Abstract
Cyclin-dependent kinases regulatory subunit 2 (CKS2) is a cyclin-dependent kinase-interacting protein, which is essential for cell cycle regulation. Elevated expression of CKS2 has been demonstrated in multiple types of human malignancies. However, the clinical significance, oncogenic functions and related mechanisms of CKS2 in colorectal cancer (CRC) remain largely unexplored. In this study, data from Oncomine database revealed that CKS2 is significantly up-regulated in CRC tissues compared with their normal counterparts. Immunohistochemical analysis of a CRC tissue microarray demonstrated that elevated CKS2 expression is closely associated with enhanced TNM stage, larger tumor size and a poor prognosis in patients with CRC. Multivariate Cox regression analysis revealed that CKS2 and TNM stage are two independent prognostic factors for CRC. Suppression of CKS2 expression resulted in decreased cell viability, increased cell apoptosis, cell cycle arrest and reduced expression of cyclins in Caco-2 and SW620 cells. Furthermore, gain and loss of function studies demonstrated that CKS2 promotes cell invasion in CRC cells through regulating claudin1. Taken together, our study reveal that CKS2 is promising prognostic indicator and contributes to tumor progression in CRC, and support that CKS2-related signaling may represent a novel target for CRC therapy.
Keywords: CKS2, colorectal cancer, prognosis, growth, invasion, claudin1
Introduction
Colorectal cancer (CRC) is one of the most common human malignancies in the world [1,2]. Due to the imperceptible symptoms of CRC, most patients are diagnosed at advanced stage at the time of diagnosis. According to the statistical analysis of American association of colorectal surgeon (ACS), the poor prognosis of patients with CRC is related to primary tumor metastasis and spread [3]. Thus, cancer metastasis is considered to be the leading causes of death in patients with CRC [4]. Meanwhile, accumulating evidences also demonstrate that alternation in cell cycle regulation and signal transduction molecules contribute to for tumor etiology and pathogenesis of CRC [5]. Therefore, it is of great significance to identify the oncogenic pathway involved in CRC progression and develop novel therapeutic approaches to treat this deadly disease.
Cyclin-dependent kinases regulatory subunit 2 (CKS2) is the member of cell cycle dependent protein kinase subunits family, which participates in cell cycle regulation [6-8]. CKS2 has an important role for early embryonic development and the process of somatic cell division [9]. Apart from its regulatory role in the transition of cell cycle, CKS2 also exhibits certain functions in tumor development [10-17]. Previous researches have demonstrated that CKS2 is up-regulated in many types of tumors, including prostate cancer, bladder cancer, breast cancer [18] and liver cancer [19] and contributes to tumor progression [20]. However, the underlying cellular functions of CKS2 and related mechanisms involved in its carcinogenicity remain largely unexplored. In a DNA microarray analysis, Menghi F et al. identified CKS2 as potential markers of cancer metastasis [21]. And in our previous study, we have demonstrated the expression and the clinical significance of CKS2 in CRC [22]. However, the fidelity is limited due to the small number of specimens studied.
In current study, by a large scale of immunohistochemical analysis, the prognostic value of CKS2 was determined in CRC. We found that up-regulated CKS2 is closely associated with tumor progression and predicts poor prognosis in CRC patients. Attenuation of CKS2 expression inhibited cell proliferation and promoted cell apoptosis. Moreover, we demonstrated that suppression of CKS2 promotes the invasive potential of CRC cells by regulating claudin1 expression.
Materials and methods
Clinical tissue samples
The specimens of 345 consecutive patients with CRC were collected from January 2003 to November 2010 at Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University. The diagnosis was confirmed based on clinical manifestation, pathological and serological examinations. The cases of CRC were selected in this study only if clinical data were available. The follow-up time was calculated from the date of surgery to the date of death, or the last known follow-up. None of them had received radiotherapy, chemotherapy, hormone therapy or other related anti-tumor therapies before surgery. All patients were well informed and the process was approved by Ethics Committee of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, China.
Cell culture and transfection
Human CRC cell lines Caco-2, SW480, SW620 and LOVO were all obtained from Cell Bank of the Chinese Academy of Sciences. The normal colonic epithelial cell line NCM460 was purchased from American Type Culture Collection (ATCC). All cells were cultured in a humidified incubator under 5% CO2 condition at 37°C and supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% antibiotics (penicillin and streptomycin) according to ATCC protocols. Small interfere RNAs (siRNA) targeting CKS2, claudin1 and a negative control were obtained from GenePharma (Shanghai, China). CKS2 and claudin1 over-expressing plasmids were purchased from GeneCopoeia (Guangzhou, China). The transfection was performed according to the manufacturer’s protocol.
Immunohistochemical staining
Immunohistochemical analysis was performed as previously described [23]. Briefly, tissue sections were deparaffinized in xylene and rehydrated with graded ethanol. After neutralization of endogenous peroxidase and antigen retrieval, the sections were blocked with 10% BSA (Sangon, Shanghai). After washing three times with phosphate-buffered saline (PBS), slides were first incubated using the antibody for CKS2 (Abcam, ab155078, US) at 4°C overnight with optimal dilution. After washing three times with PBS, slides were incubated with second antibody labeled by HRP (rabbit) (Proteintech, US) at room temperature for 1 h. Finally, the bound antibodies were visualized with 3,3’-diaminobenzidine tetrahydrochloride and counterstained by hematoxylin. Scoring was conducted according to the percent of positive cells: less than 5% scored 0; 6-25% scored 1; 25-50% scored 2; more than 50% scored 3 and staining intensity: no staining scored 0, weakly staining scored 1, moderately staining scored 2 and strongly staining scored 3, respectively. The final score was designated as low or high expression group using the percent of positive cell score × staining intensity score as follows: low expression was defined as a total score < 4 and high expression with a total score ≥ 4. These scores were determined independently by two senior pathologists.
Cell viability and apoptosis analysis
Cell viability was detected using a Cell Counting Kit (CCK-8, Dojindo, Japan). Twenty-four hours after cells were transfected with si-CKS2 and their nonspecific controls, cells were seeded into 96-well plate at 4 × 103 per well with 100 μl culture medium supplemented in the presence of 10% FBS. After cells were incubated for 24, 48, 72 or 96 h, cell viability was quantified by addition 10 µl of Cell Counting Kit-8. Then the absorbance at 450 nm was measured using a Power Wave XS microplate reader (BIO-TEK). For cell apoptosis assay, Caspase-3/7 Glo kit (promega) was used. Briefly, Caco-2 and SW620 cells transfected with si-CKS2 or their nonspecific controls were cultured in the presence of serum deprivation, after 48 h incubation, caspase-3/7 activity was measured according to the manufacturer’s protocol.
Cell cycle analysis
The influence of CKS2 on cell cycle progression was measured using propidium iodide staining and analyzed by flow cytometry. Briefly, cells were seeded in 6-well plate at 1 × 105 per well and transfected with si-CKS2 and their nonspecific controls. After 48 h incubation, cells were detached, washed with PBS, harvested and fixed in 70% ethanol for 24 h. Followed by treatment with DNase-free RNase and stained with propidium iodide, the cell cycle was measured by flow cytometric analysis of the DNA content of cell populations. Finally, the distribution of cells within G1/S and G2, and M phases was determined by using CELLQUEST software (Becton Dickinson).
Cell invasion assay
The invasive ability of Caco-2 and SW620 cells was detected by transwell model (Corning, NY) according to the manufacturer’s instructions. Briefly, 2 × 104 cells in 100 μl medium were seeded into the upper chamber of matrigel-coated filters (BD Bioscience, USA). A total of 700 μl culture medium containing 4 % (v/v) FBS was added to the bottom chamber. After incubation for 48 h, the non-invaded cells that remained on the upper chamber were removed. The invaded cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Finally, the number of invaded cells on the lower surface was counted under a light microscope in six random fields.
Quantitative real-time PCR
Total RNA from CRC cell lines and normal colonic epithelial cell line was extracted by RNA Extraction Kit (SLNco, Cinoasia, China), and cDNA was synthesized using PrimeScript RT reagent Kit (TaKaRa Biotechnology, Japan). The primers used in this study were shown in Table 1. Primers were designed with PRIMER 5.0 (ABI, Foster City, CA, USA) and synthesized by Generay (Shanghai, China). Expression of indicated genes was conducted on a Real-time Thermo Cycler (FTC3000, Funglyn, Canada) with SYBR Green Real-time PCR Master Mix (QPK-201, TOYOBO, Japan). The specificity of real-time PCR was confirmed by melting-curve analysis. Relative expressions were determined by normalizing expression of each Ct value to GAPDH Ct value and data were analyzed according to the 2-ΔΔCt formula.
Table 1.
Quantitative Real-time PCR primers used in this study
| Gene name | Primer sequence (5’ to 3’) | Amplicon size |
|---|---|---|
| CCNA1 | Forward: GCAGCCAGACATCACGGAAG | 114 bp |
| Reverse: TCCAGGAAGTTGACAGCCAGA | ||
| CCNB1 | Forward: AATAAGGCGAAGATCAACATGGC | 111 bp |
| Reverse: TTTGTTACCAATGTCCCCAAGAG | ||
| CCNB2 | Forward: GCTGGTACAAGTCCACTCCAAG | 144 bp |
| Reverse: GAAGCCAAGAGCAGAGCAGTAA | ||
| CCND1 | Forward: GCTGCGAAGTGGAAACCATC | 135 bp |
| Reverse: CCTCCTTCTGCACACATTTGAA | ||
| CCNE1 | Forward: GCCAGCCTTGGGACAATAATG | 104 bp |
| Reverse: CTTGCACGTTGAGTTTGGGT | ||
| Claudin1 | Forward: TGAGGATGGCTGTCATTGGG | 244 bp |
| Reverse: CTTGGTGTTGGGTAAGAGGTTGT | ||
| Claudin4 | Forward: GGGGCAAGTGTACCAACTG | 109 bp |
| Reverse: GACACCGGCACTATCACCA | ||
| Claudin7 | Forward: AGCTGCAAAATGTACGACTCG | 75 bp |
| Reverse: GGAGACCACCATTAGGGCTC | ||
| Occludin | Forward: AGACCCAAGAGCAGCAAAGG | 165 bp |
| Reverse: AACACCATGATGCCCAGGATA | ||
| ZO-1 | Forward: TATTCACGCAGTTACGAGCAAG | 103 bp |
| Reverse: AAGGTATCAGCGGAGGGACA | ||
| CKS2 | Forward: TTCGACGAACACTACGAGTACC | 109 bp |
| Reverse: GGACACCAAGTCTCCTCCAC | ||
| GAPDH | Forward: TGAAGGTCGGAGTCAACGGA | 225 bp |
| Reverse: CCTGGAAGATGGTGATGGGAT |
Statistical analysis
Data were presented as the means ± SD. All statistical analyses were performed using the SPSS 16.0 software and graphical representations were performed with GraphPad Prism 5 (San Diego, CA) software. Correlation of CKS2 expression with clinicopathologic parameters was analyzed by Pearson chi-square test. Overall survival rate was calculated according to the Kaplan-Meier method and the difference in survival curves was evaluated by the log-rank test. Independent prognostic factors were analyzed by the Cox proportional hazards regression model. The two-sided Student’s t test was used to assess the difference in other groups. P values less than 0.05 were considered statistically significant.
Results
CKS2 expression is significantly up-regulated in CRC
To determine the expression of CKS2 in CRC, we first analyzed its expression in Oncomine datasets. Data from two Oncomine datasets showed that CKS2 is commonly expressed in multiple cancer cells and is pronounced elevated in CRC cell lines (Figure 1). In addition, by searching Oncomine datasets, we also found that CKS2 expression was remarkably higher in tumor specimens compared with normal control specimens (Figure 2). These data suggest that CKS2 expression is deregulated in CRC and might contribute to tumor development and progression of CRC.
Figure 1.

CKS2 expression is elevated in CRC cells in Oncomine database. A. Gene expression analysis of CKS2 in a series of cancer cell lines demonstrated an increase in expression in CRC versus 15 other cancer cell types. B. CKS2 expression is significantly elevated in CRC cells compared with 7 other cancer cell types as demonstrated by analysis of Su multi-cancer dataset.
Figure 2.
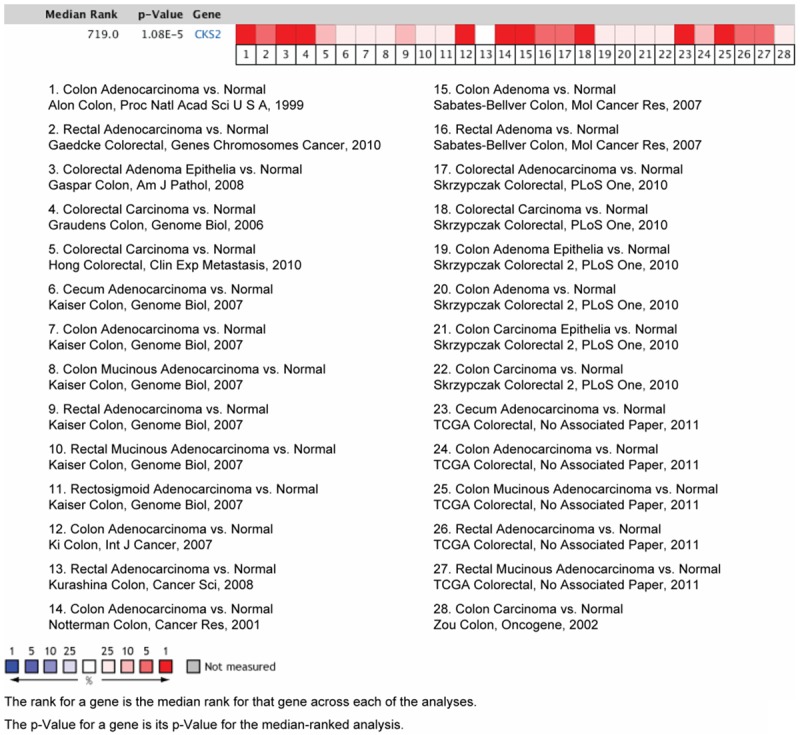
CKS2 expression is elevated in CRC tissues in Oncomine database. Oncomine heat map demonstrated a statistically significant increase in CKS2 expression in CRC tissues compared with the normal control tissues.
Elevated CKS2 expression predicts poor prognosis in patients with CRC
To further confirm the clinical significance of CKS2 in CRC, we analyzed CKS2 protein expression in a tissue microarray containing 345 cases of CRC specimens, which clinical data are available. The immunohistochemical results showed that CKS2 expression is up-regulated in 69.9% (241/345) CRC tissues (Figure 3A). By pearson chi-square test, we observed that high expression of CKS2 is positively associated with larger tumor size and enhanced TNM stage, whereas no significant difference between CKS2 expression and age, gender, tumor location or CEA levels are found (Table 2). To determine the prognostic value of CKS2, the overall survival (OS) rate of 183 CRC patients in current cohort was analyzed using Kaplan-Meier survival curves and the log-rank test. As shown in Figure 3B, patients with lower CKS2 expression had a better outcome than those with higher CKS expression (P = 0.0002). In univariate analysis, CKS2 expression level, tumor size, TNM stage and serum CEA levels were significant risk factors for OS (Table 3). And multivariate analysis revealed that CKS2 expression, tumor size and TNM stage are independent factors in the prediction of OS rate in patients with CRC (Table 3). The relative risk was 14.736 for CRC patients with a high CKS2 expression level in relative to those with a lower CKS2 expression level. Collectively, these data above indicate that up-regulated CKS2 predicts a poor prognosis in CRC patients and is closely correlated with tumor progression in CRC.
Figure 3.

Elevated CKS2 predicts a poor prognosis in CRC patients. A. Representative images of CKS2 immunoreactivity in CRC tissues. Scale bar: 50 μm. B. Kaplan-Meier curves for patients grouped based on CKS2 expression.
Table 2.
Relationship between CKS2 expression and corresponding clinicopathological features in patients with CRC
| Parameters | CKS2 (n) | |||
|---|---|---|---|---|
|
| ||||
| Low (n = 104, %) | High (n = 241, %) | P value | ||
| Age | ≤ 65 years | 61 (32.6) | 126 (67.4) | 0.276 |
| > 65 years | 43 (27.2) | 115 (72.8) | ||
| Gender | Male | 66 (33.2) | 133 (66.8) | 0.153 |
| Female | 38 (26.0) | 108 (74.0) | ||
| Tumor size | ≤ 5 cm | 68 (39.1) | 106 (60.9) | 0.000 |
| > 5 cm | 36 (21.1) | 135 (78.9) | ||
| Tumor location | Rectum | 69 (34.2) | 133 (65.8) | 0.054 |
| Colon | 35 (24.5) | 108 (75.5) | ||
| CEA level | ≤ 5 ng/ml | 59 (32.6) | 122 (67.4) | 0.297 |
| > 5 ng/ml | 45 (27.4) | 119 (72.6) | ||
| TNM stage | I | 16 (18.8) | 69 (81.2) | 0.000 |
| II | 24 (20.2) | 95 (79.8) | ||
| III | 45 (38.5) | 72 (61.5) | ||
| IV | 19 (79.2) | 5 (20.8) | ||
| Histology | Mucinous | 21 (38.9) | 33 (61.1) | 0.127 |
| Non-mucinous | 83 (28.5) | 208 (71.5) | ||
Values in parentheses indicate percentage values. The bold number represents the P-values with significant differences.
Table 3.
Univariate and multivariate analysis of prognostic parameters for survival in patients with CRC
| Parameters | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| CKS2 expression | 16.332 | 6.424-41.522 | 0.000 | 14.736 | 5.762-37.687 | 0.000 |
| Age | 1.493 | 0.969-2.300 | 0.069 | - | - | - |
| Gender | 1.256 | 0.831-1.897 | 0.279 | - | - | - |
| Tumor location | 0.800 | 0.510-1.255 | 0.332 | - | - | - |
| Tumor size | 1.713 | 1.102-2.661 | 0.017 | 1.898 | 1.215-2.964 | 0.005 |
| TNM stage | 2.057 | 1.592-2.657 | 0.000 | 1.768 | 1.381-2.263 | 0.000 |
| Serum CEA level | 1.578 | 1.030-2.418 | 0.036 | 1.290 | 0.840-1.982 | 0.245 |
HR: Hazard ratio; CI: Confidence interval. The bold number represents the P-values with significant differences.
Silencing of CKS2 inhibits cell proliferation and increases cell apoptosis in CRC cells
As CKS2 expression is closely associated with tumor size and TNM stage, we hypothesized that CKS2 contributes to the malignant biological behaviors of CRC cells. To test this hypothesis, we selected two CRC cell lines, Caco-2 and SW620, which showed higher CKS2 expression, for next investigation (Figure 4A). As shown in Figure 4B, treatment with si-RNAS targeting CKS2 resulted in significant decrease in CKS2 expression. CCK-8 assay showed that knockdown of CKS2 reduces cell viability of Caco-2 and SW620 cells (Figure 4C). Meanwhile, suppression of CKS2 also increased cell apoptosis as demonstrated by increased caspase-3/7 activity (Figure 4D). As CKS2 is a member of cell cycle dependent protein kinase subunits family, we further tested the effect of CKS2 on the distribution of cell cycle. Expectedly, silencing of CKS2 resulted in cell cycle arrest at the G1/S transition (Figure 4E). And consistent with this finding, inhibition of CKS2 also resulted in decreased expression of cyclins, including CCNA1, CCNB1, CCNB2 and CCNE1 (Figure 4F). Taken together, these data above suggest that CKS2 inhibits cell proliferation and increases cell apoptosis and this prerequisite facilitates tumor growth.
Figure 4.

Suppression of CKS2 inhibits cell proliferation and promotes cell apoptosis. (A) The mRNA expression of CKS2 in CRC cell lines and the normal control cells. (B) Western blotting analysis of the interfere efficiency of CKS2 in Caco-2 and SW620 cells. Effects of CKS2 knockdown on cell viability (C), cell apoptosis (D) and cell cycle (E) of Caco-2 and SW620 cells were analyzed by CCK8, caspase-3/7 activity and PI staining, respectively. (F). The implications of silencing of CKS2 on cyclins expression. (C-F). si-Ctrl versus si-CKS2-1 or si-CKS2-2, *P < 0.05, **P < 0.01, ***P < 0.001.
CKS2 contributes to cell invasion through regulating claudin1 expression
To further determine the role of CKS2 in tumor metastasis of CRC, we measured the invasive potential of Caco-2 and SW620 cells upon si-CKS2 treatment by Transwell model. As shown in Figure 5A, the invaded cells in si-CKS2 group were significantly reduced compared with si-Ctrl group in both Caco-2 and SW620 cells. And expectedly, over-expression of CKS2 also remarkably promoted the invasive potential of CRC cells (Figure 5B, 5C). To gain insight into the molecular mechanisms underlying CKS2-mediated pro-metastasis of CRC cells, we focused and detected the family of tight junction protein (TJP), which play critical roles in cancer metastasis [24]. The mRNA expression of five TJPs including claudin1, claudin4, claudin7, occludin1 and ZO-1 was detected after CKS2 was silenced. The resulted showed that claudin1 and claudin4 expression were reduced by si-CKS2 treatment. Notably, alternation in claudin1 expression was most significant (Figure 6A). Then we silenced claudin1 and the invasive potential of Caco-2 and SW620 cells was measured. Indeed, silencing of claudin1 suppressed the invasive ability of CRC cells and also compromised the pro-metastasis role of CKS2 (Figure 6B). And overexpression of claudin1 completely recovered the invasive potential of si-CKS2 cells (Figure 6C). Collectively, the data above indicates that CKS2 contributes to CRC cell invasion and this effect might mediate by altered expression of claudin1.
Figure 5.

Effects of CKS2 on cell invasion. A. The effect of silencing of CKS2 on cell invasion was analyzed by Transwell model (si-Ctrl versus si-CKS2-1 or si-CKS2-2, *P < 0.05, **P < 0.01). B. The over-expression efficiency of CKS2 was measured by Western blotting. C. The effect of over-expression of CKS2 on cell invasion (pcDNA3.1-Vector versus pcDNA3.1-CKS2, *P < 0.05, **P < 0.01).
Figure 6.

Inhibition of CKS2 inhibits cell invasion via down-regulating claudin1 expression. A. Alternation of tight junction protein expression was measured upon silencing of CKS2 (si-Ctrl versus si-CKS2-1 or si-CKS2-2, *P < 0.05, **P < 0.01). B. The implication of silencing of CKS2 on the invasive potential of Caco-2 and SW620 cells was detected in the presence of si-claudin1 treatment (*P < 0.05, **P < 0.01). C. The effect of silencing of CKS2 on the invasive potential of Caco-2 and SW620 cells was detected in the presence of reintroduction of Claudin1 (**P < 0.01).
Discussion
The CKS family, including two highly conserved paralogs CKS1 and CKS2, has been shown to play a regulatory role in cell cycle. Deregulation of CKS has been demonstrated in various human malignancies [15,19,25-27]. However, limited knowledge is known about CKS2 in CRC. In this study, we observed that CKS2 is commonly up-regulated in both CRC cell lines and clinical specimens compared with their normal counterparts and predicts a poor prognosis in CRC patients. Through siRNA-mediated inhibition, we demonstrated that CKS2 contributes to the malignant phenotype of CRC cells.
Previously, we have identified that CKS2 is up-regulated at both mRNA and protein levels in CRC tissues compared with the adjacent non-tumor and normal colon tissues [22]. However, due to limited clinical specimens and also follow-ups of CRC patients, the prognostic value of CKS2 was not determined. In this study, by analysis of a tissue microarray of 345 clinical samples, we found that CKS2 is closely correlated with tumor size and TNM stage. This observation is consistent with the findings in our previous reports as demonstrated by Western blotting analysis. Meanwhile, Kaplan-Meier survival analysis showed that patients with high CKS2 expression have a poor prognosis. In previous studies focused on gastric cancer [11] and esophageal squamous cell carcinoma [15], the mRNA expression of CKS2 is an independent prognostic factor. In line with this, the protein expression of CKS2 is also an independent factor in the prediction of OS rate in CRC patients by a large scale of immunohistochemical study.
In prostate cancer, aberrant expression of CKS2 contributes to tumorigenesis by enhancing cell proliferation and inhibiting programmed cell death [12]. In a papillary thyroid carcinoma, miR-26a modulates tumor growth and tumorigenesis by targeting CKS2 [13]. Consistent with this, attenuation of CKS2 in CRC cells results in decreased cell viability, increased cell apoptosis and cell cycle arrest in current study. However, further investigations are needed to explore the underlying mechanisms involved in those CKS-mediated effects. Except for tumor growth, we also found that CKS2 facilitates tumor metastasis by regulating cell tight junction protein claudin1. Altered tight junction structure and increased tight junction leakiness in cancer has implications for cancer progression [28,29]. And there is a strong association between colon neoplasia and increased claudin-1 expression [24,30]. Up-regulation of claudin1 contributes to cellular changes, including loss of cell polarity, abnormal cellular organization, as well as a decreased cell differentiation [30]. In line with this notion, silencing of claudin1 also suppresses the invasive potential of CRC cells. Meanwhile, silencing of claudin1 also compromised the pro-metastasis role of CKS2, whereas over-expression of claudin1 rescued the effect of si-CKS2. These cellular functions of CKS2 may confirm the prognostic value of CKS in CRC patients.
In conclusion, to the best of our knowledge, this is the first time to investigate the cellular functions of CKS2 in CRC cells and determine the prognostic value of CKS2 at a large scale. The results of our study provide evidences that CKS2 might be a useful predictor for malignant properties and patients’ prognosis, and suggest that targeting the oncogenic activities of CKS2 might be a novel approach for CRC treatment.
Acknowledgements
This work was supported by the grant from Science and Technology Commission of Shanghai, China (No. 13DZ1942806).
Disclosure of conflict of interest
None.
References
- 1.Fedirko V, Romieu I, Aleksandrova K, Pischon T, Trichopoulos D, Peeters PH, Romaguera-Bosch D, Bueno-de-Mesquita HB, Dahm CC, Overvad K, Chirlaque MD, Johansen C, Bidstrup PE, Dalton SO, Gunter MJ, Wark PA, Norat T, Halkjaer J, Tjonneland A, Dik VK, Siersema PD, Boutron-Ruault MC, Dossus L, Bastide N, Kuhn T, Kaaks R, Boeing H, Trichopoulou A, Klinaki E, Katsoulis M, Pala V, Panico S, Tumino R, Palli D, Vineis P, Weiderpass E, Skeie G, Gonzalez CA, Sanchez MJ, Barricarte A, Amiano P, Quiros JR, Manjer J, Jirstrom K, Ljuslinder I, Palmqvist R, Khaw KT, Wareham N, Bradbury KE, Stepien M, Duarte-Salles T, Riboli E, Jenab M. Prediagnostic anthropometry and survival after colorectal cancer diagnosis in Western European populations. Int J Cancer. 2014;135:1949–1960. doi: 10.1002/ijc.28841. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 4.Weidle UH, Birzele F, Kruger A. Molecular targets and pathways involved in liver metastasis of colorectal cancer. Clin Exp Metastasis. 2015;32:623–35. doi: 10.1007/s10585-015-9732-3. [DOI] [PubMed] [Google Scholar]
- 5.Jung Y, Lee S, Choi HS, Kim SN, Lee E, Shin Y, Seo J, Kim B, Jung Y, Kim WK, Chun HK, Lee WY, Kim J. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin Cancer Res. 2011;17:700–709. doi: 10.1158/1078-0432.CCR-10-1300. [DOI] [PubMed] [Google Scholar]
- 6.Pines J. Cell cycle: reaching for a role for the Cks proteins. Curr Biol. 1996;6:1399–1402. doi: 10.1016/s0960-9822(96)00741-5. [DOI] [PubMed] [Google Scholar]
- 7.Spruck CH, de Miguel MP, Smith AP, Ryan A, Stein P, Schultz RM, Lincoln AJ, Donovan PJ, Reed SI. Requirement of Cks2 for the first metaphase/anaphase transition of mammalian meiosis. Science. 2003;300:647–650. doi: 10.1126/science.1084149. [DOI] [PubMed] [Google Scholar]
- 8.Frontini M, Kukalev A, Leo E, Ng YM, Cervantes M, Cheng CW, Holic R, Dormann D, Tse E, Pommier Y, Yu V. The CDK subunit CKS2 counteracts CKS1 to control cyclin A/CDK2 activity in maintaining replicative fidelity and neurodevelopment. Dev Cell. 2012;23:356–370. doi: 10.1016/j.devcel.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinsson-Ahlzen HS, Liberal V, Grunenfelder B, Chaves SR, Spruck CH, Reed SI. Cyclindependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol Cell Biol. 2008;28:5698–5709. doi: 10.1128/MCB.01833-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R, Feng C, Xu Y. Cyclin-dependent kinase-associated protein Cks2 is associated with bladder cancer progression. J Int Med Res. 2011;39:533–540. doi: 10.1177/147323001103900222. [DOI] [PubMed] [Google Scholar]
- 11.Kang MA, Kim JT, Kim JH, Kim SY, Kim YH, Yeom YI, Lee Y, Lee HG. Upregulation of the cycline kinase subunit CKS2 increases cell proliferation rate in gastric cancer. J Cancer Res Clin Oncol. 2009;135:761–769. doi: 10.1007/s00432-008-0510-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lan Y, Zhang Y, Wang J, Lin C, Ittmann MM, Wang F. Aberrant expression of Cks1 and Cks2 contributes to prostate tumorigenesis by promoting proliferation and inhibiting programmed cell death. Int J Cancer. 2008;123:543–551. doi: 10.1002/ijc.23548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv M, Zhang X, Li M, Chen Q, Ye M, Liang W, Ding L, Cai H, Fu D, Lv Z. miR-26a and its target CKS2 modulate cell growth and tumorigenesis of papillary thyroid carcinoma. PLoS One. 2013;8:e67591. doi: 10.1371/journal.pone.0067591. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Mezzasoma L, Antognelli C, Del Buono C, Stracci F, Cottini E, Cochetti G, Talesa VN, Mearini E. Expression and biological-clinical significance of hTR, hTERT and CKS2 in washing fluids of patients with bladder cancer. BMC Urol. 2010;10:17. doi: 10.1186/1471-2490-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kita Y, Nishizono Y, Okumura H, Uchikado Y, Sasaki K, Matsumoto M, Setoyama T, Tanoue K, Omoto I, Mori S, Owaki T, Ishigami S, Nakagawa H, Tanaka F, Mimori K, Mori M, Natsugoe S. Clinical and biological impact of cyclin-dependent kinase subunit 2 in esophageal squamous cell carcinoma. Oncol Rep. 2014;31:1986–1992. doi: 10.3892/or.2014.3062. [DOI] [PubMed] [Google Scholar]
- 16.Shen DY, Zhan YH, Wang QM, Rui G, Zhang ZM. Oncogenic potential of cyclin kinase subunit-2 in cholangiocarcinoma. Liver Int. 2013;33:137–148. doi: 10.1111/liv.12014. [DOI] [PubMed] [Google Scholar]
- 17.Wang JJ, Fang ZX, Ye HM, You P, Cai MJ, Duan HB, Wang F, Zhang ZY. Clinical significance of overexpressed cyclin-dependent kinase subunits 1 and 2 in esophageal carcinoma. Dis Esophagus. 2013;26:729–736. doi: 10.1111/dote.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Xu L, Liu Y, Chen J, Jiang H, Yang S, Tan H. Expression of cyclin kinase subunit 2 in human breast cancer and its prognostic significance. Int J Clin Exp Pathol. 2014;7:8593–8601. [PMC free article] [PubMed] [Google Scholar]
- 19.Shen DY, Fang ZX, You P, Liu PG, Wang F, Huang CL, Yao XB, Chen ZX, Zhang ZY. Clinical significance and expression of cyclin kinase subunits 1 and 2 in hepatocellular carcinoma. Liver Int. 2010;30:119–125. doi: 10.1111/j.1478-3231.2009.02106.x. [DOI] [PubMed] [Google Scholar]
- 20.You H, Lin H, Zhang Z. CKS2 in human cancers: Clinical roles and current perspectives (Review) Mol Clin Oncol. 2015;3:459–463. doi: 10.3892/mco.2015.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menghi F, Orzan FN, Eoli M, Farinotti M, Maderna E, Pisati F, Bianchessi D, Valletta L, Lodrini S, Galli G, Anghileri E, Pellegatta S, Pollo B, Finocchiaro G. DNA microarray analysis identifies CKS2 and LEPR as potential markers of meningioma recurrence. Oncologist. 2011;16:1440–1450. doi: 10.1634/theoncologist.2010-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu M, Zhong M, Qiao Z. Expression and clinical significance of cyclin kinase subunit 2 in colorectal cancer. Oncol Lett. 2013;6:777–780. doi: 10.3892/ol.2013.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang ZS, Cong ZJ, Luo Y, Mu YF, Qin SL, Zhong M, Chen JJ. Decreased expression of interleukin-36alpha predicts poor prognosis in colorectal cancer patients. Int J Clin Exp Pathol. 2014;7:8077–8081. [PMC free article] [PubMed] [Google Scholar]
- 24.Martin TA. The role of tight junctions in cancer metastasis. Semin Cell Dev Biol. 2014;36:224–231. doi: 10.1016/j.semcdb.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 25.Khattar V, Thottassery JV. Cks1 proteasomal turnover is a predominant mode of regulation in breast cancer cells: role of key tyrosines and lysines. Int J Oncol. 2015;46:395–406. doi: 10.3892/ijo.2014.2728. [DOI] [PubMed] [Google Scholar]
- 26.Nagler RM, Ben-Izhak O, Ostrovsky D, Golz A, Hershko DD. The expression and prognostic significance of Cks1 in salivary cancer. Cancer Invest. 2009;27:512–520. doi: 10.1080/07357900802239116. [DOI] [PubMed] [Google Scholar]
- 27.Westbrook L, Manuvakhova M, Kern FG, Estes NR nd, Ramanathan HN, Thottassery JV. Cks1 regulates cdk1 expression: a novel role during mitotic entry in breast cancer cells. Cancer Res. 2007;67:11393–11401. doi: 10.1158/0008-5472.CAN-06-4173. [DOI] [PubMed] [Google Scholar]
- 28.Forster C. Tight junctions and the modulation of barrier function in disease. Histochem Cell Biol. 2008;130:55–70. doi: 10.1007/s00418-008-0424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez-Mariscal L, Dominguez-Calderon A, Raya-Sandino A, Ortega-Olvera JM, Vargas-Sierra O, Martinez-Revollar G. Tight junctions and the regulation of gene expression. Semin Cell Dev Biol. 2014;36:213–223. doi: 10.1016/j.semcdb.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Tully O, Ngo B, Zitin M, Mullin JM. Epithelial tight junctional changes in colorectal cancer tissues. ScientificWorldJournal. 2011;11:826–841. doi: 10.1100/tsw.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]


