Abstract
Introduction
Break-apart fluorescence in situ hybridization (FISH) is the FDA-approved assay for detecting anaplastic lymphoma kinase (ALK) rearrangements in non-small cell lung cancer (NSCLC), identifying patients who can gain dramatic benefit from ALK kinase inhibitors. Assay interpretation can be technically challenging, and either splitting of the 5′ and 3′ probes or loss of the 5′ probe constitute rearrangement. We hypothesized that there may be clinical differences depending upon rearrangement pattern on FISH.
Methods
An IRB-approved database of NSCLC patients at Dana-Farber Cancer Institute was queried for ALK rearrangement. Clinical characteristics and response to crizotinib were reviewed. Immunohistochemistry (IHC) and targeted next-generation sequencing (NGS) were obtained when available.
Results
Of 1,614 NSCLC patients with ALK testing, 82 (5.1%) patients had ALK rearrangement by FISH: 30 with split signals, 25 with 5′ deletion, and 27 with details unavailable. Patients with 5′ deletion were older (p=0.01) and tended to have more extensive smoking histories (p=0.08). IHC was positive for ALK rearrangement in all 27 patients with FISH split signals, while 3 of 21 patients with FISH 5′ deletion had negative IHC (p=0.05). Targeted NGS on 2 of 3 cases with discordant FISH and IHC results did not identify ALK rearrangement, instead finding driver mutations in EGFR and KRAS. Patients with 5′ deletion treated with crizotinib had a smaller magnitude of tumor response (p=0.03).
Conclusions
Patients with 5′ deletion on ALK FISH harbor features less typical of ALK-rearranged tumors, potentially indicating that some cases with this variant are false-positives. Corroborative testing with IHC or NGS may be beneficial.
Keywords: anaplastic lymphoma kinase (ALK) rearrangement, crizotinib, non-small cell lung cancer (NSCLC), fluorescence in situ hybridization (FISH)
Introduction
Rearrangement of the anaplastic lymphoma kinase (ALK) gene results in expression of a potent oncogenic driver in 3–5% of non-small cell lung cancer (NSCLC).1, 2 Patients with ALK rearrangement tend to be younger in age and have less extensive smoking histories.3 Identification of lung cancers harboring ALK-rearrangements is important clinically as these cancers have a 50–60% response rate to crizotinib, with improved progression-free survival compared to conventional chemotherapy.4, 5
Crizotinib approval by the FDA was accompanied by a commercially available diagnostic assay for ALK rearrangement – the Vysis ALK Break Apart fluorescence in situ hybridization (FISH) Probe Kit. The assay utilizes DNA probes that hybridize to the 3′ and 5′ regions of the common fusion breakpoint in ALK; rearrangement is identified by either splitting of the 3′ and 5′ signals or loss of the 5′ signal in ≥12% of nuclei (Figure 1). Interpretation of ALK break-apart FISH can be technically challenging due to subtlety of signals that fade over time and inter-observer variability. Furthermore, a proportion of cells in ALK-positive tumors may have no detectable rearrangement by FISH, while a small number of cells in normal tissue may yield patterns consistent with rearrangement.6
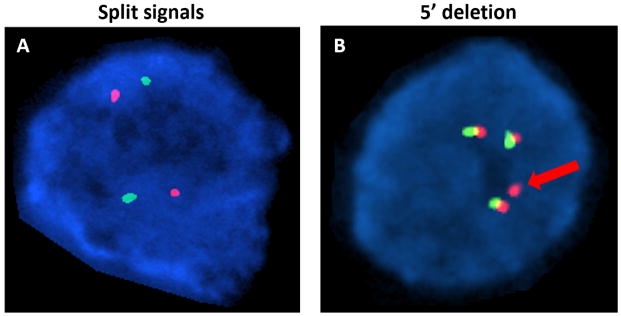
Figure 1.
ALK break-apart FISH utilizes DNA probes that hybridize to the 3′ (red signal) and 5′ (green signal) regions of the common fusion breakpoint in ALK. Rearrangement may be identified by two variant FISH patterns – splitting of signals and 5′ deletion. A, ALK rearrangement is identified by splitting of the red and green signals in both nuclei in this field. B, Rearrangement is identified by a single red signal (red arrow) with loss of the 5′ green signal in one of the four nuclei observed. An additional three nuclei demonstrate overlapping of the 3′ and 5′ probes, reflecting wildtype ALK.
Here, we study the clinical and pathologic characteristics of patients with split signals versus 5′ deletion on ALK FISH. We hypothesized that there may be differences between these two populations that might indicate an unappreciated risk of false positive results using ALK break-apart FISH.
Materials and Methods
An institutional database of NSCLC patients was queried for those identified to harbor ALK rearrangement on FISH between 2009 and 2014. In general, the predominant ALK FISH pattern was used to characterize the sample as either split signal or 5′ deletion. Patients with insufficient information regarding the specific type of ALK FISH abnormality were excluded from analysis. ALK immunohistochemistry (IHC) was performed using the monoclonal antibody 5A4 (Novocastra, Newcastle, UK), and any tumor with at least multifocal low to moderate cytoplasmic expression was considered positive.7 Next-generation sequencing (NGS) was performed via a targeted hybrid capture panel that detects mutations, insertions, deletions, copy number changes, and rearrangements within exons and key introns of 645 cancer-associated genes, including ALK. Objective tumor response was determined by Response Evaluation Criteria in Solid Tumors version 1.1, and best overall response during therapy was obtained for each patient.8 Maximal tumor shrinkage was calculated using the smallest sum of target lesions after baseline, in reference to baseline measurements. A one-sided Wilcoxon-Mann-Whitney test was used to test the hypothesis that 5′ deletion tumors are less responsive to crizotinib. Statistical analyses were performed using SAS version 9.1 (SAS Institute Inc., Cary, NC).
Results
Patient characteristics
Of 1,614 NSCLC patients who underwent ALK testing, 82 (5.1%) had ALK rearrangement identified via FISH. Of those, 30 (37%) showed split signals and 25 (30%) showed 5′ deletion. The remaining 27 (33%) patients had insufficient details regarding the specific pattern identified on FISH, with many tested using an alternative FISH assay during the early days of ALK genotyping, predating the current break-apart FISH. The median number of FISH-positive nuclei in cases with split signals was 62%, compared to 81% in cases with 5′ deletion. Relative to patients with split signals, those with 5′ deletion were older (median age 58 vs 50; p=0.01) and tended to have more extensive smoking histories (p=0.08) (Table 1). There were no significant differences between the two groups with regard to gender, race, and tumor histology (all adenocarcinoma).
Table 1.
Patient characteristics of FISH split signals vs 5′ deletion
| Split signals (n=30) | 5′ deletion (n=25) | p-value | |
|---|---|---|---|
| Age at diagnosis (years) | |||
| Median | 50 | 58 | 0.01 |
| Range | 22–82 | 28–76 | |
| Gender | |||
| Female | 18 (60%) | 17 (68%) | 0.55 |
| Male | 12 (40%) | 8 (32%) | |
| Race | |||
| Caucasian | 24 (80%) | 22 (88%) | 0.83 |
| Asian | 2 (7%) | 2 (8%) | |
| Others | 4 (13%) | 1 (4%) | |
| Smoking history | |||
| Never smoker | 19 (63%) | 11 (44%) | 0.08 |
| 0–5 pack years | 6 (20%) | 1 (4%) | |
| 5–15 pack years | 1 (3%) | 8 (32%) | |
| ≥15 pack years | 4 (13%) | 5 (20%) | |
| Immunohistochemistry | |||
| n=27 | n=21 | ||
| Positive | 27 (100%) | 18 (86%) | 0.05 |
| Negative | 0 (0%) | 3 (16%) | |
Immunohistochemistry and targeted next-generation sequencing
Tissue was available to perform ALK IHC in 27 of 30 samples demonstrating split signals on FISH and 21 of 25 samples demonstrating 5′ deletion (Table 1). All 27 samples with split signals were also positive for ALK rearrangement by IHC, whereas 3 of 21 samples with 5′ deletion were negative for ALK rearrangement by IHC. The 3 cases with positive FISH and negative IHC results had between 38–48% nuclei positive for 5′ deletion by FISH.
Targeted NGS was performed on a total of 13 available samples, including 2 of the 3 cases with discordant ALK FISH and IHC results. Neither of the 2 specimens with discordant FISH and IHC results was found to harbor ALK rearrangement by NGS. Instead, NGS identified alternate driver mutations in both cases, one in EGFR (L858R) and the other in KRAS (Q61L). An additional 8 cases with FISH 5′ deletion and 3 cases with split signals demonstrated ALK rearrangement by NGS, and all were IHC positive. Ten of the 11 cases with ALK rearrangement detected by NGS showed sequencing evidence of an EML4-ALK rearrangement. A single case contained a DCTN1-ALK rearrangement, a rarely reported fusion that has been associated with response to crizotinib therapy in ALK-rearranged inflammatory myofibroblastic tumor.9 No other driver alterations were seen in cases with ALK rearrangement detected by NGS.
Response to crizotinib
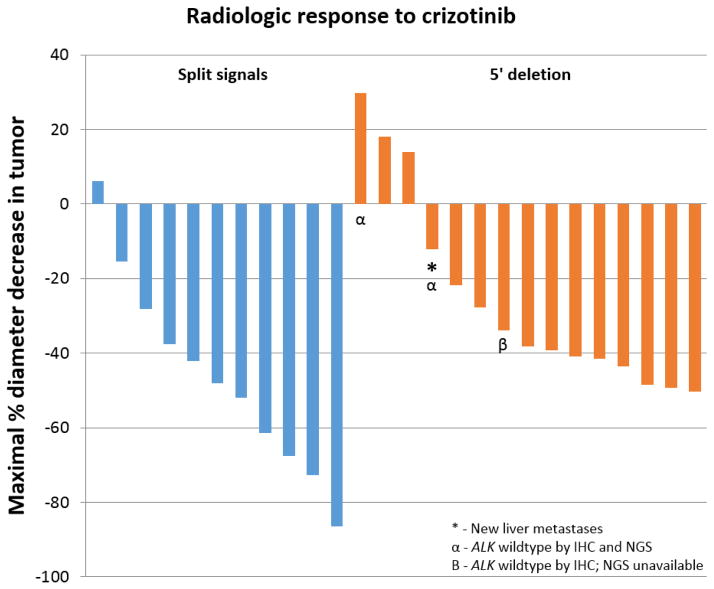
Twenty-six patients who received crizotinib had requisite radiologic follow-up for analysis of response. Of 11 patients with split signals, 8 had partial response (73%), 3 had stable disease, and none had disease progression as the overall response (Figure 3). Of 15 patients with 5′ deletion, 9 had partial response (60%), 4 had stable disease, and 2 had disease progression, including one case with new liver metastases. Patients with split signals on ALK FISH had greater median decrease in tumor diameter (48%) than patients with 5′ deletion (38%) (p=0.03).
Figure 3.
Patients with split signals on ALK FISH had a median decrease in tumor diameter of 48% compared to patients with 5′ deletion on ALK FISH (38%; p=0.03). Of 11 patients with split signals, 8 had partial response, 3 had stable disease, and none had disease progression. Of 15 patients with 5′ deletion, 9 had partial response, 4 had stable disease, and 2 had disease progression, including one case with new liver metastases. Two of the 3 cases with discordant FISH and IHC results showed disease progression – both cases were negative for ALK rearrangement by NGS.
All 3 patients with discordant ALK FISH and IHC results were treated with crizotinib, with 2 out of the 3 showing disease progression. The patient with KRAS Q61L mutation showed a 30% increase in tumor size (Figure 2), while the patient with EGFR L858R mutation showed a 12% decrease in tumor size, but presence of new liver metastases. The final case with conflicting FISH and IHC results, which had insufficient tissue for NGS, showed partial response to crizotinib, with a 34% maximal decrease in tumor size.
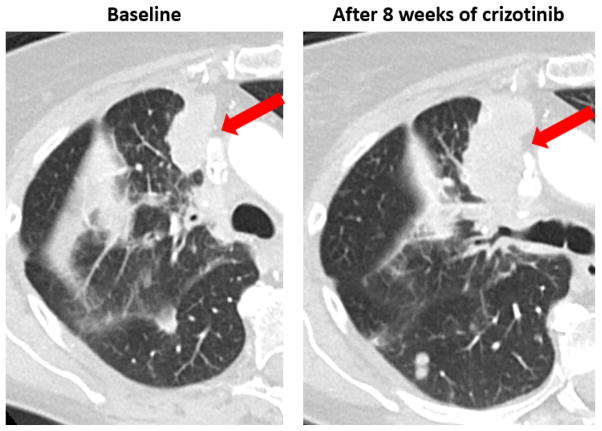
Figure 2.
Progression of disease through crizotinib in a 71-year-old patient with a heavy smoking history who was positive for ALK rearrangement by FISH 5′ deletion, but negative by IHC and NGS. NGS found the tumor to be wildtype for ALK and instead identified a Q61L KRAS mutation. Treatment with crizotinib over 8 weeks yielded no radiologic response, with an increase in the tumor size by 30%.
Discussion
ALK break-apart FISH has been the diagnostic assay used for patient selection in major trials involving ALK-targeted TKIs4, 5 and remains the only FDA-approved test for ALK rearrangement. While the incidence of various ALK FISH patterns has been previously reported,10 ours is the first study examining the clinical and pathologic characteristics of the two FISH patterns that constitute ALK rearrangement. Compared to patients with FISH split signals, those with 5′ deletion in our analysis were more likely to harbor characteristics less typical of ALK-rearranged patients, including older age and more extensive smoking history. More importantly, we found that specimens with FISH 5′ deletion were more prone to negative IHC and NGS results, identifying discordant cases in 3 of 21 samples. Our results suggest that the 5′ deletion pattern may be vulnerable to false positive results.
Both groups had high percentage of nuclei positive for ALK rearrangement – 62% in split signals and 81% in 5′ deletion. Although discordant cases had a lower percentage of nuclei positive for rearrangement compared with the rest of our samples, they each harbored between 38–48% positive nuclei, well above the lower cutoff for ALK rearrangement. Moreover, prior research has shown that the percentage of tumor cells with ALK rearrangement does not correlate with response to crizotinib.11
Our finding that ALK IHC was concordant with FISH in 45 of 48 (94%) samples is supported by prior studies demonstrating the ALK antibody 5A4 to have sensitivity and specificity in the range of 93–100% relative to FISH.7, 12 A multi-institutional Canadian study demonstrated essentially perfect performance of ALK IHC with the 5A4 antibody as compared to FISH when employing careful assay validation procedures.13 Because ALK FISH presents several limitations including high cost, requirement of specialized equipment, and technical challenges with result interpretation, alternative assays such as IHC and reverse transcription polymerase chain reaction (RT-PCR) have been evaluated.7, 12, 14 A risk of 5′ deletion on FISH representing a false positive result suggests that assays such as IHC, RT-PCR, or NGS could be beneficial as confirmatory tests prior to consideration of ALK-targeted therapy.
While 5′ deletion patients demonstrated less radiographic response to crizotinib, the majority did nonetheless exhibit a response. This finding suggests that most 5′ deletion cases do in fact represent true ALK rearrangement. Additional studies are encouraged given the relatively small sample size of patients receiving crizotinib in our study. The two cases in this study involving FISH 5′ deletion, negative IHC, and negative NGS who had poor response to crizotinib highlight the potential risks of relying solely on the 5′ deletion FISH pattern for clinical decision-making regarding ALK-targeted therapy – both cases in fact harbored an alternate oncogenic driver mutation. Indeed, one prior study of NSCLC patients found ALK FISH abnormalities to be almost mutually exclusive from EGFR and KRAS co-mutations, with all cases of co-mutations occurring in samples with KRAS mutations and loss of either the 5′ or 3′ signal.15 A recent multi-institutional study on oncogenic driver mutations in NSCLC similarly demonstrated that co-existence of ALK rearrangement with other driver mutations is rare, with two of four co-mutant cases on initial testing subsequently proven to be false-positives on IHC and repeat FISH.16 Our study similarly suggests corroborative testing with alternative assays may be helpful in cases with complex mutational findings.
Why might the 5′ deletion variant of FISH be less reliable in predicting true ALK rearrangement and therefore responsiveness to ALK-targeted therapy? The 5′ deletion pattern may reflect ALK rearrangement by either true loss of the 5′ probe binding site via rearrangement or loss of the 5′ probe within the plane of the section. However, large deletions and structural variants affecting the binding site of the 5′ probe, without ALK rearrangement, may result in an identical FISH pattern. Such findings may be expected more frequently in genomically-deranged tumors, such as smoking-related cancers. Visualization of splitting of the 5′ and 3′ probes may therefore be a more rigorous standard for ALK rearrangement than 5′ deletion and less susceptible to false positive results.
Although laboratories are encouraged to clearly communicate ALK FISH results, detailed reporting of the specific FISH pattern is not explicitly required according to published guidelines.17 Our results suggest that ALK FISH reporting should be standardized to include the variant identified. Furthermore, our findings may also extend beyond ALK rearrangement as break-apart FISH is increasingly used to identify targetable rearrangements in ROS1 and RET.18, 19 We encourage additional investigations to confirm our observations as the effectiveness of therapies targeting ALK, ROS1, and RET is highly dependent upon appropriate selection of patients with cancers harboring these relatively rare genotypes.
Acknowledgments
Sources of Support:
Wong Family Fellowship in Translational Oncology, Harold and Gail Kirstein Lung Cancer Research Fund, 1K23CA157631 (NCI), and R01CA136851.
Footnotes
LMS retains board membership with Genentech. MN receives consulting fees from Bristol-Myers Squibb. PAJ has received consulting fees from Pfizer and Chugai Pharmaceuticals. GRO has received consulting fees from AstraZeneca, Sanofi, Clovis, Novartis, Genentech, Boehringer-Ingelheim and honoraria from Chugai.
References
- 1.Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol. 2008;3:13–17. doi: 10.1097/JTO.0b013e31815e8b60. [DOI] [PubMed] [Google Scholar]
- 2.Wong DW, Leung EL, So KK, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 3.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 6.Camidge DR, Kono SA, Flacco A, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clin Cancer Res. 2010;16:5581–5590. doi: 10.1158/1078-0432.CCR-10-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sholl LM, Weremowicz S, Gray SW, et al. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol. 2013;8:322–328. doi: 10.1097/JTO.0b013e31827db604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1. 1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Subbiah V, McMahon C, Patel S, et al. STUMP un“stumped”: anti-tumor response to anaplastic lymphoma kinase (ALK) inhibitor based targeted therapy in uterine inflammatory myofibroblastic tumor with myxoid features harboring DCTN1-ALK fusion. J Hematol Oncol. 2015;8:66. doi: 10.1186/s13045-015-0160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai Z, Kelly JC, Meloni-Ehrig A, et al. Incidence and patterns of ALK FISH abnormalities seen in a large unselected series of lung carcinomas. Mol Cytogenet. 2012;5:44. doi: 10.1186/1755-8166-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camidge DR, Theodoro M, Maxson DA, et al. Correlations between the percentage of tumor cells showing an anaplastic lymphoma kinase (ALK) gene rearrangement, ALK signal copy number, and response to crizotinib therapy in ALK fluorescence in situ hybridization-positive nonsmall cell lung cancer. Cancer. 2012;118:4486–4494. doi: 10.1002/cncr.27411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLeer-Florin A, Moro-Sibilot D, Melis A, et al. Dual IHC and FISH testing for ALK gene rearrangement in lung adenocarcinomas in a routine practice: a French study. J Thorac Oncol. 2012;7:348–354. doi: 10.1097/JTO.0b013e3182381535. [DOI] [PubMed] [Google Scholar]
- 13.Cutz JC, Craddock KJ, Torlakovic E, et al. Canadian anaplastic lymphoma kinase study: a model for multicenter standardization and optimization of ALK testing in lung cancer. J Thorac Oncol. 2014;9:1255–1263. doi: 10.1097/JTO.0000000000000239. [DOI] [PubMed] [Google Scholar]
- 14.Wu YC, Chang IC, Wang CL, et al. Comparison of IHC, FISH and RT-PCR methods for detection of ALK rearrangements in 312 non-small cell lung cancer patients in Taiwan. PLoS One. 2013;8:e70839. doi: 10.1371/journal.pone.0070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res. 2013;19:4273–4281. doi: 10.1158/1078-0432.CCR-13-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sholl LM, Aisner DL, Varella-Garcia M, et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol. 2015;10:768–777. doi: 10.1097/JTO.0000000000000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol. 2013;8:823–859. doi: 10.1097/JTO.0b013e318290868f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30:4352–4359. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]