Proteins annotated as subunits of branched-chain ketoacid dehydrogenase contribute to the regulation of isoleucine, leucine, and valine homeostasis.
Abstract
The branched-chain amino acids (BCAAs) Leu, Ile, and Val are among nine essential amino acids that must be obtained from the diet of humans and other animals, and can be nutritionally limiting in plant foods. Despite genetic evidence of its importance in regulating seed amino acid levels, the full BCAA catabolic network is not completely understood in plants, and limited information is available regarding its regulation. In this study, transcript coexpression analyses revealed positive correlations among BCAA catabolism genes in stress, development, diurnal/circadian, and light data sets. A core subset of BCAA catabolism genes, including those encoding putative branched-chain ketoacid dehydrogenase subunits, is highly expressed during the night in plants on a diel cycle and in prolonged darkness. Mutants defective in these subunits accumulate higher levels of BCAAs in mature seeds, providing genetic evidence for their function in BCAA catabolism. In addition, prolonged dark treatment caused the mutants to undergo senescence early and overaccumulate leaf BCAAs. These results extend the previous evidence that BCAAs can be catabolized and serve as respiratory substrates at multiple steps. Moreover, comparison of amino acid profiles between mature seeds and dark-treated leaves revealed differences in amino acid accumulation when BCAA catabolism is perturbed. Together, these results demonstrate the consequences of blocking BCAA catabolism during both normal growth conditions and under energy-limited conditions.
The branched-chain amino acids (BCAAs) Leu, Ile, and Val are among nine amino acids essential for humans and other animals because they cannot be synthesized de novo (Harper et al., 1984). Plants synthesize BCAAs and are the main source of these essential nutrients in the diets of humans and agriculturally important animals. In addition to their nutritional value, BCAAs and BCAA-derived metabolites such as glucosinolates, fatty acids, and acyl sugars contribute to plant growth, development, defense, and flavor (Mikkelsen and Halkier, 2003; Taylor et al., 2004; Ishizaki et al., 2005; Slocombe et al., 2008; Araújo et al., 2010; Ding et al., 2012; Kochevenko et al., 2012).
The BCAA biosynthetic pathway and its regulation have been investigated in Arabidopsis (Arabidopsis thaliana) and other plants for the past two decades, in large part because of the commercial importance of herbicides that inhibit acetohydroxy acid synthase, which is the committing enzyme of BCAA biosynthesis (Singh and Shaner, 1995; Aubert et al., 1997; Singh, 1999; McCourt et al., 2006; Tan et al., 2006; Binder, 2010; Chen et al., 2010; Yu et al., 2010). Strong correlations between the levels of free BCAAs were found in wild-type Arabidopsis seeds and tomato (Solanum lycopersicum) fruits (Schauer et al., 2006; Lu et al., 2008), which suggests coregulation of biosynthesis and/or degradation. This presumably is due, at least in part, to the fact that they share four common biosynthetic enzymes and three catabolic steps. However, despite long-term interest in the desirability of optimizing the content of these essential amino acids in plants, the genes and proteins that constitute the full BCAA catabolic network are not completely characterized in Arabidopsis or any other plant, and there is much to learn about the genetic and biochemical regulation of this process.
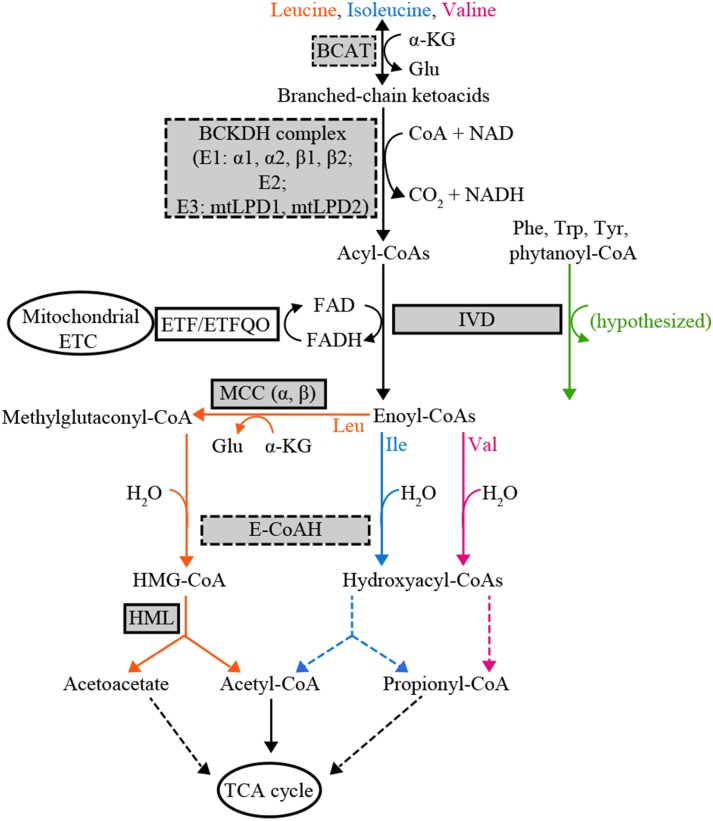
Recent observations using mutants blocked in BCAA catabolism (Fig. 1) indicate that the regulation of seed amino acid metabolism has unexpected, and potentially important, features. Previous work revealed that mutants defective in the Arabidopsis mitochondrial enzymes BCAT2, IVD, alpha (MCCA) and beta (MCCB) subunits of MCC, and HML exhibit increases in all three free BCAAs in mature seeds (Gu et al., 2010; Lu et al., 2011; Angelovici et al., 2013). Although it is logical that defects in the enzymes early in the catabolic pathway (the bcat2 and ivd1 mutants) would cause accumulation of Leu, Ile, and Val, it was not expected that mutants blocked in three enzymes specific to Leu degradation (mcca1, mccb1, and hml1) would also accumulate Ile and Val. Even more surprising is that biosynthetically unrelated amino acids, including His and Arg, accumulate to higher levels in seeds of ivd1, mcca1-1, mccb1-1, and hml1 mutants compared with the wild type (Gu et al., 2010; Lu et al., 2011). This suggests that the Arabidopsis amino acid networks are more interconnected than previously thought, and reveals that there are important gaps in our knowledge of the regulation of amino acid metabolism.
Figure 1.
Proposed Arabidopsis BCAA catabolism pathway. Enzyme names are abbreviated in rectangles with BCAA catabolic enzymes highlighted in gray. Validated BCAA catabolic enzyme activities are surrounded by solid lines, and putative BCAA catabolic enzyme activities by dashed lines. Metabolic processes directly or indirectly connected to BCAA catabolism in the mitochondrion are represented in ellipses. Reactions with one step are shown with solid arrows, and those with multiple steps with dashed arrows. Enzyme activities specific to Leu, Ile, and Val degradation are highlighted in orange, blue, and pink, respectively. Hypothesized isovaleryl-CoA dehydrogenase (IVD) enzyme activity toward non-BCAA catabolic intermediates is indicated with a green arrow. BCAT, Branched-chain aminotransferase; α-KG, α-ketoglutarate; BCKDH, branched-chain ketoacid dehydrogenase; mtLPD, mitochondrial lipoamide dehydrogenase; ETF, electron transfer flavoprotein; ETFQO, electron transfer flavoprotein ubiquinone oxidoreductase; ETC, electron transport chain; MCC, 3-methylcrotonyl-CoA carboxylase; E-CoAH, enoyl-CoA hydratase; HMG-CoA, 3-hydroxy-3-methyl-glutaryl-CoA; HML, 3-hydroxyl-3-methylglutaryl-CoA lyase; TCA, tricarboxylic acid.
Recent studies in Arabidopsis revealed that BCAA catabolism plays physiological roles beyond maintaining free amino acid homeostasis (Ishizaki et al., 2005; Araújo et al., 2010). In addition to catalyzing the third step in the degradation of BCAAs, IVD helps plants survive under energy-limited conditions by serving as a source of electrons for the mitochondrial electron transport chain via ETFα and ETFβ and the ETFQO (Fig. 1). Two lines of evidence for this role are that the ivd1-2 mutant becomes senescent faster than the wild type in prolonged darkness, and mutants defective in ETFβ and ETFQO accumulate more free BCAAs and the IVD substrate isovaleryl-CoA (Ishizaki et al., 2005, 2006; Araújo et al., 2010). In addition, the transcripts of the functionally validated BCAA catabolism genes BCAT2, IVD1, MCCA1, and MCCB1 rapidly increase following transition from light to dark, and this increase is inhibited by Suc (Fujiki et al., 2000; Che et al., 2002; Binder, 2010; Angelovici et al., 2013). These observations suggest that IVD and other BCAA catabolic enzymes contribute to plant fitness under energy-limited conditions.
Although genetic and biochemical evidence exists for the participation of Arabidopsis enzymes BCAT2, IVD, MCCA, MCCB, and HML in BCAA catabolism (Gu et al., 2010; Lu et al., 2011; Ding et al., 2012; Angelovici et al., 2013), much less is known about the genes and encoded proteins for the BCKDH complex. Published biochemical evidence demonstrated the BCKDH complex enzyme activity in isolated Arabidopsis mitochondria (Taylor et al., 2004). The better characterized mammalian BCKDH is composed of multiple copies of three proteins: the α-ketoacid dehydrogenase/carboxylase E1 (E1α and E1β), dihydrolipoyl acyltransferase E2, and dihydrolipoyl dehydrogenase E3 (also known as mtLPD; Mooney et al., 2002). This multi-megadalton molecular mass complex catalyzes the second step of BCAA degradation, converting branched-chain ketoacids (intermediates in BCAA biosynthesis and catabolism) into acyl-CoA esters (Fig. 1). The complex is homologous to mitochondrial and chloroplastic pyruvate dehydrogenase complexes and the mitochondrial α-ketoglutarate dehydrogenase complex, and shares enzyme subunits with the Gly decarboxylase complex (Oliver, 1994; Mooney et al., 2002). In addition, the BCKDH complex is hypothesized to contribute to the mitochondrial electron transport by providing NADH to complex I and the internal alternative NADH dehydrogenases (Schertl and Braun, 2014).
The size of the BCKDH complex has hindered a detailed in vitro characterization in plants, and identification of the plant BCKDH complex subunits is based upon sequence annotation rather than functional analysis. Pairs of paralogous genes are annotated as encoding Arabidopsis E1α, E1β, and E3 subunits, and one gene is annotated for E2. These assignments are based on (1) sequence similarity with proteins identified from other organisms, especially mammals, and (2) mitochondrial localization evidence using tandem mass spectrometry, rather than enzyme activities (Fujiki et al., 2000; Mooney et al., 2000; Taylor et al., 2004).
We describe results from a functional genomics analysis of genes annotated as encoding subunits of the BCKDH complex. A group of eight validated or hypothesized BCAA catabolic genes were found to be coexpressed. Mutants in these genes accumulate higher levels of BCAAs in seeds and have enhanced senescence and increased amino acid accumulation in leaves during prolonged darkness. These data provide experimental evidence for the participation of BCKDH subunit genes E1A1, E1A2, E1B1, E1B2, and E2 in BCAA catabolism, reinforcing the importance of BCAA catabolism in regulating amino acid homeostasis under day-night cycles and prolonged darkness, and during seed development. These results are consistent with the hypothesis that the Arabidopsis BCAA catabolic network interacts with energy metabolism at multiple steps.
RESULTS
Known and Hypothesized Arabidopsis BCAA Catabolism Genes Form a Coexpression Module
Transcripts of the functionally validated Arabidopsis BCAA catabolism genes BCAT2, IVD1, MCCA1, and MCCB1 were reported to increase rapidly following the transition from light to dark (Däschner et al., 2001; Fujiki et al., 2001; Schuster and Binder, 2005; Araújo et al., 2010; Angelovici et al., 2013). These results led us to hypothesize that candidate genes encoding additional enzymes in BCAA catabolism could be identified by their coexpression with these known genes. Coexpression analysis was performed with 13 genes encoding proteins proposed (based on sequence similarity) or experimentally validated to participate in BCAA catabolism (the gene list is shown in Table I). First, to determine the extent of coexpression among these genes, pairwise Pearson correlation coefficients (PCCs) were calculated, and average linkage hierarchical clustering was performed with four data sets: development, abiotic and biotic stress, and light data sets from AtGenExpress (Schmid et al., 2005; Kilian et al., 2007), and the diurnal/circadian data set from the DIURNAL database (Mockler et al., 2007; Fig. 2A; Supplemental Table S1).
Table I. List of genes encoding experimentally validated and computationally annotated BCAA catabolic enzymes.
Duplication type was derived from the physical location of the gene paralogs on chromosomes and published data (Bowers et al., 2003). α-WGD, Most recent whole genome duplication event.
| Gene | Arabidopsis Genome Initiative | Annotation | Paralogous Gene | Duplication Type |
|---|---|---|---|---|
| BCAT1 | AT1G10060 | Branched-chain aminotransferase1, putative | BCAT2 | Tandem |
| BCAT2 | AT1G10070 | Branched-chain aminotransferase2 | BCAT1 | Tandem |
| E1A1 | AT1G21400 | α-Subunit of branched-chain ketoacid dehydrogenase E1, putative | E1A2 | Before α-WGD |
| E1A2 | AT5G09300 | α-Subunit of branched-chain ketoacid dehydrogenase E1, putative | E1A1 | Before α-WGD |
| E1B1 | AT1G55510 | β-Subunit of branched-chain ketoacid dehydrogenase E1, putative | E1B2 | α-WGD |
| E1B2 | AT3G13450 | β-Subunit of branched-chain ketoacid dehydrogenase E1, putative | E1B1 | α-WGD |
| E2 | AT3G06850 | Branched-chain ketoacid dehydrogenase E2, putative | None | Not applicable |
| mtLPD1 | AT1G48030 | Branched-chain ketoacid dehydrogenase E3, putative | mtLPD2 | α-WGD |
| mtLPD2 | AT3G17240 | Branched-chain ketoacid dehydrogenase E3, putative | mtLPD1 | α-WGD |
| IVD1 | AT3G45300 | Isovaleryl-CoA dehydrogenase | None | Not applicable |
| MCCA1 | AT1G03090 | α-Subunit of 3-methylcrotonyl-CoA carboxylase | None | Not applicable |
| MCCB1 | AT4G34030 | β-Subunit of 3-methylcrotonyl-CoA carboxylase | None | Not applicable |
| HML1 | AT2G26800 | Hydroxymethylglutaryl-CoA lyase | None | Not applicable |
Figure 2.
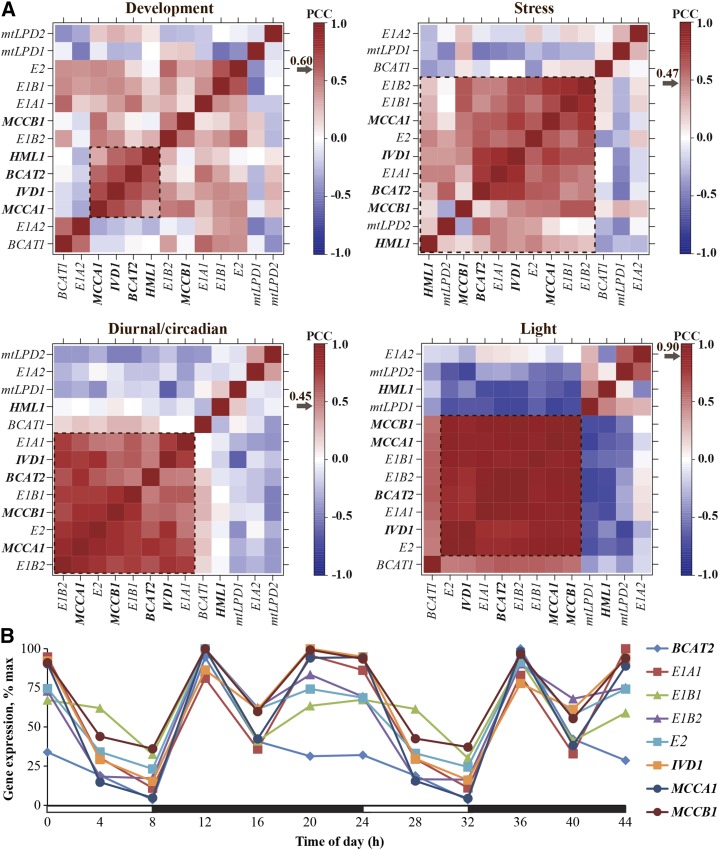
Analysis of known or proposed BCAA catabolism gene transcripts in the wild type (Col-0). A, Transcript coexpression analysis. Four data sets were tested: development (top left), stress (top right), diurnal/circadian (bottom left), and light (bottom right). Values represent the PCC for each gene pair. Arrows indicate the PCC value representing the 95th percentile of each null PCC distribution for individual data sets. Transcript names for validated genes are shown in bold and italic font, and those for proposed genes in regular italic font. Dotted rectangles represent coexpression modules in each data set, defined by clustered genes with significant expression correlations. Heat maps were generated using microarray data from AtGenExpress for stress, development, and light data sets (Schmid et al., 2005; Kilian et al., 2007) and the Diurnal database for the diurnal/circadian data set (Mockler et al., 2007). B, Expression profiles of highly coexpressed BCAA catabolism genes under short-day conditions. Rosette leaves from 4-week-old Col-0 plants were used in microarray experiments from the Diurnal database (http://diurnal.mocklerlab.org; Mockler et al., 2007). Each gene was normalized to its maximal expression. Transcript names for validated genes are emphasized in bold and italic font, and those for proposed genes in regular italic font. White bars on the x axis represent the time in the light, and black bars represent the time in the dark.
The degrees of coexpression among the five validated BCAA catabolism bait genes (BCAT2, IVD1, MCCA1, MCCB1, and HML1) were evaluated to establish the foundation for our coexpression analyses. In the stress data set, all five bait genes form a coexpression module with significant expression correlations (Fig. 2A; Supplemental Fig. S1). In the other three data sets (development, diurnal/circadian, and light), four out of the five bait genes form coexpression modules. These modules (one from each data set) contain three common bait members, BCAT2, IVD1, and MCCA1, with MCCB1 in only the diurnal/circadian and the light modules, and HML1 only in the development module (Fig. 2A; Supplemental Fig. S1). Because of the significant expression correlations among bait genes, these results indicate the feasibility of this approach for identifying candidate genes in the BCAA catabolic pathway. We next evaluated coexpression of the eight proposed BCAA catabolism genes with the five bait genes (dotted squares in Fig. 2A; Supplemental Fig. S1). The stress coexpression module contains the largest number of members: five proposed genes in addition to the five bait genes. Furthermore, four genes proposed to encode proteins of BCAA catabolism and four validated bait genes are in the light and diurnal/circadian coexpression modules. In contrast, none of the proposed genes is in the development module that contains four bait genes. In summary, highly interconnected modules containing validated and proposed BCAA catabolic enzyme genes emerged from three data sets: stress, diurnal/circadian, and light. These modules have eight members in common: the validated bait genes BCAT2, IVD1, MCCA1, and MCCB1, and proposed BCKDH genes E1A1, E1B1, E1B2, and E2 (Fig. 2A; Supplemental Fig. S1; Supplemental Table S1).
The observation that four out of five bait genes, and four out of eight proposed BCAA catabolic enzyme genes, are strongly coexpressed in the diurnal/circadian data set (Fig. 2A; Supplemental Fig. S1) suggests that these genes are coordinately regulated by light and the circadian clock. Further examination of transcript profiles confirmed common oscillation patterns among these eight validated or proposed BCAA catabolism genes under most diurnal/circadian experiments (Supplemental Fig. S2). It is notable that these gene transcript levels are reduced during the day and increased during the night in the rosette leaves of 4-week-old, short-day (SD; 8 h of light/16 h of dark)-grown plants (Fig. 2B). This common oscillation pattern is consistent with the previous observation that free BCAA levels fluctuate in day/night cycles and peak toward the end of the day (Gibon et al., 2006; Espinoza et al., 2010), suggesting that upregulation of the catabolism genes contributes to the decreased free BCAAs at night. In addition, the eight transcripts also oscillate in a similar fashion in constant light after entrainment in light/dark or hot/cold cycles (Supplemental Fig. S2), consistent with the hypothesis that BCAA catabolism is subject to regulation by the circadian clock. These results further demonstrated the coexpression of BCAA catabolic enzyme gene transcripts in diurnal/circadian conditions, especially on a day/night cycle.
Distinct Expression Patterns among BCAA Catabolic Enzyme Paralogs
There are four pairs of paralogous genes encoding known or proposed enzymes in the first two steps of BCAA catabolism (Table I). However, it is not known whether all paralogs participate in the BCAA catabolic process because BCAT activities are involved in both BCAA biosynthesis and degradation (Diebold et al., 2002), and some BCKDH subunits are similar to those of pyruvate dehydrogenases in mitochondria and chloroplasts, and mitochondrial α-ketoglutarate dehydrogenase and Gly decarboxylase (Oliver, 1994; Lutziger and Oliver, 2001; Mooney et al., 2002). We hypothesized that genes encoding these BCAA degradation enzyme paralogs would be coexpressed with the documented catabolic enzymes because of their significant expression correlations revealed from our previous coexpression analyses (Fig. 2A; Supplemental Fig. S1). Indeed, four out of eight paralogs, BCAT2 and BCKDH subunits E1A1, E1B1, and E1B2, are members of the coexpression modules identified from the light and the diurnal/circadian data sets (Fig. 2A). This observation suggested that these proteins have roles in BCAA catabolism.
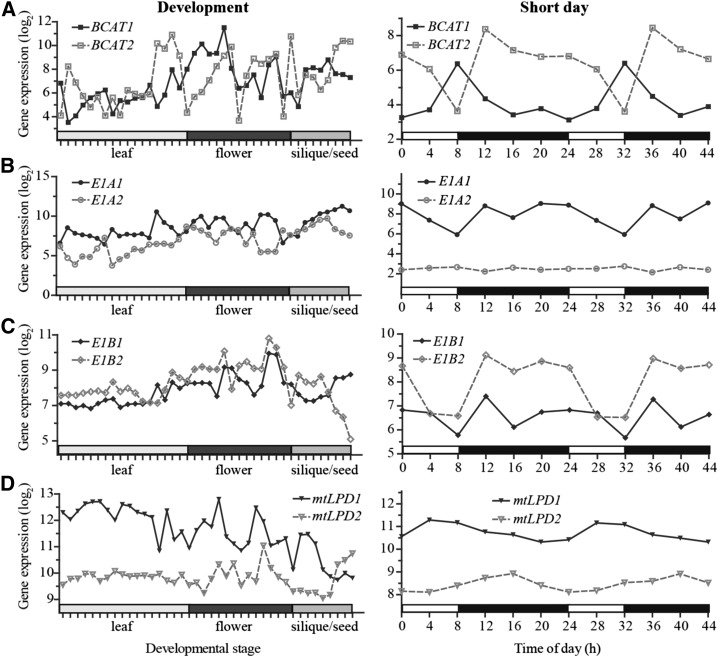
Expression divergence is often observed among paralogous gene pairs that can be due to modification of regulatory elements during evolution (Shariati and De Strooper, 2013), and likely results in function diversification (Stern and Orgogozo, 2008). To evaluate the degree of expression divergence, we compared the expression profiles of the four paralogous pairs. The flowering and seed development data sets were included in this analysis because the known BCAA catabolism genes BCAT2, IVD1, MCCA1, and HML1 have increased transcript levels during flowering and seed development (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). In addition, data for the SD growth experiments were also selected because of the highly coordinated transcript oscillations described earlier (Fig. 2B). All four pairs of paralogs were found to have divergent expression profiles in these data sets that followed two general patterns (Fig. 3). First, expression of paralogs BCAT1/2, E1A1/2, and E1B1/2 was anticorrelated during the late stages of seed development (Fig. 3, left; PCC value r = −0.98, −0.55, and −0.87, respectively, for the last four time points), suggesting more important roles in seed amino acid regulation for the isoforms encoded by the more highly expressed transcripts. Varying degrees of anticorrelation were also found for BCAT1/2, E1A1/2, and mtLPD1/2 under SD conditions (Fig. 3, right; r = −0.41, −0.82, and −0.32, respectively). The second pattern of expression divergence is illustrated by E1A1/2, E1B1/2, and mtLPD1/2 under SD growth conditions, and mtLPD1/2 during development, where one copy is expressed at a higher level than the other (Fig. 3; Ganko et al., 2007; Zou et al., 2009). Among these pairs, the expression profiles of E1B1 and E1B2 are most similar (r = 0.66). In summary, examples of divergent expression profiles of paralogs can be distinguished in the development or SD diurnal cycling data sets, indicating divergence in mRNA regulation. The differences in expression patterns suggest a possible functional divergence between the paralogs. In addition, this expression divergence under SD conditions also explains why BCAT1, E1A2, mtLPD1, and mtLPD2, genes proposed for BCAA catabolism, are not found in the coexpression module in the diurnal/circadian data set (Fig. 2; Supplemental Fig. S1).
Figure 3.
Expression analysis of paralogous genes known or proposed to be involved in BCAA catabolism. Microarray data were obtained from the AtGenExpress development data set (left column; Schmid et al., 2005) and the Diurnal database (right column; Mockler et al., 2007), respectively. White bars on the x axis under SD conditions represent the time in the light, and black bars represent the time in the dark. Gene expression values were converted to log2 and are shown on the y axis. Gene pairs indicated in each row are BCAT1 versus BCAT2 (A), E1A1 versus E1A2 (B), E1B1 versus E1B2 (C), and mtLPD1 versus mtLPD2 (D).
Double Mutants of BCKDH E1α Subunits Accumulate Increased Seed Free BCAAs
The coexpression analyses led to the hypothesis that the putative BCKDH E1α1, E1β1, E1β2, and E2 subunits, which are coregulated with the known BCAA catabolic enzymes, function in BCAA degradation (Fig. 2A). Because ivd1-2, mcca1-1, mccb1-1, hml1 (Gu et al., 2010; Lu et al., 2011), and bcat2 mutants (Angelovici et al., 2013) have increased free BCAAs in mature seeds, we hypothesized that mutants defective in bona fide BCKDH subunits would share this phenotype.
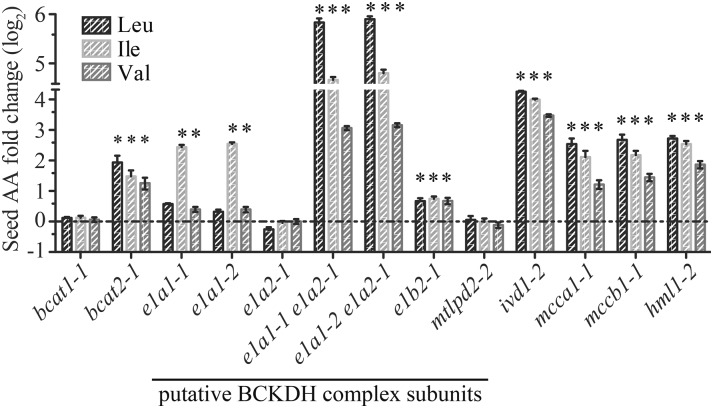
To test this idea, free BCAA content of mature dry seed was examined in homozygous mutants of proposed BCAA catabolic enzyme genes (Fig. 4). Quantitative PCR analysis revealed that e1a1-1, e1a1-2, e1a2-1, and e1b2-1 behave as null alleles, whereas mtlpd2-2 showed approximately 50% reduction of the mtLPD2 transcripts relative to the wild type (Supplemental Fig. S3). Unfortunately, no homozygous transfer DNA (T-DNA) mutant lines could be identified for E1B1, E2, or mtLPD1. The positive control mutants bcat1-1, bcat2-1, ivd1-2, mcca1-1, mccb1-1, and hml1-2 (Fig. 4; Supplemental Table S2) displayed BCAA changes similar to those previously reported (Gu et al., 2010; Lu et al., 2011; Angelovici et al., 2013). Some of the single mutants defective in the putative BCKDH subunits yielded small but significant (P < 0.05) changes in free BCAAs. For example, both e1a1 mutants and e1b2-1 had modest increases (1.5- to 5.8-fold relative to the wild type) in Ile and Val, or all three BCAAs (Fig. 4; Supplemental Table S2). We tested the hypothesis that partial genetic buffering was responsible for the modest increases in the single mutants by examining free seed BCAA levels in e1a1-1; e1a2-1 and e1a1-2; e1a2-1 double mutants, defective in both genes annotated as encoding E1α subunits. Consistent with this hypothesis, both double mutants had large and statistically significant (P < 0.05) increases in seed free BCAAs: 45-, 22-, and 9-fold increases for Leu, Ile, and Val, respectively (Fig. 4; Supplemental Table S2). These data support the hypothesis that E1α1 and E1α2 contribute to the degradation of BCAAs during seed development.
Figure 4.
Changes in free BCAA content in dry seeds of mutants relative to the wild type. The bars show the fold change of individual amino acids (AA) in each mutant compared with the wild type (horizontal dashed line) grown at the same time. Four or more biological replicates were measured for each genotype. An asterisk indicates a significant difference from the wild type, determined by the Student’s t test (P < 0.05). Error bars represent means ± se. The experiments were done at least three times with similar results obtained, and representative results are shown.
Because no null e1b1 mutants were available, constructs encoding artificial microRNAs (amiRNAs) targeting E1B1 were transformed into the e1b2-1 mutant background in an attempt to reduce expression of both paralogous E1β subunit genes (Supplemental Fig. S4). True-breeding homozygous T3 transgenic lines from two independent primary transformants had E1B1 transcript reduction to approximately 30% of the wild-type level (Supplemental Fig. S4). The free amino acid content from the seeds of homozygous T3 lines exhibited moderate but significant (P < 0.05) BCAA increases compared with the wild type (Supplemental Fig. S4). Thus, modest reduction of the E1B1 transcript in the e1b2-1 mutant background results in more seed free BCAAs. This supports the hypothesis that both E1β enzyme paralogs participate in BCAA catabolism during seed development. Genetic analysis of the role of mtLPDs was not possible since no null alleles were available for mtLPD1 or mtLPD2.
BCAA Catabolic Mutants Exhibit Early Senescence under Prolonged Darkness
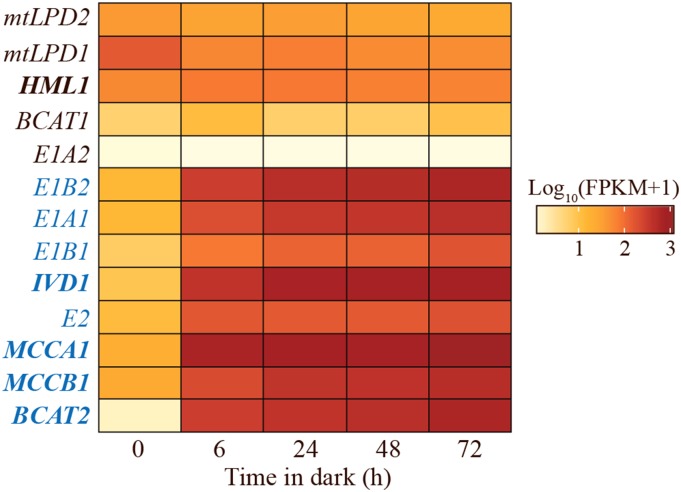
The increased seed free BCAAs in the mutant and amiRNA lines deficient in E1A1, E1A2, E1B1, and E1B2 support the hypothesis that the tested genes encode proteins of BCAA catabolism. Published studies revealed that transcripts of the known BCAA catabolism genes BCAT2, IVD1, MCCA1, and MCCB1 increase following transition from light to dark in plants grown under light-dark conditions and during prolonged darkness (Däschner et al., 2001; Fujiki et al., 2001; Schuster and Binder, 2005; Araújo et al., 2010). We used RNA sequencing (RNA-Seq) analysis to ask whether the transcripts for the proposed BCKDH subunits are also upregulated in plants subjected to prolonged darkness. These experiments were performed using rosette leaves from 5-week-old, SD-grown Columbia-0 (Col-0) wild-type plants moved to constant darkness for 6, 24, 48, and 72 h (Fig. 5). The transcripts of the eight genes found in the coexpression modules derived from stress, light, and diurnal/circadian data sets (Supplemental Fig. S1) were increased 9- to 400-fold within the first 6 h of prolonged darkness, and remained high until the last time point (gene names highlighted in blue in Fig. 5). These results are consistent with the hypothesis that BCAA catabolic enzymes, including BCKDH subunits E1A1, E1B1, E1B2, and E2, have one or more physiological roles in the dark.
Figure 5.
Heat map of log10 (FPKM+1) transcript levels of known and proposed BCAA catabolic genes in prolonged darkness in Col-0. Values are the mean of two independent biological replicates, which were grown and sequenced separately. FPKM, fragments per kilobase of transcript per million mapped reads. Genes with names in blue text showed statistically significantly increased transcript levels during prolonged darkness compared with time zero (P < 0.05, determined by exact test implemented in edgeR with Benjamini-Hochberg correction at false discovery rate < 0.05; Chen et al., 2011).
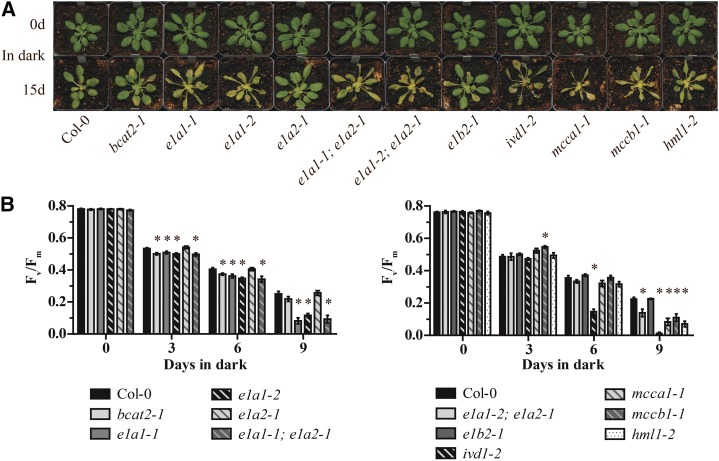
To explore this idea, BCAA catabolic mutant leaf morphology was monitored during prolonged darkness for 15 d. Prior to dark treatment, all of the lines being assessed were green and showed normal rosette leaf morphology (Fig. 6; Supplemental Fig. S5). As previously described for ivd1-2 (Araújo et al., 2010), we found that mutants defective in the other validated Leu catabolic enzyme genes, MCCA1, MCCB1, and HML1, exhibited enhanced dark-induced senescence phenotypes (Fig. 6A; Supplemental Fig. S5). These results demonstrated that early leaf senescence is observed in mutants blocked from the middle to the end of the Leu catabolism pathway. We asked whether the putative BCKDH subunit mutants had abnormal dark-induced senescence. The two mutants defective in the putative E1A1 subunit gene that is upregulated in the dark (e1a1-1 and e1a1-2) showed early dark-induced senescence (Fig. 6A; Supplemental Fig. S5). Double mutants that combine these mutations with the loss-of-function allele for the paralogous gene (e1a1-1; e1a2-1 and e1a1-2; e1a2-1 double mutants) exhibited senescence phenotypes similar to the e1a1 single mutants. These results are not surprising because, in contrast to the dark inducibility of the E1A1 transcript, E1A2 mRNA is not induced in the dark (Fig. 5). In fact, the e1a2-1 single mutant did not exhibit enhanced dark-induced senescence compared with the wild-type control, even after 15 d. Two other single mutants, bcat2-1 and e1b2-1, also did not exhibit enhanced senescence. To assess viability following the 15-d dark treatment, plants were transferred back to a 8-h light/16-h dark photoperiod and examined for new growth after 1 week. All genotypes recovered from this prolonged dark treatment except for ivd1-2, which had dry and yellow leaves at the end of the prolonged dark treatment (Fig. 6A; Supplemental Fig. S5). These results demonstrate that, in addition to IVD1, the disruption of E1A1, MCCA1, MCCB1, or HML1 also results in early senescence in prolonged darkness.
Figure 6.
Phenotypes of BCAA mutants subjected to prolonged darkness. A, Photographs of 5-week-old, SD-grown Arabidopsis plants taken prior to (0 d) and after 15 d of prolonged darkness. The leaves of e1a1-1, e1a1-2, both e1a1; e1a2 double mutants, ivd1-2, mcca1-1, mccb1-1, and hml1-2 were visibly yellowed and dehydrated following 15 d of prolonged darkness compared with the wild type. The experiments were done at least three times with similar results, and representative results are shown. B, Analysis of the maximum photochemical efficiency of PSII (Fv/Fm) to quantify the kinetics of leaf senescence. Plants were grown under the same conditions as in A. Values are means ± se of three to five biological replicates. An asterisk indicates a significant difference from the wild type, determined by the Student’s t test (P < 0.05). The two bar graphs represent data from plants grown in separate flats. Each experiment was done twice with similar results, and representative results are shown.
To complement the analysis of gross morphological changes, the maximum photochemical efficiency of PSII (Fv/Fm) was used to quantify leaf senescence caused by prolonged darkness (Oh et al., 1997). Mutant chlorophyll fluorescence was indistinguishable from that of the wild type prior to dark treatment (Fig. 6B, day 0). In contrast, on day 9, the mutants that showed early senescence also exhibited statistically significantly lower Fv/Fm values relative to the corresponding wild type (Fig. 6B).
Enhanced Leaf Free Leu, Ile, and Val Accumulation during Prolonged Darkness
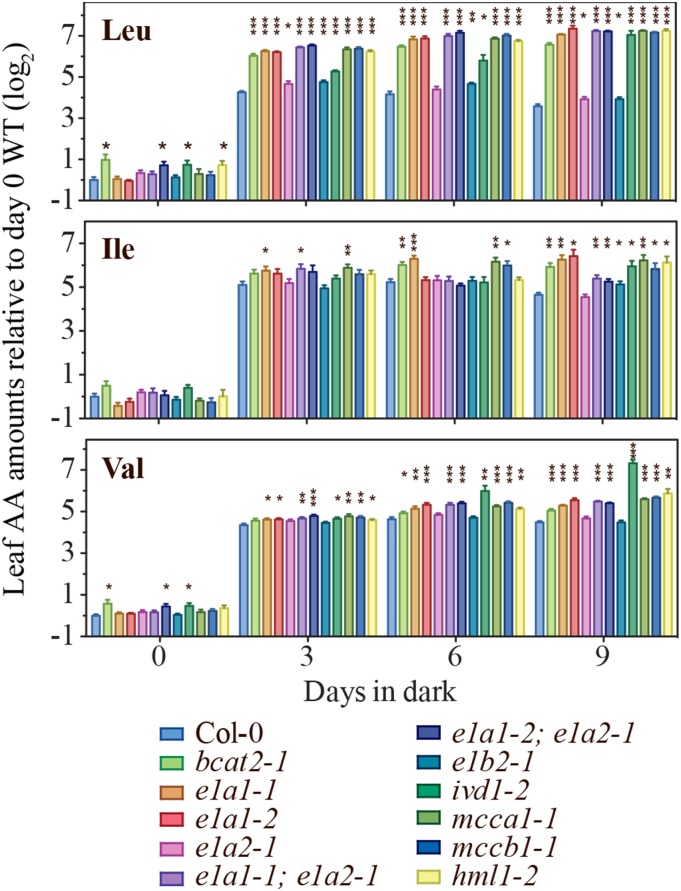
The ivd1-2 mutant was demonstrated to have increased leaf free BCAAs in prolonged darkness (Araújo et al., 2010). To test whether other mutants defective in BCAA catabolism also show enhanced BCAA accumulation, free rosette leaf amino acid content was analyzed in wild-type and catabolic mutants subjected to prolonged darkness. All three BCAAs increased in dark-treated wild-type plants at all three time points, and further increases due to blocked BCAA catabolism were seen for some combinations of mutants and amino acids (Fig. 7; Supplemental Table S3). The effects of all mutations, except for e1a2 and e1b2, were strongest and most consistent for Leu (8- to 14-fold increases compared with the wild type at day 9), with the least significant increases in Ile. Of the mutants, e1a2-1 and e1b2-1 displayed the least significant increases compared with the wild type, and this is especially obvious for Leu and Val. The small impact of these mutations is reminiscent of the results with free seed amino acid changes (Fig. 4; Supplemental Table S2). The E1B1-silenced e1b2-1 plants were also evaluated for leaf amino acid content after 6 d in prolonged darkness. The homozygous T3 lines from two independent primary transformants showed stronger BCAA increases compared with the e1b2-1 single mutant and the wild type (Supplemental Fig. S4). In summary, the BCAA increases in the mutants support the hypothesized roles of E1α1, E1β1, and E1β2 BCKDH subunits in BCAA catabolism, especially in Leu degradation, during prolonged darkness.
Figure 7.
Relative levels of leaf free BCAAs in mutants during prolonged darkness. The y axis values represent the log2 transformed amino acid (AA) levels normalized to the wild type (WT; Col-0) at day 0. Values are means ± se of five biological replicates. An asterisk indicates a significant difference from the wild type at the same time point, determined by the Student’s t test (*, P < 0.05; **, P < 0.01; and ***, P < 0.001).
Taken together, analysis of the mutants and amiRNA lines demonstrated that disruption of BCAT2, E1α1, E1β1, E1β2, IVD, MCCA, MCCB, and HML enzymes led to early senescence and/or increased leaf free BCAAs in prolonged darkness. These results further support the hypothesis that the full BCAA catabolic pathway plays a role in plant survival under carbon-limited conditions. These data also add to the evidence that E1A1, E1B1, and E1B2 encode subunits of the BCKDH complex.
Beyond Leu, Ile, and Val: Blocking BCKDH Causes Broad Changes in Leaf and Seed Amino Acids
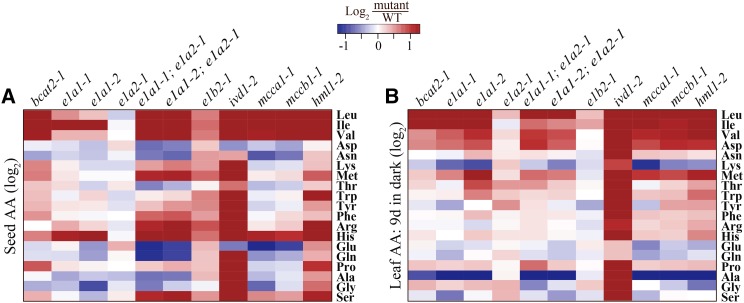
In addition to accumulating seed free BCAAs, ivd1-2, mcca1-1, mccb1-2, and hml1-2 mutants were previously shown to have increased levels of biosynthetically seemingly unrelated amino acids, with increased Arg and His common to all (Gu et al., 2010; Lu et al., 2011). We asked whether this interpathway phenomenon was observed in mutants annotated as defective in BCKDH subunit genes (Fig. 8; Supplemental Tables S3 and S4). The most obvious sign of cross pathway seed amino acid accumulation was seen in the e1a1-1; e1a2-1 and e1a1-2; e1a2-1 double mutants, which had 2- to 7-fold and statistically significant (P < 0.001) increases in Arg, His, Met, and Ser (Fig. 8A; Supplemental Table S2). The e1b2-1 mutant also had more modest, but quite widespread increases in multiple seed amino acids, despite the presence of the intact paralogous E1B1 gene. As previously reported for mutants blocked later in the pathway (Gu et al., 2010; Lu et al., 2011), high seed His was a hallmark of all the putative BCKDH subunit mutants, with the exception of e1a2-1. The positive control ivd1-2 mutant showed the expected highly pleiotropic amino acid changes in both seeds and leaves by the end of the dark treatment: it exhibited significant increases for 16 out of 19 amino acids detected in seeds, as was previously reported (Gu et al., 2010; Lu et al., 2011; Fig. 8A; Supplemental Table S2), and for 17 out of 19 leaf amino acids by the end of the 9-day dark treatment (Fig. 8B; Supplemental Table S3). The ivd1-2 mutant displayed severe dehydration and senescence when tissue was extracted after 9 d in the dark. Thus, the data from 6-d dark-treated ivd1-2 mutant plants, which are similar to the 9-d data in Figure 8B (Supplemental Table S4), are likely to be more reliable. With the possible exception of increased Asp and Met, dark-induced leaf amino acid changes were less consistent across the mutants in BCKDH and in other enzymes of BCAA catabolism.
Figure 8.
Heat map showing the effect of disrupting known or proposed BCAA catabolism genes on amino acid (AA) homeostasis. Comparisons of amino acid contents between (A) dry seeds and (B) leaves of 5-week-old plants kept in the dark for 9 d. Values represent the mean fold change (log2) in mutants compared with the wild type (WT). Four biological replicates were used for analyzing seed amino acid content, and five for leaf. Maximum color intensities correspond to −1.3 and +1.3, which are equivalent to a fold change of 0.4 and 2.5, respectively.
DISCUSSION
Advances in genomics technologies create new opportunities to identify candidate genes and proteins for complex physiological and biochemical processes. This has led to a situation where it is relatively inexpensive and straightforward to mine data and create hypotheses, whereas testing theories is often the costly and time-consuming step. Analysis of components of large multisubunit enzymes creates a special challenge requiring expression of multiple subunits, assembly of the component parts, and establishment of an in vitro activity assay that faithfully represents the in vivo process. The megadalton, multisubunit plant BCKDH enzyme is an example of such a complex. Here, we took a systems approach to hypothesize functions of putative BCKDH subunits through a combination of sequence similarity, transcript coexpression analyses, and mutant characterization. We provided evidence for the participation of putative BCKDH subunits in BCAA catabolism, and demonstrated examples of partial genetic buffering and functional divergence between enzyme paralogs. Moreover, the data revealed that BCAA catabolic mutants defective at multiple steps in the pathway exhibit early senescence with high leaf free BCAAs in prolonged darkness, supporting a previous hypothesis that BCAA degradation generates alternative sources of energy in plants under energy-limited conditions (Ishizaki et al., 2005; Araújo et al., 2010).
Considerations in Using Transcript Coexpression Analysis in Gene Functional Characterization
There are important considerations in using gene expression analysis to develop or test hypotheses regarding gene function. First, it is essential to have a set of validated bait genes for the analysis; in this study, five bait genes of BCAA catabolism were used (Table I). Second, success of the approach requires that appropriate transcript profiling data be available. In this study, we explored multiple microarray data sets (Fig. 2; Supplemental Table S1); these included a developmental data set, with a variety of tissues (development), plants grown under differing light and temperature regimes (diurnal/circadian and light), as well as abiotic and biotic stress conditions (stress). The broad choice of types of data sets was important in this study because each yielded coexpression modules of different sizes and structures (Supplemental Fig. S1). For example, analysis of the stress data set produced a module with the largest number of nodes (10), whereas the diurnal/circadian data set yielded a smaller (eight) but more highly connected module (Supplemental Fig. S1). In contrast, the development data set provided the least information, with a weakly connected module of four bait genes that had no significant expression correlation with the proposed BCAA catabolic enzyme genes (Supplemental Fig. S1). Taken together, our results demonstrate the importance of using validated bait genes and different types of expression data to identify candidate genes.
Coexpression analysis can be confounded in a number of ways. One scenario is when paralogous genes exist, with one copy that actively responds to environmental perturbations, whereas the other generally expresses at a much lower level (Keith et al., 1991; Duarte et al., 2006; Ganko et al., 2007; Székely et al., 2008; Zou et al., 2009). For example, we found that the E1α subunit gene E1A1 coexpressed with the other core BCAA catabolic genes, whereas the paralog E1A2 did not (Fig. 2A; Supplemental Fig. S1). Consistent with different roles for the two paralogous genes, the e1a1 mutants had high leaf BCAAs under prolonged dark conditions, whereas the e1a2-1 mutant did not (Fig. 7). In contrast, both E1α subunit genes are required to maintain BCAA homeostasis in seeds (Fig. 4), emphasizing that individual paralogs can have diverse roles throughout development and in response to the environment.
Using coexpression analysis to develop hypotheses regarding function also can be confounded if the protein product participates in multiple different processes. For example, mtLPD is proposed to function as a subunit of the mitochondrial pyruvate dehydrogenase, α-ketoglutarate dehydrogenase, Gly decarboxylase, and BCKDH complexes (Lutziger and Oliver, 2001). Needs for this subunit in physiologically diverse processes might be responsible for the observed lack of coregulation with other known and proposed genes of BCAA degradation (Fig. 2A; Supplemental Fig. S1). In addition, the single-copy and experimentally validated hydroxymethylglutaryl-CoA lyase gene HML1 was coexpressed with other BCAA catabolic genes in the stress and development data sets (Fig. 2A; Supplemental Fig. S1). We hypothesize that lack of membership of HML1 mRNA in the light and diurnal/circadian coexpression modules reflects that this enzyme participates in other metabolic processes (Ashmarina et al., 1994, 1999). Alternatively, perhaps this and other noncoregulated BCAA catabolic enzymes are subject to posttranscriptional regulation.
Evidence that BCAA Catabolism and Energy Metabolism Interact at Multiple Steps
In this study, we demonstrated that eight out of the 13 proposed or validated BCAA catabolism genes are coexpressed and share common transcript oscillation patterns in diurnal and circadian treatments (Fig. 2; Supplemental Fig. S2). These findings are consistent with the observed fluctuation of free BCAA levels on diel cycles (Gibon et al., 2006; Espinoza et al., 2010), and demonstrate the physiological importance of BCAA catabolism at night. BCAAs are proposed to provide their downstream catabolic products, acetoacetate, acetyl-CoA, and propionyl-CoA, to the TCA cycle for energy generation (Fig. 1; Anderson et al., 1998). In addition, Mentzen et al. (2008) identified a coexpression supermodule capable of maintaining cellular energy balance via catabolism using coexpression analysis with microarray data from 70 experiments. This supermodule contained genes encoding enzymes in the catabolism of amino acids (including BCAAs), carbohydrates, lipids, and cell wall components. Despite the proposed role of BCAA catabolism in energy generation, no vegetative or reproductive phenotype was observed for BCAA catabolic mutants grown in photoperiods with 16-, 12-, or 8-h light (Lu et al., 2011, and day 0 in Fig. 6 and Supplemental Fig. S5 of this study), in contrast to the reported aberrant reproductive development of the mcca1 and mccb1 mutants (Ding et al., 2012). Our results suggest a limited role of BCAA catabolism in providing TCA cycle substrates in day/night cycling conditions (Lu et al., 2011 and this study).
Although diurnal regulation is more physiologically relevant, the prolonged darkness assay is experimentally convenient for studying the regulation of energy metabolism and interacting metabolic processes in plants. Success using such an approach was previously demonstrated in identifying and characterizing mutants defective in genes participating in energy-related processes including autophagy, mitochondrial electron transport chain, and starch metabolism (Gibon et al., 2004; Ishizaki et al., 2005, 2006; Liu and Bassham, 2012). During energy-limited conditions, autophagy was demonstrated to promote organelle degradation including chloroplasts, partially contributing to the increase of free amino acid pools including free BCAAs in vegetative tissues (Hanaoka et al., 2002; Wada et al., 2009; Izumi et al., 2010). Consistent with this idea, we observed dramatic free amino acid increases including BCAAs, aromatic amino acids, Lys, His, Asn, and Arg in the wild type within 3 d in prolonged darkness (Supplemental Table S3). In addition, Lys metabolism was previously demonstrated to interact with plant energy metabolism: high seed Lys Arabidopsis lines with a knockout mutation in the catabolic lys-ketoglutarate reductase/saccharopine dehydrogenase and expression of a deregulated bacterial dihydrodipicolinate synthase showed reduced levels of the TCA cycle intermediates (Angelovici et al., 2011).
Our results are consistent with the notion that multiple steps of the BCAA degradation pathway provide alternative sources of energy under long-term dark treatment conditions. Prior to our study, IVD was the only enzyme in BCAA catabolism that was shown to play a role in plant survival in energy-limited conditions, because ivd1-2 exhibited enhanced senescence relative to the wild type in prolonged darkness (Araújo et al., 2010). The authors hypothesized that the early senescence of ivd1-2 resulted from deficiency in supplying electrons to the mitochondrial electron transport chain and/or providing the BCAA catabolic products to the TCA cycle (Ishizaki et al., 2005; Araújo et al., 2010; Fig. 1). In our study, BCAA catabolic mutants defective in enzymes both upstream (e1a1 single mutants and e1a1; e1a2 double mutants) and downstream (mcca1-1, mccb1-1, and hml1-2 mutants) of IVD displayed enhanced senescence in prolonged darkness (Fig. 6; Supplemental Fig. S5), supporting the hypothesized role of BCAA catabolism in providing TCA cycle substrates in energy-limited conditions. Interestingly, ivd1-2 exhibited dehydrated and yellowed rosette leaves more rapidly than the other mutants (Supplemental Fig. S5), and was the only mutant that did not recover after the 15-d dark treatment (Fig. 6). This is consistent with the hypothesis that IVD influences energy homeostasis in multiple ways, not only by providing BCAA catabolic CoA intermediates to the mitochondrial electron transport chain, but also by catabolizing additional substrates such as phytanoyl-CoA and aromatic amino acids (Araújo et al., 2010). The observation that e1a1 single mutants and e1a1; e1a2 double mutants, which are defective in subunits of the upstream BCKDH complex, showed enhanced senescence at a weaker level than ivd1-2 (Fig. 6; Supplemental Fig. S5) supports the hypothesized broader substrate range for IVD. Taken together, our results provide genetic evidence supporting the hypothesized interaction between BCAA catabolism and energy metabolism at multiple steps, and are consistent with a broader substrate range for IVD in plants.
Broader Implications
This and previously published work on amino acid catabolism have a variety of implications for our understanding of plant metabolism and attempts to improve productivity and nutritional quality of plants. First, a growing body of evidence points to major roles of amino acid catabolism and interconversion in regulating levels of six of the nine free essential amino acids (Ile, Leu, Lys, Met, Thr, and Val) in seeds of a variety of plants (Karchi et al., 1994; Zhu and Galili, 2003; Jander et al., 2004; Joshi et al., 2006; Lee et al., 2008; Gu et al., 2010; Lu et al., 2011; Angelovici et al., 2013). These results indicate that modifying catabolism has the potential to improve the nutritional quality of crop seeds and vegetative tissues. However, a variety of pleiotropic effects were seen for these examples, ranging from increased seed Cys in threonine aldolase1 mutants (Lu et al., 2008) to broader amino acid increases in Lys overproducing lines (Karchi et al., 1994; Zhu and Galili, 2003) and BCAA catabolic mutants (Gu et al., 2010; Lu et al., 2011; and this study), and defects in vegetative and seed development or seed viability (Zhu and Galili, 2004; Lee et al., 2008; Ding et al., 2012). It is not surprising that these relatively old processes are broadly connected and difficult to perturb without resultant undesirable effects (Milo and Last, 2012). Although these pleiotropies create hurdles for metabolic engineering of nutritional quality, understanding the basis for these syndromes should lead to a deeper understanding of the architecture of these and other metabolic networks.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia CS60000, T-DNA lines bcat1-1 (SALK_138630), bcat2-1 (SALK_037854), e1a1-1 (SALK_071680), e1a1-2 (WiscDsLox470G12), e1a2-1 (SAIL_113_D07), e1b2-1 (SALK_098054), and mtlpd2-2 (SALK_027039) were obtained from the Arabidopsis Biological Resource Center. Homozygous mutant lines that we obtained or validated were deposited at the Arabidopsis Biological Resource Center: bcat1-1 stock CS68922, bcat2-1 CS68923, e1a1-1 CS68924, e1a1-2 CS68925, e1a2-1 CS68926, e1b2-1 CS68927, mtlpd2-2 CS68928, double mutants e1a1-1; e1a2-1 CS68929 and e1a1-2; e1a2-1 CS68930. The ivd1-2, mcca1-1, mccb1-1, and hml1-2 mutants were described previously (Gu et al., 2010; Lu et al., 2011). All plants, including double mutants, were genotyped and confirmed to be homozygous with primers in Supplemental Table S5 prior to further analyses. Plants were grown in soil in chambers at 21°C with fluorescent lamps (100 μmol m−2 s−1) under different photoperiods (16 h for long day, 8 h for SD, and continuous darkness). Seeds for amino acid assay were harvested from mature plants grown under long day conditions. Leaves for transcript analysis and amino acid assay were harvested from plants grown under SD for 5 weeks and subjected to various lengths of prolonged darkness after the end of the night.
Transcript Coexpression Analysis
Coexpression analysis of the previously documented and hypothesized BCAA catabolic enzyme genes was performed with the following microarray data sets: stress (AtGenExpress, abiotic and biotic stress treatments in roots and shoots with time points from 0.5 to 24 h; Kilian et al., 2007), development (AtGenExpress, atlas of developmental stages consisting of a variety of tissues; Schmid et al., 2005), light (AtGenExpress, light treatments of different wavelengths, fluence, and durations; Kilian et al., 2007), and diurnal/circadian (Diurnal, combinations of light and temperature conditions; Mockler et al., 2007). The development, light, and diurnal/circadian data were downloaded in a robust multiarray average normalized form from http://jsp.weigelworld.org/expviz/expviz.jsp and ftp://www.mocklerlab.org/diurnal/. For the stress data set, the degrees of differential expression (in the form of fold change) were obtained from an earlier study (Zou et al., 2009). For all data sets, pairwise PCCs were calculated among the genes of interest using the SciPy library in Python (http://www.scipy.org/; Jones et al., 2001). To identify gene pairs with significantly higher than randomly expected PCC values, 500,000 gene pairs were selected randomly from each data set to calculate PCCs and establish a null PCC distribution. The 95th percentile of each null PCC distribution was used as the threshold for calling two genes as significantly positively correlated with a 5% false-positive rate (arrows in Fig. 2A; Supplemental Fig. S1; Supplemental Table S1). Coexpression modules were defined by clustered genes with significant correlations in each data set. Heat maps were generated with levelplot within the R lattice package (Sarkar and Sarkar, 2007).
RNA Extraction and Quantitative Reverse Transcription-PCR Analysis
Total RNA from 9th to 12th rosette leaves was extracted using the RNeasy plant mini kit (Qiagen) and digested on column with RNase-free DNase Set (Qiagen). First-strand complementary DNA was synthesized from 2 µg of total RNA with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and oligo(dT)12-18 primer (Invitrogen; Kotewicz et al., 1985). Gene-specific primers were designed to span two or more exons as listed in Supplemental Table S5. Quantitative PCR analyses were performed on a 7500 Fast Real-Time PCR System with Power or Fast SYBR green PCR master mix (Applied Biosystems), with ACTIN2 (At3G18780) transcript level used as an internal control (Czechowski et al., 2005).
Free Amino Acid Analysis by Liquid Chromatography-Tandem Mass Spectrometry
Free amino acids from dry seeds or the 10th and 11th rosette leaves were extracted and analyzed by modifying a previously described method (Gu et al., 2007, 2012; Lu et al., 2008). One micromolar of five heavy amino acids were added to the extraction buffer for a more accurate quantification: Leu-d10 for Leu and Ile, His-d3 for His, Trp-d5 for Trp, Val-d8 for Val, and Phe-d8 for all other amino acids. Details on the detection of selected ion monitoring pairs were as described previously (Angelovici et al., 2013). The amino acid quantities were normalized to the fresh weight of the harvested samples. All heavy amino acids were purchased from Cambridge Isotope Laboratories.
Generation of E1B1-Silenced e1b2-1 Mutant
To clone the amiRNA constructs, two amiRNAs targeting E1B1 were designed using the WMD3-Web MicroRNA Designer (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi): amiE1B1-1 (5′-TATGCGATTACATTAGTCCTT-3′) and amiE1B1-2 (5′-TAACTACAGATAGTACGCCTA-3′). The precursor amiRNAs were cloned by overlapping PCR following protocols provided by WMD3-Web MicroRNA Designer (http://wmd3.weigelworld.org/downloads/Cloning_of_artificial_microRNAs.pdf), amplified by Gateway-compatible primers (Supplemental Table S5), and inserted into pEarley100 (Earley et al., 2006). The resultant constructs were transformed into the e1b2-1 single mutant. All progeny lines were tested for the presence of the amiRNA and T-DNA prior to further analyses.
RNA-Seq Analysis
Leaf tissues from Col-0, ivd1-2, and hml1-2 treated for 0, 6, 24, 48, and 72 h in prolonged darkness and 72 h under SD were harvested for RNA-Seq. For each genotype under the same treatment, two replicates were grown and harvested a month apart. In total, 36 samples were sequenced.
Total RNA from 9th to 12th rosette leaves was extracted using the RNeasy plant mini kit (Qiagen) and on-column digestion performed with the RNase-free DNase Set (Qiagen). RNA quality was assessed using the Agilent 2100 Bioanalyzer with the RNA 6000 Pico Chip (Agilent Technologies). Library construction and sequencing were conducted by the Michigan State University Research Technology Support Facility using the Illumina TruSeq Stranded kit, following the manufacturer’s protocols. Six samples were multiplexed and sequenced in one lane using the Illumina HiSEquation 2500 sequencer, and 50-bp single-end reads were generated. A total of six lanes were sequenced for 36 samples.
For read processing and assembly, the sequencing adapters were removed using the following parameters in Trimmomatic version 0.30 (Bolger et al., 2014): ILLUMINACLIP: TruSeq3-SE, SLIDINGWINDOW: 4:15, and MINLEN: 35. Processed reads were filtered by fastq_quality_filter in the FASTX-Toolkit version 0.0.13.2 (http://hannonlab.cshl.edu/fastx_toolkit/index.html), satisfying the criterion that >85% bases must have a Q-score ≥20. The processed and filtered reads were mapped to the Arabidopsis reference genome (The Arabidopsis Information Resource Genome Annotation version 10) using TopHat version 1.4.1 (Trapnell et al., 2009), sorted by SAMtools version 0.1.19 (Li et al., 2009), and analyzed with Cuffdiff version 2.1.1 (Trapnell et al., 2013). The transcript levels represented as FPKM were visualized with CummeRbund version 2.6.1 (Goff et al., 2012) in R version 3.0.3 (R Core Team, 2012). The RNA-Seq data set was deposited in the National Center for Biotechnology Information Gene Expression Omnibus under GEO accession number GSE67956.
Determination of the PSII Photochemical Efficiency
All chlorophyll fluorescence experiments were performed at the Center for Advanced Algal and Plant Phenotyping at Michigan State University (http://www.prl.msu.edu/caapp) with previously described setups for the growth chambers (Attaran et al., 2014). Plants were grown under SD for 5 weeks and then subjected to prolonged darkness for 14 d. Chlorophyll fluorescence parameters were measured each day at the end of the subjective night from days 0 to 14 in prolonged darkness. Experiments were repeated twice and representative results were shown.
Sequence data from this article can be found in the GenBank/EMBL data libraries under Gene Expression Omnibus accession number GSE67956.
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Graphical representation of transcript correlation modules among the four data sets.
Supplemental Figure S2. Transcript profiles for known and proposed BCAA catabolism genes under diurnal/circadian conditions.
Supplemental Figure S3. Characterization of BCKDH complex subunit mutants.
Supplemental Figure S4. Transcript and mutant analyses of E1B1-silenced e1b2-1 lines.
Supplemental Figure S5. Phenotypes of BCAA mutants subjected to prolonged darkness.
Supplemental Table S1. Pairwise PCCs for transcripts of BCAA catabolism genes.
Supplemental Table S2. Mutant seed free amino acid profiles.
Supplemental Table S3. Mutant leaf free amino acid profiles in prolonged darkness.
Supplemental Table S4. Leaf amino acid profiles of ivd1-2 relative to Col-0 at 6 and 9 d in prolonged darkness.
Supplemental Table S5. Primers for genotyping, quantitative PCR, and amiRNA generation.
Supplementary Material
Acknowledgments
We thank multiple colleagues at Michigan State University for help with this work: David Hall and Kathleen Imre for genotyping homozygous mutants, Ruthie Angelovici for providing heavy amino acids and the mass spectrometry detection method, Lijun Chen for modifying and fine-tuning the mass spectrometry method, Ronghui Pan for providing vectors for artificial microRNA mutant generation, and Gaurav Moghe and Jeongwoon Kim for constructive suggestions on RNA-Seq analysis. We also thank Linda Savage and David Hall (Center for Advanced Algal and Plant Phenotyping) for help with all chlorophyll fluorescence experiments.
Glossary
- BCAA
branched-chain amino acid
- BCKDH
branched-chain ketoacid dehydrogenase
- TCA
tricarboxylic acid
- PCC
Pearson correlation coefficient
- amiRNA
artificial microRNA
- FPKM
fragments per kilobase of transcript per million mapped reads
- T-DNA
transfer DNA
- RNA-Seq
RNA sequencing
- Col-0
Columbia-0
- SD
short day
Footnotes
This work was supported by the U.S. National Science Foundation (grant nos. MCB–1119778 and NSF MCB–124400) and the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (grant no. DE–FG02–91ER20021 to C.P. and S.U.).
Articles can be viewed without a subscription.
References
- Anderson MD, Che P, Song J, Nikolau BJ, Wurtele ES (1998) 3-Methylcrotonyl-coenzyme A carboxylase is a component of the mitochondrial leucine catabolic pathway in plants. Plant Physiol 118: 1127–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Fait A, Fernie AR, Galili G (2011) A seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytol 189: 148–159 [DOI] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D (2013) Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 25: 4827–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al. (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashmarina LI, Pshezhetsky AV, Branda SS, Isaya G, Mitchell GA (1999) 3-Hydroxy-3-methylglutaryl coenzyme A lyase: targeting and processing in peroxisomes and mitochondria. J Lipid Res 40: 70–75 [PubMed] [Google Scholar]
- Ashmarina LI, Rusnak N, Miziorko HM, Mitchell GA (1994) 3-Hydroxy-3-methylglutaryl-CoA lyase is present in mouse and human liver peroxisomes. J Biol Chem 269: 31929–31932 [PubMed] [Google Scholar]
- Attaran E, Major IT, Cruz JA, Rosa BA, Koo AJ, Chen J, Kramer DM, He SY, Howe GA (2014) Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol 165: 1302–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Bligny R, Day DA, Whelan J, Douce R (1997) Induction of alternative oxidase synthesis by herbicides inhibiting branched-chain amino acid synthesis. Plant J 11: 649–657 [Google Scholar]
- Binder S. (2010) Branched-chain amino acid metabolism in Arabidopsis thaliana. Arabidopsis Book 8: e0137., doi/10.1199/tab.0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Che P, Wurtele ES, Nikolau BJ (2002) Metabolic and environmental regulation of 3-methylcrotonyl-coenzyme A carboxylase expression in Arabidopsis. Plant Physiol 129: 625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Saksa K, Zhao F, Qiu J, Xiong L (2010) Genetic analysis of pathway regulation for enhancing branched-chain amino acid biosynthesis in plants. Plant J 63: 573–583 [DOI] [PubMed] [Google Scholar]
- Chen Y, McCarthy D, Robinson M, Smyth GK (2011) edgeR: differential expression analysis of digital gene expression data. User’s Guide. http://www.bioconductor.org/packages/release/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf (August 13, 2013)
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Däschner K, Couée I, Binder S (2001) The mitochondrial isovaleryl-coenzyme A dehydrogenase of Arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol 126: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold R, Schuster J, Däschner K, Binder S (2002) The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol 129: 540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Che P, Ilarslan H, Wurtele ES, Nikolau BJ (2012) Genetic dissection of methylcrotonyl CoA carboxylase indicates a complex role for mitochondrial leucine catabolism during seed development and germination. Plant J 70: 562–577 [DOI] [PubMed] [Google Scholar]
- Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N, dePamphilis CW (2006) Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol 23: 469–478 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Espinoza C, Degenkolbe T, Caldana C, Zuther E, Leisse A, Willmitzer L, Hincha DK, Hannah MA (2010) Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS ONE 5: e14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Ito M, Nishida I, Watanabe A (2001) Leucine and its keto acid enhance the coordinated expression of genes for branched-chain amino acid catabolism in Arabidopsis under sugar starvation. FEBS Lett 499: 161–165 [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Sato T, Ito M, Watanabe A (2000) Isolation and characterization of cDNA clones for the e1β and E2 subunits of the branched-chain α-ketoacid dehydrogenase complex in Arabidopsis. J Biol Chem 275: 6007–6013 [DOI] [PubMed] [Google Scholar]
- Ganko EW, Meyers BC, Vision TJ (2007) Divergence in expression between duplicated genes in Arabidopsis. Mol Biol Evol 24: 2298–2309 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JH, Palacios N, Cross J, Selbig J, Stitt M (2004) A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Trapnell C, Kelley D (2012) CummeRbund: visualization and exploration of Cufflinks high-throughput sequencing data. R Package Version 2.2.0. http://www.bioconductor.org/packages/release/bioc/vignettes/cummeRbund/inst/doc/cummeRbund-manual.pdf (September 19, 2013)
- Gu L, Jones AD, Last R (2012) Rapid LC–MS/MS profiling of protein amino acids and metabolically related compounds for large-scale assessment of metabolic phenotypes. In Alterman MA, Hunziker P, eds, Amino Acid Analysis, Vol 828 Humana Press, New York, pp 1-11 [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL (2007) LC-MS/MS assay for protein amino acids and metabolically related compounds for large-scale screening of metabolic phenotypes. Anal Chem 79: 8067–8075 [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL (2010) Broad connections in the Arabidopsis seed metabolic network revealed by metabolite profiling of an amino acid catabolism mutant. Plant J 61: 579–590 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper AE, Miller RH, Block KP (1984) Branched-chain amino acid metabolism. Annu Rev Nutr 4: 409–454 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Larson TR, Schauer N, Fernie AR, Graham IA, Leaver CJ (2005) The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K, Schauer N, Larson TR, Graham IA, Fernie AR, Leaver CJ (2006) The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J 47: 751–760 [DOI] [PubMed] [Google Scholar]
- Izumi M, Wada S, Makino A, Ishida H (2010) The autophagic degradation of chloroplasts via Rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol 154: 1196–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Joshi V, Fraga M, Rugg A, Yu S, Li L, Last RL (2004) Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening; evidence that threonine aldolase plays a role in seed nutritional quality. Plant J 39: 465–475 [DOI] [PubMed] [Google Scholar]
- Jones E, Oliphant T, Peterson P (2001) SciPy: Open source scientific tools for Python. http://www.scipy.org/ (April 2, 2014)
- Joshi V, Laubengayer KM, Schauer N, Fernie AR, Jander G (2006) Two Arabidopsis threonine aldolases are nonredundant and compete with threonine deaminase for a common substrate pool. Plant Cell 18: 3564–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchi H, Shaul O, Galili G (1994) Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc Natl Acad Sci USA 91: 2577–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith B, Dong XN, Ausubel FM, Fink GR (1991) Differential induction of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase genes in Arabidopsis thaliana by wounding and pathogenic attack. Proc Natl Acad Sci USA 88: 8821–8825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kochevenko A, Araújo WL, Maloney GS, Tieman DM, Do PT, Taylor MG, Klee HJ, Fernie AR (2012) Catabolism of branched chain amino acids supports respiration but not volatile synthesis in tomato fruits. Mol Plant 5: 366–375 [DOI] [PubMed] [Google Scholar]
- Kotewicz ML, D’Alessio JM, Driftmier KM, Blodgett KP, Gerard GF (1985) Cloning and overexpression of Moloney murine leukemia virus reverse transcriptase in Escherichia coli. Gene 35: 249–258 [DOI] [PubMed] [Google Scholar]
- Lee M, Huang T, Toro-Ramos T, Fraga M, Last RL, Jander G (2008) Reduced activity of Arabidopsis thaliana HMT2, a methionine biosynthetic enzyme, increases seed methionine content. Plant J 54: 310–320 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2012) Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Ajjawi I, Imre KM, Yoder DW, Benning C, Dellapenna D, Ohlrogge JB, Osteryoung KW, Weber AP, (2008) New connections across pathways and cellular processes: industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiol 146: 1482–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Larson MD, Wilkerson CG, Last RL (2011) Chloroplast 2010: a database for large-scale phenotypic screening of Arabidopsis mutants. Plant Physiol 155: 1589–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutziger I, Oliver DJ (2001) Characterization of two cDNAs encoding mitochondrial lipoamide dehydrogenase from Arabidopsis. Plant Physiol 127: 615–623 [PMC free article] [PubMed] [Google Scholar]
- McCourt JA, Pang SS, King-Scott J, Guddat LW, Duggleby RG (2006) Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc Natl Acad Sci USA 103: 569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentzen WI, Peng J, Ransom N, Nikolau BJ, Wurtele ES (2008) Articulation of three core metabolic processes in Arabidopsis: fatty acid biosynthesis, leucine catabolism and starch metabolism. BMC Plant Biol 8: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Halkier BA (2003) Metabolic engineering of valine- and isoleucine-derived glucosinolates in Arabidopsis expressing CYP79D2 from Cassava. Plant Physiol 131: 773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Last RL (2012) Achieving diversity in the face of constraints: lessons from metabolism. Science 336: 1663–1667 [DOI] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J (2007) The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Mooney BP, Henzl MT, Miernyk JA, Randall DD (2000) The dihydrolipoyl acyltransferase (BCE2) subunit of the plant branched-chain α-ketoacid dehydrogenase complex forms a 24-mer core with octagonal symmetry. Protein Sci 9: 1334–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney BP, Miernyk JA, Randall DD (2002) The complex fate of α-ketoacids. Annu Rev Plant Biol 53: 357–375 [DOI] [PubMed] [Google Scholar]
- Oh SA, Park JH, Lee GI, Paek KH, Park SK, Nam HG (1997) Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J 12: 527–535 [DOI] [PubMed] [Google Scholar]
- Oliver DJ. (1994) The glycine decarboxylase complex from plant mitochondria. Annu Rev Plant Physiol Plant Mol Biol 45: 323–337 [Google Scholar]
- R Core Team (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Sarkar D, Sarkar MD (2007) The lattice Package. http://cran.r-project.org/web/packages/lattice/lattice.pdf (May 14, 2014)
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Schertl P, Braun HP (2014) Respiratory electron transfer pathways in plant mitochondria. Front Plant Sci 5: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schuster J, Binder S (2005) The mitochondrial branched-chain aminotransferase (AtBCAT-1) is capable to initiate degradation of leucine, isoleucine and valine in almost all tissues in Arabidopsis thaliana. Plant Mol Biol 57: 241–254 [DOI] [PubMed] [Google Scholar]
- Shariati SA, De Strooper B (2013) Redundancy and divergence in the amyloid precursor protein family. FEBS Lett 587: 2036–2045 [DOI] [PubMed] [Google Scholar]
- Singh BK. (1999) Biosynthesis of valine, leucine, and isoleucine. In Singh BK, ed, Plant Amino Acids: Biochemistry and Biotechnology. Marcel Dekker, New York, pp 227-247 [Google Scholar]
- Singh BK, Shaner DL (1995) Biosynthesis of branched chain amino acids: from test tube to field. Plant Cell 7: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocombe SP, Schauvinhold I, McQuinn RP, Besser K, Welsby NA, Harper A, Aziz N, Li Y, Larson TR, Giovannoni J, et al. (2008) Transcriptomic and reverse genetic analyses of branched-chain fatty acid and acyl sugar production in Solanum pennellii and Nicotiana benthamiana. Plant Physiol 148: 1830–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V (2008) The loci of evolution: how predictable is genetic evolution? Evolution 62: 2155–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székely G, Abrahám E, Cséplo A, Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, et al. (2008) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 53: 11–28 [DOI] [PubMed] [Google Scholar]
- Tan S, Evans R, Singh B (2006) Herbicidal inhibitors of amino acid biosynthesis and herbicide-tolerant crops. Amino Acids 30: 195–204 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH (2004) Lipoic acid-dependent oxidative catabolism of α-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiol 134: 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, Makino A (2009) Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol 149: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Han H, Vila-Aiub MM, Powles SB (2010) AHAS herbicide resistance endowing mutations: effect on AHAS functionality and plant growth. J Exp Bot 61: 3925–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Galili G (2003) Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Galili G (2004) Lysine metabolism is concurrently regulated by synthesis and catabolism in both reproductive and vegetative tissues. Plant Physiol 135: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Lehti-Shiu MD, Thomashow M, Shiu SH (2009) Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genet 5: e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.