Abstract
Growing old is our destiny. However, the mature differentiated cells making up our body can be rejuvenated to an embryo-like fate called pluripotency which is an ability to differentiate into all cell types by enforced expression of defined transcription factors. The discovery of this induced pluripotent stem cell (iPSC) technology has opened up unprecedented opportunities in regenerative medicine, disease modelling and drug discovery. In this review, we introduce the applications and future perspectives of human iPSCs and we also show how iPSC technology has evolved along the way.
Keywords: reprogramming, induced pluripotent stem cells, regenerative medicine, epigenetics, cancer
1. Introduction
The cell, the smallest unit of a living organism, which was first observed by Robert Hooke in 1665, still fascinates the scientists of today [1]. Our body consists of more than 200 committed cell types, some of which work independently, such as blood cells, whereas others form tissues and work in networks, like synapses from the brain to the end of the body. Despite their great diversity, all of the cells in our body evolve from a unicellular zygote.
A zygote, which is the earliest developmental stage of embryogenesis, transforms into a morula and then a blastocyst through mitotic cell division before implantation. The inner cell mass (ICM), which is a component of the blastocysts, matures into an epiblast of the post-implantation embryo, and then commits to one of the three germ layers: the endoderm, mesoderm or ectoderm. In other words, the ICM can differentiate into all of the cell types in the human body. This highly specialized ability is referred to as pluripotency. Pluripotency was first introduced to the culture dish as embryonic stem cells (ESCs). ESCs have made a great contribution to developmental biology through the generation of genetically engineered mice [2–4]. In combination with homologous recombination technology innovated by Smithies, Capecchi reported Hprt deficient mice, the first knockout mice [5,6]. These technologies robustly accelerated the following era of life science. Normal embryonic development, including cellular differentiation, has long been thought of as a one-way street, which may be likened to a ball rolling downhill, from an undifferentiated stem or progenitor cell state to a physiologically mature cell, as depicted by Conrad Waddington during the mid-twentieth century (figure 1) [7]. Indeed, one can think of cells rolling down this landscape into deeper, inescapable valleys that represent the determination of cell fate during development until the cells reach a stable state at the bottom. It was once believed that an unnecessary genetic code in committed cells other than germ cells was released—a principle referred to as the Weismann barrier [8]. The cloning of a frog through the nuclear transfer of embryonic cells, but not somatic cells, in 1952 by Robert Briggs and Thomas Joseph King suggested that irreversible changes took place in the somatic nuclei [9]. Thus, it seemed likely that a change of cell fate was impossible once cells were committed. However, the story took an unexpected turn with Sir John Gurdon's landmark experiments in Xenopus laevis, which first established the concept of the reprogramming of cell fate promptly after the emergence of Waddington's dogma [10]. The birth of a cloned sheep in the late-twentieth century, famously named Dolly, meant that the erasing of epigenetic memories in somatic nuclei could be achieved, even in mammals [11]. In this way, these facts suggest that an unidentified reprogramming activity exists in oocytes and that the activity is conserved beyond species (figure 1).
Figure 1.
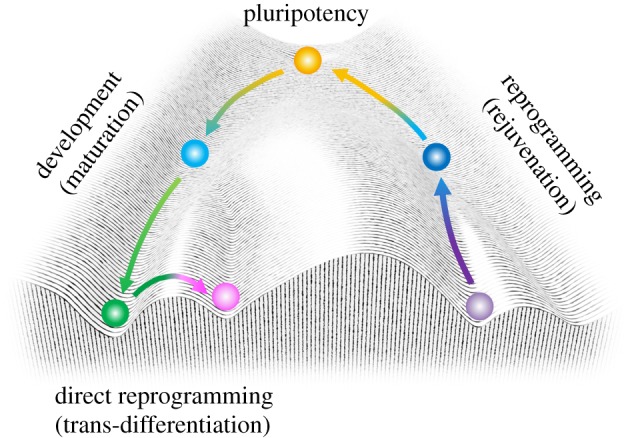
Cell fate changes on Waddington's epigenetic landscape. Pluripotent stem cells (top, yellow) can commit to various somatic lineages (bottom left, green) via a progenitor state (middle left, light blue) during embryonic development and in vitro differentiation. Direct reprogramming, or transdifferentiation, using tissue-specific transcription factors convert the fate of lineage-committed cells (bottom left, green) to another differentiated fate (bottom centre, pink), bypassing the need for a pluripotent state. There are several ways of reprogramming lineage-committed cells (bottom right, purple) toward pluripotency (top, yellow). Adapted, with permission, from Waddington [7]. (Online version in colour.)
The first discovery of defined reprogramming factors was reported in 1987 [12]. Davis et al. performed a complementary DNA subtraction and found three genes that were expressed predominantly in the proliferative myoblasts. One of them was myogenic differentiation 1 (MYOD1, also known as MYOD), which encoded a basic-helix–loop–helix transcription factor that shared homology with a transactivation domain of the c-myc proto-oncogene. The forced expression of Myod1 is alone sufficient to convert fibroblasts to myosin-expressing stable myoblasts. This precise moment was the dawn of the direct reprogramming of somatic cells into cells of another lineage—which is one of the trends in cell biology. It showed that the concept was thoroughly alive and extremely changeable (figure 1) [13].
The two scientific streams emerged as induced pluripotent stem cells (iPSCs), which bear a striking resemblance to ESCs (figure 1) [14]. Although the reprogramming process during iPSC generation is still wrapped in mystery, the products offer promise in many areas, such as drug discovery, pathological studies, toxicology, the evaluation of secondary drug effects and regenerative medicine [15]. Here, we outline the current knowledge and future prospects for induced pluripotency.
2. Applications of induced pluripotent stem cells
(a). Cell transplantation therapy
Since they were first reported in 1998, human ESCs have been strongly expected to be a key to the treatment of intractable diseases such as Parkinson's disease and spinal cord injuries [16]. In 2010, Geron Corporation pressed for the initiation of the first-ever clinical trial of ESC-derived oligodendrocyte progenitor cells for the patient with spinal cord injury. Advanced Cell Technology, another biotechnology company, reported a clinical trial using ESC-derived retinal pigment epithelium to treat dry age-related macular degeneration (AMD). However, the acceptance of human ESCs has faced several hurdles, including an ethical concern regarding the use of human embryos and immune rejection after transplantation.
iPSC technology has the potential to overcome these issues. In 2007, the proof of concept of the therapeutic usage of iPSCs was reported in a mouse model of sickle-cell anaemia, a genetic blood disorder caused by a defect in the β-globin gene [17]. Homologous recombination-mediated gene correction in mutant iPSCs allowed for the disease to be cured in donor mice. This was an example of a perfect model for iPSC-mediated regenerative medicine. Namely, an ideal and distinctive potential of iPSC technology exists in their use in made-to-order therapies with autologous cells. The iPSC-based autologous method is advantageous in comparison with allografts from other donors, because it is not associated with immunological rejection or infection with unidentified viruses or other pathogens [18–20]. The first clinical trial of the treatment of a wet AMD patient using autologous iPSCs was launched in September 2014 by Masayo Takahashi in Japan.
In spite of the potential benefits of autologous iPSC therapies, there are some associated limitations. First, the preparation of autologous iPSCs from each patient carries a high medical cost. In addition, because more than three months are needed to generate the iPSCs, it is not possible to meet the deadline for effective treatments of some disorders such as spinal cord injuries. It is therefore important to take a realistic approach to the use of allogenic iPSCs in regenerative medicine. Fortunately, the striking advantage of iPSC technology is the flexibility of establishment from any age of donor candidates in the world and the ease of access to their origins. Before the generation of clinical-grade iPSC clones, we can closely examine all of the aspects of donors such as health conditions and their human leucocyte antigen (HLA) type. In addition, it is possible to rigorously evaluate single nucleotide and copy number variation based on donor information. Another strength is the abundant availability of multiple clones from each donor. Because human pluripotent stem cells generally show clonal variations such as differentiation propensities, it allows researchers to make choices based on the purpose of each application [21,22].
Based on the experiences of bone marrow transplantation, the matching of types of three major HLA loci including A, B and DR between recipient and donor is expected to elicit immune rejection after transplantation less frequently. The most realistic method for iPSC therapy that is conceivable under the present circumstances is based on the collection of iPSC stock from various HLA-homozygous donors [23–25]. In the case of Japan, for example, 75 types of HLA-homozygous donors can theoretically cover approximately 80% of the Japanese population (127 300 000). It is calculated that 40 000 candidates could include these 75 donors [25]. This search would become more promising with the cooperation of existing cell banks making available their HLA data of stocked cells. In fact, in 2013, the Hyogo Cord blood bank, one of the non-government organizations in Japan, decided to provide 10 cord blood cells of HLA-homozygous donors for an iPSC stock project organized by Kyoto University.
Early human iPSC research has coped with problems such as the integration of viral vectors as a possible risk of tumourigenicity [26,27]. Furthermore, the culture media for human pluripotent stem cells contained animal-derived components that could pose a hurdle for therapeutic use in humans. However, most of the issues have been solved by recent technological innovations such as use of integration-free methods and xenofree culture (figure 2) [25,28–32]. Hence, we expect that, in the near future, clinical-grade iPSCs will be generated under conditions that correspond to good manufacturing practices (GMPs).
Figure 2.
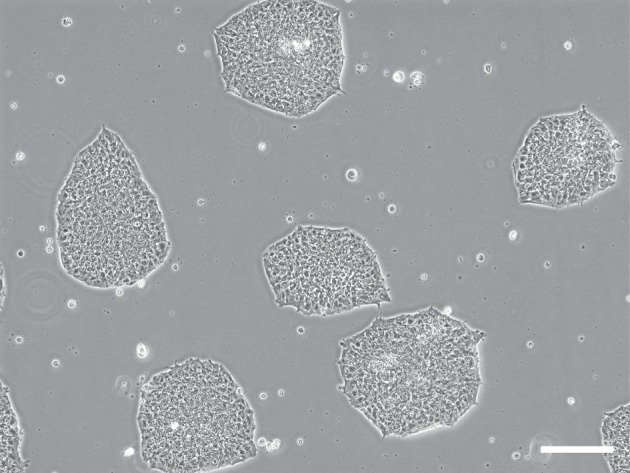
Human iPSCs for clinical use. A phase contrast image of human iPSCs maintained in chemical defined xenofree medium on laminin 511 E8-coated tissue culture plate. The scale bar indicates 100 µm.
Human iPSCs were first established from skin fibroblasts [26,27,33]. A skin biopsy for fibroblast isolation may only be a sideline issue for the surgery. However, for healthy individuals, it is important to be cautious when deciding on the harvest sites, because a biopsy may leave a visible scar, and because the procedure carries some associated risk of infection. The sources of human iPSCs should therefore be, to the maximum extent possible, obtained with minimally invasive procedures. Cord blood is an attractive source for iPSCs [34–36]. Immature cells such as CD34-positive cells, which are contained abundantly in cord blood, can be a suitable source of iPSCs, not only from the standpoint of availability, but also because of the expectation of fewer acquired genetic mutations. However, while cord blood banks should stock only the cells derived from seemingly healthy donors, it is difficult to trace whether the donors remain healthy in older age. Peripheral blood is another promising source of iPSCs [37,38]. In particular, T lymphocytes can be reproducibly converted into iPSCs through transient-expression methods [25,39,40]. Because the rearrangement patterns of T-cell receptor loci are different and readily distinguishable in each T lymphocyte, it would be easy to monitor and trace the clonality of iPSCs in the procedure under GMP. Through medical diagnosis, we are at least able to confirm the donor's health at the age of collection. Because of the minimal invasiveness associated with peripheral blood collection, there is a strong expectation that a large number of healthy donors will cooperate with collection efforts. In many nations, there are huge stocks of cord blood and large numbers of donor candidates for platelet transfusion or bone marrow transplantation. Peripheral blood donors are definitely one of the most powerful and effective sources for clinical-grade iPSC collection.
Overall, although many drawbacks of iPSC for cell therapy were pointed out, advances have overcome many of them and consequently led to the first clinical study. Researchers succeeded in eliminating oncogene from reprogramming factors, genomic integration of viral vector, and any animal-derived compounds from culture conditions. To save time and cost, some HLA-homozygous iPSC bank projects have been prepared and maintained in Japan, Europe and USA. The quality of ESC/iPSC, though it is not easy to tell with 100% accuracy, became able to be assessed in combination with genomic sequencing testing.
(b). Disease modelling and drug screening
A stereotypical conception related to iPSCs is that their best medical application is in cell transplantation therapy. In our opinion, however, disease modelling and drug screening are at least as important as cell therapy [41]. Animal models have made a tremendous contribution to the better understanding of disease mechanisms. However, we do recognize limitations of animal models in recapitulating human diseases. For example, a number of drugs have been developed that showed therapeutic effects in rodent models of amyotrophil lateral sclerosis, but which proved to be ineffective in the treatment of human patients. Such occurrences underscore the necessity of disease models using human cells.
Disease-specific iPSC lines were first reported from two groups in 2008 [42,43]. The first successful in vitro reconstruction of a disease state was the reconstruction of spinal muscular atrophy [44,45]. Patient-derived iPSCs were demonstrated to be useful for drug validating in Rett syndrome [46] and in familial dysautonomia [47]. Recently, Yamashita et al. successfully demonstrated that statin, a well-known drug for high blood pressure, could correct degraded cartilage in both chondrogenically differentiated thanatophoric dysplasia type I and achondroplasia iPSCs [48]. These results not only showed that the reproduction of disease phenotypes using patient-derived iPSCs was possible, but also the potential applications of iPSCs in drug screening including drug repositioning. To date, many patient-specific iPSC lines have been established and used for disease modelling. These are expected to facilitate the accession of rare disease studies [49]. One of the critical issues regarding patient-derived iPSC is of control. Despite the ready availability of ES cells and iPSCs derived from healthy donors, the big differences that may exist in genetic backgrounds are often a source of controversy. Healthy family members such as mothers and brothers are better targets for control donors. In addition, the recent progress of genetic editing technologies using custom-made nucleases, including zinc-finger nucleases, transcription activator-like effector nucleases and clustered regularly interspaced short palindromic repeat/Cas9 ground the gene collection in patient-specific iPSCs more in reality [50,51].
3. The challenges of induced pluripotent stem cells
(a). Diversity of induced pluripotent stem cell characteristics
Although it has been demonstrated that each ESC line has its own clonal differences [21], the iPSC lines have shown greater diversity than ESCs. The cause of the variety has been explained in several ways such as retained epigenetic memory [52,53], genetic background [54] and features newly obtained during reprogramming. Recent analysis dissecting the reprogramming process in mouse [55] and human [56] revealed that the cells in transitional phase are dramatically distinct from both original and fully reprogrammed cells. Because of that iPSC diversity could be due to the epigenetic dynamics during the process of iPSC generation from cells of somatic origin. This idea is supported by the evidence that some distinct iPSC lines exhibit features of incomplete reprogramming [57].
Many of the reported ‘incomplete’ human/mouse iPSC lines have characteristics that are similar to ESCs, such as morphology, marker gene expression and basic pluripotency represented in the teratoma formation, while they exhibit particular defects such as poor quality of differentiation, low growth rate, aberrant transcription, DNA methylation, chromatin regulation or chimeric animal contribution in mouse [58–63]. Dissecting the molecular and biological differences among the various iPSC lines has greatly helped in gaining an in-depth understanding of the mechanisms that are central to complete pluripotency.
To select completely reprogrammed iPSC lines, evidence-based key criteria are required to be defined. However, there have not been many reports that exhibited the link between biological phenotype and molecular marker of human ES/iPSCs. For example, KLF4, one of the reprogramming factors, was considered to interrupt neurogenesis of iPSCs [57,64]. XIST is also implied as a benchmark to assess human ESC/iPSC quality. The study comparing XaXi hiPSCs with and without XIST expression suggests the possibility that XIST expression influences the proliferation speed and differentiation potential of hiPSCs [65]. Like these, further studies to pursue molecular markers to evaluate ESC/iPSC quality are required in the future.
(b). Differences between embryonic stem cells and induced pluripotent stem cells
The claim, in dozens of reports, that epigenetic relics of somatic origin, including DNA methylation and gene expression, remain in iPSCs, distinguishes iPSCs from ESCs despite their shared pluripotency [66–73]. On the other hand, many other reports have demonstrated that no distinct differences (including differences in epigenetic memory) exist between ESCs and iPSCs [54,74–76]. The number of cells used in such studies may influence conclusions. Studies that used 2–6 ESCs and 2–12 iPSCs found notable differences in gene expression and/or DNA methylation between ESCs and iPSCs [66–73]. Those that investigated 20–36 ESCs and 12–68 iPSCs found otherwise [74–76].
Koyanagi-Aoi et al. performed comparison analyses of gene expression, miRNA expression and DNA methylation between 10 ESCs and 49 iPSCs [77]. Although they saw no clear differences between the two cell types, they did find that some iPSCs exhibited distinguishing expression signatures that were related to defects in differentiation. In contrast, incomplete reprogramming produced clones with distinguishable properties. A recent study has further shown that approximately 40% of the 3771 human endogenous retroviruses type-H (HERV-Hs) on the human genome are transiently activated during reprogramming [57]. An aberrant increase of HERV-H expression in human ESCs/iPSCs caused by failure of silencing induces a defective phenotype in the directed differentiation into the neural lineage [57,77]. Therefore, with the exception of incompletely reprogrammed clones, substantially mature iPSCs are thought to be indistinguishable (with regard to gene expression and epigenetic status) from ESCs. However, the variation in genetic backgrounds has made certain analyses more difficult.
Recently, the invention of human somatic cell nuclear transfer (SCNT)-ESCs has been reported [78–80], and the comparison of gene expressions, epigenetic statuses and genetic alterations between isogenic human SCNT-ESCs and iPSCs derived from the same somatic cell cultures has been discussed [81,82]. Ma et al. concluded that iPSCs have inherent abnormalities, because the similarity of SCNT-ESCs to ESCs derived from in vitro fertilized eggs is greater than that of iPSCs [82]. They found that iPSCs have aberrant DNA methylation statuses for some imprinted genes, such as DIRAS3, MEG3 and PEG3, and regions of X-chromosome inactivation. However, Johannesson et al. concluded that human SCNT-ESCs and iPSCs have no significant differences in gene expression, DNA methylation or frequency of de novo coding mutations [81]. In addition, imprinting loss was shown to occur in both SCNT-ESC and iPSC lines with a similar frequency. A better description of human pluripotency will help us to identify which of these two cell types makes the more suitable model for development.
(c). Naive pluripotency
The combination of bone morphogenic protein and leukaemia inhibitory factor (LIF) causes mouse naive pluripotent stem cells to self-renew, but into a heterogeneous population [83]. The ground state of mouse naive pluripotency, which is defined as a fundamental proliferative state with no epigenetic restriction and minimal requirements of extrinsic signals, can be achieved using chemical inhibitors for mitogen-activated protein kinase (MEK) and glycogen synthase kinase 3 (GSK3) [84]. It makes pluripotent stem cell populations homogeneous and allows for the generation of germline competent ESCs derived from non-permissive mouse strains, such as non-obese diabetic mice [85]. Thus, the ground state buffers cell characteristics acquired from genetic backgrounds that create a considerable difference in extrinsic stimuli responsiveness. It has since been demonstrated that another approach to generating a homogenous population is the continuous passaging of mouse iPSCs, which abrogates transcriptional, epigenetic and functional differences [53]. Additionally, the tetraploid complementation of mouse iPSCs has been shown to produce normal pups, suggesting that selected fully reprogrammed iPSCs have bona fide pluripotency that is indistinguishable from that of ESCs [86–88]. In conclusion, these data suggest that transcription factor-mediated reprogramming can achieve the full spectrum of mouse naive pluripotency as well as embryo-derived pluripotent stem cells in mice.
Recent technologies have allowed for the analysis of human embryos [89], and two recent papers have revealed significant differences in the global DNA methylation statuses of ESCs and blastocysts whose origins are ESCs [90,91]. Thus, the human ESCs in the dish were no longer identical to their embryonic origins. It therefore makes no sense to ask, at the present time, whether factor-induced reprogramming causes abnormalities, because we still lack a description of desirable human pluripotent stem cells.
A way to define human pluripotency is to understand the ground state of human pluripotent stem cells. In the past few years, several strategies to create human naive pluripotency have been proposed [92–97]. The published strategies for the conversion of human primed pluripotent stem cells to naive state commonly use inhibitors of MEK and GSK3. However, the use of only these inhibitors differentiates human primed pluripotent stem cells into neural stem-like cells [98]. Therefore, the published studies differ in their use of additional chemical compounds or growth factors. Notably, naive human ESCs were obtained using the inhibitors for MEK, GSK3 and protein kinase C (PKC) developed by Takashima et al. which exhibited global DNA hypomethylation (except for the imprinted gene loci), a feature similar to that of pre-implantation embryos [94]. A deeper understanding of the ground state of human pluripotency will bring a definitive end to the controversial comparisons and shed light on the goal of reprogramming.
4. Conclusion and future perspectives
Towards the practical use of human ESCs/iPSCs for clinical and industrial application, a large-scale suspension cell culture system for human ESCs/iPSCs has been proposed instead of the conventional adherent cell culture system [35,99–101]. To achieve scaling-up, uniformed quality and low cost, three-dimensional culture devices have been developed such as a spinner flask with dynamic stirring system. These efficient manufacture technologies should promote the widespread use of pluripotent cells in future [100].
iPSC generation can be used as the technology of genome-wide epigenetic resetting in the cancer research field. The stepwise accumulation of genetic mutation is a fundamental model of cancer initiation and progression [102], but the contribution of epigenetic abnormality such as aberrant methylation is still unclear. To assess the influence of abnormal epigenetic alteration on malignant cancer cell behaviour, Stricker et al. established glioblastoma-derived iPSCs and induced neural progenitors. iPSCs exhibited erasure of cancer-specific epigenome whereas re-established neural progenitors showed infiltrative behaviour upon xenotransplantation, indicating the mutated genome of glioblastoma would be a definitive cause of malignant behaviour rather than epigenetic changes [103]. On the other hand, the epigenetic instability induced after the forced expression of reprogramming factors can be also used in the field of cancer development. That the reprogramming process, in some respects, resembles the cancer initiation process [104–110] implies the involvement of common molecular mechanisms. The incomplete reprogramming by overexpression of OSKM factors drove the development of Wilms-like tumours in mouse kidney without any genomic alterations [111]. Further OSKM expression can reprogram these tumour cells into complete iPSCs in vitro, suggesting that an epigenetic change, but not genomic mutation, could be sufficient for the occurrence of some tumorigenesis. Mutual progression in both fields could facilitate an in-depth understanding of cancer development.
Most of the directed differentiation methods from ESCs/iPSCs were established models for in vivo differentiation from the embryo. The ability of pluripotent stem cells to recapitulate the developmental process in vitro makes PSCs useful in the study of developmental biology [112]. The less invasive generation of iPSCs from somatic cells of rare animals facing extinction allows us to access their developmental processes. Thus far, iPSCs have been generated from, for example, a northern white rhino, drill monkey [113], snow leopard [114], domesticated horse [115] and prairie vole [116]. Such iPSCs from various bioresources should facilitate the understanding of species-specific molecular biology. The information yielded from such research can be used for the conservation of endangered animals, in the industrial use of molecules from valuable bioresources and in the study of species specification.
In particular, the non-human primate iPSCs, which have thus far been established from a range of primates [113,117–119] are expected to serve as tools for evolutionary analysis [120]. Their iPSCs, which are derived from fibroblasts or blood cells which are relatively easy to obtain, have made it possible to analyse and compare interspecies cells in vitro. The comparison of humans and apes has revealed human-specific traits in the regulation mechanisms of LINE-1 transposons [121]. Although the iPSC-facilitated study of evolution and development has just begun, the iPSCs in those fields offer huge potential in elucidating the processes by which living things obtained diversity and complexity.
In summary, the technology of factor-mediated induced pluripotency has had a great influence upon the field of medicine such as in transplantation therapy, disease modelling and drug discovery. We have picked the recent advances both in medicine and life science triggered by the generation of iPSCs. Ongoing clinical studies of cell transplantation, improvement of the clinical-grade iPSC bank and storage, drug screening and repositioning will accelerate the achievements of iPSC-based therapy. Developments have also been made in the fundamental life sciences, such as stem cell biology, cancer research and evolution, by using iPSC techniques. These studies, though still exploratory and even challenging, have a future growth potential in these fields.
Acknowledgements
We thank Yoko Miyake, Rie Kato, Eri Minamitani, Sayaka Takeshima, Ryoko Fujiwara and Kyoko Nakahara for their administrative support.
Competing interests
No competing interests.
Funding
This work was supported in part by a grant from Japan Foundation for Applied Enzymology, and the iPS Cell Research Fund. M.O. is a research fellow of JSPS.
References
- 1.Hooke R. 1665 Micrographia: some physiological descriptions of minute bodies made by magnifying glasses with observations and inquiries thereupon. London, UK: Printed by Jo. Martyn and Ja. Allestry, Printers to the Royal Society. [Google Scholar]
- 2.Evans MJ, Kaufman MH. 1981. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. ( 10.1038/292154a0) [DOI] [PubMed] [Google Scholar]
- 3.Martin GR. 1981. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA 78, 7634–7638. ( 10.1073/pnas.78.12.7634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley A, Evans M, Kaufman MH, Robertson E. 1984. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255–256. ( 10.1038/309255a0) [DOI] [PubMed] [Google Scholar]
- 5.Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. 1987. Targeted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature 330, 576–578. ( 10.1038/330576a0) [DOI] [PubMed] [Google Scholar]
- 6.Thomas KR, Capecchi MR. 1987. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51, 503–512. ( 10.1016/0092-8674(87)90646-5) [DOI] [PubMed] [Google Scholar]
- 7.Waddington CH. 1957. The strategy of the genes. A discussion of some aspects of theoretical biology. London, UK: George Allen & Unwin. [Google Scholar]
- 8.Weismann A. 1893. The germ-plasm: a theory of heredity. (Transl. by WN Parker, H Ronnfeldt). New York, NY: Charles Scribners Sons. [Google Scholar]
- 9.Briggs R, King TJ. 1952. Transplantation of living nuclei from blastula cells into enucleated frogs eggs. Proc. Natl Acad. Sci. USA 38, 455–463. ( 10.1073/pnas.38.5.455) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurdon JB. 1962. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J. Embryol. Exp. Morphol. 10, 622–640. [PubMed] [Google Scholar]
- 11.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. 1997. Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813. ( 10.1038/385810a0) [DOI] [PubMed] [Google Scholar]
- 12.Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000. ( 10.1016/0092-8674(87)90585-X) [DOI] [PubMed] [Google Scholar]
- 13.Takahashi K. 2012. Cellular reprogramming - lowering gravity on Waddingtons epigenetic landscape. J. Cell Sci. 125, 2553–2560. ( 10.1242/jcs.084822) [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. ( 10.1016/j.cell.2006.07.024) [DOI] [PubMed] [Google Scholar]
- 15.Yamanaka S. 2009. A fresh look at iPS cells. Cell 137, 13–17. ( 10.1016/j.cell.2009.03.034) [DOI] [PubMed] [Google Scholar]
- 16.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. ( 10.1126/science.282.5391.1145) [DOI] [PubMed] [Google Scholar]
- 17.Hanna J. et al. 2007. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318, 1920–1923. ( 10.1126/science.1152092) [DOI] [PubMed] [Google Scholar]
- 18.Okita K, Nagata N, Yamanaka S. 2011. Immunogenicity of induced pluripotent stem cells. Circ. Res 109, 720–721. ( 10.1161/RES.0b013e318232e187) [DOI] [PubMed] [Google Scholar]
- 19.Araki R. et al. 2013. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature 494, 100–104. ( 10.1038/nature11807) [DOI] [PubMed] [Google Scholar]
- 20.Guha P, Morgan JW, Mostoslavsky G, Rodrigues NP, Boyd AS. 2013. Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell 12, 407–412. ( 10.1016/j.stem.2013.01.006) [DOI] [PubMed] [Google Scholar]
- 21.Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. 2008. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26, 313–315. ( 10.1038/nbt1383) [DOI] [PubMed] [Google Scholar]
- 22.Kajiwara M. et al. 2012. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc. Natl Acad. Sci. USA 109, 12 538–12 543. ( 10.1073/pnas.1209979109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima F, Tokunaga K, Nakatsuji N. 2007. Human leucocyte antigen matching estimations in a hypothetical bank of human embryonic stem cell lines in the Japanese population for use in cell transplantation therapy. Stem Cells 25, 983–985. ( 10.1634/stemcells.2006-0566) [DOI] [PubMed] [Google Scholar]
- 24.Nakatsuji N, Nakajima F, Tokunaga K. 2008. HLA-haplotype banking and iPS cells. Nat. Biotechnol. 26, 739–740. ( 10.1038/nbt0708-739) [DOI] [PubMed] [Google Scholar]
- 25.Okita K. et al. 2011. A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412. ( 10.1038/nmeth.1591) [DOI] [PubMed] [Google Scholar]
- 26.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. ( 10.1016/j.cell.2007.11.019) [DOI] [PubMed] [Google Scholar]
- 27.Yu J. et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. ( 10.1126/science.1151526) [DOI] [PubMed] [Google Scholar]
- 28.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. 2009. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn Acad. Ser. B, Phys. Biol. Sci. 85, 348–362. ( 10.2183/pjab.85.348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura K. et al. 2010. Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. J. Biol. Chem. 286, 4760–4771. ( 10.1074/jbc.M110.183780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren L. et al. 2010. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630. ( 10.1016/j.stem.2010.08.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyazaki T. et al. 2012. Laminin E8 fragments support efficient adhesion and expansion of dissociated human pluripotent stem cells. Nat. Commun. 3, 1236 ( 10.1038/ncomms2231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakagawa M. et al. 2014. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci. Rep. 4, 3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ.. 2008. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146. ( 10.1038/nature06534) [DOI] [PubMed] [Google Scholar]
- 34.Giorgetti A. et al. 2009. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell 5, 353–357. ( 10.1016/j.stem.2009.09.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haase A. et al. 2009. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell 5, 434–441. ( 10.1016/j.stem.2009.08.021) [DOI] [PubMed] [Google Scholar]
- 36.Takenaka C, Nishishita N, Takada N, Jakt LM, Kawamata S. 2010. Effective generation of iPS cells from CD34(+) cord blood cells by inhibition of p53. Exp. Hematol. 38, 154–162. ( 10.1016/j.exphem.2009.11.003) [DOI] [PubMed] [Google Scholar]
- 37.Loh YH. et al. 2009. Generation of induced pluripotent stem cells from human blood. Blood 5476–5479. ( 10.1182/blood-2009-02-204800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staerk J, Dawlaty MM, Gao Q, Maetzel D, Hanna J, Sommer CA, Mostoslavsky G, Jaenisch R. 2010. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell 7, 20–24. ( 10.1016/j.stem.2010.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seki T. et al. 2010. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell 7, 11–14. ( 10.1016/j.stem.2010.06.003) [DOI] [PubMed] [Google Scholar]
- 40.Nishishita N, Ijiri H, Takenaka C, Kobayashi K, Goto K, Kotani E, Itoh T, Mori H, Kawamata S. 2011. The use of leukemia inhibitory factor immobilized on virus-derived polyhedra to support the proliferation of mouse embryonic and induced pluripotent stem cells. Biomaterials 32, 3555–3563. ( 10.1016/j.biomaterials.2010.12.063) [DOI] [PubMed] [Google Scholar]
- 41.Yamanaka S. 2010. Patient-specific pluripotent stem cells become even more accessible. Cell Stem Cell 7, 1–2. ( 10.1016/j.stem.2010.06.009) [DOI] [PubMed] [Google Scholar]
- 42.Dimos JT. et al. 2008. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321, 1218–1221. ( 10.1126/science.1158799) [DOI] [PubMed] [Google Scholar]
- 43.Park IH. et al. 2008. Disease-specific induced pluripotent stem cells. Cell 134, 877–886. ( 10.1016/j.cell.2008.07.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ebert AD, Yu J, Rose FF Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. 2009. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457, 277–280. ( 10.1038/nature07677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebert AD, Liang P, Wu JC. 2012. Induced pluripotent stem cells as a disease modeling and drug screening platform. J. Cardiovasc. Pharmacol. 60, 408–416. ( 10.1097/FJC.0b013e318247f642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. 2010. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell 143, 527–539. ( 10.1016/j.cell.2010.10.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee G. et al. 2009. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 461, 402–406. ( 10.1038/nature08320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashita A. et al. 2014. Statin treatment rescues FGFR3 skeletal dysplasia phenotypes. Nature 513, 507–511. ( 10.1038/nature13775) [DOI] [PubMed] [Google Scholar]
- 49.Bellin M, Marchetto MC, Gage FH, Mummery CL. 2012. Induced pluripotent stem cells: the new patient? Nat. Rev. Mol. Cell Biol. 13, 713–726. ( 10.1038/nrm3448) [DOI] [PubMed] [Google Scholar]
- 50.Hockemeyer D. et al. 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27, 851–857. ( 10.1038/nbt.1562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hockemeyer D. et al. 2011. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731–734. ( 10.1038/nbt.1927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim K. et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290. ( 10.1038/nature09342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polo JM. et al. 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848–855. ( 10.1038/nbt.1667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L, Gaffney D. 2014. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 10, e1004432 ( 10.1371/journal.pgen.1004432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polo JM. et al. 2012. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151, 1617–1632. ( 10.1016/j.cell.2012.11.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi K. et al. 2014. Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat. Commun. 5, 3678 ( 10.1038/ncomms4678) [DOI] [PubMed] [Google Scholar]
- 57.Ohnuki M. et al. 2014. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc. Natl Acad. Sci. USA 111, 12 426–12 431. ( 10.1073/pnas.1413299111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lander ES. et al. 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921. ( 10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- 59.Meissner A. et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770. ( 10.1038/nature07107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mikkelsen TS. et al. 2008. Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55. ( 10.1038/nature07056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. 2009. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377. ( 10.1016/j.cell.2009.01.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu L. et al. 2010. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J. Biol. Chem. 285, 19 483–19 490. ( 10.1074/jbc.M110.131995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. 2010. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–181. ( 10.1038/nature09017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim H, Lee G, Ganat Y, Papapetrou EP, Lipchina I, Socci ND, Sadelain M, Studer L. 2011. miR-371–3 expression predicts neural differentiation propensity in human pluripotent stem cells. Cell Stem Cell 8, 695–706. ( 10.1016/j.stem.2011.04.002) [DOI] [PubMed] [Google Scholar]
- 65.Anguera MC. et al. 2012. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell 11, 75–90. ( 10.1016/j.stem.2012.03.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chin MH. et al. 2009. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5, 111–123. ( 10.1016/j.stem.2009.06.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deng J. et al. 2009. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 27, 353–360. ( 10.1038/nbt.1530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doi A. et al. 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 41, 1350–1353. ( 10.1038/ng.471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. 2009. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE 4, e7076 ( 10.1371/journal.pone.0007076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC. 2010. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS ONE 5, e8975 ( 10.1371/journal.pone.0008975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim K. et al. 2011. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat. Biotechnol. 29, 1117–1119. ( 10.1038/nbt.2052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lister R. et al. 2011. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73. ( 10.1038/nature09798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohi Y. et al. 2011. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 13, 541–549. ( 10.1038/ncb2239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. 2010. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7, 249–257. ( 10.1016/j.stem.2010.06.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Newman AM, Cooper JB. 2010. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell 7, 258–262. ( 10.1016/j.stem.2010.06.016) [DOI] [PubMed] [Google Scholar]
- 76.Bock C. et al. 2011. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439–452. ( 10.1016/j.cell.2010.12.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koyanagi-Aoi M. et al. 2013. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc. Natl Acad. Sci. USA 110, 20 569–20 574. ( 10.1073/pnas.1319061110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tachibana M. et al. 2013. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153, 1228–1238. ( 10.1016/j.cell.2013.05.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chung YG. et al. 2014. Human somatic cell nuclear transfer using adult cells. Cell Stem Cell 14, 777–780. ( 10.1016/j.stem.2014.03.015) [DOI] [PubMed] [Google Scholar]
- 80.Yamada M. et al. 2014. Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells. Nature 510, 533–536. ( 10.1038/nature13287) [DOI] [PubMed] [Google Scholar]
- 81.Johannesson B. et al. 2014. Comparable frequencies of coding mutations and loss of imprinting in human pluripotent cells derived by nuclear transfer and defined factors. Cell Stem Cell 15, 634–642. ( 10.1016/j.stem.2014.10.002) [DOI] [PubMed] [Google Scholar]
- 82.Ma H. et al. 2014. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511, 177–183. ( 10.1038/nature13551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ying QL, Nichols J, Chambers I, Smith A. 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292. ( 10.1016/S0092-8674(03)00847-X) [DOI] [PubMed] [Google Scholar]
- 84.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. 2008. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. ( 10.1038/nature06968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hanna J. et al. 2009. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell 4, 513–524. ( 10.1016/j.stem.2009.04.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. 2009. Adult mice generated from induced pluripotent stem cells. Nature 461, 91–94. ( 10.1038/nature08310) [DOI] [PubMed] [Google Scholar]
- 87.Kang L, Wang J, Zhang Y, Kou Z, Gao S. 2009. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell 5, 135–138. ( 10.1016/j.stem.2009.07.001) [DOI] [PubMed] [Google Scholar]
- 88.Zhao XY. et al. 2009. iPS cells produce viable mice through tetraploid complementation. Nature 461, 86–90. ( 10.1038/nature08267) [DOI] [PubMed] [Google Scholar]
- 89.Yan L. et al. 2013. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139. ( 10.1038/nsmb.2660) [DOI] [PubMed] [Google Scholar]
- 90.Guo H. et al. 2014. The DNA methylation landscape of human early embryos. Nature 511, 606–610. ( 10.1038/nature13544) [DOI] [PubMed] [Google Scholar]
- 91.Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. 2014. DNA methylation dynamics of the human preimplantation embryo. Nature 511, 611–615. ( 10.1038/nature13581) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan YS. et al. 2013. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13, 663–675. ( 10.1016/j.stem.2013.11.015) [DOI] [PubMed] [Google Scholar]
- 93.Gafni O. et al. 2013. Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286. ( 10.1038/nature12745) [DOI] [PubMed] [Google Scholar]
- 94.Takashima Y. et al. 2014. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254–1269. ( 10.1016/j.cell.2014.08.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Theunissen TW. et al. 2014. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15, 471–487. ( 10.1016/j.stem.2014.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Valamehr B. et al. 2014. Platform for induction and maintenance of transgene-free hiPSCs resembling ground state pluripotent stem cells. Stem Cell Rep. 2, 366–381. ( 10.1016/j.stemcr.2014.01.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ware CB. et al. 2014. Derivation of naive human embryonic stem cells. Proc. Natl Acad. Sci. USA 111, 4484–4489. ( 10.1073/pnas.1319738111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hirano K, Nagata S, Yamaguchi S, Nakagawa M, Okita K, Kotera H, Ainscough J, Tada T. 2012. Human and mouse induced pluripotent stem cells are differentially reprogrammed in response to kinase inhibitors. Stem Cells Dev. 21, 1287–1298. ( 10.1089/scd.2011.0283) [DOI] [PubMed] [Google Scholar]
- 99.Amit M, Laevsky I, Miropolsky Y, Shariki K, Peri M, Itskovitz-Eldor J. 2011. Dynamic suspension culture for scalable expansion of undifferentiated human pluripotent stem cells. Nat. Protoc. 6, 572–579. ( 10.1038/nprot.2011.325) [DOI] [PubMed] [Google Scholar]
- 100.Zweigerdt R, Olmer R, Singh H, Haverich A, Martin U. 2011. Scalable expansion of human pluripotent stem cells in suspension culture. Nat. Protoc. 6, 689–700. ( 10.1038/nprot.2011.318) [DOI] [PubMed] [Google Scholar]
- 101.Otsuji TG. et al. 2014. A 3D sphere culture system containing functional polymers for large-scale human pluripotent stem cell production. Stem Cell Rep. 2, 734–745. ( 10.1016/j.stemcr.2014.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hanahan D, Weinberg RA. 2000. The hallmarks of cancer. Cell 100, 57–70. ( 10.1016/S0092-8674(00)81683-9) [DOI] [PubMed] [Google Scholar]
- 103.Stricker SH. et al. 2013. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 27, 654–669. ( 10.1101/gad.212662.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507. ( 10.1038/ng.127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Banito A. et al. 2009. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 23, 2134–2139. ( 10.1101/gad.1811609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. 2009. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460, 1132–1135. ( 10.1038/nature08235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–1144. ( 10.1038/nature08311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460, 1136–1139. ( 10.1038/nature08290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460, 1145–1148. ( 10.1038/nature08285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. 2011. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 14, 264–271. ( 10.1016/j.cmet.2011.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohnishi K. et al. 2014. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 156, 663–677. ( 10.1016/j.cell.2014.01.005) [DOI] [PubMed] [Google Scholar]
- 112.Lancaster MA, Knoblich JA. 2014. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 ( 10.1126/science.1247125) [DOI] [PubMed] [Google Scholar]
- 113.Ben-Nun IF. et al. 2011. Induced pluripotent stem cells from highly endangered species. Nat. Methods 8, 829–831. ( 10.1038/nmeth.1706) [DOI] [PubMed] [Google Scholar]
- 114.Verma R, Holland MK, Temple-Smith P, Verma PJ. 2012. Inducing pluripotency in somatic cells from the snow leopard (Panthera uncia), an endangered felid. Theriogenology 77, 220–228, 228.e1-2 ( 10.1016/j.theriogenology.2011.09.022) [DOI] [PubMed] [Google Scholar]
- 115.Nagy K. et al. 2011. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev. 7, 693–702. ( 10.1007/s12015-011-9239-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Manoli DS, Subramanyam D, Carey C, Sudin E, Van Westerhuyzen JA, Bales KL, Blelloch R, Shah NM. 2012. Generation of induced pluripotent stem cells from the prairie vole. PLoS ONE 7, e38119 ( 10.1371/journal.pone.0038119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu H. et al. 2008. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell 3, 587–590. ( 10.1016/j.stem.2008.10.014) [DOI] [PubMed] [Google Scholar]
- 118.Wu Y, Zhang Y, Mishra A, Tardif SD, Hornsby PJ. 2010. Generation of induced pluripotent stem cells from newborn marmoset skin fibroblasts. Stem Cell Res. 4, 180–188. ( 10.1016/j.scr.2010.02.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wunderlich S. et al. 2012. Induction of pluripotent stem cells from a cynomolgus monkey using a polycistronic simian immunodeficiency virus-based vector, differentiation toward functional cardiomyocytes, and generation of stably expressing reporter lines. Cell Reprogram. 14, 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wunderlich S. et al. 2014. Primate iPS cells as tools for evolutionary analyses. Stem Cell Res. 12, 622–629. ( 10.1016/j.scr.2014.02.001) [DOI] [PubMed] [Google Scholar]
- 121.Marchetto MC. et al. 2013. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503, 525–529. ( 10.1038/nature12686) [DOI] [PMC free article] [PubMed] [Google Scholar]


