The synthesis of specialized metabolites is actively repressed in a Mediator-dependent manner in an Arabidopsis phenylpropanoid pathway mutant.
Abstract
Phenylpropanoids are phenylalanine-derived specialized metabolites and include important structural components of plant cell walls, such as lignin and hydroxycinnamic acids, as well as ultraviolet and visible light-absorbing pigments, such as hydroxycinnamate esters (HCEs) and anthocyanins. Previous work has revealed a remarkable degree of plasticity in HCE biosynthesis, such that most Arabidopsis (Arabidopsis thaliana) mutants with blockages in the pathway simply redirect carbon flux to atypical HCEs. In contrast, the ferulic acid hydroxylase1 (fah1) mutant accumulates greatly reduced levels of HCEs, suggesting that phenylpropanoid biosynthesis may be repressed in response to the loss of FERULATE 5-HYDROXYLASE (F5H) activity. Here, we show that in fah1 mutant plants, the activity of HCE biosynthetic enzymes is not limiting for HCE accumulation, nor is phenylpropanoid flux diverted to the synthesis of cell wall components or flavonol glycosides. We further show that anthocyanin accumulation is also repressed in fah1 mutants and that this repression is specific to tissues in which F5H is normally expressed. Finally, we show that repression of both HCE and anthocyanin biosynthesis in fah1 mutants is dependent on the MED5a/5b subunits of the transcriptional coregulatory complex Mediator, which are similarly required for the repression of lignin biosynthesis and the stunted growth of the phenylpropanoid pathway mutant reduced epidermal fluorescence8. Taken together, these observations show that the synthesis of HCEs and anthocyanins is actively repressed in a MEDIATOR-dependent manner in Arabidopsis fah1 mutants and support an emerging model in which MED5a/5b act as central players in the homeostatic repression of phenylpropanoid metabolism.
The phenylpropanoid pathway is required for the biosynthesis of a wide variety of soluble specialized plant metabolites, including hydroxycinnamate esters (HCEs), flavonols, and anthocyanins, as well as wall-bound hydroxycinnamic acids (HCAs) and lignin. Sinapoylmalate, the predominant HCE in Arabidopsis (Arabidopsis thaliana) leaves, accumulates in adaxial epidermal cells, where it serves as a UV light protectant (Chapple et al., 1992; Gräwe et al., 1992; Liang et al., 2006). A number of previous studies have shown that, when the phenylpropanoid pathway is blocked by mutation or through metabolic engineering, phenylpropanoid precursors are redirected from sinapoylmalate to various normally low-abundance, atypical HCEs. Thus, in place of sinapate-derived HCEs, Arabidopsis mutants with deficiencies in the phenylpropanoid biosynthetic enzymes CINNAMATE 4-HYDROXYLASE (C4H), p-COUMAROYLSHIKIMATE 3′-HYDROXYLASE (C3′H), CINNAMOYL COENZYME A REDUCTASE (CCR), or CAFFEIC ACID/5-HYDROXYFERULIC ACID O-METHYLTRANSFERASE (COMT/ATOMT1) instead accumulate HCEs derived from cinnamate, p-coumarate, ferulate, or 5-hydroxyferulate, respectively (Franke et al., 2002; Do et al., 2007; Mir Derikvand et al., 2008; Schilmiller et al., 2009).
The cytochrome P450-dependent monooxygenase FERULATE 5-HYDROXYLASE (F5H), the product of the FERULIC ACID HYDROXYLASE1 (FAH1) gene, is required for the hydroxylation of coniferaldehyde and coniferyl alcohol and for the subsequent formation of sinapoylated compounds and syringyl lignin (Humphreys et al., 1999). Despite their metabolic block, fah1 mutant plants are morphologically indistinguishable from wild-type plants and accumulate wild-type levels of total lignin, with all flux to syringyl lignin apparently redirected to coniferyl alcohol-derived guaiacyl lignin (Meyer et al., 1998). Unlike the phenylpropanoid mutants described above, and in contrast with their flexibility in lignin biosynthesis, fah1 mutants accumulate greatly reduced levels of total HCEs, suggesting that their HCE biosynthetic pathway may be actively repressed.
The anthocyanins are compounds derived from the flavonoid branch of the phenylpropanoid pathway that absorb visible light and thus function as pigments ranging in color from red to blue. They are widely distributed among the vascular plants, where they play important roles in photoprotection, plant-pollinator interactions, and responses to abiotic stress (Spaethe et al., 2001; Steyn et al., 2009; Schenke et al., 2011; Yuan et al., 2013). Variations in the spectral properties of different anthocyanins primarily arise from a small number of ring modifications on the chromophore itself, together with the combinatorial addition of peripheral moieties by various glycosyltransferases and acyltransferases (Mol et al., 1998). The predominant anthocyanin in Arabidopsis is a highly decorated cyanidin molecule with three glucosyl units and single additions of Xyl, malonate, p-coumarate, and sinapate (Tohge et al., 2005). The biochemical pathways responsible for the production and modification of the anthocyanin chromophore are relatively well understood, and substantial progress has been made toward elucidating the transcriptional mechanisms by which anthocyanin biosynthesis is regulated (Grotewold, 2006; Solfanelli et al., 2006; Gonzalez et al., 2008).
Mediator is a large, multisubunit transcriptional coregulator that physically bridges the interaction of gene-specific transcription factors with the core transcription machinery (Malik and Roeder, 2010). Recent work has established an important role for Mediator in a number of biological processes in plants, including freezing tolerance, pathogen response, and the production of small noncoding RNAs (Knight et al., 1999, 2009; Kim et al., 2011; Zhang et al., 2013). We previously showed that the paralogous Mediator subunits MED5a (At2g48110) and MED5b (At3g23590), formerly known as REDUCED EPIDERMAL FLUORESCENCE4 (REF4) and REF4-RELATED1, respectively, are required for phenylpropanoid homeostasis in Arabidopsis (Bonawitz et al., 2012). Mutants with dominant, gain-of-function mutations in MED5a show globally reduced phenylpropanoids, whereas mutants lacking MED5a and MED5b accumulate elevated levels of HCEs and HCE:hydroxycinnamyl alcohol coupling products (Stout et al., 2008; Bonawitz et al., 2012). We have also recently shown that disruption of MED5a and MED5b restores lignin biosynthesis and growth of the C3′H-deficient ref8 mutant (Bonawitz et al., 2014), demonstrating a role for Mediator in repressing lignin biosynthesis in response to a metabolic block in the phenylpropanoid pathway.
Here, we report the results of experimental efforts aimed at understanding the lack of metabolic plasticity in the HCE biosynthetic pathway of fah1 mutants. We show that, in addition to their HCE deficiency, fah1 mutants also exhibit decreased accumulation of anthocyanins. Both the HCE and anthocyanin deficiency phenotypes of fah1 are dependent upon an intact Mediator complex, and both exist despite the fact that fah1 mutants apparently possess all of the required enzymatic activities for HCE and anthocyanin biosynthesis. On the basis of these results, we propose the existence of a feedback mechanism whereby phenylpropanoid synthesis is inhibited by a Mediator-dependent process in response to specific metabolic blocks in the pathway.
RESULTS
fah1 Accumulates Low Amounts of HCEs
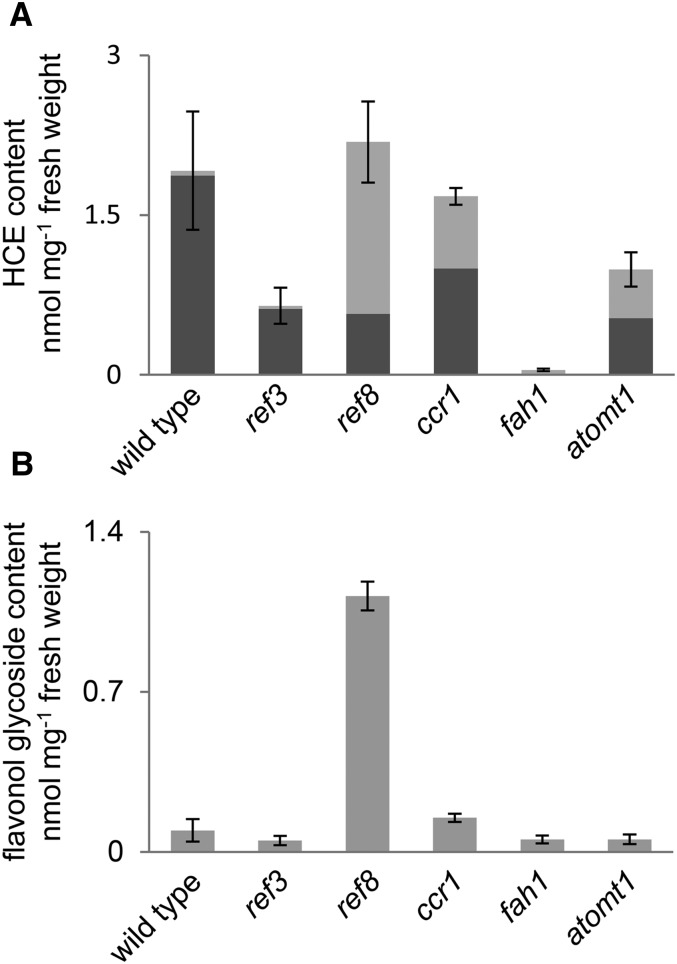
The plasticity of the HCE biosynthetic pathway has been observed in multiple Arabidopsis ref and related phenylpropanoid mutants, including ref3 (c4h), ref8, ccr1, and atomt1 (Franke et al., 2002; Do et al., 2007; Mir Derikvand et al., 2008; Schilmiller et al., 2009; for an overview of the phenylpropanoid pathway in Arabidopsis, see Supplemental Fig. S1). Consistent with these previous reports, a comparison of total HCE content of these mutants grown in parallel revealed that all accumulate substantial or nearly wild-type levels of HCEs. In each mutant, the increase of atypical cinnamate-, p-coumarate-, ferulate-, or 5-hydroxyferulate-derived HCEs substantially accounts for their reduced level of sinapate-derived HCEs (Fig. 1; Supplemental Table S1). In contrast, fah1 mutant plants accumulate less total HCEs, with the ferulate-derived HCEs feruloylglucose and feruloylmalate predominating. The accumulation of flavonol glycosides in leaves was largely unaffected in these mutants (Fig. 1), with the exception of ref8, which, as shown previously, exhibited globally elevated flavonoids (Franke et al., 2002; Weng et al., 2010).
Figure 1.
Disruption of phenylpropanoid biosynthetic genes results in the accumulation of normally low-abundance HCEs. Shown are quantifications by HPLC of total HCEs (A) and total flavonol glycosides (B) extracted from 3-week-old whole rosettes of wild-type plants and plants with the indicated mutations. Sinapoylmalate in A is represented in dark gray, and the sum of all other hydroxycinnamoyl esters is represented in light gray. Graphs show means ± sd of three biological replicates.
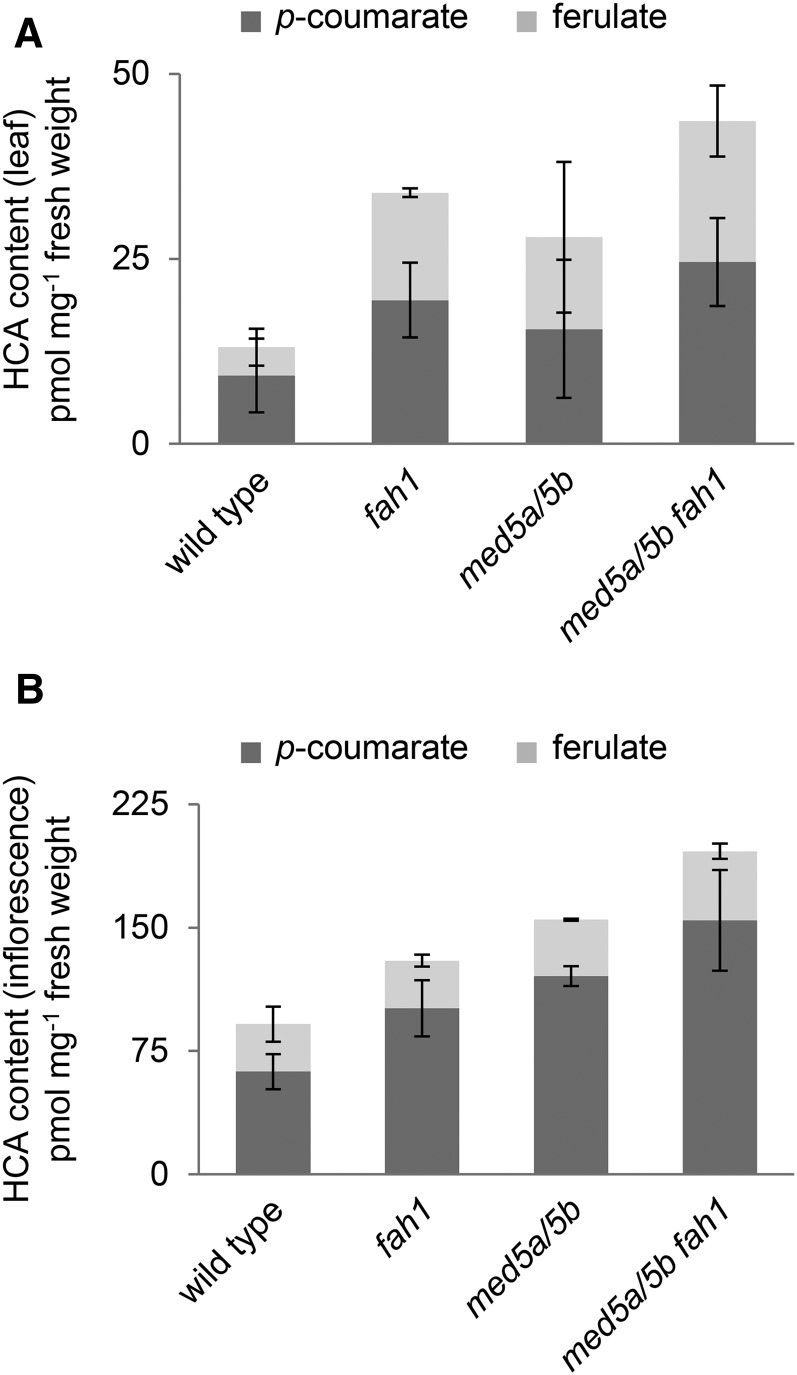
Because phenylpropanoid flux normally directed to HCE biosynthesis did not appear to be redistributed to flavonol glycosides in fah1, we tested the hypothesis that it is instead channeled to insoluble cell wall-bound lignins or HCAs. Lignin quantification using the acetyl bromide method revealed no change in total lignin accumulation in fah1 mutants compared with wild-type plants (percentage acetyl bromide-soluble lignin per extract-free cell wall: wild type, 11.2 ± 0.72; fah1, 10.3 ± 1.23), nor did fah1 mutants exhibit autofluorescence indicative of ectopic lignification of the epidermis (Zhou et al., 2009; Supplemental Fig. S2). Although significantly higher levels of cell wall-bound HCAs were observed in both leaf and inflorescence tissue of fah1 mutants (Fig. 2), this increase is far from sufficient to account for the decrease in HCE content. In leaves, for example, there is a 0.15 nmol mg−1 fresh weight increase of HCA accumulation compared with a 1.9 nmol mg−1 fresh weight decrease in HCE content.
Figure 2.
Loss of F5H does not lead to decreased levels of cell wall-bound HCAs. Shown are quantifications by HPLC of p-coumarate and ferulate released by saponification from whole rosettes (A) and inflorescence stems (B) of wild-type plants and plants with the indicated mutations. Whole rosettes were harvested 3 weeks after planting and inflorescence stems were harvested 10 weeks after planting. Multiple plants were pooled for each biological replicate. Graphs show means ± sd of three biological replicates.
The Activity of HCE-Specific Biosynthetic Enzymes Is Not Limiting for HCE Accumulation in fah1 Mutants
The HCE biosynthetic enzymes REF1 (a hydroxycinnamaldehyde dehydrogenase) and UGT84A2/bright trichomes1 (a UDP-Glc-dependent glucosyltransferase [UGT]) are required for normal sinapoylmalate accumulation in wild-type plants (Nair et al., 2004; Sinlapadech et al., 2007). REF1 is also required for the synthesis of the low levels of feruloylmalate in fah1 mutants and for a significant portion of cell wall-bound ferulic acid in wild-type plants (Nair et al., 2004). We hypothesized that the reduction in total HCE levels in fah1 mutant plants might be the result of the inability of REF1 to efficiently utilize the ferulate precursor coniferaldehyde as a substrate. To test this hypothesis, we evaluated the ability of REF1 to catalyze the oxidation of several hydroxycinnamaldehydes in vitro. Although REF1 displayed the highest catalytic efficiency toward sinapaldehyde, comparable catalytic efficiencies were observed with the other substrates tested, including coniferaldehyde (Table I). Given that REF1 displays similar activity toward a range of hydroxycinnamaldehydes, we conclude that it is unlikely that REF1 limits feruloylmalate accumulation in fah1 mutants.
Table I. Kinetic values for the REF1-catalyzed oxidation of several hydroxycinnamaldehydes.
Values shown are means ± sd of three technical replicates.
| Substrate | Vmax | Km | Vmax:Km Ratio |
|---|---|---|---|
| pkat mg−1 protein | µm | ||
| Coniferaldehyde | 560 ± 30.0 | 43.0 ± 5.90 | 13.0 |
| Sinapaldehyde | 1.70 × 103 ± 103 | 150 ± 60.0 | 11.0 |
| p-Coumaraldehyde | 310 ± 62.0 | 19.0 ± 4.80 | 16.0 |
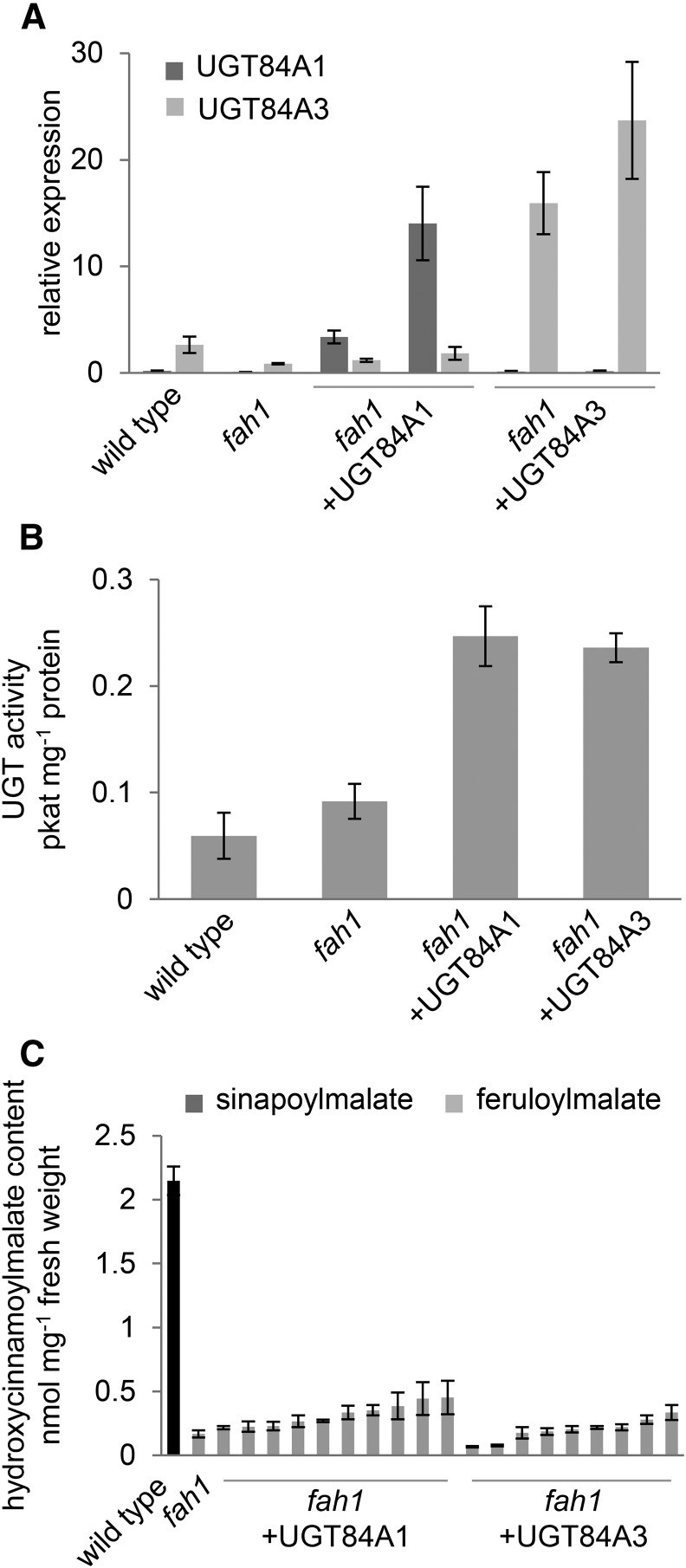
The predominant UGT in sinapoylmalate biosynthesis in Arabidopsis, UGT84A2, is expressed at high levels in leaves but exhibits little to no glucosyltransferase activity toward ferulate in vitro (Lim et al., 2001; Sinlapadech et al., 2007). In contrast, the related enzymes UGT84A1 and UGT84A3 exhibit substantial glucosyltransferase activity toward ferulate in vitro but are expressed at relatively low levels in leaves. To test the hypothesis that the low expression of a ferulate-utilizing UGT limits ferulate ester biosynthesis in fah1 mutants, we overexpressed UGT84A1 and UGT84A3 under the control of the C4H promoter in the fah1 background. Although fah1 transformants overexpressing UGT84A1 and UGT84A3 showed increased UGT transcript levels (Fig. 3) and glucosyltransferase activity toward ferulate (Fig. 3), they exhibited at best modest increases in the accumulation of feruloylmalate (Fig. 3). From these data, we conclude that UGT activity also does not limit HCE accumulation in fah1 mutants.
Figure 3.
Overexpression of neither UGT84A1 nor UGT84A3 rescues the HCE deficiency of the fah1 mutant. Shown are steady-state transcript levels of UGT84A1 and UGT84A3 (A), ferulate:UDP-Glc glucosyltransferase activity (B), and hydroxycinnamoylmalate content (C) of 3-week-old whole rosettes of wild-type plants, fah1 plants, and T1 progeny of fah1 plants transformed with either a UGT84A1 or UGT84A3 overexpression construct. Transcript abundance in A is shown relative to that of the control gene At1g13320. Graphs in A and C show means ± sd of three biological replicates of individual plants, whereas those in B show means ± sd of three technical replicates of pooled samples.
Disruption of the Mediator Subunits MED5a and MED5b Substantially Alleviates the HCE Deficiency of the fah1 Mutant
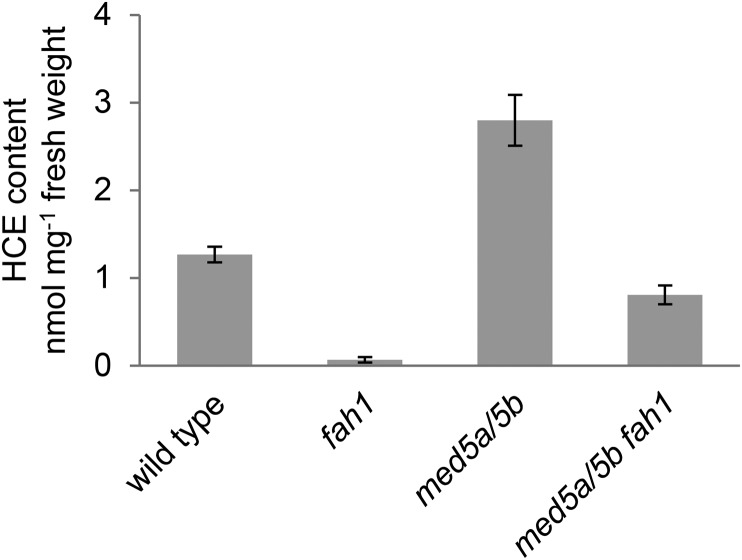
The observation that fah1 mutants fail to synthesize wild-type levels of HCEs, despite their apparent biochemical capability to do so, was reminiscent of our recent observations with ref8 mutants, which similarly fail to synthesize wild-type levels of p-hydroxyphenyl lignin despite possessing the requisite biochemical activities (Bonawitz et al., 2014). Because the Mediator subunits MED5a/5b are required for the down-regulation of lignin deposition in ref8 mutants, we hypothesized that they might play a similar role in the HCE deficiency of fah1 mutants. To test this hypothesis, we generated med5a/5b fah1 triple mutants and measured their total leaf HCE content (Fig. 4). These analyses revealed that disruption of med5a/5b in the fah1 mutant background increased total HCEs approximately 12-fold, to a level near that of wild-type plants but still approximately half that of med5a/5b mutants. The predominant HCEs observed in med5a/5b fah1 plants were feruloylmalate, feruloylglucose, and several normally low-abundance feruloyl ester:coniferyl alcohol coupling products that have been described previously (Supplemental Table S2; Rohde et al., 2004; Bonawitz et al., 2012). Disruption of MED5a/5b had no effect on cell wall-bound HCA content in either wild-type or fah1 mutant plants (Fig. 2). From these data, we conclude that MED5a/5b are required for the severe HCE deficiency of fah1 mutants, but there also appear to exist Mediator-independent mechanisms that limit the accumulation of HCEs in plants lacking functional F5H.
Figure 4.
Disruption of MED5a and MED5b substantially alleviates the HCE deficiency of the fah1 mutant. Shown is the quantification by HPLC of total methanol-soluble HCEs extracted from rosette leaves of fah1, med5a/5b, and med5a/5b fah1 mutants 3 weeks after planting. The graph shows means ± sd of three biological replicates.
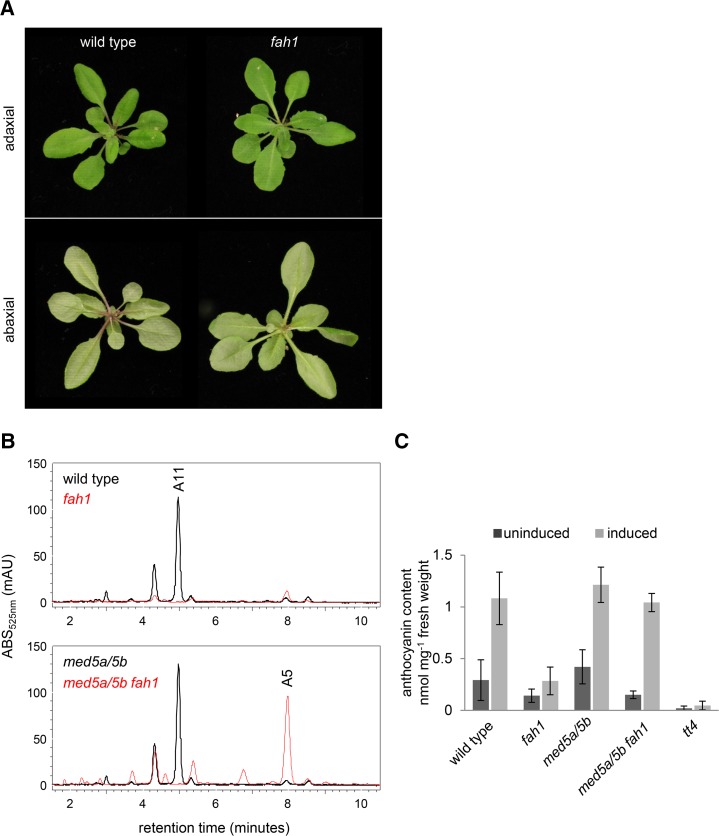
Anthocyanin Accumulation Is Repressed in a Mediator-Dependent Manner in fah1 Mutants
Although F5H is required for the formation of the sinapate moiety that decorates the most abundant anthocyanin in Arabidopsis leaves (A11 according to the nomenclature of Tohge et al. [2005]), this addition is not required for anthocyanin accumulation (Fraser et al., 2007). Nevertheless, we repeatedly noticed a clear reduction in visible purple pigmentation of soil-grown fah1 plants, particularly near the hypocotyl and at the base of rosette leaf petioles (Fig. 5), an observation that was recently reported elsewhere (Maruta et al., 2014). Consistent with this observation, we found that fah1 mutant seedlings also accumulated markedly lower levels of anthocyanins than wild-type seedlings in response to Suc stress (Fig. 5). Parallel analysis of med5a/5b fah1 seedlings showed that the inhibition of anthocyanin accumulation in fah1, like the inhibition of HCE accumulation, is also Mediator dependent (Fig. 5). As expected, med5a/5b fah1 mutants did not exhibit restored synthesis of A11 or other sinapoylated anthocyanins but instead primarily accumulated the nonsinapoylated anthocyanin A5 (Fig. 5). Importantly, and in contrast to our measurements of HCEs, the disruption of MED5a/5b did not lead to increased anthocyanin biosynthesis in plants with functional F5H. This observation strongly suggests that the increased anthocyanin accumulation of med5a/5b fah1 plants is the result of the derepression of phenylpropanoid biosynthesis and not a global increase unrelated to FAH1.
Figure 5.
Inhibition of anthocyanin biosynthesis in fah1 mutants is dependent on the Mediator subunits MED5a and MED5b. A, Soil-grown wild-type and fah1 plants 3 weeks after planting. B, Representative HPLC chromatograms at 525 nm of seedlings after the induction of anthocyanin biosynthesis with Suc stress. For clarity, chromatograms from wild-type and med5a/5b samples are overlaid at top and those from fah1 and med5a/5b fah1 samples are overlaid at bottom. C, Quantification by HPLC of anthocyanins in Suc-stressed seedlings. Each bar shows the mean of three biological replicates for each genotype, with error bars indicating sd.
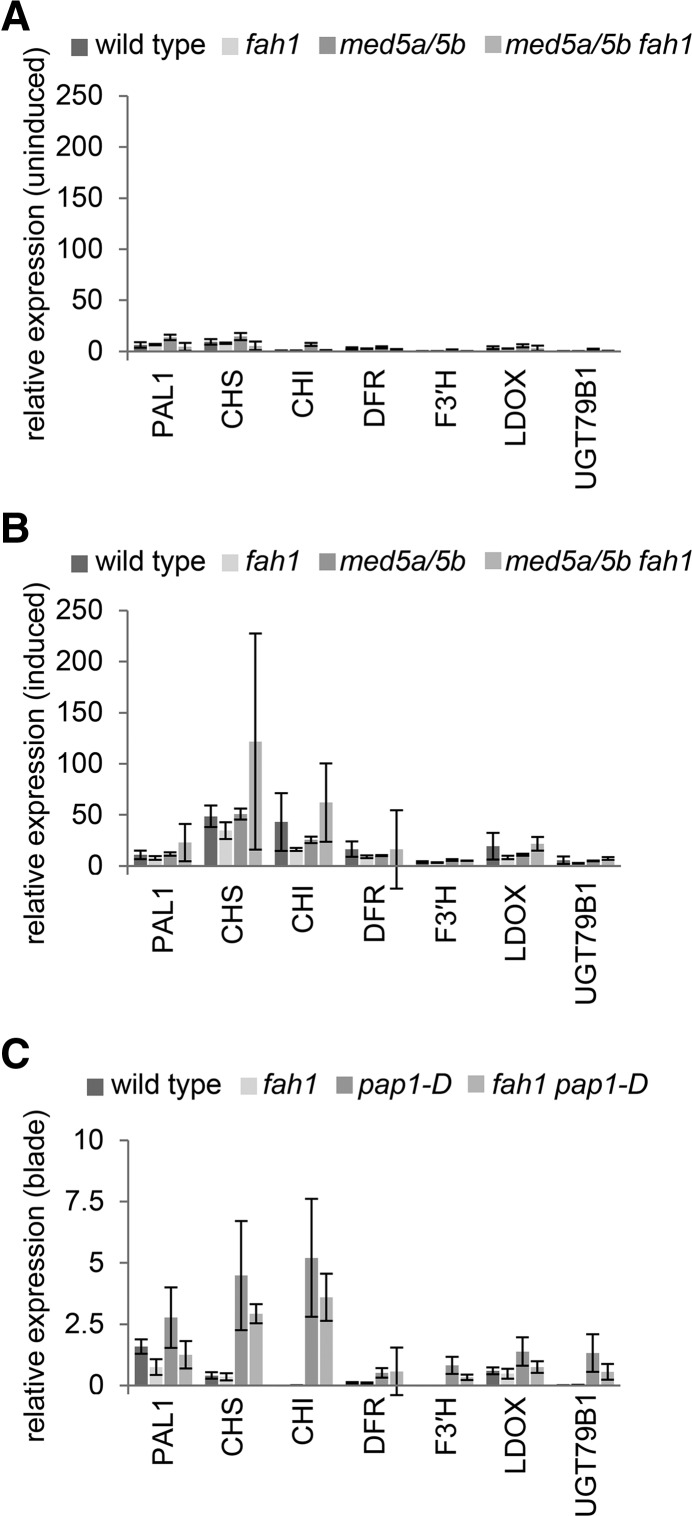
To test the possibility that the restoration of anthocyanin accumulation in med5a/5b fah1 mutants is a result of the transcriptional derepression of anthocyanin biosynthesis, we measured the expression of anthocyanin biosynthetic genes in wild-type, fah1, med5a/5b, and med5a/5b fah1 seedlings 24 and 48 h after induction with Suc (Fig. 6), when their expression in wild-type plants was highest (Supplemental Fig. S3). In fact, fah1 plants exhibited up-regulation of anthocyanin biosynthetic genes similar to that of wild-type plants, and the disruption of MED5a/5b had no effect on this induction in either background. Although we cannot rule out the repression of anthocyanin biosynthetic genes in fah1 in a small subset of cells or during a short temporal window, the most straightforward interpretation of these data is that derepression of anthocyanin accumulation in med5a/5b fah1 does not occur at the level of transcription of anthocyanin biosynthetic genes.
Figure 6.
Transcriptional induction of anthocyanin biosynthetic genes in fah1 mutants is indistinguishable from that of wild-type plants. A and B, Transcript levels of the anthocyanin biosynthetic genes PAL1, CHS, CHI, DFR, F3ʹH, LDOX, and UGT79B1 in wild-type seedlings and seedlings with the indicated mutations grown in either 0.5% Suc (A) or 3% Suc (B). C, Transcript levels of the same genes in rosette leaf blades of soil-grown wild-type, fah1, pap1-D, and fah1 pap1-D plants 3 weeks after planting. Transcript abundances shown are relative to those of the control gene At1g13320 for all experiments. Graphs show means ± sd of three biological replicates. CHI, Chalcone isomerase; CHS, chalcone synthase; DFR, dihydroflavonol reductase; F3′H, flavonoid 3′-hydroxylase; LDOX, leucoanthocyanidin dioxygenase; PAL1, phenylalanine ammonia lyase1.
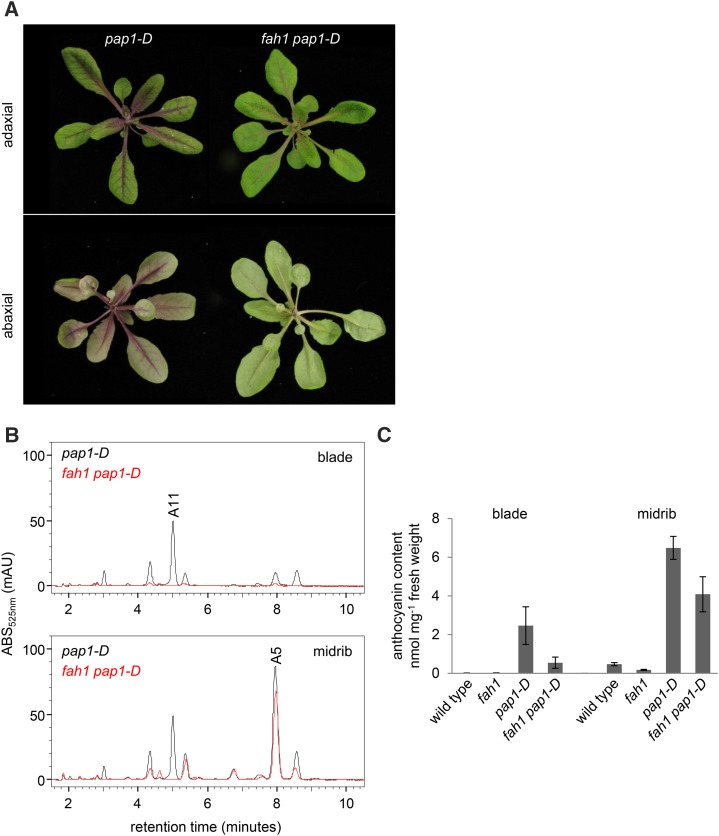
The Effectiveness of PRODUCTION OF ANTHOCYANIN PIGMENT1/MYB75 Overexpression Is Limited by Tissue in fah1 Mutants
Overexpression of the anthocyanin-specific transcription factor PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1)/MYB75 and the consequent increased expression of anthocyanin biosynthetic genes leads to anthocyanin hyperaccumulation in the Arabidopsis pap1-D mutant (Borevitz et al., 2000). To test whether overexpression of PAP1 can restore anthocyanin biosynthesis in plants lacking functional F5H, we generated fah1 pap1-D double mutants. Initial inspection of fah1 pap1-D plants revealed reduced visible pigmentation compared with that of pap1-D (Fig. 7). Anthocyanin quantification by HPLC was consistent with this observation, as rosette leaves of fah1 pap1-D plants were found to contain significantly lower levels of total anthocyanins than leaves of pap1-D plants (Fig. 7). Dissection of leaves into blade and midrib sections revealed that the majority of anthocyanins in pap1-D leaves are found in the midrib (Fig. 7). The most abundant anthocyanin in the blade is the sinapoylated anthocyanin A11, whereas in the midrib it is the nonsinapoylated anthocyanin A5 (Fig. 7). Notably, the relative abundance of sinapoylated versus nonsinapoylated anthocyanins in pap1-D leaf blades and midribs mirrors that of anthocyanin deficiency in fah1 pap1-D leaves, which was significantly more pronounced in the leaf blade than in the midrib. These data suggest that deficiencies in anthocyanin accumulation in fah1 pap1-D are only observed in tissues where F5H is normally expressed. Steady-state transcript levels of anthocyanin biosynthetic genes were similarly up-regulated in both pap1-D and fah1 pap1-D mutants (Fig. 6), even in leaf blades, where the anthocyanin deficiency of fah1 pap1-D mutants is profound (Fig. 7).
Figure 7.
Overexpression of the anthocyanin-specific transcription factor PAP1 fails to overcome the anthocyanin repression of fah1. A, Soil-grown pap1-D and fah1 pap1-D plants 3 weeks after planting. B, Representative chromatograms of dissected leaf blades (top) and midribs (bottom) of pap1-D and fah1 pap1-D plants. C, Quantification of total anthocyanin levels in the leaf sections shown in B together with wild-type and fah1 controls. Each bar shows the mean of three samples, with error bars indicating sd.
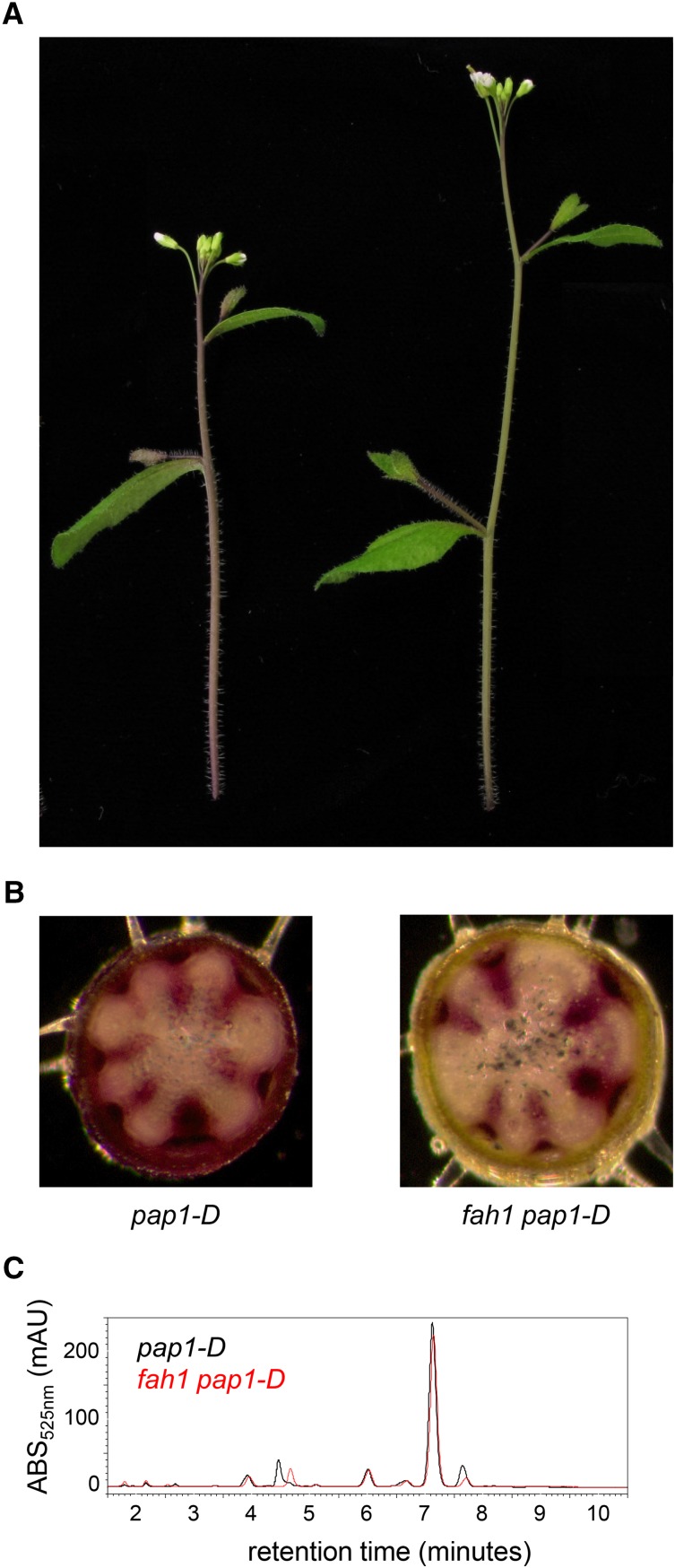
To extend these observations, we investigated anthocyanin content and distribution in pap1-D and fah1 pap1-D inflorescence stems (Fig. 8). Initial inspection of intact inflorescence stems revealed a subtle, yet highly reproducible, reduction in visible pigmentation in fah1 pap1-D stems compared with pap1-D stems (Fig. 8); however, we were unable to detect any significant difference in the total anthocyanin content by HPLC (Fig. 8). Interestingly, microscopic examination of hand sections of pap1-D inflorescence stems revealed that anthocyanins in pap1-D plants are found in the stem cortex, interfascicular region, and vascular bundles and are especially pronounced in the phloem (Fig. 8). In contrast, fah1 pap1-D plants showed little pigmentation in the cortical region of the stem and interfascicular region but relatively normal pigmentation in vascular bundles. Thus, it appears that the majority of anthocyanins in the inflorescence stem of pap1-D mutants are nonsinapoylated, localized to vascular bundles, and unaffected by the loss of F5H. In contrast, a relatively small quantity of predominantly sinapoylated anthocyanins accumulate in the epidermis and interfascicular regions, cell types that synthesize sinapoylmalate and syringyl lignin, respectively, and express F5H (García et al., 2014). The F5H-dependent accumulation of these anthocyanins is responsible for most of the visible pigmentation of intact pap1-D stems and is deficient in fah1 pap1-D stems. Taken together, these experiments show that overexpression of PAP1 is able to drive the synthesis of high levels of anthocyanins in the fah1 mutant background only in tissues in which F5H is not normally expressed. In tissues that would otherwise express it, the absence of F5H results in a lack of anthocyanin accumulation, apparently the result of Mediator-dependent feedback inhibition associated with a block in phenylpropanoid metabolism.
Figure 8.
Anthocyanin accumulation is inhibited in the cortex of fah1 pap1-D inflorescence stems but is unaffected in vascular bundles. A, Representative inflorescence stems of pap1-D and fah1 pap1-D. B, Semithin sections of pap1-D and fah1 pap1-D inflorescence stems taken from the bottom of the second internode as viewed without staining using a light microscope. C, Representative chromatograms showing anthocyanins in pap1-D and fah1 pap1-D inflorescence stems.
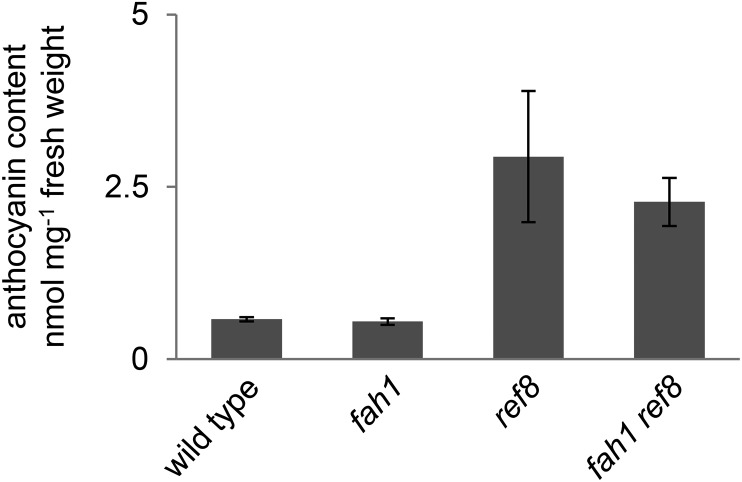
ref8 Is Epistatic to fah1 with Respect to Anthocyanin Accumulation
Given the observation that anthocyanin accumulation is specifically inhibited in tissues in which F5H is normally expressed, we hypothesized that the accumulation of a pathway product upstream of F5H might be responsible for the feedback inhibition of phenylpropanoid metabolism. To test this hypothesis, we quantified total anthocyanin accumulation in fah1 ref8 double mutants, which have been described previously (Weng et al., 2010). These analyses revealed that fah1 ref8 double mutants accumulate levels of total anthocyanins indistinguishable from those of ref8 mutants with functional F5H (Fig. 9). As expected, fah1 ref8 plants contained no detectable sinapoylated anthocyanins. These results further demonstrate that sinapoylation is dispensable for anthocyanin accumulation and indicate that disruption of C3′H prevents the repression of anthocyanin accumulation in the fah1 mutant background, suggesting that the accumulation of a biosynthetic intermediate between C3′H and F5H may contribute to the inhibition of anthocyanin accumulation in fah1 mutants.
Figure 9.
ref8 is epistatic to fah1 with respect to anthocyanin inhibition. Shown is the quantification by HPLC of anthocyanins in whole rosettes of wild-type plants and plants with the indicated mutations 3 weeks after planting. Each bar shows the mean of three biological replicates, with error bars indicating sd.
DISCUSSION
Although most core phenylpropanoid biosynthetic genes are thought to be known and characterized (Boerjan et al., 2003; Fraser and Chapple, 2011; Vanholme et al., 2013) and many important transcription factors have been identified (Mele et al., 2003; Mehrtens et al., 2005; Tohge et al., 2005; Zhong et al., 2007; Zhou et al., 2009; Umezawa, 2010; Zhao and Dixon, 2011), a predictive understanding of the phenylpropanoid pathway remains elusive, and additional regulatory and even biosynthetic components of the pathway continue to be discovered. The observation that fah1 mutants do not redirect carbon from sinapoylmalate to feruloylmalate biosynthesis revealed a gap in our understanding of phenylpropanoid metabolism and suggested the possible existence of a repressive mechanism responding to the loss of F5H activity. The experiments we have described here provide important new insight into this repression and suggest that a common mechanism may underlie both the lignin deficiency and dwarfing of ref8-1 mutants (Bonawitz et al., 2014) and the HCE and anthocyanin deficiency of fah1 mutants.
In most Arabidopsis phenylpropanoid mutants, intermediates that are normally used for the synthesis of sinapoylmalate are redistributed to other HCEs as a result of blockage of the pathway. This plasticity is the result of HCE biosynthetic enzymes having relatively relaxed substrate specificity (Lim et al., 2001; Shirley and Chapple, 2003; Mittasch et al., 2013). The data presented here show that neither REF1 nor UGT activity limits ferulate ester biosynthesis in fah1 mutants, thus demonstrating that the failure of these plants to accumulate wild-type levels of HCEs is not due to the lack of the required enzymatic activities. Furthermore, carbon flux is not diverted from the synthesis of HCEs to HCAs, lignin, or flavonoid biosynthesis in fah1. Instead, we have shown that the HCE deficiency of fah1 mutants is dependent on the transcriptional coregulator Mediator, as elimination of the Mediator subunits MED5a and MED5b restores total HCE accumulation to nearly wild-type levels.
Our experiments with soil-grown and Suc-stressed fah1 plants demonstrate that the repression of phenylpropanoid metabolism in fah1 mutants extends to the anthocyanin branch of the pathway and that this repression is also dependent on Mediator. In contrast with HCEs, anthocyanin accumulation is only affected by the disruption of MED5a/5b in the absence of FAH1, strengthening the conclusion that phenylpropanoids are derepressed and not simply globally increased in med5a/5b fah1 mutants. These observations are consistent with those of Maruta et al. (2014), who recently showed that fah1 seedlings fail to accumulate wild-type levels of anthocyanins in response to methylviologen-induced oxidative stress. In contrast with the conclusions of Maruta et al. (2014), however, we have found evidence that the anthocyanin deficiency of fah1 plants is not a result of expression changes at the transcriptional level of anthocyanin biosynthetic genes. Not only do fah1 mutants show wild-type transcriptional induction of anthocyanin biosynthesis genes in response to Suc stress, but also the rescue of anthocyanin accumulation in med5a/5b fah1 mutants is not associated with any changes in the expression of these genes. Moreover, we have shown that overexpression of the MYB75/PAP1 transcription factor leads to similar induction of anthocyanin biosynthetic genes in leaf blades of pap1-D and fah1 pap1-D mutants, despite the striking anthocyanin deficiency of the latter. Although we cannot formally exclude the possibility that Mediator regulates anthocyanin biosynthetic genes in fah1 mutants in a limited spatial or temporal window, the most straightforward interpretation of our data is that the repression of anthocyanin biosynthesis in fah1 is independent of changes in the transcription of the anthocyanin biosynthetic genes. Indeed, these results are similar to those we have reported for med5a/5b ref8-1 mutants, in which the rescue of lignin biosynthesis is not accompanied by changes in the expression of the lignin biosynthetic genes (Bonawitz et al., 2014).
Finally, multiple experimental observations reported here support a model in which the Mediator-dependent repression of anthocyanin biosynthesis (and, by extension, of HCE biosynthesis) is the result of a homeostatic feedback mechanism triggered in cells with an active phenylpropanoid pathway blocked at F5H (Supplemental Fig. S1). First, ref8 is epistatic to fah1 with respect to anthocyanin biosynthesis, such that elimination of FAH1 in the ref8 mutant background has no effect on total anthocyanin levels. Again in disagreement with Maruta et al. (2014), this result shows that sinapate and, indeed, FAH1 itself are entirely dispensable for high levels of anthocyanin biosynthesis. Second, the pattern of sinapoylated and nonsinapoylated anthocyanins in pap1-D leaves closely mirrors the pattern of anthocyanin deficiency in fah1 pap1-D leaves, with fah1 pap1-D showing the strongest inhibition in the portion of the leaf where anthocyanins are primarily sinapoylated. Finally, anthocyanin deficiency in fah1 pap1-D inflorescence stems is most striking in the epidermis and interfascicular fibers, precisely the same tissues in which F5H is normally expressed. The simplest model that accounts for all of these observations is that the accumulation in fah1 mutants of one or more pathway intermediates upstream of F5H and downstream of C3′H (or derivatives thereof) results in the initiation of a Mediator-dependent feedback mechanism that represses phenylpropanoid metabolism specifically in those cells that normally express F5H and therefore express all of the pathway genes leading up to this block.
The role of Mediator as a transcriptional coregulator involved in processes essential for the growth, development, and perception of environmental conditions is starting to be defined in plants (Kidd et al., 2011; Hemsley et al., 2014; Molloy, 2014; Yang et al., 2014), as is the role of MED5 in the regulation of the phenylpropanoid pathway (Bonawitz et al., 2012, 2014). Consistent with our previous observations on the MED5-dependent repression of lignin biosynthesis in ref8-1 mutants, the repression of phenylpropanoid biosynthesis in fah1 plants appears to be independent of changes in the steady-state level of the biosynthetic genes themselves. Nevertheless, given the established role of Mediator as a transcriptional coregulator and the sophisticated regulatory mechanisms controlling metabolic pathways, it remains a strong possibility that Mediator inhibits phenylpropanoid accumulation in fah1 via its influence on transcription. For instance, MED5a/5b-dependent changes in the transcription of one or more genes that influence the translation, stability, or activity of one or more phenylpropanoid biosynthetic genes, or the transport or turnover of pathway products or intermediates, could lead to the phenotypes observed in fah1 mutants. Determining the signaling components upstream and downstream of Mediator responsible for the repression of soluble phenylpropanoid metabolism in fah1 mutants and determining the extent to which they are shared with the Mediator-dependent pathway repressing lignin biosynthesis in ref8 remain exciting questions for future investigation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown under a 16-h-light/8-h-dark photoperiod in Redi-Earth Plug and Seedling Mixture (Sun Gro Horticulture) supplemented with Scotts Osmocote Plus controlled-release fertilizer (Hummert International) at 22°C. For seedling plant material to be used in the analysis of anthocyanin accumulation, seeds were surface sterilized for 30 min in a 2:1 mixture of 0.1% (v/v) Triton X-114 and household bleach. Seeds were rinsed thoroughly with sterile water and plated onto sterile filter paper discs wetted with 0.5% (v/v) ammonia-free Murashige and Skoog medium (Murashige and Skoog, 1962; Fraser et al., 2007). Plates were kept in the dark at 4°C for 2 d and then transferred to continuous light at 23°C. On day 4, the medium from each plate was decanted and replaced with new ammonia-free Murashige and Skoog medium containing 3% (w/v) Suc. Seedlings were analyzed for gene expression and anthocyanin content at various stages of development after high-Suc induction as described below.
Transgenic Arabidopsis
To generate the UGT overexpression constructs, the open reading frames for UGT84A1 and UGT84A3 were amplified from genomic Columbia wild-type DNA using CC2382/CC2383 and CC2384/CC2385, respectively (Supplemental Table S3). pCC0996 (Bonawitz et al., 2012) was used as the Gateway destination vector for both genes. Plant transformation using transformed Agrobacterium tumefaciens was conducted as described by Weigel and Glazebrook (2002).
Methanolic Soluble Secondary Metabolite Analysis
Flavonol glycosides and HCEs from whole rosettes and inflorescence tissue were analyzed as follows. Three-week-old whole rosettes and 8-week-old inflorescence tissue, stripped of siliques and cauline leaves, were harvested. Samples were extracted in solvent at 100 mg fresh weight mL−1 for 2 h at 65°C in 50% (v/v) methanol in triplicate from three independent biological populations. Compounds were quantified using a kaempferol standard for the flavonol glycosides and the standards cinnamic, p-coumaric, ferulic, 5-hydroxy ferulic, or sinapic acid (Sigma-Aldrich) for HCEs. Extracts were separated on a Shim-pack XR-ODS column (Shimadzu; column dimensions, 3 × 75 mm; bead size, 2.2 µm) using a gradient of increasing acetonitrile from 2% to 25% (v/v) for 29.5 min in 0.1% (v/v) formic acid at a flow rate of 0.9 mL min−1. Anthocyanins were extracted for 2 h at 65°C in a 50% (v/v) methanol and 4% (v/v) acetic acid solution (100 mg fresh weight mL−1) in triplicate from three independent biological populations. Extracts were separated on a Shim-pack XR-ODS column (Shimadzu; column dimensions, 3 × 75 mm; bead size, 2.2 µm) using an increasing acetonitrile gradient from 10% to 20% (v/v) for 10.5 min in 10% (v/v) formic acid at a flow rate of 0.7 mL min−1 and quantified with respect to a cyanidin standard (Sigma-Aldrich).
Cell Wall-Bound Secondary Metabolite Analysis
To analyze cell wall-bound p-coumaric and ferulic acid, triplicate samples from three independent biological populations were ground in liquid nitrogen and washed and dried to remove soluble metabolites (Meyer et al., 1998). The residual cell wall tissue was saponified using 1 n NaOH and incubated for 24 h at 37°C with constant agitation. The solution was acidified with 1 m HCl and extracted using ethyl acetate. Extracts were then dried, redissolved in 50% methanol, and analyzed by HPLC. Extracts were separated by HPLC using the same method described above for HCEs, and metabolites were quantified using authentic standards obtained from Sigma-Aldrich. Lignin content was determined using the acetyl bromide method as described by Fukushima and Hatfield (2004), using the extinction coefficient 23.35 g L−1 cm−1 as determined by Chang et al., (2008).
Heterologous Expression in Escherichia coli and Purification of REF1
Purified Arabidopsis REF1 protein was produced in BL21DE3 pLysS E. coli carrying a pET30a-REF1 expression vector (Nair et al., 2004). Cells were first grown to an optical density at 600 nm of 0.6 at 37°C, induced using 1 mm isopropyl-β-d-1-thiogalactopyranoside, and then grown overnight at 18°C. Cells were harvested by centrifugation 10,000g for 10 min at 4°C, resuspended in lysis buffer (50 mm NaCl and 20 mm Tris-HCl, pH 8), and 1 mg of each lysozyme and DNase1 was added. Samples were then incubated at room temperature for 30 min and frozen at −70°C. Samples were separated into soluble and insoluble fractions by centrifugation at 5,000g for 10 min at 4°C. The soluble fraction was affinity purified using a nickel-charged HiTrap Chelating HP column (http://www.gelifesciences.com) integrated in an FPLC system (http://www.gelifesciences.com) and washed using 20 mm Tris-HCl, pH 8, containing 50 mm NaCl and 20 mm imidazole. Protein was eluted using the same buffer containing 250 mm imidazole. The purity of purified protein was evaluated using SDS-PAGE (data not shown). Samples were stored at −70°C until use.
Enzyme Assays
REF1 activity assays were conducted according to Nair et al. (2004) with several modifications. Assays were conducted in 50 mm MOPS-KOH, pH 7, 5 mm DTT, 10% (v/v) glycerol, 50 mm NaCl, 1 mm NAD+, and varying concentrations of hydroxycinnamaldehyde substrate (Sigma-Aldrich) in a final volume of 500 µL. The buffer was prewarmed to 30°C, and 100 µL of diluted purified enzyme was added to initiate the reaction. Protein concentration was determined by the bicinchoninic acid assay method (www.piercenet.com) using bovine serum albumin (Sigma-Aldrich) as a standard for quantification. The assay was stopped after 20 min by the addition of 100 µL of 1 m HCl containing 10 µm 3,4,5-trimethoxy cinnamic acid (Sigma-Aldrich) as an internal standard. Samples were extracted using 1 mL of ethyl acetate and dried in vacuo. Samples were redissolved in 50% (v/v) methanol and analyzed by HPLC using the same method as described for HCEs above. UGT enzyme assays were conducted essentially as reported by Sinlapadech et al. (2007) using extracts of 3-week-old leaf tissue.
Quantitative Reverse Transcription-PCR
Quantitative reverse transcription-PCR was carried out as described by Li et al. (2010) using complementary DNA reverse transcribed from RNA extracted from 3-week-old leaf tissue from soil-grown plants or from seedlings harvested at different time points and exposed to different conditions. At1g13320 was used as a reference gene for all of the experiments, since the expression of this gene is stable over a wide range of conditions. The list of primers used can be found in Supplemental Table S3.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL database under the following accession numbers: C3ʹH, At2g40890; C4H, At2g30490; CCR, At1g15950; CHI, At5g05270; CHS, At5g13930; COMT, At5g54160; DFR, At5g42800; F3′H, At5g07990; F5H, At4g36220; LDOX, At4g22880; MED5a, At2g48110; MED5b, At3g23590; PAL1, At2g37040; PAP1, At1g56650; REF1, AT3G24503; UGT79B1, At5g54060; UGT84A1, At4g15480; UGT84A2, At3g21560; and UGT84A3, At4g15490.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic representation of the phenylpropanoid pathway in wild-type Arabidopsis and in the F5H-deficient fah1 mutant.
Supplemental Figure S2. fah1 mutants do not show ectopic lignification.
Supplemental Figure S3. Time course of transcript abundance of anthocyanin biosynthetic genes with or without induction with Suc stress.
Supplemental Table S1. Hydroxycinnamoyl ester and flavonol glycoside metabolic profile of fah1 plants and other plants with disrupted lignin biosynthesis.
Supplemental Table S2. Hydroxycinnamoyl ester and hydroxycinnamic acid derivative metabolic profile of leaf tissue.
Supplemental Table S3. List of relevant primers.
Supplementary Material
Glossary
- HCE
hydroxycinnamate ester
- HCA
hydroxycinnamic acid
Footnotes
This work was supported by the U.S. Department of Energy (Office of Science, Basic Energy Sciences grant no. DE–SC0000997 and Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences grant no. DE–FG02–07ER15905).
Articles can be viewed without a subscription.
References
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M, et al. (2014) Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509: 376–380 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Soltau WL, Blatchley MR, Powers BL, Hurlock AK, Seals LA, Weng JK, Stout J, Chapple C (2012) REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J Biol Chem 287: 5434–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang XF, Chandra R, Berleth T, Beatson RP (2008) Rapid, microscale, acetyl bromide-based method for high-throughput determination of lignin content in Arabidopsis thaliana. J Agric Food Chem 56: 6825–6834 [DOI] [PubMed] [Google Scholar]
- Chapple CC, Vogt T, Ellis BE, Somerville CR (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CT, Pollet B, Thévenin J, Sibout R, Denoue D, Barrière Y, Lapierre C, Jouanin L (2007) Both caffeoyl coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C (2002) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J 30: 47–59 [DOI] [PubMed] [Google Scholar]
- Fraser CM, Chapple C (2011) The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book 9: e0152, doi/10.1199/tab.0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Thompson MG, Shirley AM, Ralph J, Schoenherr JA, Sinlapadech T, Hall MC, Chapple C (2007) Related Arabidopsis serine carboxypeptidase-like sinapoylglucose acyltransferases display distinct but overlapping substrate specificities. Plant Physiol 144: 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima RS, Hatfield RD (2004) Comparison of the acetyl bromide spectrophotometric method with other analytical lignin methods for determining lignin concentration in forage samples. J Agric Food Chem 52: 3713–3720 [DOI] [PubMed] [Google Scholar]
- García JR, Anderson N, Le-Feuvre R, Iturra C, Elissetche J, Chapple C, Valenzuela S (2014) Rescue of syringyl lignin and sinapate ester biosynthesis in Arabidopsis thaliana by a coniferaldehyde 5-hydroxylase from Eucalyptus globulus. Plant Cell Rep 33: 1263–1274 [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Gräwe W, Bachhuber P, Mock HP, Strack D (1992) Purification and characterization of sinapoylglucose:malate sinapoyltransferase from Raphanus sativus L. Planta 187: 236–241 [DOI] [PubMed] [Google Scholar]
- Grotewold E. (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57: 761–780 [DOI] [PubMed] [Google Scholar]
- Hemsley PA, Hurst CH, Kaliyadasa E, Lamb R, Knight MR, De Cothi EA, Steele JF, Knight H (2014) The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26: 465–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys JM, Hemm MR, Chapple C (1999) New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA 96: 10045–10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Cahill DM, Manners JM, Schenk PM, Kazan K (2011) Diverse roles of the Mediator complex in plants. Semin Cell Dev Biol 22: 741–748 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Mugford SG, Ulker B, Gao D, Thorlby G, Knight MR (2009) Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J 58: 97–108 [DOI] [PubMed] [Google Scholar]
- Knight H, Veale EL, Warren GJ, Knight MR (1999) The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bonawitz ND, Weng JK, Chapple C (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YS, Kim HK, Lefeber AWM, Erkelens C, Choi YH, Verpoorte R (2006) Identification of phenylpropanoids in methyl jasmonate treated Brassica rapa leaves using two-dimensional nuclear magnetic resonance spectroscopy. J Chromatogr A 1112: 148–155 [DOI] [PubMed] [Google Scholar]
- Lim EK, Li Y, Parr A, Jackson R, Ashford DA, Bowles DJ (2001) Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J Biol Chem 276: 4344–4349 [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG (2010) The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 11: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta T, Noshi M, Nakamura M, Matsuda S, Tamoi M, Ishikawa T, Shigeoka S (2014) Ferulic acid 5-hydroxylase 1 is essential for expression of anthocyanin biosynthesis-associated genes and anthocyanin accumulation under photooxidative stress in Arabidopsis. Plant Sci 219-220: 61–68 [DOI] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138: 1083–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake S (2003) The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev 17: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C (1998) Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc Natl Acad Sci USA 95: 6619–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir Derikvand M, Sierra JB, Ruel K, Pollet B, Do CT, Thévenin J, Buffard D, Jouanin L, Lapierre C (2008) Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227: 943–956 [DOI] [PubMed] [Google Scholar]
- Mittasch J, Böttcher C, Frolov A, Strack D, Milkowski C (2013) Reprogramming the phenylpropanoid metabolism in seeds of oilseed rape by suppressing the orthologs of reduced epidermal fluorescence1. Plant Physiol 161: 1656–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3: 212–217 [Google Scholar]
- Molloy S. (2014) Oomycetes: attacking the mediator. Nat Rev Microbiol 12: 74–75 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue. Physiol Plant 15: 473–497 [Google Scholar]
- Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C (2004) The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al. (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenke D, Böttcher C, Scheel D (2011) Crosstalk between abiotic ultraviolet-B stress and biotic (flg22) stress signalling in Arabidopsis prevents flavonol accumulation in favor of pathogen defence compound production. Plant Cell Environ 34: 1849–1864 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C (2009) Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Shirley AM, Chapple C (2003) Biochemical characterization of sinapoylglucose:choline sinapoyltransferase, a serine carboxypeptidase-like protein that functions as an acyltransferase in plant secondary metabolism. J Biol Chem 278: 19870–19877 [DOI] [PubMed] [Google Scholar]
- Sinlapadech T, Stout J, Ruegger MO, Deak M, Chapple C (2007) The hyper-fluorescent trichome phenotype of the brt1 mutant of Arabidopsis is the result of a defect in a sinapic acid:UDPG glucosyltransferase. Plant J 49: 655–668 [DOI] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaethe J, Tautz J, Chittka L (2001) Visual constraints in foraging bumblebees: flower size and color affect search time and flight behavior. Proc Natl Acad Sci USA 98: 3898–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJ, Jacobs G, Rosecrance RC, Roberts SC (2009) Evidence for a photoprotective function of low-temperature-induced anthocyanin accumulation in apple and pear peel. Physiol Plant 136: 461–472 [DOI] [PubMed] [Google Scholar]
- Stout J, Romero-Severson E, Ruegger MO, Chapple C (2008) Semidominant mutations in reduced epidermal fluorescence 4 reduce phenylpropanoid content in Arabidopsis. Genetics 178: 2237–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Umezawa T. (2010) The cinnamate/monolignol pathway. Phytochem Rev 9: 1–17 [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Weng JK, Akiyama T, Bonawitz ND, Li X, Ralph J, Chapple C (2010) Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ou B, Zhang J, Si W, Gu H, Qin G, Qu LJ (2014) The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J 77: 838–851 [DOI] [PubMed] [Google Scholar]
- Yuan YW, Sagawa JM, Young RC, Christensen BJ, Bradshaw HD Jr (2013) Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics 194: 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yao J, Zhang Y, Sun Y, Mou Z (2013) The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J 75: 484–497 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Dixon RA (2011) Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci 16: 227–233 [DOI] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19: 2776–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH (2009) MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.