Expressing glucan branching enzymes from different plants and bacteria in branching enzyme-deficient Arabidopsis plants reveals marked differences in their specificity, yielding unique starch structures.
Abstract
The major component of starch is the branched glucan amylopectin, the branching pattern of which is one of the key factors determining its ability to form semicrystalline starch granules. Here, we investigated the functions of different branching enzyme (BE) types by expressing proteins from maize (Zea mays BE2a), potato (Solanum tuberosum BE1), and Escherichia coli (glycogen BE [EcGLGB]) in Arabidopsis (Arabidopsis thaliana) mutant plants that are deficient in their endogenous BEs and therefore, cannot make starch. The expression of each of these three BE types restored starch biosynthesis to differing degrees. Full complementation was achieved using the class II BE ZmBE2a, which is most similar to the two endogenous Arabidopsis isoforms. Expression of the class I BE from potato, StBE1, resulted in partial complementation and high amylose starch. Expression of the glycogen BE EcGLGB restored only minimal amounts of starch production, which had unusual chain length distribution, branch point distribution, and granule morphology. Nevertheless, each type of BE together with the starch synthases and debranching enyzmes were able to create crystallization-competent amylopectin polymers. These data add to the knowledge of how the properties of the BE influence the final composition of starch and fine structure of amylopectin.
Starch is composed of two glucan polymers: amylopectin and amylose. Amylopectin constitutes around 80% of the mass of most starches and is a large, branched polymer with a tree-like architecture. The positioning and frequency of branch points together with the distribution of chain lengths are thought to be critical factors allowing amylopectin to adopt a semicrystalline state. Within amylopectin molecules, clusters of unbranched chain segments align, and adjacent chains form double helices. These pack into crystalline lamellae that alternate with amorphous regions containing the branch points. Longer chain segments span from one cluster to the next (Zeeman et al., 2010).
Amylopectin is synthesized by three enzyme activities. First, starch synthases (SSs) transfer the glucosyl part of ADP-Glc to the nonreducing end of existing glucan chains, forming new α-1,4 glucosidic bonds. Second, branching enzymes (BEs) cleave part of an α-1,4-linked chain and through an inter- or intramolecular transfer reaction, reattach it, creating α-1,6-branch points. This reaction creates additional nonreducing ends on which SSs can act. Third, debranching enzymes (DBEs) hydrolyze some of these branches, tailoring the structure of the polymer to promote its crystallization.
Several SS and BE isoforms are involved in starch synthesis in plants. There are five conserved classes of SSs (granule-bound starch synthase [GBSS] and SS1–SS4) and two conserved classes of BEs (classes I and II; also referred to as classes B and A, respectively; Nougué et al., 2014). In addition, plants contain two classes of DBEs: isoamylases (ISAs) and limit dextrinases (LDAs; also called pullulanases). One ISA, a multimeric enzyme composed of either a mixture of ISA1 and ISA2 subunits or just ISA1 subunits, is primarily involved in amylopectin synthesis (James et al., 1995; Mouille et al., 1996; Nakamura et al., 1996; Delatte et al., 2005). The other DBEs (i.e. ISA3 and LDA) are primarily involved in starch degradation (Wattebled et al., 2005; Delatte et al., 2006).
Based on the in vitro analysis of purified or recombinant proteins and the phenotypes of mutant plants, the different SS isoforms are proposed to have distinct, albeit overlapping, functions. SS1 is thought to preferentially elongate short chains produced by the branching reactions to between 8 and 12 Glc units (Delvallé et al., 2005; Fujita et al., 2006). SS2 is proposed to elongate such chains farther to about 20 Glc units, optimal for cluster formation (Edwards et al., 1999; Umemoto et al., 2002; Zhang et al., 2008). The precise role of SS3 is less clear, although it has been proposed to generate long, cluster-spanning chains (Fujita et al., 2007). SS4 has a distinct role in initiating and/or coordinating granule formation (Roldán et al., 2007; Crumpton-Taylor et al., 2013).
The two different BE classes are also proposed to have distinct functions in amylopectin synthesis. In vitro analyses of maize (Zea mays), rice (Oryza sativa), and potato (Solanum tuberosum) enzymes suggest that the class I enzymes preferentially act on amylose and transfer longer chains, whereas class II enzymes preferentially act on branched substrates, such as amylopectin, and transfer shorter chains (Guan and Preiss, 1993; Rydberg et al., 2001; Nakamura et al., 2010). This knowledge derives largely from experiments where linear or branched substrates were provided to recombinant or purified enzymes and the increased degree of branching was monitored. Similar conclusions were gained by recombinant protein expression in Escherichia coli and yeast (Saccharomyces cerevisiae) strains deficient in their endogenous glycogen BEs (Guan et al., 1995; Seo et al., 2002), where chain elongation by glycogen synthases occurred concurrently with branching.
Models have been proposed in which both BE classes help create the final cluster structure of amylopectin: class I BEs initiate branching by transferring long or branched chains, which are subsequently acted on by class II BEs to create more numerous shorter chains. These shorter chains are then elaborated by the SSs to create the clusters (Nakamura et al., 2010). After the branching reactions, a degree of debranching occurs, which is thought to control branch number and positioning and thereby, facilitate amylopectin crystallization (Myers et al., 2000; Zeeman et al., 2010). Several studies have shown that isa1-deficient mutants produce starch with an altered amylopectin, accumulate a related soluble polymer (phytoglycogen), or both (James et al., 1995; Mouille et al., 1996; Nakamura et al., 1996; Delatte et al., 2005).
Despite the wide conservation of the two BE classes, major alterations in starch properties are only observed when genes encoding class II enzymes are mutated or repressed. Loss of class I BE activity in maize endosperm, rice endosperm, or potato tuber did not alter starch content and caused only minor differences in amylopectin structure (e.g. the distribution of chain lengths and branch points) and/or starch properties (e.g. gelatinization or digestibility; Safford et al., 1998; Blauth et al., 2002; Satoh et al., 2003; Xia et al., 2011). In contrast, loss of class II BE results in significant changes, such as decreased starch content and a high apparent amylose content. This has been observed in several species, including maize (Stinard et al., 1993), potato (Jobling et al., 1999), pea (Pisum sativum; Bhattacharyya et al., 1990), rice (Mizuno et al., 1993), barley (Hordeum vulgare; Regina et al., 2010), and wheat (Triticum aestivum; Regina et al., 2006). The high apparent amylose content was caused at least in part by the accumulation of less-frequently branched amylopectin that stains with a higher wavelength of maximal absorption (λmax) than that of the wild type (Boyer et al., 1976). In potato, this phenotype was enhanced by the simultaneous suppression of BE1 (Schwall et al., 2000), a result also shown recently in barley (Carciofi et al., 2012).
Arabidopsis (Arabidopsis thaliana) has three genes annotated as BEs, At3g20440 (BE1), At5g03650 (BE2), and At2g36390 (BE3), but it seems that only BE2 and BE3 are active. Both BE2 and BE3 are class II BEs, making Arabidopsis somewhat unusual in not possessing a class I BE. The gene annotated as BE1 encodes a related protein that falls into a separate clade to either class I or II BEs (Dumez et al., 2006; Han et al., 2007; Wang et al., 2010). It was initially suggested that plants with mutations in this gene had a wild-type phenotype (Dumez et al., 2006), but subsequent work indicated that homozygous be1 mutation causes embryo lethality (hence, its alternative name EMBRYO DEFECTIVE2729; Wang et al., 2010). Thus, the function of the protein encoded at At3g20440 is currently unknown but unlikely to be a functional BE.
The be2 and be3 single mutants have phenotypes that closely resemble the wild type, indicating that there is a high degree of redundancy between the enzymes. However, be2be3 double mutants lack starch (Dumez et al., 2006). Instead, the plants accumulate large amounts of maltose and other linear malto-oligosaccharides (MOSs). This is presumably because linear chains produced by the SSs are cleaved by starch-degrading enzymes (α- and β-amylases; Dumez et al., 2006). The altered metabolism of these double-mutant plants impedes growth, and they are smaller and paler than the wild type. The precise reason for this is unclear.
In addition to mutagenesis, there have been several studies where BEs were overexpressed in transgenic plants. Overexpression of the E. coli glycogen BE (EcGLGB) in potato tubers or rice endosperm resulted in an increased degree of branching of amylopectin (Shewmaker et al., 1994; Kortstee et al., 1996; Kim et al., 2005). Overexpression of endogenous plant BE2 genes has also been performed in both rice and potato, increasing the proportion of shorter amylopectin chains (Tanaka et al., 2004; Brummell et al., 2015), and rice, leading to the accumulation of highly branched, water-soluble polysaccharides (Tanaka et al., 2004). Transgenic expression of genes from different photosynthetic organisms has also shown the degree of functional conservation within the plant BE classes. Sawada et al. (2009) showed that class II BE from Chlorella kessleri could rescue the BE2b-deficient phenotype in rice endosperm.
The aim of this work was to investigate the capacity of different types of BEs to mediate starch granule formation by assessing their ability to function in the context of an otherwise intact starch biosynthesis pathway. To do this, we used the Arabidopsis be2be3 double mutants as a line in which to express three types of BEs. We chose BE2a from maize (required for leaf starch synthesis and similar to the endogenous Arabidopsis proteins; Yandeau-Nelson et al., 2011), BE1 from potato (represents the plant class I BEs that Arabidopsis lacks; Safford et al., 1998), and GLGB (the BE from E. coli involved in glycogen biosynthesis). This approach differs from previous investigations, because the activity of each BE type (working in planta with the same set of SSs and DBEs) can be assessed, and the results can be directly compared. In addition, we sought to address whether a glycogen BE was sufficient for starch production—in other words, whether the remaining starch biosynthetic enzymes are capable of generating a crystallization competent polymer, even when partnered with a BE with a different specificity. In previously described transgenic plants expressing E. coli GLGB, the endogenous plant BEs were still present (Shewmaker et al., 1994; Kortstee et al., 1996; Kim et al., 2005).
In the transgenic lines generated here, we analyzed glucan synthesis, starch structure, and composition. Our results show that all three BE types can mediate starch granule production but to differing degrees. In each case, the structure of amylopectin and the amylose content depend on the type of BE present, as does starch granule morphology. We discuss the reasons for these differences in relation to previously reported BE properties.
RESULTS
Transformation of the be2be3 Double Mutant with ZmBE2a, StBE1, and EcGLGB Genes
Three genes were used to transform the be2be3 double mutant: BE2a from maize, BE1 from potato, and GLGB (the BE from E. coli). In the constructs, each gene was driven by the constitutive cauliflower mosaic virus 35S promoter (Supplemental Fig. S1A). For the maize and potato genes, we used the full-length coding sequences, including their plastid transit peptides. For the E. coli gene, the coding sequence was cloned such that the expressed protein was fused to the C terminus of the yellow fluorescent protein (YFP), which was itself preceded by transit peptide of the Arabidopsis Rubisco small subunit gene (At5g38430 encoding amino acids 1–57). We confirmed the correct targeting of this fusion protein to the chloroplasts by confocal fluorescence microscopy (Supplemental Fig. S1B). For each of the constructs, multiple independent transgenic lines were obtained, which were initially screened for the presence of branched glucans by qualitative iodine staining. We selected three independent lines for each transgene for further analysis.
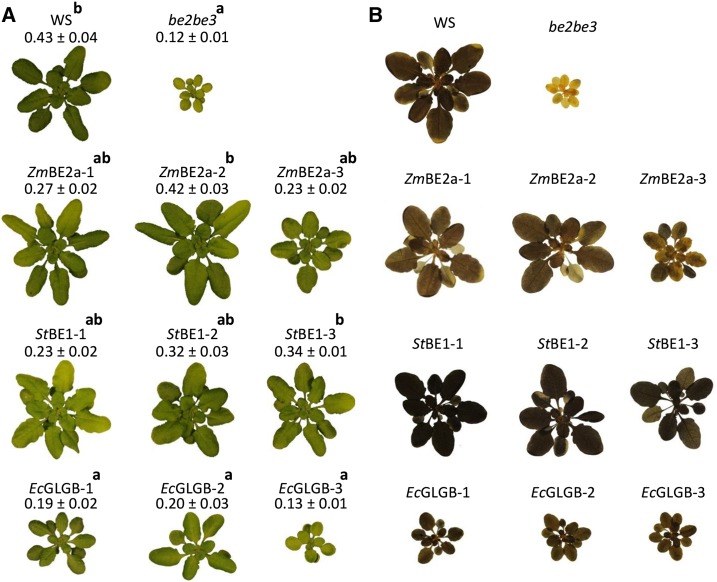
Previous studies showed that be2be3 double mutants were pale green, slow growing, and contained no starch but accumulated a pool of MOSs, primarily maltose (Dumez et al., 2006; Fig. 1). We evaluated the transformed plants for each of these phenotypes. All of the plants transformed with ZmBE2a or StBE1 were darker green and had increased growth rates compared with the be2be3 parental line (Fig. 1A). Most lines were still significantly smaller than the wild type, but a few were comparable in size (e.g. ZmBE2a-2 and StBE1-3). Of the transgenic lines, those complemented with the E. coli GLGB grew most poorly. They were darker green than be2be3, and some appeared larger, but overall, there was no statistically significant size difference. Most plants were still visibly paler than the wild type. Staining the plants for starch at the end of the day confirmed that the be2be3 parental line was starchless (Fig. 1B). Consistent with the initial screening, all of the transformed plants stained more darkly than be2be3 plants, indicating the presence of starch. However, they did not all stain the same color; the plants expressing ZmBE2a and EcGLGB stained brown, quite similarly to the wild type, whereas the plants expressing StBE1 stained even darker, suggesting the presence of more starch, longer amylopectin chains, or a higher amylose content. However, iodine staining alone can be difficult to interpret given that it is influenced by both the amount and the structure of glucans.
Figure 1.
Growth and starch staining of be2be3 mutants expressing different BE types. The class II BE from maize (ZmBE2a), the class I BE from potato (StBE1), and the glycogen BE from E. coli (EcGLGB) were expressed in the Arabidopsis be2be3 double mutant. For each transformation, three independent lines are shown (designated 1–3 in each case). A, Thirty days after germination, plants were harvested at the end of the day, weighed, and photographed. Representative plants for each transgenic line are shown. The rosette fresh weight for each line is given (mean ± sem of between four and eight plants). Significant differences in growth rate, as determined by ANOVA analysis (P < 0.05), are designated by the letters a and b. B, Plants, like in A, were decolorized in hot ethanol and stained for starch with iodine-potassium iodide solution. After rinsing in water, the plants were photographed. Note that the be2be3 parental line does not stain and the differences in starch staining between the lines transformed with different genes.
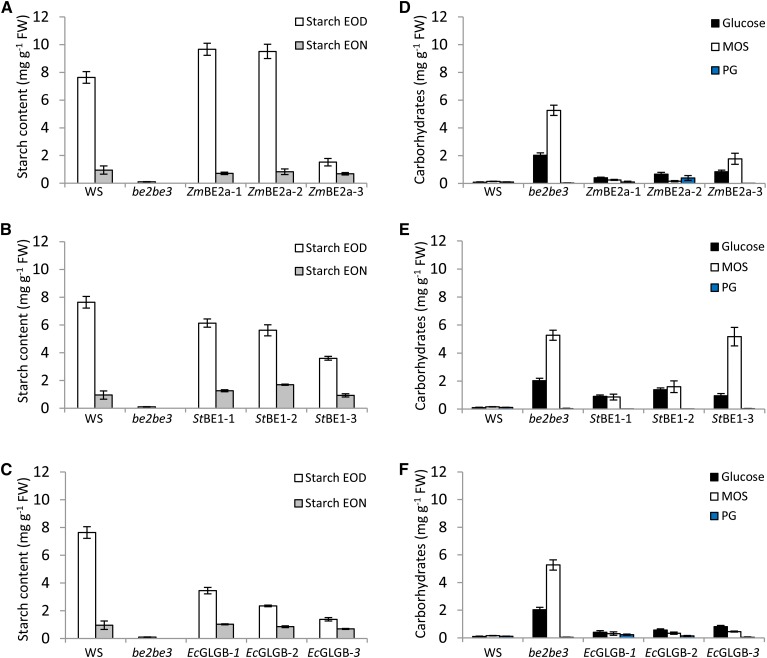
To obtain precise information, we quantified the starch content of the transformed plants at the end of the day and the end of the subsequent night (Fig. 2). Consistent with the iodine staining and previous reports (Dumez et al., 2006), the be2be3 mutant had no detectable starch. In contrast, we observed normal starch levels in two of the three lines expressing ZmBE2a. The third line (ZmBE2a-3) contained about 85% less starch than the wild type (Fig. 2A). Despite the dark-staining rosettes (Fig. 1B), plants expressing StBE1 contained less starch than the wild type (Fig. 2B), although there was again variation in the amounts of starch synthesized between the independent transformants. Low starch levels were observed in the plants expressing EcGLGB (Fig. 2C), although this was not apparent from the iodine staining. In all cases, starch levels at the end of the night were close to that of the wild type.
Figure 2.
Starch and soluble glucan content of be2be3 mutants expressing different BE types. Whole rosettes from plants were harvested at the end of the day (EOD) or the end of the night (EON) and immediately frozen in liquid N2. Starch and soluble glucans (free Glc, MOSs, and phytoglycogen [PG]) were extracted using perchloric acid, and glucans were quantified after enzymatic digestion to Glc. Each value is the mean ± se (n = 6). The data for the wild type (WS) and the be2be3 parental line are the same in A to C and in D to F. A to C, Starch contents of plants expressing ZmBE2a (A), StBE1 (B), or EcGLGB (C) are compared with the wild type and be2be3 at EOD and EON. D to F, Soluble glucan contents of plants expressing ZmBE2a (D), StBE1 (E), or EcGLGB (F) are compared with the wild type and be2be3 at EOD. FW, Fresh weight.
We measured water-soluble glucans, including free Glc, MOSs, and soluble polyglucans, such as phytoglycogen, in plants harvested at the end of the day. As expected, we detected only low levels of MOSs, soluble polyglucans, and Glc in the wild type (Fig. 2D). In contrast, be2be3 contained very high levels of MOSs and high levels of free Glc but no soluble polyglucans. The plants expressing ZmBE2a had low levels of MOSs, with the exception of ZmBE2a-3, which had intermediate levels between the wild type and the be2be3 parental line (Fig. 2D). Glc levels were slightly higher than those in the wild type, especially in ZmBE2a-3. None of the ZmBE2a-expressing plants contained significant levels of soluble polyglucans. In the plants expressing StBE1, MOSs were detectable in all lines, and the amounts correlated inversely with the amount of starch. Glc levels were also high, but again, very few soluble polyglucans were present (Fig. 2E). Interestingly, in all of the plants expressing EcGLGB, MOS levels were very low, regardless of the amount of starch that they produced. Glc levels were higher than in the wild type, but soluble polyglucan levels were again low (Fig. 2F).
The MOS in be2be3 was reported to be predominantly maltose (Dumez et al., 2006), with smaller amounts of longer oligosaccharides, such as maltotriose. We analyzed all of the transgenic lines to determine the nature of the MOS. In all cases, maltose accounted for 95% or more of the MOSs, with the remainder present as longer oligosaccharides (Supplemental Fig. S2).
Detection of the Activities of ZmBE2a, StBE1, and EcGLGB
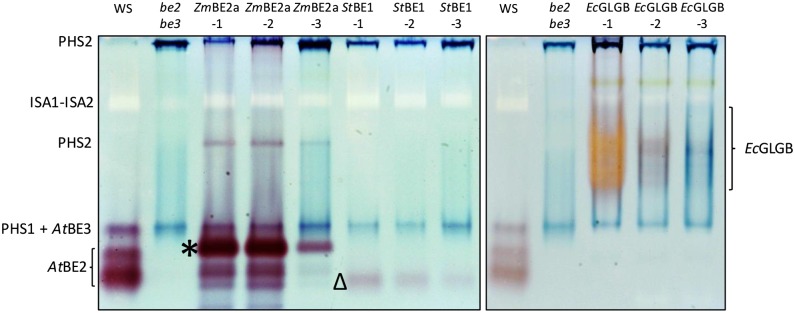
To detect the activities of the different BE types, we used a zymogram approach and quantitative assays based on the stimulation of rabbit muscle phosphorylase-a by the presence of BE. For the zymograms, extracts were made from fresh leaf material, and the soluble proteins were separated on native PAGE gels containing small amounts of glycogen as a primer. After electrophoresis, the gels were incubated in a solution containing phosphorylase-a. The gels were then stained with iodine to detect bands of BE-stimulated phosphorylase activity (Fig. 3). In extracts of the be2be3 parental line, we observed only the blue-staining activities of the endogenous α-glucan phosphorylases (PHS1 and PHS2), whereas in extracts of the wild type, we detected three additional bands of red-staining activity. Two fast-migrating bands corresponded to the BE2 activity, whereas one band, which frequently comigrated with the plastidial α-glucan phosphorylase PHS1, corresponded to BE3 (see Supplemental Fig. S3, where these activities are resolved). In plants expressing ZmBE2a, we observed a major red-staining band of activity migrating in a similar part of the gel as the endogenous BE2. Additional minor bands were observed running slightly faster than the major band. The activity was strongest in lines ZmBE2a-1 and ZmBE2a-2 and much weaker in line ZmBE2a-3. In lines expressing StBE1, a faint, fast-migrating, red-staining band of activity was detected. The band was strongest in line StBE1-1 and weakest in line StBE1-3. In lines expressing EcGLGB, a slow-migrating and poorly resolved band of activity was detected. This activity was strongest in line EcGLGB-1 and stained bright orange. In line EcGLGB-2, the activity was weaker, and the band was red stained. In the line EcGLGB-3, the activity was weakest, and the staining was faint blue. We attribute the differences in staining color to the balance between chain elongation by phosphorylase-a and the branching by GLGB. Thus, the in-gel product of EcGLGB-1 extracts is more highly branched than that of EcGLGB-3 extracts.
Figure 3.
Native PAGE (zymograms) of be2be3 mutants expressing different BE types. Soluble proteins were extracted from the indicated lines and separated by native PAGE in gels containing 0.02% (w/v) glycogen. BE activity was detected by incubating the gels in a medium containing phosphorylase-a and its substrate, Glc-1-P. The identifications of the endogenous BE2 and BE3 activities are based on comparable analysis of the wild type, the be2 and be3 single mutants, and the be2be3 double mutant (see supplemental data in Pfister et al., 2014). Dark bands represent the endogenous cytosolic (PHS2) and plastidial (PHS1) α-glucan phosphorylases. In these gels, BE3 comigrates with PHS1. A pale band corresponds to the heteromultimeric ISA1-ISA2 DBE. Unique activities attributable to ZmBE2a (*), StBE1 (Δ), and EcGLGB are indicated.
In addition to BE activities, other enzyme activities were also revealed on the zymograms. The blue-staining bands of PHS1 and PHS2 were stronger in be2be3 parental lines than in the wild type as previously reported (Dumez et al., 2006). Below the cytosolic α-glucan phosphorylase (PHS2), a white band could be identified as ISA1 and ISA2 activity, which showed no significant difference between lines. We performed further zymograms to detect SS activities (Supplemental Fig. S4), which revealed bands of activity previously attributed to SS1 and SS3 (Pfister et al., 2014). Again, there were no major differences between the transgenic lines in these activities.
These zymograms suggested that the lines with the highest BE activity showed the strongest phenotypic changes compared with be2be3 plants (Figs. 1 and 2). In each case, the line with the lowest detectable BE activity (assigned as line 3) also had the weakest starch-accumulating phenotype. To support this observation, we performed a quantitative assay also based on phosphorylase-a stimulation, in which the incorporation of 14C-labeled Glc-1-P into phosphorylase-a products is measured precisely. The results (Table I) confirmed that there were quantitative differences in BE activity among the transgenic lines expressing each type of BE, which was apparent from the zymograms. We emphasize, however, that these numbers should be treated with caution when comparing between the sets of transgenic lines, because different BE types are known to give different absolute activities in the phosphorylase-a stimulation assay (Guan and Priess, 1993; Guan et al., 1997). The reasons for this are unclear but probably relate to differences in BE substrate specificities—a factor that likely limits other types of BE assays as well.
Table I. Quantification of BE activity in plants expressing different BE types.
For each sample, six leaves from six plants of each line were harvested in the middle of day and pooled. Extracts of soluble proteins (10 µg) were added to an assay mixture containing 50 mm Glc-1-P, including [U-14C]Glc-1-P. The reactions were started by adding 2 units of phosphorylase-a, and they were incubated at 25°C for 3 h. The activity of the BE from each line was calculated from the rate obtained between 60 and 180 min (mean ± se; n = 3).
| Genotype | BE Activity |
|---|---|
| nmol Glc-1-P incorporation min−1 mg−1 protein | |
| Wild type (WS) | 476 ± 47 |
| be2be3 | 27 ± 50 |
| ZmBE2a-1 | 1,740 ± 6 |
| ZmBE2a-2 | 1,591 ± 79 |
| ZmBE2a-3 | 318 ± 24 |
| StBE1-1 | 230 ± 39 |
| StBE1-2 | 232 ± 32 |
| StBE1-3 | 54 ± 15 |
| EcGLGB-1 | 344 ± 39 |
| EcGLGB-2 | 223 ± 14 |
| EcGLGB-3 | 216 ± 56 |
Differences in Amylopectin Structure in Starches Made by ZmBE2a, StBE1, and EcGLGB
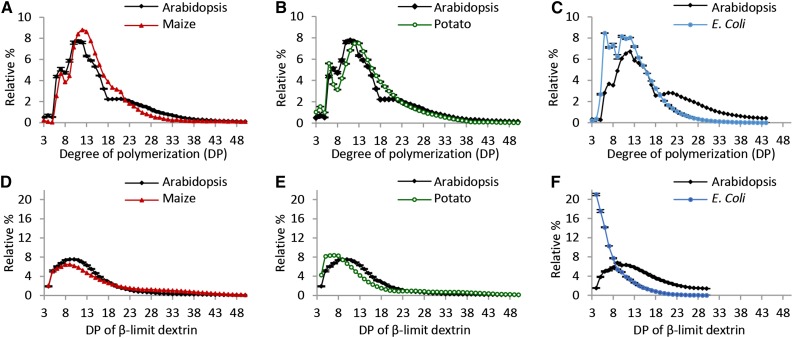
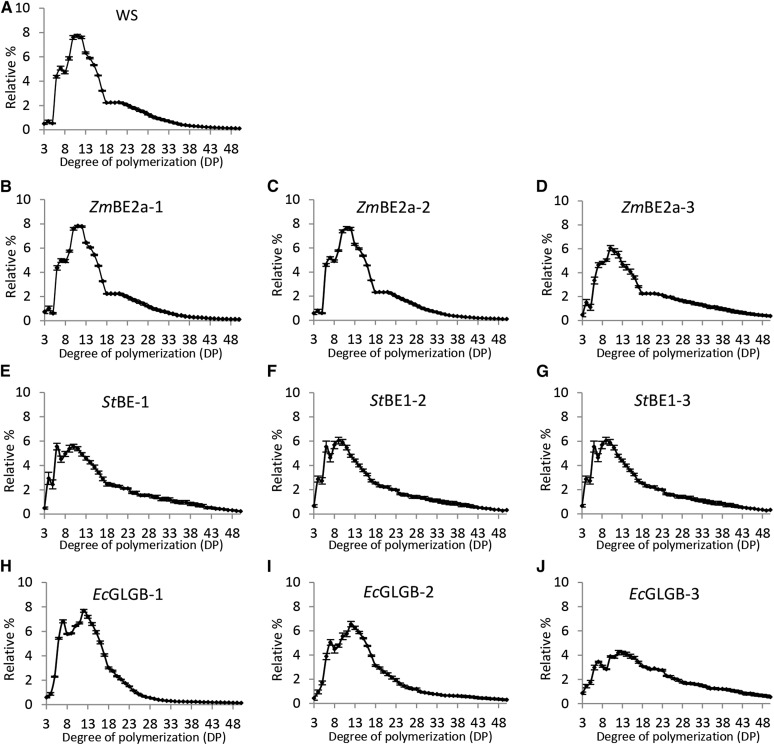
We investigated the structural characteristics of the starch made with the contribution of each BE class. We analyzed the chain length distribution (CLD) of amylopectin after gelatinizing and completely debranching the starch. For comparison, we analyzed the CLDs of wild-type Arabidopsis leaf starch, maize leaf starch (because ZmBE2a is the predominant BE expressed in the maize leaf; Blauth et al., 2001), potato tuber amylopectin, and E. coli glycogen (Fig. 4). The CLD of Arabidopsis wild-type starch, when analyzed this way, has three subpopulations of chains: short chains (with a degree of polymerization [DP] of between 6 and 8), intermediate chains (DP of between 8 and 18), including the cluster-forming chains, and long chains (DP of between 18 and 50) that include chains spanning one or more clusters (Fig. 4A). Both maize leaf starch and potato tuber amylopectin also had subpopulations of short chains. The distinction between intermediate and longer chains around DP of 18 was apparent for maize leaf starch (albeit less pronounced than for Arabidopsis) but not for potato tuber amylopectin (Fig. 4, A and B).
Figure 4.
CLDs and β-CLDs of starch from Arabidopsis leaves, maize leaves, and potato tuber and glycogen from E. coli. Starch/glycogen was debranched either without (A–C) or with (D–F) prior treatment with an excess of β-amylase. The resultant linear chains were separated and detected by HPAEC-PAD. Peak areas in the ranges given were summed, and the relative peak area for each chain length was calculated. Values are the means ± se from three or four biological replicates. A and D, CLDs and β-CLDs of Arabidopsis and maize leaf starches. B and E, CLDs and β-CLDs of Arabidopsis leaf and potato tuber starches. The data for Arabidopsis (the WS wild type) are the same as in A and D. C and F, CLDs and β-CLDs of Arabidopsis leaf starch and E. coli glycogen. The data for Arabidopsis are from a different batch of WS wild-type plants that were analyzed at the same time as the E. coli glycogen.
Plants expressing ZmBE2a had a CLD identical to that of the Arabidopsis wild type (Fig. 5), with the exception of ZmBE2a-3, the CLD of which had fewer intermediate-length chains and relatively more long chains (DP of 25–45; Fig. 5, B–D). Nevertheless, the three subpopulations of chain lengths were still apparent with the CLD of ZmBE2a-3 (Fig. 5D). Plants expressing StBE1 produced starch with a distinct CLD (Fig. 5, E–G). The profile contained short chains of DP of six to seven and a continuous distribution of intermediate to long chains. Compared with wild-type Arabidopsis starch, there was a depletion in chains with a DP of 10 to 17 and an enrichment of very short chains (DP of 4–6) and long chains (DP of 28–50). The CLDs of the three independent transformants were similar, although StBE1-3, which had the least starch and the least detectable BE activity, had a CLD with more long chains, fewer intermediate chains, and more very short chains (Fig. 5G).
Figure 5.
CLD of starch from be2be3 mutants expressing different BE types. Starch in the insoluble material derived from perchloric acid extracts from the given lines was debranched, and the resulting linear chains were analyzed using HPAEC-PAD. Peak areas from DP of 3 to DP of 50 were summed, and the relative peak area for each chain length was calculated. Values are the means ± se from three or four biological replicates. A, CLD of starch from the wild type (WS). B to D, CLD of starch from independent transgenic lines expressing ZmBE2a. E to G, CLD of starch from independent transgenic lines expressing StBE1. H to J, CLD of starch from independent transgenic lines expressing EcGLGB.
The CLD of E. coli glycogen is quite distinct from that of amylopectin (Fig. 4C). The glycogen has subpopulations of short chains (DP of between 5 and 9) and intermediate chains (DP of between 9 and 30). Interestingly, the starch from plants expressing EcGLGB had glycogen-like characteristics in that there was a subpopulation of DP of five to nine short chains together with a population of intermediate to long chains (Fig. 5, H–J). The CLDs from the three independent lines differed significantly. EcGLGB-1, which had the highest starch level (Fig. 2C) and the highest detectable BE activity (Fig. 3), had many short chains and intermediate chains with DP of 10 to 18 but fewer chains more than a DP of 20 (Fig. 5H). EcGLGB-3 showed the opposite (Fig. 5J).
We repeated the analyses on high Mr amylopectin fractions derived from Sepharose CL2B gel-permeation chromatography (GPC). Although this approach does not necessarily capture all of the amylopectin in the starch, it rules out the possibility that short branches of the amylose fraction interfere in the amylopectin CLD analysis. However, overall, the results were very similar to those obtained when analyzing the whole-starch preparations (Supplemental Fig. S5).
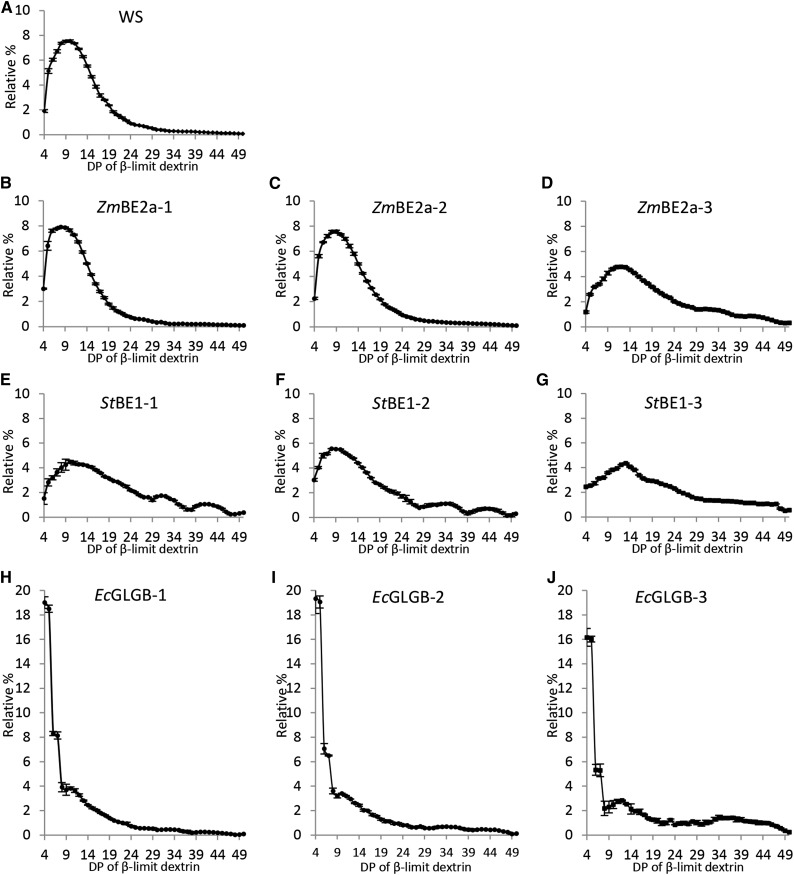
The CLD is an important architectural feature of branched glucans but does not provide information on the relative positioning of the branch points. Therefore, we analyzed the chain length distribution of the β-limit (β-CLD) dextrin of each of the starches and E. coli glycogen (Figs. 4 and 6). During β-amylase treatment, external chains that do not carry other branches (so-called A chains) are shortened to stubs of DP of two or three as the exoamylase sequentially removes maltosyl groups from the exposed nonreducing ends. For chains carrying other chains through a branch point (so-called B chains), their internal segments are protected from degradation, because β-amylase cannot work past a branch point, stopping one or two Glc residues before. By debranching the β-limit dextrin, information about the lengths of the internal B-chain segments can be obtained. It is important to note that the method does not discriminate between singly and doubly branched chains. Thus, the β-CLDs reflects the length between the initiation of a B-chain at a branch point and the outermost branch that it carries (plus one or two Glc units).
Figure 6.
β-CLDs of starch from be2be3 mutants expressing different BE types. Starch in the insoluble material derived from perchloric acid extracts from the given lines was treated with an excess of β-amylase. The resultant β-limit glucan was debranched to obtain linear chains that were analyzed by HPAEC-PAD. To compare the lengths of internal chain segments, peak areas from DP of 4 to DP of 50 were summed, and the relative peak area for each chain length was calculated. Values are the means ± se from three or four biological replicates. A, β-CLD of starch from the wild type (WS). B to D, β-CLD of starch from independent transgenic lines expressing ZmBE2a. E to G, β-CLD of starch from independent transgenic lines expressing StBE1. H to J, β-CLD of starch from independent transgenic lines expressing EcGLGB.
The β-CLDs of the three starch control samples were similar but not identical (Fig. 4, D and E). Arabidopsis and maize leaf starches had populations of internal chain segments ranging in DP of from 4 to >25, with modal values of DP of 10 and 9, respectively. The β-CLD of potato tuber amylopectin had more short chains (DP of five to eight) than the leaf starches, with a modal value of DP of seven. The β-CLDs of starch from the transgenic lines expressing ZmBE2a were nearly identical to those of Arabidopsis (Fig. 6, A–D). The exception was ZmBE2a-3, which had the lowest starch levels and BE activity. The β-CLD of this line was broader and shifted toward longer chains, which could indicate that branches are more widely spaced (Fig. 6D). All lines expressing StBE1 had broad β-CLDs, with fewer chains between 5 and 15 in length compared with wild-type Arabidopsis starch (Fig. 6, E–G). Again, the line with the lowest BE activity (StBE1-3) had the β-CLD profile most shifted toward longer chains (Fig. 6G). The β-CLD of E. coli glycogen was radically different from those of the starches. The most abundant chains by far were those of DP of 4 and 5, and the frequency of longer chains decreased steadily until DP of 20 (Fig. 4F). Remarkably, in this respect, the β-CLD of the starches from transgenic lines expressing EcGLGB strongly resembled E. coli glycogen rather than any of the other starches (Fig. 6, H–J). However, whereas the abundance of longer chains in the β-CLD of E. coli glycogen decreased smoothly, that of the starch from lines expressing EcGLGB had a sharp decline in abundance and then a second peak in the region of DP of 9 to 11 (Fig. 6, H–J).
Collectively, these data show that specificities of each BE type present are clearly reflected in the CLD and the branch point positioning of amylopectin produced in combination with the other starch biosynthetic enzymes. Our data also show that the amount of BE activity relative to the other enzymes of starch metabolism influences both the quantity of starch made and to an extent, its structure. This supports previous studies where the rice BE2 mutant was complemented by overexpression, giving rise to a range of lines with different BE2 levels (Tanaka et al., 2004). This dosage relationship presumably reflects the fact that, on the one hand, the SSs are dependent on BE to create substrates in the form of nonreducing ends; on the other hand, when BEs are limiting, the SSs will continue to elongate the available chains, thus altering the CLD and the substrate for the BE reaction, leading to a different amylopectin product.
Composition of Starch Granules Produced by the Different BE Classes
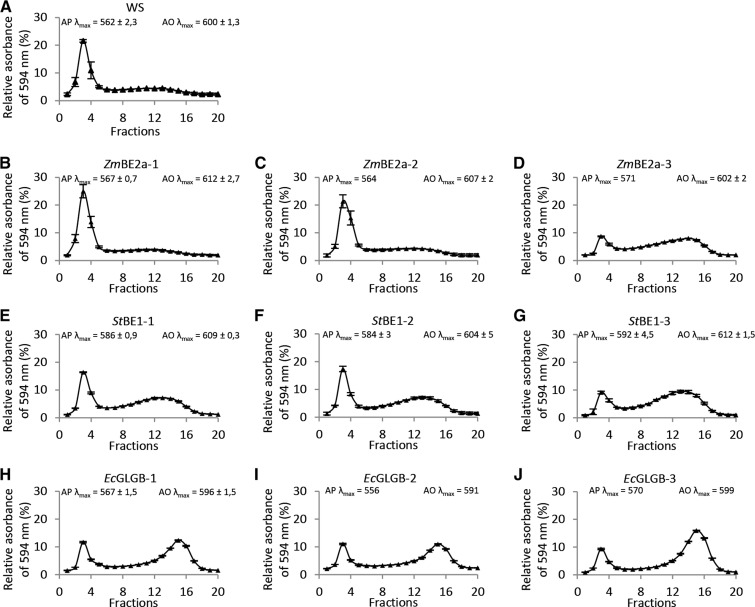
We used Sepharose CL2B GPC to analyze the composition of the starches made in the transgenic lines (Fig. 7). This medium is effective in separating larger amylopectin molecules, which elute early from the column, from the smaller amylose molecules that elute later (Zeeman et al., 2002a, 2002b). Wild-type starch separated into a sharp peak of amylopectin (fractions 1–5; λmax with iodine of 562 nm) followed by a broad amylose peak (fractions 6–19; λmax with iodine of 600 nm; Fig. 7A). The composition of the starch from plants expressing ZmBE2a was quite similar to the wild type, except for ZmBE2a-3, which had relatively more amylose than the other two lines (Fig. 7, B– D). The composition of the starch from plants expressing StBE1 differed, with all three lines having higher amylose contents than the wild type. Furthermore, the λmax of the amylopectin fraction complexed with iodine was between 584 and 592 nm—a significantly longer wavelength than for wild-type amylopectin. This suggests that the amylopectin from these lines contains more long chains than wild-type amylopectin and hence, an overall staining intermediate between that of amylopectin and amylose (Fig. 7, E and F). The starch from plants expressing EcGLGB also contained a high proportion of amylose, although the staining of the amylopectin fraction was, in this case, similar to the wild type (Fig. 7, H–J). As with the ZmBE2a-expressing plants, the EcGLGB-expressing line with the lowest activity (EcGLGB-3) had the highest amylose content. Interestingly, the amylose fractions in all EcGLGB plants had slightly lower λmax values than the other lines (between 591 and 599 nm) and eluted slightly later from the GPC column. This could mean that the amylose fraction contained more branch points and was lower in Mr.
Figure 7.
Amylose and amylopectin contents of starch from be2be3 mutants expressing different BE types. Starch extracted from the given lines was dissolved in 0.5 m NaOH and separated by Sepharose CL-2B gel filtration chromatography. The eluate was collected in fractions, which were neutralized, mixed with an iodine-potassium iodide solution, and analyzed for their absorbance at wavelengths between 400 and 700 nm. The interpolated line is the absorption at 594 nm. The λmax values for the amylopectin and amylose peaks are given. Values are the means ± se from two to three experimental replicates, expect for those for EcGLGB-2 and EcGLGB-3, which were from single replicates conducted on starch extracted from a pool of plants. A, CL-2B elution profile of starch from the wild type (WS). B to D, CL-2B elution profile of starch from independent transgenic lines expressing ZmBE2a. E to G, CL-2B elution profile of starch from independent transgenic lines expressing StBE1. H to J, CL-2B elution profile of starch from independent transgenic lines expressing EcGLGB. AO, Amylose; AP, amylopectin.
Given the differences in amylose content, we evaluated whether the abundance of GBSS varied in the different transgenic lines. Immunoblots of total protein extracts probed with GBSS-specific antibodies revealed differences in GBSS abundance (Supplemental Fig. S6), which correlated with starch amount but not with amylose content. For example, the wild type, ZmBE2a-1, and StBE1-1 all had comparable amounts of starch and GBSS but of the three, only StBE1-1 had high amylose. These data suggest that GBSS abundance is not the primary cause of the differences in amylose content.
Formation of Starch Granules in the Mesophyll of be2be3 Double Mutants by Expression of Different BE Classes
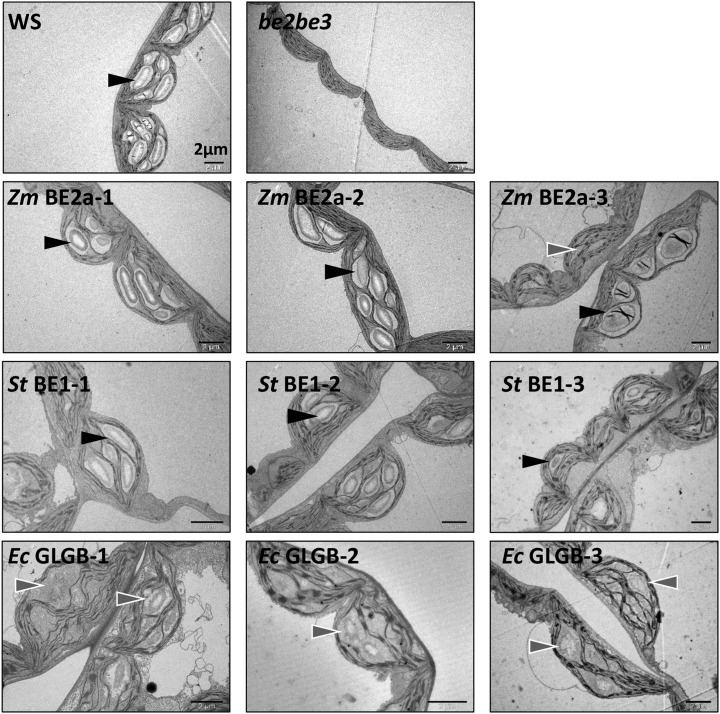
We analyzed leaf mesophyll cells by transmission electron microscopy (TEM) to observe the appearance of the starch produced in the transgenic lines (Fig. 8). Normal lenticular granules were observed in the chloroplast of the wild type, whereas no granules were visible in the chloroplasts of the be2be3 parental line as expected (Dumez et al., 2006). The expression of ZmBE2a in be2be3 restored the production of normal looking granules, although in ZmBE2a-3, there were fewer granules per chloroplast and in a few cases, these granules were highly irregular in appearance (Fig. 8; Supplemental Fig. S7). We observed normal-looking granules in all three lines expressing StBE1. However, in the transgenic lines expressing EcGLGB, the appearance of most of the granules was abnormal, having irregular shapes and poorly defined boundaries to the chloroplast stroma.
Figure 8.
Appearance of starch granules within leaf mesophyll chloroplasts of be2be3 mutants expressing different BE types. Leaves from the indicated lines were harvested close to the end of day, fixed with glutaraldehyde, and embedded in resin, and chloroplasts in the mesophyll cells were imaged by TEM. Normal-looking starch granules (black arrows) and aberrant granule-like structures (gray arrows) are indicated. Additional images are presented in Supplemental Figure S7. Bars = 2 µm.
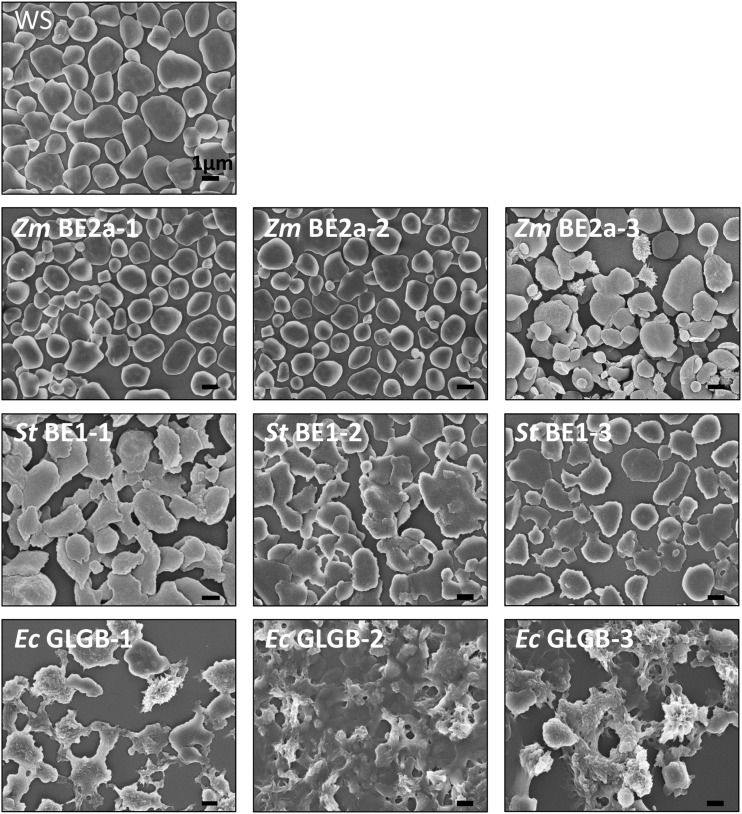
To observe directly the starch granule morphology, we extracted and purified starch from each of the lines and examined it using scanning electron microscopy (SEM; Fig. 9). This analysis revealed that the starch granules from ZmBE2a-expressing plants were indeed very similar to those of the wild type. In the case of ZmBE2a-3, most granules appeared normal, but they were variable in size, and among them were granules with highly irregular surfaces. The starch granules of the StBE1- and EcGLGB-expressing plants appeared different from the wild type. All three StBE1-expressing lines had flat granules with smooth surfaces, like those of the wild type, but with irregular margins. In some cases, the granules appeared to have fused with each other. However, the granules from the EcGLGB-expressing plants were radically different in appearance, having very rough surfaces and highly irregular shapes, with frequent protrusions.
Figure 9.
Morphology of starch granules from be2be3 mutants expressing different BE types. Starches extracted from the given lines were analyzed by SEM. Bars = 1 µm.
DISCUSSION
The data presented here provide valuable information about the importance of BE specificity in starch biosynthesis. Our work complements previous in vitro enzymatic studies, mutant phenotypic studies, and transgenic approaches, because it allows the direct comparison of each BE type in the same plant background, in which the other types of starch biosynthetic enzymes (SSs and DBEs) are present. Most other mutant and gene repression lines that have been analyzed still contain more than one functional BE or a degree of residual activity. Furthermore, different plant species and/or tissue types have different amounts of the other starch biosynthetic enzymes, which also influence the final starch structure and make cross comparisons problematic.
Our work confirms how important BE specificity is for starch biosynthesis in not only terms of the branch lengths that they transfer but also, how they position branch points relative to each other. The resulting glucans have very different structures, compositions, and granule morphologies. Some aspects of the phenotypes that we observe are very intriguing. In particular, it is not fully clear why MOSs accumulate in the be2be3 double mutant and in some—but not all—of the transgenic lines derived from it.
ZmBE2a Can Fully Complement the Arabidopsis be2be3 Mutant
Because Arabidopsis contains two class II BEs, which are largely redundant with each other, it is perhaps not surprising that, of the three BEs examined, the maize class II BE provides the most convincing complementation of the be2be3 double-mutant phenotype. Two of the three ZmBE2a-expressing lines (ZmBE2a-1 and ZmBE2a-2), which have the highest BE activity, are wild type-like in terms of their appearance, growth rate, and starch/soluble glucan contents. Furthermore, their starch granules appear normal by SEM and TEM, have a normal composition, have a wild type-like CLD, and differ only slightly in β-CLD. These results argue for a conservation of function among class II BEs types, particularly when these results are juxtaposed with the phenotypes observed with the other BE types. This idea is consistent with the previous observation—for example, that C. kessleri class II BE was able to complement the be2b mutant of rice (Sawada et al., 2009). Furthermore, our observations show that the differences in CLDs between Arabidopsis and maize leaf amylopectin are caused by the other starch synthetic enzymes. We suggest that this is primarily because of a different complement of SSs isoforms, because these are known to vary in relative abundance from one starch-synthesizing system to another. It is less likely to be caused by a difference in ISA specificity. It was recently reported that the expression of maize or rice ISA1 in the Arabidopsis isa1 mutant resulted in a very similar amylopectin structure to that of the wild type (Facon et al., 2013; Streb and Zeeman, 2014).
The third line, ZmBE2a-3, displays only partial complementation, presumably because the transgene inserted in a chromosomal location that does not permit high levels of expression (reflected in the lower BE activity than the other two transgenic lines; Fig. 3; Table I). In this line, BE activity presumably limits starch production, explaining why there is only around 15% of the wild-type starch level. This is surprising given that the phosphorylase-a stimulation assay indicates that it has two thirds of the activity of the endogenous Arabidopsis BE. It is possible that ZmBE2a does not work efficiently with the other Arabidopsis starch biosynthetic enzymes. Alternatively, our BE assay might underestimate the Arabidopsis activity—an idea that is supported by work suggesting that BE3 activity is difficult to measure this way (Dumez et al., 2006). Whatever the cause, the limitation in BE activity in ZmBE2a-3 results in some important differences in starch content, amylopectin structure, and granule morphology compared with the other ZmBE2a-expressing lines. For example, the CLD shows that average chain lengths of amylopectin are longer, and the β-CLD suggests that the branch points are more widely spaced—features consistent with a limitation in branching relative to chain elongation activity.
Interestingly, the MOS-accumulating phenotype characteristic of the be2be3 parental line is still apparent in ZmBE2a-3. It was suggested that, in be2be3, linear chains can still be synthesized and that these are degraded by chloroplast-localized amylases resulting in MOS (Dumez et al., 2006). Normally during starch degradation, β-amylases produce β-maltose, which after export from the plastid, is metabolized by the cytosolic glucosyltransferase disproportionating enzyme2 (DPE2; Chia et al., 2004; Lu and Sharkey, 2004; Niittylä et al., 2004; Weise et al., 2004; Fulton et al., 2008). In be2be3, the type of MOS accumulating was reported to be primarily α-maltose. It was further suggested that DPE2 can only metabolize this maltose after it has slowly mutorotated to the β-configuration. As yet, the enzyme(s) generating α-maltose and the precise nature of the substrates on which they act have not been established, but it seems that the aberrant, long chain-enriched amylopectin made in the presence of limiting ZmBE2a activity is susceptible to the same process, whereas wild-type amylopectin made in the presence of sufficient ZmBE2a activity is not.
Interestingly, ZmBE2a-3 also has relatively more amylose than the other ZmBE2a lines and the wild type. This does not seem to be because of an increased abundance of GBSS, the levels of which reflected the amount of starch present. Rather, it may be because the limitation of amylopectin synthesis renders more ADP-Glc substrate available for amylose synthesis by GBSS. It is also possible that GBSS activity is stimulated by the presence of MOS in the chloroplast stroma (Denyer et al., 1999; Zeeman et al., 2002a, 2002b).
Partial Complementation of be2be3 by StBE1 Results in High Amylose Starch
Although Arabidopsis does not possess a class I BE, the expression of StBE1 restores starch biosynthesis and partially complements the be2be3 phenotype. The StBE1-expressing plants grew faster than the be2be3 parental line and contained 60% to 80% as much starch as the wild type. However, there are some interesting differences in the phenotypes of these plants compared with those complemented with ZmBE2a.
Despite having less starch than the wild type, the StBE1-complemented plants show dark iodine staining. Our data show that this is because of the high amylose content of the starch but also because the amylopectin-iodine complex has a high λmax (glucans with high λmax-values have a higher affinity for iodine and therefore, generally stain more strongly). The CLDs and β-CLD show that the amylopectin differs from that of the wild type, having relatively more long chains, fewer intermediate chains, and branch points spaced farther apart. Thus, some long chains may adopt single helical structures like amylose, whereas others form double helices like amylopectin, leading to intermediate staining characteristics. Similar λmax values have been reported for endosperm amylopectin from be2b or amylose extender mutants of maize, where much of the remaining BE activity could be attributable to BE1 (Boyer et al., 1976). These aspects of the starch phenotype are broadly consistent with the reported action of class I BEs in transferring longer chains than class II BEs (Guan and Preiss, 1993; Rydberg et al., 2001; Nakamura et al., 2010).
Interestingly, the CLDs of StBE1 amylopectin also contained very short chains of DP of four to six, which is surprising given that in vitro studies showed that class I BEs both transfer chain segments longer than DP of six (usually much longer) and also, leave residual chains of DP of six or longer. The presence of shorter chains could be explained by the interference of glucan-degrading enzymes in the biosynthetic process. This is supported by the presence of high MOS levels in the StBE1 lines, even in those lines with near wild-type levels of starch (Fig. 2). Thus, it seems that the aforementioned degradative process that generates MOS affects long chain-containing amylopectin produced by StBE1 far more than the wild type-like amylopectin produced by ZmBE2a (e.g. compare the starch and MOS levels for ZmBE2a-3 with those of the StBE1 lines; Fig. 2, D and E). Again, the MOS could contribute to the high amylose phenotype of the StBE1 lines by providing primers for GBSS, which itself was similar in abundance to that in wild-type plants (Supplemental Fig. S6). It would be interesting to see whether MOSs are present in other situations where starch made predominantly by BE1 was found to have high amylose (Bhattacharyya et al., 1990; Regina et al., 2006, 2010).
The β-CLDs of the StBE1-expressing lines with the highest starch levels (StBE1-1 and StBE1-2) have a distinctive pattern, with three subpopulations of chains of DPs of 4 to 28, 29 to 38, and 39 to 48. It is possible that the longer populations may be formed when a BE acts on internal chains rather than external chains (i.e. transferring an already branched or doubly branched fragment rather than a linear one). Such activity has been proposed for class I BEs from studies of the recombinant rice enzyme (Nakamura et al., 2010). The longer subpopulations were not apparent in the ZmBE2a lines with the highest starch levels (ZmBE2a-1 and ZmBE2a-2), consistent with the idea that class II BEs preferentially transfer linear chain segments by acting on external chains (Nakamura et al., 2010). The subpopulations could also reflect a relative increase in chains that spans two or more amylopectin clusters. However, it is important to bear in mind that there are different ways in which the CLDs can interpreted, because different models for amylopectin architecture have been proposed (Pérez and Bertoft, 2010). Most models have radially oriented cluster-spanning chains, but it is plausible that there are long chains in the amorphous lamellae that are perpendicular to the clusters. Furthermore, it is generally overlooked that a significant fraction of the glucan within a starch granule is not necessarily organized in the often-presented cluster configuration.
Finally, although the starch granule morphologies of the StBE1-expressing lines looked normal in TEM sections, they were distinct from those of the wild type and lines complemented with ZmBE2a when viewed by SEM, because they were larger and more irregular in shape. This could be a feature caused by the aberrant biosynthesis, the susceptibility of the glucan to degrading enzymes, or both.
Expression of EcGLGB in be2be3 Results in Highly Abnormal Starch
Perhaps the most intriguing results come from the be2be3 plants expressing the E. coli GLGB. Despite synthesizing starch, the be2be3 growth phenotype was poorly complemented in these plants. They grew only marginally better than the parental lines, and some plants were still pale. The starch content was generally much lower than for the lines expressing the plant BEs, even though large amounts of EcGLGB activity were detected on the zymograms (e.g. line EcGLGB-1; Fig. 3).
Using some methods, the starch made with EcGLGB does not appear too different from the other lines. Although the amylopectin CLD is clearly distinct from those of the ZmBE2a- and StBE1-derived amylopectins, it is no more different to them than they are from each other (Fig. 5). The distinctive features of the EcGLGB amylopectin CLDs are the abundance of short chains and the lack of longer chains. However, using other methods, it is clear that the starch made with EcGLGB is radically different. First, the β-CLDs of EcGLGB amylopectin are dominated by extremely short chains of DPs of four and five, suggesting that branches are introduced extremely close to other branches (a chain of DP of four suggests branches within two or three glucosyl residues of one another). In this way, the β-CLDs resembled those of E. coli glycogen much more than the ones of starch. Second, whereas the starch fractionates into peaks corresponding to amylopectin and amylose, the amylose peak differs, eluting late (indicating a low Mr) and having a shorter λmax when complexed with iodine (indicating more branches). Third, the appearance of the starch by TEM and SEM is very unusual, with highly irregular shapes and poorly defined granules. Nevertheless, the analysis of these plants shows that the remaining starch biosynthetic enzymes are indeed capable of working together with this unusual BE to generate a structure capable of crystallization. It will be interesting to explore further the crystallinity and functional properties of this unique starch. This will reveal if the higher order structures observed in wild-type starches are present and whether the unusual structure imparts useful characteristics. The previous use of GLGB to increase the branching of potato tuber and rice endosperm amylopectin altered starch properties, although in these cases, it acted together with the endogenous BEs (Shewmaker et al., 1994; Kortstee et al., 1996, 1998; Kim et al., 2005).
No soluble, glycogen-like polymers accumulated in these EcGLGB-expressing lines. It is possible that a fraction of the synthesized polymers is soluble but does not accumulate, because they are simultaneously degraded. Sundberg et al. (2013) showed that the heteromultimeric ISA ISA1/ISA2 that normally facilitates amylopectin crystallization actually suppressed glycogen accumulation when expressed in E. coli. It was suggested that E. coli glycogen cannot be debranched into a form that will crystallize, because the glycogen synthase and glycogen BE do not generate a suitable substrate. A similar explanation was recently proposed to explain the nearly starchless phenotype of the ss2ss3 double mutant of Arabidopsis. In that case, SS1 and SS4 are present, but almost no starch is made. Instead, soluble glucans and MOS were detectable. The additional loss of ISA1/ISA2 in this line resulted in the accumulation of large amounts of soluble glucan, showing that SS1 and SS4 together are capable of glucan synthesis and suggesting that accumulation is suppressed by ISA1/ISA2 (Pfister et al., 2014).
However, despite having low starch contents, the MOS content of the lines expressing EcGLGB is much lower than for the lines expressing StBE1. This suggests that, if simultaneous degradation does occur, the process differs from what is happening in be2be3 and the StBE1-expressing lines that generate long chain-rich amylopectin. Further experiments are needed to resolve these issues.
CONCLUSION
Some of our findings are in agreement with in vitro studies of the different BE types, which have investigated factors including the lengths of chains transferred and the position at which the transferred chain segment is reattached to the acceptor chain (Guan and Preiss, 1993; Rydberg et al., 2001; Nakamura et al., 2010; Sawada et al., 2014). Our data are consistent with the idea that BEs can be similar in terms of the lengths of chains that they transfer, but differ in the location where they reattach it. This is clear for EcGLGB, which transfers chains of similar lengths to the plant BEs but positions them extremely close to other existing branch points (Sawada et al., 2014). In contrast, the plant BEs seem to place branches farther apart, which has been proposed to be important for the correct alignment of double helices within the crystalline lamellae of amylopectin (O’Sullivan and Perez, 1999).
The fact that the Arabidopsis SSs and DBEs can form an insoluble glucan even working with BEs of distinct origins that have quite different specificities suggests that their combined specificities are integral to the starch biosynthetic process. However, it also seems that the resulting enzyme combinations are not equally efficient in every case. The major structural differences in the resultant insoluble polymers reinforce just how important the different specificities of the BEs are in determining the branch point distribution. Although the ISA1/ISA2 DBE also plays an important role in this process in Arabidopsis, as illustrated by the phytoglycogen accumulation the isa1 and isa2 mutants (Zeeman et al., 1998; Delatte et al., 2005), the data here indicate that it cannot correct the unusual branching patterns generated by StBE1 and EcGLGB. Nevertheless, introgressing these transgenes into an isa mutant background could be useful and help to establish the contribution of the debranching reaction to the structure and amount of glucan made in each case.
Finally, it should be pointed out that our data support the idea that the amount of BE activity relative to the other enzymes also influences the final amount and structure of the glucan. This is apparent in all three sets of transgenic plants, where differences in the amount and structure of the starch correlated with the amount of activity that we detected on our zymogram analyses and in quantitative BE assays. However, it is important to restate that comparisons of the absolute activity of the different BE types is problematic, because the nature of each assay (e.g. phosphorylase stimulation assay as done here or the amylose branching assay) is liable to favor one BE type over another. Nevertheless, it seems that the differences in glucan structure resulting from expression of each BE type were greater than the differences between plants with different levels of a given BE. Therefore, we conclude that manipulating BE types rather than amounts (combined with SS types) may represent the better approach to significantly alter the structure and properties of amylopectin in starch crops.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) Wassilewskija (WS) ecotype and its mutant be2be3, which lacks both BEs (Dumez et al., 2006), were used. Sown Arabidopsis seeds were stratified at 4°C for 2 to 3 d in the dark, and the plants were then grown in Percival Growth Chambers (CLF Plant Climatics) under a 12-h-light/12-h-dark regime. The temperature was 20°C, the relative humidity was 70%, and the light intensity was 150 mol quanta m−2 s−1. Maize (Zea mays) plants were grown in the greenhouse at between 20°C and 23°C, with a minimum 12-h photoperiod. Potato (Solanum tuberosum) tuber amylopectin for structural analysis was purchased from Sigma-Aldrich.
Production of Transgenic Lines
The full lengths of coding sequences of starch BE2a of maize (ZmBE2a) and starch BE1 of potato (StBE1) were separately amplified from complementary DNA using primers given in Supplemental Table S1 and cloned by recombinant cloning into the vector pH7WG2,0 (Invitrogen). The full-length Escherichia coli GLGB coding sequence was amplified from genomic DNA of strain DH5α and cloned by recombinant cloning into the vector pH7WGY2,0, which was modified to contain a chloroplast transit peptide (from the Arabidopsis Rubisco small subunit B) upstream of the YFP that precedes the LR recombinant cloning site (designated pH7WGY2-ctp). Resulting constructs are shown in Supplemental Figure S1A. Each construct was transformed into Arabidopsis be2be3 plants by Agrobacterium tumefaciens-mediated transformation (strain GV3101; Zhang et al., 2008). The expression of each of the recombinant genes was under the control of the cauliflower mosaic virus 35S promoter. Multiple transgenic T1 plants expressing ZmBE2a, StBE1, or EcGLGB were isolated by hygromycin resistance screening on 1% (w/v) Murashige and Skoog medium plates. Three lines of each were selected for further analysis. T2 plants carrying the transgenes were identified individually by PCR-based genotyping for hygromycin resistance at the time of harvesting using primers shown in Supplemental Table S1.
Starch and Water-Soluble Polysaccharide Extraction and Quantification
Whole rosettes from 30-d-old Arabidopsis plants or leaves from 45-d-old maize plants were harvested at the end of the day or the end of the night, weighed, and frozen in liquid N2. Extraction was done as described previously (Pfister et al., 2014). Briefly, plant material was homogenized in 1.12 m perchloric acid. The homogenate was subject to centrifugation (10 min at 3,000g) to separate the starch-containing insoluble fraction from the soluble fraction (containing sugars and soluble glucans). The insoluble fraction was washed one time with water and four times with 80% (v/v) ethanol, dried, resuspended in water, and stored at −20°C. The soluble fraction was adjusted to pH 6 with 2 m KOH, 0.4 m MES, and 0.4 m KCl. Precipitated KClO4 was removed by centrifugation (3,000g for 10 min). Phytoglycogen in the supernatant was precipitated at −20°C for 2 h by adding methanol (final concentration of 75% [v/v]). After centrifugation (3,000g for 10 min), the pellet was washed one time with 75% (v/v) methanol, dried, and resuspended in water. Glucans in the insoluble fraction, the soluble fraction, and the precipitated phytoglycogen fractions were quantified as described (Hostettler et al., 2011). Total nonprecipitatable MOSs were calculated by subtracting the amount of precipitated phytoglycogen and the free Glc from the glucan in the soluble fraction.
To analyze MOSs, the neutralized soluble fractions from the perchloric acid extraction (above) were mixed with cellobiose (5 nmol per 20 mg of original plant material) as an internal standard. Neutral sugars were eluted from sequential ion exchange columns of Dowex 50W and Dowex 1 (Sigma-Aldrich) and analyzed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) as described by Egli et al. (2010). Standard solutions containing known amounts of maltose (5 µm to 0.8 mm) and MOS DPs of three to seven (25 nm to 0.8 µm) were used to identify and quantify MOSs. Analyses were performed on four to six biological replicates, each comprising an individual plant.
To purify starch granules, whole rosettes from 35-d-old Arabidopsis plants were homogenized in 50 mm Tris-HCl, pH 8, 0.2 mm EDTA, and 0.5% (v/v) Triton X-100 using a Waring Blender. The homogenate was filtered through a nylon mesh (pore size of 100 µm), and the insoluble fraction was sedimented by centrifugation (2,500g for 15 min), then resuspended in the same medium, and sequentially filtered through nylon meshes with 30- and 15-µm pore sizes. Starch granules were separated from the filtrate at 2,500g for 25 min over a 95% (v/v) Percoll cushion (Sigma-Aldrich). The starch pellet was then washed in 0.5% (w/v) SDS. Excess SDS was removed by extensively washing the pellet in water. The pellet was then washed one time in 80% (v/v) ethanol and dried under vacuum overnight. A fraction of each starch granule preparation (0.5 mg) was stored in water without ethanol washing for analysis by SEM. Glycogen from E. coli was obtained as described previously (Sundberg et al., 2013).
Structural Analysis of Starch and Glycogen
For iodine staining, whole rosettes of 30-d-old plants were harvested at the end of the day, decolorized in hot 80% (v/v) ethanol, stained with Lugol’s Solution (Sigma-Aldrich), and briefly destained in water.
To obtain amylopectin and glycogen CLDs and β-CLDs, the insoluble fractions from the perchloric acid extraction (above) were prepared for and analyzed by HPAEC-PAD (Dionex) as described (Streb et al., 2008). In each case, starch from individual plants was analyzed. In the case of E. coli glycogen, the methanol-KCl precipitate was used as the starting material (Sundberg et al., 2013).
Analysis of Amylopectin and Amylose Content
Purified starch granules were analyzed for amylose content as described previously (Zeeman et al., 2002a, 2002b). Briefly, 3 to 5 mg of starch granules were dissolved in 0.1 mL of 0.5 m NaOH. The amylose and amylopectin fractions were separated over a Sepharose CL-2B Column (30 cm long, 10 mm i.d.) using 50 mm NaOH as the eluent (0.2-mL min−1 flow rate). Twenty 1-mL fractions containing the eluted glucans were collected. An aliquot of each fraction was mixed with the same volume of 50 mm HCl with 200 mm MES to neutralize it before mixing with 10× diluted Lugol’s Solution (Sigma-Aldrich). The A594 was recorded. To obtain the λmax values of amylopectin and amylose, aliquots of fractions of 2 to 6 and 7 to 17 were pooled, respectively. These pooled samples were neutralized and mixed with 10× diluted Lugol’s Solution, and the absorption spectrum between 400 and 800 nm was recorded. To obtain the CLD of isolated amylopectin, fractions 2 to 6 from the CL-2B columns were pooled and adjusted to pH 7.0 by the addition of 50 mm HCl. Amylopectin was precipitated for 2 h at 4°C with 75% (v/v) ethanol and collected by centrifugation (3,000g for 10 min). The amylopectin pellet was washed with 75% (v/v) ethanol and resuspended in water. A 200-µg amylopectin sample was dissolved by boiling, debranched, and analyzed as described above.
Electron Microscopy
For TEM, mature leaves from 32-d-old plants were cut with a razor blade. The leaf sections were fixed for 4 h at 4°C (2% [v/v] glutaraldehyde in 0.05 m sodium cacodylate buffer, pH 7.4) for 16 h, washed three times (0.1 m sodium cacodylate buffer, pH 7.4), and incubated for 16 h in 1% (w/v) osmium tetroxide (in 0.1 m sodium cacodylate buffer, pH 7.4) at 4°C. After three washes in ice cold 0.1 m sodium cacodylate buffer (pH 7.4) and one time with water, samples were dehydrated in a series of aqueous ethanol solutions from 50% (v/v) to 100% (v/v) ethanol and then 100% acetone. The leaf sections were then incubated for 2 h in 25% (v/v) epoxy resin (in acetone; Spurr’s; Agar Scientific), 2 h in 50% (v/v) epoxy resin, 16 h in 75% (v/v) epoxy resin, and 7 h in 100% (v/v) epoxy resin. Embedding was completed in fresh 100% epoxy resin by incubating at 70°C for 48 h. Ultrathin (70 µm) sections were cut with a diamond knife, placed on formvar carbon-coated copper grids, stained with 2% (w/v) uranyl acetate and Reynold’s lead citrate, and imaged with an FEI Morgagni 268 Electron Microscope. Pictures are representative for the analysis of sections from two individual plants per genotype.
For SEM, purified starch granules were coated with a 3-nm platinum layer using a Balzers MED010 Coating Device and visualized in a Leo 1530 Gemini (Zeiss) Microscope.
Analysis of Enzyme Activity by Native PAGE and BE Assays
Rosette material from three to five 30-d-old plants was harvested in the middle of a 12-h day, pooled, and frozen in liquid N2. Proteins were extracted using an all-glass homogenizer in 100 mm MOPS, pH 7.0, 1 mm EDTA, 5 mm dithiothreitol (DTT), and 10% (v/v) glycerol containing protease inhibitors (Complete EDTA-Free; Roche). To detect BE activity, extracts (50 µg of soluble protein) were separated on a 7.5% (w/v) native polyacrylamide gel containing 0.02% (w/v) oyster glycogen (Sigma-Aldrich) and run at 10 V cm−1 for 3 h at 4°C. The gels were washed for 30 min at 4°C in 50 mm HEPES-NaOH (pH 7.0) and 10% (v/v) glycerol and then incubated for 16 h at 20°C with gentle shaking in 50 mm HEPES-NaOH, 10% (v/v) glycerol, 2.5 mm AMP (Sigma-Aldrich), 50 mm Glc-1-P, and 28 units (per gel) phosphorylase-a (from rabbit muscle; Sigma-Aldrich). The BE activity bands were visualized by staining with Lugol’s Solution. To detect SS activities, extracts (50 µg of soluble protein) were separated as above in gels containing 0.3% (w/v) oyster glycogen. The gel was incubated for 16 h in a solution containing 100 mm HEPES-NaOH (pH 7.5), 2 mm DTT, 10% (v/v) glycerol, 0.5 mm EDTA, 0.5 m trisodium citrate, and 0.8 mm ADP-Glc. The SS activity bands were revealed by Lugol’s staining.
Quantitative assays for BE activity were done using the phosphorylase-a stimulation method. Soluble proteins were extracted in 100 mm MOPS (pH 7.0), 1 mm DTT, and 10% (v/v) glycerol containing protease inhibitors. For each assay, soluble proteins (10 µg) were added to a 300-µL reaction mixture containing 200 mm MES (pH 6.6), 50 mm Glc-1-P (including 3.38 kBq of [U-14C]Glc-1-P), and 1 mm AMP. The reactions were started by adding 2 units of phosphorylase-a and incubated at 25°C. Aliquots (50 μL) were taken at intervals of 0, 30, 60, 90, 120, and 180 min. Each was boiled for 2 min, mixed with 50 μL of amylopectin (10 mg mL−1), and adjusted to 75% (v/v) methanol and 1% (w/v) KCl. Glc polymers were precipitated for 3 h at 4°C and then collected by centrifugation (2,000g for 5 min). The pellets were washed with 75% (v/v) methanol and 1% (w/v) KCl, dried, and resuspended in water. The radioactivity incorporated in to the glucan was measured by liquid scintillation counting. The activity of BE was calculated from the changes in incorporation observed between 60 and 180 min.
Immunoblotting to Detect the GBSS Protein
Rosette leaves (300 mg) were harvested in the middle of day and homogenized in 1 mL of extraction medium containing 50 mm Tris-HCl (pH 7.0), 5 mm DTT, 2% (w/v) SDS, 10% (v/v) glycerol, and protease inhibitor (Complete EDTA-Free; Roche). A sample of the homogenate was mixed with an equal volume of SDS loading solution (50 mm Tris-HCl, 200 mm DTT, 4% [w/v] SDS, 10% [v/v] glycerol, and 0.1% [w/v] bromophenol blue) and boiled 10 min to release proteins from starch. As positive and negative controls, 1 mg of purified starch from the wild type (WS) and a mutant lacking GBSS (GBSS and STARCH EXCESS4-deficient mutant), respectively, were used. These were dissolved in 1 mL of extraction solution and boiled. After centrifugation (5 min at 20,000g), 25 μL of supernatant from each line was loaded in a 10% (w/v) acrylamide SDS-PAGE gel. The immunoblotting was as described in Seung et al., 2015.
Arabidopsis Genome Initiative gene codes for the Arabidopsis genes discussed in this study are as follows: BE2, At5g03650; and BE3, At2g36390. Accession numbers for the coding sequences of the BEs from other species are EG10378 (EcoGene accession no.; E. coli glycogen BE), U65948 (GRMZM2G073054; maize type II BE), and Y08786 (Sotub07g025820; potato type I BE).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Constructs used for the expression of different BE types in the Arabidopsis be2be3 double mutant and the correct localization of E. coli GLGB.
Supplemental Figure S2. MOS composition from be2be3 mutants expressing different BE types.
Supplemental Figure S3. Native PAGE (Zymograms) of wild-type Arabidopsis (WS) and be2be3 mutants to detect BE activities.
Supplemental Figure S4. Native PAGE (Zymograms) of wild-type Arabidopsis (WS) and be2be3 mutants expressing different BE types to detect SS activities.
Supplemental Figure S5. CLD of amylopectin from wild-type Arabidopsis (WS) and be2be3 mutants expressing different BE types.
Supplemental Figure S6. GBSS abundance in total protein extracts of wild-type Arabidopsis (WS) and be2be3 mutants expressing different BE types.
Supplemental Figure S7. Various granule morphology in line ZmBE2a-3 and different EcGLGB-expressing lines.
Supplemental Table S1. Primer sequences used for amplification of full-length complementary DNA of ZmBE2a, StBE1, and EcGLGB for genotyping the Arabidopsis be2 and be3 mutations and genotyping with respect to the hygromycin resistance gene associated with recombinant BE expression.
Acknowledgments
We thank Andrea Ruckle and David Seung (ETH Zurich) for assistance with plant cultivation and for technical assistance with separation of amylopectin and amylose, respectively; and Christophe D'Hulst (Université des Sciences et Technologies de Lille, France) for the seeds of the BE mutants.
Glossary
- β-CLD
chain length distribution of the β-limit
- CLD
chain length distribution
- DP
degree of polymerization
- DTT
dithiothreitol
- GPC
gel-permeation chromatography
- HPAEC-PAD
high-performance anion-exchange chromatography with pulsed amperometric detection
- λmax
wavelength of maximal absorption
- MOS
malto-oligosaccharide
- SEM
scanning electron microscopy
- TEM
transmission electron microscopy
- WS
Wassilewskija
Footnotes
This work was supported by the Swiss National Science Foundation (grant no. 31003A–153144/1) and ETH Zurich.
K.-J.L. performed most of the experiments; S.S. generated the transgenic lines; F.M. and B.P. assisted with the experimental works; S.C.Z. supervised the work and completed the writing.
Articles can be viewed without a subscription.
References
- Bhattacharyya MK, Smith AM, Ellis THN, Hedley C, Martin C (1990) The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell 60: 115–122 [DOI] [PubMed] [Google Scholar]
- Blauth SL, Kim KN, Klucinec J, Shannon JC, Thompson D, Guiltinan M (2002) Identification of Mutator insertional mutants of starch-branching enzyme 1 (sbe1) in Zea mays L. Plant Mol Biol 48: 287–297 [DOI] [PubMed] [Google Scholar]
- Blauth SL, Yao Y, Klucinec JD, Shannon JC, Thompson DB, Guilitinan MJ (2001) Identification of Mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiol 125: 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer CD, Garwood DL, Shannon JC (1976) Interaction of amylose-extender and waxy mutants of maize (Zea Mays L) – fine structure of amylose-extender waxy starch. Stärke 28: 405–410 [Google Scholar]
- Brummell DA, Watson LM, Zhou J, McKenzie MJ, Hallett IC, Simmons L, Carpenter M, Timmerman-Vaughan GM (2015) Overexpression of STARCH BRANCHING ENZYME II increases short-chain branching of amylopectin and alters the physicochemical properties of starch from potato tuber. BMC Biotechnol 15: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carciofi M, Blennow A, Jensen SL, Shaik SS, Henriksen A, Buléon A, Holm PB, Hebelstrup KH (2012) Concerted suppression of all starch branching enzyme genes in barley produces amylose-only starch granules. BMC Plant Biol 12: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia T, Thorneycroft D, Chapple A, Messerli G, Chen J, Zeeman SC, Smith SM, Smith AM (2004) A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J 37: 853–863 [DOI] [PubMed] [Google Scholar]
- Crumpton-Taylor M, Pike M, Lu KJ, Hylton CM, Feil R, Eicke S, Lunn JE, Zeeman SC, Smith AM (2013) Starch synthase 4 is essential for coordination of starch granule formation with chloroplast division during Arabidopsis leaf expansion. New Phytol 200: 1064–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte T, Trevisan M, Parker ML, Zeeman SC (2005) Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J 41: 815–830 [DOI] [PubMed] [Google Scholar]
- Delatte T, Umhang M, Trevisan M, Eicke S, Thorneycroft D, Smith SM, Zeeman SC (2006) Evidence for distinct mechanisms of starch granule breakdown in plants. J Biol Chem 281: 12050–12059 [DOI] [PubMed] [Google Scholar]
- Delvallé D, Dumez S, Wattebled F, Roldán I, Planchot V, Berbezy P, Colonna P, Vyas D, Chatterjee M, Ball S, et al. (2005) Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J 43: 398–412 [DOI] [PubMed] [Google Scholar]
- Denyer K, Waite D, Motawia S, Møller BL, Smith AM (1999) Granule-bound starch synthase I in isolated starch granules elongates malto-oligosaccharides processively. Biochem J 340: 183–191 [PMC free article] [PubMed] [Google Scholar]
- Dumez S, Wattebled F, Dauvillee D, Delvalle D, Planchot V, Ball SG, D’Hulst C (2006) Mutants of Arabidopsis lacking starch branching enzyme II substitute plastidial starch synthesis by cytoplasmic maltose accumulation. Plant Cell 18: 2694–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards A, Fulton DC, Hylton CM, Jobling SA, Gidley M, Rössner U, Martin C, Smith AM (1999) A combined reduction in activity of starch synthases II and III of potato has novel effects on the starch of tubers. Plant J 17: 251–261 [Google Scholar]
- Egli B, Kölling K, Köhler C, Zeeman SC, Streb S (2010) Loss of cytosolic phosphoglucomutase compromises gametophyte development in Arabidopsis. Plant Physiol 154: 1659–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facon M, Lin Q, Azzaz AM, Hennen-Bierwagen TA, Myers AM, Putaux JL, Roussel X, D’Hulst C, Wattebled F (2013) Distinct functional properties of isoamylase-type starch debranching enzymes in monocot and dicot leaves. Plant Physiol 163: 1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y (2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park JH, Jane JL, et al. (2007) Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol 144: 2009–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, Gil M, Reinhold H, Eicke S, Messerli G, et al. (2008) Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Kuriki T, Sivak M, Preiss J (1995) Maize branching enzyme catalyzes synthesis of glycogen-like polysaccharide in glgB-deficient Escherichia coli. Proc Natl Acad Sci USA 92: 964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Li P, Imparl-Radosevich J, Preiss J, Keeling P (1997) Comparing the properties of Escherichia coli branching enzyme and maize branching enzyme. Arch Biochem Biophys 342: 92–98 [DOI] [PubMed] [Google Scholar]
- Guan HP, Preiss J (1993) Differentiation of the properties of the branching isozymes from maize (Zea Mays). Plant Physiol 102: 1269–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Sun FJ, Rosales-Mendoza S, Korban SS (2007) Three orthologs in rice, Arabidopsis, and Populus encoding starch branching enzymes (SBEs) are different from other SBE gene families in plants. Gene 401: 123–130 [DOI] [PubMed] [Google Scholar]
- Hostettler C, Kölling K, Santelia D, Streb S, Kötting O, Zeeman SC (2011) Analysis of starch metabolism in chloroplasts. Methods Mol Biol 775: 387–410 [DOI] [PubMed] [Google Scholar]
- James MG, Robertson DS, Myers AM (1995) Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling SA, Schwall GP, Westcott RJ, Sidebottom CM, Debet M, Gidley MJ, Jeffcoat R, Safford R (1999) A minor form of starch branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: cloning and characterisation of multiple forms of SBE A. Plant J 18: 163–171 [DOI] [PubMed] [Google Scholar]
- Kim WS, Kim J, Krishnan HB, Nahm BH (2005) Expression of Escherichia coli branching enzyme in caryopses of transgenic rice results in amylopectin with an increased degree of branching. Planta 220: 689–695 [DOI] [PubMed] [Google Scholar]
- Kortstee AJ, Suurs LCJM, Vermeesch AMG, Keetels CJAM, Jacobsen E, Visser RGF (1998) The influence of an increased degree of branching on the physicochemical properties of starch from genetically modified potato. Carbohydr Polym 37: 173–184 [Google Scholar]
- Kortstee AJ, Vermeesch AMS, de Vries BJ, Jacobsen E, Visser RGF (1996) Expression of Escherichia coli branching enzyme in tubers of amylose-free transgenic potato leads to an increased branching degree of the amylopectin. Plant J 10: 83–90 [DOI] [PubMed] [Google Scholar]
- Lu Y, Sharkey TD (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218: 466–473 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Kawasaki T, Shimada H, Satoh H, Kobayashi E, Okumura S, Arai Y, Baba T (1993) Alteration of the structural properties of starch components by the lack of an isoform of starch branching enzyme in rice seeds. J Biol Chem 268: 19084–19091 [PubMed] [Google Scholar]
- Mouille G, Maddelein ML, Libessart N, Talaga P, Decq A, Delrue B, Ball S (1996) Preamylopectin processing: a mandatory step for starch biosynthesis in plants. Plant Cell 8: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG (2000) Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol 122: 989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Umemoto T, Takahata Y, Komae K, Amano E, Satoh H (1996) Changes in structure of starch and enzyme activities affected by sugary mutations in developing rice endosperm: possible role of starch debranching enzyme (R-enzyme) in amylopectin biosynthesis. Physiol Plant 97: 491–498 [Google Scholar]
- Nakamura Y, Utsumi Y, Sawada T, Aihara S, Utsumi C, Yoshida M, Kitamura S (2010) Characterization of the reactions of starch branching enzymes from rice endosperm. Plant Cell Physiol 51: 776–794 [DOI] [PubMed] [Google Scholar]
- Niittylä T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89 [DOI] [PubMed] [Google Scholar]
- Nougué O, Corbi J, Ball SG, Manicacci D, Tenaillon MI (2014) Molecular evolution accompanying functional divergence of duplicated genes along the plant starch biosynthesis pathway. BMC Evol Biol 14: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan AC, Perez S (1999) The relationship between internal chain length of amylopectin and crystallinity in starch. Biopolymers 50: 381–390 [DOI] [PubMed] [Google Scholar]
- Pérez S, Bertoft E (2010) The molecular structures of starch components and their contribution to the architecture of starch granules: a comprehensive review. Stärke 62: 389–420 [Google Scholar]
- Pfister B, Lu KJ, Eicke S, Feil R, Lunn JE, Streb S, Zeeman SC (2014) Genetic evidence that chain length and branch point distributions are linked determinants of starch granule formation in Arabidopsis. Plant Physiol 165: 1457–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li Z, Rahman S, Morell M (2006) High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc Natl Acad Sci USA 103: 3546–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A, Kosar-Hashemi B, Ling S, Li Z, Rahman S, Morell M (2010) Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb. J Exp Bot 61: 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán I, Wattebled F, Mercedes Lucas M, Delvallé D, Planchot V, Jiménez S, Pérez R, Ball S, D’Hulst C, Mérida A (2007) The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J 49: 492–504 [DOI] [PubMed] [Google Scholar]
- Rydberg U, Andersson L, Andersson R, Aman P, Larsson H (2001) Comparison of starch branching enzyme I and II from potato. Eur J Biochem 268: 6140–6145 [DOI] [PubMed] [Google Scholar]
- Safford R, Jobling SA, Sidebottom CM, Westcott RJ, Cooke D, Tober KJ, Strongitharm BH, Russell AL, Gidley MJ (1998) Consequences of antisense RNA inhibition of starch branching enzyme activity on properties of potato starch. Carbohydr Polym 35: 155–168 [Google Scholar]
- Satoh H, Nishi A, Yamashita K, Takemoto Y, Tanaka Y, Hosaka Y, Sakurai A, Fujita N, Nakamura Y (2003) Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol 133: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada T, Francisco PB Jr, Aihara S, Utsumi Y, Yoshida M, Oyama Y, Tsuzuki M, Satoh H, Nakamura Y (2009) Chlorella starch branching enzyme II (BEII) can complement the function of BEIIb in rice endosperm. Plant Cell Physiol 50: 1062–1074 [DOI] [PubMed] [Google Scholar]
- Sawada T, Nakamura Y, Ohdan T, Saitoh A, Francisco PB Jr, Suzuki E, Fujita N, Shimonaga T, Fujiwara S, Tsuzuki M, et al. (2014) Diversity of reaction characteristics of glucan branching enzymes and the fine structure of α-glucan from various sources. Arch Biochem Biophys 562: 9–21 [DOI] [PubMed] [Google Scholar]
- Schwall GP, Safford R, Westcott RJ, Jeffcoat R, Tayal A, Shi YC, Gidley MJ, Jobling SA (2000) Production of very-high-amylose potato starch by inhibition of SBE A and B. Nat Biotechnol 18: 551–554 [DOI] [PubMed] [Google Scholar]
- Seo BS, Kim S, Scott MP, Singletary GW, Wong KS, James MG, Myers AM (2002) Functional interactions between heterologously expressed starch-branching enzymes of maize and the glycogen synthases of Brewer’s yeast. Plant Physiol 128: 1189–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D, Soyk S, Coiro M, Maier BA, Eicke S, Zeeman SC (2015) PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol 13: e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker CK, Boyer CD, Wiesenborn DP, Thompson DB, Boersig MR, Oakes JV, Stalker DM (1994) Expression of Escherichia coli glycogen synthase in the tubers of transgenic potatoes (Solanum tuberosum) results in a highly branched starch. Plant Physiol 104: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinard PS, Robertson DS, Schnable PS (1993) Genetic isolation, cloning, and analysis of a Mutator-induced, dominant antimorph of the maize amylose extender1 locus. Plant Cell 5: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Delatte T, Umhang M, Eicke S, Schorderet M, Reinhardt D, Zeeman SC (2008) Starch granule biosynthesis in Arabidopsis is abolished by removal of all debranching enzymes but restored by the subsequent removal of an endoamylase. Plant Cell 20: 3448–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streb S, Zeeman SC (2014) Replacement of the endogenous starch debranching enzymes ISA1 and ISA2 of Arabidopsis with the rice orthologs reveals a degree of functional conservation during starch synthesis. PLoS One 9: e92174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg M, Pfister B, Fulton D, Bischof S, Delatte T, Eicke S, Stettler M, Smith SM, Streb S, Zeeman SC (2013) The heteromultimeric debranching enzyme involved in starch synthesis in Arabidopsis requires both isoamylase1 and isoamylase2 subunits for complex stability and activity. PLoS One 8: e75223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Fujita N, Nishi A, Satoh H, Hosaka Y, Ugaki M, Kawasaki S, Nakamura Y (2004) The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol J 2: 507–516 [DOI] [PubMed] [Google Scholar]
- Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y (2002) Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor Appl Genet 104: 1–8 [DOI] [PubMed] [Google Scholar]
- Wang X, Xue L, Sun J, Zuo J (2010) The Arabidopsis BE1 gene, encoding a putative glycoside hydrolase localized in plastids, plays crucial roles during embryogenesis and carbohydrate metabolism. J Integr Plant Biol 52: 273–288 [DOI] [PubMed] [Google Scholar]
- Wattebled F, Dong Y, Dumez S, Delvallé D, Planchot V, Berbezy P, Vyas D, Colonna P, Chatterjee M, Ball S, et al. (2005) Mutants of Arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol 138: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise SE, Weber AP, Sharkey TD (2004) Maltose is the major form of carbon exported from the chloroplast at night. Planta 218: 474–482 [DOI] [PubMed] [Google Scholar]
- Xia H, Yandeau-Nelson M, Thompson DB, Guiltinan MJ (2011) Deficiency of maize starch-branching enzyme I results in altered starch fine structure, decreased digestibility and reduced coleoptile growth during germination. BMC Plant Biol 11: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandeau-Nelson MD, Laurens L, Shi Z, Xia H, Smith AM, Guiltinan MJ (2011) Starch-branching enzyme IIa is required for proper diurnal cycling of starch in leaves of maize. Plant Physiol 156: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM (2010) Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 61: 209–234 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM (2002a) The priming of amylose synthesis in Arabidopsis leaves. Plant Physiol 128: 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Tiessen A, Pilling E, Kato KL, Donald AM, Smith AM (2002b) Starch synthesis in Arabidopsis: granule synthesis, composition, and structure. Plant Physiol 129: 516–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Umemoto T, Lue WL, Au-Yeung P, Martin C, Smith AM, Chen J (1998) A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10: 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Szydlowski N, Delvallé D, D’Hulst C, James MG, Myers AM (2008) Overlapping functions of the starch synthases SSII and SSIII in amylopectin biosynthesis in Arabidopsis. BMC Plant Biol 8: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]