Abstract
Like other cancers, renal cell carcinoma (RCC) derives the essential energy for proliferation and survival from high rates of glycolysis rather than from oxidative phosphorylation of the mitochondrial respiration pathway. NDUFA4 (NADH Dehydrogenase (Ubiquinone) 1 Alpha Subcomplex, 4) is encoding a protein belonging to the respiratory chain of mitochondria. For a better understanding of the tumor biology and for identification of a potential new biomarker, we analyzed the regulation of NDUFA4 in RCC compared to normal tissue cells. Downregulation of NDUFA4 mRNA and protein was detected in RCC compared to normal renal tissues in quantitative real-time PCR as well as in western blot and immunohistochemical staining. Histological analysis revealed higher NDUFA4 expression in the distal tubules compared to the proximal tubules and the loop of Henle. A higher molecular weight of the NDUFA4 protein was discovered in RCC samples, possibly indicating a posttranslational modification. Moreover, NDUFA4 protein expression was predictive for cancer-specific survival. Our analysis revealed a potential new biomarker, but future studies are warranted to investigate the prognostic value of NDUF4A expression.
Keywords: Renal cell carcinoma, NDFUA4, biomarker, mitochondria, electron transport chain
Introduction
Renal Cell Carcinoma accounts for approximately 2-3% of all cancers and is the twelfth most common malignancy in the world [1]. The incidence has increased over time by 1.5-5.9% each year [2]. In 2012, approximately 2.4% new cases of RCC and 1.7% kidney cancer-related deaths were counted in the world (European Union: incidence 4.0%, mortality-rate 3.2%) [3]. It is the most common malignancy of all urological tumors for females and, after prostate and bladder cancer, on third position for men.
Due to improved imaging techniques (ultrasound, computed tomography and magnetic resonance imaging) and their widespread use, the number of incidentally diagnosed small renal lesions has significantly increased over the last decades. About one third of all renal incidentalomas are of benign origin, but often malignancy can not be ruled out using imaging alone. To identify malignant and potentially aggressive tumors, novel non-invasive biomarkers are needed.
NDUFA4 is encoding a protein belonging to the respiratory chain of mitochondria [4,5]. Mitochondria cell metabolism-especially the regulation of cell death pathways - had become a major research focus over the last decades, given the fact that most tumor cells are resistant to apoptosis [6]. This resistance could be associated with special attributes of mitochondria in cancer cells compared to normal tissue cells. It is already known that cancer cells display high rates of glycolysis to produce their adenosine triphosphate (ATP) and some investigations suggest this is due to alterations of signaling pathways that govern glucose uptake and utilization [7,8]. Stimulation of glycolysis in tumor cells could also occur through downregulation of NDUFA4, an important protein-coding gene for the mitochondrial respiration pathway to produce ATP.
NDUFA4 downregulation was observed in an earlier microarray study by our group [9]. The present study is devoted to analyze the regulation of NDUFA4 in RCC compared to normal tissue and to reveal a novel biomarker not only to help to identify RCC but to monitor treatment response.
Material and methods
Patients
We determined the expression of NDUFA4 mRNA in each 51 clear cell renal cell carcinoma (ccRCC) and normal renal tissue samples. Fresh-frozen tissue samples were collected from patients undergoing radical or partial nephrectomy at the Department of Urology at the University Hospital Bonn. Furthermore, formalin-fixed and paraffin embedded tissue samples from 143 patients with ccRCC were used for the construction of a tissue microarray [10]. The clinical-pathological parameters are shown in Table 1. The collection of tissue samples was performed within the framework of the Biobank initiative of the CIO Köln-Bonn. All patients provided written informed consent for the collection of biomaterials. The study was approved by the local ethic committee (number: 280/12).
Table 1.
Clinical-pathological parameters of the study cohorts
| Quantitative real-time PCR | Immunohistochemistry | |||
|---|---|---|---|---|
|
|
||||
| Cancer n=51 (%) | Normal n=51 (%) | Cancer n=143 (%) | Normal n=21 (%) | |
| Sex | ||||
| male | 34 (66.6) | 35 (68.6) | 93 (65.0) | 16 (76.1) |
| female | 17 (33.3) | 16 (31.4) | 50 (35.0) | 5 (23.9) |
| Age | ||||
| mean | 62.5 | 62.0 | 63.2 | 55.7 |
| min-max | 36-86 | 36-86 | 33-85 | 28-77 |
| Pathological stage | ||||
| pT1 | 28 (54.9) | n.a. | 53 (37.1) | n.a. |
| pT2 | 7 (13.7) | n.a. | 28 (19.6) | n.a. |
| pT3 | 16 (31.4) | n.a. | 47 (32.9) | n.a. |
| pT4 | 0 (0) | n.a. | 2 (1.4) | n.a. |
| lymph node metastasis | 2 (3.9) | n.a. | 8 (5.6) | n.a. |
| distant metastasis | 7 (13.7) | n.a. | 17 (11.9) | n.a. |
| Grading | ||||
| grade 1 | 4 (7.8) | n.a. | 38 (26.6) | n.a. |
| grade 2 | 39 (76.5) | n.a. | 89 (62.2) | n.a. |
| grade 3 | 7 (13.7) | n.a. | 3 (2.1) | n.a. |
| grade 4 | 1 (1.9) | n.a. | 0 (0) | n.a. |
n.a., not applicable.
RNA isolation and quantitative real-time PCR
The RNA isolation procedure was performed as described earlier [11]. In brief, total RNA was isolated using the mirVana miRNA Isolation Kit and treated with the DNA-free Kit (both: Ambion, Foster City, CA, USA). The RNA quantity and quality was analyzed at a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Gel electrophoresis was used to exclude RNA degradation. cDNA was synthesized using 1 µg total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio, Saint-Germain-en-Laye, France). Quantitative real-time PCR was performed using 5 ng/µl cDNA, Takara Bio SYBR Premix Ex Taq II with ROX Plus and 10 pmol/µl forward/reverse primer. The primer sequences were: NDUFA4 (forward AACCTCTAAGAACAGGGTTTCTCA; reverse GGATGGTCTGCTAAACCAAACT), ACTB (forward CCAACCGCGAGAAGATGA; reverse CCAGAGGCGTACAGGGATAG) and TPB (forward GAACATCATGAATCAGAACAACA; reverse ATAGGGATTCCGGGAGTCAT). PCR was performed on an ABIPrism 7900 HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Data were analyzed using Qbase+ (Biogazelle, Ghent, Belgium) with ACTB and TBP as reference genes in the 2-∆∆CT algorithm. Statistical analysis was done using the t-test (SPSS Statistics v21 (IBM, Ehningen, Germany).
Western blot
Corresponding fresh-frozen tumor and normal renal tissue of 8 patients with ccRCC was used for Western Blot. Approximately 50 mg tissue was homogenized in a Precellys 24 (Peqlab, Erlangen, Germany) with 400 µl Cell Lysis Buffer (Cell Signaling, Cambridge, United Kingdom) with Complete Mini EDTA-free protease inhibitor (Roche, Basel, Switzerland). The protein concentration was determined (BCA Protein Assay Kit, Pierce Biotechnology, Rockford, IL, USA), 30 ng protein per lane was loaded into a NuPAGE 4-12% denaturating PAA Gel (Life Technologies, Carlsbad, CA, USA) and separated in a XCell4 SureLock electrophoresis system (Life Technologies). The samples were transferred on 0.2 µm nitrocellulose (XCell II, Life Technologies) and immunostaining was performed with an antibody against NDUFA4 (#ab133698, Abcam, Cambridge, United Kingdom). The detection was carried out with horseradish peroxidase conjugated to secondary antibodies (anti-rabbit-POD, #170-6515, Biorad, Hercules, CA, USA; anti-biotin-POD, #7075, Cell Signaling). SuperSignal West Femto Kit (Thermo Scientific, Waltham, MA, USA) showed the evolvement of the chemiluminescent signal, which was recorded by the LAS 3000 Image Reader (Fujifilm, Tokyo, Japan).
Immunohistochemistry
Immunohistochemistry was performed to specify the expression of NDUFA4 in ccRCC (n=143) and normal renal tissue samples (n=21) using a tissue microarray [10]. The NDUFA4 expression was quantified for cells in the proximal/distal tubules and the loop of Henle separately. Paraffin sections of 5 µm thickness were cut from the tissue block and stained at the Ventana Benchmark automated staining system (Ventana Medical System, Tuscon, AZ, USA). After incubation of the slides with the primary NDUFA4 antibody (dilution 1:100) for 40 minutes at room temperature, signal detection was performed with the Ventana DABMap Kit combined with Universal Secondary Antibody. The slides were counterstained with haematoxylin and bluing reagent, dehydrated and mounted.
The slides were scanned (Panoramic Midi, 3D HISTECH, Budapest, Hungary), and Definiens Tissue Studio v2.1 (Munich, Germany) image analysis software was used to obtain a continuous spectrum of average staining intensity. SPSS Statistics v21 was used to correlate NDUFA4 expression with normal/tumor tissue and clinical-pathological parameters (Mann-Whitney-U test) and to determine the relevance of NDUFA4 expression for progression-free, cancer-specific and overall survival (Kaplan Meier estimates and Cox regression analyses).
Results
mRNA expression of NDUFA4
We first investigated the mRNA expression of NDUFA4 in each 51 clear cell renal cell carcinoma (ccRCC) and normal renal tissue samples using qPCR. The NDUFA4 expression was significantly decreased in ccRCC tissue compared to normal renal tissue (mean: 0.668 vs. 1.282; P=0.004). NDUFA4 mRNA levels in ccRCC tissue did not correlate to clinical-pathological parameters (i.e. TNM-stage, age, gender; all P>0.1). See Figure 1.
Figure 1.
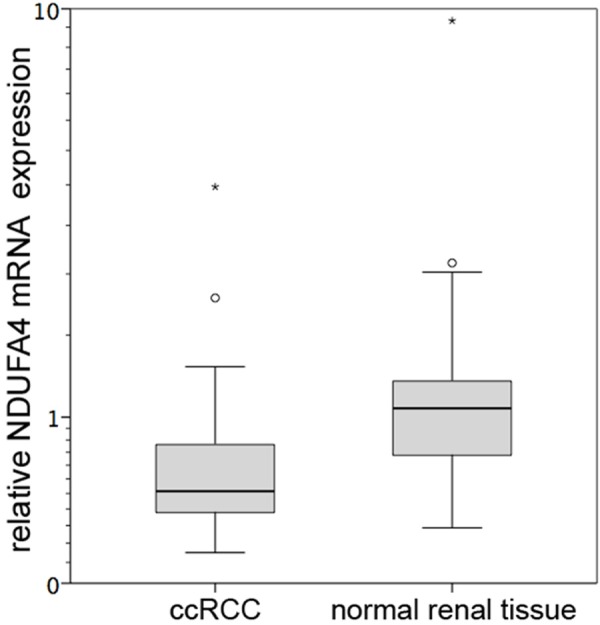
NDUFA4 mRNA expression. Relative NDUFA4 mRNA expression levels were determined in each 51 clear cell renal cell carcinoma (ccRCC) and normal renal tissue samples; NDUFA4 mRNA expression was significantly decreased in ccRCC samples (P=0.004).
Protein expression of NDUFA4
In order to confirm the differential mRNA expression on the protein level, we next performed Western Blot with 8 corresponding normal renal and ccRCC tissue lysates. As expected, the NDUFA4 protein level was lower in ccRCC samples. Interestingly, the protein in ccRCC samples had a somewhat higher molecular weight, indicating that the protein may undergo posttranslational modification. See Figure 2.
Figure 2.
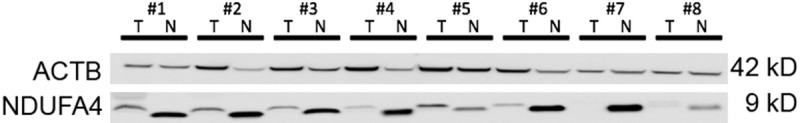
Western blot. Western blot experiments were performed to determine the protein expression in 8 corresponding normal (N) and clear cell renal cell carcinoma (T) tissue. NDUFA4 protein expression was lower in tumor than in normal renal tissue (ACTB: actin, beta).
Immunohistochemistry
In order to investigate the NDUFA4 protein in a validation cohort allowing meaningful statistical analyses, we performed immunohistochemical staining with a NDUFA4 antibody in a tissue microarray cohort which included 143 ccRCC samples as well as 21 normal renal tissue samples. The immunohistochemical analysis confirmed the decrease of mean NDUFA4 expression in ccRCC compared to normal renal tissue (see Figure 3A, 3B). In detail, mean immunostaining scores were 0.167 in ccRCC, 0.464 in distal tubules (compared to ccRCC: P<0.001), 0.215 in proximal tubules (P=0.002) and 0.052 in the loop of Henle (P<0.001).
Figure 3.
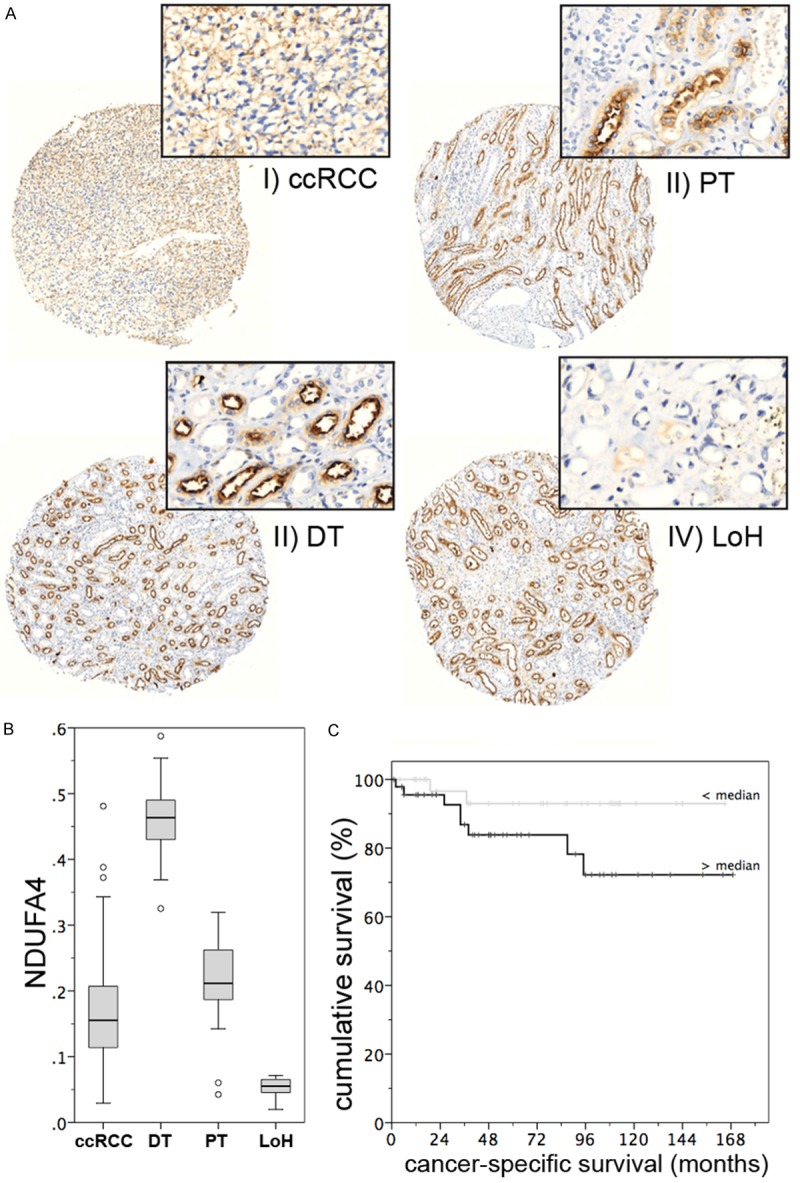
Protein expression of NDUFA4 in ccRCC. A. Representative immunohistochemical images from tissue of ccRCC (I), PT (II), DT (III) and LoH (IV) analysis of NDUFA4 protein expression. B. NDUF4A expression is lower in clear cell renal cell carcinoma (ccRCC) than in proximal (PT) and distal tubules (DT), whereas the cells of the loop of Henle (LoH) have a very low expression of NDUFA4. C. The expression of NDUFA4 is predictive of the period of cancer-specific survival in patients with ccRCC.
NDUFA4 protein levels were not related to any clinical pathological parameter (i.e. pT-stage, grade, lymph node involvement, distant metastasis, age, sex; all P>0.2). However, univariate Cox regression analysis indicated that NDUFA4 was predictive for cancer-specific survival in a subset of 83 patients with available follow-up information (P=0.046; hazard ratio 967.7, 95% confidence interval 1.13-825.500). The Kaplan Meier estimate for cancer-specific survival is shown in Figure 3C. As expected, established prognostic parameters like pT-stage (P=0.041), grade (P=0.005), lymph node and distant metastasis (both P<0.001) were also predictive for cancer-specific survival. NDUF4A lost its predictive value (P=0.134) in the multivariate model which included TNM-stage and grade, however, the small cohort size (n=83) limits powerful statistical analyses.
Discussion
For many malignancies there are biomarkers that help to identify cancer cells in doubt of an unknown lesion. Unfortunately this is not the case for the renal cell carcinoma. In order to detect a novel biomarker, we analyzed the NDUFA4 expression in ccRCC. As a result, downregulation of NDUFA4 mRNA and protein was detected in RCC compared to normal renal tissues in quantitative real-time PCR as well as in western blot and immunohistochemistry. Thereby, our data confirm the results of an earlier microarray screening study [9]. NDUFA4 mRNA levels in ccRCC tissue did not correlate to clinical-pathological parameters. Thus, NDUFA4 expression could be used for diagnostic purposes. Interestingly, the detailed histological analysis revealed a higher NDUFA4 expression in the distal tubules compared to the proximal tubules and the loop of Henle, whereby NDUF4A was also lower in ccRCC cells compared to proximal tubules. These findings are indicating that downregulation of NDUFA4 is not only accounting for the derivation of ccRCC from the cells of the proximal tubules. Besides that, we detected a higher molecular weight of the NDUFA4 protein in ccRCC samples, indicating that the protein may undergo posttranslational modification, leading to a possible impairment of the protein function.
NDUFA4 protein expression was predictive for cancer-specific survival in a univariate cox regression analysis, however it lost its predictive value in the multivariate model, which included established predictive parameters like TNM-stage and grade. However, the small size of the cohort (n=83) limits the statistical power and future studies are warranted to investigate the potential prognostic value of NDUF4A expression.
Already in the early 1930s, researchers observed that cancer cells have an increase in glycolysis and lactate production simultaneously with a decrease of oxidative phosphorylation [12,13]. The mechanisms of the mitochondrial respiratory chain to produce energy in form of ATP are lowered; tumor cells derive their energy from glycolysis. It has already been reported that the expression level of some mitochondrial transcripts is lower in RCC than in normal tissue [14]. Our analysis in ccRCC shows a decrease of NDUFA4, which is implicated in the mitochondrial respiratory chain and therefore in the oxidative phosphorylation. Furthermore, the protein seems to be modified in ccRCC as the molecular weight is different in tumor and normal renal tissue, which may further lead to an impairment of the protein.
In ccRCC respiratory chain complex activities and subunit amounts are severely diminished [15]. We could proof that the transcription of genes involved in the structure and biogenesis of these complexes are significantly decreased - especially NDUFA4.
Acknowledgements
The tissue samples were collected within the framework of the Biobank of the Center for Integrated Oncology Cologne Bonn at the University Hospital Bonn. Isabella Syring is supported by a grant of the Ferdinand Eisenberger Foundation of the German Association of Urology. Sven Perner is supported by a grant of the Rudolf Becker Foundation.
Disclosure of conflict of interest
None.
References
- 1.Porta C, Paglino C, Grünwald V. Sunitinib rechallenge in advanced renal-cell carcinoma. Br J Cancer. 2014;111:1047–53. doi: 10.1038/bjc.2014.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLaughlin JK, Lipworth L, Tarone RE. Epidemiologic aspects of renal cell carcinoma. Semin Oncol. 2006;33:527–33. doi: 10.1053/j.seminoncol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012, Cancer Incidence and Mortality Worldwide. Lyon, France: International Agency for Research on Cancer; 2014. [Google Scholar]
- 4.Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landázuri MO, Enríquez JA. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16:378–86. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Garbian Y, Ovadia O, Dadon S, Mishmar D. Gene expression patterns of oxidative phosphorylation complex I subunits are organized in clusters. PLoS One. 2010;5:e9985. doi: 10.1371/journal.pone.0009985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fulda S. Tumor resistance to apoptosis. Int J Cancer. 2009;124:511–5. doi: 10.1002/ijc.24064. [DOI] [PubMed] [Google Scholar]
- 7.Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria in cancer cells: what is so special about them? Trends Cell Biol. 2008;18:165–73. doi: 10.1016/j.tcb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Frezza C, Gottlieb E. Mitochondria in cancer: not just innocent bystanders. Semin Cancer Biol. 2009;19:4–11. doi: 10.1016/j.semcancer.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Deng M, Blondeau JJ, Schmidt D, Perner S, Müller SC, Ellinger J. Identification of novel differentially expressed lncRNA and mRNA transcripts in clear cell renal cell carcinoma by expression profiling. Genomics Data n.d. doi: 10.1016/j.gdata.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellinger J, Kahl P, Mertens C, Rogenhofer S, Hauser S, Hartmann W, Bastian PJ, Büttner R, Müller SC, von Ruecker A. Prognostic relevance of global histone H3 lysine 4 (H3K4) methylation in renal cell carcinoma. Int J Cancer. 2010;127:2360–6. doi: 10.1002/ijc.25250. [DOI] [PubMed] [Google Scholar]
- 11.Blondeau JJ, Deng M, Syring I, Schrödter S, Schmidt D, Perner S, Müller SC, Ellinger J. Identification of novel long non-coding RNAs in clear cell renal cell carcinoma. Clin Epigenetics. 2015;7:10. doi: 10.1186/s13148-015-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warburg O, Wind F, Neglers E. Metabolism of Tumors. Constable Co Lond. 1930:254–70. [Google Scholar]
- 13.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 14.Faure Vigny H, Heddi A, Giraud S, Chautard D, Stepien G. Expression of oxidative phosphorylation genes in renal tumors and tumoral cell lines. Mol Carcinog. 1996;16:165–72. doi: 10.1002/(SICI)1098-2744(199607)16:3<165::AID-MC7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 15.Hervouet E, Simonnet H, Godinot C. Mitochondria and reactive oxygen species in renal cancer. Biochimie. 2007;89:1080–8. doi: 10.1016/j.biochi.2007.03.010. [DOI] [PubMed] [Google Scholar]


