43], measurements [40,43] and the treatment of missing data [23] can be found elsewhere.
Figure 1.
Cranial measurements. The approximate locations of the 27 cranial measurements used in the analyses (defined in electronic supplementary material, table S1) superimposed as dark grey lines on lateral, anterior and inferior views of a human cranium. Note that when the endpoints of a measurement are not visible, the line is projected into a plane situated in front of the cranium.
(b). Split-time analyses
If the morphological measurements of two groups have diverged neutrally (by genetic drift and mutation alone), then PTD, a morphological split-time estimator, is expected to be equal to how many generations in the past the groups shared a common ancestor [17] (electronic supplementary material, appendix A1). We calculated PTD for the split between Neandertals and modern humans, and between the western subspecies and the central and eastern subspecies of common chimpanzee, assuming mutation drift equilibrium (balance between the addition of variation by mutation and removal of variation by genetic drift; V0 = (V1 + V2)/2 in equation (1.1) of electronic supplementary material, appendix A1) or no additive genetic variance in the last common ancestor (V0 = 0 in equation (1.1) of electronic supplementary material, appendix A1). The former assumption gives a minimum estimate unless there was a decline in effective population size; the latter gives a maximum estimate [17,44,45].
For each split, we calculated PTD along each of the 27 eigenvectors of the human within-group variance–covariance matrix, taking the mean as a point estimate and resampling with replacement 10 000 times from the individual estimates to produce bootstrap confidence limits [17]. We added 25 000 years to each Neandertal versus modern human split-time estimate to account for the fact (averaging dates) that Neandertals lived approximately 50 000 years ago [17]. For consistency with Weaver et al.'s [17] analyses, we used the same values for the (narrow-sense) heritability, mutation constant and generation length (table 1) for our PTD calculations. Heritability allows within-group morphological variances to be converted into within-group additive genetic variances; the mutation constant and heritability allow within-group morphological variances to be converted into mutational variances; and generation length allows split times in generations to be converted into split times in years (see electronic supplementary material, appendix A3 for a numerical example).
Table 1.
Parameter values for split-time calculations.
| heritability (h2) | mutation constant (m) | generation length (g) |
|---|---|---|
| 0.37 | 1.20 × 10−4 | 25 years |
Weaver et al. [17] used patterns of DNA sequence and cranial variation in recent modern human groups to estimate the heritability and mutation constant values. More specifically, this previous study used two ‘known’ reference points: the split time between sub-Saharan African and other recent modern human populations, and the effective population size for sub-Saharan African populations, picking values for heritability and the mutation constant that made the morphological estimates of these reference points match those from microsatellite (short-tandem repeat) markers. We explore potential variation in these, and other parameters in additional, mutational variance analyses. We performed all of the split-time analyses in Matlab (Mathworks).
(c). Mutational variance analyses
If assumptions are made about the split time between two groups and generation length (table 2), then it is possible to calculate the mutational variance required, if the measurements diverged neutrally, to produce the observed morphological divergence (equation (2.2) of electronic supplementary material, appendix A2). Similarly, if assumptions are made about heritability, effective population size of the two groups (considered together) and among-region differentiation (FST) (table 2), then it is possible to calculate the mutational variance required, if the measurements evolved neutrally, to produce the observed within-group morphological variance (equation (2.5) of electronic supplementary material, appendix A2). Under neutrality, these two mutational variance estimates should match, because both between- and within-group patterns of variation are expected to depend on the same mutation constant [54–56].
Table 2.
Assumed parameter ranges for mutational variance analyses.
| parameter | range | sources |
|---|---|---|
| heritability (h2) | (0.3, 0.6) | [25,46,47] |
| generation length (g) | (25, 30) | [15,48] |
| human among-region differentiation (FST) | (0.10, 0.15) | [49,50] |
| common chimpanzee among subspecies differentiation (FST) | (0.20, 0.25) | [32,49] |
| Neandertal versus modern human split time (ty) | (270 000, 790 000) | [14,15] |
| western versus other common chimpanzee split time (ty) | (420 000, 850 000) | [31–33] |
| human effective population size (Ne) | (4000, 17 000) | [31,51,52] |
| common chimpanzee effective population size (Ne) | (21 000, 62 000) | [31,53] |
For consistency with the PTD calculations, we conducted the split-time mutational variance analyses in the space of the eigenvectors of the human within-group variance–covariance matrix; the results are similar if the common chimpanzee within-subspecies variance–covariance matrix is used instead. The results of the within-group mutational variance analyses are identical regardless of whether the calculations are done in the space of the original variables, the eigenvectors of the human within-group variance–covariance matrix or the eigenvectors of the common chimpanzee within-group variance–covariance matrix. We performed all of the mutational variance analyses in Matlab.
3. Results
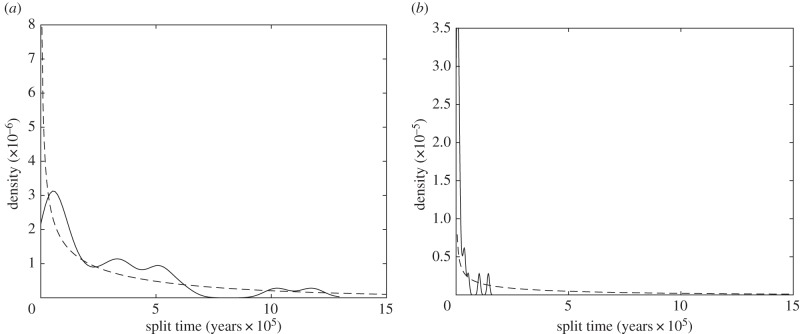
Morphological split-time estimates for Neandertals versus modern humans based on 27 measurements—281 or 405 ka—are slightly lower than (but similar to) those based on 37 measurements—311 or 435 ka (table 3) [17]. By contrast, analogous estimates for western versus central common chimpanzees—16 or 140 ka—or western versus eastern common chimpanzees—26 or 151 ka—are much lower (table 3). Evidence from DNA sequences indicates that all three of these splits occurred at approximately 500 ka [2,14,15,31–33] (table 3), so the morphological estimates for Homo correspond, at least broadly, with DNA sequence estimates, whereas those for Pan are too young (figure 2). Using a shorter generation length for common chimpanzees would only increase the contrast between Homo and Pan by lowering the common chimpanzee split-time estimates.
Table 3.
Split-time estimates.
| assumption | Neandertal versus modern human | western versus central common chimpanzee | western versus eastern common chimpanzee |
|---|---|---|---|
| mutation drift equilibrium | 281a (181, 400)b | 16 (9, 24) | 26 (11, 47) |
| no initial within-group variance (V0 = 0) | 405 (304, 522) | 140 (133, 148) | 151 (135, 171) |
| molecular estimates | 270–790c | 420–850d | 420–850d |
Figure 2.
Split-time estimate distributions. (a) Neandertal versus modern human. (b) Pan troglodytes verus versus other subspecies. In both panels, the solid curve summarizes the distribution of the split-time estimates along each of the 27 eigenvectors of the human within-group variance–covariance matrix, and the dashed curve gives the expected distribution under neutrality, if the split occurred at 500 ka. The y-axis (density) is scaled so that each curve integrates to one.
Under neutrality and mutation drift equilibrium, the rate of morphological divergence will not depend on effective population size. Changes in effective population size will impact the rate, but PTD can account for some such changes, as long as the divergence was neutral. Roughly, if the average effective population size of the daughter groups is larger than the effective population size of the ancestral group, the actual split time will be in between the mutation drift equilibrium (lower) and V0 = 0 (higher) estimates; if the effective population sizes at both time points are roughly the same, the mutation drift equilibrium estimate will be close to the actual split time; and if the average effective population size of the daughter groups is smaller than the effective population size of the ancestral group, even the mutation drift equilibrium estimate will be too high.
Based on these considerations and estimates of the effective population sizes of common chimpanzees, Neandertals, modern humans and their ancestors [2,31], the most appropriate model for the splits between the western and central subspecies of common chimpanzee, and between Neandertals and modern humans, appear to be somewhere in between the mutation drift equilibrium and V0 = 0 models. That is, if the divergence was neutral, the mutation drift equilibrium and V0 = 0 estimates would be expected to bracket the actual split time. For the split between the western and eastern subspecies of common chimpanzee, it appears that a small effective population size in the western subspecies is not compensated for by a large effective population size in the eastern subspecies, resulting in a reduction, on average, in effective population size relative to the last common ancestor of common chimpanzees [31]. In this case, even the mutation drift equilibrium estimate would be expected to be too high under neutrality.
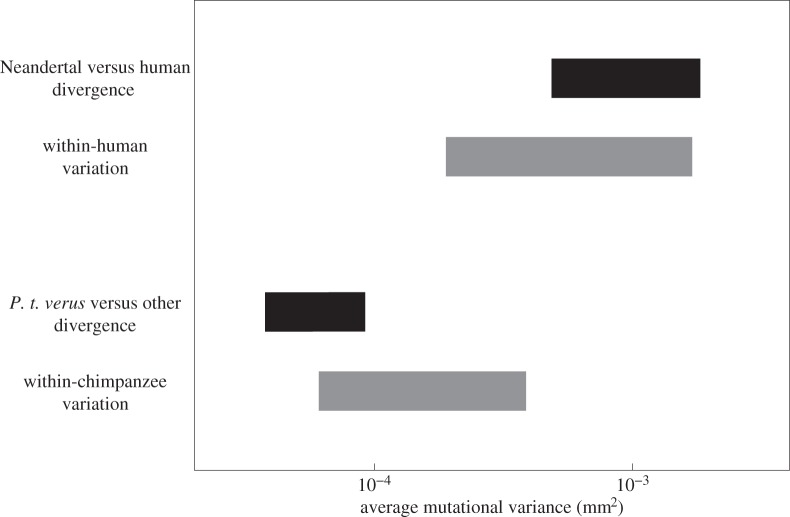
All of these split-time estimates, however, depend on assumptions about mutation; in this case, how much additive genetic variance is introduced by mutation per zygote per generation (mutational variance, which depends on the values of the mutation constant and heritability). Therefore, to investigate potential variation in these and other parameters, we performed analyses of the mutational variance required to explain between- and within-group patterns of variation, given a range of possible parameter values (table 2). As expected based on the young common chimpanzee morphological split-time estimates, the mutational variance required, if the measurements diverged neutrally, to produce the observed morphological divergence is lower for common chimpanzees than for Neandertals and modern humans (figure 3). In other words, to be consistent with neutral morphological divergence, the mutational input in common chimpanzees would need to be lower than in Neandertals and modern humans, because less morphological divergence occurred in roughly the same amount of time.
Figure 3.
Mutational variances consistent with between-group divergence and within-group variation. The black bars give the range of average mutational variance values consistent with between-group divergence in cranial form and the generation length and split-time ranges given in table 2. The grey bars give the range of average mutation variance values consistent with within-group variation in cranial form and the heritability, among-region differentiation (FST), and effective population size ranges given in table 2.
The ranges of mutational variance values consistent with neutrality and within-group morphological variation for common chimpanzees overlap with those for modern humans, but the lower end of the common chimpanzee range is lower and the higher end of the human range is higher (figure 3). For both Homo and Pan, the between-group and within-group mutational variance ranges overlap (figure 3), which is consistent with both aspects of variation being shaped by neutral evolutionary processes.
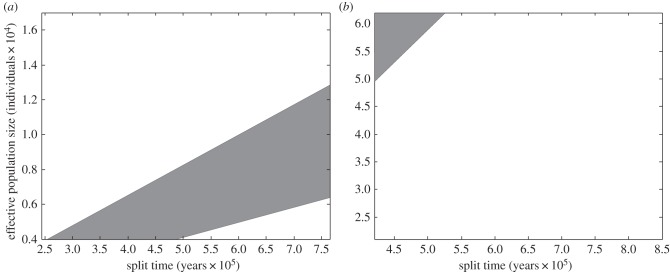
Assuming a generation length of 25 years and the minimum FST given in table 2, the modern human effective population size would have to be on the lower end of DNA sequence estimates for patterns of morphological variation in Neandertals and modern humans to be consistent with neutrality (table 4 and figure 4a); for common chimpanzees, effective population sizes on the higher end and split times on the lower end of DNA sequence estimates would be needed (table 4 and figure 4b).
Table 4.
Effective population sizes and split times consistent with between-group divergence and within-group variation.
| taxa analysed | effective population size (Ne) | split time (ty) |
|---|---|---|
| Neandertals and modern humansa | (4000, 13 000)b | (270 000, 790 000) |
| common chimpanzeesc | (50 000, 62 000) | (420 000, 525 000) |
aCalculations assume g = 25, FST = 0.10 and h2 = (0.3, 0.6).
bEffective population size is for modern humans only.
cCalculations assume g = 25, FST = 0.20 and h2 = (0.3, 0.6).
Figure 4.
Combinations of effective population size and split time consistent with patterns of cranial variation, neutral cranial evolution and DNA sequence estimates. (a) Neandertal versus modern human divergence and within-human variation. (b) Pan troglodytes verus versus other subspecies divergence and within common chimpanzee variation. The shaded area encompasses the space of consistent parameter combinations. The axes span the split time and effective population size ranges given in table 2.
4. Discussion and conclusion
Both the split-time (table 3) and mutational variance (figure 3) analyses demonstrate that cranial divergence was accelerated in Neandertals and modern humans relative to common chimpanzees. Furthermore, the correspondence between the cranial and DNA sequence split-time estimates for Neandertals versus modern humans is consistent with neutral divergence along the evolutionary lineages leading to these two groups. Importantly, directional natural selection may still have shifted both Neandertal and modern human cranial form relative to their last common ancestor's morphology (e.g. parallel increases in brain size relative to body size), because these shifts would maintain similarity between the two lineages, and our analyses focus on differences [17].
Harvati et al.'s [57] investigation of cranial variation in Neandertals, modern humans, African apes and Old World monkeys is one of the only other studies to compare morphological divergence in Neandertals and modern humans with morphological divergence in common chimpanzees. They found that Mahalanobis squared distances calculated from three-dimensional coordinate data were greater between Neandertals and modern humans than between the western subspecies and either the central or the eastern subspecies of common chimpanzee, which is consistent with accelerated cranial divergence in Neandertals and modern humans compared with common chimpanzees. With neutral divergence at mutation drift equilibrium, Mahalanobis squared distances are expected to be proportional to split times [38], but they would need to be calibrated in some way to make quantitative comparisons, as was done here, with split times from DNA sequences.
Using a dataset overlapping with this study, Weaver [43] found that while FST estimates from cranial measurements and DNA sequences were similar for recent modern humans, the cranial estimates were lower than the DNA sequence estimates for common chimpanzees and bonobos. FST measures the degree of departure of two or more groups from a single randomly mating group, with smaller values indicating less differentiation between the groups. The close match of the cranial and DNA sequence FST estimates in modern humans is consistent with neutral cranial divergence, and if morphological divergence in humans was neutral, then it follows that there have been constraints on cranial divergence in chimpanzees [43].
Results of this study and those of Harvati et al. [57] and Weaver [43], taken together, support the idea that stabilizing natural selection has acted more strongly on the common chimpanzee cranium than on Neandertal and modern human crania [58]. It may be that stabilizing selection was stronger in Pan than in Homo because culture (i.e. technology) buffered Homo, at least to a certain extent, from selection on cranial form [38,58,59]. Consistent with this idea, morphological divergence appears to have been constrained by stabilizing selection in a variety of mammalian taxa (including among great ape species) [38]. Nevertheless, based solely on the overlap of the between-group and within-group mutational variance ranges (figure 3), both the common chimpanzee and the Neandertal and modern human patterns of variation are consistent with neutrality. With neutral cranial evolution for both Homo and Pan, the accelerated cranial divergence in Neandertals and modern humans would be the result of more within-group variance (relative to effective population size) available for genetic drift to act on, perhaps because of stronger genetic and/or developmental constraints in common chimpanzees [43,58]. Both the ‘selection-limited’ and ‘variation-limited’ [43] explanations are consistent with the mutational variance results, in part, because we currently have fairly imprecise split time and effective population size estimates. As more DNA sequence data become available, the split time and effective population size ranges estimated from these data should contract, making it possible to more definitely distinguish between selection-limited and variation-limited explanations for the different rates of cranial divergence in Neandertals and modern humans in comparison with common chimpanzees.
Supplementary Material
Supplementary Material
Acknowledgements
We thank I. Cuthill, P. Donoghue, M. dos Reis, T. Steele and an anonymous reviewer for helpful comments; H. Hoekstra, J. Chupasko, D. Lieberman, M. Morgan, J. Rousseau, O. Herschensohn, R. Thorington, D. Lunde, E. Westwig, K. Botha, A. Gill, B. Wilkey, P. Jenkins, L. Tomsett, R. Portella-Miguez, J.-J. Hublin, C. Boesch, U. Schwartz, E. Gilissen and W. Wendelen for access to and assistance with collections; the Ivory Coast authorities for allowing fieldwork in Taï National Park and the export of common chimpanzee skeletal remains; the late W. Howells for generously sharing his data; M. v. Harling for the CT scans used to create figure 1; and the L. S. B. Leakey Foundation, the Calleva Foundation and the Human Origins Research Fund for funding.
Endnotes
Here, ‘modern human’ includes recent (present-day) humans and Holocene and Late Pleistocene fossils that document the evolutionary lineage leading to recent humans, as separate from the lineages leading to Neandertals and other extinct human groups.
With isolation between groups and complete neutrality (i.e. natural selection is not acting at all), genetic drift provides the mechanism and mutation provides the raw material for evolutionary divergence.
Data accessibility
The data required to replicate the analyses can be found in the electronic supplementary material (common chimpanzee and Neandertal measurements), reference [23] (Neandertal measurement means) and on Dr Benjamin M. Auerbach's website (recent modern human measurements): http://web.utk.edu/~auerbach/HOWL.htm.
Authors' contributions
T.D.W. designed the study, collected the common chimpanzee data, analysed the data and wrote the manuscript; C.B.S. collected the Neandertal data and wrote the manuscript. Both authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
T.D.W. received support from the L. S. B. Leakey Foundation to collect the common chimpanzee data. C.B.S.'s research is supported by the Calleva Foundation and the Human Origins Research Fund.
References
- 1.Reich D, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060. ( 10.1038/nature09710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prüfer K, et al. 2013. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49. ( 10.1038/nature12886) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rink WJ, Schwarcz HP, Smith FH, Radovcic J. 1995. ESR ages for Krapina hominids. Nature 378, 24 ( 10.1038/378024a0) [DOI] [PubMed] [Google Scholar]
- 4.Hublin J-J. 2009. The origin of Neandertals. Proc. Natl Acad. Sci. USA 106, 16 022–16 027. ( 10.1073/pnas.0904119106) [DOI] [Google Scholar]
- 5.Higham T, et al. 2014. The timing and spatiotemporal patterning of Neanderthal disappearance. Nature 512, 306–309. ( 10.1038/nature13621) [DOI] [PubMed] [Google Scholar]
- 6.Krause J, et al. 2007. Neanderthals in central Asia and Siberia. Nature 449, 902–904. ( 10.1038/nature06193) [DOI] [PubMed] [Google Scholar]
- 7.Deacon HJ. 1995. Two Late Pleistocene–Holocene archaeological depositories from the southern Cape, South Africa. South Afr. Archaeol. Bull. 50, 121–131. ( 10.2307/3889061) [DOI] [Google Scholar]
- 8.White TD, Asfaw B, DeGusta D, Gilbert H, Richards GD, Suwa G, Howell FC. 2003. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature 423, 742–747. ( 10.1038/nature01669) [DOI] [PubMed] [Google Scholar]
- 9.McDougall I, Brown FH, Fleagle JG. 2005. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature 433, 733–736. ( 10.1038/nature03258) [DOI] [PubMed] [Google Scholar]
- 10.Pearson OM. 2000. Postcranial remains and the origin of modern humans. Evol. Anthropol. 9, 229–247. () [DOI] [Google Scholar]
- 11.Trinkaus E. 2005. Early modern humans. Annu. Rev. Anthropol. 34, 207–230. ( 10.1146/annurev.anthro.34.030905.154913) [DOI] [Google Scholar]
- 12.Henn BM, Cavalli-Sforza LL, Feldman M. 2012. The great human expansion. Proc. Natl Acad. Sci. USA 109, 17 758–17 764. ( 10.1073/pnas.1212380109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hershkovitz I, et al. 2015. Levantine cranium from Manot Cave (Israel) foreshadows the first European modern humans. Nature 520, 216–520. ( 10.1038/nature14134) [DOI] [PubMed] [Google Scholar]
- 14.Green RE, et al. 2010. A draft sequence of the Neanderthal genome. Science 328, 710–722. ( 10.1126/science.1188021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langergraber KE, et al. 2012. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl Acad. Sci. USA 109, 15 716–15 721. ( 10.1073/pnas.1211740109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stringer CB, Hublin J-J. 1999. New age estimates for the Swanscombe hominid, and their significance for human evolution. J. Hum. Evol. 37, 873–877. ( 10.1006/jhev.1999.0367) [DOI] [PubMed] [Google Scholar]
- 17.Weaver TD, Roseman CC, Stringer CB. 2008. Close correspondence between quantitative- and molecular-genetic divergence times for Neandertals and modern humans. Proc. Natl Acad. Sci. USA 105, 4645–4649. ( 10.1073/pnas.0709079105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arsuaga JL, et al. 2014. Neandertal roots: cranial and chronological evidence from Sima de los Huesos. Science 344, 1358–1363. ( 10.1126/science.1253958) [DOI] [PubMed] [Google Scholar]
- 19.Endicott P, Ho SYW, Stringer C. 2010. Using genetic evidence to evaluate four paleoanthropological hypotheses for the timing of Neanderthal and modern human origins. J. Hum. Evol. 59, 87–95. ( 10.1016/j.jhevol.2010.04.005) [DOI] [PubMed] [Google Scholar]
- 20.Currat M, Excoffier L. 2011. Strong reproductive isolation between humans and Neanderthals inferred from observed patterns of introgression. Proc. Natl Acad. Sci. USA 108, 15 129–15 134. ( 10.1073/pnas.1107450108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weaver TD. 2009. The meaning of Neandertal skeletal morphology. Proc. Natl Acad. Sci. USA 106, 16 028–16 033. ( 10.1073/pnas.0903864106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson OM. 2013. Hominin evolution in the Middle-Late Pleistocene: fossils, adaptive scenarios, and alternatives. Curr. Anthropol. 54(Suppl 8), S221–S233. ( 10.1086/673503) [DOI] [Google Scholar]
- 23.Weaver TD, Roseman CC, Stringer CB. 2007. Were Neandertal and modern human cranial differences produced by natural selection or genetic drift? J. Hum. Evol. 53, 135–145. ( 10.1016/j.jhevol.2007.03.001) [DOI] [PubMed] [Google Scholar]
- 24.Lynch M. 1989. Phylogenetic hypotheses under the assumption of neutral quantitative-genetic variation. Evolution 43, 1–17. ( 10.2307/2409160) [DOI] [PubMed] [Google Scholar]
- 25.Relethford JH. 1994. Craniometric variation among modern human populations. Am. J. Phys. Anthropol. 95, 53–62. ( 10.1002/ajpa.1330950105) [DOI] [PubMed] [Google Scholar]
- 26.Holton NE, Franciscus RG. 2008. The paradox of a wide nasal aperture in cold-adapted Neandertals: a causal assessment. J. Hum. Evol. 55, 942–951. ( 10.1016/j.jhevol.2008.07.001) [DOI] [PubMed] [Google Scholar]
- 27.O'Connor CF, Franciscus RG, Holton NE. 2005. Bite force production capability and efficiency in Neandertals and modern humans. Am. J. Phys. Anthropol. 127, 129–151. ( 10.1002/ajpa.20025) [DOI] [PubMed] [Google Scholar]
- 28.Clement AF, Hillson SW, Aiello LC. 2012. Tooth wear, Neanderthal facial morphology and the anterior dental loading hypothesis. J. Hum. Evol. 62, 367–376. ( 10.1016/j.jhevol.2011.11.014) [DOI] [PubMed] [Google Scholar]
- 29.Hublin J-J. 1998. Climatic changes, paleogeography, and the evolution of the Neandertals. In Neandertals and modern humans in Western Asia (eds Akazawa T, Aoki K, Bar-Yosef O), pp. 295–310. New York, NY: Plenum Press. [Google Scholar]
- 30.Hublin J-J. 2014. How to build a Neandertal? Science 344, 1338–1339. ( 10.1126/science.1255554) [DOI] [PubMed] [Google Scholar]
- 31.Prado-Martinez J, et al. 2013. Great ape genetic diversity and population history. Nature 499, 471–475. ( 10.1038/nature12228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becquet C, Patterson N, Stone AC, Przeworski M, Reich D. 2007. Genetic structure of chimpanzee populations. PLoS Genet. 3, e66 ( 10.1371/journal.pgen.0030066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caswell J, Mallick S, Richter DJ, Neubauer J, Schirmer C, Gnerre S, Reich D. 2008. Analysis of chimpanzee history based on genome sequence alignments. PLoS Genet. 4, e1000057 ( 10.1371/journal.pgen.1000057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gingerich PD. 1983. Rates of evolution: effects of time and temporal scaling. Science 222, 159–161. ( 10.1126/science.222.4620.159) [DOI] [PubMed] [Google Scholar]
- 35.Gingerich PD. 2001. Rates of evolution on the time scale of the evolutionary process. Genetica 112–113, 127–144. ( 10.1023/A:1013311015886) [DOI] [PubMed] [Google Scholar]
- 36.Gingerich PD. 2009. Rates of evolution. Annu. Rev. Ecol. Syst. 40, 657–675. ( 10.1146/annurev.ecolsys.39.110707.173457) [DOI] [Google Scholar]
- 37.Kinnison MT, Hendry AP. 2001. The pace of modern life II: from rates of contemporary microevolution to pattern and process. Genetica 112–113, 145–164. ( 10.1023/A:1013375419520) [DOI] [PubMed] [Google Scholar]
- 38.Lynch M. 1990. The rate of morphological evolution in mammals from the standpoint of neutral expectation. Am. Nat. 136, 727–741. ( 10.1086/285128) [DOI] [Google Scholar]
- 39.Roopnarine PD. 2003. Analysis of rates of morphologic evolution. Annu. Rev. Ecol. Syst. 34, 605–632. ( 10.1146/annurev.ecolsys.34.011802.132407) [DOI] [Google Scholar]
- 40.Howells WW. 1973. Cranial variation in man: a study by multivariate analysis of patterns of difference among recent human populations. Cambridge, MA: Harvard University Press. [Google Scholar]
- 41.Howells WW. 1989. Skull shapes and the map: craniometric analyses in the dispersion of modern Homo. Cambridge, MA: Harvard University Press. [Google Scholar]
- 42.Howells WW. 1995. Who's who in skulls: ethnic identification of crania from measurements. Cambridge, MA: Harvard University Press. [Google Scholar]
- 43.Weaver TD. 2014. Brief communication: quantitative- and molecular-genetic differentiation in humans and chimpanzees: implications for the evolutionary processes underlying cranial diversification. Am. J. Phys. Anthropol. 154, 615–620. ( 10.1002/ajpa.22538) [DOI] [PubMed] [Google Scholar]
- 44.Zhivotovsky LA. 2001. Estimating divergence time with the use of microsatellite genetic distances: impacts of population growth and gene flow. Mol. Biol. Evol. 18, 700–709. ( 10.1093/oxfordjournals.molbev.a003852) [DOI] [PubMed] [Google Scholar]
- 45.Zhivotovsky LA, Rosenberg NA, Feldman MW. 2003. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am. J. Hum. Genet. 72, 1171–1186. ( 10.1086/375120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carson EA. 2006. Maximum likelihood estimation of human craniometric heritabilities. Am. J. Phys. Anthropol. 131, 169–180. ( 10.1002/ajpa.20424) [DOI] [PubMed] [Google Scholar]
- 47.Martínez-Abadías N, Esparza M, Sjøvold T, Gonzáles-José R, Santos M, Hernández M. 2009. Heritability of human cranial dimensions: comparing the evolvability of different cranial regions. J. Anat. 214, 19–35. ( 10.1111/j.1469-7580.2008.01015.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fenner JN. 2005. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 128, 415–423. ( 10.1002/ajpa.20188) [DOI] [PubMed] [Google Scholar]
- 49.Fischer A, Pollack J, Thalmann O, Nickel B, Pääbo S. 2006. Demographic history and genetic differentiation in apes. Curr. Biol. 16, 1133–1138. ( 10.1016/j.cub.2006.04.033) [DOI] [PubMed] [Google Scholar]
- 50.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. 2008. Natural selection has driven population differentiation in modern humans. Nat. Genet. 40, 340–345. ( 10.1038/ng.78) [DOI] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475, 403–407. ( 10.1038/nature10215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melé M, et al. 2011. Recombination gives a new insight in the effective population size and the history of the old world human populations. Mol. Biol. Evol. 29, 25–30. ( 10.1093/molbev/msr213) [DOI] [PubMed] [Google Scholar]
- 53.Yu N, Jensen-Seaman MI, Chemnick L, Ryder O, Li W-H. 2004. Nucleotide diversity in gorillas. Genetics 166, 1375–1383. ( 10.1534/genetics.166.3.1375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217, 624–626. ( 10.1038/217624a0) [DOI] [PubMed] [Google Scholar]
- 55.Kimura M. 1989. The neutral theory of molecular evolution and the world view of the neutralists. Genome 31, 24–31. ( 10.1139/g89-009) [DOI] [PubMed] [Google Scholar]
- 56.Lynch M, Hill WG. 1986. Phenotypic evolution by neutral mutation. Evolution 40, 915–935. ( 10.2307/2408753) [DOI] [PubMed] [Google Scholar]
- 57.Harvati K, Frost SR, McNulty KP. 2004. Neanderthal taxonomy reconsidered: implications of 3D primate models of intra- and interspecific differences. Proc. Natl Acad. Sci. USA 101, 1147–1152. ( 10.1073/pnas.0308085100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weaver TD. 2011. Rates of cranial evolution in Neandertals and modern humans. In Computational paleontology (ed. Elewa AMT.), pp. 165–178. Berlin, Germany: Springer. [Google Scholar]
- 59.Ackermann RR, Cheverud JM. 2004. Detecting genetic drift versus selection in human evolution. Proc. Natl Acad. Sci. USA 101, 17 946–17 951. ( 10.1073/pnas.0405919102) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data required to replicate the analyses can be found in the electronic supplementary material (common chimpanzee and Neandertal measurements), reference [23] (Neandertal measurement means) and on Dr Benjamin M. Auerbach's website (recent modern human measurements): http://web.utk.edu/~auerbach/HOWL.htm.