The synthesis of pharmaceutically active acylphloroglucinols during strawberry fruit ripening is catalyzed by dual-function chalcone synthases/valerophenone synthases.
Abstract
Phenolics have health-promoting properties and are a major group of metabolites in fruit crops. Through reverse genetic analysis of the functions of four ripening-related genes in the octoploid strawberry (Fragaria × ananassa), we discovered four acylphloroglucinol (APG)-glucosides as native Fragaria spp. fruit metabolites whose levels were differently regulated in the transgenic fruits. The biosynthesis of the APG aglycones was investigated by examination of the enzymatic properties of three recombinant Fragaria vesca chalcone synthase (FvCHS) proteins. CHS is involved in anthocyanin biosynthesis during ripening. The F. vesca enzymes readily catalyzed the condensation of two intermediates in branched-chain amino acid metabolism, isovaleryl-Coenzyme A (CoA) and isobutyryl-CoA, with three molecules of malonyl-CoA to form phlorisovalerophenone and phlorisobutyrophenone, respectively, and formed naringenin chalcone when 4-coumaroyl-CoA was used as starter molecule. Isovaleryl-CoA was the preferred starter substrate of FvCHS2-1. Suppression of CHS activity in both transient and stable CHS-silenced fruit resulted in a substantial decrease of APG glucosides and anthocyanins and enhanced levels of volatiles derived from branched-chain amino acids. The proposed APG pathway was confirmed by feeding isotopically labeled amino acids. Thus, Fragaria spp. plants have the capacity to synthesize pharmaceutically important APGs using dual functional CHS/(phloriso)valerophenone synthases that are expressed during fruit ripening. Duplication and adaptive evolution of CHS is the most probable scenario and might be generally applicable to other plants. The results highlight that important promiscuous gene function may be missed when annotation relies solely on in silico analysis.
Phenolic compounds constitute one of the most numerous and ubiquitous groups of plant secondary metabolites, and they have attracted much attention due to their reputed beneficial effects on human health protection (Scalbert et al., 2005; Maher et al., 2006; Saito and Matsuda, 2010; De Luca et al., 2012). Garden strawberry (Fragaria × ananassa) fruits are consumed in high quantities worldwide and have health-promoting potential, as they are a valuable source of phenolics such as flavonoids, anthocyanins, flavonols, and flavan-3-ols, including proanthocyanidins, phenolic acids, and ellagitannins (Seeram et al., 2006). These compounds play several important functions in plants, as the red-colored anthocyanins may attract frugivores that help to disperse seeds, flavonols act as protective UV-B-absorbing chemicals in fruit skin, and proanthocyanidins contribute to defense and stress resistance (Cheynier et al., 2013). The Glc esters of phenylpropanoic acids serve as energy-rich substrates in plant secondary metabolism, and ellagic acid may play a role in protection from predation and plant growth regulation (Cheynier et al., 2013). It is now generally accepted that phytochemicals are also responsible for the health-promoting effect of Fragaria spp. fruits (Hannum, 2004).
Phenolic compounds originate from the shikimate, phenylpropanoid, flavonoid, and the lignin pathways (Vogt, 2010). In most plants, the biosynthesis of the phenolics starts with 4-coumaric acid formation from the primary metabolite Phe. Genes and enzymes of the basic biosynthetic pathway leading to anthocyanins are known (Ververidis et al., 2007), and remarkable progress has been made in understanding the regulation of this pathway (Allan et al., 2008), although the regulation of their accumulation and flux through the pathway is not that well established. In a recent study, an examination of the transcriptome of different Fragaria spp. fruit genotypes coupled with targeted metabolite profiling was undertaken to reveal genes whose expression levels correlate with an altered composition of phenolics (Ring et al., 2013). This led to the identification of candidate genes that might control accumulation of phenolic compounds in Fragaria spp. fruit.
This work was undertaken to confirm the relationship between the expression pattern of the candidate genes and the accumulation of phenolics using reverse genetics approaches and to structurally identify unique metabolites whose levels are affected by the transcript levels of the candidate genes. Biologically active acylphloroglucinol (APG) glucosides, which have been only detected in a limited number of plants (Bohr et al., 2005; Crispin et al., 2013), were unambiguously uncovered as natural metabolites of Fragaria spp. fruit. In hops (Humulus lupulus), the APG aglycones, phlorisovalerophenone (PIVP), and phlorisobutyrophenone (PIBP) are generated by the gene product of valerophenone synthase (VPS; Paniego et al., 1999; Okada et al., 2004). Because this gene has not been annotated in the genome of the diploid woodland strawberry Fragaria vesca (Shulaev et al., 2011), we needed to rationalize the biosynthesis of PIVP and PIBP in Fragaria spp. fruit. Based on the basic catalytic mechanisms of VPS and chalcone synthase (CHS) and untargeted metabolite profiling analysis, we hypothesized that a CHS may have dual functionality and also act as VPS in Fragaria spp. fruit. Thus, detailed enzymatic characterization of three recombinant CHS enzymes was performed. Their dual CHS/VPS function was confirmed by activity assays and by suppression of CHS catalytic activity in transient CHS-silenced Fragaria spp. fruit and stable antisense chs transgenic lines, as well as by tracer experiments using isotopically labeled precursor amino acids.
RESULTS
Selection of Candidate Genes and Gain- or Loss-of-Function Phenotypes
The relative levels of mRNA of 13 candidate genes that might affect the accumulation of flavonoids and anthocyanins in Fragaria spp. fruit during ripening (Ring et al., 2013) were determined to select genes that show a ripening-related expression pattern (Supplemental Fig. S1; Supplemental Table S1). Gene21343 (expansin-A8-like) transcripts are highly abundant in the ripe fruit, and the level strongly increases during ripening. Gene33865 (ephrin-A1-like) mRNA is found at low levels in the different tissues, but the relative transcript level increases during fruit development. By contrast, transcript levels of the gene10776 (senescence-related gene1 [SRG1]-like) are at the highest in vegetative tissue, and the level decreases with the development of the fruit. Similarly, gene00897 (defensin-like) mRNA levels are highest in stem tissue, but in fruit, the level falls during ripening. These genes were chosen for further analyses.
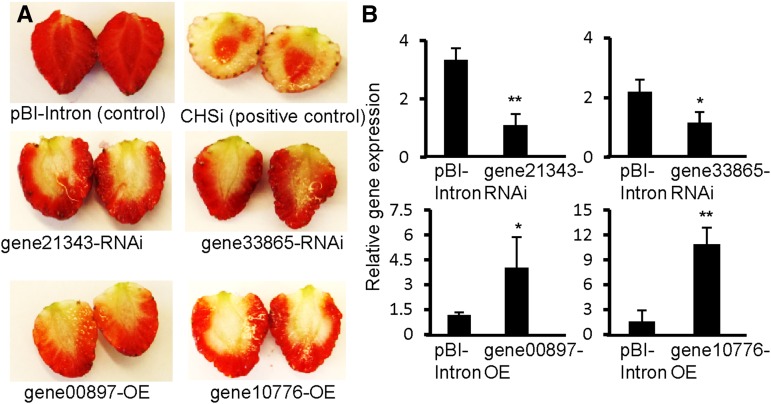
To determine whether the transcription of the four genes affects the accumulation of phenolic compounds in Fragaria spp. fruit, gain- and loss-of-function phenotypes were generated by transient overexpression or silencing of the candidate genes by agroinfiltration (Supplemental Fig. S2). A previously reported chalcone synthase gene from strawberry CHS (Lunkenbein et al., 2006), whose product catalyzes one of the first steps in the flavonoid pathway, was chosen as a positive reporter gene (Vogt, 2010). Down-regulation of CHS transcript levels rapidly leads to lower levels of pigmentation in Fragaria spp. fruit and is thus easy to detect (Fig. 1; Hoffmann et al., 2006). Transcript levels of the gene21343 and gene33865 were successfully down-regulated and resulted in loss-of-function phenotypes, as exhibited by white regions (Fig. 1), a clear sign of impaired anthocyanin accumulation. On the other hand, up-regulation of gene10776 and gene00897 mRNA levels produced gain-of-function phenotypes with white regions, indicative that gene10776 and gene00897 might be negative regulators of the anthocyanin pathway.
Figure 1.
Fruit phenotypes and relative mRNA levels of candidate genes in the transgenic fruits. Fruit phenotypes (A) and relative mRNA levels of candidate genes (B) in agroinfiltrated fruits. Infiltration with pBI-Intron alone resulted in no color change. Fruits agroinfiltrated with pBI-CHSi served as positive control. Pigmentation was decreased in fruits agroinfiltrated with gene-silencing (RNAi) and overexpression (OE) constructs of defined genes. Transcript levels decreased in gene21343- and gene33865-RNAi agroinfiltrated fruit, and transcript levels of gene00897 and gene10776 increased in fruits agroinfiltrated with overexpression constructs relative to pBI intron controls. Data represent means ± se for triplicate technical repetitions from five cDNA preparations. Asterisks indicate significant differences (*P = 0.05 and **P = 0.01).
Targeted and Untargeted Metabolite Profiling Analysis
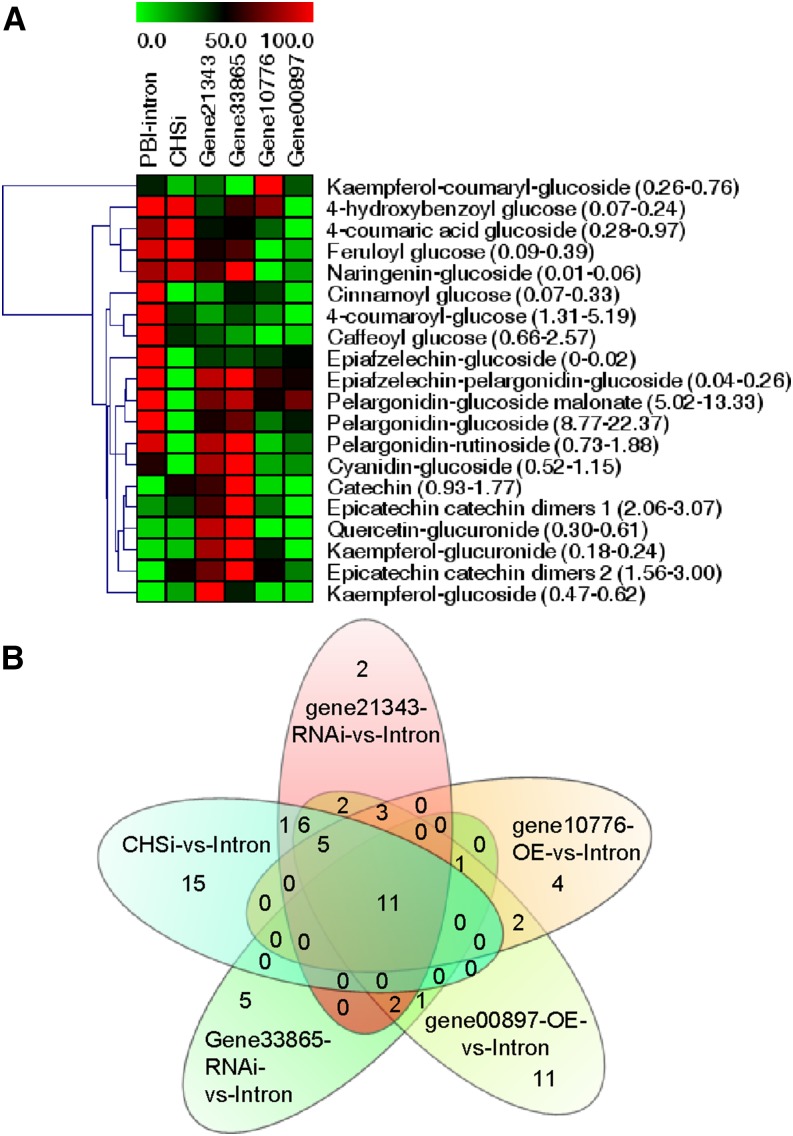
To understand how metabolism is altered when mRNA levels are up- or down-regulated we compared the metabolite profiles of the transgenic fruits with those of the control fruits. Targeted analysis showed that the levels of anthocyanins (pelargonidin and cyanidin derivatives), flavonoids (naringenin, kaempferol, quercetin, afzelechin, and [epi]catechin derivatives), and phenylpropanoids (cinnamoyl, 4-coumaroyl, caffeoyl, and feruloyl Glc) were differently affected in the transgenic fruit compared with the control, pBI-intron (Fig. 2). Consistent with the gain- and loss-of-function phenotypes (Fig. 1), it is evident that all four selected candidate genes are involved in the accumulation of phenolics in Fragaria spp. fruit, as changes in their transcript levels alter the pool sizes of metabolites in the different branches of the phenolics pathway. In addition to pelargonidin glucoside, the major pigment in Fragaria spp. fruit, the levels of cinnamoyl Glc, 4-coumaroyl Glc, caffeoyl Glc, and epiafzelechin glucoside were significantly reduced in all transgenic fruits.
Figure 2.
Targeted metabolite analysis and Venn diagram. Targeted metabolite analysis (A) and Venn diagram (B) showing the results of the second order analysis of untargeted LC-MS data. Metabolites were extracted from Fragaria spp. fruits in which the candidate genes were differentially expressed due to agroinfiltration of RNAi and overexpression (OE) constructs, using chalcone synthase RNAi constructs (CHSi) as positive control. Heat map also shows the range of concentration of the individual metabolites in parenthesis. Second order (meta-)analysis was performed according to Tautenhahn et al. (2011).
Pairwise untargeted comparisons provide physiologically relevant data but often result in hundreds of differences (Patti et al., 2012). To facilitate the extraction of interesting metabolites from our large untargeted liquid chromatography (LC)-mass spectrometry (MS) data sets, we performed second order (meta-)analysis using metaXCMS (Tautenhahn et al., 2011; Gowda et al., 2014). In this investigation, second order comparison was applied using a tolerance of 0.01 mass-to-charge ratio (m/z) and 60-s retention time to find metabolite features that were down- or up-regulated in the transgenic fruits. In the negative MS ion model, CHS-silenced transgenic plants showed the most unique metabolite features (15 unique metabolites whose levels were not affected in the other transgenic fruits), followed by the gene00897-, gene33865-, gene10776-, and gene21343-silenced transgenic plants, which showed 11, five, four, and two unique metabolites in this analysis, respectively (Fig. 2). Eleven features were found to be differentially accumulating in all five samples. Of these, nine metabolites could be confirmed manually and assigned to LC-MS signals (Supplemental Table S2). All confirmed metabolites were previously unknown to occur in Fragaria spp. The structures of these confirmed metabolites were unknown, so they were isolated and their chemical structures were determined.
APGs Are Unique Fragaria spp. Metabolites That Differentially Accumulate in Transgenic Plants
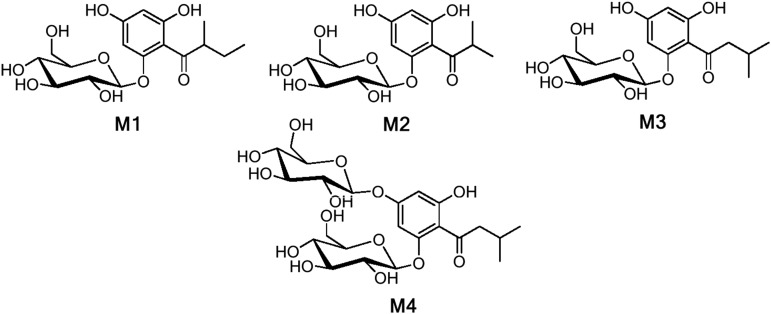
Metabolites M1 and M3 are unique to CHS-RNA interference (CHSi) plants, while M2 is differentially accumulated in all transgenic plants compared with the control (Supplemental Fig. S3; Supplemental Tables S2 and S3). M1 was identified as 1-[(2-methylbutyryl)-phloroglucinyl]-2-O-β-d-glucopyranoside by comparison of its 1H-NMR, heteronuclear multiple quantum coherence (HMQC), and heteronuclear multiple-bond correlation (HMBC) data (Supplemental Table S4) with those of known phloroglucinol glucosides (Kosasi et al., 1989; Tsukamoto et al., 2004; Bohr et al., 2005). The 1H-NMR spectrum shows one methyl doublet at δ 1.15 ppm, d (3H), one methyl triplet at δ 0.90 ppm, t (3H), a methine signal at δ 3.93 ppm, m (H-2′), and a geminally coupled methylene spin system (δ 1.81 ppm, m, H-3′), which indicates a 2-methylbutyryl moiety (Kosasi et al., 1989). In addition, the 1H-NMR spectrum reveals two meta-coupled aromatic doublets (δ 5.98 ppm and δ 6.20 ppm, each 1H). The HMQC data suggests that these hydrogens are connected to carbons at δ 99.3 ppm (C-4) and δ 96.4 ppm (C-6), indicating an asymmetric substituted phloroglucinol moiety. Therefore, the sugar residue must be attached to C-1 of the phloroglucinol, and this is supported by the upfield resonances of C-4 and C-6 (Supplemental Table S4). The sugar moiety was identified by comparison of its 1H-NMR, HMQC, and HMBC data with those of known phloroglucinol glucosides (Kosasi et al., 1989; Tsukamoto et al., 2004; Bohr et al., 2005).
The 1H-NMR spectrum of M3 (Supplemental Table S5) differed only in the signals of the acyl side chain from M1, supporting the structure of 1-[(3-methylbutyryl)-phloroglucinyl]-2-O-β-d-glucopyranoside; Kosasi et al., 1989; Gao et al., 2004; Bohr et al., 2005). Similarly, M2 was identified as 1-[(2-methylpropanoyl)-phloroglucinyl]-2-O-β-d-glucopyranoside (Okada et al., 2004; Bohr et al., 2005) by comparison of its LC-MS and high-resolution MS data with those of authentic reference material (Intelmann et al., 2011). Metabolites of strawberry ‘Senga Sengana’ were also analyzed to identify additional phloroglucinol derivatives in Fragaria spp. fruit, revealing the presence of 1-[(3-methylbutyryl)-phloroglucinyl]-2,4-di-O-β-d-glucopyranoside, denoted as metabolite M4 (m/z 533 [M-H]–, Ms2 m/z 323; Fig. 3). M4 was identified by comparing its retention time and mass spectra (Ms and Ms2) and high-resolution mass spectra with those of the authentic compound (Intelmann et al., 2011).
Figure 3.
Structures of APGs identified in strawberry fruit. APGs were identified in strawberry ‘Elsanta,’ ‘Senga Sengana,’ ‘Mara des Bois,’ and ‘Calypso’ fruit. M1, 1-[(2-Methylbutyryl)-phloroglucinyl]-2-O-β-d-glucopyranoside; M2, 1-[(2-methylpropanoyl)-phloroglucinyl]-2-O-β-d-glucopyranoside; M3, 1-[(3-methylbutyryl)-phloroglucinyl]-2-O-β-d-glucopyranoside; M4, 1-[(3-methylbutyryl)-phloroglucinyl]-2,4-di-O-β-d-glucopyranoside.
Except for M3, which was found as a cytochrome P450 inhibitor (Tsukamoto et al., 2004), APGs have not been reported from Fragaria spp. fruit. APGs show various pharmacological activities in vitro and in vivo and have been found in only a limited number of plants (Suksamrarn et al., 1997; Zhang et al., 2002). The altered concentrations of APGs in response to changed expression levels of the candidate genes provide further evidence that these four genes, along with CHS, are involved in the biosynthesis of phenolic compounds. Metabolites M1 to M4 were detected in fruit of strawberry ‘Elsanta’ and ‘Senga Sengana’ and M1 to M3 in strawberry ‘Mara des Bois’ and ‘Calypso.’ In comparison, the white-fruited diploid F. vesca var HI 4 (HW4) genotype, which lacks anthocyanin pigments, also lacks the APG metabolites (Table I).
Table I. Relative concentration of APG glucosides in relation to the internal standard (Biochanin A) in Fragaria spp. fruit of different varieties.
cv Elsanta, Senga Sengana, Mara des Bois, and Calypso are cultivars of the strawberry hybrid, whereas HW4 (HI 4) is a white-fruited variety of diploid F. vesca. Relative concentration is expressed in milligram equivalent g–1 lyophilized Fragaria spp. fruit powder (mean ± sd). N.D., Not detected by LC-MS.
| Compound | Elsanta (Red) | Senga Sengana (Red) | Mara des Bois (Red) | Calypso (Red) | HW4 (White) |
|---|---|---|---|---|---|
| M1 | 0.134 ± 0.052 | 0.094 ± 0.021 | 0.036 ± 0.007 | 0.220 ± 0.103 | N.D. |
| M2 | 0.084 ± 0.023 | 0.379 ± 0.023 | 0.233 ± 0.023 | 0.766 ± 0.296 | N.D. |
| M3 | 0.287 ± 0.081 | 0.208 ± 0.085 | 0.018 ± 0.006 | 0.332 ± 0.111 | N.D. |
| M4 | 0.002 ± 0.001 | 0.003 ± 0.001 | N.D. | N.D. | N.D. |
APGs Are Synthesized by Dual Functional CHS/VPS Enzymes in Vitro
The presence of APGs suggests a key role for enzymes with VPS activity (Paniego et al., 1999; Okada et al., 2004) in Fragaria spp. fruit. The diploid woodland strawberry F. vesca and the cultivated octoploid strawberry share a common ancestor (Shulaev et al., 2011); however, a VPS gene has not been annotated in the F. vesca reference genome sequence (Shulaev et al., 2011). Untargeted metabolite profiling analysis (Supplemental Table S2) showed that the levels of M1, M2, and M3 all declined due to silencing of CHS (including its homologs in strawberry due to RNA interference (RNAi) off-target effects; Supplemental Table S3), suggesting that CHS might also act as VPS and play a role in the biosynthesis of APGs. To verify that CHS can perform the function of VPS in Fragaria spp. fruit, we characterized the enzymatic activities of CHS proteins from F. vesca.
Eight putative CHS genes have been annotated in the F. vesca genome sequence, although two of these appear to be truncated. Primers (Supplemental Table S1) were designed based on complementary DNA (cDNA) sequences of F. vesca gene26825 and gene26826, which are highly expressed in Fragaria spp. tissue during early stages of development (Supplemental Fig. S4) and are annotated as CHS2-like. Both have close homology to FaCHS (accession no. AI795154.1), which is highly expressed in strawberry fruit (Lunkenbein et al., 2006). Three full-length CHS amplicons (FvCHS2-1, FvCHS2-2, and FvCHS2-3) were obtained from F. vesca (Supplemental Figs. S5 and S6) and heterologously expressed in Escherichia coli.
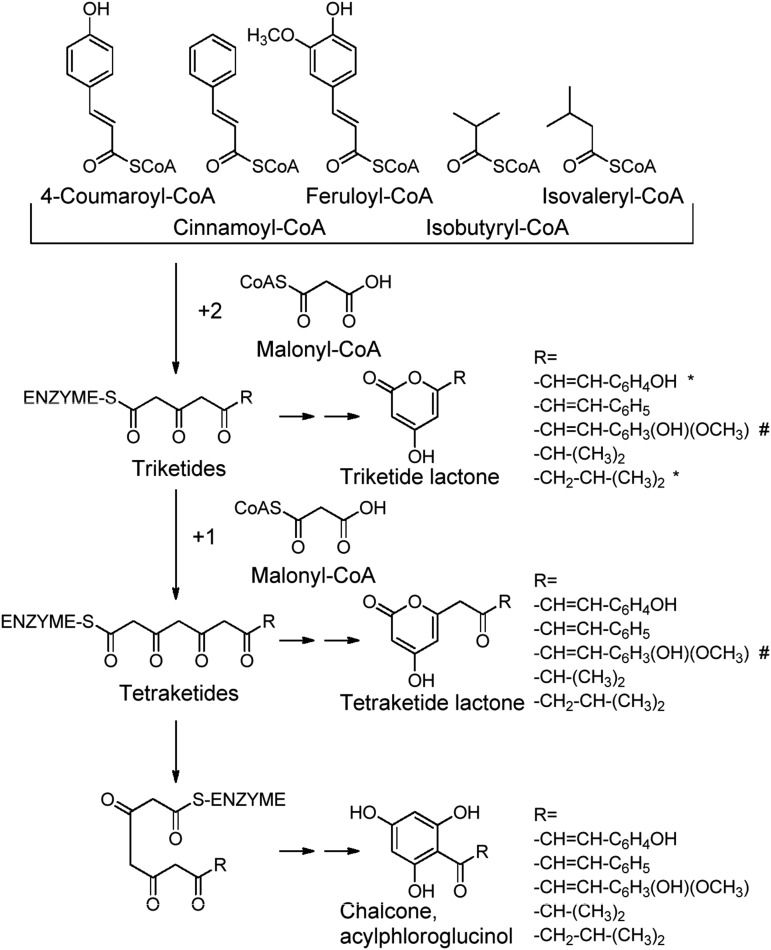
In the presence of 4-coumaroyl-CoA, all three recombinant enzymes FvCHS2-1, FvCHS2-2, and FvCHS2-3 produced 4-coumaroyl tetraketide lactone and naringenin-chalcone (Fig. 4; Table II; Supplemental Fig. S7), indicating that all three enzymes function as CHS. When isovaleryl-CoA and isobutyryl-CoA were used as starter substrates instead of 4-coumaroyl-CoA, APGs PIVP and PIBP were readily produced, respectively, by all three FvCHSs (Fig. 4; Table II; Supplemental Figs. S8 and S9). The product profiles formed in the presence of isovaleryl-CoA or isobutyryl-CoA were identical to those produced by VPS (Yamazaki et al., 2001; Okada et al., 2004). Because APGs PIVP and PIBP were the major products formed by CHS2-1 and CHS2-2 (Table I), we concluded that CHS2-1 and CHS2-2 act as VPS in vitro.
Figure 4.
Proposed formation mechanism and in vitro products. Products formed by FvCHS2-1, FvCHS2-2, and FvCHS2-3 when 4-coumaroyl-CoA, cinnamoyl-CoA, feruloyl-CoA, isobutyryl-CoA, and isovaleryl-CoA were used as starter molecules. A number sign indicates that the product was not detected by LC-MS, and an asterisk indicates that the product represents less than 10% of total products. -SCoA and CoAS- denote Coenzyme A.
Table II. LC-MS analysis of products formed and product ratio.
Products formed by CHS2-1, CHS2-2, and CHS2-3 and obtained product ratio using negative electrospray ionization mode. N.D., Not detected.
| Starter CoA | Product | Elemental Formula | Ms | Ms2 | Product Ratio |
||
|---|---|---|---|---|---|---|---|
| CHS2-1 | CHS2-2 | CHS2-3 | |||||
|
m/z |
% |
||||||
| Isovaleryl-CoA | APG PIVP | C11H14O4 | [M-H]– 209 | 165 and 125 | 40 to 45 | 70 to 80 | 10 to 30 |
| Tetraketide lactone | C11H14O4 | [M-H]– 209 | 165 and 125 | 25 to 30 | 10 to 15 | <10 | |
| Triketide lactone | C9H12O3 | [M-H]– 167 | 123 | 30 to 35 | 15 to 20 | 65 to 85 | |
| Isobutyryl-CoA | APG PIBP | C10H12O4 | [M-H]– 195 | 151 and 131 | 40 to 45 | 70 to 80 | <10 |
| Tetraketide lactone | C10H12O4 | [M-H]– 195 | 151 and 125 | 40 to 45 | 20 to 30 | 90 to 95 | |
| Triketide lactone | C8H10O3 | [M-H]– 153 | 109 | 10 to 15 | <10 | <10 | |
| 4-Coumaroyl-CoA | Naringenin | C15H12O5 | [M-H]– 271 | 177,151, and 107 | 25 to 30 | 70 to 75 | 80 to 90 |
| Tetraketide lactone | C15H12O5 | [M-H]– 271 | 227,201, and 125 | 70 to 75 | 25 to 30 | 15 to 20 | |
| Cinnamoyl-CoA | Chalcone | C15H12O4 | [M-H]– 255 | 211 and 151 | 80 to 85 | 80 to 90 | 10 to 20 |
| Tetraketide lactone | C15H12O4 | [M-H]– 255 | 211 and 187 | 5 to 15 | 10 | 10 to 20 | |
| Triketide lactone | C13H10O3 | [M-H]– 213 | 169 | <10 | <10 | 70 to 75 | |
| Feruloyl-CoA | Triketide lactone | C16H14O6 | [M-H]– 259 | 215 | 100 | N.D. | 100 |
The relative ratio of isovaleryl-CoA/malonyl-CoA and isobutyryl-CoA/malonyl-CoA affects the composition of the products and thus the PIVP and PIBP content (Supplemental Fig. S10). In addition to isovaleryl-CoA, isobutyryl-CoA, and 4-coumaroyl-CoA, FvCHS2-1 and FvCHS2-3 also accept feruloyl-CoA as starter molecule but form only the triketide lactone (Table II; Supplemental Fig. S11). With cinnamoyl-CoA, all three enzymes yield varying amounts of the tetraketide and triketide lactone and the chalcone (Table II; Supplemental Fig. S12). No products are detected when caffeoyl-CoA was used as starter substrate.
The highest activity of FvCHS2-1, FvCHS2-2, and FvCHS2-3 was detected at pH 6.0, 7.0, and 7.0, respectively, and the optimal temperature was 40°C for all enzymes using either 4-coumaroyl-CoA or isovaleryl-CoA as the starter substrate (Supplemental Fig. S13). To compare starter-CoA preference of these three FvCHS enzymes, kinetic properties were determined in the linear range of the enzymatic reaction (Supplemental Figs. S14 and S15). Isovaleryl-CoA is the preferred substrate of FvCHS2-1 (specificity constant [kcat/Km, where kcat is the turnover number] 21,700 m–1 s–1), whereas a 5-fold and 4-fold lower kcat/Km value was calculated for this starter substrate in the case of FvCHS2-2 and FVCHS2-3, respectively (Table III). The kcat value for FvCHS2-1 with isobutyryl-CoA is 5-fold and 4-fold higher than the corresponding values for FvCHS2-2 and FvCHS2-3, respectively, yielding a maximum specificity constant kcat/Km value for FvCHS2-1 of 847 m–1 s–1.
Table III. Steady-state kinetic constants for FvCHS2-1, FvCHS2-2, and FvCHS2-3 of F. vesca with different starter substrates.
Values shown are means (n = 3).
| Enzyme | Starter Substrate | Km | kcat | kcat/Km |
|---|---|---|---|---|
| µm | min–1 | m–1 s–1 | ||
| CHS2-1 | Isovaleryl-CoA | 14.7 ± 2.3 | 19.1 ± 1.5 | 21,700 |
| Isobutyryl-CoA | 86.6 ± 11.6 | 4.4 ± 0.5 | 847 | |
| 4-Coumaroyl-CoA | 6.4 ± 1.7 | 0.8 ± 0.1 | 2,021 | |
| CHS2-2 | Isovaleryl-CoA | 19.7 ± 6.3 | 5.1 ± 0.5 | 4,355 |
| Isobutyryl-CoA | 88.6 ± 14.5 | 1.0 ± 0.1 | 179 | |
| 4-Coumaroyl-CoA | 10.4 ± 3.8 | 1.3 ± 0.2 | 2,045 | |
| CHS2-3 | Isovaleryl-CoA | 18.0 ± 2.7 | 5.8 ± 0.2 | 5,431 |
| Isobutyryl-CoA | 62.3 ± 13.4 | 1.2 ± 0.1 | 313 | |
| 4-Coumaroyl-CoA | 10.2 ± 2.5 | 2.42 ± 0.12 | 3,957 | |
| Cinnamoyl-CoA | 77.5 ± 17.2 | 4.6 ± 0.5 | 981 | |
| Feruloyl-CoA | 16.1 ± 3.3 | 2.3 ± 0.1 | 2,364 |
Down-Regulation of FvCHS Enzymatic Activity in a Transient System and a Stable Transgenic Line
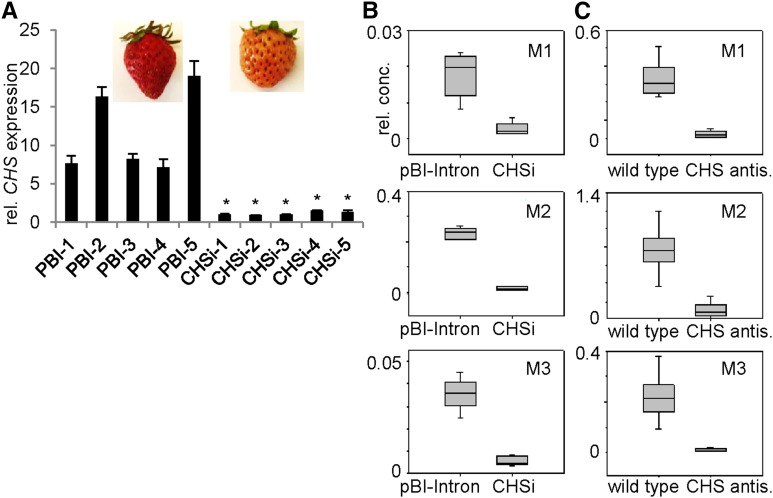
To corroborate that FvCHS genes play a role in the biosynthesis of APGs in vivo, metabolite profiling analyses of transiently silenced FaCHS fruits (Fig. 5) and fruits of a stable transgenic FaCHS antisense line (Lunkenbein et al., 2006) were performed. Fruits infiltrated with the control vector ripened normally to red, whereas fruits agroinfiltrated with plasmid binary (pBI)-CHSi became only slightly red (orange) colored (Fig. 5). Metabolite analyses revealed that FaCHS-silenced receptacles produced significantly lower levels not only of anthocyanins (Fig. 2), but also of M1 (10% of control), M2 (8%), and M3 (14%) when compared with the levels in controls (Fig. 5). M4 was not detected in fruit of strawberry ‘Mara de Bois’ (Table I). Similarly, levels of APG derivatives (M1–M3) in fruits of a stable transgenic antisense FaCHS line (Lunkenbein et al., 2006), which shows a CHS transcript level less than 5% of control fruit, were significantly reduced in response to the down-regulation of the CHS function (Fig. 5), confirming the biochemical role of CHS enzymes in the biosynthesis of APGs in Fragaria spp. fruit. Hardly any APG glucoside was detected in CHS-silenced fruit, suggesting that CHS2 proteins are the only enzymes that catalyze the biosynthesis of APGs in Fragaria spp. fruit.
Figure 5.
APGs decrease in both transient and stable CHS-silenced fruits. Fragaria spp. fruit phenotypes and CHS gene expression levels in agroinfiltrated fruits (A), and effect of CHS gene down-regulation on APGs (M1–M3) in transiently CHS-silenced fruits (B; CHSi, strawberry ‘Mara des Bois’) and a stable CHS antisense (antis.) transgenic line (C; CHS antis.; strawberry ‘Calypso’). Fourteen days after pollination, green Fragaria spp. fruit was infiltrated with Agrobacterium tumefaciens transformed with a construct encoding CHSi. Levels of metabolites were determined by LC-MS 14 d after infiltration in CHS-silenced fruits (CHSi; n = 5) and fruits infiltrated with A. tumefaciens containing the control vector (PBI, pBI-Intron; n = 5). Down-regulation of the CHS genes results in a decrease of APGs in the CHSi fruits and fruits of the stable transgenic line (CHS antis.; strawberry ‘Calypso’) compared with the control fruits (pBI-Intron and the wild type; for each, n = 5). Relative concentration (rel. conc.) is expressed in milligram equivalent internal standard g–1. Asterisks indicate significant differences (*P = 0.01).
Production of Volatiles Is Enhanced in Transient and Stable FaCHS-Silenced Fruits
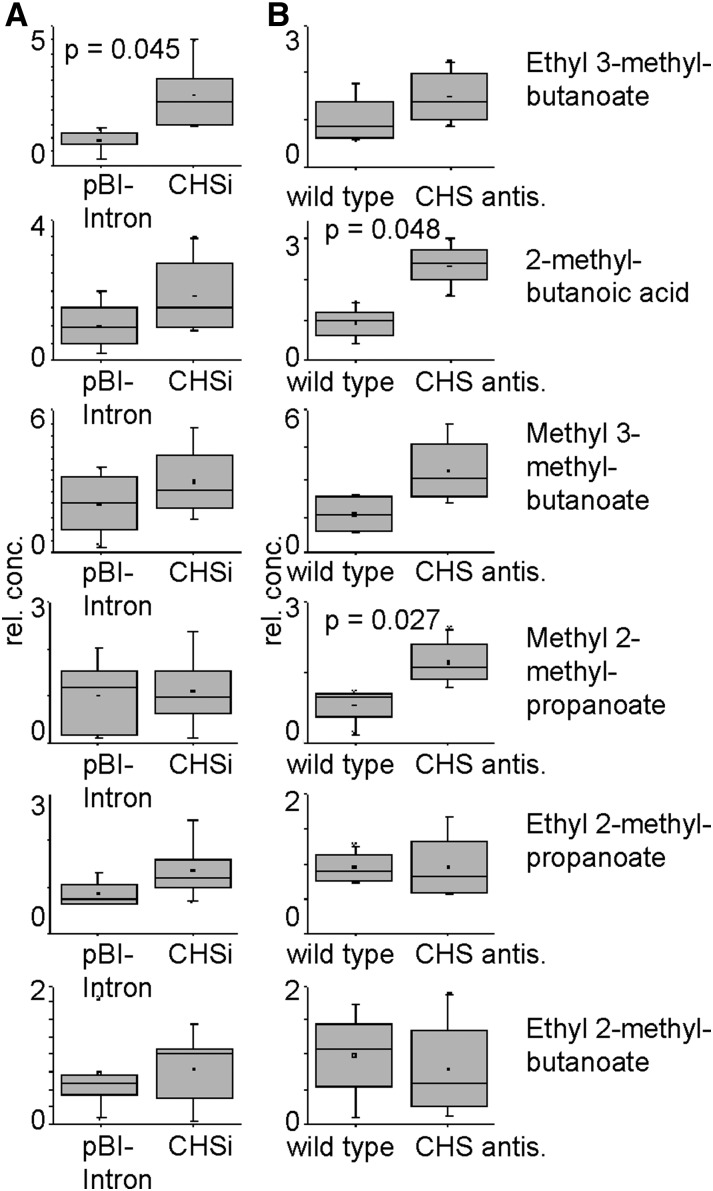
Because the precursor molecules of APGs, isovaleryl-CoA and isobutyryl-CoA, also act as substrates for alcohol acyl-CoA transferase enzymes that form aroma esters during Fragaria spp. fruit ripening (Aharoni et al., 2000; Cumplido-Laso et al., 2012), we analyzed the production of volatile esters by solid-phase microextraction gas chromatography (GC)-MS in RNAi-mediated CHS-silenced fruits and in fruits of the stable CHS antisense transgenic line. The relative content of ethyl 3-methylbutanoate was significantly increased by 2.2-fold in fruits after transient silencing of FaCHS, whereas levels of 2-methylbutanoic acid and methyl 2-methylpropanoate were significantly enhanced by 2.5- and 2.1-fold, respectively, in fruits of the stable transgenic line compared with the values of the wild-type fruits (Fig. 6). Most of the methyl-branched esters show higher concentrations in fruits in which the CHS transcript levels have been down-regulated, but the differences were not statistically significant due to the high biological variation of the values, indicated by the sizes of the boxes.
Figure 6.
Ester production in both transient and stable CHS-silenced fruits. Relative concentrations of esters in CHSi agroinfiltrated fruits (A; strawberry ‘Elsanta’) and a stable transgenic CHS antisense (CHS antis.) line (B; strawberry ‘Calypso’). Metabolite levels were determined by GC-MS 14 d after infiltration with A. tumefaciens transformed with a construct encoding CHS-inverted hairpin RNA or with pBI-Intron. Metabolite levels in fruits of the CHS antisense line and wild-type strawberry ‘Calypso’ fruits were determined at the mature ripening stage (n = 5–7). Identity of the compounds was confirmed by authentic references. Relative concentration (rel. conc.) is expressed in in milligram equivalent internal standard kg–1.
Stable Isotope Labeling Experiments
The APG pathway was traced by injection of stable isotope-labeled l-Ile (the biogenic precursor of 2-methylbutyryl-CoA), l-Leu (precursor of isovaleryl-CoA), and l-Val (precursor of isobutyryl-CoA) throughout attached Fragaria spp. fruits in the turning stage of ripening, which is just before APGs start to accumulate (Supplemental Fig. S16). The isotopically labeled APG products were analyzed by LC-MS, and the levels were calculated as a percentage of the unlabeled naturally occurring metabolites (Table IV; Supplemental Figs. S17 and S18). One day after the application of 10 mm l-Leu-13C6, l-Ile-13C6, and l-Val-2,3,4,4,4,5,5,5-D8, the level of isotopically labeled M3, M1, and M2 accounted for 20%, 16%, and 23% of the unlabeled metabolites, respectively. These values increased 4-, 2.5-, and 2.1-fold when the amounts of the labeled precursors were enhanced by a factor of 5 (50 mm solution), indicating that Fragaria spp. fruit readily form APGs from amino acids. The effect of longer incubation periods, up to 7 d, on the accumulation of labeled APGs was also studied. The contents of isotopically labeled APG products reached the highest levels at 4 d after injection and decreased thereafter (Supplemental Figs. S17D and S18D).
Table IV. Detection of isotopically labeled APGs.
Isotopically labeled APGs were detected 1 d after injection of different concentrations of l-Ile-13C6, l-Leu-13C6, and l-Val-2,3,4,4,4,5,5,5-D8 into strawberry ‘Mara des Bois’ fruits.
| Concentration Applied | Substrates | Compound | Products |
|
|---|---|---|---|---|
| Unlabeled | Labeled | |||
| mm | % | |||
| 10 | l-Leu | M3 | 100a | 0b |
| 50 | l-Leu | M3 | 100 | 0 |
| 10 | l-Leu-13C6 | M3 | 100 | 19.7 ± 1.3 |
| 50 | l-Leu-13C6 | M3 | 100 | 78.7 ± 6.3 |
| 10 | l-Ile | M1 | 100 | 0 |
| 50 | l-Ile | M1 | 100 | 0 |
| 10 | l-Ile-13C6 | M1 | 100 | 15.9 ± 5.3 |
| 50 | l-Ile-13C6 | M1 | 100 | 40.4 ± 0.3 |
| 10 | l-Val | M2 | 100 | 0 |
| 50 | l-Val | M2 | 100 | 0 |
| 10 | l-Val-2,3,4,4,4,5,5,5-D8 | M2 | 100 | 22.9 ± 4.7 |
| 50 | l-Val-2,3,4,4,4,5,5,5-D8 | M2 | 100 | 48.0 ± 24.9 |
Integrated peak areas of pseudomolecular ions m/z –371, –371, and –357 (after application of l-Leu, l-Ile, and l-Val, respectively) were set to 100%. b Percentages of labeled products were calculated from the integrated peak areas of the pseudomolecular ions of isotopically labeled products m/z –376, –376, and –364 (after application of l-Leu-13C6, l-Ile-13C6, and l-Val-2,3,4,4,4,5,5,5-D8, respectively).
DISCUSSION
Plants produce numerous structurally diverse metabolites that have beneficial effects on human health (Saito and Matsuda, 2010; De Luca et al., 2012). Phenolic compounds are the most widely distributed secondary metabolites in the plant kingdom, and there is increasing evidence that consumption of a variety of phenolic compounds may lower the risk of serious health disorders (Visioli et al., 2011). Although the basic reactions of the biosynthetic pathways for phenolics in plants have been intensively analyzed, the regulation of their accumulation and flux through the pathway is not that well established. By comparing the transcript patterns and metabolic profiles of different Fragaria spp. genotypes, recent research revealed unique candidate genes that might affect accumulation of flavonoids and anthocyanins in Fragaria spp. fruit (Ring et al., 2013).
Candidate Genes Function in Anthocyanin Accumulation
Four of these candidate genes (expansin-A8-like, SRG1-like, ephrin-A1-like, and defensin-like) were selected for further analysis, as they show a ripening-related expression pattern (Supplemental Fig. S1). Transient up- or RNAi-mediated down-regulation of the candidate genes clearly confirmed the correlation of transcript abundance with flavonoid and anthocyanin accumulation (Fig. 2). Loss-of-function phenotypes of the expansin-A8-like and ephrin-A1-like gene as well as the gain-of-function phenotypes of the SRG1-like and defensin-like gene showed impaired phenylpropanoid and anthocyanin accumulation, indicating that the first two genes might be positive and the last two genes negative regulators in the biosynthesis pathway.
Expansins were originally identified as cell wall-loosening proteins and are now considered as key regulators of cell wall breakdown and softening in processes such as fruit ripening, pollination, germination, and abscission (Li et al., 2003). SRG1 in Arabidopsis (Arabidopsis thaliana) is senescence related (Callard et al., 1996) and is a member of the Fe(II)/ascorbate oxidase superfamily. Some members of this family have been shown to catalyze the oxidation of intermediates of the flavonoid pathway (Almeida et al., 2007). Ephrins are receptor protein-Tyr kinases and have been implicated in mediating developmental events in animals (Wilkinson, 2000). They regulate cellular responses, and their ectodomain is an eight-stranded β-barrel with topological similarity to plant nodulins and phytocyanins (Toth et al., 2001). Plant defensins are cationic peptides that belong to a large superfamily of antimicrobial peptides found in several types of organisms (Carvalho and Gomes, 2011). They have numerous biological activities and are involved in inhibiting protein synthesis, mediating abiotic stress, and altering the ascorbic acid redox state.
During fruit ripening, flavonoids and anthocyanins are synthesized by highly complex but coordinated pathways that are regulated at the levels of signal reception (Jia et al., 2011), signal transduction (Jia et al., 2013), transcription factors (Medina-Puche et al., 2014), and structural genes (Griesser et al., 2008). In addition, complex feedback mechanisms controlling anthocyanin synthesis have been described (Muñoz et al., 2010). Although the four candidate genes studied are probably not directly involved in flavonoid formation, their function may be required for the coordinate progression of the ripening process, and they may have different roles for the different classes of phenolics (Fig. 2). Further investigations are needed to elucidate the precise functions of these candidate genes that are essential for pigment formation in the ripe fruit.
Untargeted Analysis Revealed Unique Fragaria spp. Metabolites
MS-based untargeted metabolite analysis of the data sets was performed, followed by second order comparisons to identify shared disturbances among the phenotypes of the transgenic plants. This led to the identification of four APGs, three of which have not previously been reported from Fragaria spp. fruit (Tsukamoto et al., 2004). APGs are prominent secondary metabolites of the genera Hypericum (Hypericaceae; Crispin et al., 2013) and Humulus (Cannabinaceae; Bohr et al., 2005) and have been detected in Phyllanthus emblica (Zhang et al., 2002), Jatropha multifida (Kosasi et al., 1989), and Curcuma comosa (Suksamrarn et al., 1997) but are rarely found in other plant species.
The structural diversity among APGs leads to various pharmacological activities in vitro and in vivo. APGs show significant antibacterial activity, as well as cytotoxic, antiproliferative, and antiangiogenic effects (Schmidt et al., 2012). The altered concentrations of APGs in response to changed expression levels of the candidate genes provide further evidence that the five genes studied are involved in the biosynthesis of phenolic compounds, but the role of the bioactive APGs in Fragaria spp. fruit remains unclear.
CHS Genes Are Involved in the Biosynthesis of APGs
APGs are generated by VPS, a key enzyme in the bitter acid biosynthesis pathway of hops (Zuurbier et al., 1998; Paniego et al., 1999; Okada et al., 2004). As there are no VPS genes annotated in the F. vesca genome sequence (Shulaev et al., 2011), we needed to rationalize the biosynthesis of PIVP and PIBP in Fragaria spp. fruit. Based on the basic catalytic mechanisms of VPS and CHS and untargeted metabolite profiling analysis, we hypothesized that a CHS may have dual functionality and also act as VPS in Fragaria spp. fruit. Eight CHS genes were detected in the F. vesca genome sequence, only two of which (gene26825 and gene26826) are transcribed during early Fragaria spp. fruit development and are annotated as CHS2 (Supplemental Fig. S4; Kang et al., 2013). Three CHS2-like sequences were cloned, including a unique CHS sequence, named FvCHS2-3. Comparison of the deduced amino acid sequences (Supplemental Fig. S5) showed that the N-terminal part of FvCHS2-3 is identical with gene26825 (FvCHS2-1), whereas the C terminus contains the sequence of gene26826 (FvCHS2-2). Thus, we assume that FvCHS2-3 is an artifact formed during PCR.
Recent structural and functional studies have elucidated the basic chemical mechanism for polyketide formation in CHS (Ferrer et al., 1999; Funa et al., 1999; Jez et al., 2000). Three essential catalytic amino acids, Cys-164, His-303, and Asn-336 (Jez et al., 2000), and two Phe residues (Phe-215 and Phe-265) important in determining the substrate specificity (Jez et al., 2002) are all well conserved in the FvCHS2 proteins (Supplemental Fig. S5). CHS2-1, CHS2-2, and CHS2-3 from F. vesca form naringenin chalcone from 4-coumaroyl-CoA with similar efficiency (Table III), but FvCHS2-1 shows superior VPS activity with isovaleryl-CoA as a starter molecule. Thus, CHS2-2 and, in particular, CHS2-1 were shown to efficiently catalyze the formation of the aglycones of M1 to M4 in vitro. Dual functional CHS/VPS have rarely been reported. CHS-hops1 (H1) from hops (HlCHS-H1) and CHS from Pinus sylvestris, Pinus strobus, and Sinapis alba can perform the function of VPS, although the majority of the products were released from the polyketide synthases after only two condensation reactions (Zuurbier et al., 1998; Novák et al., 2006).
In Planta Functional Analysis of CHS2 Genes
In planta function of CHS2 was analyzed by RNAi-mediated down-regulation of CHS2 transcript abundance in receptacles of strawberry ‘Mara des Bois’ and in fruits of a stable transgenic CHS antisense line (strawberry ‘Calypso’). In both cases, CHS homologous genes were also down-regulated. In addition to lower levels of flavonoids and anthocyanins and higher levels of some phenylpropanoids (4-coumaroyl and feruloyl Glc; Fig. 2) in CHS2-silenced fruits, our results show that down-regulation of CHS2 also resulted in significantly reduced levels of APGs (M1–M3; Fig. 5). Plants of a stable transgenic CHS antisense line (Lunkenbein et al., 2006) also contained significantly lower concentrations of APGs than Fragaria spp. fruit of wild-type plants (Fig. 5). The results of both reverse genetic approaches clearly confirm the biochemical function of the CHS2 enzymes in the anthocyanin and in the APG biosynthesis pathway in vivo.
APG Pathway in Fragaria spp. Fruit
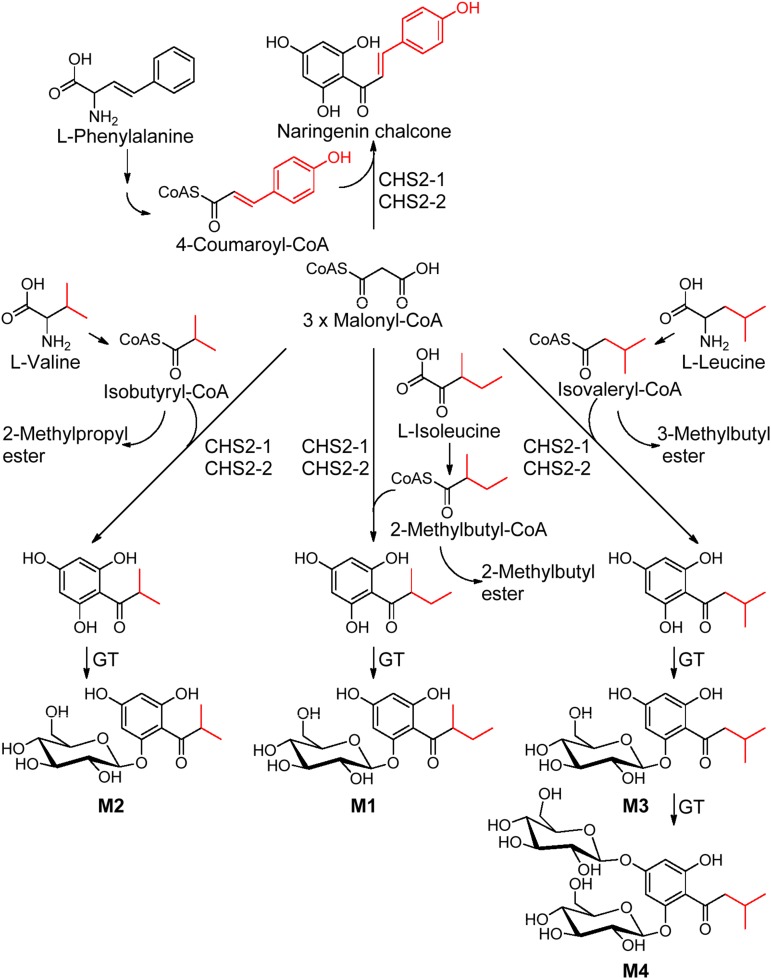
Polyketide synthases play major roles in the biosynthesis of diverse secondary metabolites, as they generate the backbones of chalcones, stilbenes, phloroglucinols, resorcinols, benzophenones, biphenyls, dibenzyls, chromones, acridones, pyrones, and curcuminoides (Abe and Morita, 2010). CHS is the best studied plant-specific polyketide synthase and shows extremely broad substrate promiscuity (Helariutta et al., 1996; Zuurbier et al., 1998). Similarly, CHS2-1, CHS2-2, and CHS2-3 can use both aromatic and methyl branched-chain aliphatic CoAs derived from the transformation of aromatic and branched-chain amino acids (Table II; Xu et al., 2013) as starter substrates. Isovaleryl-CoA, isobutyryl-CoA, and 2-methylbutyryl-CoA are produced from Leu, Val, and Ile, respectively, by the oxidative decarboxylation of the ketoacid intermediates (Xu et al., 2013). Thus, we propose Ile, Val, and Leu as biogenetic precursors of M1, M2, and M3, respectively, with M4 being formed by glucosylation of M3 (Fig. 7). The transformation of branched-chain amino acids to M1 to M3 was confirmed by stable isotope tracer experiments in which 40% to 79% of the APG products were labeled with the isotopes after 1 d when solutions of labeled precursor amino acids were injected into fruits (Table IV). The degree of labeling was already high in the APG products after 1 d; however, calculations showed that the amount of labeled Val, Leu, and Ile (2.9, 3.3, and 3.3 mg per 10 g fresh fruit, respectively) that was injected into fruit exceeded the amount of the naturally unlabeled amino acids (1.0–2.0, 0.4, and 1.1–1.4 mg per 100 g fresh weight of Val, Leu, and Ile, respectively; Pérez et al., 1992; Keutgen and Pawelzik, 2008) in the fruit by at least a factor of 15 (1,500%). Thus, substantial amounts of the labeled precursors are probably utilized by other pathways or were not yet transformed. In fact, maximum labeling was achieved after 4 d (Supplemental Figs. S17 and S18), peaking at 1,100% in the case of Ile.
Figure 7.
The proposed APG biosynthesis pathway catalyzed by CHS2-1 and CHS2-2 in Fragaria spp. fruit. GT, Glucosyl transferase; -CoAS, Coenzyme A.
One of the main sources of substrates for volatile ester production is the metabolism of amino acids, generating alcohols and acids, either aliphatic, branched chain, or aromatic (Aharoni et al., 2000; Pérez et al., 2002; Cumplido-Laso et al., 2012). These esters contribute, and in some cases are determining, to the primary aroma of many fruits. Feeding of Ile resulted in a substantial increase in 2-methylbutanoate esters and 2-methylbutyl esters (Pérez et al., 2002). Similarly, levels of volatile branched-chain aliphatic esters were increased in CHSi fruits compared with controls (Fig. 6), demonstrating that higher concentrations of precursor CoA thioesters are available for ester formation due to the down-regulation of the CHS function. Thus, the pathways producing branched-chain aliphatic esters and APGs compete for their common CoA thioester substrates.
Evolutionary Relevance
Gene duplication has been proposed to be an important process in the generation of evolutionary novelty (Hughes, 2005). Neofunctionalization, as an adaptive process where one copy of the original gene mutates into a function that was not present in the ancestral gene, is one mechanism that can lead to the retention of both gene copies. In the case of FvCHS2-1 and FvCHS2-2 (CHS2-3 is considered an artifact), duplication of an ancestral CHS2 gene appears to have occurred only recently on evolutionary scales, as the sequences of CHS2-1 and CHS2-2 are highly similar (95% and 98% nucleotide and amino acid identity, respectively), and both genes show identical expression profiles (Supplemental Fig. S4). However, enzymatic activity of FvCHS2-1 for the starter substrate isovaleryl-CoA is much higher than that of FvCHS2-2 (Table II). Similarly, VPS and HICHS-H1 from hops show very different substrate specificity toward the substrates 4-coumaroyl-CoA and isovaleryl-CoA (Novák et al., 2006), but protein sequence comparison reveals that all three essential catalytic amino acids (Jez et al., 2000) and known residues important in determining the substrate specificity (Jez et al., 2002) are well conserved in CHS2-1, CHS2-2, VPS, and HlCHS-H1 (Supplemental Fig. S19). Thus, with the present knowledge, it is not possible to define what properties of these proteins is conferring the increased VPS activity. Site-directed mutagenesis experiments and modeling analyses are needed to identify the essential amino acids.
Subfunctionalization, as a neutral process where a paralog specializes in one (here, VPS function) of several ancestral functions, best describes the fact that both CHS2 copies are retained in the genome of F. vesca (Lynch and Force, 2000). The results of the studies on FvCHS2 confirm that when multifunctionality precedes gene duplication, it is straightforward for duplicates to specialize by sharing the ancestral function (Jensen, 1976; Hughes, 2005).
CONCLUSION
Global epidemiological studies confirm an inverse relationship between the consumption of fruit and the incidence of diverse diseases. There is convincing evidence that the considerable health benefits of fruits are due to their specific chemical compositions, particularly to compounds of nutritional relevance. Although the major components of fruits have been frequently quantified, detailed analysis by more sensitive analytical methods shows that plants have a much higher capacity to synthesize secondary metabolites than previously thought. To our knowledge, we demonstrate for the first time that Fragaria spp. plants are able to synthesize and accumulate APGs, compounds with exciting pharmacological properties (Singh et al., 2010), due to the catalytic properties of bifunctional chalcone synthase/valerophenone synthase enzymes. APGs might be important physiologically active metabolites in Fragaria spp. fruit that contribute to its health-promoting activity. Additionally, the findings show that duplication of a dually functioning CHS2 and subsequent specialization for VPS function might explain the occurrence of paralogous CHS enzymes with overlapping but not identical catalytic activities in Fragaria spp. fruit.
MATERIAL AND METHODS
Plant Material
The octoploid strawberry (Fragaria × ananassa) cultivars used for this study (cv Elsanta, Mara des Bois, Senga Sengana, and Calypso and a stable transgenic FaCHS antisense Calypso line; Lunkenbein et al., 2006) were cultured in a greenhouse in Freising, Germany. Growing conditions were maintained at 25°C and a 16-h photoperiod providing 120 µmol m–2 s–1 irradiance from Osram Fluora lamps. For genetic and molecular analyses, fruits were injected 14 d after pollination and harvested 10 to 14 d after agroinfiltration. Seasonal flowering short-day diploid strawberry Fragaria vesca ‘Reine de Vallees’ and perpetual-flowering F. vesca accession HI-4 were used for cloning and were also cultured in the greenhouse. F. vesca HI-4 was kindly provided by Vladimir Shulaev (University of North Texas).
Chemicals and Reagents
Isovaleryl-CoA, isobutyryl-CoA, malonyl-CoA, l-Ile-13C6,15N (98+ atom% 13C and 98+ atom% 15N), l-Leu-13C6, 15N (98 atom% 13C and 98 atom% 15N), and l-Val-2,3,4,4,4,5,5,5,-D8 (98 atom% D) were purchased from Sigma. All chemicals and solvents were obtained from Sigma, Fluka, and Aldrich, Carl Roth, and VWR International, unless otherwise noted. Authentic phloroglucinol derivatives were kindly provided by the Chair of Food Chemistry and Sensory Analysis (Technische Universität München).
Plasmid Construction
Fragments (200–400 bp) corresponding to the candidate genes expansin-A8-like (gene21343) and ephrin-A1-like (gene33865) were PCR amplified from strawberry ‘Elsanta’ DNA using primers shown in Supplemental Table S1 and cloned into the p9U10 vector (DNA-Cloning Services e.K.). DNA corresponding to the full-length SRG1-like (gene10776) and defensin-like (gene00897) genes was PCR amplified from strawberry ‘Elsanta’ DNA using the primers listed in Supplemental Table S1 and cloned into the pBI121 vector. PBI with an intron-containing GUS gene, pBI-Intron, was used as control.
Transfection of Fragaria spp. Fruit by Agroinfiltration
The Agrobacterium tumefaciens strain AGL0 containing the p9U10-gene21343, p9U10-gene33865, pBI121-gene10776, pBI121-gene00897, pBI-Intron-GUS, and pBI-CHS was grown at 28°C in Luria-Bertani (LB) medium with appropriate antibiotics. When the culture reached an optical density at 600 nm of about 0.8, cells were harvested and resuspended in a modified modified MacConkey agar medium (Murashige and Skoog salts, 10 mm MES, pH 5.6, and 20 g L–1 Suc). The A. tumefaciens suspension was evenly infiltrated throughout the entire attached fruit (strawberry ‘Mara des Bois’ and ‘Elsanta’) about 14 d after pollination using a sterile 1-mL hypodermic syringe according to Hoffmann et al. (2006).
Isolation of Nucleic Acids and Quantitative PCR Analysis
For candidate gene expression analysis, wild-type fruit (strawberry ‘Elsanta’) of different ripening stages (green, white, and red), root, stem (petiole), and leaves were harvested and freeze dried. Agroinfiltrated fruit was harvested 10 to 14 d after injection and individually freeze dried. After grinding to a fine powder, 200 mg of the powder from each sample was used for total RNA extraction according to Liao et al. (2004), followed by DNase I (Fermentas) treatment and reverse transcription with oligo(dT) primer (Promega). Real-time PCR was performed with a StepOnePlus real-time PCR system (Applied Biosystems) using Fast SYBR Green Master Mix (Applied Biosystems) to monitor double-stranded cDNA synthesis according to Ring et al. (2013). At least five biological replicates for each sample were used for the quantitative reverse transcription-PCR analysis, and at least two technical replicates were analyzed for each biological replicate. The interspacer 26S-18S strawberry RNA housekeeping gene FaRib413 was used as an internal control for normalization, and data were analyzed as described in Ring et al. (2013). Gene-specific primers used to detect the transcripts are listed in Supplemental Table S1.
Metabolite Analysis
Metabolites in individual Fragaria spp. fruits were analyzed about 14 d after injection in the ripe stage. The materials were ground in liquid nitrogen and kept at –80°C prior to analyses. For metabolite analysis, an aliquot of 50 mg was used for each of the three biological replicates. Sample preparation and analysis by LC-MS was performed as described previously (Ring et al., 2013).
Preparative Isolation of Unknown Metabolites
Frozen strawberry ‘Elsanta’ fruits (2.0 kg) were extracted with 1 L of methanol. After vortexing and sonication for 10 min, the sample was centrifuged at 16,000g for 10 min. The supernatant was removed, and the residue was reextracted with 500 mL of methanol. The supernatants were combined, concentrated to dryness in a vacuum concentrator, and redissolved in 10 mL of water. After 1-min vortexing, 10-min sonication, and 10-min centrifugation at 16,000g, the clear supernatant was used for preparative fractionation. Separations were carried out using an RP18 column (25 cm × 46 mm; particle size, 5 µm; Phenomenex) at room temperature, connected to a Jasco PU-1580 LC system and Jasco UV-1575 detector. Metabolites were separated by the following gradient of water containing 1% (v/v) formic acid (A) and methanol containing 1% (v/v) formic acid (B): 0 to 5 min, 0% B; 5 to 30 min, 0% to 40% B; and 30 to 35 min, 100% B. The injection volume was 2 mL, and the flow rate was 10 mL min–1. One fraction was collected every minute. Fractions containing the target compounds were further purified on the RP18 column with a gradient of 0 to 5 min, 20% B, and 5 to 30 min, 20% to 40% B. A normal phase column (SeQuant ZIC-HILIC, 200 Å, 5 µm, 250 × 10 mm, Merck) was further used with a gradient consisting of water with 1% (v/v) formic acid (A) and acetonitrile with 1% (v/v) formic acid (C): 0 to 30 min, 95% to 80% C; 30 to 35 min, 80% to 0% C; and 35 to 45 min, 0% C. Final purification was achieved by analytical LC (RP18 column; 25 cm × 4.0 mm; particle size, 5 µm; Phenomenex) applying the following gradient: 0 to 5 min, 0% B; 5 to 35 min, 0% to 100% B; and 35 to 50 min, 100% B. Purity was analyzed by LC-MS as described previously (Ring et al., 2013). High-resolution mass spectra of the compounds were measured on a Bruker Micro-TOF (Bruker Daltronics) mass spectrometer and referenced to sodium formate (Intelmann et al., 2011).
NMR Spectroscopy
The samples were evaporated and dissolved in methanol-d4 (99.8%) containing 0.03% (v/v) trimethylsilane. 1H-NMR spectra were recorded at 500.13 MHz with a Bruker DRX 500 spectrometer. The chemical shifts were referred to the solvent signal. The one- and two-dimensional correlation spectroscopy, HMQC, and HMBC spectra were acquired and processed with standard Bruker software (XWIN-NMR).
Cloning the Full-Length CHS Genes
Total RNA was isolated from mature fruit of F. vesca using the method described previously (Liao et al., 2004). First strand cDNA was synthesized from 1 µg of DNase I (Fermentas)-treated total RNA using Moloney murine leukemia virus reverse transcriptase H2 (Promega) and an oligo(dT) primer. CHS2 was amplified by PCR from the F. vesca fruit cDNA using a pair of gene-specific primers (Supplemental Table S1), designed according to the two F. vesca mRNA sequences annotated as CHS2 (gene26825 and gene26826) at Genome Database for Rosaceae and Strawberry Genomic Resources (Darwish et al., 2013; Jung et al., 2014). The full-length coding sequences were amplified using proof-reading Pfu DNA polymerase (Promega). The PCR was carried out in a 25-µL total reaction volume, and the temperature program used was 5 min at 95°C, one cycle; 45 s at 95°C, 45 s at 55°C, and 2 min at 72°C, 35 cycles; and final extension at 72°C for 10 min. The PCR products were A tailed and cloned into the pGEM-T Easy (Promega) vector, and the ligation product transformed into Escherichia coli NEB10 β (New England Biolabs). The identity of the cloned genes was confirmed by sequencing the complete insert (MWG Biotech) from both sides and by restriction enzyme digest with BamH1 and EcoRI.
Construction of the Expression Vector pGEX-4T1-CHS
To obtain recombinant glutathione S-transferase (GST) fusion proteins for functional characterization, the amplified CHS sequences were digested with EcoRI and BamH1, and the resulting gene fragments were cloned into the expression vector pGEX-4 T-1 (Amersham Biosciences). The recombinant plasmid (pGEX-4T1-CHS) was then introduced into E. coli NEB10 β. Plasmid DNA was purified from the bacteria and sequenced to check for correct insertion.
Heterologous Expression and Partial Purification of the Recombinant Protein
Expression constructs were transformed into E. coli strain BL21(DE3) pLysS (Promega). A single colony of E. coli strain BL21(DE3) pLysS cells harboring the pGEX-4T1-CHS plasmid was cultured overnight at 37°C in LB liquid medium containing ampicillin (50 µg mL–1) and chloramphenicol (50 µg mL–1). The following day, the culture was diluted 1:40 with LB medium containing the antibiotics and grown under the same conditions as above until optical density at 600 nm of the cultured cells reached 0.8. Expression was induced with isopropyl-β-d-thiogalactopyranoside (1 mm), and the culture was incubated at 16°C and 150 rpm overnight. Cells were harvested by centrifugation (5,000g, 10 min), and recombinant protein was purified by GST binding (Novagen), following the manufacturer’s protocol with slight modifications. All steps were performed at 4°C with prechilled buffers to maintain enzyme activity. The harvested cells were frozen at –80°C for 15 min, and the pellet resuspended in 10 mL of GST wash buffer (43 mm Na2HPO4, 14.7 mm KH2PO4, 1.37 m NaCl, and 27 mm KCl, pH 7.3) followed by sonication in three intervals of 30 s at 50% power (Sonopuls UW2200, Bandelin Electronic). The lysate was centrifuged (10,000g, 20 min) and incubated with gentle shaking for at least 60 min with GST binding resin previously equilibrated with GST wash buffer. The resin was washed two times with 5 mL of GST wash buffer and then incubated 5 min at room temperature with 200 µL of 1× elution buffer (50 mm Tris-Cl, pH 8.0, and 10 mm reduced glutathione). This step was repeated three times. The resulting fractions were analyzed by SDS-PAGE. BL21 and BL21 harboring the pEXT-4T1 empty vector were induced and subjected to the same procedure and served as controls. Protein concentration was determined by the method of Bradford (1976).
Preparation of Starter CoA Esters
Enzymatic synthesis of cinnamoyl-CoA, 4-coumaroyl-CoA, caffeoyl-CoA, and feruloyl-CoA was carried out with purified 4-coumarate:CoA ligase, according to Beuerle and Pichersky (2002). 4-Coumarate:CoA ligase was a gift from Till Beuerle (Universität Braunschweig). To purify the hydroxycinnamoyl-CoAs, 0.8 g of ammonium acetate was added to the reaction mixture. The reaction mixture was loaded onto a 6-mL Isolute C8 (extraction cartridge) solid-phase extraction column (Biotage), which had been previously washed successively with methanol, distilled water, and 4% ammonium acetate solution. After loading, the column was washed with 4% ammonium acetate solution until free CoA was detectable spectrophotometrically at 259 nm. The CoA esters were then obtained by elution with Milli-Q water. Fractions (2 mL) were collected and lyophilized. The reaction products were dissolved in Milli-Q water after freeze drying, and the concentration of the CoAs was determined spectrophotometrically by measuring the absorbance at the absorption maxima of the products (Beuerle and Pichersky, 2002).
Enzyme Assay
The reactions were performed as described previously (Zuurbier et al., 1998) with modifications. The standard assay for determining CHS activity (2 µg of protein) was conducted in a total volume of 100 µL containing 100 mm potassium phosphate buffer (pH 7.0), 300 µm malonyl-CoA, and 100 µm starter-CoA. The mixtures were incubated at 30°C for 30 min. Reactions were initiated by addition of enzyme and were quenched with 5% (v/v) acetic acid. The products were extracted twice with 200 µL of ethyl acetate and analyzed by LC-MS. Optimum reaction temperature was determined in assays carried out in the range of 10°C to 50°C at pH 7.0. Before adding the enzyme and substrate, the mixture was equilibrated to the tested temperature. The pH optimum was tested in the range from pH 3 to 10. Citric acid, sodium phosphate, and Tris-HCl buffers were used for pH 3 to 6, pH 6 to 8, and pH 8 to 10, respectively. At least two biological replicates (different preparations) were carried out, and the total product formation was measured by LC-MS analysis. LC-MS was performed using a Nucleosil C18 (15 cm × 4.0 mm; particle size, 4 µm; Macherey Nagel) column with a flow rate of 0.1 mL min–1 at UV 280-nm detection. Gradient elution was performed with water (A) and methanol (B), both containing 0.1% (v/v) formic acid: 0 to 5 min, 0% to 70% B; 5 to 25 min, 70% to 100% B; and 25 to 30 min, 100% B. Products were identified by comparison of the retention times and MS data with those of authentic references (Akiyama et al., 1999; Jez et al., 2001). The amount of each product was calculated as milligram equivalent of naringenin using a standard curve. Steady-state kinetic constants were determined from initial velocity measurements (Jez et al., 2000). Kinetic experiments were conducted with 2 µg of protein in 100 µL of enzyme assay buffer at pH 7.0 with 300 µm malonyl-CoA and various concentrations of CoA starter molecules, and the reactions were incubated at 40°C for 10 min. Reactions were initiated and quenched as above. Then, the products were extracted twice with 200 µL of ethyl acetate for 2 min, and the top layer was analyzed by LC-MS. At least seven different substrate concentrations covering the range of 1 to 300 µm were used. Data were fitted to the Michaelis-Menten equation using a nonlinear regression program (Enzyme Kinetics, SigmaPlot) to calculate Vmax and Km values. Protein concentration was determined using the protein assay dye reagent (Bio-Rad) with bovine serum albumin as standard.
Analyses of Volatiles by GC-MS
Volatiles released by fruits of CHS-silenced transgenic lines (agroinfiltrated fruits and fruits of a stable CHS transgenic line; Lunkenbein et al., 2006) and wild-type Fragaria spp. were sampled by solid-phase microextraction. Intact fruits of approximately the same size at mature stages were placed under a glass funnel with caps to create a headspace as described previously (Aharoni et al., 2000). The volatile compounds collected from the headspace were analyzed by a Thermo Finnigan Trace DSQ mass spectrometer coupled to a BPX520M-fused silica capillary column with a 30-m × 0.25-mm i.d. Helium (1.1 mL min–1) was used as carrier gas. The injector temperature was 250°C, and the ion source and interface temperatures were kept at 250°C and 280°C, respectively. The temperature program was 40°C for 3 min and increased to 280°C at a rate of 5°C min–1. The electron impact-MS ionization voltage was 70 eV (electron impact ionization). Mass data were acquired in the range of 50 to 650 m/z. Compounds were identified by comparing their mass spectra and retention indices to the National Institute of Standards and Technology mass spectra library and authentic reference compounds. Relative concentration of volatile esters was determined as described previously (Sinz and Schwab, 2012).
Stable Isotope Labeling
l-Ile-13C6, l-Leu-13C6, and l-Val-2,3,4,4,4,5,5,5,-D8 were injected into the tops of strawberry ‘Mara des Bois’ fruit still attached to the plant with a 1-mL sterile hypodermic syringe similarly as described (Wein et al., 2001). Five mature fruits of similar sizes were selected for the applications. Solutions (500 µL) containing the isotopically labeled compounds (10–50 mm in water) were injected. As a control experiment, 500 µL of an aqueous solution containing 10 to 50 mm of unlabeled l-Ile, l-Leu, and l-Val was injected into Fragaria spp. fruits. The experiments were repeated at least twice. The fruits were harvested after 1 and 4 d and stored at –20°C until they were analyzed by LC-MS. In a separate set of experiments, equal amounts of l-Leu-13C6 (10 mm) and l-Ile-13C6 (10 mm) were administered to strawberry ‘Mara des Bois’ fruit of the same ripening stage and similar weight. After 1, 2, 4, 5, and 7 d, individual fruits were harvested and stored at –20°C until analysis by LC-MS.
Untargeted Metabolite Analysis
Analysis by metaXCMS (Gowda et al., 2014) was performed at https://xcmsonline.scripps.edu/ as described previously (Tautenhahn et al., 2011).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FvCHS2-1 (gene26825) gi|470141648|ref|XP 004306542.1| and FvCHS2-2 (gene26826) gi|470141650|ref|XP 004306543.1|.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Relative mRNA expression level of candidate genes in different tissues.
Supplemental Figure S2. Constructs used for this study.
Supplemental Figure S3. Extracted ion chromatogram (m/z 357 and 371, superimposed), Ms and Ms2 spectra of metabolites M1, M2, and M3.
Supplemental Figure S4. Relative expression levels of putative chalcone synthase genes in strawberry (F. vesca ‘Hawaii-4’) fruit tissue.
Supplemental Figure S5. Comparison of the deduced amino acid sequences of CHS enzymes from F. vesca.
Supplemental Figure S6. Phylogenetic tree of CHS/VPS enzymes.
Supplemental Figure S7. LC-MS analysis of products formed by the empty vector control and FvCHS2-3 from the starter molecule 4-coumaroyl-CoA.
Supplemental Figure S8. LC-MS analysis of products formed by the empty vector control and FvCHS2-1 from the starter molecule isovaleryl-CoA.
Supplemental Figure S9. LC-MS analysis of products formed by the empty vector control and FvCHS2-1 from the starter molecule isobutyryl-CoA.
Supplemental Figure S10. Products formed by FvCHS2-1 using 300 μM malonyl-CoA and different concentrations of isovaleryl-CoA.
Supplemental Figure S11. LC-MS analysis of the product formed by the empty vector control and FvCHS2-1 from the starter molecule feruloyl-CoA.
Supplemental Figure S12. LC-MS analysis of products formed by the empty vector control and FvCHS2-3 from the starter molecule cinnamoyl-CoA.
Supplemental Figure S13. The pH and temperature optima.
Supplemental Figure S14. The effect of different amounts of protein (0.5–10 μg) and incubation time (5–60 min) on the product formation of FvCHS2-3 using cinnamoyl-CoA as the starter substrate.
Supplemental Figure S15. Michaelis-Menten plots of purified FvCHS2-3 with the substrate isobutyryl-CoA, 4-coumaroyl-CoA, isovaleryl-CoA, feruloyl-CoA, and cinnamoyl-CoA.
Supplemental Figure S16. Relative content of M1, M2, and M3 measured by LC-MS (n = 3–5) and relative CHS expression (gene 26825 and gene 26826) determined by quantitative PCR.
Supplemental Figure S17. Ms and Ms2 of unlabeled M1 and isotopically labeled M1 after application of l-Ile-13C6, proposed pathway, as well as the effect of the incubation period on the accumulation of labeled product.
Supplemental Figure S18. Ms and Ms2 of unlabeled M3 and isotopically labeled M3 after application of l-Leu-13C6, proposed pathway, as well as the effect of the incubation period on the accumulation of labeled product.
Supplemental Figure S19. Comparison of the deduced amino acid sequences of CHS enzymes from F. vesca, FvCHS2-1, and FvCHS2-2, as well as VPS and HICHSH1 from hops.
Supplemental Table S1. List of primers used in this study.
Supplemental Table S2. Metabolites (m/z negative ionization mode and retention time) that were found to be differentially accumulated in Fragaria spp. fruit after down-regulation and overexpression of different genes.
Supplemental Table S3. Metabolites (m/z negative ionization and retention time) that were found to be differentially accumulated in Fragaria spp. fruit after down-regulation of CHS (CHSi).
Supplemental Table S4. 1H- and 13C-NMR data derived from HSQC and HMBC data of isolated metabolite M1 and data from literature.
Supplemental Table S5. 1H-NMR data derived from HSQC and HMBC data of isolated metabolite M3 and 1H-NMR data from literature.
Supplementary Material
Acknowledgments
We thank Ludwig Friedrich (Chair of Food Chemistry and Molecular Sensory Science of Technische Universität München) for providing authentic reference material from hop and the high-resolution mass spectrometry analysis.
Glossary
- APG
acylphloroglucinol
- PIVP
phlorisovalerophenone
- PIBP
phlorisobutyrophenone
- LC
liquid chromatography
- MS
mass spectrometry
- m/z
mass-to-charge ratio
- cDNA
complementary DNA
- GC
gas chromatography
- GST
glutathione S-transferase
- LB
Luria-Bertani
- kcat/Km
specificity constant
- HMBC
heteronuclear multiple-bond correlation
- HMQC
heteronuclear multiple quantum coherence
- RNAi
RNA interference
Footnotes
This work was supported by the China Scholarship Council (grant no. 2011630188), Plant-KBBE FraGenomics, and Deutsche Forschungsgemeinschaft (grant no. DFG–SFB924).
References
- Abe I, Morita H (2010) Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat Prod Rep 27: 809–838 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Keizer LC, Bouwmeester HJ, Sun Z, Alvarez-Huerta M, Verhoeven HA, Blaas J, van Houwelingen AM, De Vos RC, van der Voet H, et al. (2000) Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell 12: 647–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T, Shibuya M, Liu HM, Ebizuka Y (1999) p-Coumaroyltriacetic acid synthase, a new homologue of chalcone synthase, from Hydrangea macrophylla var. thunbergii. Eur J Biochem 263: 834–839 [DOI] [PubMed] [Google Scholar]
- Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13: 99–102 [DOI] [PubMed] [Google Scholar]
- Almeida JRM, D’Amico E, Preuss A, Carbone F, de Vos CHR, Deiml B, Mourgues F, Perrotta G, Fischer TC, Bovy AG, et al. (2007) Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria × ananassa). Arch Biochem Biophys 465: 61–71 [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichersky E (2002) Enzymatic synthesis and purification of aromatic coenzyme a esters. Anal Biochem 302: 305–312 [DOI] [PubMed] [Google Scholar]
- Bohr G, Gerhäuser C, Knauft J, Zapp J, Becker H (2005) Anti-inflammatory acylphloroglucinol derivatives from Hops (Humulus lupulus). J Nat Prod 68: 1545–1548 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Callard D, Axelos M, Mazzolini L (1996) Novel molecular markers for late phases of the growth cycle of Arabidopsis thaliana cell-suspension cultures are expressed during organ senescence. Plant Physiol 112: 705–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AdeO, Gomes VM (2011) Plant defensins and defensin-like peptides: biological activities and biotechnological applications. Curr Pharm Des 17: 4270–4293 [DOI] [PubMed] [Google Scholar]
- Cheynier V, Comte G, Davies KM, Lattanzio V, Martens S (2013) Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol Biochem 72: 1–20 [DOI] [PubMed] [Google Scholar]
- Crispin MC, Hur M, Park T, Kim YH, Wurtele ES (2013) Identification and biosynthesis of acylphloroglucinols in Hypericum gentianoides. Physiol Plant 148: 354–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumplido-Laso G, Medina-Puche L, Moyano E, Hoffmann T, Sinz Q, Ring L, Studart-Wittkowski C, Caballero JL, Schwab W, Muñoz-Blanco J, et al. (2012) The fruit ripening-related gene FaAAT2 encodes an acyl transferase involved in strawberry aroma biogenesis. J Exp Bot 63: 4275–4290 [DOI] [PubMed] [Google Scholar]
- Darwish O, Slovin JP, Kang C, Hollender CA, Geretz A, Houston S, Liu Z, Alkharouf NW (2013) SGR: an online genomic resource for the woodland strawberry. BMC Plant Biol 13: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca V, Salim V, Atsumi SM, Yu F (2012) Mining the biodiversity of plants: a revolution in the making. Science 336: 1658–1661 [DOI] [PubMed] [Google Scholar]
- Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP (1999) Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat Struct Biol 6: 775–784 [DOI] [PubMed] [Google Scholar]
- Funa N, Ohnishi Y, Fujii I, Shibuya M, Ebizuka Y, Horinouchi S (1999) A new pathway for polyketide synthesis in microorganisms. Nature 400: 897–899 [DOI] [PubMed] [Google Scholar]
- Gao S, Feng N, Yu S, Yu D, Wang X (2004) Vasodilator constituents from the roots of Lysidice rhodostega. Planta Med 70: 1128–1134 [DOI] [PubMed] [Google Scholar]
- Gowda H, Ivanisevic J, Johnson CH, Kurczy ME, Benton HP, Rinehart D, Nguyen T, Ray J, Kuehl J, Arevalo B, et al. (2014) Interactive XCMS Online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal Chem 86: 6931–6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesser M, Hoffmann T, Bellido ML, Rosati C, Fink B, Kurtzer R, Aharoni A, Muñoz-Blanco J, Schwab W (2008) Redirection of flavonoid biosynthesis through the down-regulation of an anthocyanidin glucosyltransferase in ripening strawberry fruit. Plant Physiol 146: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum SM. (2004) Potential impact of strawberries on human health: a review of the science. Crit Rev Food Sci Nutr 44: 1–17 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Kotilainen M, Elomaa P, Kalkkinen N, Bremer K, Teeri TH, Albert VA (1996) Duplication and functional divergence in the chalcone synthase gene family of Asteraceae: evolution with substrate change and catalytic simplification. Proc Natl Acad Sci USA 93: 9033–9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, Kalinowski G, Schwab W (2006) RNAi-induced silencing of gene expression in strawberry fruit (Fragaria × ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J 48: 818–826 [DOI] [PubMed] [Google Scholar]
- Hughes AL. (2005) Gene duplication and the origin of novel proteins. Proc Natl Acad Sci USA 102: 8791–8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intelmann D, Haseleu G, Dunkel A, Lagemann A, Stephan A, Hofmann T (2011) Comprehensive sensomics analysis of hop-derived bitter compounds during storage of beer. J Agric Food Chem 59: 1939–1953 [DOI] [PubMed] [Google Scholar]
- Jensen RA. (1976) Enzyme recruitment in evolution of new function. Annu Rev Microbiol 30: 409–425 [DOI] [PubMed] [Google Scholar]
- Jez JM, Austin MB, Ferrer J, Bowman ME, Schröder J, Noel JP (2000) Structural control of polyketide formation in plant-specific polyketide synthases. Chem Biol 7: 919–930 [DOI] [PubMed] [Google Scholar]
- Jez JM, Bowman ME, Noel JP (2001) Structure-guided programming of polyketide chain-length determination in chalcone synthase. Biochemistry 40: 14829–14838 [DOI] [PubMed] [Google Scholar]
- Jez JM, Bowman ME, Noel JP (2002) Expanding the biosynthetic repertoire of plant type III polyketide synthases by altering starter molecule specificity. Proc Natl Acad Sci USA 99: 5319–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Lu D, Sun JH, Li CL, Xing Y, Qin L, Shen YY (2013) Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J Exp Bot 64: 1677–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Ficklin SP, Lee T, Cheng CH, Blenda A, Zheng P, Yu J, Bombarely A, Cho I, Ru S, et al. (2014) The Genome Database for Rosaceae (GDR): year 10 update. Nucleic Acids Res 42: D1237–D1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z (2013) Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25: 1960–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keutgen AJ, Pawelzik E (2008) Contribution of amino acids to strawberry fruit quality and their relevance as stress indicators under NaCl salinity. Food Chem 111: 642–647 [Google Scholar]
- Kosasi S, Van der Sluis WG, Labadie RP (1989) Multifidol and multifidol glucoside from the latex of Jatropha multifida. Phytochemistry 28: 2439–2441 [Google Scholar]
- Li Y, Jones L, McQueen-Mason S (2003) Expansins and cell growth. Curr Opin Plant Biol 6: 603–610 [DOI] [PubMed] [Google Scholar]
- Liao Z, Chen M, Guo L, Gong Y, Tang F, Sun X, Tang K (2004) Rapid isolation of high-quality total RNA from taxus and ginkgo. Prep Biochem Biotechnol 34: 209–214 [DOI] [PubMed] [Google Scholar]
- Lunkenbein S, Coiner H, de Vos CHR, Schaart JG, Boone MJ, Krens FA, Schwab W, Salentijn EMJ (2006) Molecular characterization of a stable antisense chalcone synthase phenotype in strawberry (Fragaria × ananassa). J Agric Food Chem 54: 2145–2153 [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher P, Akaishi T, Abe K (2006) Flavonoid fisetin promotes ERK-dependent long-term potentiation and enhances memory. Proc Natl Acad Sci USA 103: 16568–16573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Puche L, Cumplido-Laso G, Amil-Ruiz F, Hoffmann T, Ring L, Rodríguez-Franco A, Caballero JL, Schwab W, Muñoz-Blanco J, Blanco-Portales R (2014) MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. J Exp Bot 65: 401–417 [DOI] [PubMed] [Google Scholar]
- Muñoz C, Hoffmann T, Escobar NM, Ludemann F, Botella MA, Valpuesta V, Schwab W (2010) The strawberry fruit Fra a allergen functions in flavonoid biosynthesis. Mol Plant 3: 113–124 [DOI] [PubMed] [Google Scholar]
- Novák P, Krofta K, Matoušek J (2006) Chalcone synthase homologues from Humulus lupulus: some enzymatic properties and expression. Biol Plant 50: 48–54 [Google Scholar]
- Okada Y, Sano Y, Kaneko T, Abe I, Noguchi H, Ito K (2004) Enzymatic reactions by five chalcone synthase homologs from hop (Humulus lupulus L.). Biosci Biotechnol Biochem 68: 1142–1145 [DOI] [PubMed] [Google Scholar]
- Paniego NB, Zuurbier KW, Fung SY, van der Heijden R, Scheffer JJ, Verpoorte R (1999) Phlorisovalerophenone synthase, a novel polyketide synthase from hop (Humulus lupulus L.) cones. Eur J Biochem 262: 612–616 [DOI] [PubMed] [Google Scholar]
- Patti GJ, Tautenhahn R, Siuzdak G (2012) Meta-analysis of untargeted metabolomic data from multiple profiling experiments. Nat Protoc 7: 508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez AG, Olías R, Luaces P, Sanz C (2002) Biosynthesis of strawberry aroma compounds through amino acid metabolism. J Agric Food Chem 50: 4037–4042 [DOI] [PubMed] [Google Scholar]
- Pérez AG, Rios JJ, Sanz C, Olías JM (1992) Aroma components and free amino acids in strawberry variety Chandler during ripening. J Agric Food Chem 40: 2232–2235 [Google Scholar]
- Ring L, Yeh SY, Hücherig S, Hoffmann T, Blanco-Portales R, Fouche M, Villatoro C, Denoyes B, Monfort A, Caballero JL, et al. (2013) Metabolic interaction between anthocyanin and lignin biosynthesis is associated with peroxidase FaPRX27 in strawberry fruit. Plant Physiol 163: 43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Matsuda F (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61: 463–489 [DOI] [PubMed] [Google Scholar]
- Scalbert A, Manach C, Morand C, Rémésy C, Jiménez L (2005) Dietary polyphenols and the prevention of diseases. Crit Rev Food Sci Nutr 45: 287–306 [DOI] [PubMed] [Google Scholar]
- Schmidt S, Jürgenliemk G, Skaltsa H, Heilmann J (2012) Phloroglucinol derivatives from Hypericum empetrifolium with antiproliferative activity on endothelial cells. Phytochemistry 77: 218–225 [DOI] [PubMed] [Google Scholar]
- Seeram NP, Lee R, Scheuller HS, Heber D (2006) Identification of phenolic compounds in strawberries by liquid chromatography electrospray ionization mass spectroscopy. Food Chem 97: 1–11 [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh IP, Sidana J, Bharate SB, Foley WJ (2010) Phloroglucinol compounds of natural origin: synthetic aspects. Nat Prod Rep 27: 393–416 [DOI] [PubMed] [Google Scholar]
- Sinz Q, Schwab W (2012) Metabolism of amino acids, dipeptides and tetrapeptides by Lactobacillus sakei. Food Microbiol 29: 215–223 [DOI] [PubMed] [Google Scholar]
- Suksamrarn A, Eiamong S, Piyachaturawat P, Byrne LT (1997) A phloracetophenone glucoside with choleretic activity from Curcuma comosa. Phytochemistry 45: 103–105 [Google Scholar]
- Tautenhahn R, Patti GJ, Kalisiak E, Miyamoto T, Schmidt M, Lo FY, McBee J, Baliga NS, Siuzdak G (2011) metaXCMS: second-order analysis of untargeted metabolomics data. Anal Chem 83: 696–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth J, Cutforth T, Gelinas AD, Bethoney KA, Bard J, Harrison CJ (2001) Crystal structure of an ephrin ectodomain. Dev Cell 1: 83–92 [DOI] [PubMed] [Google Scholar]
- Tsukamoto S, Tomise K, Aburatani M, Onuki H, Hirorta H, Ishiharajima E, Ohta T (2004) Isolation of cytochrome P450 inhibitors from strawberry fruit, Fragaria ananassa. J Nat Prod 67: 1839–1841 [DOI] [PubMed] [Google Scholar]
- Ververidis F, Trantas E, Douglas C, Vollmer G, Kretzschmar G, Panopoulos N (2007) Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: Chemical diversity, impacts on plant biology and human health. Biotechnol J 2: 1214–1234 [DOI] [PubMed] [Google Scholar]
- Visioli F, De La Lastra CA, Andres-Lacueva C, Aviram M, Calhau C, Cassano A, D’Archivio M, Faria A, Favé G, Fogliano V, et al. (2011) Polyphenols and human health: a prospectus. Crit Rev Food Sci Nutr 51: 524–546 [DOI] [PubMed] [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Wein M, Lewinsohn E, Schwab W (2001) Metabolic fate of isotopes during the biological transformation of carbohydrates to 2,5-dimethyl-4-hydroxy-3(2h)-furanone in strawberry fruits. J Agric Food Chem 49: 2427–2432 [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. (2000) Eph receptors and ephrins: regulators of guidance and assembly. Int Rev Cytol 196: 177–244 [DOI] [PubMed] [Google Scholar]
- Xu H, Zhang F, Liu B, Huhman DV, Sumner LW, Dixon RA, Wang G (2013) Characterization of the formation of branched short-chain fatty acid:CoAs for bitter acid biosynthesis in hop glandular trichomes. Mol Plant 6: 1301–1317 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Suh DY, Sitthithaworn W, Ishiguro K, Kobayashi Y, Shibuya M, Ebizuka Y, Sankawa U (2001) Diverse chalcone synthase superfamily enzymes from the most primitive vascular plant, Psilotum nudum. Planta 214: 75–84 [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Abe T, Tanaka T, Yang CR, Kouno I (2002) Two new acylated flavanone glycosides from the leaves and branches of Phyllanthus emblica. Chem Pharm Bull (Tokyo) 50: 841–843 [DOI] [PubMed] [Google Scholar]
- Zuurbier KWM, Leser J, Berger T, Hofte AJP, Schröder G, Verpoorte R, Schröder J (1998) 4-Hydroxy-2-pyrone formation by chalcone and stilbene synthase with nonphysiological substrates. Phytochemistry 49: 1945–1951 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.