A plastidial glycolytic enzyme modulates the metabolism of carbon and nitrogen in nonphotosynthetic cells and affects plant organ development.
Abstract
This study functionally characterizes the Arabidopsis (Arabidopsis thaliana) plastidial glycolytic isoforms of glyceraldehyde-3-phosphate dehydrogenase (GAPCp) in photosynthetic and heterotrophic cells. We expressed the enzyme in gapcp double mutants (gapcp1gapcp2) under the control of photosynthetic (Rubisco small subunit RBCS2B [RBCS]) or heterotrophic (phosphate transporter PHT1.2 [PHT]) cell-specific promoters. Expression of GAPCp1 under the control of RBCS in gapcp1gapcp2 had no significant effect on the metabolite profile or growth in the aerial part (AP). GAPCp1 expression under the control of the PHT promoter clearly affected Arabidopsis development by increasing the number of lateral roots and having a major effect on AP growth and metabolite profile. Our results indicate that GAPCp1 is not functionally important in photosynthetic cells but plays a fundamental role in roots and in heterotrophic cells of the AP. Specifically, GAPCp activity may be required in root meristems and the root cap for normal primary root growth. Transcriptomic and metabolomic analyses indicate that the lack of GAPCp activity affects nitrogen and carbon metabolism as well as mineral nutrition and that glycerate and glutamine are the main metabolites responding to GAPCp activity. Thus, GAPCp could be an important metabolic connector of glycolysis with other pathways, such as the phosphorylated pathway of serine biosynthesis, the ammonium assimilation pathway, or the metabolism of γ-aminobutyrate, which in turn affect plant development.
Glycolysis is an essential primary metabolic pathway in most living organisms whose main function is to oxidize hexoses to provide ATP, reducing power and pyruvate, and to produce precursors for anabolism (Plaxton, 1996). In plants, glycolysis is particularly important because it is considered the predominant pathway that fuels the tricarboxylic acid cycle in mitochondria and is an important source of precursors for secondary metabolism, amino acids, and fatty acid biosynthesis (Plaxton, 1996). Plant glycolysis possesses some differences with respect to other organisms, which complicates its understanding (Plaxton, 1996). There are two glycolytic pathways operating in parallel in the cytosol and plastids, and both interact through highly selective transporters present in the inner plastid membrane (Weber et al., 2005), which may suggest that glycolytic intermediates are fully equilibrated in both compartments. However, the characterization of plastidial glycolytic mutants suggests that this may not always be the case, at least for some glycolytic intermediates in certain cellular types (Muñoz-Bertomeu et al., 2009, 2010). Furthermore, plants possess autotrophic photosynthetic cells and heterotrophic nonphotosynthetic cells, where the functions of the glycolytic pathway could be completely different. Besides, some of the glycolytic reactions in the chloroplast are identical to those in the Calvin-Benson cycle but operate in the opposite direction. For this reason, the functional significance of plastidial glycolysis has been questioned and still remains controversial, especially in photosynthetic cells, where chloroplasts may lack one or several glycolytic enzymes (e.g. enolase and phosphoglycerate mutase; Van der Straeten et al., 1991; Andriotis et al., 2010; Prabhakar et al., 2010). For instance, plastidial glycolytic enolase has been demonstrated to have poor or no expression in chloroplasts, and mutant plants do not display drastic visible phenotypes (Prabhakar et al., 2010). Since the double mutant of enolase and the phosphoenolpyruvate (PEP) transporter show a lethal phenotype and the PEP transporter single mutant phenotype is not lethal, it can be assumed that the plastidial enolase is functional in some cell types. However, Andriotis et al. (2010) concluded that, although plastids in developing Arabidopsis (Arabidopsis thaliana) embryos have an active lower part of the glycolytic pathway, the cytosolic glycolytic pathway suffices to support the flux from 3-phosphoglycerate (3-PGA) to PEP required for oil production. Other plastidial glycolytic enzymes of the upper and lower parts of the pathways have also been characterized. For instance, it has been demonstrated that pyruvate kinases, in the lower part of the glycolytic pathway, play an important role in seed oil biosynthesis (Andre and Benning, 2007; Andre et al., 2007; Baud et al., 2007), while triose-phosphate isomerase, in the middle part, is essential for the postembryonic transition from heterotrophic to autotrophic growth (Chen and Thelen, 2010). All these results highlight the specific functions of plastidial glycolytic enzymes in plants. However, our understanding of the general relevance of plastidial and cytosolic glycolysis in the plant is far from complete. Thus, it is a challenge to elucidate which enzyme isoforms are functionally important, in which cells there is a complete plastidial pathway, whether there is equilibrium of metabolic intermediates between the cytosol and the plastids, and in particular the exact role, regulation, and relative importance of different glycolytic isoforms in specific cell types and under different environmental conditions.
According to Arabidopsis genome databases (https://www.arabidopsis.org/), the number of annotated genes coding for different glycolytic enzyme isoforms varies greatly. Some enzymes are coded by few genes, such as triose-phosphate isomerase, which is represented by only two annotations in the databases, while others, such as phosphoglycerate mutase, is represented by more than 40 annotations. The Arabidopsis Information Resource database describes seven genes encoding phosphorylating glyceraldehyde 3-phosphate dehydrogenase (GAPDH), four of which participate in glycolysis and three in the Calvin-Benson cycle. Two of the glycolytic isoforms (GAPC1 and GAPC2) are cytosol localized and two (GAPCp1 and GAPCp2) are localized in the plastid (Muñoz-Bertomeu et al., 2009). This may indicate the complexity of the GAPDH reaction in terms of the integration and regulation of enzyme activity. By following an antisense approach in potato (Solanum tuberosum), it was concluded that decreased GAPC activities exhibited no major changes in either whole-plant or tuber morphology and that the enzyme plays only a minor role in the regulation of metabolism (Hajirezaei et al., 2006). By contrast, an Arabidopsis mutant in the GAPC1 isoform displayed delayed growth and several morphological alterations in siliques and seeds (Rius et al., 2008). The Arabidopsis double knockout gapc mutants, however, did not display any visible phenotype under normal growth conditions, but lack of enzyme activity decreased the oil content in developing seeds (Guo et al., 2012). Additional nonglycolytic functions for cytosolic GAPDHs have also been attributed in both plants and mammals (Kim and Dang, 2005; Zaffagnini et al., 2013). In Arabidopsis, GAPCs have been documented to be involved in the response to stress, for instance, participating in hydrogen peroxide signal transduction, in the response to cadmium toxicity, or in the immunity response (Guo et al., 2012; Vescovi et al., 2013; Han et al., 2015).
The plastidial GAPCps have been demonstrated to be critical for primary root growth and essential for microspore development (Muñoz-Bertomeu et al., 2009, 2010). It was hypothesized that the main function of GAPCps in roots is to supply 3-PGA to the phosphorylated pathway of serine biosynthesis (PPSB; Muñoz-Bertomeu et al., 2009). This hypothesis was later confirmed by the characterization of the PPSB (Cascales-Miñana et al., 2013). Double GAPCp1 and GAPCp2 mutants (gapcp1gapcp2) also display a drastic reduced growth of the aerial part (AP; Muñoz-Bertomeu et al., 2009), suggesting a role of the enzyme in this organ. Petersen et al. (2003) suggested that GAPCp is absent in the chloroplasts of angiosperms. However, Backhausen et al. (1998) postulated that GAPCp is essential for starch metabolism during the dark period in green and nongreen plastids. In this scenario, it would be involved along with phosphoglycerate kinase in the ATP production needed for starch metabolism (Backhausen et al., 1998).
In this work, we deepen the functional characterization of GAPCps in photosynthetic and heterotrophic organs by performing metabolomic and transcriptomic studies in gapcp mutants and in mutant lines in which the enzyme is specifically expressed in heterotrophic or in photosynthetic cells. We provide new insights concerning how GAPCp activity affects other metabolic pathways. We also identify genomic and metabolic targets responding to GAPCp activity in both roots and the AP. We conclude that GAPCp is an important link that connects metabolism with mineral nutrition and development in plants.
RESULTS
Specific Expression of GAPCp under the Control of the Rubisco Small Subunit Promoter Does Not Restore AP Growth But Complements gapcp1gapcp2 Sterility
The phenotypic analysis of gapcp1gapcp2 indicated that they display a drastic reduction not only of root growth but also of the AP when grown both on plates and in greenhouse conditions (Supplemental Fig. S1; Muñoz-Bertomeu et al., 2009). This phenotype is observed in double homozygous mutants only. Single mutants (gapcp1 or gapcp2) or mutant plants homozygous for one of the genes and heterozygous for the other are phenotypically indistinguishable from the wild-type plants (Supplemental Fig. S1), which demonstrates that GAPCp1 and GAPCp2 are redundant to one another. All the phenotypes of gapcp1gapcp2 could also be complemented with constructs that carry the GAPCp1 genomic sequence or the complementary DNA (cDNA) under the control of its native promoter (Muñoz-Bertomeu et al., 2009, 2010), which corroborates that GAPCp1 can compensate the lack of GAPCp2. The metabolite profile of the gapcp1gapcp2 AP indicated that 67% of analyzed metabolites were significantly altered and 45% were changed by more than 40% as compared with the wild type (Supplemental Table S1). These results point to the importance of GAPCp for metabolism in the AP. In this sense, mRNA for both GAPCp1 and GAPCp2 was clearly found in leaves, shoots, and reproductive organs (Muñoz-Bertomeu et al., 2009). Western blot using antibodies raised against an epitope of GAPCp isoforms further confirmed that the enzyme is present in the AP, although in lower amount than in roots (Supplemental Fig. S1).
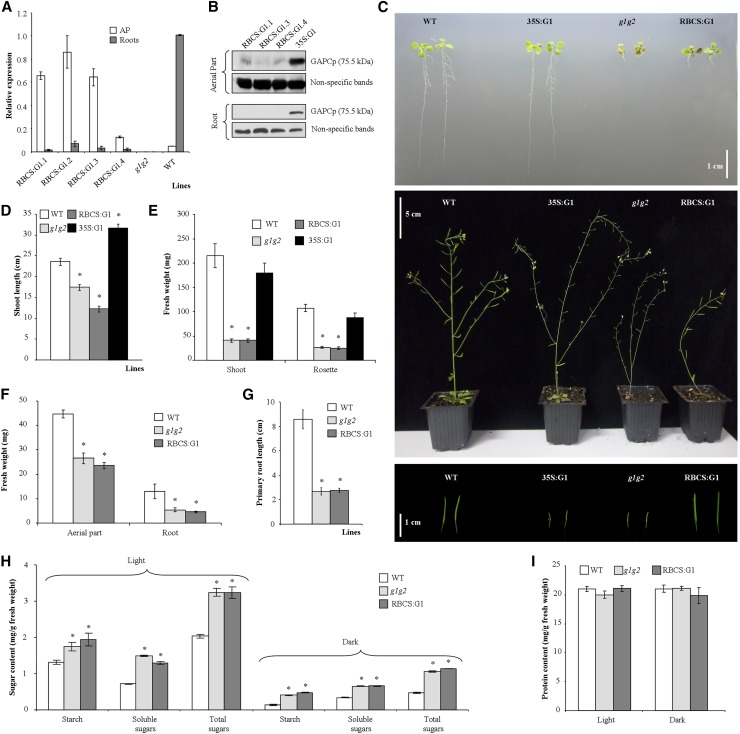
In order to dissect the function of GAPCp in the AP, we expressed GAPCp1-GFP in gapcp1gapcp2 under the control of the Rubisco small subunit RBCS2B promoter (RBCS:GAPCp1). According to the Bio-Analytic Resource for Plant Biology database (http://bar.utoronto.ca/), RBCS2B is mostly expressed in photosynthetic tissues in both dark and light conditions. It is also expressed in the shoot apex, shoots, seeds, and flowers but poorly expressed or nonexpressed in roots. GAPCp1 expression in the AP of different gapcp1gapcp2 RBCS:GAPCp1 lines was confirmed by quantitative real-time (qRT)-PCR, western blot using anti-GFP antibodies, and confocal microscopy, and those lines with the lowest expression in roots (RBCS:G1.1, RBCS:G1.3, and RBCS:G1.4) were further characterized (Fig. 1, A and B; Supplemental Fig. S2). At the adult stage, GAPCp1 expression in the AP was able to complement the sterile phenotype of gapcp1gapcp2, resulting in plants with siliques and fertile seeds (Fig. 1C). The developmental pattern of gapcp1gapcp2 RBCS:GAPCp1 was also altered as compared with gapcp1gapcp2, probably as a consequence of the fertile phenotype, displaying shorter shoots than gapcp1gapcp2 (Fig. 1, C and D). A similar developmental pattern alteration was observed in the sterile gapcp1gapcp2 35S:GAPCp1 lines (Muñoz-Bertomeu et al., 2010) as compared with the wild type (Fig. 1, C and D). These results corroborate that GAPCp1 expression in reproductive organs is necessary for Arabidopsis fertility and confirm that GAPCp1 is active in gapcp1gapcp2 RBCS:GAPCp1 lines.
Figure 1.
Molecular and phenotypical analysis of gapcp1gapcp2 (g1g2) expressing GAPCp1-GFP under the control of the RBCS promoter (RBCS:G1). A, qRT-PCR analysis of GAPCp1 in the APs and roots of several 18-d-old RBCS:G1 lines as compared with the wild type (WT) and g1g2. B, Immunoblot showing GAPCp1 expression in the APs and roots of three representative RBCS:G1 lines as compared with g1g2 expressing GAPCp1 under the control of the 35S promoter (35S:G1). Protein gel-blot analysis was performed using anti-GFP antibodies. Nonspecific bands are shown as a sample loading control at bottom. C, Morphology of seedlings (top), adult plants (middle), and siliques (bottom) from wild-type, 35S:G1, g1g2, and RBCS:G1 plants. D and E, Shoot length (D) and fresh weight of shoots and rosette leaves (E) of plants grown in the greenhouse. F to I, Fresh weight (F), root length (G), starch and total soluble sugar contents (H), and protein content (I) of wild-type, g1g2, and RBCS:G1 lines grown for 18 d on plates. In A, H, and I, values are means ± se of three independent biological replications. In D to G, values are means ± se of three independent transgenic lines (n ≥ 36 plants). Asterisks indicate significant differences as compared with the wild type (P < 0.05).
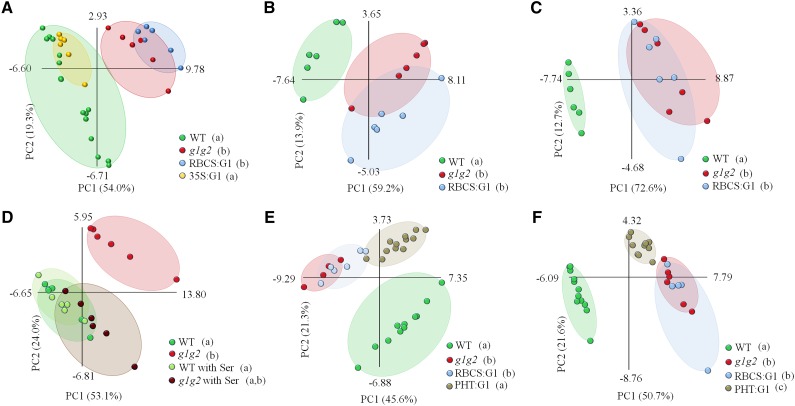
The presence of GAPCp1 in the photosynthetic cells did not recover the growth retardation of the rosette leaves and roots when plants were cultured in pots or on plates (Fig. 1, E–G). Similarly, the metabolite profile of the AP at the end of the light period in gapcp1gapcp2 RBCS:GAPCp1 lines overlapped that of gapcp1gapcp2 according to principal component analysis (PCA; Fig. 2A; Supplemental Table S1). Other metabolites measured in gapcp1gapcp2 RBCS:GAPCp1 lines, such as starch, total soluble sugars, and proteins, were also not significantly different from gapcp1gapcp2 (Fig. 1, H and I). In order to investigate whether GAPCp1 expression performed a function in photosynthetic cells under growth conditions where glycolysis is more important, for instance at night or when photosynthetic activity is inhibited, we analyzed the metabolite profile under dark conditions and in the presence of high concentrations of Suc, which inhibits photosynthesis (Fig. 2, B and C; Supplemental Table S2). Similar to samples obtained at the end of the light period, gapcp1gapcp2 RBCS:GAPCp1 lines displayed a metabolite profile that overlapped with gapcp1gapcp2 in both darkness and under growth medium containing Suc (Fig. 2, B and C; Supplemental Table S2). Lack of metabolic and growth complementation in the rosette of gapcp1gapcp2 RBCS:GAPCp1 lines suggested that GAPCp1 expression in photosynthetic cells is not functionally important in spite of the drastic phenotypes observed in this organ in gapcp1gapcp2.
Figure 2.
PCA of the metabolite profiles of different GAPCp mutant lines. Data from the gas chromatography-mass spectrometry analysis were evaluated using PCA, with the two first components accounting for at least 67% of total metabolic variance. Values in parentheses give the relative contribution of each component to the total variance observed in the data set. The first principal component was also used for the ANOVA of different lines, with Tukey’s posthoc tests to differentiate between the lines. A, The AP at the end of the light period. According to Tukey’s test, two different subsets are established: subset 1 (wild type [WT] and gapcp1gapcp2 35S:GAPCp1 [35S:G1] [a]) and subset 2 (gapcp1gapcp2 [g1g2] and g1g2 RBCS:GAPCp1 [RBCS:G1] [b]). B, The AP at the end of the dark period. According to Tukey’s test, two different subsets are established: subset 1 (wild type [a]) and subset 2 (g1g2 and RBCS:G1 [b]). C, The AP at the end of the light period in medium supplemented with 2% (w/v) Suc. According to Tukey’s test, two different subsets are established: subset 1 (wild type [a]) and subset 2 (g1g2 and RBCS:G1 [b]). D, The AP at the end of the light period in medium supplemented with 0.1 mm Ser. According to Tukey’s test, two different subsets are established: subset 1 (g1g2 with Ser, wild type, and wild type with Ser [a]) and subset 2 (g1g2 and g1g2 with Ser [b]). E, The AP at the middle of the light period. According to Tukey’s test, two different subsets are established: subset 1 (wild type and g1g2 PHT:GAPCp1 [PHT:G1] [a]) and subset 2 (g1g2 and RBCS:G1 [b]). F, Roots at the middle of the light period. According to Tukey’s test, three different subsets are established: subset 1 (wild type [a]), subset 2 (g1g2 and RBCS:G1 [b]), and subset 3 (PHT:G1 [c]). Each ellipse includes all the data points of the same line being analyzed (cover hull).
According to Tukey’s posthoc test of the ANOVA of the first principal component, the metabolite profile of the AP of gapcp1gapcp2 lines expressing GAPCp1-GFP under the control of 35S (35S:GAPCp1) is similar to that of the wild type but differs from that of gapcp1gapcp2 and gapcp1gapcp2 RBCS:GAPCp1 (Fig. 2A; Supplemental Table S1). In gapcp1gapcp2 35S:GAPCp1 lines, 14 of the 28 metabolites altered in gapcp1gapcp2 recovered the wild-type levels and seven changed in the opposite direction to that in gapcp1gapcp2, which is suggestive of an overexpression effect. Since 35S:GAPCp1 is expressed in both photosynthetic and nonphotosynthetic cells (Fig. 1B), we postulated that lack of GAPCp activity in the latter cells could affect metabolism in the AP.
Expression of GAPCp under the Control of the Phosphate Transporter Promoter Modifies the AP and Root Metabolite Pattern and Partly Complements gapcp1gapcp2 Phenotypes
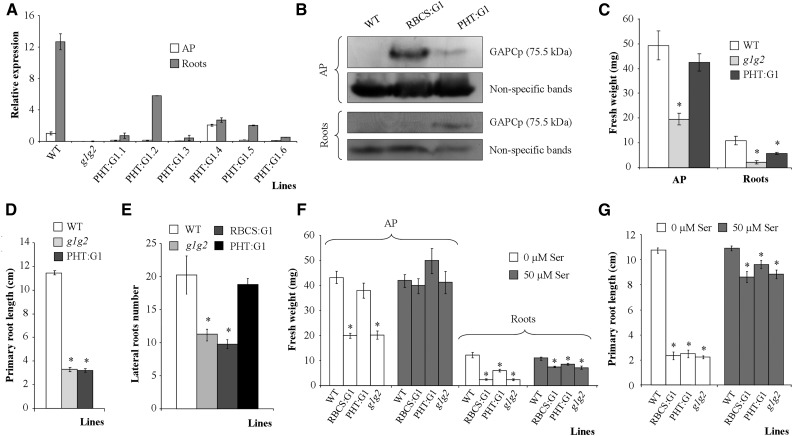
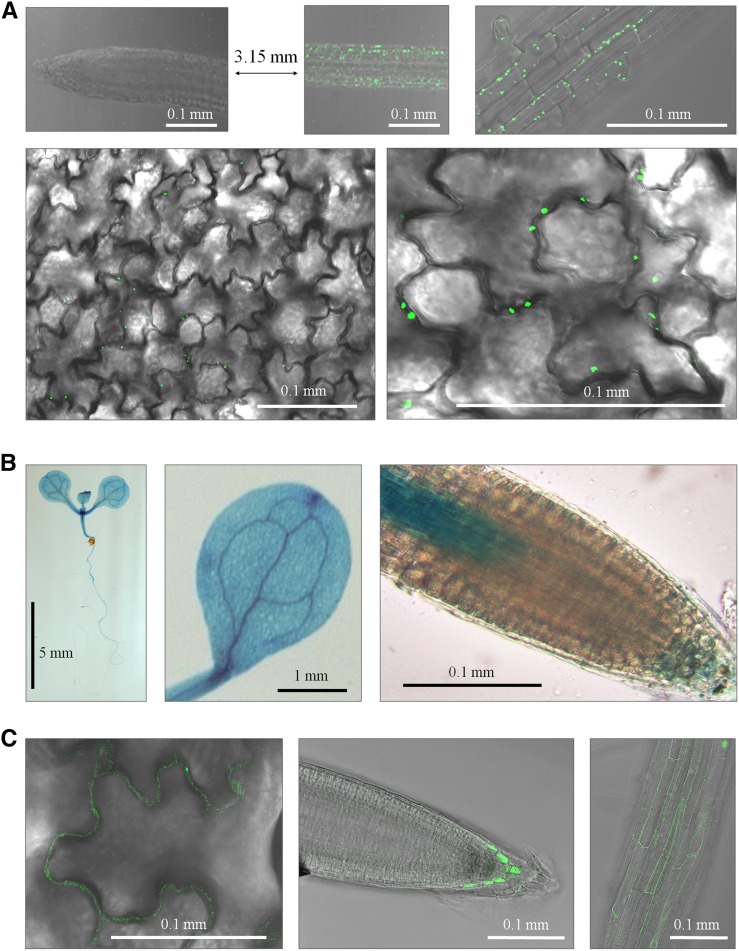
According to the microarray databases and the literature, the phosphate transporter PHT1.2 (PHT) promoter directs gene expression in root hair, epidermal, and cortex cells, with weak expression in rosette leaves (Mudge et al., 2002; Nussaume et al., 2011; Kirchsteiger et al., 2012; http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). For this reason, we next expressed GAPCp1-GFP under the control of the PHT1.2 promoter (PHT:GAPCp1). Several lines with different GAPCp1 mRNA expression levels in roots and the AP were obtained (Fig. 3A). Two of these lines (PHT:G1.2 and PHT:G1.5) displaying high GAPCp1 expression in roots and low expression in the AP were selected for further characterization. Western blots using anti-GFP antibodies confirmed that GAPCp1 was mainly expressed in roots, although it could also be detected in the AP (Fig. 3B). In any case, the GAPCp content in the AP was much lower than in the selected gapcp1gapcp2 RBCS:GAPCp1 lines (Fig. 3B). The PHT:GAPCp1 construct complemented the delayed AP growth of gapcp1gapcp2 (Fig. 3C). Analysis of GAPCp1 expression under the control of the PHT promoter indicated that the enzyme was located in leaf epidermal cell plastids (Fig. 4A), suggesting that it is necessary in these cell types. The expression analysis of the native GAPCp1 promoter in GUS and GFP fusions corroborated that, in addition to its expression in vascular cells, the GAPCp1 promoter directs gene expression in leaf epidermal cells (Fig. 4, B and C). The PHT:GAPCp1 construct increased the root weight (Fig. 3C) by an increase of lateral root formation (Fig. 3E) but did not recover the growth of primary roots (Fig. 3D). This may be related to the lack of expression of PHT:GAPCp1 in specific cell types of the root tip (Mudge et al., 2002). Analysis of the native GAPCp1 promoter in roots indicated that it was expressed in root vascular cells (Muñoz-Bertomeu et al., 2009), in epidermal cells, in the proximal part of meristems, in the columella, and in the lateral root cap cells (Fig. 4, B and C). PHT:GAPCp1 started to be expressed in the root epidermis at about 1.5 to 2 mm from the apex, having maximal expression at about 3 to 4 mm, but was not expressed in the meristem or the root cap (Fig. 4A). These results may suggest that GAPCp activity in root meristems is necessary for proper primary root growth.
Figure 3.
Molecular and phenotypical analyses of gapcp1gapcp2 (g1g2) expressing GAPCp1-GFP under the control of the PHT promoter (PHT:G1). A, qRT-PCR analysis of GAPCp1 in the APs and roots of several 21-d-old PHT:G1 lines as compared with g1g2 and the wild type (WT). B, Immunoblot showing GAPCp1 expression in the APs and roots of representative RBCS:G1 and PHT:G1 lines as compared with the wild type. Protein gel-blot analysis was performed using anti-GFP antibodies. Nonspecific bands are shown as a sample loading control at bottom. C to E, Fresh weight (C), primary root length (D), and lateral root number (E) of 18-d-old wild-type, g1g2, and PHT:G1 plants. F and G, Fresh weight (F) and primary root growth (G) of wild-type, g1g2, RBCS:G1, and PHT:G1 lines in the absence or presence of 50 µm Ser. In A, values are means ± se of three independent biological replications. In C to E, values are means ± se of two independent transgenic lines (n ≥ 36 plants). In F and G, values are means ± se (n ≥ 40 plants). Asterisks indicate significant differences as compared with the wild type (P < 0.05).
Figure 4.
Cellular expression of GAPCp1 under the control of its native promoter or the PHT promoter. A, GAPCp1-GFP expression under the control of the PHT promoter at the root tip (top left), at 3.15 mm from the root tip (top middle and right), and in epidermal leaf cells (bottom). B, GUS expression under the control of the GAPCp1 promoter (ProG1:GUS) in seedlings (left), cotyledons (middle), and root meristem (right). C, GFP expression under the control of the GAPCp1 promoter (ProG1:GFP) in leaf epidermal cells (left), root columella (middle), and root epidermal cells (right).
The analysis of gapcp1gapcp2 PHT:GAPCp1 lines confirmed that their metabolite profile in the AP is different from gapcp1gapcp2 and similar to that of the wild type, according to Tukey’s posthoc test of the ANOVA of the first principal component (Fig. 2E; Supplemental Table S3). Thus, in the gapcp1gapcp2 PHT:GAPCp1 AP, 62% of metabolites (24 out of 39) were significantly changed as compared with gapcp1gapcp2, and most of them (14 out of 24) reached wild-type levels (Supplemental Table S3). In quantitative terms, the most important changes in gapcp1gapcp2 PHT:GAPCp1 metabolites in relation to gapcp1gapcp2 were the reductions in the levels of some amino acids (Asn and Gln) and sugars (Fru, maltose, Glc, and trehalose; Supplemental Table S3).
In roots, the metabolite profile of gapcp1gapcp2 PHT:GAPCp1 was different from that of the wild type but also from that of gapcp1gapcp2 and gapcp1gapcp2 RBCS:GAPCp1, according to Tukey’s posthoc test of the ANOVA of the first principal component (Fig. 2F). The relative levels of 68% of metabolites analyzed in these lines (25 out of 37 metabolites) were significantly different from those of gapcp1gapcp2 mutants, but only six of them reached wild-type levels (Supplemental Table S3). The intermediate metabolite profile of gapcp1gapcp2 PHT:GAPCp1 between the wild type and gapcp1gapcp2 indicates a partial complementation of the gapcp1gapcp2 phenotype and may reflect the lack of recovery of primary root growth (Fig. 3D). We previously proposed that one of the main functions of GAPCp in roots is to supply the 3-PGA precursor of Ser through the so-called PPSB (Muñoz-Bertomeu et al., 2009; Cascales-Miñana et al., 2013). The primary root growth of the gapcp1gapcp2 mutant was 21% of that of the wild type in the absence of Ser and 81% of that of the wild type in its presence in the growth medium (Fig. 3G). In addition, growth of the AP was completely reestablished in the presence of Ser (Fig. 3F). The root growth of gapcp1gapcp2 RBCS:GAPCp1 lines was similar to that of gapcp1gapcp2 in the presence of Ser, and that of gapcp1gapcp2 PHT:GAPCp1 roots was still significantly lower than that of the wild type (Fig. 3, F and G). As shown by PCA, the metabolite profile of the AP of Ser-grown gapcp1gapcp2 plants overlaps that of the wild type (Fig. 2D). However, Tukey’s posthoc test of the ANOVA of the first principal component indicates that gapcp1gapcp2 grown in the presence of Ser can be grouped within both subsets, that of the wild type and that of gapcp1gapcp2 grown in the absence of Ser. These results indicate that, although Ser restores part of the gapcp1gapcp2 metabolite values that come close to those found in the wild type, some metabolite changes still occur compared with the controls (Supplemental Table S2). This suggests that GAPCp may have other functions in addition to providing substrates for Ser biosynthesis via the PPSB in specific cell types.
Metabolic and Transcriptomic Pathways Affected by GAPCp
To assess the metabolic pathways affected by GAPCp activity in the AP and in roots, we followed a metabolomic and transcriptomic approach. Lack of GAPCp expression clearly affected amino acid metabolism, as shown previously (Muñoz-Bertomeu et al., 2009), but also the carbon metabolism of sugars and organic acids (Tables I and II; Supplemental Tables S1–S3). Since metabolism is a dynamic process, we reasoned that a search for those metabolites that systematically changed under all different assayed conditions (end of the night, middle of the light period, end of the light period, and in the presence of Suc) may help narrow the identification of important pathways affected by GAPCp activity. We concentrated only on those metabolites that changed in gapcp1gapcp2 independently of the growth conditions (Table I; Supplemental Tables S1–S4). In the AP, two amino acids related to Glu metabolism were increased, γ-aminobutyrate (GABA) and Gln. There was also an increase in many sugars and their derivatives (Fru, myoinositol, Fuc, galactinol, Glc, trehalose, and Xyl) in gapcp1gapcp2 as compared with the wild type. However, the sugar phosphates Glc-6-P and Fru-6-P decreased in gapcp1gapcp2 in all the experiments in which they could be quantified as compared with controls. Two organic acids of the tricarboxylic acid cycle, citrate and succinate, also systematically increased in the AP of gapcp1gapcp2, which suggests that the respiratory pathway is affected by the lack of GAPCp activity. Glycerate, a metabolite that can be synthesized from 3-PGA through the reaction catalyzed by 3-PGA phosphatase or, alternatively, used to synthesize 3-PGA through the reaction catalyzed by glycerate kinase, was also increased in gapcp1gapcp2 when compared with the controls. In all instances, phosphoric acid was drastically reduced in gapcp1gapcp2 (more than 90% under some conditions; Table I; Supplemental Tables S1–S3).
Table I. List of metabolites that significantly increased (I) or decreased (D) in all assayed conditions in the AP of gapcp1gapcp2 (g1g2) when compared with wild-type plants.
Numerical values of g1g2 (means ± se) are relative values normalized to the mean response calculated for the wild type of one representative experiment. Significant changes (I or D) in g1g2 RBCS:GAPCp1 (RBCS:G1), g1g2 PHT:GAPCp1 (PHT:G1), and g1g2 35S:GAPCp1 (35S:G1) lines as well as in g1g2 plants grown in the presence of 100 µm Ser (g1g2 Ser) when compared with the wild type are also reported. ND and NC stand for not determined and not changed, respectively. The full data sets are shown in Supplemental Tables S1 to S4.
| Sample | g1g2 | RBCS:G1 | PHT:G1 | 35S:G1 | g1g2 Ser | |
|---|---|---|---|---|---|---|
| Metabolites | ||||||
| Citrate | I (2.57 ± 0.23) | I | ND | NC | NC | |
| Fru | I (2.71 ± 0.16) | I | I | I | I | |
| Fru-6-P | D (0.86 ± 0.03) | D | D | ND | ND | |
| Fuc | I (1.77 ± 0.10) | I | ND | NC | I | |
| GABA | I (1.31 ± 0.06) | I | ND | D | I | |
| Galactinol | I (8.86 ± 0.46) | I | D | D | NC | |
| Glc | I (2.92 ± 0.13) | I | NC | NC | I | |
| Glc-6-P | D (0.68 ± 0.06) | D | D | I | NC | |
| Gln | I (1.94 ± 0.09) | I | NC | NC | I | |
| Glycerate | I (3.18 ± 0.30) | I | NC | I | NC | |
| Myoinositol | I (2.61 ± 0.13) | I | NC | NC | I | |
| Phosphoric acid | D (0.13 ± 0.01) | D | D | NC | D | |
| Succinate | I (1.20 ± 0.07) | I | ND | I | NC | |
| Trehalose | I (1.47 ± 0.09) | I | I | NC | NC | |
| Xyl | I (2.38 ± 0.13) | I | ND | I | I | |
| No. of metabolites changed as compared with the wild type | ||||||
| Changed in the same direction as g1g2 | 15 | 15 | 5 | 4 | 8 | |
| Not changed/opposite direction to g1g2 | 0 | 0 | 5 | 10 | 6 | |
| Not determined | 0 | 0 | 5 | 1 | 1 |
Table II. List of metabolites that significantly increased (I) or decreased (D) in all assayed experiments in roots of gapcp1gapcp2 (g1g2), g1g2 RBCS: GAPCp1 (RBCS:G1), and g1g2 PHT: GAPCp1 (PHT:G1) when compared with wild-type plants.
Numerical values of g1g2 (means ± se) are relative values normalized to the mean response calculated for the wild type of one representative experiment. Significant changes in RBCS:G1 and PHT:G1 when compared with g1g2 are also reported. Only metabolites that displayed more than a 1.5-fold change in at least one experiment are listed. NC stands for not changed. The full data sets are shown in Supplemental Tables S3 and S4.
| Sample | Compared with the Wild Type |
Compared with g1g2 |
|||
|---|---|---|---|---|---|
| g1g2 | RBCS:G1 | PHT:G1 | RBCS:G1 | PHT:G1 | |
| Metabolites | |||||
| Ala | I (1.94 ± 0.07) | I | I | NC | NC |
| Ala, β | I (3.05 ± 0.13) | I | I | NC | D |
| Asn | I (2.71 ± 0.29) | I | I | NC | D |
| Fru | I (2.38 ± 0.06) | I | I | I | D |
| GABA | I (1.86 ± 0.04) | I | I | NC | D |
| Galactinol | I (2.69 ± 0.07) | I | I | NC | D |
| Gln | I (1.91 ± 0.11) | I | I | NC | NC |
| Glycerate | I (3.89 ± 0.17) | I | I | I | D |
| Myoinositol | I (1.75 ± 0.07) | I | I | NC | D |
| Raffinose | I (1.87 ± 0.05) | I | I | I | NC |
| Threitol | D (0.20 ± 0.02) | D | D | NC | I |
| Trehalose | I (3.58 ± 0.16) | I | I | NC | D |
| No. of metabolites changed as compared with the wild type or g1g2 | |||||
| Changed | 12 | 12 | 12 | 3 | 9 |
| Not changed | 0 | 0 | 0 | 9 | 3 |
The 15 metabolites that systematically changed in the AP of gapcp1gapcp2 all changed in the same direction in the gapcp1gapcp2 RBCS:GAPCp1 lines (Table I). However, most of these changes were reverted in 35S:GAPCp1, and some were reverted in gapcp1gapcp2 PHT:GAPCp1 or gapcp1gapcp2 grown in the presence of Ser. These results corroborate the lack of gapcp1gapcp2 complementation when the enzyme is expressed in photosynthetic cells.
In roots, only two different experiments were studied in either gapcp1gapcp2 mutants or conditional gapcp1gapcp2 mutants (Supplemental Tables S3 and S4). In order to filter the results, we chose the metabolites that changed in both conditions and displayed more than a 1.5-fold change in at least one experiment (Table II). In general, amino acids tended to increase in gapcp1gapcp2, but only Ala, Gln, Asn, and GABA fulfilled the chosen criterion. Regarding sugars, Fru, myoinositol, galactinol, raffinose, and trehalose increased more than 2-fold, while threitol decreased more than 50%, as compared with the controls. Glycerate increased more than 3-fold in gapcp1gapcp2. As in the AP, phosphoric acid was also depleted in roots of gapcp1gapcp2. In this case, the reduction was only about 40% (Supplemental Tables S3 and S4).
Changes in all these metabolites were maintained in gapcp1gapcp2 RBCS:GAPCp1 lines as compared with the wild type (Table II). However, the levels of some of them (nine out of 12) were partly reverted in gapcp1gapcp2 PHT:GAPCp1 and differed significantly from those in gapcp1gapcp2, which confirms the idea that GAPCp expression under the control of the PHT promoter partly complements gapcp1gapcp2 (Table II). Seven out of 12 metabolites whose levels changed in gapcp1gapcp2 roots (Fru, galactinol, glycerate, Gln, GABA, myoinositol, and trehalose) were also modified in the AP (Tables I and II), which suggest an effect of the PHT:GAPCp1 construct in this organ.
A transcriptomic approach in the APs and roots showed that several functional categories of genes were up- or down-regulated in gapcp1gapcp2 as compared with the wild type (Table III; Supplemental Table S5; Supplemental Materials S1). Changes in gene expression were also confirmed by qRT-PCR (Supplemental Fig. S3). According to the Munich Information Center for Protein Sequences Funcat Functional Annotation program (http://mips.gsf.de/proj/funcatDB; P < 0.005), only genes related to phosphate transport were significantly induced in the gapcp1gapcp2 AP compared with the wild type (Table III). Specifically, two genes coding for phosphate transporters were up-regulated in the AP of gapcp1gapcp2 (At2g38940 [PHT1.4] and At5g43370 [PHT1.2]; Supplemental Table S5). In order to further specify important genes that might be affected by the lack of GAPCp activity, we compared misregulated genes in all microarrays obtained in this study and also with a previous microarray obtained by our group in which whole seedlings were analyzed (Muñoz-Bertomeu et al., 2009). We found that only two genes displayed common changes in all microarrays (wild-type versus gapcp1gapcp2 whole seedling, wild-type versus gapcp1gapcp2 AP, wild-type versus gapcp1gapcp2 roots, and gapcp1gapcp2 versus gapcp1gapcp2 HS:GAPCp1 induced in APs and roots; see below), At5g06300 (LONELY GUY7 [LOG7]) and At1g73010 (PYROPHOSPHATE-SPECIFIC PHOSPHATASE1 [PPsPase1]; Supplemental Table S5). Both genes were up-regulated in gapcp1gapcp2 in all microarrays studied, and their induction was confirmed by qRT-PCR (Supplemental Fig. S3). Since PPsPase1 is postulated to play an important role in the inorganic phosphate (Pi) starvation response (May et al., 2011) and the metabolomic analysis of the AP of gapcp1gapcp2 indicated a drastic reduction in the phosphoric acid level in the mutant, we searched in its transcriptome for additional misregulated genes involved in phosphate metabolism and homeostasis (Supplemental Table S5). In microarrays of gapcp1gapcp2 AP, we found four up-regulated genes (At1g23140, At3g17790 [PURPLE ACID PHOSPHATASE17; PAP17], At5g20150, and At2g38940 [PHT1.4]) whose expression is also up-regulated in the shoots of the Ca2+/H+ transporter cax1/cax3 mutants. The activity of this transporter is required for systemic phosphate homeostasis involving shoot-to-root signaling in Arabidopsis (Liu et al., 2011). Two additional genes, encoding a purple acid phosphatase (PAP8; At2g01890) and a phosphate transporter (PHT1.2; At5g43370), were also induced in the gapcp1gapcp2 AP. Finally, from 16 common misregulated genes in the APs and roots of gapcp1gapcp2, three were related to phosphate homeostasis. Apart from the previously mentioned PPsPase1 (At1g73010), a second gene (At5g03545), known to be expressed in response to phosphate starvation, was induced in gapcp1gapcp2 as compared with the controls. The third gene, At2g33770, encodes a ubiquitin-conjugating E2 enzyme, PHOSPHATE2, which was the only phosphorus-related gene repressed in both roots and AP of gapcp1gapcp2.
Table III. Functional categories of genes differentially expressed in the APs and roots of gapcp1gapcp2 as compared with the wild type.
Significantly enriched categories (P < 0.005) are shown.
| Functional Category | P |
|---|---|
| APs | |
| Induced | |
| Phosphate transport | 0.00441 |
| Repressed | |
| Secondary metabolism | 0.00457 |
| Metabolism of glucosinolates and derivatives | 0.00467 |
| Stress response | 0.00489 |
| Roots | |
| Induced | |
| Nitrogen, sulfur, and selenium metabolism | 0.00301 |
| Nucleotide-sugar metabolism | 0.00061 |
| Lipid binding | 0.00182 |
| Cellular transport, transport facilities, and transport routes | 0.00288 |
| Transported compounds (substrates) | 0.00369 |
| Lipid/fatty acid transport | 0.00110 |
| Detoxification | 0.00300 |
| Oxygen and radical detoxification | 0.00277 |
| Response to biotic stimulus | 0.00078 |
| Repressed | |
| Monoterpene metabolism | 0.00074 |
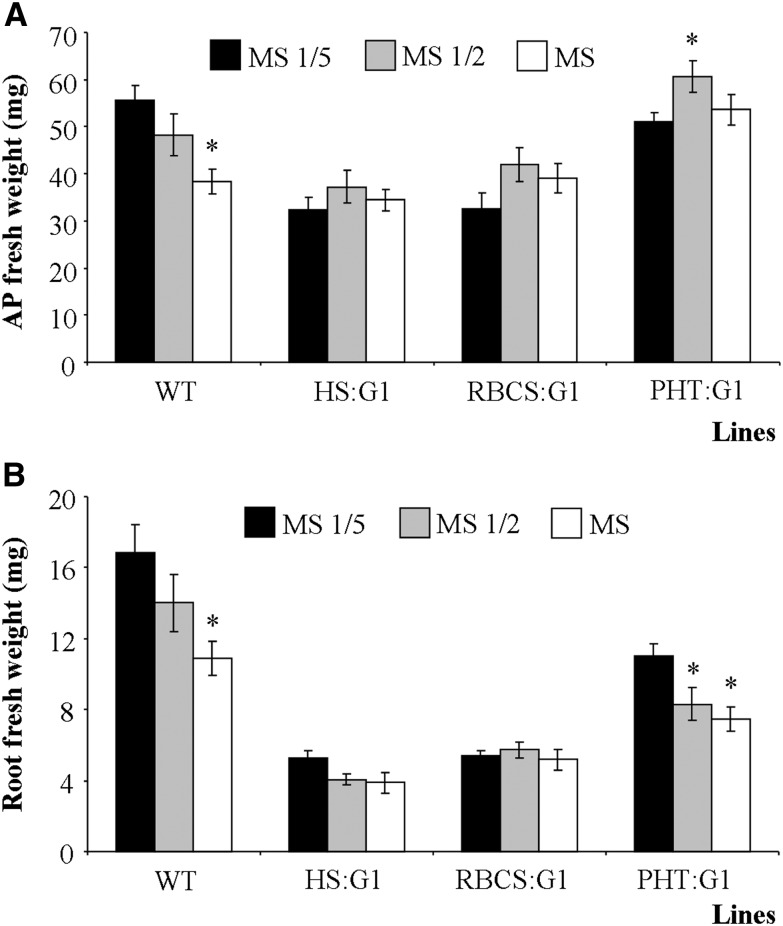
Since metabolomic and transcriptomic analyses indicated an alteration of Pi homeostasis, we measured the mineral content of gapcp1gapcp2. Several minerals were reduced in the gapcp1gapcp2 AP, but the main nutrient deficiency observed was Pi (Supplemental Table S6). In gapcp1gapcp2 roots, although some nutrients were reduced (potassium, copper, and iron), no Pi deficiency was observed, and some nutrients (nitrogen, manganese, and zinc) were even increased (Supplemental Table S6). In order to investigate whether mineral deficiency could affect gapcp1gapcp2 development, we assayed the mutant growth in medium with increased mineral supply (Fig. 5). As compared with a one-fifth-strength Murashige and Skoog (MS) medium, growth in a one-half-strength MS medium only improved the AP growth in the gapcp1gapcp2 PHT:GAPCp1 line, having no significant effect in the other lines and even decreasing root growth in the PHT:GAPCp1 line. These results indicated that nutrient deficiency is not the main cause of the growth defects in gapcp1gapcp2.
Figure 5.
Growth parameters of gapcp1gapcp2 lines grown in medium with increasing MS medium concentrations. Fresh weights of the AP (A) and roots (B) of 21-d-old wild type (WT) and gapcp1gapcp2 plants expressing GAPCp1 (G1) under the control of the HS (HS:G1), the RBCS (RBCS:G1), and the PHT (PHT:G1) promoters are shown. Values are means ± se of two independent transgenic lines (n ≥ 36 plants). Asterisks indicate significant differences as compared with plants grown in one-fifth-strength MS medium (P < 0.05).
Metabolite and Transcripts Responding to GAPCp Activity
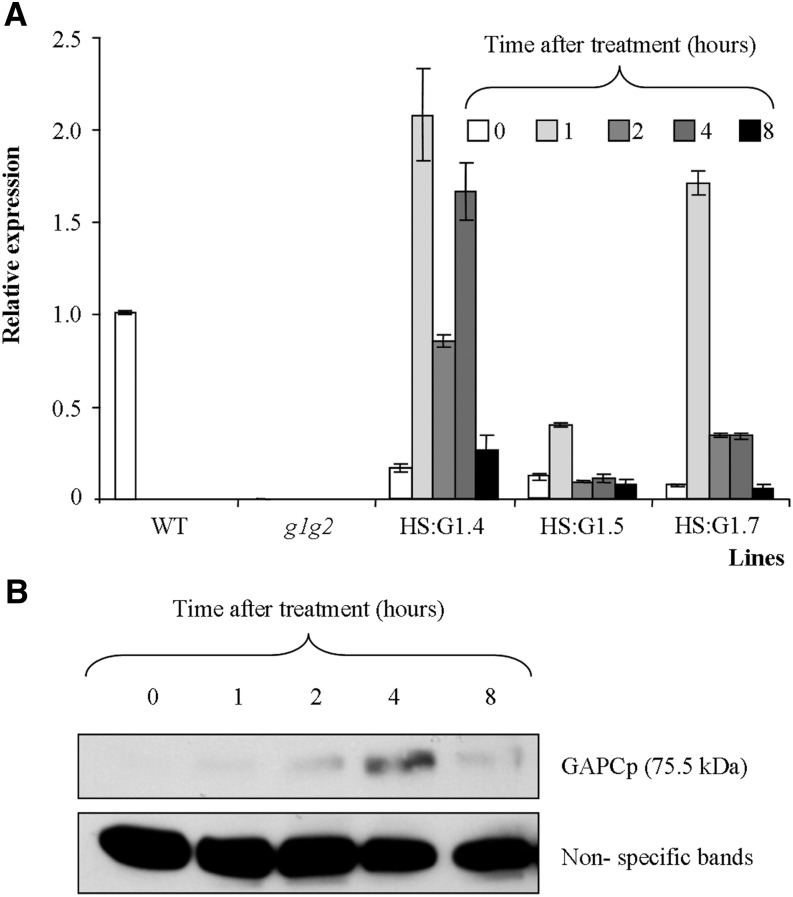
In order to identify targets responding to GAPCp activity, we performed a metabolomic and transcriptomic study of conditional gapcp1gapcp2 mutants. For this purpose, we used gapcp1gapcp2 transformed with a GAPCp1-GFP cDNA under the control of the heat-shock promoter HSP18.2 (HS:GAPCp1). Seven conditional lines were obtained, and two or three of them (depending on the experiment) were selected for further characterization, taking into account their basal GAPCp1 level, their induction level, and their complementation of the gapcp1gapcp2 root growth and sterile phenotype (Fig. 6; Supplemental Fig. S4). First, we performed a time-course experiment to decide the best time to perform the analyses. All selected gapcp1gapcp2 HS:GAPCp1 lines displayed a peak of GAPCp1 mRNA 1 h after induction (Fig. 6A). The GAPCp1 protein peak was reached 4 h after induction and was still present 8 h after induction (Fig. 6B; Supplemental Fig. S4). We assumed that translation is mostly resumed in Arabidopsis 1 h after a heat-shock treatment at 37°C (M. Castellanos, personal communication). Since GAPCp1 had to be translated and to participate in glycolysis before having an effect on transcription, we chose 4 h after induction for the realization of transcriptomic experiments and 8 h after induction for the metabolomic experiments. In order to discard any effect of heat shock on the metabolite profile of conditional mutants, we additionally studied the metabolite profile of gapcp1gapcp2 8 h after a heat-shock treatment. Only one metabolite (Tyr) out of 42 measured changed in the AP, and four (Pro, Glc-6-P, Fru-6-P, and trehalose) out of 37 changed in the roots, of heat shock-treated gapcp1gapcp2 as compared with untreated plants (Supplemental Table S7). Changes in these metabolites were not interpreted as such if the observed changes in the conditional mutants were in the same direction after induction.
Figure 6.
Molecular and phenotypical analyses of gapcp1gapcp2 (g1g2) expressing GAPCp1-GFP (G1) under the control of the HS promoter (HS:G1). A, Time-course analysis of GAPCp1 mRNA expression in three 18-d-old HS:G1 lines as compared with the wild type (WT) and g1g2. B, Time-course analysis of GAPCp1 protein expression in HS:G1 lines. Protein gel-blot analysis was performed using anti-GFP antibodies. Nonspecific bands are shown as a sample loading control at bottom. In A, values means ± se of three independent biological replications.
The metabolomics analysis indicated that 54% (21 out of 39) and 22% (eight out of 37) of measured metabolites recovered wild-type levels in the APs and roots of induced gapcp1gapcp2 HS:GAPCp1, respectively (Supplemental Table S4). In general, lower amino acid and sugar levels were observed in these lines, which indicates the crucial role of GAPCp activity in nitrogen and carbon metabolism. Tables IV and V list the metabolites whose levels changed by more than 20% following GAPCp1 induction in APs and roots, respectively. Three of them, Gln, glycerate, and galactinol, are particularly interesting because they were the only ones altered in all experimental conditions assayed in both roots and AP of gapcp1gapcp2 and those that displayed the most important changes after GAPCp1 induction. In the AP, Gln was the metabolite whose levels changed the most after GAPCp1 induction (31% reduction). Galactinol and glycerate were reduced by 24% and 22%, respectively. In roots, the levels of these three metabolites lowered even more drastically than in the AP after GAPCp1 induction, with glycerate reduced the most of all metabolites measured (68% reduction), followed by Gln (45%) and galactinol (33%). To provide additional insights that could explain changes in glycerate levels, we next measured the activity of phosphoglycerate kinase in roots and in the AP. The enzyme activity was higher in the AP of gapcp1gapcp2 than in the wild type (264 ± 6 versus 246 ± 2 nmol NADH g−1 fresh weight min−1 in gapcp1gapcp2 and the wild type, respectively) and especially in roots (60 ± 2 versus 34 ± 2 nmol NADH g−1 fresh weight min−1 in gapcp1gapcp2 and the wild type, respectively), where the highest levels of glycerate in the mutant were found.
Table IV. Metabolites in AP of gapcp1gapcp2 HS:GAPCp1 plants that significantly changed more than 20% following GAPCp1 induction.
Data for gapcp1gapcp2 HS:GAPCp1 induced and noninduced plants (means ± se) are relative values normalized to the mean response calculated for wild-type plants. The full data set is shown in Supplemental Table S4.
| Metabolite | Noninduced | Induced |
|---|---|---|
| Arg | 1.01 ± 0.06 | 0.80 ± 0.05 |
| Galactinol | 1.19 ± 0.05 | 0.91 ± 0.04 |
| Glc | 1.38 ± 0.07 | 1.05 ± 0.07 |
| Gln | 1.53 ± 0.11 | 1.05 ± 0.08 |
| Glycerate | 1.44 ± 0.04 | 1.13 ± 0.06 |
| Gly | 0.79 ± 0.03 | 0.98 ± 0.03 |
| Maltose | 0.92 ± 0.08 | 1.33 ± 0.04 |
| Tyr | 1.04 ± 0.11 | 0.56 ± 0.05 |
| Urea | 0.66 ± 0.03 | 0.94 ± 0.03 |
Table V. Metabolites in roots of gapcp1gapcp2 HS:GAPCp1 plants that significantly changed more than 20% following GAPCp1 induction.
Data for gapcp1gapcp2 HS:GAPCp1 induced and noninduced plants (means ± se) are relative values normalized to the mean response calculated for wild-type plants. The whole metabolite set is presented in Supplemental Table S4.
| Metabolite | Noninduced | Induced |
|---|---|---|
| Ala | 2.01 ± 0.20 | 1.47 ± 0.11 |
| Ala, β | 2.33 ± 0.15 | 1.43 ± 0.10 |
| Asn | 2.79 ± 0.40 | 1.82 ± 0.21 |
| Asp | 1.35 ± 0.06 | 0.88 ± 0.01 |
| Fru-6-P | 0.99 ± 0.05 | 0.72 ± 0.03 |
| Galactinol | 2.25 ± 0.15 | 1.50 ± 0.07 |
| Glc-6-P | 0.96 ± 0.05 | 0.65 ± 0.02 |
| Glu | 1.17 ± 0.09 | 0.84 ± 0.05 |
| Gln | 2.30 ± 0.23 | 1.26 ± 0.12 |
| Glycerate | 4.40 ± 0.06 | 1.42 ± 0.07 |
| Myoinositol | 1.52 ± 0.07 | 1.21 ± 0.05 |
| Malate | 1.18 ± 0.04 | 0.92 ± 0.03 |
| Raffinose | 1.96 ± 0.13 | 1.51 ± 0.08 |
| Trehalose | 2.29 ± 0.20 | 1.71 ± 0.12 |
The high levels of Gln in gapcp1gapcp2 could be related to the arrest in PPSB activity, which has been shown to affect the ammonium assimilation pathway (Benstein et al., 2013). According to this, the ammonium content in gapcp1gapcp2 roots was significantly higher than in the wild type (Fig. 7), which could contribute to growth impairment in the mutant. Ammonium toxicity is currently associated with acidification of the medium (Li et al., 2014). To study such acidification, we used the visual assay for H+ efflux from root cells based on plates containing the pH indicator Bromocresol Purple. As indicated in Figure 7, the yellow color surrounding the roots of gapcp1gapcp2 roots, indicative of moderate acidification, was much greater than in the wild type.
Figure 7.
Ammonium content and proton extrusion increase in roots of gapcp1gapcp2. Two-week-old Arabidopsis gapcp1gapcp2 (g1g2) and wild-type (WT) plants were grown for 6 h in one-fifth-strength MS medium at pH 5.7 without MES and containing 0.003% (w/v) Bromocresol Purple. The yellow color of the pH indicator around g1g2 roots indicates increased proton extrusion in the mutant. The table at bottom presents data of ammonium content (means ± se; n = 3) in roots of g1g2 and the wild type. The asterisk indicates a significant difference as compared with the wild type (P < 0.05). FW, Fresh weight.
The transcriptomic study revealed that, in the AP, several functional categories of genes were significantly up- or down-regulated following the induction of GAPCp1 (Supplemental Table S8). In order to narrow the identification of putative targets responding to GAPCp1 activity, we searched for those genes that were altered in gapcp1gapcp2 as compared with the wild type and whose expression was reverted by the induction of GAPCp1 activity in the mutant. We considered that these genes could be mediating the response to altered GAPCp1 activity. Twenty-four genes in the AP and three genes in roots whose expression was misregulated in gapcp1gapcp2 were then reverted after the induction of GAPCp1 (Table VI). Hormone (At5g06300, At1g67100, At1g80340, At3g23150, and At4g37150) and sugar signaling (At3g30720), cell cycle regulation (At2g29680), and metabolism (At5g17330, At5g52570, and At5g67520) associated genes fitted these criteria (Table VI). Two of them are particularly interesting (At5g06300 [LOG7] and At5g17330, which encodes one of two isoforms of Glu decarboxylase [GAD]) as development and metabolic mediators of the response to altered GAPCp1 activity. LOG7, one of the two common misregulated genes in all gapcp1gapcp2 microarrays, was up-regulated in gapcp1gapcp2 APs and roots and down-regulated in both organs upon GAPCp1 induction. LOG7, which encodes an enzyme participating in cytokinin biosynthesis, has been implicated in the maintenance of the shoot apical meristem and plays a role in normal primary root growth (Tokunaga et al., 2012). Similarly, GAD was up-regulated in the gapcp1gapcp2 AP and down-regulated after GAPCp1 activity induction. This enzyme, which plays an important role in balancing carbon and nitrogen metabolism, catalyzes the irreversible decarboxylation of Glu to form GABA (Fait et al., 2011).
Table VI. List of genes in APs and roots whose expression was misregulated in gapcp1gapcp2 (g1g2) as compared with the wild-type plants and then reverted after GAPCp1 induction in g1g2 HS:GAPCp1 (HS:G1).
| Gene | Fold Change (Log 2) |
Gene Description | |
|---|---|---|---|
| Wild Type versus g1g2 | g1g2 versus HS:G1 | ||
| APs | |||
| At1g14880 | −2.6 | 1.7 | Plant Cadmium Resistance1 (PCR1) |
| At1g23840 | −1.4 | 1.3 | Unknown function |
| At1g53480 | −2.0 | 1.6 | Encodes MRD1 (mto1 responding down) |
| At1g67100 | −1.9 | 0.9 | LOB domain-containing protein40 (LBD40) |
| At3g23150 | −0.8 | 1.0 | Involved in ethylene perception in Arabidopsis |
| At3g23440 | −1.3 | 0.7 | Embryo Sac Development Arrest6 (EDA6) |
| At3g30720 | −0.6 | 1.0 | Qua-Quine Starch (QQS) |
| At4g13345 | −0.9 | 2.3 | Maternal Effect Embryo Arrest55 (MEE55) |
| At4g22280 | −0.8 | 0.9 | F-box/RNI-like superfamily protein |
| At5g13270 | −0.6 | 0.8 | Encodes RARE1 (Required for accD RNA Editing1) |
| At5g41620 | −0.6 | 1.9 | Unknown function |
| At5g67520 | −1.5 | 0.7 | Provides activated sulfate for the sulfation of secondary metabolites, including the glucosinolates |
| At1g59930 | 2.1 | −1.1 | Encodes a maternally expressed imprinted gene |
| At1g80090 | 2.5 | −1.4 | Cystathionine β-synthase (CBS) family protein |
| At1g80340 | 1.0 | −0.9 | Encodes a protein with GA3 β-hydroxylase activity |
| At2g01890 | 1.0 | −1.1 | Encodes a purple acid phosphatase (PAP) |
| At2g29680 | 1.4 | −0.7 | Encodes cell division control protein6 (CDC6) |
| At3g11580 | 2.5 | −0.6 | APETALA2/B3-like transcriptional factor family protein |
| At3g59700 | 1.7 | −0.7 | Member of a receptor kinase-like protein family |
| At4g01985 | 0.6 | −0.9 | Unknown function |
| At4g37150 | 1.2 | −1.1 | Methyl Esterase9 (MES9) |
| At5g06300 | 3.0 | −1.3 | Putative Lys decarboxylase family protein (LOG7) |
| At5g17330 | 0.7 | −0.8 | Encodes one of two isoforms of Glu decarboxylase (GAD1) |
| At5g52570 | 1.3 | −0.9 | Converts β-carotene to zeaxanthin via cryptoxanthin (BCH2) |
| Roots | |||
| At1g78070 | −1.2 | 0.7 | Transducin/WD40 repeat-like superfamily protein |
| At5g03090 | −4.5 | 1.8 | Unknown function |
| At5g06300 | 1.4 | −2.5 | Putative Lys decarboxylase family protein (LOG7) |
DISCUSSION
GAPCp Activity in Nonphotosynthetic Cells Restores the Metabolite Profile and Growth of the AP
Knowledge concerning the specific roles of glycolysis in photosynthetic and heterotrophic cells is scarce. Given that gapcp1gapcp2 displays a drastic growth reduction of the AP, we explored the specific effect of GAPCp in photosynthetic cells by expressing the enzyme under the control of the RBCS promoter. Morphological and metabolomic analysis indicated that RBCS:GAPCp1 expression did not complement the reduced growth phenotype either in roots or in the AP. The metabolite profile of the AP in gapcp1gapcp2 and gapcp1gapcp2 RBCS:GAPCp1 was similar under both photosynthetic and nonphotosynthetic conditions. These results indicate that plastidial glycolysis, or at least GAPCp activity, is not functionally important in photosynthetic cells. GAPCp1 expression under the control of the 35S promoter, however, restored both the growth phenotype and the metabolite profile of gapcp1gapcp2, suggesting that metabolic alterations in the AP could be caused by a lack of GAPCp activity in roots or in other AP cell types. In this sense, GAPCp1 expression under the control of the PHT promoter improved the growth of the AP in gapcp1gapcp2 and modified its metabolite profile. We found that PHT1.2 was not only expressed in roots, as described elsewhere (Mudge et al., 2002; Nussaume et al., 2011; Kirchsteiger et al., 2012), but also in the leaf epidermal cells of gapcp1gapcp2. PHT:GAPCp1 expression in the AP may be related to the Pi-deficient phenotype of gapcp1gapcp2, as this promoter is induced by Pi starvation in wild-type roots (Mudge et al., 2002), although not in the AP, as was observed in gapcp1gapcp2. Given that GAPCp1 expression in photosynthetic cells did not complement gapcp1gapcp2 AP phenotypes, we postulate that improved AP metabolism and development by PHT:GAPCp1 expression could be the result of GAPCp expression in AP heterotrophic cells, such as epidermal or vascular cells, where GAPCp is currently expressed under the control of its native promoter. This would explain why most metabolites whose levels changed in gapcp1gapcp2 roots were also modified in the AP, why some of the metabolites that directly respond to GAPCp activity, such as Gln and glycerate, increased in both gapcp1gapcp2 APs and roots, and why these metabolites reached wild-type levels in the gapcp1gapcp2 PHT:GAPCp1 AP. However, an indirect effect of GAPCp1 expression in roots on AP metabolism cannot completely be ruled out.
The partial complementation of gapcp1gapcp2 when expressing PHT:GAPCp1 may be due to the enzyme not being present in specific root cell types. Confocal microscopy indicated that GAPCp1 expression under the control of the PHT promoter was observed in root differentiated cells but not in meristematic and root cap cells, where GAPCp (this work) and PPSB genes are normally expressed (Cascales-Miñana et al., 2013; Toujani et al., 2013). It has been postulated that PPSB is an important source of Ser in specific cell types, such as the anther tapetum and meristematic cells (Ros et al., 2014). Lack of expression of PHT:GAPCp1 in gapcp1gapcp2 root meristematic cells and root cap cells would prevent the supply of Ser to these cells and thus prevent the recovery of primary root growth. This idea is supported by the recovery of gapcp1gapcp2 primary root growth by Ser supplementation. Regarding this observation, it would be interesting to know why the AP growth is reduced in gapcp1gapcp2, since this organ is not Ser deficient (Muñoz-Bertomeu et al., 2009; this work). It could be that the AP growth inhibition is due to mineral deficiency, and specifically Pi deficiency, as a consequence of root growth inhibition in the mutant. Organic and mineral Pi deficiency was measured in the AP of gapcp1gapcp2. However, an increased supply of mineral nutrients did not significantly increase the AP growth or the primary root growth of gapcp1gapcp2 or gapcp1gapcp2 RBCS:GAPCp1 lines. Besides, GAPCp1 expression in roots directed by the PHT promoter did not increase organic or mineral Pi content but significantly improved AP growth. Thus, Pi deficiency is probably not the primary cause of AP growth inhibition in gapcp1gapcp2. However, it remains possible that this deficiency could potentiate the phenotypic alterations observed in the mutant.
Lack of GAPCp Activity Affects Nitrogen and Carbon Metabolism and Mineral Nutrition
Several genes coding for proteins involved in Pi transport and signaling were up-regulated in the AP of gapcp1gapcp2. This observation is in accordance with the Pi deficiencies observed in the gapcp1gapcp2 AP, which could limit its growth. PPsPase1 deserves a special mention, since it was commonly induced in all the gapcp1gapcp2 microarrays. PPsPase1 encodes a pyrophosphate-specific phosphatase (PPsPase1) that has been suggested to play a role in the Pi starvation response by providing Pi from phosphorylated substrates (May et al., 2011). However, the expression pattern of eight out of nine misregulated Pi-related genes in gapcp1gapcp2, including PPsPase1, was not modified by GAPCp1 activation, which indicates that they are not genes primarily responding to GAPCp1 induction. This would corroborate that Pi deficiency is the consequence rather than the cause of the inhibition of primary root growth in gapcp1gapcp2 mutants.
In roots, misregulated functional categories included nitrogen and sulfur metabolism and transport of amino acids and ions. Unlike in the AP, genes involved in stress responses were induced in gapcp1gapcp2 roots, although in both roots and the AP metabolite markers related to stress (raffinose and galactinol) were increased. The metabolomic analysis also indicated that not only carbon metabolism, including the tricarboxylic acid cycle, but also amino acid metabolism was seriously affected in gapcp1gapcp2. These results indicate that GAPCp is required to maintain the normal interactions between carbon and nitrogen metabolism that, in turn, affect plant development. The dramatic phenotypic and metabolic alterations in GAPCp mutants, therefore, may be related to multiple requirements of the products of GAPCp in plant metabolism rather than to a specific role in glycolysis itself. Indeed, the effects of a lack of GAPCp activity in the metabolite profile were much less severe when plants were grown in the presence of Ser, which partially rescues primary root growth, indicating that the main role of GAPCp could be the supply of precursors for PPSB rather than its role in glycolysis per se.
Metabolite and Gene Targets of GAPCp Activity
Three metabolites (galactinol, glycerate, and Gln) systematically changed in both roots and the AP of gapcp1gapcp2 and tend to recover wild-type levels upon GAPCp1 induction. Galactinol (and myoinositol) increased in gapcp1gapcp2 and decreased after GAPCp induction in conditional mutants. Raffinose also increased in gapcp1gapcp2 in most of the assayed experimental conditions. These metabolites participate as substrates (galactinol and myoinositol) or final products (raffinose) in the biosynthesis of the raffinose family oligosaccharides (Nishizawa et al., 2008) and have been shown to play a role in the plant response to environmental stresses (Taji et al., 2002; Kaplan et al., 2007). It has been postulated that galactinol and raffinose could increase tolerance to chilling, salinity, or drought stress by protecting plants against oxidative damage (Nishizawa et al., 2008). The high level of these metabolites in gapcp1gapcp2 is probably an indicator of oxidative stress in gapcp1gapcp2. This could be a secondary consequence of metabolic perturbations resulting from the lack of GAPCp. Alternatively, GAPCp could be involved in plant protection against oxidative stress, a nonmetabolic function already suggested for the plant cytosolic GAPCs (Baek et al., 2008; Guo et al., 2012).
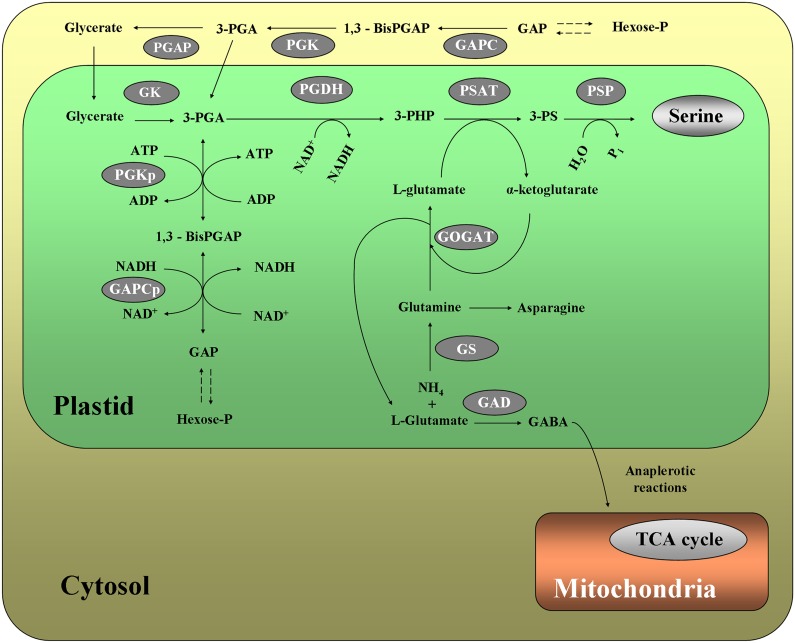
Glycerate accumulated in gapcp1gapcp2, especially in roots, where the levels clearly lowered after GAPCp1 activation in conditional mutants. Glycerate can be converted into 3-PGA in a reaction catalyzed by glycerate kinase in plastids (Fig. 8). Kleczkowski and Randall (1986) proposed that glycerate kinase, in addition to its role in photorespiration in leaves, could serve in C4 species as part of a facilitated diffusion system for the intercellular transport of 3-PGA. Glycerate accumulation in gapcp1gapcp2 roots may form part of the plant response to maintain 3-PGA levels in plastids. The high phosphoglycerate kinase activity measured in gapcp1gapcp2 would be anticipated to provide an increased 3-PGA concentration that would be converted into glycerate by the phosphoglycerate phosphatase (Fig. 8). If this is correct, the increased glycerate concentration in gapcp1gapcp2 should be related to the activity of the cytosolic phosphoglycerate kinase isoform, since the activity of the plastidial isoform should be compromised in the mutant due to the lack of substrate provided by GAPCp in vivo. Glycerate accumulated in the cytosol could then be transported into the plastid, where glycerate kinase would convert glycerate into 3-PGA (Fig. 8). A glycerate transporter was recently identified in Arabidopsis chloroplasts (Pick et al., 2013). However, both glycerate and the triose-phosphate transporters, which can efficiently exchange 3-PGA and glycerate between the cytosol and the plastid, are poorly expressed or nonexpressed in roots (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) and probably in nonphotosynthetic cells in general. We propose that the absence of GAPCp constrains plastid 3-PGA production, making plastids dependent on a cytosolic source of this precursor. This could be supplied either as 3-PGA or glycerate. Since the capacities of both Triose Phosphate Translocator and glycerate transporter are likely to be limited in nonphotosynthetic cells, 3-PGA may be imported via the Glc-P/Pi translocator (albeit inefficiently); alternatively, glycerate, which is present at high levels in gapcp1gapcp2, may also enter by diffusion. If the latter, then plastidic glycerate kinase activity could be a limiting factor.
Figure 8.
Proposed model for metabolic pathways affected by GAPCp activity. Lack of GAPCp activity reduces the 3-PGA precursor for the PPSB, which provides Ser to specific nonphotosynthetic cells but also α-ketoglutarate required for ammonium assimilation through the Gln synthetase/Glu synthase (GS/GOGAT) pathway and/for anaplerotic reactions into the tricarboxylic acid (TCA) cycle. The reduced activity of the GS/GOGAT pathway would increase Gln and would divert l-Glu to the biosynthesis of GABA, which could play a role in balancing carbon and nitrogen metabolism in gapcp1gapcp2 by contributing to the anaplerotic flux of Gln-derived carbon into the tricarboxylic acid cycle when PPSB is restricted. The reduced 3-PGA levels in plastids could be compensated in part by the transport of glycerate from the cytosol and subsequent conversion into 3-PGA by glycerate kinase (GK). The enzymes participating in each biosynthetic pathway are as follows: for the phosphorylated pathway (PPSB), 3-phosphoglycerate dehydrogenase (PGDH), 3-phospho-Ser aminotransferase (PSAT), and 3-phospho-Ser phosphatase (PSP); and for the ammonium assimilation pathway, GS/GOGAT. Other enzymes are as follows: PGAP, phosphoglycerate phosphatase; and PGK, phosphoglycerate kinase; p stands for plastidial. Metabolites are as follows: 1,3-bis-PGAP, 1,3-bis-phosphoglycerate; GAP, glyceraldehyde 3-phosphate; 3-PHP, 3-phosphohydroxypyruvate; and 3-PS, 3-phospho-Ser.
Gln levels were also significantly altered in both roots and the AP following the induction of the enzyme in conditional mutants. The connection between GAPCp, PPSB, and the ammonium assimilation pathway (GS/GOGAT; Muñoz-Bertomeu et al., 2009; Benstein et al., 2013; Cascales-Miñana et al., 2013) could provide clues about the possible mechanism involved in the modification of Gln content. The second enzyme of the PPSB, PSAT, uses l-Glu as cosubstrate in the formation of α-ketoglutarate (Fig. 8). If PSAT activity is restricted, α-ketoglutarate would not be regenerated, and then the second reaction of the GS/GOGAT pathway (GOGAT), which uses α-ketoglutarate as cosubstrate along with Gln, would be inhibited. Thus, Gln levels would increase in the cell. Consequently, the high levels of Gln found in the roots and the AP of gapcp1gapcp2 may be a direct consequence of the arrest in PPSB activity, which in turn affects the GS/GOGAT pathway. The facts that NH+4 levels are increased in roots of gapcp1gapcp2 and that one of the main changes of activating GAPCp1 was the reduction of Gln levels in both roots and the AP would support this idea. In addition, levels of Gln were not reduced in the AP of gapcp1gapcp2 supplied with Ser when the PPSB would be anticipated to be inactive. This would support the high Gln levels in the mutant coming from the reduction of PSAT activity. Gln is the primary product of nitrogen assimilation in plants and a central metabolite of nitrogen metabolism. Although as yet unknown, a Gln-sensing mechanism similar to that recently identified in other plant families (Chellamuthu et al., 2014) could play a pivotal role in coordinating nitrogen metabolism in response to the general metabolic state of the cell. In this regard, Gln along with Asn are the main organic nitrogen forms transported between roots and the AP. Indeed, Gln is the most abundant amino acid in the xylem and phloem sap (Zhang et al., 2010). As such, Gln could be a good candidate for communication between the root and the AP in order to induce the required metabolic adjustment in gapcp1gapcp2.
Concerning gene targets responding to altered GAPCp1 activity, many functional categories were significantly affected by the induction of the enzyme. Many hormone-signaling genes (including those involved in cytokinin, GA, ethylene, and jasmonate responses) responded to the activation of the enzyme, which suggests that they participate in the recovery of plant development. One of them, known as LOG7, is a Lys decarboxylase (cytokinin riboside 5′-monophosphate phosphoribohydrolase) that was up-regulated in both the APs and roots of gapcp1gapcp2 and down-regulated upon GAPCp1 induction. LOG7 has been described as performing a regulatory function in the shoot and root apical meristem and could play an important role in the control of plant development through the cytokinin activation pathway (Tokunaga et al., 2012). Transfer DNA (T-DNA) insertion mutants in which the LOG-dependent pathway is impaired displayed severe retardation of shoot and root growth (Tokunaga et al., 2012). We found that LOG7 was not repressed, but rather induced, in gapcp1gapcp2, which may suggest that LOG7 up-regulation is a consequence of the mutant’s reduced growth rather than a cause of it.
Another gene candidate that could mediate the lack of GAPCp activity is At5g17330, which codes for one of the isoforms of GAD. This enzyme participates in Glu metabolism by catalyzing its irreversible decarboxylation to form GABA (Fait et al., 2011; Fig. 8). The increased GABA levels measured in roots, and especially in the AP, of the gapcp1gapcp2 mutant could well be the consequence of a higher GAD expression in the mutant. GAD could metabolize l-Glu to avoid the accumulation of Gln through the GS reaction. In this sense, GAD expression is repressed after GAPCp1 activation and Gln levels are lowered. On the other hand, GABA catabolism produces succinic semialdehyde, which is converted into succinate by succinic semialdehyde dehydrogenase. We can speculate that this mechanism might contribute to the anaplerotic flux of Gln-derived carbon into the tricarboxylic acid cycle. Increased GAD activity could form part of a metabolic adjustment in gapcp1gapcp2 to restore metabolite homeostasis. Fait et al. (2011) proposed that GAD-mediated conversion of l-Glu into GABA plays a role in balancing carbon and nitrogen metabolism. In this vein, overexpression of GAD during seed maturation increases the nitrogen-to-carbon ratio. As gapcp1gapcp2 mutant roots have an increased nitrogen-to-carbon ratio (Muñoz-Bertomeu et al., 2009), GAD could be a mediator of the response to altered GAPCp1 activity connecting carbon and nitrogen metabolism. We propose a model in which a lack of GAPCp1 activity reduces the PPSB flux that provides Ser to specific heterotrophic cells but also supplies α-ketoglutarate required for the GS/GOGAT pathway (Fig. 8). Inhibition of the GS/GOGAT pathway would increase Gln and divert l-Glu to the biosynthesis of GABA. This metabolite may play a role in balancing carbon and nitrogen metabolism in gapcp1gapcp2 by contributing to the anaplerotic flux of Gln-derived carbon into the tricarboxylic acid cycle (Fig. 8). The 3-PGA deficit in the plastids of specific heterotrophic cells, therefore, would be compensated by increased glycerate and 3-PGA biosynthesis in the cytosol, which we believe would be inefficiently transported to the plastid lumen. Further studies are needed, however, to fully understand how these metabolic and transcriptomic networks are connected and how they interact through GAPCp to regulate plant metabolism and development.
CONCLUSION
The results presented here show that GAPCps do not have a major function in photosynthetic cells but play a crucial role in nonphotosynthetic cells of both photosynthetic and nonphotosynthetic organs, where they could provide essential metabolites for plant survival. The very specific expression pattern of GAPCp in primary root meristems and the root cap could be the key for its important role in this organ. Our results indicate that GAPCp interacts with essential metabolic pathways that affect nitrogen and carbon metabolism and in turn interfere with plant development. Then, GAPCp seems to be an important metabolic connector of essential pathways of primary metabolism in plants. A prerequisite of metabolic engineering is the precise understanding of plant metabolic networks and how they are interconnected. The results here open new directions for research on the mechanisms connecting plant metabolic networks.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) initial seed stocks (ecotype Columbia-0) were supplied by the European Arabidopsis Stock Centre (Scholl et al., 2000). The plastidial gapcp1gapcp2 is a T-DNA insertional double mutant (SAIL_390_G10 and SALK_137288) of genes At1g79530 and At1g16300, respectively, which was obtained previously by our group (Muñoz-Bertomeu et al., 2009). Unless stated otherwise, seeds were vertically sown on 0.8% (w/v) agar plates containing one-fifth-strength MS medium with Gamborg vitamins, as described previously (Muñoz-Bertomeu et al., 2009). Some plates were supplemented with Ser, additional MS salts, or 0.003% (w/v) Bromocresol Purple as indicated in the figure legends. For acidification studies, plates were prepared without MES and adjusted to pH 5.7 with KOH. For the selection of transgenic plants, one-half-strength MS plates supplemented with 0.5% (w/v) Suc and appropriate selection markers were used. Some plantlets were also grown under greenhouse conditions as described elsewhere (Muñoz-Bertomeu et al., 2009). Wherever indicated, conditional gapcp1gapcp2 was treated for 1 h at 37°C several hours before sampling.
Primers
All primers used in this work are listed in Supplemental Table S9.
Cloning and Plant Transformation
The cDNA corresponding to GAPCp1 (At1g79530) was placed under the control of four different promoters: 35S promoter, AP-specific promoter of gene At5g38420 (RBCS2B; Kim et al., 2005), promoter of gene At5g43370 (PHT1.2; Mudge et al., 2002), and heat shock-inducible promoter of gene At5g59720 (HSP18.2; Matsuhara et al., 2000), giving constructs 35S:GAPCp1, RBCS:GAPCp1, PHT:GAPCp1, and HS:GAPCp1, respectively.
Construct 35S:GAPCp1 carrying GAPCp1 fused in frame with GFP at the C-terminal position (GAPCp1-GFP) in plasmid pMDC83 (Curtis and Grossniklaus, 2003) was obtained previously as described (Muñoz-Bertomeu et al., 2009). This plasmid was used to obtain constructs RBCS:GAPCp1, PHT:GAPCp1, and HS:GAPCp1. The 35S promoter of 35S:GAPCp1 was exchanged with the 1,335-bp RBCS promoter previously PCR amplified from plasmid pSBright (Kim et al., 2005), kindly provided by Dr. D. Alabadí (Instituto de Biología Molecular y Celular de Plantas-Consejo Superior de Investigaciones Científicas), using primers described in Supplemental Table S9 to introduce HindIII and PacI sites, giving RBCS:GAPCp1. For construct PHT:GAPCp1, the 35S promoter of 35S:GAPCp1 was exchanged with the 2,012-bp PHT promoter previously PCR amplified from plasmid pGWB1 (Nakagawa et al., 2007), in which PHT1.2 had been subcloned (Kirchsteiger et al., 2012), using primers described in Supplemental Table S9 to introduce HindIII and SpeI sites, giving PHT:GAPCp1. For construct HS:GAPCp1, the 35S promoter of 35S:GAPCp1 was exchanged for an 852-bp fragment corresponding to the promoter region of gene HSP18.2, giving the vector HS:GAPCp1. The HSP18.2 promoter was obtained by digestion of the PTT101 plasmid (Matsuhara et al., 2000) with enzymes SpeI and HindIII. For GUS-GFP gene promoter-reporter fusions, a 1.5-kb fragment corresponding to the native GAPCp1 promoter (−1,521 to +18 relative to the GAPCp1 translation start) was placed before the 5′-GUS-GFP-3′ gene fusion in pCAMBIA1303 as described previously (Muñoz-Bertomeu et al., 2009). All PCR-derived constructs were verified by DNA sequencing.
Arabidopsis plants were transformed with the different constructs by the floral dipping method (Clough and Bent, 1998) with Agrobacterium tumefaciens carrying pSOUP. As gapcp1gapcp2 is sterile, the progeny of heterozygous plants (heterozygous for GAPCp1 and homozygous for the mutant allele of GAPCp2) were transformed with the different constructs. Transformants were selected by antibiotic selection, while homozygous gapcp1gapcp2, heterozygous (homozygous for GAPCp1 and heterzygous for GAPCp2), and the wild type were identified by PCR genotyping using gene-specific primers (RP and LP) and left border primers of the T-DNA insertions described in Supplemental Table S9. Several independent single-insertion homozygous T3 lines were obtained for all the different constructs. After characterization by qRT-PCR, two to three different lines were selected for further analyses depending on the experiment. Syngenic wild-type lines were used as controls for our studies.
qRT-PCR and Microarrays
qRT-PCR was performed as described previously (Cascales-Miñana et al., 2013). Each reaction was performed in triplicate. Data are means of three biological samples. PCR amplification specificity was confirmed with a heat dissociation curve (from 60°C to 95°C). The efficiency of the PCR was calculated, and different internal standards were selected (Czechowski et al., 2005) depending on the primers’ efficiency. Relative mRNA abundance was calculated using the comparative cycle threshold method according to Pfaffl (2001). Primers used for PCRs are listed in Supplemental Table S9.
For microarray experiments, total RNAs from three pools of 21-d-old roots and AP (gapcp1gapcp2 and the wild type or gapcp1gapcp2 and gapcp1gapcp2 HS:GAPCp1), vertically grown on one-fifth-strength MS plates as described above, were extracted using the total RNA isolation kit (Macherey-Nagel). For the induction experiments, gapcp1gapcp2 and gapcp1gapcp2 HS:GAPCp1 were treated for 1 h at 37°C 4 h before sampling. RNA integrity was determined using RNA 6000 Nano Labchips in an Agilent 2100 Bioanalyzer following the manufacturer’s protocol. RNA amplification, labeling, and hybridization were done as described by Bueso et al. (2007). The Arabidopsis oligonucleotide microarrays were obtained from David Galbraith (http://www.ag.arizona.edu/microarray/). The chosen design for the microarray experiments was a balanced block design, with three to four biological replicates and two dye swaps. The microarray data analysis was done as described by Bissoli et al. (2012). Genes were selected as differentially expressed when expression was at least 1.5-fold higher or lower than the controls and the false discovery rate was lower than 10% when applying the significance analysis. Significant functional categories were selected using the Munich Information Center for Protein Sequences software (http://mips.helmholtz-muenchen.de/proj/funcatDB/). Microarray data were submitted to the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) with the accession number E-MTAB-3262 and are listed in Supplemental Table S9.
Enzyme Assays, Metabolite Determination, and Ion Analysis
APs and roots of 18- to 21-d-old wild-type and different gapcp1gapcp2 transgenic lines were used for the determination of metabolite content. Starch and total soluble sugars were determined with the ENZYTEC starch kit (R-Biopharm). Protein content was quantified using the Bio-Rad protein assay kit. The levels of other metabolites were determined in derivatized methanol extracts by gas chromatography-mass spectrometry following the protocol defined by Lisec et al. (2006). Macronutrients and micronutrients were determined according to Quiñones et al. (2011). Ammonium content was determined according to Sarasketa et al. (2014). Phosphoglycerate kinase activity was determined according to Brandina et al. (2006).
Western Blots
Crude protein extracts from whole wild-type and different gapcp1gapcp2 transgenic lines were obtained by harvesting 0.5 g of plant material in liquid nitrogen and grinding in 1 mL of ice-cold homogenization buffer as described previously (Cascales-Miñana et al., 2013). Proteins were electrotransferred onto Immune-Blot nitrocellulose membranes (Bio-Rad) using the Mini Tran-Blot Cell (Bio-Rad) for 1 h at 100 V with the transfer buffer (17 mm Tris, 192 mm Gly, and 20% [v/v] methanol). Immunoblots were probed with anti-GFP antibodies (Molecular Probes; A-11122) or a specific antibody raised against a 16-amino acid specific epitope (Ac-KGSINVIDDSTLEINC-amide) of GAPCp1. Ponceau S-stained membranes or nonspecific bands are shown as loading controls. Cross-reacting bands were detected using the ECL Select Western-Blotting Detection Reagent Kit (Amersham Biosciences).
Microscopy
For GUS activity assays, roots were processed as described (Muñoz-Bertomeu et al., 2009). Images were acquired with a Leica DM1000 microscope and a Leica DC350 digital camera. GFP fluorescence was observed with a confocal microscope (Leica TCS-SP).
Statistics
Experimental values represent means and se, and n represents the number of independent samples. P values were calculated with Student’s t test (two tailed) using Microsoft Excel. The level of significance was fixed at 5% (0.05). PCA of metabolomic data was performed using the Excel add-in Multibase package (Numerical Dynamics). For PCA, relative values normalized to the mean response calculated for the wild type were scaled by sd prior to the analysis. The first principal component was used for the ANOVA of different lines, with Tukey’s posthoc tests to differentiate between the lines.
Sequence data from this article can be found in the Arabidopsis Genome Initiative data libraries under accession numbers At1g79530 (GAPCp1) and At1g16300 (GAPCp2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Molecular and physiological analysis of GAPCp isoforms.
Supplemental Figure S2. Cellular expression of GAPCp1 under the control of the RBCS and the 35S promoters.
Supplemental Figure S3. Quantification of changes in transcript level using qRT-PCR.
Supplemental Figure S4. Molecular and phenotypical analyses of gapcp1gapcp2 expressing GAPCp1-GFP under the control of the HS promoter.
Supplemental Table S1. Metabolite levels in the aerial parts of wild-type, gapcp1gapcp2 (g1g2), g1g2 RBCS:GAPCp1, and g1g2 35S:GAPCp1 plants sampled at the end of the light period.
Supplemental Table S2. Metabolite levels in the aerial parts of wild-type, gapcp1gapcp2 (g1g2), and g1g2 RBCS:GAPCp1 plants grown in standard medium or supplemented with either 2% Suc or 0.1 mm Ser.
Supplemental Table S3. Metabolite levels in aerial parts and roots of wild-type, gapcp1gapcp2 (g1g2), g1g2 RBCS:GAPCp1, and g1g2 PHT:GAPCp1 sampled at the middle of the light period.
Supplemental Table S4. Metabolite levels in aerial parts and roots of wild-type and gapcp1gapcp2 HS:GAPCp1 plants.
Supplemental Table S5. List of genes mentioned in this work whose expression was misregulated in gapcp1gapcp2 (g1g2) or g1g2 HS:GAPCp1 following microarray experiments.
Supplemental Table S6. Macronutrient and micronutrient concentrations in aerial parts and roots of wild-type, gapcp1gapcp2 (g1g2), and g1g2 PHT:GAPCp1 plants.
Supplemental Table S7. Effect of 1-h treatment at 37°C on metabolite levels in aerial parts and roots of gapcp1gapcp2 plants.
Supplemental Table S8. Functional categories of genes differentially expressed in the aerial parts and roots of gapcp1gapcp2 HS:GAPCp1 after GAPCp1 induction as compared with gapcp1gapcp2.
Supplemental Table S9. List of primers used in this work.
Supplemental Materials S1. Genes differentially expressed in transgenic and mutant lines.
Acknowledgments
We thank Servei Central de Suport a la Investigació Experimental and Unitat Central de Investigació de Medicina of the Universitat de València for technical assistance.
Glossary
- PEP
phosphoenolpyruvate
- 3-PGA
3-phosphoglycerate
- PPSB
phosphorylated pathway of serine biosynthesis
- AP
aerial part
- cDNA
complementary DNA
- RT
reverse transcription
- PCA
principal component analysis
- GABA
γ-aminobutyrate
- qRT
quantitative reverse transcription
- Pi
inorganic phosphate
- MS
Murashige and Skoog
- T-DNA
transfer DNA
- qRT
quantitative real-time
Footnotes
This work was supported by the Spanish Government and the European Union (Fondo Europeo de Desarrollo Regional grant no. BFU2012–31519 to J.M.-B., Formación del Personal Investigador fellowship to S.R.-T., and Agencia Española de Cooperación Internacional fellowship to A.D.A.), by the Valencian Regional Government (PROMETEO grant no. II/2014/052), and by the University of Valencia (Atracció de Talent fellowship to M.F.-T.).
References
- Andre C, Benning C (2007) Arabidopsis seedlings deficient in a plastidic pyruvate kinase are unable to utilize seed storage compounds for germination and establishment. Plant Physiol 145: 1670–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19: 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriotis VM, Kruger NJ, Pike MJ, Smith AM (2010) Plastidial glycolysis in developing Arabidopsis embryos. New Phytol 185: 649–662 [DOI] [PubMed] [Google Scholar]
- Backhausen JE, Vetter S, Baalmann E, Kitzmann C, Scheibe R (1998) NAD-dependent malate dehydrogenase and glyceraldehyde 3-phosphate dehydrogenase isoenzymes play an important role in dark metabolism of various plastid types. Planta 205: 359–366 [Google Scholar]
- Baek D, Jin Y, Jeong JC, Lee HJ, Moon H, Lee J, Shin D, Kang CH, Kim DH, Nam J, et al. (2008) Suppression of reactive oxygen species by glyceraldehyde-3-phosphate dehydrogenase. Phytochemistry 69: 333–338 [DOI] [PubMed] [Google Scholar]
- Baud S, Wuillème S, Dubreucq B, de Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C (2007) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52: 405–419 [DOI] [PubMed] [Google Scholar]
- Benstein RM, Ludewig K, Wulfert S, Wittek S, Gigolashvili T, Frerigmann H, Gierth M, Flügge UI, Krueger S (2013) Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25: 5011–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissoli G, Niñoles R, Fresquet S, Palombieri S, Bueso E, Rubio L, García-Sánchez MJ, Fernández JA, Mulet JM, Serrano R (2012) Peptidyl-prolyl cis-trans isomerase ROF2 modulates intracellular pH homeostasis in Arabidopsis. Plant J 70: 704–716 [DOI] [PubMed] [Google Scholar]
- Brandina I, Graham J, Lemaitre-Guillier C, Entelis N, Krasheninnikov I, Sweetlove L, Tarassov I, Martin RP (2006) Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim Biophys Acta 1757: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Bueso E, Alejandro S, Carbonell P, Perez-Amador MA, Fayos J, Bellés JM, Rodriguez PL, Serrano R (2007) The lithium tolerance of the Arabidopsis cat2 mutant reveals a cross-talk between oxidative stress and ethylene. Plant J 52: 1052–1065 [DOI] [PubMed] [Google Scholar]
- Cascales-Miñana B, Muñoz-Bertomeu J, Flores-Tornero M, Anoman AD, Pertusa J, Alaiz M, Osorio S, Fernie AR, Segura J, Ros R (2013) The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell 25: 2084–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellamuthu VR, Ermilova E, Lapina T, Lüddecke J, Minaeva E, Herrmann C, Hartmann MD, Forchhammer K (2014) A widespread glutamine-sensing mechanism in the plant kingdom. Cell 159: 1188–1199 [DOI] [PubMed] [Google Scholar]
- Chen M, Thelen JJ (2010) The plastid isoform of triose phosphate isomerase is required for the postgerminative transition from heterotrophic to autotrophic growth in Arabidopsis. Plant Cell 22: 77–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Nesi AN, Angelovici R, Lehmann M, Pham PA, Song L, Haslam RP, Napier JA, Galili G, Fernie AR (2011) Targeted enhancement of glutamate-to-γ-aminobutyrate conversion in Arabidopsis seeds affects carbon-nitrogen balance and storage reserves in a development-dependent manner. Plant Physiol 157: 1026–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Devaiah SP, Narasimhan R, Pan X, Zhang Y, Zhang W, Wang X (2012) Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24: 2200–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajirezaei MR, Biemelt S, Peisker M, Lytovchenko A, Fernie AR, Sonnewald U (2006) The influence of cytosolic phosphorylating glyceraldehyde 3-phosphate dehydrogenase (GAPC) on potato tuber metabolism. J Exp Bot 57: 2363–2377 [DOI] [PubMed] [Google Scholar]
- Han S, Wang Y, Zheng X, Jia Q, Zhao J, Bai F, Hong Y, Liu Y (2015) Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27: 1316–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL (2007) Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J 50: 967–981 [DOI] [PubMed] [Google Scholar]
- Kim JW, Dang CV (2005) Multifaceted roles of glycolytic enzymes. Trends Biochem Sci 30: 142–150 [DOI] [PubMed] [Google Scholar]
- Kim JY, Rim Y, Wang J, Jackson D (2005) A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev 19: 788–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchsteiger K, Ferrández J, Pascual MB, González M, Cejudo FJ (2012) NADPH thioredoxin reductase C is localized in plastids of photosynthetic and nonphotosynthetic tissues and is involved in lateral root formation in Arabidopsis. Plant Cell 24: 1534–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski LA, Randall DD (1986) Thiol-dependent regulation of glycerate metabolism in leaf extracts: the role of glycerate kinase in C4 plants. Plant Physiol 81: 656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li G, Kronzucker HJ, Baluška F, Shi W (2014) Ammonium stress in Arabidopsis: signaling, genetic loci, and physiological targets. Trends Plant Sci 19: 107–114 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Liu TY, Aung K, Tseng CY, Chang TY, Chen YS, Chiou TJ (2011) Vacuolar Ca2+/H+ transport activity is required for systemic phosphate homeostasis involving shoot-to-root signaling in Arabidopsis. Plant Physiol 156: 1176–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhara S, Jingu F, Takahashi T, Komeda Y (2000) Heat-shock tagging: a simple method for expression and isolation of plant genome DNA flanked by T-DNA insertions. Plant J 22: 79–86 [DOI] [PubMed] [Google Scholar]
- May A, Berger S, Hertel T, Köck M (2011) The Arabidopsis thaliana phosphate starvation responsive gene AtPPsPase1 encodes a novel type of inorganic pyrophosphatase. Biochim Biophys Acta 1810: 178–185 [DOI] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Irles-Segura A, Mateu I, Nunes-Nesi A, Fernie AR, Segura J, Ros R (2010) The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol 152: 1830–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J, Cascales-Miñana B, Mulet JM, Baroja-Fernández E, Pozueta-Romero J, Kuhn JM, Segura J, Ros R (2009) Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol 151: 541–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC (2011) Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Brinkmann H, Cerff R (2003) Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. J Mol Evol 57: 16–26 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick TR, Bräutigam A, Schulz MA, Obata T, Fernie AR, Weber AP (2013) PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc Natl Acad Sci USA 110: 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC. (1996) The organization and regulation of plant glycolysis. Annu Rev Plant Physiol Plant Mol Biol 47: 185–214 [DOI] [PubMed] [Google Scholar]
- Prabhakar V, Löttgert T, Geimer S, Dörmann P, Krüger S, Vijayakumar V, Schreiber L, Göbel C, Feussner K, Feussner I, et al. (2010) Phosphoenolpyruvate provision to plastids is essential for gametophyte and sporophyte development in Arabidopsis thaliana. Plant Cell 22: 2594–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones A, Martínez-Alcántara B, San-Francisco S, García-Mina JM, Legaz F (2011) Methyl xanthine as a potential alternative to gibberellic acid in enhancing fruit set and quality in clementine citrus trees in Spain. Exp Agric 47: 159–171 [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez-Casati DF (2008) Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol 148: 1655–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros R, Muñoz-Bertomeu J, Krueger S (2014) Serine in plants: biosynthesis, metabolism, and functions. Trends Plant Sci 19: 564–569 [DOI] [PubMed] [Google Scholar]
- Sarasketa A, González-Moro MB, González-Murua C, Marino D (2014) Exploring ammonium tolerance in a large panel of Arabidopsis thaliana natural accessions. J Exp Bot 65: 6023–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Tokunaga H, Kojima M, Kuroha T, Ishida T, Sugimoto K, Kiba T, Sakakibara H (2012) Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J 69: 355–365 [DOI] [PubMed] [Google Scholar]
- Toujani W, Muñoz-Bertomeu J, Flores-Tornero M, Rosa-Téllez S, Anoman AD, Alseekh S, Fernie AR, Ros R (2013) Functional characterization of the plastidial 3-phosphoglycerate dehydrogenase family in Arabidopsis. Plant Physiol 163: 1164–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Straeten D, Rodrigues-Pousada RA, Goodman HM, Van Montagu M (1991) Plant enolase: gene structure, expression, and evolution. Plant Cell 3: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vescovi M, Zaffagnini M, Festa M, Trost P, Lo Schiavo F, Costa A (2013) Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed Arabidopsis roots. Plant Physiol 162: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AP, Schwacke R, Flügge UI (2005) Solute transporters of the plastid envelope membrane. Annu Rev Plant Biol 56: 133–164 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P (2013) Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci 4: 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tan Q, Lee R, Trethewy A, Lee YH, Tegeder M (2010) Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22: 3603–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]