Carotenoid biosynthesis is regulated by plant proximity (shade) signals through specific sets of positive and negative transcriptional regulators targeting the first gene of the pathway.
Abstract
Carotenoids are photosynthetic pigments essential for the protection against excess light. During deetiolation, their production is regulated by a dynamic repression-activation module formed by PHYTOCHROME-INTERACTING FACTOR1 (PIF1) and LONG HYPOCOTYL5 (HY5). These transcription factors directly and oppositely control the expression of the gene encoding PHYTOENE SYNTHASE (PSY), the first and main rate-determining enzyme of the carotenoid pathway. Antagonistic modules also regulate the responses of deetiolated plants to vegetation proximity and shade (i.e. to the perception of far-red light-enriched light filtered through or reflected from neighboring plants). These responses, aimed to adapt to eventual shading from plant competitors, include a reduced accumulation of carotenoids. Here, we show that PIF1 and related photolabile PIFs (but not photostable PIF7) promote the shade-triggered decrease in carotenoid accumulation. While HY5 does not appear to be required for this process, other known PIF antagonists were found to modulate the expression of the Arabidopsis (Arabidopsis thaliana) PSY gene and the biosynthesis of carotenoids early after exposure to shade. In particular, PHYTOCHROME-RAPIDLY REGULATED1, a transcriptional cofactor that prevents the binding of true transcription factors to their target promoters, was found to interact with PIF1 and hence directly induce PSY expression. By contrast, a change in the levels of the transcriptional cofactor LONG HYPOCOTYL IN FAR RED1, which also binds to PIF1 and other PIFs to regulate shade-related elongation responses, did not impact PSY expression or carotenoid accumulation. Our data suggest that the fine-regulation of carotenoid biosynthesis in response to shade relies on specific modules of antagonistic transcriptional factors and cofactors.
During their lifetime, plants are exposed to large variations in the quantity and quality of the incoming light that eventually determine their growth and development. Strong sunlight sometimes overwhelms the photosynthetic capacity of plants and, hence, leads to the production of highly reactive oxygen species that can potentially damage photosynthetic and cell structures. To cope with this danger, plant chloroplasts accumulate carotenoids, photoprotective compounds that channel energy away from chlorophylls and protect against reactive oxygen species and free radicals (Niyogi, 1999; Domonkos et al., 2013). On the other hand, photosynthesis and growth can be heavily compromised by the shading of nearby plants that compete for light. Phytochromes, a family of plant photoreceptors, are able to perceive changes in light quality associated with crowded (i.e. high-density) plant environments and to rapidly transduce them into changes in gene expression aimed to anticipate and avoid shading by overgrowing neighboring plants (i.e. promoting elongation growth), readjusting photosynthetic metabolism (i.e. decreasing the production of chlorophylls and carotenoids), or launching reproductive development (Franklin, 2008; Martinez-Garcia et al., 2010; Casal, 2013; Gommers et al., 2013).
Although regulating carotenoid biosynthesis and accumulation is central for plants to successfully adapt to changes in light quantity and quality, our understanding of how light cues regulate the accumulation of these essential metabolites is still limited (Ruiz-Sola and Rodríguez-Concepción, 2012). A first level of regulation of carotenoid accumulation in plants is the control of the transcription of biosynthetic genes. Recent reports have shown that the expression of the Arabidopsis (Arabidopsis thaliana) gene encoding PHYTOENE SYNTHASE (PSY), the first and main rate-determining enzyme of the carotenoid pathway (Ruiz-Sola and Rodríguez-Concepción, 2012), is under the direct control of two transcription factors involved in the transduction of light signals: PHYTOCHROME-INTERACTING FACTOR1 (PIF1) and LONG HYPOCOTYL5 (HY5; Toledo-Ortiz et al., 2010, 2014). PIF1 is a basic helix-loop-helix (bHLH) protein that, like other members of the so-called PIF quartet (formed by PIF1, PIF3, PIF4, and PIF5, collectively referred to as PIFq), accumulates in the dark and is degraded in the light (Leivar et al., 2008a; Leivar and Quail, 2011). By contrast, HY5 belongs to the basic Leu zipper (bZIP) family, accumulates in the light, and is degraded in the dark (Lau and Deng, 2010). PIFq and HY5 act antagonistically for a broad set of responses (Kami et al., 2010; Lau and Deng, 2010; Leivar and Quail, 2011; Chen et al., 2013), including the control of PSY expression and carotenoid biosynthesis (Toledo-Ortiz et al., 2010, 2014). PIF1 (repressor) and HY5 (activator) were demonstrated to bind to the same G-box motif in the promoter of PSY, forming a dynamic repression-activation transcriptional module that provides robustness in response to light but also to temperature cues (Toledo-Ortiz et al., 2014).
The combination of positive and negative regulators also appears to be instrumental in the responses of plants to shade (i.e. to light signals generated by the presence of nearby vegetation; Franklin, 2008; Martinez-Garcia et al., 2010; Casal, 2013). When sunlight is filtered through leaves or reflected from the surface of neighboring plants, the red light wavelengths of the spectrum are preferentially absorbed and the resulting light becomes enriched in far-red light. The reduction in the red light-far-red light ratio (R:FR) caused by plant proximity or canopy shade displaces the photoequilibrium toward the inactive form of phytochromes, hence inducing the so-called shade-avoidance syndrome (SAS) in shade-intolerant species like Arabidopsis and most crops. The four PIFq members and the photostable PIF7 protein have been demonstrated to directly contribute to the SAS (Lorrain et al., 2008; Hornitschek et al., 2012; Leivar et al., 2012; Li et al., 2012; Sellaro et al., 2012). Exposure to low R:FR promotes PIFq protein accumulation (Lorrain et al., 2008; Leivar et al., 2012) and binding of PIF7 to its target promoters (Li et al., 2012). Based on their role in promoting elongation, all PIFs are considered as positive regulators of the SAS (Lorrain et al., 2008; Leivar et al., 2012; Li et al., 2012). A decrease in the R:FR also triggers the production of negative regulators of hypocotyl elongation, including PHYTOCHROME-RAPIDLY REGULATED1 (PAR1), its paralog PAR2, and LONG HYPOCOTYL IN FAR RED1 (HFR1). These are helix-loop-helix proteins that lack a proper DNA-binding domain and act as transcriptional cofactors (Hornitschek et al., 2009; Galstyan et al., 2011, 2012): that is, they regulate gene expression by heterodimerizing with true bHLH transcription factors. For example, both PAR1 and HFR1 interact with PIFs and prevent their binding to their target promoters (Hornitschek et al., 2009; Hao et al., 2012; Shi et al., 2013). This dynamic balance of positive (PIFs) and negative (PAR1 and HFR1) regulators of the SAS was proposed to function as a gas-and-brake mechanism that prevents an excessive elongation in response to the initial low R:FR signal (Sessa et al., 2005; Roig-Villanova et al., 2007). Although a role for HY5 in the SAS has been proposed in plants grown under prolonged low R:FR (Sellaro et al., 2011; Ciolfi et al., 2013), the relevance of the antagonistic PIFq-HY5 module in the response to plant proximity remains to be determined.
Whereas the main output response of the SAS studied to date is elongation growth in seedlings, low R:FR also triggers a reduction in the levels of photosynthetic pigments (including carotenoids) in seedlings and adult plants (Roig-Villanova et al., 2007; Patel et al., 2013). Here, we investigated to what extent PIFs and their known antagonists in the control of light responses regulate carotenoid biosynthesis, and particularly PSY expression, in response to plant proximity signals.
RESULTS AND DISCUSSION
PIFq Proteins Repress PSY Gene Expression and Eventually Carotenoid Biosynthesis under Simulated Shade
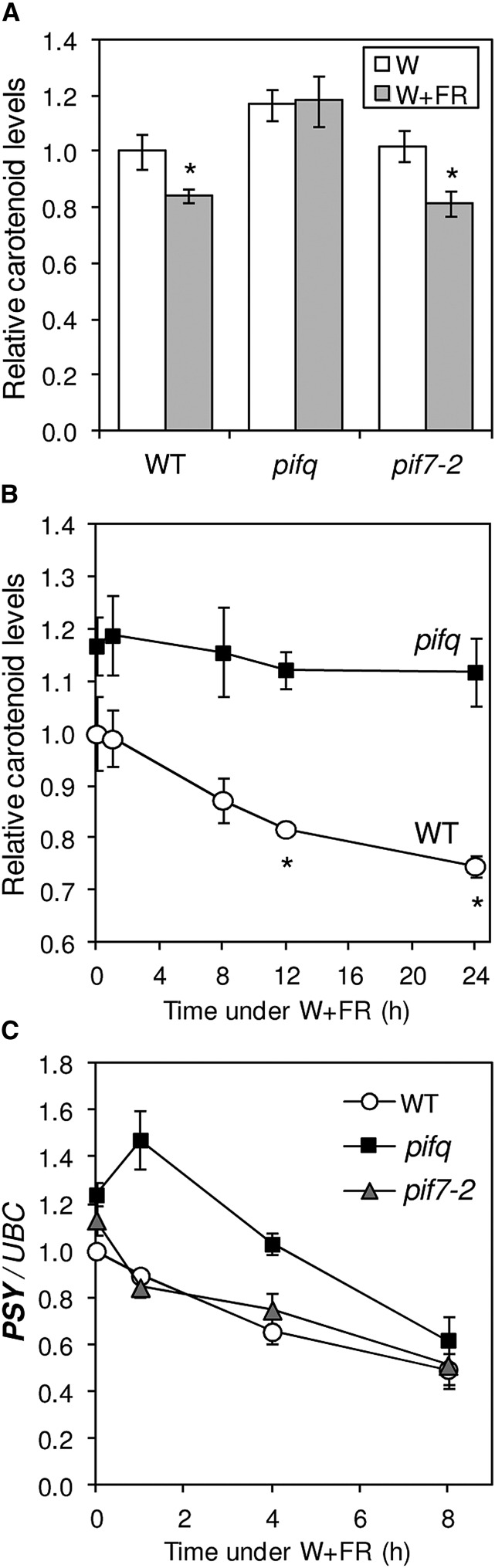
Treatment of Arabidopsis plants with low R:FR results in a fast increase in the level of PIFq proteins (Lorrain et al., 2008; Leivar et al., 2012) and a decreased accumulation of chlorophyll and carotenoid pigments (Roig-Villanova et al., 2007; Patel et al., 2013; Supplemental Fig. S1). To investigate whether PIFs participated in the control of carotenogenesis in response to shade, we used Arabidopsis wild-type (ecotype Columbia) plants, a knockout allele (pif7-2) impaired in PIF7 function (Leivar et al., 2008a), and a quadruple mutant line (pifq) defective in all four PIFq proteins (Leivar et al., 2008b). After germination and growth for 2 d under continuous white light (W), seedlings were either left under W or exposed to far-red light-supplemented white light (W+FR; a treatment referred to as simulated shade) for another 5 d. At day 7, W and W+FR samples were collected to quantify the accumulation of total carotenoids (Fig. 1). Treatment with simulated shade led to decreased carotenoid accumulation in wild-type plants but had no effect on the pifq mutant (Fig. 1A). By contrast, the pif7-2 mutant showed a virtually wild-type phenotype in terms of carotenoid accumulation under both W and W+FR (Fig. 1A). The higher carotenoid content in W-grown pifq plants is consistent with the proposed role for PIFq proteins as repressors of carotenoid biosynthesis (Toledo-Ortiz et al., 2010), whereas the lack of response to simulated shade indicates that PIFq activity is required to repress carotenoid accumulation in response to low R:FR (W+FR). These results together suggest that PIFq proteins, but not PIF7, are negative regulators of carotenoid biosynthesis both before and after the shade signal is perceived.
Figure 1.
Carotenoid and PSY transcript levels in wild-type and PIF-defective plants exposed to simulated shade. A, Carotenoids quantified in wild-type (WT), pifq, and pif7-2 seedlings grown for 2 d under W and then either left under W (white bars) or transferred to W+FR (gray bars) for an additional 5 d. B, Carotenoids measured in wild-type and pifq seedlings grown for 7 d under W and then treated with W+FR for the indicated times. C, Quantitative reverse transcription-PCR (qPCR) analysis of PSY transcript levels in wild-type and pifq seedlings grown as described in B. The UBIQUITIN-CONJUGATING ENZYME (UBC) gene (At5g25760) was used for normalization. Values are shown relative to those in W-grown wild-type samples and correspond to means and sd of biological triplicates (n = 3). The mean values for total carotenoid levels in W-grown wild-type samples were 111.03 µg g−1 fresh weight (A) and 119.14 µg g−1 fresh weight (B). Asterisks in A and B mark statistically significant differences (P < 0.05) relative to W-grown plants. In C, W+FR exposure led to statistically significant differences (P < 0.05) in transcript levels relative to untreated (0-h) samples for all the genes and genotypes tested.
Our previous work indicated that one of the mechanisms by which PIFq proteins control carotenogenesis is the direct repression of the transcriptional activity of the PSY gene (Toledo-Ortiz et al., 2010). To investigate the relevance of PIFq-mediated changes in PSY expression for the regulation of carotenoid biosynthesis in response to plant proximity light signals, the levels of both PSY transcripts and total carotenoids were quantified in W-grown wild-type and pifq seedlings at different time points after treatment with W+FR at day 7. Consistent with Figure 1A, a decline in carotenoid levels was observed in wild-type plants after exposure to simulated shade, whereas no significant (P < 0.05) changes occurred in the pifq mutant even after 24 h of W+FR illumination (Fig. 1B). PSY transcripts also declined in wild-type plants after transferring them to simulated shade (Fig. 1C). A virtually identical profile of PSY gene expression was observed in the pif7-2 mutant (Fig. 1C). By contrast, pifq mutant plants showed higher levels of PSY transcripts under W, as anticipated (Toledo-Ortiz et al., 2010), and a reproducible increase in PSY expression early (1 h) after W+FR exposure. At later time points, PSY transcript accumulation decreased to levels similar to those of wild-type plants (Fig. 1C), an unexpected finding based on the absence of changes in the levels of carotenoids observed in pifq seedlings (Fig. 1B). A possible explanation is that PSY protein stability is increased or its degradation rate is decreased in a PIFq-defective background. Another possibility is that high PIFq activity in wild-type plants grown under low R:FR eventually results in a reduced accumulation of carotenoids in a PSY-independent manner. Consistently, PIFq proteins are known to repress the development of chloroplasts and photosynthetic structures (Leivar et al., 2008b; Leivar and Quail, 2011; Zhang et al., 2013), hence decreasing carotenoid storage capacity. We conclude that PIFq (but not PIF7) proteins repress PSY early after treatment with simulated shade, whereas at later stages, they repress carotenoid accumulation by mechanisms that do not necessarily rely on controlling PSY expression.
HY5 Is Not Required for the Shade-Triggered Down-Regulation of Carotenoid Biosynthesis
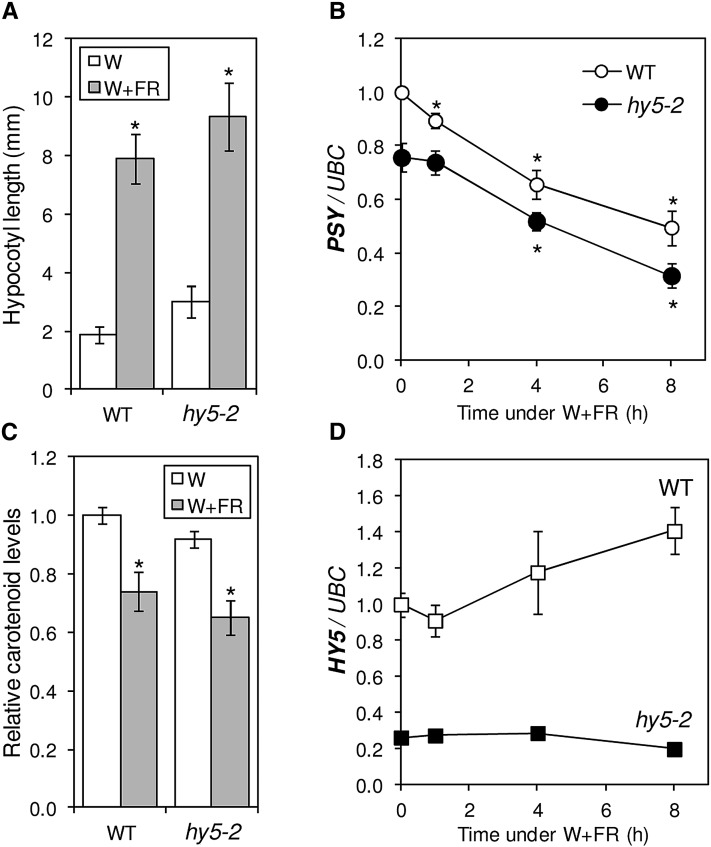
Because the combination of negative and positive regulators has been found to be instrumental in the SAS responses (Franklin, 2008; Martinez-Garcia et al., 2010; Casal, 2013), we next aimed to identify positive regulators of carotenoid biosynthesis that could counterbalance the negative role of PIFq proteins. The first selected candidate was HY5, because this transcription factor has been shown to act antagonistically with PIFq proteins in a number of physiological processes, including light-regulated elongation growth and carotenoid biosynthesis (Kami et al., 2010; Lau and Deng, 2010; Leivar and Quail, 2011; Chen et al., 2013; Toledo-Ortiz et al., 2014). In agreement with the described role of HY5 in repressing hypocotyl elongation in the light, wild-type plants grown under W showed shorter hypocotyls than the HY5-defective SALK_056405 line (Oh and Montgomery, 2013), here referred to as hy5-2 (Fig. 2). Also consistent with the proposed role of HY5 as an inducer of PSY expression and carotenoid biosynthesis, mutant hy5-2 plants showed significantly (P < 0.05) reduced levels of PSY transcripts (Fig. 2B) and carotenoids (Fig. 2C) under both W and W+FR compared with the wild type. But since wild-type and hy5-2 plants had very similar responses to simulated shade in terms of hypocotyl elongation, PSY expression, and carotenoid accumulation (Fig. 2), we conclude that HY5 is not required for the developmental (i.e. hypocotyl elongation) or the metabolic (i.e. carotenoid biosynthesis) responses to our W+FR treatment. In particular, the described repression-activation module formed by PIFq and HY5 proteins might not be relevant to regulate PSY expression in response to W+FR.
Figure 2.
Contribution of HY5 to shade-triggered responses. A, Hypocotyl elongation response in wild-type (WT) and hy5-2 seedlings grown for 2 d under W and then either kept in W (white bars) or transferred to W+FR (gray bars) for another 5 d. Hypocotyl length was measured using the ImageJ software (http://rsb.info.nih.gov/) on digital images. Columns represent means and sd of n = 2 independent experiments, each containing more than 30 seedlings of every genotype per treatment. B, qPCR analysis of PSY transcript levels in wild-type and hy5-2 seedlings grown for 7 d under W and then treated with W+FR for the indicated times. C, Carotenoid levels in seedlings grown as described in A. The mean amount in W-grown wild-type samples was 105.74 µg g−1 fresh weight. D, HY5 transcript levels in the samples described in B. Values in B to D are shown relative to those in W-grown wild-type samples and correspond to means and sd of biological triplicates (n = 3). Asterisks mark statistically significant differences (P < 0.05) relative to W-grown plants.
PAR1 Is a Positive Regulator of PSY Gene Expression and Carotenoid Accumulation during the SAS
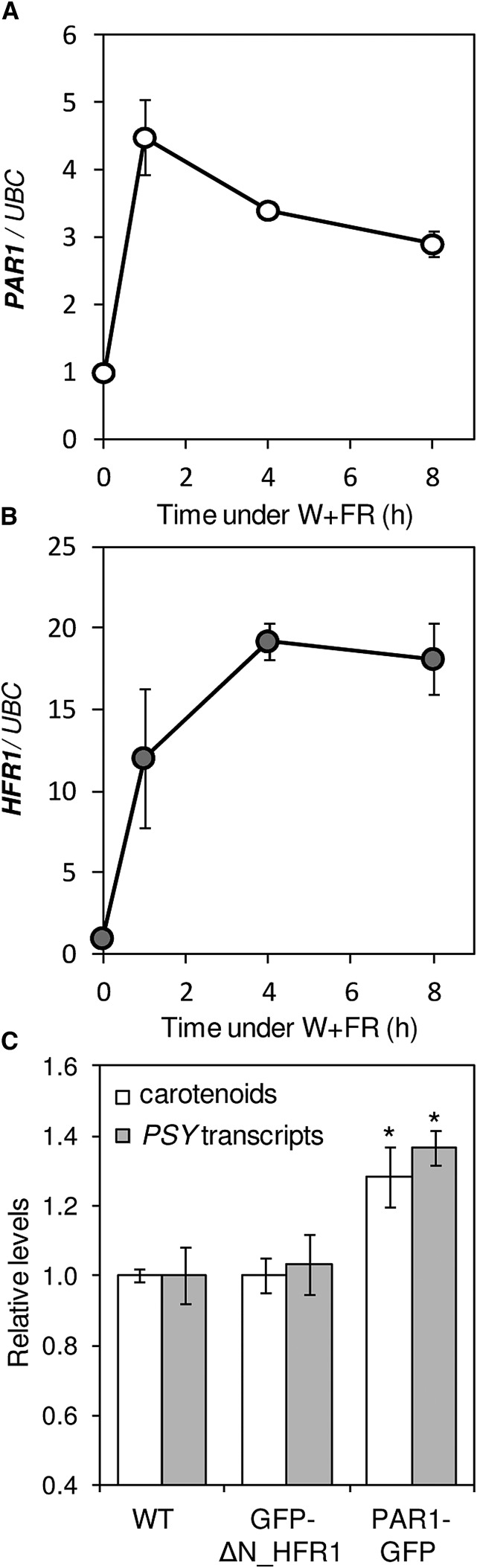
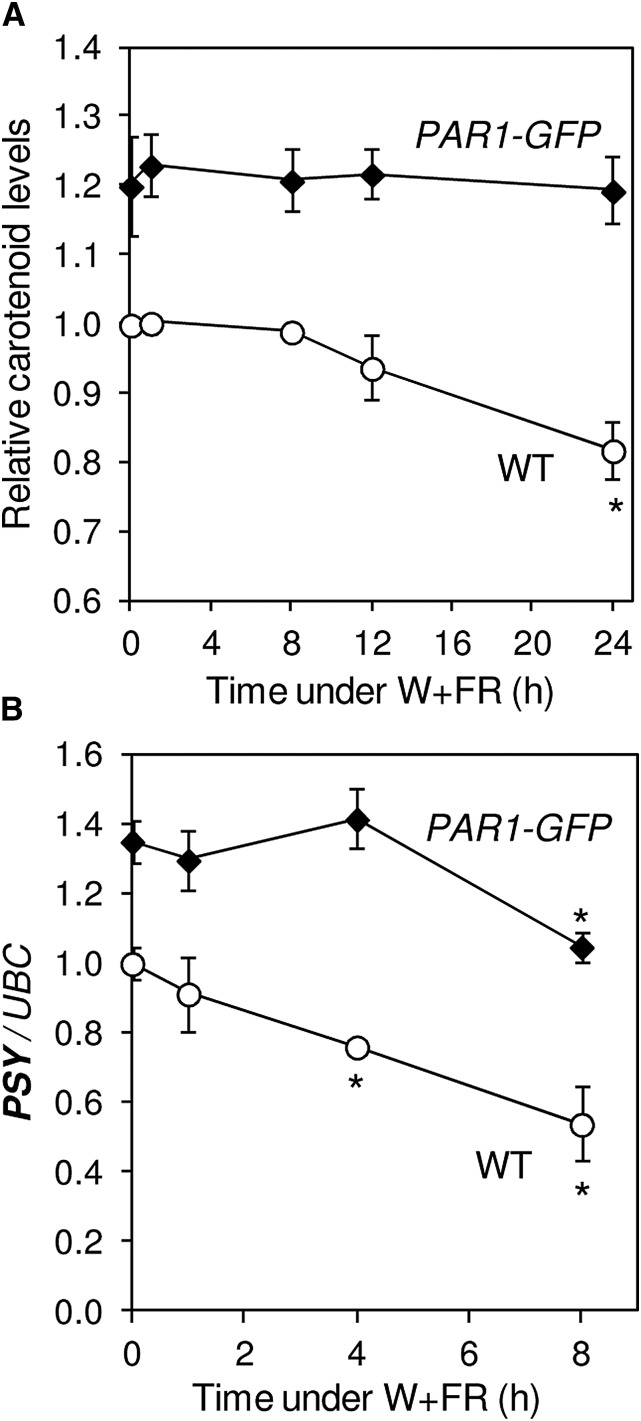
Besides HY5, other negative (i.e. antagonistic) regulators of PIF activity and SAS elongation responses are PAR1 and HFR1 (Sessa et al., 2005; Roig-Villanova et al., 2006, 2007; Hornitschek et al., 2009). But unlike HY5 expression, which only slightly increases at late time points (8 h) following W+FR treatment (Ciolfi et al., 2013; Fig. 2D), the expression of PAR1 and HFR1 is induced soon (1 h) after exposure to low R:FR (Fig. 3). Under our experimental conditions, PAR1 transcript levels peak early (1 h) after the W+FR treatment (Fig. 3A), similar to that observed for PSY transcripts in the pifq mutant (Fig. 1C), whereas HFR1 expression increases steadily when plants are transferred to simulated shade (Fig. 3B). Interestingly, an increase in PAR1 activity in transgenic plants producing the PAR1-GFP fusion protein (Roig-Villanova et al., 2007) results in increased PSY transcript levels compared with untransformed plants (Fig. 3C). By contrast, no changes in PSY expression were observed when HFR1 function was up-regulated in lines producing a GFP-tagged version of a truncated HFR1 protein (here referred to as GFP-ΔN_HFR1) previously shown to be more stable than the full-length protein and highly active in vivo (Galstyan et al., 2011). Consistent with these results, analysis of carotenoid levels showed increased contents in plants with enhanced PAR1 levels, but not in those with higher HFR1 activity, relative to untransformed controls (Fig. 3C). Therefore, we conclude that PSY gene expression and carotenoid biosynthesis are positively regulated by PAR1 but not by HFR1. When exposed to simulated shade, lines overproducing PAR1-GFP showed no changes in carotenoid levels and a strongly attenuated down-regulation of PSY transcript levels (Fig. 4). These results confirm a positive role for PAR1 in the regulation of PSY gene expression and carotenoid accumulation during the SAS. Moreover, the similar carotenoid-related phenotypes observed in lines with down-regulated PIFq levels (pifq mutant) or up-regulated PAR1 levels (PAR1-GFP-overexpressing lines) compared with the wild-type controls (compare Figs. 1 and 4) further supports the conclusion that PAR1 and PIFq proteins antagonistically contribute to the control of carotenoid accumulation both under normal light conditions and in response to shade. This antagonistic effect, however, was not observed in the dark (Supplemental Fig. S1). Comparison of carotenoid levels in etiolated seedlings of wild-type, pifq, and PAR1-GFP-overexpressing lines showed wild-type levels in the transgenic seedlings, whereas the absence of PIFq proteins in the mutant resulted in increased carotenoid levels, as reported previously (Toledo-Ortiz et al., 2010). A possible explanation is that, opposite to PIFq proteins, PAR1-GFP is degraded in darkness (Zhou et al., 2014).
Figure 3.
Contribution of PAR1 and HFR1 to the control of PSY expression and carotenoid biosynthesis. A, Levels of PAR1 transcripts quantified by qPCR analysis of RNA samples from wild-type seedlings grown for 7 d under W and then treated for the indicated times with W+FR. B, Levels of HFR1 transcripts in the samples described in A. Values in A and B are represented relative to those before the simulated shade treatment (0 h). Average and sd values of n = 3 independent samples are shown. C, Carotenoid levels (white bars) and PSY transcript abundance (gray bars) in plants producing the fusion proteins GFP-ΔN_HFR1 and PAR1-GFP and in wild-type controls (WT) grown under W for 7 d. Values are shown relative to those in wild-type samples (the mean carotenoid amount was 112.84 µg g−1 fresh weight) and correspond to means and sd of n = 3 independent samples. Asterisks mark statistically significant differences (P < 0.05) relative to wild-type plants.
Figure 4.
Carotenoid accumulation and PSY expression in plants with increased PAR1 levels. A, Carotenoids measured in wild-type (WT) plants and transgenic lines overexpressing a GFP-tagged PAR1 protein (PAR1-GFP). Plants were grown for 7 d under W and then treated with W+FR for the indicated times. The mean carotenoid level in W-grown wild-type plants was 105.17 µg g−1 fresh weight. B, PSY transcript levels quantified by qPCR analysis of RNA samples from the plants described in A. Values are shown relative to those in W-grown wild-type samples and correspond to means and sd of n = 3 experiments. Asterisks mark statistically significant differences (P < 0.05) relative to W-grown plants.
PAR1 Can Interact with PIF1 and Prevent Its Binding to the PSY Promoter to Directly Induce Gene Expression
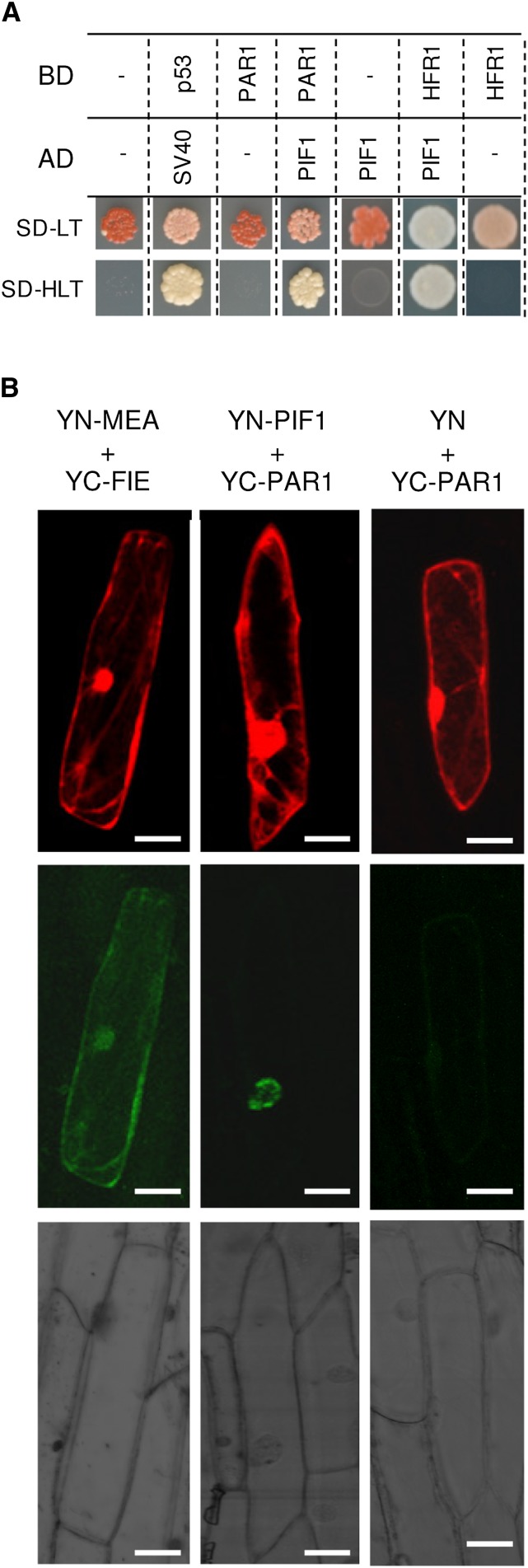
Both PAR1 and HFR1 directly regulate gene expression by heterodimerizing with true bHLH transcription factors such as PIFs to prevent their binding to the promoters of target genes (Hornitschek et al., 2009; Carretero-Paulet et al., 2010; Galstyan et al., 2011; Hao et al., 2012; Cifuentes-Esquivel et al., 2013; Shi et al., 2013). In particular, HFR1 has been shown to interact with PIF1 and PIF3 in vitro and in the yeast two-hybrid (Y2H) assay (Fairchild et al., 2000; Bu et al., 2011), whereas coimmunoprecipitation and bimolecular fluorescence complementation (BiFC) assays demonstrated its interaction with PIF1, PIF4, and PIF5 in vivo (Hornitschek et al., 2009; Shi et al., 2013). PAR1 was recently shown to bind to PIF4 in vivo to prevent its binding to DNA for transcriptional activity (Hao et al., 2012). To test whether PAR1 could also interact with PIF1, the main repressor of PSY (Toledo-Ortiz et al., 2010), we carried out two complementary assays, Y2H and BiFC, using HFR1 as a positive control (Fig. 5; Supplemental Fig. S2). The Y2H assay showed a clear interaction between PAR1 and PIF1 (Fig. 5A), whereas BiFC experiments performed by microbombarding leek (Allium ampelloprasum) epidermal cells confirmed that PAR1 binds PIF1 in the nucleus (Fig. 5B). Together, these results show that PAR1 can physically interact with PIF1 in vivo.
Figure 5.
Interaction of PAR1 with PIF1 in vivo. A, Y2H assay of the indicated proteins fused to the activation domain (AD) or the binding domain (BD) of GAL4. Mating cells (positive transformants) were selected on synthetically defined medium (SD) lacking Leu and Trp (SD-LT), whereas protein-protein interactions were assessed on medium that also lacked His (SD-HLT). Proteins p53 and SV40 are known to interact and were used as a positive control. B, BiFC assay of the indicated proteins fused to the N-terminal domain (YN) or the C-terminal domain (YC) of yellow fluorescent protein (YFP). Leek cells were cobombarded with plasmids to express the indicated fusion proteins and an additional vector producing the DsRed protein as a marker of cells expressing the microbombarded constructs. The top row shows the DsRed fluorescence, the middle row shows the fluorescence of reconstituted YFP (indicative of a positive interaction of YN and YC fusions), and the bottom row shows the bright-field images of the same area. Proteins MEDEA (MEA) and FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), known to interact in both the nucleus and the cytoplasm of plant cells (Bracha-Drori et al., 2004), were used as a positive control. A negative empty plasmid control is also shown in the right column. Additional controls are shown in Supplemental Figure S2. Bars = 20 μm.
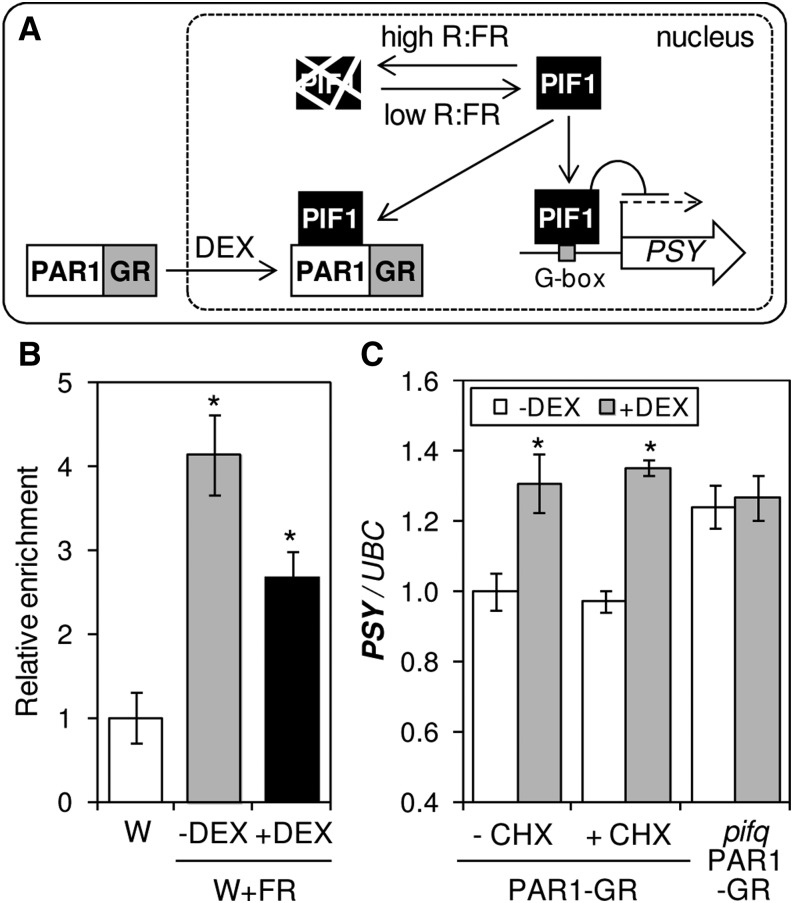
To next determine whether the interaction of PAR1 and PIF1 could modulate PSY expression in response to shade, chromatin immunoprecipitation assays were carried out using double transgenic Arabidopsis lines constitutively producing the fusion proteins PAR1-GR and TAP-PIF1 (Fig. 6). PAR1-GR, a fusion of PAR1 to the glucocorticoid receptor (GR), was previously shown to be biologically active when targeted to the nucleus upon treatment of transgenic plants with dexamethasone (DEX), a synthetic glucocorticoid (Roig-Villanova et al., 2007; Fig. 6A). The TAP-PIF1 protein contains a tandem affinity purification (TAP) tag with nine copies of the MYC repeat that allows immunoprecipitation of PIF1-bound chromatin regions using an anti-MYC serum (Toledo-Ortiz et al., 2010, 2014). After germination and growth for 2 d under W, double transgenic seedlings were either left under W or exposed to W+FR for another 5 d in the presence or absence of DEX. At day 7, samples were collected and protein-DNA complexes formed by TAP-PIF1 were immunoprecipitated. Enriched DNA sequences were then quantified by qPCR using primers that amplified the PIF1 target sequence on the PSY promoter (Toledo-Ortiz et al., 2010). As shown in Figure 6B, W+FR treatment resulted in a higher (P < 0.05) proportion of immunoprecipitated PIF1-bound PSY promoter sequences. Therefore, it is likely that an enhanced accumulation of the PIF1 protein under simulated shade conditions results in enhanced binding to the PSY promoter. Moreover, treatment with both W+FR and DEX caused a reduction in the level of immunoprecipitated PSY promoter sequences compared with samples treated only with W+FR (Fig. 6B). These results show that increased PAR1 levels in the nucleus interfere with PIF1 binding to the PSY promoter (Fig. 6A). Because PIF1 is a repressor of PSY expression (Toledo-Ortiz et al., 2010), PAR1 would then act as a positive regulator of PSY expression, consistent with the increased levels of PSY transcripts observed in light-grown PAR1-GFP-overexpressing lines (Figs. 3C and 4B).
Figure 6.
PAR1 is a direct positive regulator of PSY expression. A, Proposed model for the direct regulation of PSY gene expression by PIF1 and PAR1. PIF1 is unstable under high R:FR (W) but accumulates under low R:FR achieved by simulated shade (W+FR) treatment, eventually binding to the PSY promoter to repress it. However, when PAR1 levels are high (e.g. when nuclear translocation of the chimeric PAR1-GR protein is promoted by DEX treatment), interaction with PIF1 prevents its binding to the promoter and PSY expression increases. B, Chromatin immunoprecipitation assay of the influence of PAR1 on PIF1 binding to the PSY promoter. Double transgenic lines producing the fusion proteins TAP-PIF1 and PAR1-GR were grown for 2 d under W and then either left under W (white column) or exposed to W+FR for another 5 d in the presence (black column) or absence (gray column) of DEX. Chromatin immunoprecipitation was carried out using an anti-MYC antibody recognizing the tandem affinity purification tag. A nonantibody control sample was processed in parallel in each case. Immunoprecipitated DNA was analyzed by qPCR using specific primers covering the PIF1-binding motif in the PSY promoter. Results are represented relative to W-grown samples after normalization with the nonantibody control. Columns represent means and sd of biological triplicates. C, Single transgenic plants expressing only the PAR1-GR fusion in a wild-type or a PIF-defective (pifq) background were grown for 7 d under W and then treated (+) with DEX, cycloheximide (CHX), and/or mock solutions (−). Samples were collected 4 h after treatment to quantify PSY transcript levels. Values are shown relative to those in mock-treated PAR1-GR (wild-type) samples and correspond to means and sd of biological triplicates (n = 3). Asterisks mark statistically significant differences (P < 0.05) relative to mock samples.
To further confirm that PAR1 directly activates PSY expression by preventing the binding of transcriptional repressors like PIF1 and likely other PIFq proteins to the promoter, we used transgenic Arabidopsis lines producing the PAR1-GR protein in a genetic background in which PIFq proteins were either present (wild type) or absent (pifq). As shown in Figure 6C, treatment of W-grown PAR1-GR seedlings with DEX for 4 h resulted in a significant (P < 0.05) increase in PSY transcript levels compared with untreated plants, whereas no changes were detected in pifq PAR1-GR seedlings. This result confirms that PSY expression can be up-regulated when PAR1 levels increase in the nucleus and that this response requires the presence of PIFq proteins. Moreover, treatment of PAR1-GR plants with the protein synthesis inhibitor cycloheximide had no effect on the DEX-mediated increase of PSY transcript levels (Fig. 6C). Together, our data support the conclusion that PAR1 directly activates PSY gene expression by forming heterodimers with PIF1 and likely other PIFq proteins that repress PSY expression, hence preventing the binding of these transcription factors to their target sequences in the PSY promoter (Fig. 6A).
Specific Interactions of PAR1 and HFR1 with Distinct Subsets of Transcription Factors Might Control Different SAS Responses
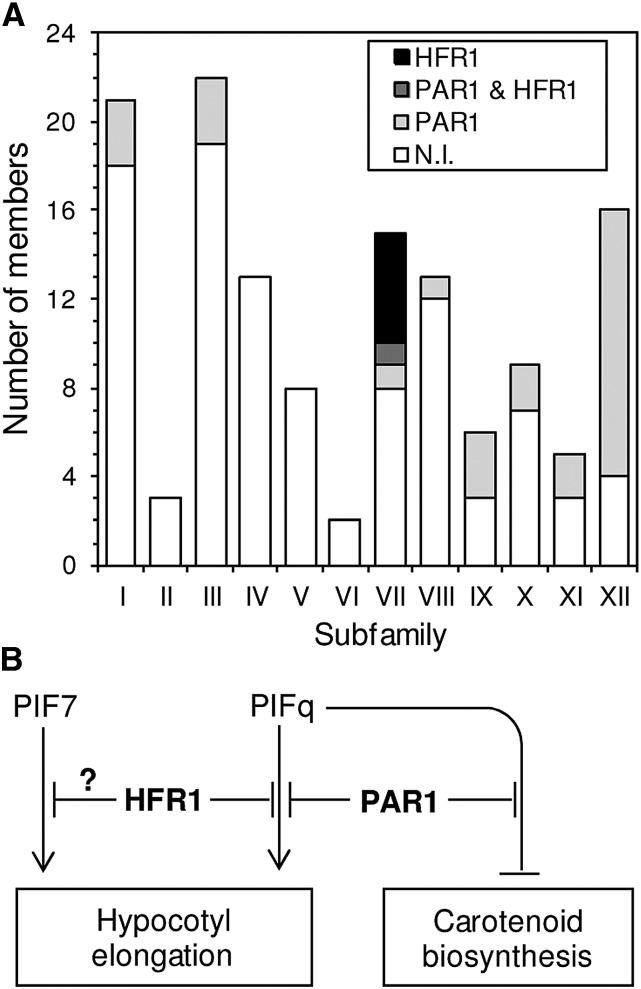
Because both PAR1 and HFR1 can bind to PIF1 (Fig. 5; Supplemental Fig. S2) and other PIFq proteins but only PAR1 regulates PSY gene expression and carotenoid biosynthesis in plants (Fig. 3C), it can be speculated that the PAR1-PIF and HFR1-PIF interaction modules might somehow unleash different signaling pathways that eventually regulate common sets of responses (such as those causing hypocotyl elongation) but also specific ones (like carotenoid biosynthesis). It is also possible that PAR1 and HFR1 might differentially interact with other bHLH transcription factors that could further contribute to provide the observed specificity in terms of SAS responses. As a first step to understand the molecular mechanism behind the differential role of PAR1 and HFR1 on the regulation of PSY expression and carotenoid biosynthesis, we aimed to compare the profiles of transcription factors that could potentially interact with these transcriptional cofactors. First, we searched for bHLH family proteins among those identified in a previous Y2H screening for PAR1-interacting partners (Cifuentes-Esquivel et al., 2013). As shown in Supplemental Table S1, 28 partners of PAR1 were found in eight of the 12 subfamilies described for bHLH proteins (Heim et al., 2003; Fig. 7). The interaction of PAR1 with individual members of subfamilies VII (PIF4 and HFR1), VIII (PAR1 and PAR2), and XII (BRASSINOSTEROID ENHANCED EXPRESSION1 [BEE1], BEE2, and BEE3) has been experimentally confirmed (Carretero-Paulet et al., 2010; Hao et al., 2012; Cifuentes-Esquivel et al., 2013). Additionally, previous work showed that PAR1 can also interact with other bHLH proteins not identified in this screen, including the atypical non-DNA-binding protein PACLOBUTRAZOL RESISTANCE1 (PRE1; Hao et al., 2012) and the subfamily V member BES1-INTERACTING MYC-LIKE1 (BIM1; Cifuentes-Esquivel et al., 2013), suggesting that the variety of bHLH targets of PAR1 action might be even wider than that unveiled by the Y2H screen.
Figure 7.
Differential roles for PAR1 and HFR1 during the SAS. A, Number of members of each bHLH subfamily found to interact with PAR1, HFR1, both transcription cofactors, and none of them (N.I.) in Y2H experiments. B, Model for antagonistic transcriptional modules regulating SAS responses. Low R:FR up-regulates the activity of transcription factors like PIFq and PIF7 and the expression of transcription cofactors like PAR1 and HFR1 (in boldface). Repression-activation modules formed by the indicated proteins thus modulate the induction of genes required for elongation growth and the repression of genes involved in carotenoid accumulation (including PSY).
A similar Y2H screening for HFR1-interacting partners identified only six bHLH proteins, all of them belonging to subfamily VII (Supplemental Table S2). They include all four PIFq proteins, previously shown to be able to heterodimerize with HFR1 (Fairchild et al., 2000; Hornitschek et al., 2009; Bu et al., 2011; Shi et al., 2013). Y2H screenings identified only one bHLH protein (PIF5) as a common target for PAR1 and HFR1 (Fig. 7A). However, other PIFq proteins (PIF1 and PIF4) have been found to also be able to interact with both transcriptional cofactors (Hornitschek et al., 2009; Hao et al., 2012; Shi et al., 2013; Fig. 5; Supplemental Fig. S2). Nevertheless, the results suggest that HFR1 might be much more target specific than PAR1. The observation that HFR1 appears to preferentially bind PIFs (i.e. members of its own subfamily) whereas PAR1 interacts with PIFs but also with a wide variety of other bHLH proteins (Fig. 7A) suggests that the impact on PSY expression and carotenoid accumulation caused by increasing the levels of PAR1 (but not HFR1) might be further mediated by non-PIF bHLH transcription factors that interact with PAR1 but not with HFR1. Consistent with this conclusion, the peak in PAR1 gene expression observed early (1 h) after W+FR treatment (Fig. 3A) correlates with a similar up-regulation of PSY transcript levels in the absence of PIFq proteins (i.e. in the pifq mutant; Fig. 1C). Identifying specific non-PIFq PSY repressors antagonized by PAR1 upon heterodimerization, however, will require further work.
CONCLUSION
Our data show that PIFq proteins (but not PIF7) are repressors of PSY expression and carotenoid biosynthesis both under W and in response to shade (Fig. 1). Besides stimulating PIF activity, low R:FR perception results in a strong up-regulation of genes coding for SAS antagonists, including the transcription cofactors PAR1 and, later, HFR1 (Sessa et al., 2005; Roig-Villanova et al., 2006, 2007; Hornitschek et al., 2009). HFR1 does not appear to regulate PSY expression or carotenoid biosynthesis (Fig. 3). By contrast, PAR1 promotes carotenoid biosynthesis and directly induces PSY expression (Fig. 4), most likely by forming nonfunctional heterodimers with PIFq proteins such as PIF1 (Figs. 5 and 6) and, perhaps, with other bHLH transcription factors that might act as PSY repressors (Fig. 7A). The fast and transient up-regulation of PAR1 expression that takes place after simulated shade exposure (Fig. 3A), therefore, might act as an early break to prevent an excessive reduction in PSY expression, allowing the plant to rapidly resume carotenoid biosynthesis if the low R:FR signal disappears (e.g. if a commitment to the shade-avoidance lifestyle is unnecessary). If the low R:FR signal persists, carotenoid accumulation decreases independently of PSY expression (Fig. 1). Our results with the hy5-2 mutant suggest that the antagonistic PIFq-HY5 module previously described to control PSY expression and carotenoid biosynthesis in response to environmental (light and temperature) signals (Toledo-Ortiz et al., 2014) might not be relevant during the SAS. Consistently, PAR1 and HY5 have been shown to act in separate pathways to inhibit hypocotyl elongation under different light conditions (Zhou et al., 2014).
Our data further suggest that specific modules of bHLH family members might control different sets of SAS-associated responses (Fig. 7B). Thus, PIF7 and HFR1 are key regulators of shade-induced elongation responses (Sessa et al., 2005; Li et al., 2012) but have no apparent role in regulating carotenoid biosynthesis, whereas PIFq and PAR1 proteins regulate both SAS responses. We propose that activation-suppression transcriptional modules formed by PAR1-PIFq proteins likely exert a dynamic control of early events in the regulation of both hypocotyl growth and carotenoid biosynthesis in response to low R:FR, whereas HFR1-PIFq and potentially HFR1-PIF7 modules might specifically control elongation responses (Fig. 7B). The lack of correlation between shade-induced hypocotyl elongation and flowering initiation in over 100 Arabidopsis ecotypes (Botto and Smith, 2002) previously supported the conclusion that different signaling pathways control distinct SAS responses (Martinez-Garcia et al., 2010). The results presented here, however, go a step beyond by highlighting the coexistence of different shade signaling pathways controlling several SAS responses (hypocotyl elongation and carotenoid accumulation) at the very same stage of development (photosynthetic seedlings).
In summary, we conclude that different modules formed by activators and repressors are recruited to regulate PSY expression and eventually carotenoid biosynthesis in response to low R:FR. Some of them regulate other responses to the same signal (e.g. the PAR1-PIFq module also controls hypocotyl elongation in response to shade). Most strikingly, individual module components do not always regulate carotenoid biosynthesis in other developmental stages (e.g. PAR1 does not appear to control carotenoid production in etiolated seedlings; Supplemental Fig. S1) or in response to different stimuli (e.g. the burst in PSY transcript accumulation that takes place in Arabidopsis roots exposed to a salt stress does not require the activity of PIFq proteins; Ruiz-Sola et al., 2014). Future work should help to unveil the complexity underlying the control of carotenoid biosynthesis in order to better understand and manipulate the carotenoid profiles of particular tissues in response to specific treatments.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All the Arabidopsis (Arabidopsis thaliana) lines used in this work are in the Columbia background. Transgenic lines producing PAR1-GFP, PAR1-GR, and GFP-ΔN_HFR1 have been described previously (Roig-Villanova et al., 2007; Galstyan et al., 2011). Double transgenic lines producing PAR1-GR and TAP-PIF1 proteins were generated by crossing the corresponding single transgenic lines (Roig-Villanova et al., 2007; Toledo-Ortiz et al., 2010). Single pif7-2 (Leivar et al., 2008a) and quadruple pifq (Leivar et al., 2008b) mutants were also available in the laboratory. The pifq PAR1-GFP lines were constructed by transforming pifq plants with the PAR1-GR construct (Roig-Villanova et al., 2007). A DEX-responsive homozygous pifq PAR1-GFP line displaying PAR1-GR transcript levels similar to those in the original PAR1-GR line (in a wild-type background) was selected for the experiments. The SALK_056405 line (here referred to as hy5-2), previously shown to be a knockout mutant (Oh and Montgomery, 2013), was obtained from the Nottingham Arabidopsis Stock Centre.
Seeds were surface sterilized and sown on petri dishes with solid growth medium without Suc (Roig-Villanova et al., 2007). When comparing different lines (e.g. wild type versus mutant), they were grown together on the same plate instead of growing each line on a different plate. After stratification for at least 3 d at 4°C in the dark, the plates were incubated in growth chambers at 22°C under continuous W (25 μmol m–2 s–1 photosynthetically active radiation, R:FR of 2.1–3.5). Simulated shade (W+FR; 25 μmol m–2 s–1 photosynthetically active radiation, R:FR of 0.05) was generated by enriching W with supplementary far-red light provided by QB1310CS-670-735 light-emitting diode hybrid lamps (Quantum Devices). Fluence rates were measured using an EPP2000 spectrometer (StellarNet) as described (Sorin et al., 2009). For experiments involving pharmacological treatments, seeds were germinated and grown under W on sterile filter paper placed on top of solid growth medium in petri dishes. At the indicated time, the filter paper with the seedlings was transferred to fresh plates containing cycloheximide and/or DEX as described (Roig-Villanova et al., 2007). For seed production, plants were grown in the greenhouse under long-day conditions.
Quantification of Transcript and Carotenoid Levels
Whole seedlings were ground in liquid nitrogen, and part of the resulting powder was used for the quantification of carotenoid levels by HPLC or spectrometric methods as described (Rodríguez-Villalón et al., 2009). HPLC separation and quantification of carotenoids and chlorophylls were performed in an initial set of experiments. Once confirmed that all individual carotenoids showed similar responses to simulated shade (Supplemental Fig. S1), total carotenoid levels were estimated spectrophotometrically (Lichtenthaler, 1987). Concentration was calculated relative to fresh weight. The rest of the powder was used for RNA extraction using the RNeasy Plant Mini Kit (Qiagen). The synthesis of complementary DNA (cDNA) from DNase-treated RNA was carried out using the Transcriptor First-Strand cDNA Synthesis Kit (Roche). The qPCR experiments were performed as described (Rodríguez-Villalón et al., 2009) using Fast Start Universal SYBR Green Master Mix (Roche) on a Light Cycler 480 apparatus (Roche). The UBC (At5g25760) gene was used for normalization (Czechowski et al., 2005). Primer sequences for qPCR are listed in Supplemental Table S3. Two-tailed Student’s t tests were performed to analyze statistical differences.
Protein-DNA and Protein-Protein Interaction Analyses
Chromatin immunoprecipitation assays were carried out as described (Toledo-Ortiz et al., 2014). The Y2H screen using HFR1 as a bait was performed by Hybrigenics (www.hybrigenics-services.com) as described previously for PAR1 (Cifuentes-Esquivel et al., 2013). Briefly, the coding sequence for HFR1 was cloned into pB27 as a C‐terminal fusion to LexA, and the resulting construct (pB27-LexA‐HFR1) was used as a bait to screen a random‐primed Arabidopsis cDNA library from 7-d-old wild-type seedlings grown at 24°C under a long-day photoperiod. Screening of 76.6 million clones was carried out using a mating approach, and positive clones were used to sequence the corresponding prey inserts by PCR. Supplemental Tables S1 and S2 summarize the results for bHLH proteins found to interact with PAR1 and HFR1, respectively, including a confidence score (predicted biological score) attributed to each interaction.
Individual Y2H assays were performed by cell mating with the Matchmaker system (Clontech). Yeast (Saccharomyces cerevisiae) cells of two different strains, the MATa Leu auxotroph YM4271a (Liu et al., 1993) and the MATɑ Trp auxotroph PJ694α (Uetz et al., 2000), were transformed with constructs to express fusion proteins to either the GAL4 activation domain (AD) or binding domain (BD), respectively, and selected in appropriate medium. PIF1 was fused to the activation domain by subcloning the full-length PIF1 cDNA into the pGADT7 plasmid, yielding pJB62 [AD-PIF1]. A BD-PAR1 fusion in plasmid pCL1 was available (Galstyan et al., 2011), whereas a similar construct was generated for HFR1 by subcloning the full-length HFR1 cDNA into the pGBKT7 plasmid to yield pJB37 [BD-HFR1]. Independent transformants were selected on SD lacking either Leu (pJB62) or Trp (pCL1 and pJB37), grown in liquid medium, and then allowed to mate by mixing equal volumes of the two types of transformed yeast cells. Mated cells were selected on SD-Leu-Trp and then transferred to SD-His-Leu-Trp to test for protein-protein interaction. Mating was repeated at least twice, with identical results.
For BiFC experiments, cDNA sequences encoding PIF1, PAR1, and HFR1 proteins were fused to the N-terminal or C-terminal regions of YFP using appropriate vectors (Bracha-Drori et al., 2004). The full-length PIF1 cDNA was cloned into pSY736 to yield pJB72 [YN-PIF1]. Plasmid pSY735 was used to construct vectors pMS2 [YC-PAR1] and pCN6 [YC-HFR1] as described (Cifuentes-Esquivel et al., 2013). Leek (Allium ampelloprasum) epidermal peels were microbombarded with DNA-coated gold microcarriers using a Biolistic PDS-1000/helium system (Bio-Rad) and incubated at 22°C in the dark for 24 h prior to observation with an Olympus FV1000-ASW confocal laser scanning microscope.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. HPLC profiles and quantification of carotenoid and chlorophyll pigments.
Supplemental Figure S2. BiFC assay of the interaction of HFR1 and PIF1.
Supplemental Table S1. PAR1-interacting bHLH proteins identified in the Y2H screen.
Supplemental Table S2. HFR1-interacting bHLH proteins identified in the Y2H screen.
Supplemental Table S3. Primers used in this work for qPCR analyses.
Supplementary Material
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre for seeds from mutant lines, Dr. Luis Oñate for Y2H yeast strains, and M. Rosa Rodríguez-Goberna and members of the Centre for Research in Agricultural Genomics Services for technical support.
Glossary
- bHLH
basic helix-loop-helix
- SAS
shade-avoidance syndrome
- R:FR
red light-far-red light ratio
- W
white light
- W+FR
far-red light-supplemented white light
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- DEX
dexamethasone
- qPCR
quantitative reverse transcription-PCR
- cDNA
complementary DNA
- SD
synthetically defined medium
Footnotes
This work was supported by the Spanish Dirección General de Investigación (grant nos. BIO2011–23680 [including an FPI fellowship for M.O.-A.] and BIO2011–23489), the Consejo Superior de Investigaciones Científicas (JAE-DOC contract for G.T.-O.), the Gobierno de Chile (predoctoral fellowship to N.C.-E.), the Generalitat de Catalunya (grant nos. 2014SGR1434 and 2014SGR447), the Programa Iberoamericano de Ciencia y Tecnologia para el Desarrollo (IBERCAROT), the U.K. Biotechnology and Biological Sciences Research Council (ROBuST, grant no. BB/F005237/1 to K.J.H.), and the European Union FP7 (TiMet, contract no. 245143) and Marie Curie (grant no. PCIG11–GA–2012–321649) programs.
Articles can be viewed without a subscription.
References
- Botto JH, Smith H (2002) Differential genetic variation in adaptive strategies to a common environmental signal in Arabidopsis accessions: phytochrome-mediated shade avoidance. Plant Cell Environ 25: 53–63 [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Bu Q, Castillon A, Chen F, Zhu L, Huq E (2011) Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol Biol 77: 501–511 [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153: 1398–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Esquivel N, Bou-Torrent J, Galstyan A, Gallemí M, Sessa G, Salla Martret M, Roig-Villanova I, Ruberti I, Martínez-García JF (2013) The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J 75: 989–1002 [DOI] [PubMed] [Google Scholar]
- Ciolfi A, Sessa G, Sassi M, Possenti M, Salvucci S, Carabelli M, Morelli G, Ruberti I (2013) Dynamics of the shade-avoidance response in Arabidopsis. Plant Physiol 163: 331–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domonkos I, Kis M, Gombos Z, Ughy B (2013) Carotenoids, versatile components of oxygenic photosynthesis. Prog Lipid Res 52: 539–561 [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Franklin KA. (2008) Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Galstyan A, Bou-Torrent J, Roig-Villanova I, Martínez-García JF (2012) A dual mechanism controls nuclear localization in the atypical basic-helix-loop-helix protein PAR1 of Arabidopsis thaliana. Mol Plant 5: 669–677 [DOI] [PubMed] [Google Scholar]
- Galstyan A, Cifuentes-Esquivel N, Bou-Torrent J, Martinez-Garcia JF (2011) The shade avoidance syndrome in Arabidopsis: a fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J 66: 258–267 [DOI] [PubMed] [Google Scholar]
- Gommers CM, Visser EJ, St Onge KR, Voesenek LA, Pierik R (2013) Shade tolerance: when growing tall is not an option. Trends Plant Sci 18: 65–71 [DOI] [PubMed] [Google Scholar]
- Hao Y, Oh E, Choi G, Liang Z, Wang ZY (2012) Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol Plant 5: 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH (2008a) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008b) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH (2012) Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 148: 351–382 [Google Scholar]
- Liu H, Styles CA, Fink GR (1993) Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262: 1741–1744 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Galstyan A, Salla-Martret M, Cifuentes-Esquivel N, Gallemi M, Bou-Torrent J (2010) Regulatory components of shade avoidance syndrome. Adv Bot Res 53: 65–116 [Google Scholar]
- Niyogi KK. (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50: 333–359 [DOI] [PubMed] [Google Scholar]
- Oh S, Montgomery BL (2013) Phytochrome-induced SIG2 expression contributes to photoregulation of phytochrome signalling and photomorphogenesis in Arabidopsis thaliana. J Exp Bot 64: 5457–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D, Basu M, Hayes S, Majláth I, Hetherington FM, Tschaplinski TJ, Franklin KA (2013) Temperature-dependent shade avoidance involves the receptor-like kinase ERECTA. Plant J 73: 980–992 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M (2009) Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J 60: 424–435 [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martínez-García JF (2006) Identification of primary target genes of phytochrome signaling: early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol 141: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M, Martínez-García JF (2007) Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J 26: 4756–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sola MA, Rodríguez-Concepción M (2012) Carotenoid biosynthesis in Arabidopsis: a colorful pathway. The Arabidopsis Book 10: e0158, doi/10.1199/tab.0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sola MA, Rodriguez-Villalon A, Rodriguez-Concepcion M (2014) Light-sensitive Phytochrome-Interacting Factors (PIFs) are not required to regulate phytoene synthase gene expression in the root. Plant Signal Behav 9: e29248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R, Pacín M, Casal JJ (2012) Diurnal dependence of growth responses to shade in Arabidopsis: role of hormone, clock, and light signaling. Mol Plant 5: 619–628 [DOI] [PubMed] [Google Scholar]
- Sellaro R, Yanovsky MJ, Casal JJ (2011) Repression of shade-avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J 68: 919–928 [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW (2013) HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25: 3770–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martínez-García JF (2009) ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J 59: 266–277 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Rodríguez-Concepción M (2010) Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci USA 107: 11626–11631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodríguez-Concepción M, Halliday KJ (2014) The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Song M, Yang Q, Su L, Hou P, Guo L, Zheng X, Xi Y, Meng F, Xiao Y, et al. (2014) Both PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiol 164: 841–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.