Abstract
Background
Promoter methylation of O6-methylguanine-DNA methyltransferase (MGMT) is an important predictive biomarker in glioblastoma. The T variant of the MGMT promoter-enhancer single nucleotide polymorphism (SNP; rs16906252) has been associated with the presence of MGMT promoter methylation in other cancers. We examined the association of the T allele of rs16906252 with glioblastoma development, tumor MGMT methylation, MGMT protein expression, and survival outcomes.
Methods
Two independent temozolomide-treated glioblastoma cohorts—one Australian (Australian Genomics and Clinical Outcomes of Glioma, n = 163) and the other American (University of California Los Angeles/Kaiser Permanente Los Angeles, n = 159)—were studied. Allelic bisulphite sequencing was used to determine if methylation was specific to the T allele. Additionally, we compared the incidence of the T allele between glioblastoma cases and matched controls to assess whether it was a risk factor for developing MGMT methylated glioblastoma.
Results
Carriage of the T allele of the rs16906252 SNP was associated with both MGMT methylation and low MGMT protein expression and predicted significantly longer survival in temozolomide-treated patients with both MGMT methylated and nonmethylated glioblastoma. Methylation was linked to the T allele, inferring that the T variant plays a key role in the acquisition of MGMT methylation. Carriage of the T allele was associated with a significantly elevated risk of developing glioblastoma (adjusted odds ratio, 1.96; P = .013), increasing further when glioblastoma was classified by the presence of MGMT methylation (adjusted odds ratio, 2.86; P = .001).
Conclusions
The T allele of the rs16906252 SNP is a key determinant in the acquisition of MGMT methylation in glioblastoma. Temozolomide-treated patients with the rs16906252 T genotype have better survival, irrespective of tumor methylation status.
Keywords: MGMT methylation, SNP, survival
Glioblastoma (GBM), the most common malignant brain tumor, is highly fatal, with a median survival of less than 15 months. In terms of years of life lost, the population burden from GBM is the highest of all the malignant cancers.1 GBM often affects both people at the peak of their work and child-rearing responsibilities and the elderly. The addition of concomitant and adjuvant temozolomide (TMZ) to radiation as a standard of care (Stupp protocol) for patients with newly diagnosed GBM has significantly increased survival from this uniformly fatal disease.2
Promoter methylation of O6-methylguanine-DNA methyltransferase (MGMT) is an important biomarker in neuro-oncology, with many studies now confirming that patients with GBM, in which MGMT is absent, derive the greatest benefit from TMZ therapy.3,4 In a study by Hegi et al,5 a survival benefit was seen in patients with MGMT methylated tumors: median survival was 23.4 months compared with 12.6 months in those with nonmethylated tumors receiving concurrent radiation therapy and TMZ.5 Nevertheless, 14.8% of patients with nonmethylated MGMT treated with TMZ still survived at least 2 years.5 This latter observation suggests that methylation testing alone may not be sensitive enough to detect all responders to TMZ and/or that there are other mechanisms, independent of methylation, leading to MGMT repression.
MGMT transcription and its downstream protein expression can be affected by inherent genetic factors such as single nucleotide polymorphisms (SNPs) in the promoter/enhancer region of the gene.6 In fact, it is likely that a combination of SNPs and differential MGMT promoter methylation plays a role in sensitivity to alkylating agents. Genome-wide association studies (GWAS) comprising large sample sizes and matched controls have provided a powerful tool for identifying glioma-risk SNPs.7–10 We wanted to study SNPs directly associated with the MGMT promoter region, and for this reason we used a candidate gene approach to identify the SNP—rs16906252:C > T (minor allele frequency: 0.077). This SNP has been linked with MGMT methylation in colorectal carcinoma,11,12 pleural mesothelioma,13 diffuse large B-cell lymphoma,14 lung cancer,15 and GBM.16 The SNP is located within the coding sequence of exon 1 and resides 92 base pairs downstream of the transcriptional start site within a cis-acting enhancer element of the MGMT gene. The variant T allele has been associated with low levels of MGMT promoter methylation in normal somatic tissues, including the normal colorectal mucosa of colorectal carcinoma cases and healthy controls,11 as well as the peripheral blood lymphocytes of healthy individuals,17 suggesting that healthy individuals possessing the T allele may be predisposed to somatic methylation at the MGMT promoter.
We previously assessed the frequency and functional relevance of the MGMT SNP rs16906252:C > T in a small cohort of 78 GBM cases.16 Methylation was associated with the variant T allele (P = .030), with a 24.4% incidence of the SNP and a 14.1% frequency of the T allele. The SNP was associated with favorable survival (the unadjusted hazard ratio [HR] for death for carriers of the T allele compared with wild-type C allele patients was 0.39 [95% CI: 0.21–0.73]; P = .003). A more recent study in GBM demonstrated an association between MGMT promoter methylation and the rs16906252 genotype, also showing a higher methylation level in T allele–bearing GBM.18 The variant T allele is likely to be functionally significant in MGMT transcriptional regulation, since promoter reporter assays performed in both lung cancer and GBM cell lines have shown that the haplotype bearing the T allele has reduced promoter activity compared with the wild-type sequence.15,16
The objectives of this study were to validate our previously reported findings of an association between the SNP rs16906252:C > T and MGMT methylation and survival benefit in larger independent GBM cohorts, and to show definitively whether MGMT methylation is linked to the variant T allele. Additionally, we compared the incidence of the MGMT SNP rs16906252 genotypes among GBM cases with a demographically matched control population to assess whether the SNP is a risk factor for developing an MGMT methylated GBM.
Materials and Methods
Ethics approval for use of the Australian Genomics and Clinical Outcomes of Glioma (AGOG) cohort was obtained from the Human Research Ethics Committee (South Eastern Sydney Local Health District). DNA extracted from a total of 163 cases of formalin-fixed paraffin-embedded (FFPE) tumor tissue was provided from the AGOG biobank (www.agogbio.org.au). Newly diagnosed GBM was surgically removed by gross total resection (no biopsies were included), and all patients subsequently received concurrent radiotherapy plus TMZ followed by adjuvant TMZ (chemoradiotherapy). Cases with mutations in the isocitrate dehydrogenase 1 gene were excluded from the study (n = 5). Medical history was reviewed for all patients and follow-up data collected. These data included sex, age, surgical procedure, radiotherapy and chemotherapy, and overall survival. MGMT expression was assessed by immunohistochemistry as determined by a trained neuropathologist (Prof Peter C. Burger, Johns Hopkins University), using a scoring system published by Lalezari and colleagues.19 The clinical characteristics of the AGOG patient cohort are listed in Table 1. All participants provided written informed consent.
Table 1.
Demographic characteristics and MGMT molecular features of the Australian GBM cohort compared with the UCLA GBM cohort and Australian healthy control group
| Characteristics | Australian Cohort (AGOG) |
American Cohort (UCLA) |
Australian Healthy Control Group |
|||||
|---|---|---|---|---|---|---|---|---|
| No. Patients | (%) | No. Patients | (%) | P | No. Controls | (%) | P | |
| 163 | 159 | 451 | ||||||
| Age, y | 160 | |||||||
| Mean (SD) | 58.3 (12.4) | 57.8 (10.3) | .670 | 45.88 (14.08) | .000 | |||
| Range | 60.0 (25.0–85.0) | 62 (22.3–84.3) | 61.0 (18.0–79.0) | |||||
| <50 | 35 | 21.5 | 34 | 21.4 | 252 | 55.9 | ||
| ≥50 | 125 | 76.7 | 125 | 78.6 | 199 | 44.1 | ||
| Sex | 156 | 159 | .212 | |||||
| Male | 100 | 61.3 | 91 | 57.2 | 263 | 58.3 | .204 | |
| Female | 56 | 34.4 | 68 | 42.8 | 188 | 41.7 | ||
| MGMT methylation | 144 | 159 | .000 | |||||
| Methylated | 92 | 56.4 | 58 | 36.5 | ||||
| Nonmethylated | 52 | 31.9 | 101 | 63.5 | ||||
| MGMT protein | 134 | 129 | .254 | |||||
| <30% | 81 | 49.7 | 69 | 43.4 | ||||
| ≥30% | 53 | 32.5 | 60 | 37.7 | ||||
| SNP genotype | 155 | 155 | .107 | |||||
| CT/TT combined | 34 | 20.9 | 23 | 14.5 | 54 | 12.0 | .002 | |
| CC (wild-type) | 121 | 74.2 | 132 | 83.0 | 397 | 88.0 | ||
| Overall survival, mo | 160 | |||||||
| Median | 13.05 | 19.13 | .000 | |||||
| Range (min–max) | 86.9 (2.9–89.8) | 125.79 (2.63–128.42) | ||||||
A second, published cohort with MGMT promoter methylation status (determined by methylation-specific PCR [MSP]) and MGMT protein expression data available was accessed from the University of California Los Angeles (UCLA) and Kaiser Permanente Los Angeles (KPLA).19 DNA extracted from FFPE specimens (n = 159) was selected based on the same criteria as the AGOG cohort, and researchers were blinded to MGMT status and patient outcome.19 Clinical data for the 155 UCLA/KPLA cases are summarized in Table 1. All patients received chemoradiotherapy.
Blood specimen DNA from 451 healthy control subjects was obtained from consenting donors recruited via the Australian Red Cross Blood Bank (Australian Red Cross Blood Service Ethics Approval 2003#08). The controls were de-identified, with sex and age data available (Table 1). The healthy control subjects and the AGOG GBM cohort were both sampled in New South Wales, Australia.
Single Nucleotide Polymorphism Genotyping
Genotyping for the rs16906252:C > T SNP was carried out on blood (healthy controls) and tumor DNA (FFPE from AGOG and UCLA/KPLA). Briefly, DNA was PCR-amplified using the primers (5′-3′) GCCCCTAGAACGCTTTGCGTC and AGACACTCACCAAGTCGCAACG and an annealing temperature of 65°C to produce a 74-bp product that spanned the MGMT enhancer region and the rs16906252 SNP. The PCR product was then incubated with the restriction enzyme HhaI (New England Biolabs), which recognizes and cuts the wild-type C allele at the rs16906252 position to produce 48-bp and 26-bp fragments, but not the variant T allele.16
Methylation of the MGMT Promoter
Bisulfite conversion of tumor DNA followed by pyrosequencing of cytosine–phosphate–guanine (CpG) was used to assess the methylation status across the MGMT promoter for AGOG specimens where status was not known. Chemically methylated and nonmethylated genomic DNA was used as positive and negative controls, respectively (Millipore). Extracted tumor DNA (500 ng) was converted with sodium bisulfite using the EZ Methylation-Gold kit as per the manufacturer's instructions (Zymo Research). Methylation analyses were performed using 100 ng of bisulfite-converted DNA as PCR template. The methylation-unbiased pyrosequencing assay was performed using the PyroMark MGMT kit (Qiagen) on the PSQ96 MA system (Qiagen) and interrogated 5 individual CpG sites within exon 1 near the MGMT transcription start site for methylation.20 PyroMark CpG software (Qiagen) was used to quantify the levels of methylation. GBM samples were scored as methylation positive by pyrosequencing if all 5 CpG sites had methylation values of 9% or higher.16
Allelic Bisulfite Sequencing
To determine the allelic methylation patterns in patient samples heterozygous for the rs16906252:C > T SNP, clonal sequencing was performed on bisulfite-converted tumor DNA. The primers GTTTGTAGGATTATTYGAGGTTGTTAT and CCCCRAATATACTAAAACAACCC were used to PCR-amplify a 171-bp fragment, which spanned 15 CpG sites and contained the rs16906252 SNP. These primers were specific to the antisense strand of bisulfite-converted DNA, to allow visualization of the SNP as a G > A change on the reverse complement strand. As methylation levels in some tumors were relatively low, the PCR cycling conditions were designed to preferentially amplify methylated DNA templates, reducing the amplification of uninformative nonmethylated DNA. An annealing temperature of 72°C was used for the first 7 cycles, then reduced by 1°C per cycle until an annealing temperature of 65°C was reached, whereupon 30 further cycles were conducted at this temperature. PCR-amplified products were cloned into the vector, pCR 4-TOPO, using the TOPO TA Cloning Kit for Sequencing (Invitrogen), and transformed into One Shot TOP10 Chemically Competent Escherichia coli cells (Invitrogen). A minimum of 12 colonies was isolated and the plasmid inserts sequenced using vector-specific primers.
Statistical Analysis
The primary endpoint of this study was overall survival; survival curves were generated using Kaplan–Meier analysis. Cox proportional hazards regression analysis was performed to analyze survival, adjusted for age (decades) and sex. In addition, survival was analyzed with the variant T allele of the rs16906252 SNP and MGMT promoter methylation as covariates, as well as for the interaction between the rs16906252 genotype and MGMT promoter methylation. P-values obtained for variables that were below .10 were considered for multivariable analysis, as marginally insignificant effects could be potential confounders. Associations among the rs16906252 genotype, MGMT promoter methylation, and MGMT protein expression were sought using 2-sided chi-square analyses on dichotomized variables. P ≤ .05 was considered significant. Where an association was found to be significant on univariate analyses, multivariate logistic regression was used to adjust for age and sex. The median k and Mann–Whitney U nonparametric tests were used to compare the median and distribution, respectively, of methylation levels detected by pyrosequencing in patients segregated by SNP genotype. A multinomial logit model was used to determine the risk association between the SNP genotype and development of GBM for all AGOG cases and for subgroups segregated by MGMT methylation status. The SPSS statistics package version 22 was used.
Results
MGMT Promoter Methylation Is Associated With Improved Survival
The frequency of MGMT promoter methylation in 163 Australian AGOG patients was 56.4% (missing data, 19). Survival data were available for 160 of 163 patients. As expected, patients with MGMT promoter methylated tumors showed a longer median survival, of 15.77 months (95% CI: 11.27–20.26), compared with those with nonmethylated tumor (median survival, 10.70 mo, 95% CI: 8.91–12.49). This difference was significant by the log-rank test (P < .001) and remained significant when adjusted for age and sex (Table 2).
Table 2.
Association between MGMT promoter methylation and rs16906252 and overall survival of GBM patients
| Genetic Features | Univariate Analysis |
Multivariate Analysisa |
|||
|---|---|---|---|---|---|
| Survival Times, mo, median (95% CI) | Log-rank P |
HR (95% CI) |
HR (95% CI) |
P | |
| Australian cohort (AGOG) | |||||
| Methylation | |||||
| Nonmethylated | 10.70 (8.91–12.49) | .000 | 1.00 (Ref) | 1.00 (Ref) | .000 |
| Methylated | 15.77 (11.27–20.26) | 0.42 (0.28–0.62) | 0.42 (0.28–0.63) | ||
| SNP | |||||
| CC (wild-type) | 12.26 (10.39–14.13) | .010 | 1.00 (Ref) | 1.00 (Ref) | .005 |
| CT/TT combined | 19.96 (14.07–25.85) | 0.56 (0.36–0.87) | 0.52 (0.33–0.82) | ||
| American cohort (UCLA/KPLA) | |||||
| Methylation | |||||
| Nonmethylated | 17.10 (15.52–18.68) | .001 | 1.00 (Ref) | 1.00 (Ref) | .000 |
| Methylated | 24.69 (19.04–30.34) | 0.56 (0.39–0.79) | 0.52 (0.36–0.73) | ||
| SNP | |||||
| CC (wild-type) | 18.20 (15.60–20.48) | .054 | 1.00 (Ref) | 1.00 (Ref) | .040 |
| CT/TT combined | 21.11 (17.80–24.42) | 0.63 (0.39–1.01) | 0.61 (0.38–0.98) | ||
Adjusted for age and sex.
MGMT SNP rs16906252:C > T Is Associated With MGMT Methylation and Improved Overall Survival Irrespective of Methylation Status
Tumor DNA from 163 AGOG cases was genotyped at the MGMT SNP rs16906252:C > T. Thirty-four cases (20.9%) were heterozygous carriers of the variant T allele with no homozygous TT carriers detected. Therefore, the minor T allele frequency in this cohort was 10.97% and this was in Hardy–Weinberg equilibrium (P = .125).
Both tumor methylation and SNP genotype data were available for 138 of the 163 cases. In univariate analyses, patients carrying the T allele were more likely to have an MGMT-methylated tumor than wild-type CC patients (odds ratio [OR], 2.64; 95% CI: 1.05–6.63, P = .035; Table 3). This was consistent with our previous findings.16 The association between carriage of the T allele and presence of MGMT methylation remained significant when adjusted for age and gender in a multivariate logistic regression model (OR, 2.56; 95% CI: 1.01–6.51, P = .049). Furthermore, the median level of MGMT methylation was significantly higher among carriers of the T allele compared with wild-type patients (P = .011), as was the distribution of methylation levels (P = .040).
Table 3.
Association between the T genotype of the rs16906252 SNP and MGMT methylation in GBM
| SNP (Genotype) | Methylation |
|||
|---|---|---|---|---|
| No. Nonmethylated | No. Methylated | OR (95% CI) | P | |
| Australian cohort (AGOG) | ||||
| CC (wild-type) | 45 | 61 | 1.00 (Ref) | |
| CT/TT combined | 7 | 25 | 2.64 (1.05–6.63) | .035 |
| American cohort (UCLA/KPLA) | ||||
| CC (wild-type) | 88 | 44 | 1.00 (Ref) | |
| CT/TT combined | 11 | 12 | 2.18 (0.89–5.34) | .083 |
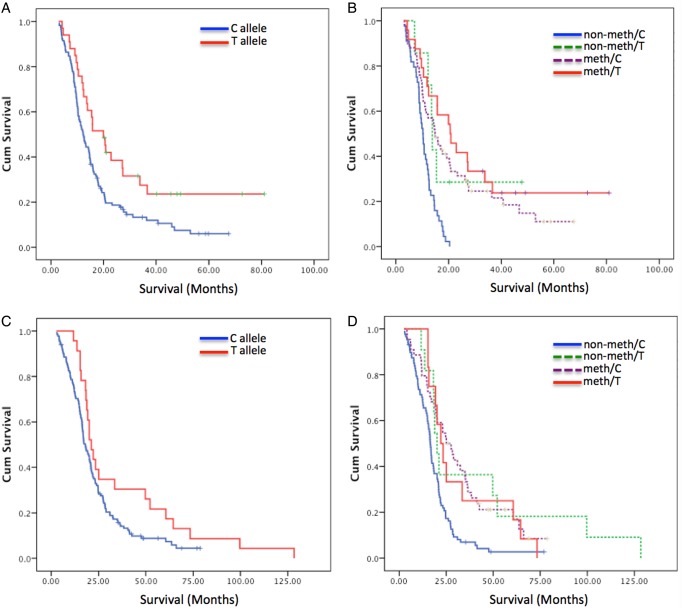
A significant survival benefit was demonstrated for carriers of the T allele, irrespective of MGMT methylation status (Fig. 1A). The median survival for carriers of the T allele was 19.96 months (95% CI: 14.07–25.85) compared with wild-type patients (12.26 mo; 95% CI: 10.39–14.13, P = .010). Multivariate analyses adjusting for age (continuous variable) and sex showed that MGMT promoter methylation status (HR = 0.42, P < .001) was the most powerful single factor associated with overall survival, followed by the MGMT SNP rs16906252:C > T (HR = 0.52, P = .005; Table 2).
Fig. 1.
Kaplan–Meier survival curves based on classification by (A) c.-56C > T SNP genotype for the AGOG cohort, (B) interaction between the c.-56C > T SNP genotype and MGMT methylation assessed by pyrosequencing for the AGOG cohort, (C) c.-56C > T SNP genotype for the UCLA/KPLA cohort, and (D) interaction between the c.-56C > T SNP genotype and MGMT methylation assessed by MSP for the UCLA/KPLA cohort. Abbreviations: Meth, methylation of the MGMT promoter; Non-meth, nonmethylated MGMT promoter; T, variant T allele of the SNP rs16906252; C, wild-type C allele.
Significantly improved survival was noteworthy in a small group of patients with nonmethylated tumors (n = 7) in whom carriage of the variant T allele was detected. The median survival of these patients was 13.86 months, significantly better than patients with the wild-type C allele and nonmethylated tumors (n = 44; median survival: 10.233 mo; P = .029).
Reduced MGMT Expression by Immunohistochemistry Correlates With Improved Survival, MGMT Methylation, and SNP Genotype
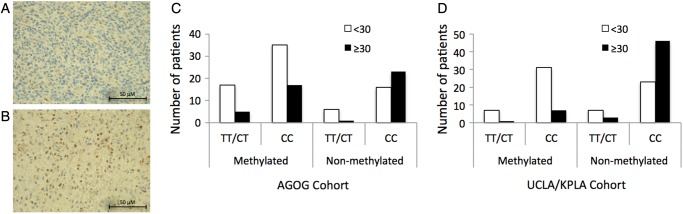
We assessed MGMT protein expression levels using the scoring system published by Lalezari and colleagues,19 separating patients into a low expression group (<30%) and a high expression group (≥30%) (Fig. 2A and B). Approximately half of the cases (49.7%) scored <30% (Fig. 2B). The low expression group demonstrated a significantly longer survival of 17.03 months, whereas patients with high tumor MGMT protein expression demonstrated a median overall survival of 10.00 months (log-rank P < .001) (data not shown). In univariate analyses, MGMT promoter methylation was significantly associated with low tumor MGMT protein expression (OR, 2.46; 95% CI: 1.16–5.21, P = .019), as was carriage of the T allele (OR, 3.13; 95% CI: 1.17–8.33, P = .019; Table 4). However, in multivariate analysis, only the association between MGMT promoter methylation and reduced protein expression remained significant (OR, 2.96; 95% CI: 1.25–7.01, P = .014). A marginal independent effect remained between the SNP genotype and reduced MGMT protein expression (P = .056; Table 4). We found no evidence for an interaction between the SNP genotype and promoter methylation on MGMT protein expression level (Table 4); however, this study had insufficient power to detect anything other than a large size effect.
Fig. 2.
MGMT protein expression. Representative photomicrograph of a GBM specimen (A) graded <30% (low MGMT protein) and (B) graded ≥30% (high MGMT protein). Scale bar, 50 µm. Bar charts summarize the number of cases (GBM patients) as either <30% or ≥30%, stratified by the presence of MGMT methylation and the SNP. (C) Australian cohort and (D) UCLA cohort. Abbreviations: T/T or T/C, variant T allele; C, wild-type C allele.
Table 4.
Associations of MGMT methylation, the T genotype of the rs16906252 SNP, and their lack of interaction, with reduced MGMT protein expression in GBM
| SNP (genotype) | MGMT Protein Expression |
Univariate Analysis |
Multivariate Analysis |
|||
|---|---|---|---|---|---|---|
| No. (<30%) | No. (≥30%) | OR (95% CI) | P | OR (95% CI) | P | |
| Australian cohort (AGOG) | ||||||
| SNP | ||||||
| CC (wild-type) | 55 | 43 | 1.00 (Ref) | 1.00 (Ref) | ||
| CT/TT combined | 24 | 6 | 3.13 (1.17–8.33) | .023 | 8.63 (0.95–78.71) | .056 |
| Methylation | ||||||
| Nonmethylated | 22 | 24 | 1.00 (Ref) | 1.00 (Ref) | ||
| Methylated | 54 | 24 | 2.46 (1.16–5.21) | .019 | 2.96 (1.25–7.00) | .014 |
| SNPa methylation | 0.19 (0.02–2.32) | .194 | ||||
| American cohort (UCLA/KPLA) | ||||||
| SNP | ||||||
| CC (wild-type) | 54 | 53 | 1.00 (Ref) | 1.00 (Ref) | ||
| CT/TT combined | 14 | 4 | 3.44 (1.06–11.11) | .039 | 4.67 (1.10–19.74) | .036 |
| Methylation | ||||||
| Nonmethylated | 30 | 51 | 1.00 (Ref) | 1.00 (Ref) | ||
| Methylated | 39 | 9 | 7.37 (3.14–17.30) | .000 | 8.86 (3.39–23.15) | .000 |
| SNPa methylation | 0.339 (0.02–4.90) | .427 | ||||
SNPa methylation, interaction between SNP and methylation.
MGMT Methylation and the Predictive Value of SNP rs16906252:C > T in an Independent Series of GBM Cases
To validate these findings, we obtained DNA extracted from a subset of 159 cases with GBM from a published UCLA/KPLA study.19 Lalezari and colleagues found in their full cohort of 418 newly diagnosed GBM that patients with tumors with tandem promoter methylation and low MGMT protein expression (<30%) demonstrated improved overall survival and progression-free survival.
Fifty-eight of these 159 tumors (36.5%) were MGMT methylated as assessed by MSP. Twenty-three of the 159 patients (14.5%) in the UCLA cohort were carriers of the T allele of MGMT SNP rs16906252:C > T, among whom 19 were heterozygous and 4 were homozygous TT. Therefore, the minor T allele frequency in this cohort was 8.7%; however, it was not in Hardy–Weinberg equilibrium, P = .004, with an overrepresentation of subjects homozygous for the T allele (n = 4).
In univariate analyses, patients who carried the T allele were more likely to have an MGMT methylated tumor than wild-type CC patients (OR, 2.18; 95% CI: 0.89–5.34, P = .083); however, significance was not reached in this USA cohort (Table 3). Nevertheless, consistent with the Australian cohort, patients who were carriers of the T allele were significantly more likely to show reduced MGMT protein expression (<30%) than wild-type CC patients (OR, 3.44; 95% CI: 1.06–11.11, P = .031; Table 3, Fig. 2D). In both univariate and multivariate analyses, MGMT promoter methylation and carriage of the T allele were significantly associated with low tumor MGMT protein expression (Table 4). Again, no evidence for an interaction between MGMT promoter methylation and SNP genotype was found on MGMT protein expression levels in this cohort (Table 4).
Carriers of the T allele of this SNP showed a trend toward better survival (21.11 mo; 95% CI: 17.80–24.42) than wild-type patients (18.20 mo; 95% CI: 15.60–20.48; log-rank P = .054; Fig. 1C). Nonmethylated T allele carriers demonstrated a significantly longer median survival time (20.05 mo) compared with nonmethylated and wild-type patients (16.54 mo; P = .018), consistent with the observations from the AGOG cohort (Fig. 1D). In fact, survival in the nonmethylated/T allele carrier group (20.05 mo; 95% CI: 17.10–23.00) did not significantly differ from that of the patients with MGMT promoter methylation irrespective of SNP genotype (24.69 mo; 95% CI: 19.14–30.24, P = .631). Therefore, patients who carried the T allele but did not have methylation in their tumors received the same survival benefit as patients with a methylated tumor.
Allelic Bisulfite Sequencing of the MGMT Enhancer Region in Selected GBM
To investigate the relationship between the rs16906252:C > T SNP and methylation at the molecular level, we performed allelic bisulfite sequencing across the enhancer region containing the SNP in the GBM from heterozygous patients from the AGOG cohort. Frozen tumor tissue was available for 7 AGOG patients who tested positive for the T allele (heterozygous CT). Loss of heterozygosity (LOH) of the chromosome arm 10q is a common event in GBM,21 thus we tested these 7 samples for LOH. Retention of heterozygosity was detected in 3 of the 7 samples, allowing an informative investigation of methylation of the individual alleles for any genotype specificity (LOH was detected in 4 samples, which were omitted from this aspect of the study for being uninformative). As illustrated in Fig. 3 and Supplementary Fig. 1, methylation was found to be specific to the T allele in all 3 tumors.
Fig. 3.
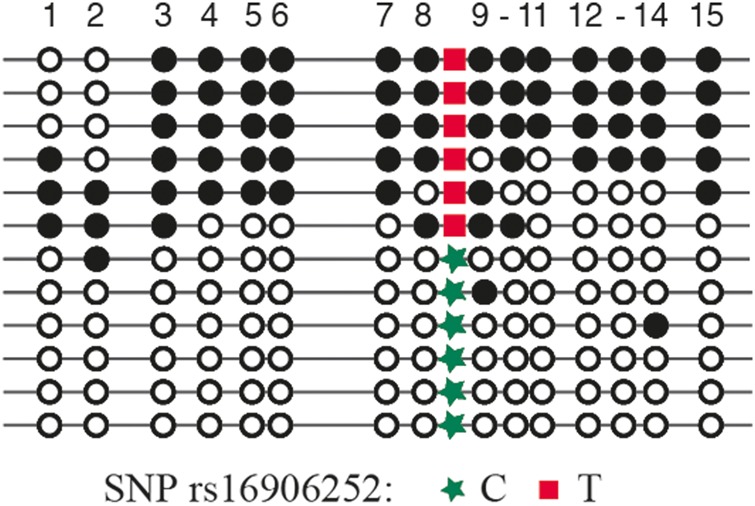
Representative pattern of allele bisulfite sequencing spanning 15 CpG sites across the MGMT enhancer region. The patient is 30% methylated (as assessed by pyrosequencing), is heterozygous for the rs16906252 SNP, and demonstrates selective methylation of the T allele. Horizontal lines indicate individual alleles, circles denote individual CpG dinucleotides, with black indicating a methylated CpG and white indicating a nonmethylated CpG.
MGMT SNP rs16906252 Is Associated With a Significantly Increased Risk of Developing MGMT Methylated GBM
Given the propensity for MGMT methylation and reduced MGMT protein expression in tumors of patients carrying the T allele, we sought to determine if carriage of the T allele is associated with an increased risk of developing GBM, specifically MGMT methylated GBM. We genotyped 451 healthy controls who were matched geographically to the AGOG cohort and compared the frequency of carriage of the T allele between the case and control groups. Among the healthy controls, 54 (12%) were carriers of the T allele, and the T allele frequency was 6.08%, conforming to Hardy–Weinberg equilibrium (P = .58).
There was a significant association between carriers of the T allele and risk for GBM compared with wild-type subjects in univariate analyses (OR, 2.07; 95% CI: 1.29–3.32, P = .003), which remained significant in multivariate logistic regression adjusted for age (OR, 1.96; 95% CI: 1.16–3.32, P = .013).
When the cases were stratified by tumor MGMT methylation status and compared with healthy controls, an even more significant association was found between carriers of the T allele and risk of developing an MGMT methylated tumor (age-adjusted OR, 2.86; 95% CI: 1.57–5.21, P = .001) compared with wild-type individuals. No significant association was found between the SNP genotype and risk of developing an MGMT a nonmethylated tumor (age-adjusted OR, 1.14; 95% CI: 0.48–2.73; P = .770). These findings suggest that carriage of the T allele confers an overall elevated risk of developing GBM but that the proportional increase is in GBM exhibiting MGMT methylation.
Discussion
Epigenetic silencing of MGMT by promoter methylation is a known predictive biomarker for therapeutic response to TMZ in GBM. This study provides compelling evidence for the cis-acting genetic variant, rs16906252, as a key determinant for MGMT methylation in GBM.
We assessed the DNA from 2 independent GBM cohorts: Australian (AGOG, n = 163) and American (UCLA/KPLA, n = 159). All patients were treated with concurrent radiation and TMZ and adjuvant TMZ. In both cohorts, as anticipated, MGMT methylation was significantly associated with better survival. One of the current issues with MGMT methylation status, determined by MSP or pyrosequencing techniques, is that it does not strictly correlate with TMZ sensitivity in all patients. For example, good response is seen in MGMT methylated patients: median survival of 23.4 months compared with 12.6 months in nonmethylated patients.5 Nevertheless, 15% of patients treated with TMZ survived 2 years or more despite having MGMT nonmethylated tumors,22 clearly indicating additional response/survival mechanisms that remain to be determined. Hence, clinically, nonmethylated MGMT does not preclude treatment with TMZ in patients who are fit for standard therapy. In our current datasets, a subset of nonmethylated GBM tumors demonstrated survival times in excess of 2 years.
An association between the presence of MGMT methylation and carriage of the T allele has been observed in numerous cancers.11–17 We tested patient DNA for the rs16906252 SNP and found carriage of the T allele at an incidence of 20% in the AGOG cohort and 15% in the UCLA/KPLA cohort. Carriage of the T allele was more frequently observed in patients with MGMT methylated tumor in the AGOG cohort. The overall risk of a T allele carrier and MGMT methylation was 2.56 (95% CI: 1.01–6.51, P = .049), independently confirming our previous finding.16 While the same trend was observed in the UCLA/KPLA cohort, this did not reach statistical significance. This difference in significance between the 2 cohorts may simply reflect the different methods used to determine methylation status, since other associations with the T allele were concordant. For example, in both cohorts, carriers of the T allele were more likely to demonstrate reduced tumor MGMT protein expression (<30%), likely representing a secondary consequence of promoter methylation. Questions regarding the sensitivity and specificity of MGMT methylation testing methods have been raised. For the Australian cohort, pyrosequencing, which is quantitative, was used. The cutoff value for dichotomizing MGMT methylated and nonmethylated gliomas was 9%, set as the nadir of the logarithmic distribution curve of quantitative methylation values. However, there is a “gray zone” around this “technical cutoff,” and perhaps tumors with percentage values close to the cutoff should not be assigned to either the methylated or the nonmethylated category. When pyrosequencing data were analyzed as a continuous variable, the T allele was also associated with increased levels of methylation compared with the C allele. MSP was used in the UCLA/KPLA cohort. MSP is a simple but nonquantitative assay that is commonly used; however, the sensitivity and specificity of this assay for detecting methylation depends upon the specific PCR protocol used. MSP typically has a high analytical sensitivity, which may detect very low levels of methylation of questionable clinical significance. We suggest that pyrosequencing may provide a more reliable methylation testing method, given that well-designed assays are quantitative and have an internal quality control measure for sodium bisulfite conversion, and this technique has generally been found to be robust for use on both formalin-fixed and fresh-frozen tissues.
An intriguing finding, consistent in both cohorts, was the significantly better overall survival in patients who were T allele carriers but had nonmethylated tumors. The median survivals for T allele/MGMT nonmethylated patients in the Australian and UCLA cohorts were 15.5 months and 25.0 months, respectively. When compared with the C allele (wild-type)/MGMT nonmethylated subset, carriage of the T allele conferred a 5-month (Australian cohort) and 8.5-month (UCLA cohort) survival advantage. In fact, survival of T allele carriers was equivalent to that of patients with MGMT methylated tumors in the respective cohorts. Although the reason for this remains uncertain, we speculate that the T allele may have direct effects on transcriptional activity, leading to lower MGMT protein expression levels. We previously used a standard luciferase promoter reporter assay to assess the effect of the T allele on MGMT promoter-enhancer activity.16 Constructs carrying either the wild-type promoter sequence or the equivalent sequence but with the T nucleotide substituted at the rs16906252 SNP position were transfected into 2 GBM cell lines, U251 and U87. A reduction of ∼30% in normalized promoter reporter activity was observed in both cell lines transfected with the haplotype carrying the T allele. If the nonmethylated T allele is indeed transcribed at a lower level than the C allele in vivo in GBM, lower levels of MGMT would be produced, thereby sensitizing cells to TMZ. This difference in transcriptional output between the 2 genetic alleles may be brought about by the altered binding of a nuclear factor or complex to the MGMT promoter enhancer, which in turn would likely alter nucleosome occupancy at the transcription initiation site. Irrespective of the underlying mechanism, our results suggest that a simple genotype test at the rs16906252 SNP may be a useful adjunctive test to MGMT methylation testing of GBM, with the utility to identify the subset of patients with unmethylated MGMT status who could also benefit from TMZ.
Questions surrounding why MGMT becomes epigenetically silenced have not been resolved. This study provides strong evidence that the T allele is a key determinant in the acquisition of MGMT methylation in GBM tumorigenesis. Since the first GWAS, several loci associated with glioma risk have been discovered, including the rs6010620 SNP in the RTEL gene (regulator of telomere elongation helicase 1) and rs2736100 in the TERT gene (telomerase reverse transcriptase).23 These 2 genetic variants of telomere regulation genes associate with glioma risk in older patients.9 In this current study, the frequency of carriage of the minor T allele of the rs16906252 variant in healthy controls was 13% (minor allele frequency: 0.065). Significantly elevated risk of GBM development was associated with carriage of the T allele. This risk increased when GBM was classified by the presence of MGMT methylation. This SNP is not included on the SNP arrays that were employed for previous GWAS, and furthermore GWAS were not performed on subsets of tumors stratified by methylation status, explaining the novelty of this finding despite prior GWAS.
MGMT silencing due to methylation has been suggested as an early event in the development of both colorectal and gastric adenocarcinoma, with MGMT promoter methylation found in adjacent normal tissues.24,25 Furthermore, the presence of low levels of MGMT promoter methylation in normal tissues from both healthy and cancer-affected individuals has been associated with the T allele of the rs16906252 SNP.11,17 In one study, the investigators detected low levels of MGMT methylation in the peripheral blood DNA of healthy individuals and found a significant association with the minor T allele. Moreover, they demonstrated that this low-level methylation in the peripheral blood leukocyte DNA of heterozygous patients was linked to the T allele. They reasoned that the T allele might affect the propensity to methylate the MGMT promoter in normal individuals.17 We found that monoallelic methylation was consistently linked to the T allele in the GBM of 4 heterozygous patients. We suggest that carriers of the T allele are at increased risk of developing MGMT methylated GBM due to the increased likelihood of epigenetic silencing of this allele within normal tissues.
In summary, our 2 key findings are that the rs16906252 variant in the MGMT promoter-enhancer region is a significant risk factor for GBM, particularly MGMT promoter methylated GBM, and that carriage of the T allele is associated with better overall survival in TMZ-treated patients, irrespective of tumor MGMT methylation status. The identification of T allele carriage as a predictive biomarker has significant and immediate translational implications in clinical practice through simple detection of the SNP in blood DNA and clinical stratification for treatment outcome with TMZ.
Supplementary Material
Funding
The work was supported by the Cancer Council NSW and the Cure Brain Cancer Foundation.
Conflict of interest statement. None declared.
Supplementary Material
References
- 1.Burnet NG, Jefferies SJ, Benson RJ, et al. , Years of life lost (YLL) from cancer is an important measure of population burden—and should be considered when allocating research funds. Br J Cancer. 2005;92(2):241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3.Reifenberger G, Hentschel B, Felsberg J, et al. Predictive impact of MGMT promoter methylation in glioblastoma of the elderly. Int J Cancer. 2012;131(6):1342–1350. [DOI] [PubMed] [Google Scholar]
- 4.Stummer W, Nestler U, Stockhammer F, et al. Favorable outcome in the elderly cohort treated by concomitant temozolomide radiochemotherapy in a multicentric phase II safety study of 5-ALA. J Neurooncol. 2011;103(2):361–370. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 6.Xu M, Nekhayeva I, Cross CE, et al. Influence of promoter/enhancer region haplotypes on MGMT transcriptional regulation: a potential biomarker for human sensitivity to alkylating agents. Carcinogenesis. 2014;35(3):564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shete S, Lau CC, Houlston RS, et al. Genome-wide high-density SNP linkage search for glioma susceptibility loci: results from the Gliogene Consortium. Cancer Res. 2011;71(24):7568–7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh KM, Codd V, Smirnov IV, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46(7):731–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh KM, Rice T, Decker PA, et al. Genetic variants in telomerase-related genes are associated with an older age at diagnosis in glioma patients: evidence for distinct pathways of gliomagenesis. Neuro Oncol. 2013;15(8):1041–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkins NJ, Lee JH, Wong JJ, et al. MGMT methylation is associated primarily with the germline C>T SNP (rs16906252) in colorectal cancer and normal colonic mucosa. Mod Pathol. 2009;22(12):1588–1599. [DOI] [PubMed] [Google Scholar]
- 12.Ogino S, Hazra A, Tranah GJ, et al. MGMT germline polymorphism is associated with somatic MGMT promoter methylation and gene silencing in colorectal cancer. Carcinogenesis. 2007;28(9):1985–1990. [DOI] [PubMed] [Google Scholar]
- 13.Kristensen LS, Nielsen HM, Hager H, et al. Methylation of MGMT in malignant pleural mesothelioma occurs in a subset of patients and is associated with the T allele of the rs16906252 MGMT promoter SNP. Lung Cancer. 2011;71(2):130–136. [DOI] [PubMed] [Google Scholar]
- 14.Kristensen LS, Treppendahl MB, Asmar F, et al. Investigation of MGMT and DAPK1 methylation patterns in diffuse large B-cell lymphoma using allelic MSP-pyrosequencing. Sci Rep. 2013;32:789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leng S, Bernauer AM, Hong C, et al. The A/G allele of rs16906252 predicts for MGMT methylation and is selectively silenced in premalignant lesions from smokers and in lung adenocarcinomas. Clin Cancer Res. 2011;17(7):2014–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald KL, Rapkins RW, Olivier J, et al. The T genotype of the MGMT C>T (rs16906252) enhancer single-nucleotide polymorphism (SNP) is associated with promoter methylation and longer survival in glioblastoma patients. Eur J Cancer. 2013;49(2):360–368. [DOI] [PubMed] [Google Scholar]
- 17.Candiloro IL, Dobrovic A. Detection of MGMT promoter methylation in normal individuals is strongly associated with the T allele of the rs16906252 MGMT promoter single nucleotide polymorphism. Cancer Prev Res (Phila). 2009;2(10):862–867. [DOI] [PubMed] [Google Scholar]
- 18.Oberstadt MC, Bien-Moller S, Weitmann K, et al. Epigenetic modulation of the drug resistance genes MGMT, ABCB1 and ABCG2 in glioblastoma multiforme. BMC Cancer. 2013;13:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lalezari S, Chou AP, Tran A, et al. Combined analysis of O6-methylguanine-DNA methyltransferase protein expression and promoter methylation provides optimized prognostication of glioblastoma outcome. Neuro Oncol. 2013;15(3):370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christians A, Hartmann C, Benner A, et al. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PloS One. 2012;7(3):e33449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tada K, Shiraishi S, Kamiryo T, et al. Analysis of loss of heterozygosity on chromosome 10 in patients with malignant astrocytic tumors: correlation with patient age and survival. J Neurosurg. 2001;95(4):651–659. [DOI] [PubMed] [Google Scholar]
- 22.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 23.Egan KM, Thompson RC, Nabors LB, et al. Cancer susceptibility variants and the risk of adult glioma in a US case-control study. J Neurooncol. 2011;104(2):535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svrcek M, Buhard O, Colas C, et al. Methylation tolerance due to an O6-methylguanine DNA methyltransferase (MGMT) field defect in the colonic mucosa: an initiating step in the development of mismatch repair-deficient colorectal cancers. Gut. 2010;59(11):1516–1526. [DOI] [PubMed] [Google Scholar]
- 25.Zou XP, Zhang B, Zhang XQ. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Human Pathology. 2009;40(11):1534–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.