Abstract
Dysregulation of NEAT1 plays critical oncogenic roles and facilitates tumorigenesis on various human tumor entities. However, little information is available about the expression pattern of NEAT1 in esophageal squamous cell carcinoma (ESCC). The contributions of this lncRNA to tumorigenesis and progression of ESCC aslo remains unclear. By performing quantitative real-time polymerase chain reaction (qRT-PCR) in 96 cases of ESCC, we found that the expression of NEAT1 was higher in ESCC tissues and cells compared with the normal counterparts. Pearson analysis showed that elevated NEAT1 levels were extraordinarily correlated with the tumor size (P=0.026), lymph node metastasis (P=0.035) and clinical stage (P=0.004). Moreover, Kaplan–Meier curves with the log-rank test showed that higher expression of NEAT1 led to a significantly poorer survival and multivariate Cox proportional hazards analysis revealed that NEAT1 was an independent risk factor of overall survival (OS). We also assessed the function of NEAT1 in vitro by gain-/loss-of-function studies. Results showed that enhanced expression of NEAT1 stimulated the proliferation of ESCC cells, and promoted their ability of forming foci, migration, and invasion. Conversely, knockdown of NEAT1 showed the opposite effect. Overall, our study indicated that the inappropriate activation of NEAT1 predicts poor prognosis and has a crucial regulatory role in in ESCC. Targeting NEAT1 could be a novel therapeutic choice for treating ESCC patients.
Keywords: NEAT1, esophageal squamous carcinoma, prognosis, proliferation, metastasis
Introduction
Human esophageal cancer (EC) is the third malignancies of digestive system with a high mortality worldwide [1]. Its high recurrence and metastasis leads to a total crude 5-year survival [2,3]. Esophageal squamous cell carcinoma (ESCC) accounts for more than 90% of EC cases, which showed distinct aetiological and pathological characteristics compared to esophageal adenocarcinoma [4]. Searching the prognostic predictor and key molecular mechanism of ESCC could be not only helpful with the clinical prevention and monitoring but also suggestive of therapeutic schemes.
Nuclear paraspeckle assembly transcript 1 (NEAT1), a long non-coding RNA (lncRNA) retained in the nucleus where it forms the core structural component of the paraspeckle sub-organelles [5], has earned the reputation as a transcriptional regulator for numerous genes. NEAT1 was firstly transcribed from the multiple endocrine neoplasia locus [6], suggesting that this lncRNA can exert its functions in cancer progression, which was subsequently confirmed by series of researches in multiple cancers [7-13]. However, whether the expression of NEAT1 in ESCC is associated with malignancy or prognosis remains unknown.
In the present study, we investigated whether NEAT1 is detectable and altered in ESCC tissues compared with adjacent normal tissues. Then, we analyzed the potential relationship between NEAT1 levels and clinicopathological features of ESCC. We also assessed whether NEAT1 influence the ESCC cell proliferation, migration or invasion in vitro by gain-/loss-of-function studies.
Materials and methods
Patients collections
A total of 96 patients (aged 34~79 years; mean, 60.8 years) with ESCC were selected from the First Affiiated Hospital of Henan University of Science and Technology from 2009-2010. All the specimens were immediately frozen in liquid nitrogen and stored at -80°C until use. None of the patients had received preoperative local or systemic treatment. The collected clinicopathological characteristics included tumor size, location, differentiation, lymph node metastasis, distant metastasis, and TNM stage. The follow-up interval was from the date of surgery to the date of death or the last clinical investigation. This study was approved by The Clinical Research Ethics Committee of First Affiiated Hospital of Henan University of Science and Technology. Written informed consent was obtained from all participants.
Cell lines and culture conditions
The immortalized human esophageal epithelial cell line (SHEE) and the malignant transformed esophageal carcinoma cell line (SHEEC) were gifts from Professor Yi Ceng (Institute of Virology, Chinese Academy of Preventive Medicine). The human ESCC cell line KYSE150 cells were purchased from Shanghai Ruilu Technology Co., Ltd. KYSE30, KYSE70, and KYSE140 were purchased from Public Health England, UK. All cells were grown and maintained in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA, USA) and 1% penicillin and streptomycin (Invitrogen, Carlsbad, CA, USA), maintain at 37°C in a humidified atmosphere with 5% CO2.
Total RNA isolation and RT-qPCR
Total RNA was isolated from tissues and cells by Trizol (Invitrogen, Life technology, USA). The RT and qPCR reactions were performed as previously described [14], and β-Actin was used as an endogenous control to normalize the data. Primer sequences used in this study were the following: 5’-ATGCCACAACGCAGATTGAT-3’ (forward) and 5’-CGAGAAACGCACAAGAAGG-3’ (reverse) for NEAT1, 5’-AAAGACCTGTACGCCAACAC-3’ (forward) and 5’-GTCATACTCCTGCTTGCTGAT-3’ (reverse) for β-Actin.
Reagents and cell transfection
For reagents used: Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). The pcDNA3.1-NEAT1 and empty vector (used as a negative control) was purchased from Invitrogen, Shanghai, China. The siRNAs of NEAT1 (5’-UGGUAAUGGUGGAGGAAGAUU-3’, 5’-GUGAGAAGUUGCUUAGAAAUU-3’) and scramble siRNA were purchased from Ribobio, Guangzhou, China. Cells were seeded in 96- or 6-well plates 24 h before the experiment. KYSE150 cells were transfected with pcDNA3.1-NEAT1 or negative control; EC9706 cells were transfected with siNEAT1 or scramble siRNA).
Cell proliferation assays
Proliferation was measured using the CCK-8 kit (Dojindo, Japan). Approximately 3 × 103 transfected cells in 100 μl were incubated in triplicate in 96-well plates. The CCK-8 reagent (10 μl) was added to each well and incubated at 37°C for 2 h at various time points. The optical density at 450 nm was measured using an automatic microplate reader (Synergy4; BioTek, Winooski, VT, USA).
Colony formation assays
Transfected cells were plated into 6-well plates (800 cells per hole) and cultured in media containing 10% FBS, which was replaced every 5 days. After 10~14 days, colonies were washed with PBS, fixed with ethanol for 30 min, and stained with 0.1% crystal violet (Sigma, St. Louis, MO) for 30 min. The visible colonies were then manually counted. For each treatment group, wells were counted in triplicate.
Cell migration and invasion detection
The Transwell assay was used to assess cell migration and invasion with the transwell system of Corning co. Ltd., USA. Inserts containing 8-μm pore filters were uncoated for the migration assays or coated with Matrigel (Becton-Dickinson Labware, Bedford, MA) for the invasion assays. A total of 1~3 × 105 cells transfected with pcDNA3.1-NEAT1 or pcDNA3.1 (KYSE150 cells) or with siNEAT1 or scramble siRNA (EC9706 cells) in 100 μl of serum-free medium were added to each well. After 24 h of incubation at 37°C, the cells that had migrated through the filters were fixed with methanol, stained with Giemsa and counted from 10 random fields under a microscope at 200 × magnification. Each experiment was performed in triplicate.
Statistical analysis
All statistical analyses were performed using SPSS 20.0 (IBM, SPSS, Chicago, IL, USA). Statistical tests for data between groups analysis included the χ2-test and Student’s two-tailed t-test. Two-tailed Student’s t-test was employed for analyzing in vitro data. Pearson analysis was used to calculate the correlation between clinicopathological features and the expression of NEAT1. OS curves were calculated with the Kaplan-Meier method and were analyzed with the log-rank test. Univariate analysis and multivariate models were fit using a Cox proportional hazards regression model. A threshold of P<0.05 was defined as statistically significant.
Results
NEAT1 was upregulated in ESCC
To investigate whether NEAT1 could be detected and whether its expression was altered in ESCC, we conducted QRT-PCR in 96 patients with ESCC. As shown in Figure 1A and 1B, comparative analyses demonstrated that NEAT1 was elevated in human ESCC samples compared with the adjacent non-neoplastic tissues (P=0.005), and NEAT1 was upregulated in all of the 6 analysed ESCC cell lines compared with the normal esophageal epithelial cell line (SHEE) (all P<0.05). These results suggested that NEAT1 was overexpressed in ESCC. Furthermore, NEAT1 expression levels were analyzed in cancer tissue samples according to lymph node status and TNM stage. As shown in Figure 1C and 1D, the expression of NEAT1 was positively correlated with the lymph node metastasis (P=0.002) and TNM stage (P=0.034) in ESCC patients, suggesting that NEAT1 up regulation is associated with the progression of human ESCC.
Figure 1.
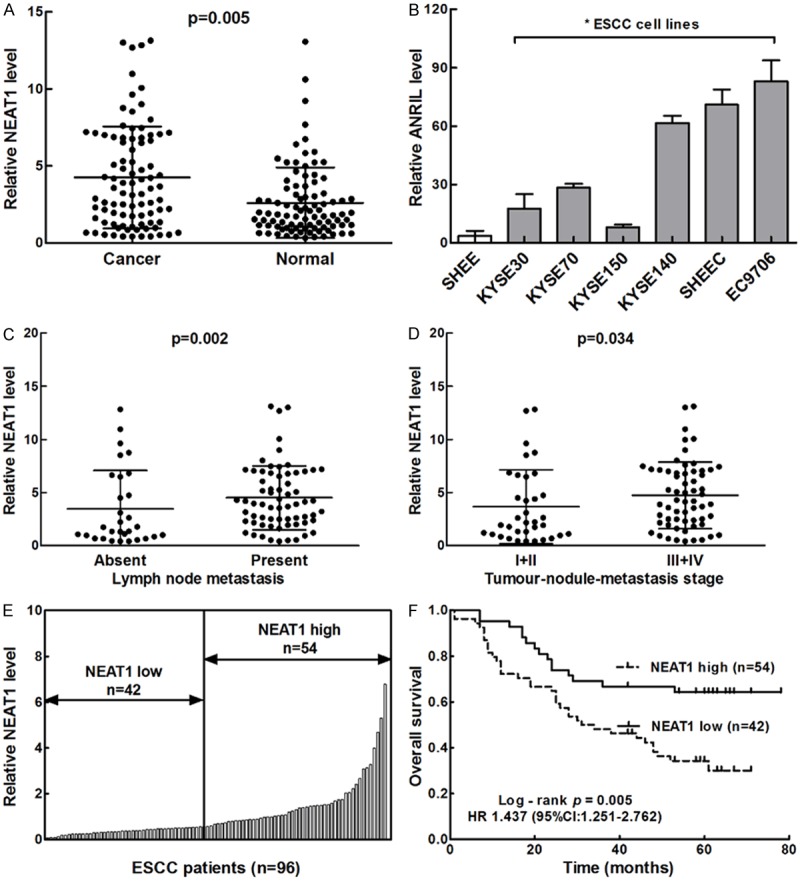
NEAT1 was highly expressed and predicted poor prognosis in ESCC. A. Representative qRT-PCR results of NEAT1 in ESCC tissues and adjacent normal tissues showed that the majority of cases exhibited elevated levels of NEAT1 in tumours compared with levels observed in noncarcinoma tissues (P=0.005). B. Representative qRT-PCR results of NEAT1 in 6 EC cell lines and one normal human esophageal epithelial cell line *P<0.05. C. NEAT1 levels categorised according to lymph node status (P=0.002). D. NEAT1 levels categorised according to tumour-nodule-metastasis stage (P=0.034). E. The total of 96 ESCC patients included in this study were divided into a low-NEAT1 expression group (n=42) and a high-NEAT1 expression group (n=54) according to the Youden index. F. Kaplan-Meier overall survival (OS) curve of patients with different expressions of NEAT1 in ESCC (Low vs. High).
NEAT1 upregulation correlats with clinicopathologic characteristics and the survival of patients with ESCC
Based on the expression levels of NEAT1 obtained by QRT-PCR, we divided the 95 ESCC patients into a high-NEAT1 expression group (n=54) and a low-NEAT1 expression group (n=42) according to the Youden index (Figure 1E). Then we analyzed the correlation of NEAT1 expression levels with the clinicopathological characteristics of patients with ESCC (Table 1). The expression levels of NEAT1 was associated with the tumor size (P=0.026), lymph node metastasis (P=0.035), as well as the TNM stage of ESCC (P=0.004) (Table 1). However, there was no significant correlation between the expression of NEAT1 with age, differentiation or remote metastasis.
Table 1.
The relationship between NEAT1 expression and clinicopathologic parameters
| Characteristics | No. of patients | Low expression | High expression | P value |
|---|---|---|---|---|
| Age (years) | 0.198 | |||
| ≤60 | 47 | 18 | 29 | |
| >60 | 49 | 24 | 25 | |
| Gender | 0.076 | |||
| Male | 72 | 35 | 37 | |
| Female | 24 | 7 | 17 | |
| Location | 0.257 | |||
| Upper | 21 | 11 | 10 | |
| Middle | 45 | 13 | 32 | |
| Lower | 30 | 18 | 12 | |
| Differentiation | 0.067 | |||
| Well | 18 | 11 | 7 | |
| Moderate | 41 | 11 | 30 | |
| Poor | 37 | 21 | 15 | |
| Tumor size | 0.026* | |||
| ≤4.0 cm | 30 | 18 | 12 | |
| >4.0 cm | 66 | 24 | 42 | |
| Lymph node metastasis | 0.035* | |||
| Absent | 46 | 26 | 20 | |
| Present | 50 | 18 | 32 | |
| Distant metastasis | 0.108 | |||
| Absent | 73 | 35 | 38 | |
| Present | 23 | 7 | 16 | |
| TNM stage | 0.004* | |||
| I+II | 35 | 22 | 13 | |
| III+IV | 61 | 20 | 41 |
P value is statistically significant.
When linked to the prognosis, the Kaplan-Meier analysis showed that patients with high levels of NEAT1 expression had significantly shorter OS (P=0.005, log-rank test; Figure 1F) than those with low levels. A univariate Cox analysis showed that tumor size, lymph node metastasis, distant metastasis, TNM stage, and NEAT1 expression were correlated with the survival (Table 2). Multivariate analysis using the Cox proportional hazard model demonstrated that NEAT1 expression was an independent risk factors for OS (P=0.033, Table 2) in addition to TNM stage (P=0.000, Table 2). These results identified that overexpression of NEAT1 seemed to be a predictive factor of poor survival of ESCC, suggesting that overexpression of NEAT1 may contribute to ESCC pathogenesis and can be employed as a powerful independent prognostic factor.
Table 2.
Univariate and multivariate analysis of different prognostic factors for OS in 96 patients with ESCC
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Prognostic factors | HR | 95% CI | P value | HR | 95% CI | P value |
| Age (>60/≤60) | 0.772 | 0.445-1.338 | 0.356 | |||
| Gender (Male/Female) | 1.083 | 0.585-2.003 | 0.800 | |||
| Tumor size (≤4 cm/>4 cm) | 1.752 | 1.005-3.055 | 0.048* | |||
| Location (Upper/Middle+Lower) | 0.819 | 0.593-1.132 | 0.227 | |||
| Differentiation (Poor/Well+Moderate) | 1.303 | 0.702-2.418 | 0.402 | |||
| Lymph node metastasis (Presence/Absence) | 5.165 | 2.195-7.154 | 0.000* | |||
| Distant metastasis (Presence/Absence) | 3.554 | 1.988-6.352 | 0.004* | |||
| TNM stage (I+II/III+IV) | 7.247 | 3.072-9.100 | 0.000* | 5.854 | 2.399-7.287 | 0.000* |
| NEAT1 (High/Low) | 2.383 | 1.302-4.361 | 0.005* | 1.919 | 1.399-6.489 | 0.033* |
P value is statistically significant.
Identification of the efficiencies of NEAT1 overexpression and knockdown in ESCC cells
KYSE150 cells transfected with pcDNA3.1-NEAT1 or negative control, and EC9706 cells with siNEAT1 or scramble siRNA transfection for 48 h were followed by qRT-PCR. The mRNA levels of NEAT1 were significantly increased in pcDNA3.1-NEAT1 transfecting KYSE150 cells compared with the vector-transfecting counterparts (Figure 2A). While in EC9706 cells the NEAT1 mRNA levels was decreased in the cells transfected with siNEAT1 (Figure 2B).
Figure 2.
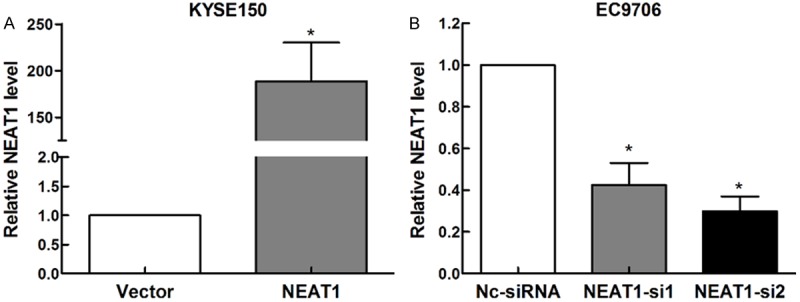
Efficiencies of overexpression and knockdown of NEAT1 in esophageal squamous carcinoma. A. The expressions of NEAT1 mRNA in KYSE150 cells transfected with pcDNA3.1- NEAT1 and vector detected by RT-qPCR. B. The mRNA levels of NEAT1 in EC9706 cells transfected with siNEAT1 or and siRNA detected by RT-qPCR.
NEAT1 promotes esophageal tumor cell proliferation
To observe the influence of NEAT1 on tumor cell proliferation, we conducted the CCK-8 cell counting assays. Results showed that the tumor cell growth of the NEAT1-overexpressing KYSE150 cells was significantly accelerated compared with the control (Figure 3A). While in EC9706 cells, knockdown of NEAT1 by siRNA effectively inhibited the cell proliferation (Figure 3B). Similarly, the colony formation assay revealed that clonogenic survival was significantly improved in NEAT1-overexpressed KYSE150 cell lines (Figure 3C). And knockdown of NEAT1 could obviously reduce the clonogenic survival (Figure 3D). These results suggested that NEAT1 could promote cell proliferation in ESCC in vitro.
Figure 3.
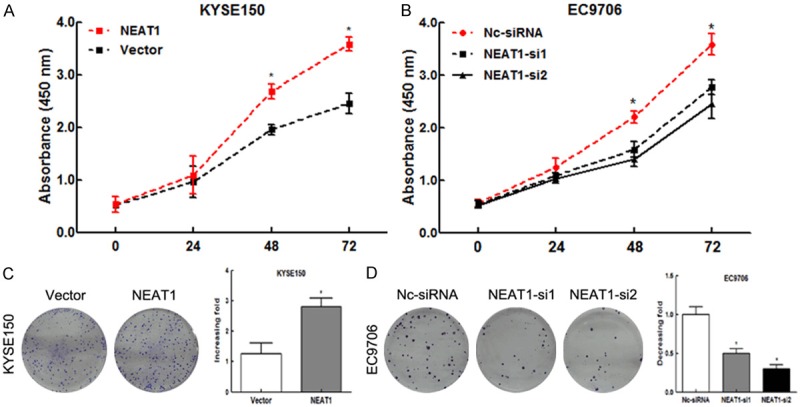
NEAT1 stimulated tumor cell proliferation in ESCC. A. The CCK8 cell counting assays revealed cell growth curves of KYSE150 cells. B. The CCK8 cell counting assays revealed cell growth curves of EC9706 cells. C. The representative pictures (left) and quantification (right) of Giemsa-stained KYSE150 cells in colony formation assay. *P<0.05. D. The representative pictures (left) and quantification (right) of Giemsa-stained EC9706 cells in colony formation assay. *P<0.05.
NEAT1 enhanced tumor cell migration and invasion in vitro
To investigate whether NEAT1 has a direct functional role in tumor cell invasion and migration in ESCC, we performed Transwell assays. Overexpression of NEAT1 in KYSE150 cells resulted in a significant increase in the migration (Figure 4A, P<0.01) and invasion ability through an extracellular matrix (Figure 4C, P<0.01). While knockdown of NEAT1 effectively abolished the mobility (Figure 4B, P<0.01) and invasiveness (Figure 4D, P<0.01) of EC9706 cells. Collectively, these data suggested that NEAT1 induced tumor cell migration and invasion in ESCC.
Figure 4.
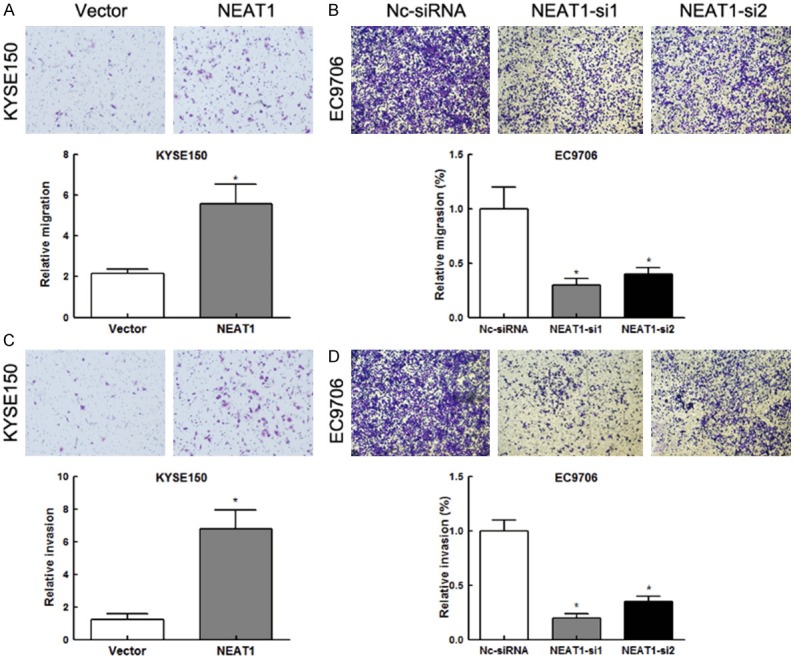
NEAT1 promoted cell invasion and migration in ESCC. A. Representative images (up) and quantification (down) of transwell migration assays for KYSE150 cells. (Scale bars=50 μm). *P<0.01. B. Representative images (up) and quantification (down) of transwell migration assays for EC9706 cells. (Scale bars=50 μm). *P<0.01. C. Representative images (up) and quantification (down) of transwell invasion assays for KYSE150 cells. (Scale bars=50 μm). *P<0.01. D. Representative images (up) and quantification (down) of transwell invasion assays for EC9706 cells. (Scale bars=50 μm). *P<0.01.
Discussion
In the present study, we explored the expression of NEAT1 and its biological functions in ESCC. We confirmed that NEAT1 was overexpressed in ESCC. Clinically, high expression levels of NEAT1 correlated with worse survival of ESCC patients, indicating that NEAT1 might be a novel prognostic indicator for patients with ESCC. In addition, NEAT1 contributes to the malignant characters of ESCC through involvement in proliferation, migration and invasion.
NEAT1 was firstly found in the multiple endocrine neoplasia locus [6], assumed as a cancer related regulator. In the past five years researchers gradually found that overexpression of NEAT1 exits in multiple malignancies, such as acute promyelocytic leukemia [7], prostate cancer [8,15], glioma [9], lung cancer [10,12], and ovarian carcinoma [13]. The present study showed that higher levels of NEAT1 expression were observed in ESCC tissues compared with adjacent normal tissues, suggesting that the up regulation of NEAT1 may also be an important molecular event in ESCC.
The correlation of NEAT1 up regulation with a higher tumor burden including larger tumor size, presence of lymph node metastasis and advanced TNM stage in our study, suggesting that NEAT1 may accelerate tumor progression in ESCC mainly via stimulating malignant biological behavior of tumor cells, which was supported by the results of overexpression of NEAT1 in KYSE150 cells. Considering that the clinical stage was majorly concerned about tumor growth and metastasis, it was not surprising that the expression of NEAT1 got intense along with the advance of stages and was tightly correlated with TNM stage. Indeed, NEAT1 might push the advance of clinical stage by inducing tumor migration and invasion in ESCC. Therefore, it was reasonable that overexpression of NEAT1 suggested a poorer survival time in ESCC patients. Taken together, monitoring the levels of NEAT1 might be an effective biomarker for the prognosis monitoring of ESCC.
Furthermore, knocking down of NEAT1 potently suppressed the tumor-promoting effects of NEAT1 in EC9706 cells. These results on one hand inversely supported that overexpression of NEAT1 should be responsible for the tumor progression in ESCC, and on the other hand suggested that suppression of NEAT1 could be an effective therapeutic choice in clinical practice, which shed a light on developing effective NEAT1-targeting chemotherapy in ESCC. Further studies might consider NEAT1 as a major molecular target in future chemotherapy regimens designing.
Previous work has identified that as an oestrogen receptor alpha specific lncRNA, NEAT1 trigger the oncogenicity of prostate cancer by altering the epigenetic landscape of target gene promoters [8]. Besides, overexpression of miR-449a could significantly downregulating NEAT1 and inhibits the cell growth and apoptosis [10]. Moreover, it can also facilitate cellular growth by modulating HIF-2α under a hypoxia environment in breast cancer [11]. However, little was known whether the dysregulation of these tumorigensis-related genes in ESCC can explain the underlying mechanisms of overexpression of NEAT1 facilitate tumor progression. Future studies might focus on the specific mechanisms of NEAT1 in the development of ESCC.
Conclusion
Our study suggests that NEAT1 predicts poor prognosis and has a vital role in the development and progression of esophageal squamous cell carcinoma. NEAT1 might be applied as a biomarker to monitor the progression of tumor and to predict the prognosis of patients. Understanding the precise role of NEAT1 in the pathogenesis of ESCC may lead to a development of therapeutic strategy for ESCC.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Abate E, DeMeester SR, Zehetner J, Oezcelik A, Ayazi S, Costales J, Banki F, Lipham JC, Hagen JA, DeMeester TR. Recurrence after Esophagectomy for Adenocarcinoma: Defining Optimal Follow-Up Intervals and Testing. J Am Coll Surg. 2010;210:428–435. doi: 10.1016/j.jamcollsurg.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Lou F, Sima CS, Adusumilli PS, Bains MS, Sarkaria IS, Rusch VW, Rizk NP. Esophageal cancer recurrence patterns and implications for surveillance. J Thorac Oncol. 2013;8:1558. doi: 10.1097/01.JTO.0000437420.38972.fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 5.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guru SC, Agarwal SK, Manickam P, Olufemi SE, Crabtree JS, Weisemann JM, Kester MB, Kim YS, Wang Y, Emmert-Buck MR, Liotta LA, Spiegel AM, Boguski MS, Roe BA, Collins FS, Marx SJ, Burns L, Chandrasekharappa SC. A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res. 1997;7:725–735. doi: 10.1101/gr.7.7.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, Chen S, Li Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. doi: 10.1186/1471-2407-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, MacDonald TY, Fontugne J, Erho N, Vergara IA, Ghadessi M, Davicioni E, Jenkins RB, Palanisamy N, Chen Z, Nakagawa S, Hirose T, Bander NH, Beltran H, Fox AH, Elemento O, Rubin MA. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Q, Sun S, Yu W, Jiang J, Zhuo F, Qiu G, Xu S, Jiang X. Altered expression of long non-coding RNAs during genotoxic stress-induced cell death in human glioma cells. J Neurooncol. 2015;122:283–292. doi: 10.1007/s11060-015-1718-0. [DOI] [PubMed] [Google Scholar]
- 10.You J, Zhang Y, Liu B, Li Y, Fang N, Zu L, Li X, Zhou Q. MicroRNA-449a inhibits cell growth in lung cancer and regulates long noncoding RNA nuclear enriched abundant transcript 1. Indian J Cancer. 2014;51(Suppl 3):e77–81. doi: 10.4103/0019-509X.154055. [DOI] [PubMed] [Google Scholar]
- 11.Choudhry H, Albukhari A, Morotti M, Hider S, Moralli D, Smythies J, Schodel J, Green CM, Camps C, Buffa F, Ratcliffe P, Ragoussis J, Harris AL, Mole DR. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4546. doi: 10.1038/onc.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan LJ, Zhong TF, Tang RX, Li P, Dang YW, Huang SN, Chen G. Upregulation and Clinicopathological Significance of Long Non-coding NEAT1 RNA in NSCLC Tissues. Asian Pac J Cancer Prev. 2015;16:2851–2855. doi: 10.7314/apjcp.2015.16.7.2851. [DOI] [PubMed] [Google Scholar]
- 13.Kim YS, Hwan JD, Bae S, Bae DH, Shick WA. Identification of differentially expressed genes using an annealing control primer system in stage III serous ovarian carcinoma. BMC Cancer. 2010;10:576. doi: 10.1186/1471-2407-10-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Gao S, Liu G, Jia R, Fan D, Feng X. Microarray expression profile analysis of long non-coding RNAs in human gastric cardiac adenocarcinoma. Cell Physiol Biochem. 2014;33:1225–1238. doi: 10.1159/000358692. [DOI] [PubMed] [Google Scholar]
- 15.Hirata Y, Ogasawara N, Sasaki M, Mizushima T, Shimura T, Mizoshita T, Mori Y, Kubota E, Wada T, Tanida S, Kataoka H, Kamiya T, Higashiyama S, Joh T. BCL6 degradation caused by the interaction with the C-terminus of pro-HB-EGF induces cyclin D2 expression in gastric cancers. Br J Cancer. 2009;100:1320–1329. doi: 10.1038/sj.bjc.6605010. [DOI] [PMC free article] [PubMed] [Google Scholar]


