Homologs of hydroxycinnamoyltransferase, involved in lignin biosynthesis, interact directly with leucine-rich receptor proteins to suppress the hypersensitive response.
Abstract
In plants, most disease resistance genes encode nucleotide binding Leu-rich repeat (NLR) proteins that trigger a rapid localized cell death called a hypersensitive response (HR) upon pathogen recognition. The maize (Zea mays) NLR protein Rp1-D21 derives from an intragenic recombination between two NLRs, Rp1-D and Rp1-dp2, and confers an autoactive HR in the absence of pathogen infection. From a previous quantitative trait loci and genome-wide association study, we identified a single-nucleotide polymorphism locus highly associated with variation in the severity of Rp1-D21-induced HR. Two maize genes encoding hydroxycinnamoyltransferase (HCT; a key enzyme involved in lignin biosynthesis) homologs, termed HCT1806 and HCT4918, were adjacent to this single-nucleotide polymorphism. Here, we show that both HCT1806 and HCT4918 physically interact with and suppress the HR conferred by Rp1-D21 but not other autoactive NLRs when transiently coexpressed in Nicotiana benthamiana. Other maize HCT homologs are unable to confer the same level of suppression on Rp1-D21-induced HR. The metabolic activity of HCT1806 and HCT4918 is unlikely to be necessary for their role in suppressing HR. We show that the lignin pathway is activated by Rp1-D21 at both the transcriptional and metabolic levels. We derive a model to explain the roles of HCT1806 and HCT4918 in Rp1-mediated disease resistance.
Plants have developed sophisticated mechanisms to inhibit infection by pathogens. One of the most important of these is mediated by dominant, major effect, resistance (R) genes that directly or indirectly recognize the presence of specific pathogen-derived molecules called effectors (Bent and Mackey, 2007). Interaction between R genes and their cognate effectors generally triggers a series of defense responses, including induction of the hypersensitive response (HR), a rapid localized cell death at the point of pathogen penetration, production of reactive oxygen species, pathogenesis-related (PR) gene expression, production of secondary metabolites, and lignification of cell walls (Heath, 2000; Mur et al., 2008).
In vascular plants, the phenylpropanoid secondary metabolism pathway is required for the biosynthesis of many metabolites, including lignin, which provides mechanical strength to vascular tissues and defense against biotic stresses (Lewis and Yamamoto, 1990; Li et al., 2010). The phenylpropanoid pathway starts with Phe, which is converted to cinnamic acid by Phe ammonia-lyase (PAL; Supplemental Fig. S1). Cinnamic acid can be further converted to either salicylic acid (SA), a hormone that plays important roles in plant defense (Dempsey et al., 2011), or p-coumaroyl CoA, the precursor of flavonoids, isoflavanoids, anthocyanins, and lignin (Boerjan et al., 2003). For lignin biosynthesis, p-coumaroyl CoA either is converted by cinnamoyl-CoA reductase (CCR) and cinnamyl-alcohol dehydrogenase (CAD) to p-hydroxyphenyl lignin (H monolignol) or undergoes a series of reactions catalyzed by different enzymes, including hydroxycinnamoyltransferase (HCT) and caffeoyl CoA O-methyltransferase (CCoAOMT), to produce guaiacyl lignin (G monolignol) and syringyl lignin (S monolignol). HCT catalyses the conversion from coumaroyl CoA to coumaroyl shikimate or coumaroyl quinate and also, from caffeoyl shikimate or caffeoyl quinate to caffeoyl CoA (Hoffmann et al., 2003). Lignin itself is a heterogeneous tridimensional phenolic polymer resulting from the oxidative polymerization of S, G, and H monolignols with hydrogen peroxide (Boerjan et al., 2003).
Most R genes encode proteins carrying a nucleotide-binding, APAF-1 (for Apoptotic protease activation factor1), R proteins and CED (for Cell death protein)-4 (NB-ARC) domain and a Leu-rich repeat (LRR) domain. The so-called nucleotide binding Leu-rich repeat (NLR) proteins (Ellis et al., 2000; Dangl and Jones, 2001) can be subdivided into two main classes according to their N-terminal secondary structure: one is a dicot-specific class that carries a Toll-IL1 receptor (TIR) domain (TIR receptor-nucleotide binding LRR [TNL]) and one, which is found in both dicots and monocots, carries a putative coiled-coil (CC) domain (coiled coil-nucleotide binding Leu-rich repeat [CNL]). The maize (Zea mays) R gene Rp1-D encodes a CNL, which confers resistance to specific races of the biotrophic fungal pathogen Puccinia sorghi, the causal agent of maize common rust (Hu et al., 1996). The locus that carries Rp1-D, Rp1, is complex, and many haplotypes have been characterized (Smith et al., 2004). The Rp1-D haplotype consists of nine NLR paralogs, Rp1-dp1 to Rp1-dp8, which have no known function, and Rp1-D (Sun et al., 2001). Rp1-D21, a mutant conferring an autoactive HR lesion phenotype in the absence of pathogen infection, was formed by unequal crossing over at the Rp1 locus and subsequent intragenic recombination between Rp1-D and Rp1-dp2 (Sun et al., 2001; Smith et al., 2010). Several other hallmarks of the pathogen-induced immune response, including hydrogen peroxide accumulation and increased expression of the defense-related genes PR1, PRms, and WIP1 (for wound-induced protein1), are also associated with Rp1-D21-mediated lesion phenotype in maize (Chintamanani et al., 2010).
The severity of Rp1-D21 HR lesion phenotype is affected by temperature, light, and importantly, genetic background (Chintamanani et al., 2010; Negeri et al., 2013). We have used Rp1-D21 as a tool to identify loci associated with modulation of the strength of HR segregating in several mapping populations (Chintamanani et al., 2010; Chaikam et al., 2011; Olukolu et al., 2013, 2014). One of the populations used, called the nested association mapping (NAM) population, is an extremely large and powerful mapping population that allows the mapping of quantitative trait loci with high precision (McMullen et al., 2009). In a previous study (Olukolu et al., 2014), we were able to identify single-nucleotide polymorphism (SNP) markers at 44 loci that were highly associated with variation in the HR phenotype. Because the markers were mapped with extremely high precision, we were also able to tentatively identify candidate genes, variation in the structure or expression of which might underlie variation in the HR phenotype. Two HCT homologous genes identified among these candidate genes are the focus of this study.
We have recently shown that transient expression of Rp1-D21 in Nicotiana benthamiana confers an HR, whereas transient expression of its nonautoactive parental alleles Rp1-D and Rp1-dp2 does not (Wang and Balint-Kurti, 2015; Wang et al., 2015b). Furthermore, we also recapitulated in N. benthamiana the HR phenotype conferred by a synthetic Rp1-D derivative gene construct, Hd2, that had previously been assessed in stably transformed maize (Smith et al., 2010) and showed that 10 of 12 Rp1-D21 suppressor mutants isolated in maize similarly abolished or reduced autoactivity in N. benthamiana (Wang et al., 2015b).
In this study, we provide evidence to validate the role of the two HCT homologs in modulating Rp1-D21-induced HR. Coexpression of either homolog with Rp1-D21 in N. benthamiana suppresses Rp1-D21-induced HR but not HR caused by other autoactive CNLs. Furthermore, the HCT proteins physically associate with Rp1 proteins in coimmunoprecipitation (Co-IP) and bimolecular fluorescence complementation (BiFC) assays. We furthermore show that nearly all of the genes required for lignin biosynthesis are differentially expressed in Rp1-D21 mutants compared with isogenic wild-type lines. We discuss the importance of the lignin pathway in modulating the defense response and generate models for NLR protein function.
RESULTS
HCT Proteins Suppress Rp1-D21-Induced HR in N. benthamiana
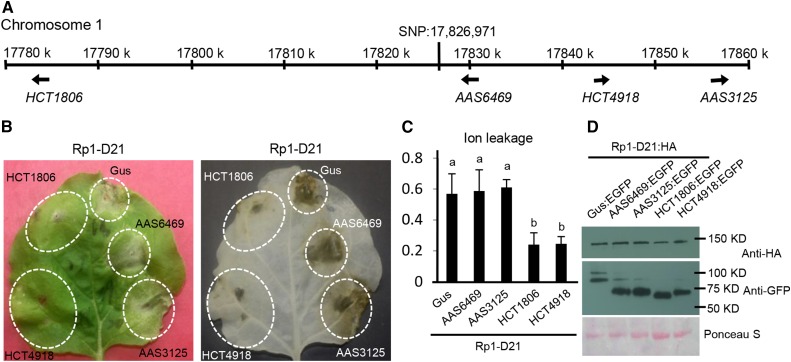
From our previous genome-wide association study (GWAS) analysis in maize using the maize NAM population (McMullen et al., 2009), we identified 44 SNPs associated with variation in Rp1-D21-induced HR (Olukolu et al., 2014). The strongest hit, with a resample model inclusion probability of 0.95 (of a maximum of 1), was at 17,826,971 bp on chromosome 1. Four genes map within 50 kb of this SNP in the annotated B73 maize genome (Schnable et al., 2009). Two of these genes, GRMZM2G061806 (hereafter called HCT1806) and GRMZM2G114918 (hereafter HCT4918), are HCT homologs that flank and are 42.4 kb distal and 16.4 kb proximal to the associated SNP, respectively. The other two genes encode an aromatic aldehyde synthase (AAS), GRMZM2G056469 (hereafter AAS6469) and GRMZM2G093125 (hereafter AAS3125), and are 2.3 kb and 28.7 kb proximal to the SNP, respectively (Fig. 1A).
Figure 1.
HCT1806 and HCT4918 suppressed Rp1-D21-induced HR. A, The positions of genes (HCT1806, HCT4918, AAS6469, and AAS3125) relative to the SNP on chromosome 1 were shown to be highly associated with variation in Rp1-D21-induced HR from a previous study (Olukolu et al., 2014). Arrows indicate the direction of transcription of the genes. B, The phenotypes resulting from transient coexpression of HCT1806, HCT4918, AAS6469, AAS3125, or Gus with Rp1-D21. The pictures were taken at 3 d after inoculation. The two pictures show the same leaf before (left) and after (right) ethanol clearing. C, Ion leakage conductivity was measured at 60 h after coexpression of HCT1806, HCT4918, AAS6469, AAS3125, or Gus with Rp1-D21. Significant differences (P < 0.05) between samples are indicated by different letters (a and b). D, Total protein was extracted from agro-infiltrated leaves at 30 h post inoculation (hpi). Anti-HA was used to detect the expression of Rp1-D21, and anti-GFP was used to detect HCT1806, HCT4918, AAS6469, AAS3125, or Gus. Equal loading of protein samples is shown by Ponceau S staining. Each experiment was performed three times with similar results.
To explore which of these four genes might be associated with variation in the severity of Rp1-D21-induced HR, we first studied the linkage disequilibrium (LD) relationships in the NAM population in this region (Supplemental Fig. S2). We detected strong LD between the most highly associated SNP at 17,826,971 bp on chromosome 1 (AGPv2 map) and SNPs within or close to all four of the annotated genes in this region. The strongest LD relationships between the associated SNP and SNPs within coding or 5′ regions of genes (where most regulatory features are found) were with the two HCT homologous genes, particularly HCT1806. Although this analysis did not rule out the possibility of any of these four genes being the causal gene, it suggested that the two HCT genes were the more likely candidates.
We then used a N. benthamiana-Agrobacterium tumefaciens-mediated transient expression system, a model system widely used for functional analysis of R gene-mediated HR for both dicots and monocots NLRs (van Ooijen et al., 2008; Slootweg et al., 2010; Bai et al., 2012; Qi et al., 2012). We have previously shown that this is an appropriate system for the functional analysis of Rp1-D21 (Wang et al., 2015b). When Rp1-D21:HA (Rp1-D21 fused with a 3× HA tag at the C terminus) was transiently coexpressed with Gus:EGFP (a GUS gene fused with enhanced GFP at the C terminus), an HR phenotype was observed 3 d postinfiltration. This was similar to previous observations of the phenotype conferred by Rp1-D21:HA alone (Wang et al., 2015b). Transient coexpression of HCT1806:EGFP or HCT4918:EGFP but not AAS6469:EGFP or AAS3125:EGFP with Rp1-D21:HA suppressed Rp1-D21-induced HR (Fig. 1B). Ion leakage conductivity data confirmed our visual observations; coexpression of either HCT protein with Rp1-D21:HA significantly reduced levels of ion leakage compared with coexpression of the Gus:EGFP control or the two AAS proteins with Rp1-D21:HA (Fig. 1C). Western-blot results showed that Rp1-D21 had similar levels of expression when coexpressed with Gus:EGFP, HCT:EGFP, or AAS:EGFP and that all of the EGFP-tagged proteins were expressed at substantial and broadly comparable levels (Fig. 1D).
Rp1-D21 Regulates the Lignin Pathway at Transcriptional and Metabolic Levels
To investigate whether HCTs and other key enzymes in lignin pathway are differentially expressed in hybrids carrying Rp1-D21 compared with wild-type isolines, we performed RNA-sequencing (RNA-Seq) analysis in two maize hybrid backgrounds: B73 × H95 and Mo17 × H95. For each background, we compared isogenic lines that differed at the Rp1 locus. Wild-type lines were heterozygous for the H95 and B73 or Mo17 haplotypes, whereas mutant lines carried the Rp1-D21 haplotype instead of the H95 Rp1 haplotype (Chaikam et al., 2011).
All of the four genes (HCT1806, HCT4918, AAS6469, and AAS3125) adjacent to the SNP associated with Rp1-D21-induced HR were more than 100-fold up-regulated in mutant compared with the wild-type hybrids in both B73 × H95 and Mo17 × H95 backgrounds (Supplemental Table S1). In Arabidopsis (Arabidopsis thaliana), there is only one HCT family member (AtHCT: AT5G48930). Using AtHCT as a query to search the maize database (www.phytozome.net), we identified 24 HCT homologs in maize (based on a cutoff of E value of 3.8E-35; Supplemental Table S1). Of the HCT homologs, we found that GRMZM2G127251 (HCT7251), GRMZM2G030436 (HCT0436), GRMZM2G158083 (HCT8083), and GRMZM2G035584 (HCT5584) were all up-regulated in the Rp1-D21 mutant lines in both backgrounds compared with the wild-type isolines. The HCT homologs AC215260.3, GRMZM2G178769, and GRMZM2G131165 were down-regulated in mutant backgrounds, whereas other HCT homologs were either not differentially expressed or not detected in the RNA-Seq data (Supplemental Table S1).
We further observed that the genes encoding homologs of nearly all key enzymes required for lignin biosynthesis, including 4-hydroxycinnamoyl-CoA ligase (4CL), p-coumarate 3-hydroxylase (C3H), cinnamate 4-hydroxylase (C4H), CAD, CCoAOMT, CCR, caffeic/5-hydroxyferulic acid O-methyltransferase (COMT), ferulate 5-hydroxylase (F5H), and PAL (Supplemental Fig. S1; Supplemental Table S1), were up-regulated in mutant compared with the wild type in both B73 × H95 and Mo17 × H95 backgrounds.
To investigate the metabolic changes in the Rp1-D21 lines, we performed liquid chromatography-mass spectrometry-based metabolite profiling on isolines with or without Rp1-D21. We found that accumulation of p-coumaroyl quinate and p-coumaroyl shikimate but not chlorogenic acid (3-O-caffeoylquinate) was significantly higher in Rp1-D21 than wild-type isolines in both B73 × H95 and Mo17 × H95 backgrounds (Supplemental Fig. S3). We also found that the lignin content was significantly higher in the Rp1-D21 isolines compared with their wild-type counterparts (Supplemental Fig. S3).
Other Maize HCT Homologs Do Not Confer the Same Level of Suppression of Rp1-D21-Induced HR
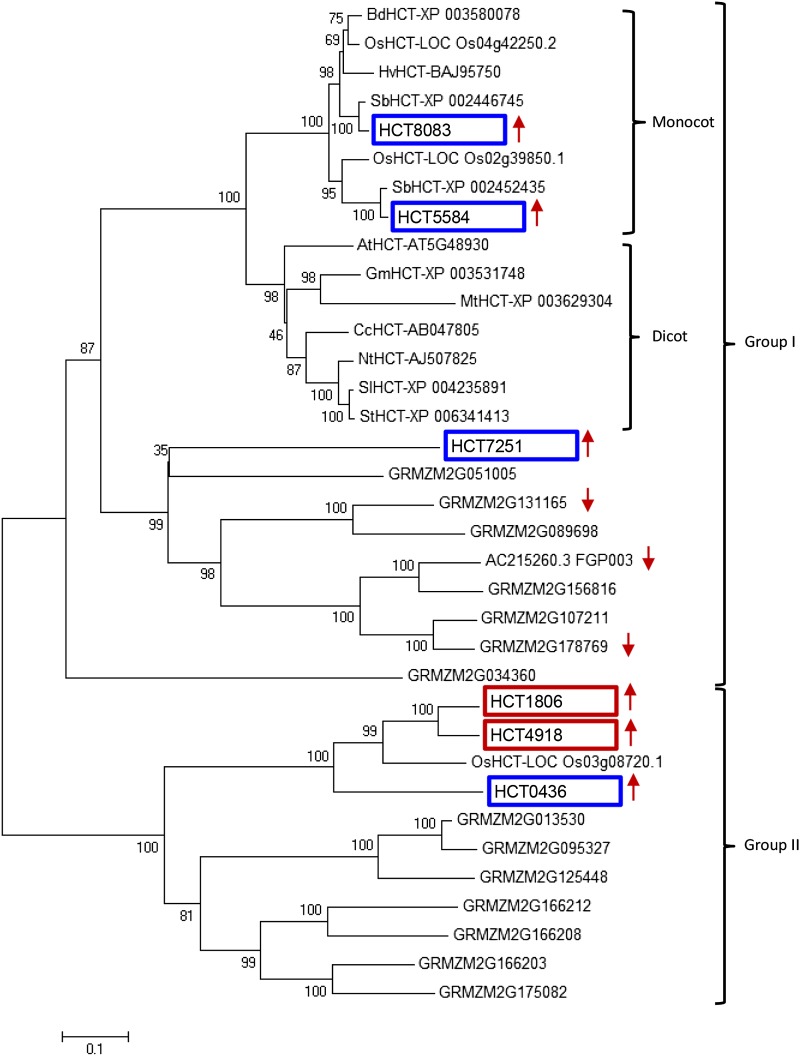
To further investigate the relationship between HCT1806/HCT4918 and other HCT homologs from maize and other plant species, we performed a phylogenetic analysis (Fig. 2). The HCTs were divided into two different groups that we termed groups 1 and 2 (Fig. 2). Both groups contained two major clades. Of the six maize HCT homologs with expressions that were up-regulated in the isolines carrying Rp1-D21 (Supplemental Table S1), HCT8083 and HCT5584 were in a monocot-specific subclade in group 1, closely related to other monocot HCTs from rice (Oryza sativa), barley (Hordeum vulgare), sorghum, and Brachypodium distachyon. HCT7251 was in the other clade in group 1, whereas HCT1806, HCT4918, and HCT0436 were closely related in group 2, forming a subclade with a rice HCT (Os03g08720; Fig. 2).
Figure 2.
Phylogenetic analysis of HCTs from diverse plant species. The two maize HCTs (HCT1806 and HCT4918) colocalized with the SNP associated with Rp1-D21-induced HR are indicated in red boxes, and the other Rp1-D21-induced HCTs used for transient expression in N. benthamiana are indicated in blue boxes. The maize HCTs with expression that is up-regulated or down-regulated by the presence of Rp1-D21 (Supplemental Table S1) are indicated by up or down arrows, respectively. All gene names starting with GRMZM and AC215260.3 are derived from maize. At, Arabidopsis; Bd, B. distachyon; Cc, Coffea canephora; Gm, Glycine max; Hv, barley; Mt, Medicago truncatula; Nt, Nicotiana tabacum; Os, rice; Sb, sorghum (Sorghum bicolor); Sl, Solanum lycopersicum; St, Solanum tuberosum.
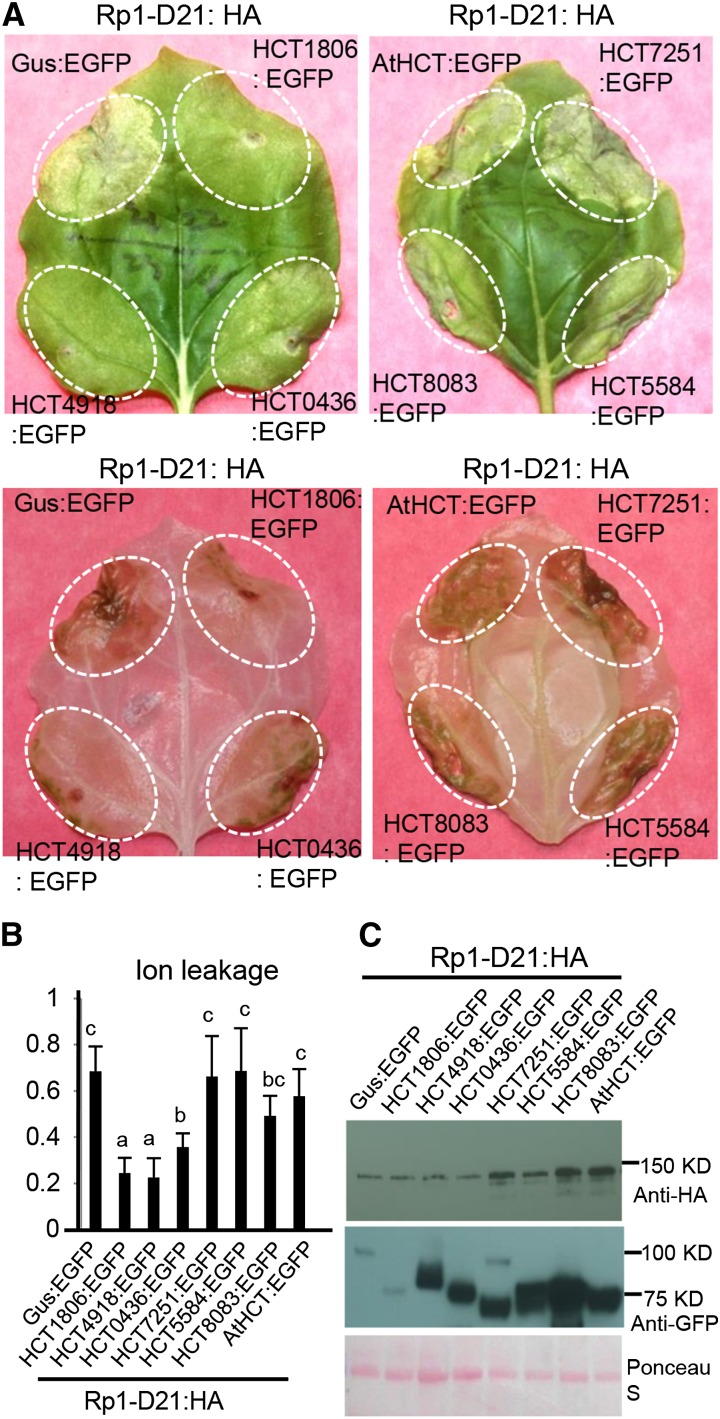
To further investigate whether the other HCTs with expressions that were induced in Rp1-D21 mutants (Supplemental Table S1) might be associated with Rp1-D21-induced HR, we coexpressed HCT0436, HCT7251, HCT5584, or HCT8083 with Rp1-D21:HA in N. benthamiana. HCT0436, which was closely related to HCT1806 and HCT4918, partially suppressed Rp1-D21-induced HR (Fig. 3A). None of the other three HCT homologs suppressed Rp1-D21-induced HR (Fig. 3A). Ion leakage conductivity data confirmed our visual observations (Fig. 3B). Our data suggest that the suppression function in Rp1-D21-induced HR is specific to the phylogenetic subclade containing HCT1806 and HCT4918 (Fig. 2). To investigate whether HCT from other species might have a suppressive role in Rp1-D21-induced HR, we coexpressed AtHCT with Rp1-D21 and found that AtHCT had no obvious suppressive effect (Fig. 3).
Figure 3.
Investigating the function of maize HCT homologs in Rp1-D21-induced HR. A, The up-regulated HCT homologs were transiently coexpressed with Rp1-D21 into N. benthamiana. The representative leaves were photographed at 3 d after inoculation (upper), and the same leaves were cleared by ethanol (lower). B, Ion leakage conductivity was measured at 60 h after coexpression of Gus or HCT homologs with Rp1-D21. Significant differences (P < 0.05) between samples are indicated by different letters (a–c). C, Total protein was extracted from agro-infiltrated leaves at 30 hpi. Anti-HA was used to detect the expression of Rp1-D21, and anti-GFP was used to detect the expression of Gus or HCT homologs. Equal loading of protein samples is shown by Ponceau S staining. The experiments were repeated three times with similar results.
HCT1806 and HCT4918 Specifically Suppress the Autoactivity of Rp1-D21 But Not That of Other Autoactive NLRs
To determine whether HCT1806 and HCT4918 could also suppress HR induced by other NLR proteins, we coexpressed HCT1806 or HCT4918 with barley MLA10(D502V) and Arabidopsis RPM1(D505V), two autoactive CNL proteins that confer an HR phenotype when transiently expressed in N. benthamiana (Gao et al., 2011; Bai et al., 2012). Neither of the HCT homologs substantially suppressed either MLA10(D502V)- or RPM1(D505V)-induced HR (Supplemental Fig. S4). HCT0436 also did not suppress MLA10(D502V)-induced HR (Supplemental Fig. S4A). We further coexpressed AtHCT with RPM1(D505V) and found that AtHCT did not inhibit RPM1(D505V)-induced HR (Supplemental Fig. S4B). The data suggest that HCT1806 and HCT4918 specifically suppress the activity of Rp1-D21.
Mutations in the Conserved Amino Acids Required for HCT Activity Do Not Affect Its Suppression of Rp1-D21-Induced HR
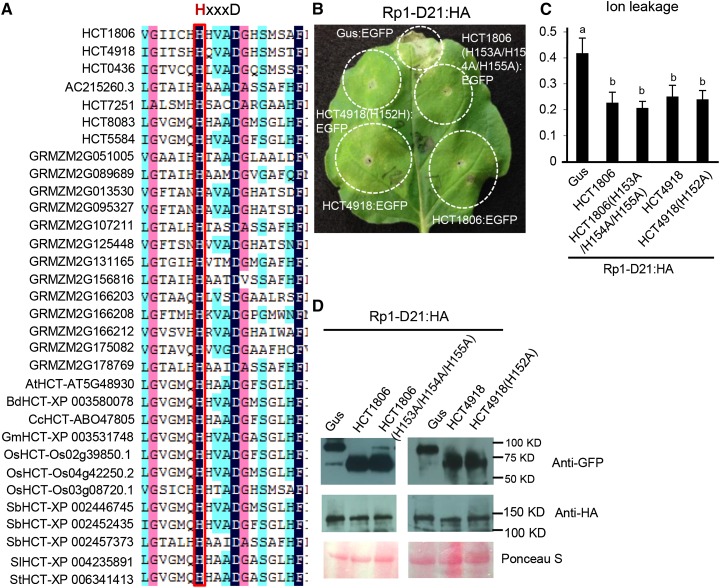
The conserved motif HxxxD, which is found in BAHD family proteins, is observed in all plant HCTs examined (Fig. 4A). The mutation in the His residue of sorghum or coffee (Coffea arabica) HCTs greatly reduced their enzymatic activities (Lallemand et al., 2012; Walker et al., 2013). We thus generated the corresponding mutation in the maize HCT4918: HCT4918(H152A). In HCT1806, there are His residues on either side of the conserved His residue (Fig. 4); we, therefore, generated the triple mutation of HCT1806(H153A/H154A/H155A). When coexpressed with Rp1-D21, both mutants still suppressed Rp1-D21-induced HR as determined by both visual examination and ion leakage assays (Fig. 4, B and C). The data suggested that enzymatic activity of these HCTs is not required for their function in regulating Rp1-D21-induced HR.
Figure 4.
Mutations in the conserved residues required for HCT activity did not affect the suppressive role of HCT on Rp1-D21-induced HR. A, Multiple sequence alignment of plant HCTs. The His residue in the conserved HxxxD motif is boxed. B, The HR phenotypes resulting from transient coexpression of HCT1806, HCT4918, and their mutant derivatives with Rp1-D21. C, Ion leakage conductivity was measured at 60 h after leaf infiltration. Significant differences (P < 0.05) between samples are indicated by different letters (a and b). D, Total protein was extracted from agro-infiltrated leaves at 30 hpi. Anti-HA was used to detect the expression of Rp1-D21, and anti-GFP was used to detect the expression of HCT homologs or Gus. Equal loading of protein samples is shown by Ponceau S staining. The experiments were repeated three times with the same results.
HCT1806 and HCT4918 Physically Associate with Rp1-D21, Rp1-D, and Rp1-dp2
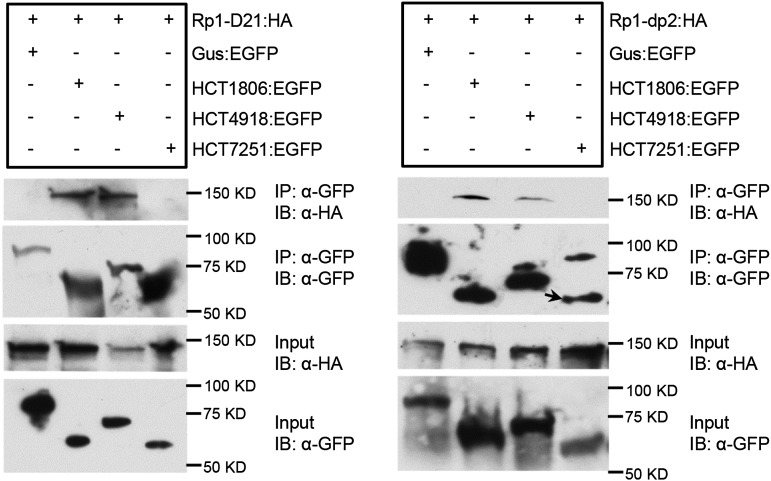
Because both HCT1806 and HCT4918 suppressed Rp1-D21-induced HR, we wanted to test whether there are physical associations between these two HCT homologs and Rp1 proteins. We performed a Co-IP assay by coexpression of HA-tagged Rp1 proteins and EGFP-tagged HCTs and found that both HCT1806 and HCT4918 coimmunoprecipated with Rp1-D21 and Rp1-dp2 (Fig. 5). However, we did not observe Co-IP between Rp1-D21/Rp1-dp2 and HCT7251, an HCT homolog that was induced in an Rp1-D21 background but did not suppress Rp1-D21-induced HR (Figs. 2 and 3A; Supplemental Table S1). We could not detect the interaction between Rp1-D and HCT1806/HCT4918 by Co-IP (data not shown), likely because of the low expression of Rp1-D:HA in the N. benthamiana system, which has been reported previously (Wang et al., 2015b).
Figure 5.
Investigating the interactions between Rp1-D21/Rp1-dp2 and HCT1806/HCT4918/HCT7251 by Co-IP assay. EGFP- and 3× HA-tagged constructs were transiently coexpressed in N. benthamiana, and samples were collected at 30 hpi for Co-IP assay. Protein extracts were immunoprecipitated (IP) by anti-GFP (α-GFP) microbeads and detected (immunblotted [IB]) by anti-GFP and anti-HA (α-HA) antibodies. In the rightmost lane of the IP:α-GFP/IB:α-GFP, an arrow indicates the target band. The experiments were repeated three times with the same results.
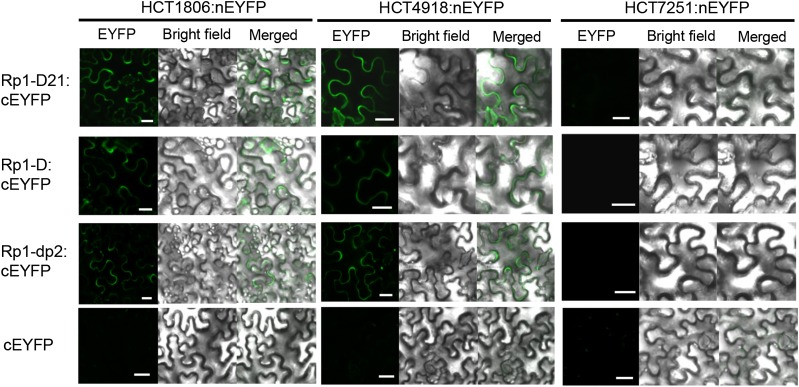
To further verify the physical interaction, the HCT homologs and Rp1 proteins were incorporated into split enhanced yellow fluorescent protein (EYFP) vectors; the HCT homologs were fused to the N-terminal of EYFP (nEYFP), and the Rp1 proteins were fused to the C-terminal of EYFP (cEYFP). These were analyzed by BiFC assay, in which a physical interaction between the proteins leads to observable fluorescence. Both HCT1806 and HCT4918 but not HCT7251 physically associated with Rp1-D21, Rp1-dp2, and Rp1-D in vivo (Fig. 6) as indicated by the fluorescence observed when the split EYFP vectors were coexpressed.
Figure 6.
Investigating the interactions between Rp1 proteins and HCT1806/HCT4918/HCT7251 by BiFC assay in N. benthamiana. Rp1-D21/Rp1-D/Rp1-dp2 was fused with cEYFP, and HCT homologs were fused with nEYFP vectors. The cEYFP- and nEYFP-derived constructs were transiently agro-infiltrated into N. benthamiana. The pictures were taken at 40 h postinfiltration under a confocal microscope. Green fluorescence indicates interaction between constructs tagged with nEYFP and constructs tagged with cEYFP. The experiments were repeated three times with the similar results. Bar = 20 µm.
Rp1-D21 contains an N-terminal CC domain, a middle NB-ARC domain, and a C-terminal LRR domain. We previously showed that the CC domain of Rp1-D21 (CCD21) but not the NB or LRR domain induced HR when fused with EGFP (Wang et al., 2015b). To determine whether HCT homologs can inhibit CCD21-induced HR, we coexpressed CCD21:EGFP and HCT1806, HCT4918, or HCT7251 in N. benthamiana and found that both HCT1806 and HCT4918 but not HCT7251 suppressed CCD21-induced HR (Supplemental Fig. S5A). Consistent with the data, we found that HCT1806 and HCT4918 but not HCT7251 associated with CCD21 as detected by Co-IP assays (Supplemental Fig. S5B).
DISCUSSION
The results presented here combined with the mapping results presented in our previous study (Olukolu et al., 2014) provide strong evidence that certain maize homologs of HCT, an enzyme required for lignin biosynthesis, interact with and regulate the activity of Rp1-D21, an autoactive NLR resistance protein. This regulation seems to be achieved through physical interaction between the proteins rather than by the enzymatic activity of HCT. To our knowledge, this is the first identification of HCT as a cofactor of NLRs.
Our previous GWAS analysis identified an SNP locus on chromosome 1 that was highly associated with variation in the severity of HR induced by Rp1-D21 (Olukolu et al., 2014). There were four genes annotated within 50 kb of this locus: two highly related HCT homologs HCT1806 and HCT4918 and two AAS homologous genes AAS6469 and AAS3125. The regions around both HCT homologous genes were in high LD with the associated SNP (Supplemental Fig. S2), indicating that both genes were strong candidates to underlie the causal variation. We have shown here that both HCT1806 and HCT4918 suppress Rp1-D21-induced HR in N. benthamiana, whereas AAS6469 and AAS3125 do not. In maize, the expression of both HCT1806 and HCT4918 and four other HCT homologs (HCT0436, HCT7251, HCT8083, and HCT5584) is induced in the presence of Rp1-D21. Except for HCT0436, which had a partially suppressive role, none of the other HCT homologs suppressed Rp1-D21-induced HR. This was consistent with the GWAS analysis and showed that control of Rp1-D21-mediated HR is associated with specific HCT homologs. HCT1806 and HCT4918 did not substantially suppress the HR phenotype caused by the other two CNL autoactive mutants: RPM1(D505V) and MLA10(D502V). These data suggest that these genes specifically regulate the activity of Rp1 proteins rather than being general suppressors of HR induced by CNLs.
HCT belongs to the BAHD family of plant acyl-CoA-dependent acyltransferases (Ma et al., 2005; D’Auria, 2006). BAHD family members share relatively low sequence identity, but nearly all members possess a conserved HxxxD motif (Burhenne et al., 2003). The His in the HxxxD motif is important for the catalytic activity of HCT, and mutation of this residue greatly reduces its activity (Lallemand et al., 2012; Walker et al., 2013). Mutations at this conserved His motif in HCT1806 and HCT4918 did not reduce their ability to inhibit Rp1-D21-induced HR, suggesting that the catalytic activity of HCT1806 and HCT4918 may not be required for their function in regulating Rp1-D21.
Maize homologs of all of the key genes involved in the lignin pathway were significantly up-regulated in Rp1-D21 backgrounds compared with their isogenic wild-type hybrids (Supplemental Fig. S1; Supplemental Table S1). Consistent with these transcriptional changes, we found that lignin content is increased in Rp1-D21 mutants compared with their isogenic wild-type backgrounds (Supplemental Fig. S3). These results are consistent with the previous observation that HR induces the phenylpropanoid pathway (Etalo et al., 2013). Interestingly, p-coumaroyl quinate and p-coumaroyl shikimate but not chlorogenic acid were hyperaccumulated in Rp1-D21 mutants, suggesting that the increases in C3H activity could not keep up with those in HCT and became a limiting step in the pathway (Supplemental Figs. S1 and S3).
Down-regulation of the lignin biosynthetic HCT gene in both Arabidopsis and alfalfa (Medicago sativa) resulted in a dwarf phenotype, constitutive activation of defense responses, increased PR gene expression, elevated SA levels (Hoffmann et al., 2004; Li et al., 2010; Gallego-Giraldo et al., 2011a, 2011b), and in alfalfa, enhanced resistance to the fungus Colletotrichum trifolii, the causal agent of alfalfa anthracnose (Gallego-Giraldo et al., 2011b). Reducing SA levels to wild-type levels in HCT down-regulated Arabidopsis plants restored a normal growth phenotype (Gallego-Giraldo et al., 2011a). Silencing HCT in N. benthamiana also led to a dwarf phenotype (Hoffmann et al., 2004). These data suggest a strong and general link, possibly mediated by SA, between HCT activity and the plant defense response.
The activity of NLR R genes must be very tightly controlled. On the one hand, inappropriate activation, as seen in the Rp1-D21 mutant, can lead to severe stunting and loss of fitness (Negeri et al., 2013). On the other hand, when the corresponding pathogen is present, activation of the defense pathway needs to be rapid for it to be effective. The precise control of the activation of NLRs has been investigated in several systems. Although there is significant apparent variation in the precise mechanisms involved in each case, in all cases, NLR proteins seem to be held in a precarious inactive state because of a combination of self-inhibitory intramolecular interactions, formation of homomers and heteromers with other NLRs, and interactions with other cofactors, including so-called guardees (see below). In a functioning wild-type system, the presence of specific pathogen-derived effectors is sufficient to disturb this balance and cause activation, leading to HR and the defense response. Furthermore, after activated, it is important that the HR be restricted to a small region, sufficient to confer resistance to the pathogen but not large enough to substantially damage the plant. Autophagy has been shown to be important in restricting the spread of HR (Liu et al., 2005).
Effectors are molecules secreted by the pathogen that facilitate infection and pathogenesis. In many cases, the effectors interact with and disrupt the function of specific plant proteins involved in the defense response. The guard hypothesis suggests that, in many cases, recognition of the effector by the NLR protein does not occur by direct interaction between the NLR and the effector but rather, occurs by the virulence target of the effector (Van der Biezen and Jones, 1998). Under this scenario, the NLR resistance protein guards the virulence target (the guardee), and a defense response is activated when disruption of the structure of the guardee by the effector is detected by the NLR. An elaboration of this model is the decoy model (van der Hoorn and Kamoun, 2008), which hypothesizes that, in some cases, the gene encoding the effector target may undergo duplication: one copy subsequently evolves to better interact with the effector to function as a bait (and possibly, lose its original metabolic function), whereas the other copy evolves to elude interaction with the effector, which allows it to better perform its original function in the defense response.
Many proteins that physically interact with and modulate the innate immunity mediated by NLRs have been identified, and several of these seem to participate in guard/guardee-type interactions. These interactions are summarized in Table I. HCTs, identified as negative regulators of Rp1-D21 in this study, have not previously been identified as cofactors of plant NLRs in any system. Among the different cofactors identified, most interact with the N-terminal CC or TIR domains of NLRs, with a few exceptions (Table I). Here, we found that HCT1806 and HCT4918 interacted with full-length Rp1 proteins and also, CCD21 to regulate their activity.
Table I. Summary of the state of knowledge of the cofactors and interaction domains of plant NLRs.
Abbreviations are as follows: Resistance to Pseudomonas syringae pv maculicola1 (RPM1), Resistance to Pseudomonas Syringae2 (RPS2), Suppressor of npr1-1, constitutive1 (SNC1), HopZ-Activated Resistance1 (ZAR1), Mildew A Locus10 (MLA10), Bacterial spot2 (Bs2), Panicle blast1 (Pb1), Constitutive expresser of PR genes1 (CPR1), basic Helix-loop-Helix84 (bHLH84), Enhanced disease susceptibility1 (EDS1), avrPphB susceptible1 (PBS1), HopZ-ETI-deficient1 (ZED1), Squamosa promoter binding protein-like6 (SPL6), suppressor of G2 allele of skp1 (SGT1), Ran GTPase activating protein2 (RanGAP2), and Bsu-like protein1 (BSL1). FL, Full length; TF, transcription factor; XNL, no typical CC or TIR domain was found in the N terminus; TNL, Toll-IL1 receptor-nucleotide binding LRR; —, no data available.
| Host Species and Gene | Protein Type | Pathogen | Effector | Cofactors | Types of Cofactors | Interaction Domains with Cofactors | Source |
|---|---|---|---|---|---|---|---|
| Arabidopsis | |||||||
| RPM1 | CNL | Pseudomonas syringae | AvrRpm1, AvrB | RIN4 | Unknown | CC, FL | Mackey et al. (2002) |
| RPS2 | CNL | P. syringae | AvrRpt2 | RIN4 | Unknown | CC, FL | Axtell and Staskawicz (2003); Mackey et al. (2003) |
| CPR1 | F-box | FL | Cheng et al. (2011) | ||||
| RPS4 | TNL | P. syringae | AvrRps4 | bHLH84 | TF | FL | Xu et al. (2014) |
| EDS1 | Lipase-like | FL | Bhattacharjee et al. (2011); Heidrich et al. (2011) | ||||
| RPS5 | CNL | Peronospora parasitica | AvrPphB | PBS1 | Kinase | CC, FL | Ade et al. (2007) |
| RPS6 | TNL | P. syringae | AvrHopA1 | EDS1 | Lipase-like | FL | Bhattacharjee et al. (2011) |
| SNC1 | TNL | P. syringae | — | bHLH84 | TF | FL | Xu et al. (2014) |
| CPR1 | F-box | FL | Cheng et al. (2011) | ||||
| EDS1 | Lipase-like | FL | Bhattacharjee et al. (2011) | ||||
| ZAR1 | CNL | P. syringae | AvrHopZ1a | ZED1 | Pseudokinase | CC, FL | Lewis et al. (2013) |
| Barley | |||||||
| MLA10 | CNL | Blumeria graminis | AvrA10 | WRKY1, WRKY2 | TF | CC, FL | Shen et al. (2007) |
| MYB6 | TF | CC, FL | Chang et al. (2013) | ||||
| Maize | |||||||
| Rp1-D | CNL | P. sorghi | — | HCT homologs | Lignin biosynthesis | FL | This study; Wang et al. (2015b) |
| Rp1-dp2 | CNL | — | — | HCT homologs | Lignin biosynthesis | CC, FL | This study; Wang et al. (2015b) |
| Rp1-D21 | CNL | — | — | HCT homologs | Lignin biosynthesis | CC, FL | This study; Wang et al. (2015b) |
| Nicotiana tabacum | |||||||
| N | TNL | Tobacco mosaic virus | P50 | SPL6 | TF | TIR, LRR, FL | Padmanabhan et al. (2013) |
| Pepper (Capsicum annuum) | |||||||
| Bs2 | XNL | Xanthomonas campestris pv vesicatoria | AvrBs2 | SGT1 | Molecular cochaperone | LRR, FL | Leister et al. (2005) |
| Potato (Solanum tuberosum) | |||||||
| Rx | CNL | Potato virus X | Coat protein | RanGAP2 | Small GTPase | CC, FL | Tameling and Baulcombe (2007) |
| R2 | CNL | Phytophthora infestans | AVR2 | BSL1 | Phosphatase | FL | Saunders et al. (2012) |
| Rice | |||||||
| Pb1 | CNL | Magnaporthe oryzae | — | WRKY45 | TF | CC, FL | Inoue et al. (2013) |
| Pit | CNL | M. oryzae | — | Rac1 | Small GTPase | NB-ARC, FL | Kawano et al. (2010) |
| Tomato (Solanum lycopersicum) | |||||||
| Prf | XNL | P. syringae | AvrPto, AvrPtoB | Pto | Kinase | CC, FL | Mucyn et al. (2006) |
The best characterized guard/guardee system is probably the RPM1-interacting protein4 (RIN4) system in Arabidopsis. RIN4 seems to be an important regulator of plant basal immunity. The balance between different phosphorylation states of RIN4 is believed to influence the switch between repression and activation of the basal defense response. At least four different bacterial effectors are known to target RIN4. This model (based on substantial experimental data) suggests that the balance between two RIN4 phosphorylation states, RIN4 pS141 and RIN4 pT166, is crucial. Upon detection of bacterial-derived microbe-associated molecular patterns (MAMPs) by plant receptors, enhanced levels of RIN4 pS141 lead to the activation of the basal defense response known as MAMP-triggered immunity. Bacterial effectors, including Avirulence protein B (AvrB) and AvrRpm1, target RIN4 and cause increased levels RIN4 pT166, which lead to repression of MAMP-triggered immunity, allowing infection. Excessive levels of RIN4 pT166 cause the activation of the NLRs RPM1, leading to HR and so-called effector-triggered immunity (Chung et al., 2014).
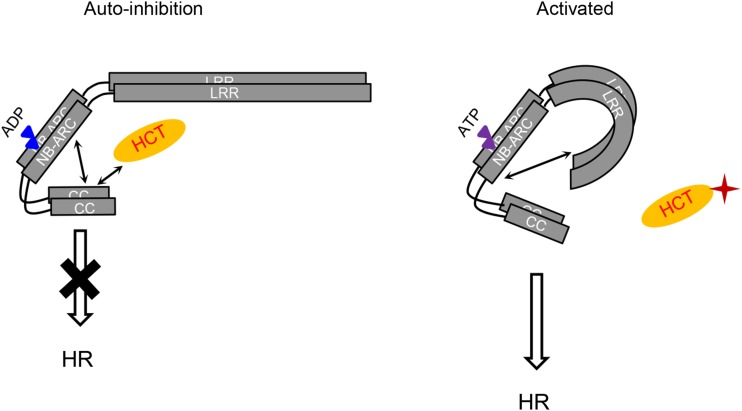
Rp1-D confers resistance to P. sorghi, the causal agent of maize common rust (Hu et al., 1996). The cognate effector of Rp1-D is not known. In our previous work using a variety of approaches to elucidate structure/function relationships, we derived a model for the self-activation/repression of Rp1 proteins with the following features (Wang et al., 2015b). (1) Self-association and heteromer formation between Rp1 proteins are important for their activity. (2) The CC domain is the signaling-competent region of the Rp1 proteins. (3) The NB-ARC domain of Rp1 proteins inhibits CC-induced HR by intramolecular interactions. (4) In Rp1-D21 and various other derived autoactive recombinants, the LRR domain (especially N1182 and the last 16 amino acids) destabilizes the interaction between the CC and NB-LRR domains through direct interaction with the NB-ARC. This destabilization allows for the exchange of bound ADP for ATP, causing activation.The data presented here support an elaboration of this model, in which Rp1 proteins are maintained in a suppressed state through the combination of intramolecular interactions and interaction with other Rp1 proteins described above as well as through interaction with other molecules, including HCT1806 and HCT4918. Because HCT and the lignin pathway are important for the defense response, it is reasonable to speculate that HCT1806 and HCT4918 might be effector targets and guarded by Rp1 proteins. Given the substantial number of HCT homologs in maize, it is further reasonable to hypothesize that HCT1806 and/or HCT4918 may have evolved to function primarily as decoys. In a wild-type situation, it may be that modification of one or more of these HCT homologs by a rust effector is sufficient to trigger the switch between the inactive and active states of the Rp1 proteins (summarized in Fig. 7).
Figure 7.
Model for the suppression and activation of Rp1 proteins. Rp1 proteins form homo- and heteromers (Wang et al., 2015b). The inhibited state of Rp1 proteins (left) is maintained through autoinhibitory intramolecular interaction and interactions between HCT1806/HCT4918 and the CC domain. In the activated state (right), pathogen effectors modify HCT1806/HCT4918, disrupting the interaction between Rp1 proteins and HCT1806/HCT4918. This, in turn, disrupts the autoinhibitory intramolecular interactions; the interaction between the LRR and NB-ARC domains is strengthened, and the CC/NB-ARC interaction is weakened, leading to activation and HR. Blue and purple triangles indicate ADP and ATP, respectively. Arrows indicate intra- and intermolecular interactions. The putative pathogen effector is labeled with a red star. The rationale behind the models is explained in the text.
This model is clearly inadequate to explain the full range of observed phenomena. For instance, Rp1-D, when transiently expressed by itself in N. benthamiana, does not induce HR, despite the absence of maize HCT1806 and HCT4918, whereas Rp1-D21 does induce HR in maize, despite the presence of the HCTs. These apparent contradictions may be resolved as we identify and characterize other proteins involved in the R gene complex. It is also likely that, as with the RIN4 model detailed above, it is not just the presence or absence per se of the HCTs that are important but rather, their relative concentrations with respect to Rp1 proteins and their modification status. For instance, it is likely that the relative levels of HCT are much higher when coexpressed with Rp1-D21 in N. benthamiana than in a maize line carrying Rp1-D21, and this may be the reason that suppression of HR is observed in N. benthamiana but not in maize. Furthermore, because activation of Rp1-D21-induced HR leads to the transcriptional up-regulation of HCT1806 and HCT4918 in maize (Supplemental Table S1), we hypothesize that the transcription levels of HCT1806 and HCT4918 will be highly increased after being infected by corresponding incompatible pathogen. We further speculate that the increased levels of both HCTs will produce large amounts of free HCT1806 and HCT4918, which can serve as a reservoir to shield Rp1 and HCT1806/HCT4918 from predicted modification and disassociation and restrict the HR spreading.
In summary, two HCT homologs were previously identified as candidate genes for modulating the severity of the HR associated with the autoactive NLR resistance protein Rp1-D21. In this study, we provide evidence that validates the roles of these genes, show the first example, to our knowledge, of HCT homologs that are cofactors of NLRs, and provide important information of the involvement of the lignin pathway in defense response. To our knowledge, this is also one of few examples of the functional validation of genes identified from GWAS in maize. These data have allowed us to generate models and hypotheses for NLR and specifically, Rp1 protein function that will form the bases of future studies.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type Nicotiana benthamiana plants were grown at 23°C in a 16-h-light/8-h-dark cycle. Maize (Zea mays) line B73 was used for isolating the complementary DNA (cDNA) or genomic DNA sequences of HCT and AAS homologous genes. Wild-type and isogenic Rp1-D21 mutant plants in B73 × H95 and Mo17 × H95 backgrounds were used for RNA extraction for RNA-Seq analysis. The only substantial genetic difference between the isogenic pairs was at the Rp1 locus, where the wild-type isoline was heterozygous for the H95 and B73 or Mo17 haplotypes, whereas mutant lines carried the Rp1-D21 haplotype instead of the H95 Rp1 haplotype (Chaikam et al., 2011). All of the maize lines were grown in constant 22°C in a 12-h-light/12-h-dark cycle.
LD Analysis
Pairwise R2 estimates (Hill and Robertson, 1968) of LD between the associated SNP marker (position 17,826,971 bp on chromosome 1) and all other markers (1,649 SNPs) within a 100-kb region (50 kb on either side of the SNP) were computed in TASSEL, v4.0 (Bradbury et al., 2007). LD was estimated based on 4,635 NAM recombinant inbred lines as well as the NAM founder lines that represent the global maize diversity, including temperate (stiff stalk and nonstiff stalk) and tropical lines, using the same marker set with which the associated SNP was originally detected (19).
RNA-Seq Library Construction and Data Processing
The third true leaves of five individual seedlings collected from 18-d-old wild-type or Rp1-D21 mutant plants from B73 × H95 or Mo17 × H95 backgrounds were pooled for total RNA isolation. The procedures for RNA-Seq assay was performed according to our previous study (Olukolu et al., 2014). Briefly, two biological replicates, each consisting of five individual plants, were performed with 100-nucleotide single-end reads. RNA-Seq libraries were constructed according to the TruSeq RNA Sample Prep v2 LS Protocol (Illumina) and sequenced by the Illumina HiSEquation 2000. The sequences were aligned to maize genome sequence v2 (www.maizegdb.org) by TopHat 2.0 (Trapnell et al., 2009) using all of the default parameter settings. Differentially expressed genes were identified using the software package edgeR from the Bioconductor suite (Robinson et al., 2010). To account for multiple testing, the procedure by Benjamini and Hochberg (1995) for controlling the false discovery rate was applied using a threshold of q ≤ 0.05 to determine significance.
Metabolite Analysis
Metabolites were extracted by 50% (v/v) methanol at 60°C for 30 min from the fourth true leaves of 22-d-old plants. Metabolite profiling experiments were performed using an Agilent 6530 Accurate-Mass Q-TOF LC/MS Instrument in Negative AJS Electrospray Ionization Mode. Samples were injected onto a ZORBAX Eclipse Plus C18 Column (3 × 100 mm, 1.8 μm; Agilent Technologies; http://www.agilent.com). A gradient from 2% acetonitrile in 0.1% formic acid to 45% acetonitrile in 0.1% formic acid at a flow rate of 0.6 mL min−1 was used for HPLC separation. Mass data were acquired with the following parameters: drying gas temperature of 300°C, drying gas flow of 7 L min−1, nebulizer pressure of 40 psi, sheath gas temperature of 350°C, sheath gas flow of 10 L min−1, capillary voltage of 3,500 V, nozzle voltage of 500 V, fragmentor voltage of 150 V, skimmer voltage of 65 V, and octopole radio frequency peak voltage of 750 V.
Lignin Analysis
Cell walls were prepared from the fourth true leaves of 22-d-old plants by sequential extractions with sodium phosphate buffer (0.1 m, pH 7.2) at 37°C, 70% ethanol at 80°C (repeated multiple times), and acetone at room temperature. Lignin content was measured by the acetyl bromide method. Briefly, 2 to 5 mg of cell wall material was dissolved in 2.5 mL of acetyl bromide:acetic acid (1:3, v/v) solution for 24 h at room temperature. The sample was qualitatively transferred to a 10-mL volumetric flask. After adding 0.35 mL of 0.5 m hydroxylamine hydrochloride, the flask was filled to volume with acetic acid. Absorbance at 280 nm was measured, and an extinction coefficient of 23.077 g−1 L cm−1 was used to calculate acetyl bromide lignin content (Fukushima and Kerley, 2011).
Plasmid Construction
All primers used in this study are listed in Supplemental Table S2. Rp1-D21, Rp1-dp2, Rp1-D, and the CC domain of Rp1-D21 (CCD21) were constructed into pGWB14 (with a 3× HA epitope tag in the C terminus) as reported in our previous study (Wang et al., 2015b). Gus:EGFP and CCD21:EGFP were generated previously (Wang et al., 2015b). Because of low expression of HCT4918, which made it difficult to clone the cDNA, the genomic DNA rather than the cDNA was cloned for transient expression in N. benthamiana. All other HCT and AAS genes were amplified from cDNA and cloned into pDONR207 vector by BP reactions. After sequencing, they were transferred into pSITEII-N1-EGFP vector (Martin et al., 2009) by LR reactions.
Site-Directed Mutagenesis
Overlapping extension PCR primers (Supplemental Table S2) were designed for generating the site-directed mutations: HCT1806(H153A/H154A/H155A) and HCT4918(H152A). The site-directed mutations were cloned into pDONR207 vector by BP reactions and verified by sequencing. Then, they were moved to gateway vector pSITEII-N1-EGFP by LR reactions.
Sequence Alignment and Phylogenetic Analysis
HCT amino acids sequences were aligned by ClustalW (www.ebi.ac.uk) and edited by BioEdit software. The phylogenetic tree was constructed with the MEGA 6.0 software (Tamura et al., 2013). The specific algorithms used for tree building were neighbor joining according to previous studies (Wang et al., 2011, 2015a).
Agrobacterium tumefaciens-Mediated Transient Expression
A. tumefaciens strain GV3101 (pMP90) transformed with binary vector constructs was grown at 28°C overnight in 5 mL of L-broth medium supplemented with appropriate antibiotics. The detailed procedures were performed according to our previous study (Wang et al., 2015b). EGFP- and 3× HA-tagged constructs were transiently coexpressed in N. benthamiana. Unless otherwise specified, agrobacterium carrying each construct was diluted to a final concentration of an optical density of 600 nm (OD600) = 0.4 plus p19 with OD600 = 0.2.
Ion Leakage Measurement
Ion leakage was measured according to a previous study (Wang and Balint-Kurti, 2015). At least five leaf discs (1.2-cm diameter) from different plants were collected and put in 4 mL of sterile water in a 15-mL polypropylene tube. The samples were shaken for 3 h at room temperature, and the conductivity (C1) was measured by a conductivity meter (model 4403; Markson Science, Inc.). Subsequently, samples were boiled for 15 min, and the total conductivity (C2) was measured again. The ion leakage was calculated as C1:C2 ratio.
Protein Analysis
For protein expression analysis, three leaf discs (1.2-cm diameter) from different single plants were collected at 30 hpi. The samples were ground with prechilled plastic pestles in liquid nitrogen, and total protein was extracted in 160 µL of extraction buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, pH 8.0, 1% Triton X-100, 0.1% SDS, 10 mm dithiothreitol, 40 µm MG132, and 1× plant protein protease inhibitor mixture; Sigma-Aldrich). For Co-IP assay, EGFP- and 3× HA-tagged constructs were transiently coexpressed in N. benthamiana. Samples were collected at 30 hpi, and proteins were extracted by grinding 0.8 g of leaf tissues in 2.4 mL of extraction buffer (50 mm HEPES, pH 7.5, 50 mm NaCl, 10 mm EDTA, pH 8.0, 0.5% Triton X-100, 4 mm dithiothreitol, and 1× plant protein protease inhibitor mixture; Sigma-Aldrich). The detailed procedures for protein analysis were conducted according to our previous study (Wang et al., 2015b).
BiFC Assay
The Rp1-D21, Rp1-D, and Rp1-dp2 genes were cloned into the gateway EYFP-split vectors and DEST-cEYFP, and HCT1806, HCT4918, and HCT7251 genes were cloned into DEST-nEYFP by LR reactions. Agrobacteria-carrying constructs derived from EYFP-split vectors were equally mixed, and each construct was diluted to a final concentration of OD600 = 0.4 plus p19 with OD600 = 0.2. The GFP fluorescence was excited at 488 nm and observed between 495 and 550 nm under a confocal microscope at 30 h after transiently coexpressed of the appropriate EYFP-split vectors into N. benthamiana.
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: nucleotide sequences: Rp1-D21 (KF951062), Rp1-dp2 (KF951063), and Rp1-D (AF107293); protein sequences Rp1-D21 (AIW65617), Rp1-dp2 (AIW65618), and Rp1-D (AAD47197); and RNA-Seq data SRP060286.
Supplementary Data
The following supplemental materials are available.
Supplemental Figure S1. The phenylpropanoid pathway, modified according to prevous studies (Hoffmann et al., 2004; Li et al., 2010).
Supplemental Figure S2. Linkage disequilibrium between the SNP at chromosome 1, 17,826,971 bp, indicated by a black line, and adjacent SNPs.
Supplemental Figure S3. Differential accumulation of hydroxycinnamic acid esters and lignin between isolines with or without Rp1-D21.
Supplemental Figure S4. HCT1806 and HCT4918 did not substantially suppress MLA10(D502V)- and RPM1(D505V)-induced HR in N. benthamiana.
Supplemental Figure S5. Investigating the function of HCT1806/HCT4918/HCT7251 in CCD21-induced HR.
Supplemental Table S1. Expression level changes of maize lignin pathway genes comparing isolines differing for the presence/absence of the Rp1-D21 gene.
Supplemental Table S2. The primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Jeff Dangl for providing strains of RPM1(D505V) and discussion of the article; Dr. Qian-Hua Shen for providing MLA10(D502V) plasmid; Dr. Marc T. Nishimura for providing pDONR207 vector; Dr. Michael Goodin for providing the vectors of pSITEII-N1-EGFP, DEST-nEYFP, and DEST-cEYFP; Dr. Shannon Sermons for help with obtaining reagents; Eva Johannes for help with microscopy observation; Drs. Guri Johal, Qin Yang, Tiffany Jamann, Farid EI-Kasmi, Marc T. Nishimura, and Jim Holland for helpful discussions; the Genomic Sciences Laboratory (North Carolina State University) for providing DNA sequencing services; and the North Carolina State University phytotron for providing controlled environment facilities.
Glossary
- BiFC
bimolecular fluorescence complementation
- cDNA
complementary DNA
- Co-IP
coimmunoprecipitation
- HR
hypersensitive response
- LD
linkage disequilibrium
- MAMP
microbe-associated molecular pattern
- NAM
nested association mapping
- RNA-Seq
RNA-sequencing
- SA
salicylic acid
- SNP
single-nucleotide polymorphism
Footnotes
This work was supported by the National Science Foundation (grant nos. 0822495 and 1444503) and the U.S. Department of Agriculture Agricultural Research Service.
References
- Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Bai S, Liu J, Chang C, Zhang L, Maekawa T, Wang Q, Xiao W, Liu Y, Chai J, Takken FL, et al. (2012) Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog 8: e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S, Halane MK, Kim SH, Gassmann W (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Burhenne K, Kristensen BK, Rasmussen SK (2003) A new class of N-hydroxycinnamoyltransferases. Purification, cloning, and expression of a barley agmatine coumaroyltransferase (EC 2.3.1.64). J Biol Chem 278: 13919–13927 [DOI] [PubMed] [Google Scholar]
- Chaikam V, Negeri A, Dhawan R, Puchaka B, Ji J, Chintamanani S, Gachomo EW, Zillmer A, Doran T, Weil C, et al. (2011) Use of Mutant-Assisted Gene Identification and Characterization (MAGIC) to identify novel genetic loci that modify the maize hypersensitive response. Theor Appl Genet 123: 985–997 [DOI] [PubMed] [Google Scholar]
- Chang C, Yu D, Jiao J, Jing S, Schulze-Lefert P, Shen QH (2013) Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25: 1158–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, Li X (2011) Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci USA 108: 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintamanani S, Hulbert SH, Johal GS, Balint-Kurti PJ (2010) Identification of a maize locus that modulates the hypersensitive defense response, using mutant-assisted gene identification and characterization. Genetics 184: 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EH, El-Kasmi F, He Y, Loehr A, Dangl JL (2014) A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immune receptors. Cell Host Microbe 16: 484–494 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- D’Auria JC. (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. Arabidopsis Book 9: e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Dodds P, Pryor T (2000) Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol 3: 278–284 [DOI] [PubMed] [Google Scholar]
- Etalo DW, Stulemeijer IJE, van Esse HP, de Vos RCH, Bouwmeester HJ, Joosten MHAJ (2013) System-wide hypersensitive response-associated transcriptome and metabolome reprogramming in tomato. Plant Physiol 162: 1599–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima RS, Kerley MS (2011) Use of lignin extracted from different plant sources as standards in the spectrophotometric acetyl bromide lignin method. J Agric Food Chem 59: 3505–3509 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Escamilla-Trevino L, Jackson LA, Dixon RA (2011a) Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc Natl Acad Sci USA 108: 20814–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA (2011b) Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol 190: 627–639 [DOI] [PubMed] [Google Scholar]
- Gao Z, Chung EH, Eitas TK, Dangl JL (2011) Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci USA 108: 7619–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath MC. (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L, Parker JE (2011) Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334: 1401–1404 [DOI] [PubMed] [Google Scholar]
- Hill WG, Robertson A (1968) Linkage disequilibrium in finite populations. Theor Appl Genet 38: 226–231 [DOI] [PubMed] [Google Scholar]
- Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M (2004) Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M (2003) Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278: 95–103 [DOI] [PubMed] [Google Scholar]
- Hu G, Richter TE, Hulbert SH, Pryor T (1996) Disease lesion mimicry caused by mutations in the rust resistance gene rp1. Plant Cell 8: 1367–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Hayashi N, Matsushita A, Xinqiong L, Nakayama A, Sugano S, Jiang CJ, Takatsuji H (2013) Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc Natl Acad Sci USA 110: 9577–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J, Yao A, Nakashima A, Takahashi H, Yoshida H, Wong HL, et al. (2010) Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7: 362–375 [DOI] [PubMed] [Google Scholar]
- Lallemand LA, Zubieta C, Lee SG, Wang Y, Acajjaoui S, Timmins J, McSweeney S, Jez JM, McCarthy JG, McCarthy AA (2012) A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiol 160: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ (2005) Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17: 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Lee AH, Hassan JA, Wan J, Hurley B, Jhingree JR, Wang PW, Lo T, Youn JY, Guttman DS, et al. (2013) The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc Natl Acad Sci USA 110: 18722–18727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NG, Yamamoto E (1990) Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol 41: 455–496 [DOI] [PubMed] [Google Scholar]
- Li X, Bonawitz ND, Weng JK, Chapple C (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP (2005) Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Ma X, Koepke J, Panjikar S, Fritzsch G, Stöckigt J (2005) Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J Biol Chem 280: 13576–13583 [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF III, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- McMullen MD, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, et al. (2009) Genetic properties of the maize nested association mapping population. Science 325: 737–740 [DOI] [PubMed] [Google Scholar]
- Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP (2006) The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18: 2792–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E (2008) The hypersensitive response; the centenary is upon us but how much do we know? J Exp Bot 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Negeri A, Wang GF, Benavente L, Kibiti CM, Chaikam V, Johal G, Balint-Kurti P (2013) Characterization of temperature and light effects on the defense response phenotypes associated with the maize Rp1-D21 autoactive resistance gene. BMC Plant Biol 13: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olukolu BA, Negeri A, Dhawan R, Venkata BP, Sharma P, Garg A, Gachomo E, Marla S, Chu K, Hasan A, et al. (2013) A connected set of genes associated with programmed cell death implicated in controlling the hypersensitive response in maize. Genetics 193: 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olukolu BA, Wang GF, Vontimitta V, Venkata BP, Marla S, Ji J, Gachomo E, Chu K, Negeri A, Benson J, et al. (2014) A genome-wide association study of the maize hypersensitive defense response identifies genes that cluster in related pathways. PLoS Genet 10: e1004562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan MS, Ma S, Burch-Smith TM, Czymmek K, Huijser P, Dinesh-Kumar SP (2013) Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog 9: e1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, DeYoung BJ, Innes RW (2012) Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol 158: 1819–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders DG, Breen S, Win J, Schornack S, Hein I, Bozkurt TO, Champouret N, Vleeshouwers VG, Birch PR, Gilroy EM, et al. (2012) Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum immune receptor R2 to mediate disease resistance. Plant Cell 24: 3420–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW, et al. (2010) Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell 22: 4195–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Pryor AJ, Hulbert SH (2004) Allelic and haplotypic diversity at the rp1 rust resistance locus of maize. Genetics 167: 1939–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Steinau M, Trick HN, Hulbert SH (2010) Recombinant Rp1 genes confer necrotic or nonspecific resistance phenotypes. Mol Genet Genomics 283: 591–602 [DOI] [PubMed] [Google Scholar]
- Sun Q, Collins NC, Ayliffe M, Smith SM, Drake J, Pryor T, Hulbert SH (2001) Recombination between paralogues at the Rp1 rust resistance locus in maize. Genetics 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Baulcombe DC (2007) Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19: 1682–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Biezen EA, Jones JD (1998) Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci 23: 454–456 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RAL, Kamoun S (2008) From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G, Mayr G, Albrecht M, Cornelissen BJ, Takken FL (2008) Transcomplementation, but not physical association of the CC-NB-ARC and LRR domains of tomato R protein Mi-1.2 is altered by mutations in the ARC2 subdomain. Mol Plant 1: 401–410 [DOI] [PubMed] [Google Scholar]
- Walker AM, Hayes RP, Youn B, Vermerris W, Sattler SE, Kang C (2013) Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme A-dependent transferases and synthases. Plant Physiol 162: 640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF, Balint-Kurti PJ (2015) Cytoplasmic and nuclear localizations are important for the hypersensitive response conferred by maize autoactive Rp1-D21 protein. Mol Plant Microbe Interact 28: 1023–1031 [DOI] [PubMed] [Google Scholar]
- Wang GF, Fan R, Wang X, Wang D, Zhang X (2015a) TaRAR1 and TaSGT1 associate with TaHsp90 to function in bread wheat (Triticum aestivum L.) seedling growth and stripe rust resistance. Plant Mol Biol 87: 577–589 [DOI] [PubMed] [Google Scholar]
- Wang GF, Ji J, El-Kasmi F, Dangl JL, Johal G, Balint-Kurti PJ (2015b) Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog 11: e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF, Wei X, Fan R, Zhou H, Wang X, Yu C, Dong L, Dong Z, Wang X, Kang Z, et al. (2011) Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytol 191: 418–431 [DOI] [PubMed] [Google Scholar]
- Xu F, Kapos P, Cheng YT, Li M, Zhang Y, Li X (2014) NLR-associating transcription factor bHLH84 and its paralogs function redundantly in plant immunity. PLoS Pathog 10: e1004312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.